Abstract
Patients with type 1 or type 2 diabetes have an insufficiency in their functional β cell mass. To advance diabetes treatment and to work toward a cure, a better understanding of how to protect the pancreatic β cells against autoimmune or metabolic assaults (e.g., obesity, gestation) will be required. Over the past decades, β cell protection has been extensively investigated in rodents both in vivo and in vitro using isolated islets or rodent β cell lines. Transferring these rodent data to humans has long been challenging, at least partly for technical reasons: primary human islet preparations were scarce and functional human β cell lines were lacking. In 2011, we described a robust protocol of targeted oncogenesis in human fetal pancreas and produced the first functional human β cell line, and in subsequent years additional lines with specific traits. These cell lines are currently used by more than 150 academic and industrial laboratories worldwide. In this Review, we first explain how we developed the human β cell lines and why we think we succeeded where others, despite major efforts, did not. Next, we discuss the use of such functional human β cell lines and share some perspectives on their use to advance diabetes research.
Introduction
The only hormone that can decrease circulating glucose levels is insulin. Insulin is produced in β cells, and its secretion into the bloodstream is tightly regulated to control glucose homeostasis. β cells reside in the pancreas and are organized in clusters of endocrine cells called islets of Langerhans. A human pancreas weights 91.8 g (ranging between 40.9 and 182 g based on analysis of 30 samples) (1). It contains approximately 1 billion β cells, which corresponds to 1 g of tissue and 10 mg of insulin — sufficient to control blood glucose levels for 2 weeks (2). A single β cell is estimated to produce 1 million molecules of insulin per minute (3) and to contain 10 pg of insulin packed into 10,000 granules (4). In people with type 1 diabetes, β cells are selectively destroyed by an autoimmune attack, creating an absolute insulin deficiency. In people with type 2 diabetes, there is a relative insulin deficiency, which is mainly due to an insulin secretory defect associated with insulin resistance (5).
Over the past century — mainly since 1921, when Banting and Best started treating diabetic dogs with pancreas extracts — our understanding of how β cells develop, grow, function, survive, and die in physiological and pathological conditions has improved impressively. Much of our understanding comes from rodent models in which the phenotype of β cells from wild-type and transgenic mice and rats was analyzed first in vivo and, once islet isolation procedures were established (6), expanded upon in vitro. A series of rodent β cell lines developed since the 1970s have further benefited β cell biology. These cell lines offered a robust and reproducible supply of β cells, allowing the use of technologies that require large amounts of cells or cell products. In recent years, major efforts have been aimed at comparing rodent β cells with their human counterparts (7–10). An increase in human islet preparations available for research has improved our understanding of what a human β cell is, but the material remains scarce, its availability is unpredictable, and the quality is variable.
In 2011, we published the generation of a new cell line, EndoC-βH1, which could be used to study human β cells (11). The work was highlighted in a Commentary in the JCI entitled “Finally! A human pancreatic β cell line” (12) and also drew the attention of the Innovative Medicines Initiative (https://www.imi.europa.eu/) (13). Here, we discuss the usefulness of human β cell lines to increase our understanding of human islet biology and diabetes.
Challenges in human islet biology
Human β cells represent 1%–2% of the total pancreatic mass. Their study has been limited for years because of the scarcity of human donor pancreata and the difficulty of accessing islet preparations. Over the past 20 years, our knowledge of human β cell biology sharply increased with the rise in human pancreata and islets available to researchers. Initiatives such as the Network for Pancreatic Organ Donors with Diabetes (https://www.jdrfnpod.org/) provide an efficient platform to procure human pancreata from diabetic and nondiabetic donors (14). Islet preparations were used to study the transcriptome and function of human β cells and to test whether knowledge derived from rodent experiments was applicable to human β cells (15–17). For example, comparisons between mouse and human islets revealed major differences regarding the deleterious effects of glucose or lipid excess on insulin secretion (15–17). In addition, islets from young and adult human donors were compared (18–22) and showed substantial differences in terms of growth factor sensitivity (18), β cell proliferation (19), and function (21). Finally, islets from healthy controls and people with type 1 (23–26), type 2 (27–31), monogenic (32), or gestational diabetes (33) were compared. Islets from type 2 diabetes have, for example, provided a transcriptional signature of the human β cells during disease (27–31).
While our knowledge surely increased, we also learned that interpreting data from human islets is often complicated. Indeed, human islets are prepared from deceased donors who differ in several aspects, including sex, age, cause of death, body mass index, comorbidity, and treatments received (34). Following pancreas procurement, the islets need to be isolated, which involves mechanical and enzymatic digestion of the exocrine pancreas without affecting the integrity of the islets (35). Next, islets are cultured at the isolation center, sent out worldwide for either clinical transplantation or research purposes, and then again cultured by the receiving institute before manipulation. While some studies have shown that human islets in culture lose their functionality with time (36), others have kept islets in culture for more than 9 months while preserving their function (37). These differences may depend on a range of variables, of which some are known, e.g., the quality of the islet preparation and the culture conditions, while others likely remain unknown. Notably, the frequency of β cells from one islet preparation to another is highly variable. Of 13 islet preparations from donors without diabetes, it was shown that the relative proportion of β cells varied between about 30% and 75% of the total endocrine cell population (38). This most likely underestimates the overall variability in β cell content of human islet preparations (percentage of the total cell number), as even expert islet isolation centers report high variability of contamination with nonendocrine cells (mainly acinar and duct cells) (39). Finally, human islet function remains highly variable from one preparation to another, even in the leading islet isolation centers (40). One of our key aims should thus be to rigorously phenotype as many human islet preparations as possible. This important issue was discussed and put forward in a recent review and will change, for the better, our ability to interpret data on human β cells (10). In a recent joint statement, the official journals of the American Diabetes Association and the European Association for the Study of Diabetes, Diabetes and Diabetologia, respectively, have highlighted the need for improved reporting of human islets used for research and proposed a checklist (http://diabetes.diabetesjournals.org/content/human-islet-policy) to facilitate standardized reporting.
Human β cell lines: from rodent to human
The first rodent β cell lines were developed in the 1970s. They have been generated in different species: Rattus norvegicus (RIN and INS1 cells), derived from x-ray–induced insulinomas in rats (41, 42); Cricetus cricetus (HIT cells), obtained by simian virus 40 (SV40) transformation of adult hamster islet cells (43); and Mus musculus (βTCs and Min6 cells), produced by targeted oncogenesis in transgenic mice expressing an insulin promoter SV40 early region that contained the coding information for SV40 large T (SV40LT) and SV40 small t tumor antigens (44–46). These cell lines have been very useful for detailed study of rodent β cells. In contrast, the generation of a functional human β cell line proved to be more complicated, and despite major investments, the undertaking remained frustrating. For example, the BetaLox-5 cell line, published in 1999, was derived from purified β cells of a human fetal pancreas at 24 weeks of development by transduction with retroviral vectors expressing the SV40LT antigen, the oncogene Hrasval12, and the human telomerase reverse transcriptase (hTERT) (47–49). However, this line expressed low levels of insulin (INS) and was unstable, with INS expression decreasing even further with passages, which explains why it was not frequently used by the scientific community. In 2005, there was a seemingly major breakthrough in the field as a multinational team published a reversibly transformed human β cell line. The NAKT-15 cell line was derived by Moloney murine leukemia virus (MoMLV) long terminal repeat–driven expression of SV40LT and hTERT in islet cells. NAKT-15 cells were highly similar to primary human β cells in terms of expression of key transcription factors, insulin production, and glucose responsiveness. It was even possible to excise the SV40LT and hTERT oncogenes in order to obtain postmitotic human β cells (50). At the time of publication, the NAKT-15 cell line drew the attention of the scientific community, as it represented a powerful tool for diabetes research (51). However, some points in the work remain unclear. First, NAKT-15 cells were generated upon transduction of adult human islets with γ-retroviral vectors. It is well established that these vectors can only stably transduce proliferating cells (52). However, adult human β cells are mostly if not exclusively postmitotic. A recent study found no β cell proliferation, as determined by Ki-67 immunostaining, among a total of 37,845 β cells analyzed on pancreata from 18 nondiabetic subjects (53). Second, when expressed by adult human β cells, SV40LT is inefficient in activating markers of proliferation (11, 54). Finally, to our knowledge, the NAKT-15 cell line has only been used in two original publications (55, 56), and it did not find its way into diabetes laboratories around the world.
In the early 2000s, we decided to try to generate human β cell lines. Our protocol was based on the successes and failures of previous attempts (44–46, 49, 50). We chose human fetal pancreata as starting material because they were thought to be more prone to immortalization than adult pancreas. We reasoned that a higher proliferation rate in fetal cells would render them more susceptible to both transduction and the pro-proliferative effect of the introduced oncogenes. Lentiviral vectors were used for transduction, and SV40LT and hTERT were chosen for immortalization. At that time this was a risky choice, since human cell transformation was considered to require the combined expression of SV40 early region, hTERT, and an oncogenic mutant of H-RAS (Hrasval12) (57). In some settings, however, hTERT alone has been used successfully to immortalize neural progenitor cells derived from human fetal spinal cord (58). In our approach, we included a 405-bp fragment of the rat insulin-2 promoter (RIP) to drive the expression of SV40LT and hTERT. This promoter fragment was well characterized and displays specificity and efficiency in both rodent and human pancreatic β cells (59–61). Overall, our approach based on targeted oncogenesis in fetal pancreatic tissues resembled the one used to derive the βTC and Min6 cell lines from transgenic mice. As access to human fetal pancreatic fragments is limited, we validated each step of the process using rat fetal pancreata as starting material (62).
We next used this protocol to generate an initial human β cell line. Drawings of this protocol can be seen in ref. 8 and ref. 12. First, we transduced fragments of human fetal pancreata (7–11 weeks of gestation) with a lentiviral vector expressing SV40LT. At this early stage of development, the human pancreas is mainly composed of progenitors and few insulin-containing cells (63–65). Our hypothesis was that the SV40LT oncogene would integrate randomly into the pancreatic cells, but that it would only be expressed when those progenitors differentiated into INS-expressing β cells (the oncogene being under the control of the insulin promoter). Next, we transplanted the transduced human pancreatic fragments into immune-compromised (SCID) mice, which formed a permissive environment for human fetal pancreatic progenitor growth and differentiation (66, 67). In the case of transduction of a β cell progenitor, this resulted in the development of an insulinoma. Next, the insulinomas were transduced with a lentiviral vector expressing hTERT and again grafted into SCID mice to amplify the proliferating β cells. Finally, the insulinomas were surgically removed, dissociated into single cells, and expanded in culture as cell lines. Different candidate cell lines were obtained, all of which had a remarkably similar expression of many key β cell transcription factors, an almost constant expression of SV40LT, but a variable expression of INS. One of these cell lines was characterized in more detail and became the first human β cell line, EndoC-βH1 (11). EndoC-βH1 cells resemble human β cells in their expression of many specific genetic and epigenetic markers (68), in the limited expression of markers of other pancreatic cell types (some rare EndoC-βH1 cells contain somatostatin, but none contain glucagon; ref. 11), and in their ability to secrete insulin in response to glucose, the glucagon-like peptide 1 analog exendin-4, sulfonylureas such as glibenclamide, and branched-chain amino acids such as leucine (11). Electrophysiological properties of EndoC-βH1 also conform with human β cells (69).
The first generation of EndoC-βH cells was followed by a second generation. In these EndoC-βH2 cells, two loxP sites flank the integrated oncogenes, which allows their excision upon Cre recombinase expression (70). Next, we generated a third-generation EndoC-βH3 by stable integration of a tamoxifen-inducible form of Cre into the EndoC-βH2 cells, which makes them an easy-to-use excisable human β cell line (71). We also used the EndoC-βH2 line to generate human Fucci β cells, in which two fluorescent reporters alternate as the cells progress through the cell cycle (72). These human Fucci β cells can be used to study the human β cell cycle and differentiation. More recently, we used a similar approach to generate a new human β cell line, ECN90, derived not from fetal but from neonatal pancreas (4 months old) (73). There, we presumably directly transduced the β cells that are present at this early postnatal age and have a substantial replicative potential (74, 75). We are convinced that our approach does not work on postmitotic adult human β cells, even when effectively transduced by the lentiviral vectors (11). This situation is reminiscent of the outcome of experiments aimed at reactivating proliferation in postmitotic muscle cells or human β cells, namely a cell cycle block due to severe DNA damage, despite an initial triggering of DNA synthesis (76, 77). Accordingly, SV40LT and hTERT may be unable to reactivate proliferation in adult human β cells as they trigger an abortive cell cycle reentry; only replicating progenitor cells or immature neonatal β cells can be immortalized using our strategy.
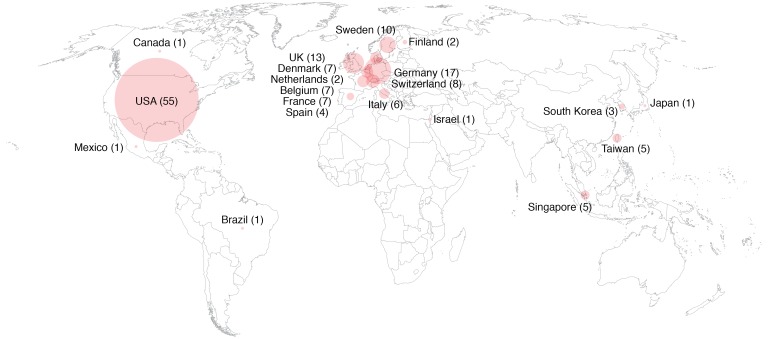
Cell line misidentification, contamination, and poor annotation are recurrent problems that affect scientific reproducibility (78, 79). To allow the detection of such problems, we analyzed short tandem repeats on each cell line at early passage for authentication, and these data are available for comparison. We have now shared the EndoC-βH cell lines with more than 150 laboratories worldwide (Figure 1). Based on feedback of colleagues and on their use in more than 100 original publications, we think that they represent useful tools for the academic scientific community. A regularly updated list of publications that have used the human β cell lines we developed can be found at https://www.humanbetacelllines.com/ The cell lines have already proved to be useful in generating new knowledge on human β cells. Data from rat islets and rat β cell lines had shown, for example, that inflammatory cytokines induce iNOS expression and thereby increase the production of nitric oxide, resulting in β cell death (80). A similar mechanism was presumed in humans, but a recent study, using EndoC-βH1 cells, demonstrated that cytokines do not induce iNOS expression in human β cells (81). The EndoC-βH1 cells were also used to show that in human β cells the methyltransferases mixed-lineage leukemia 3 and 4 (MLL3/4) bind to the transcription factors MAFA and MAFB, and that the complexes formed thereby are necessary for proper glucose-induced insulin secretion (82). Another example for which the EndoC-βH1 cells were crucial is the demonstration of physical contact between the insulin promoter and diabetes susceptibility loci and the insulin promoter’s involvement in the regulation of insulin transport and metabolism in human β cells (83). Peptidomic-based approaches have also used the human β cell lines to identify target epitopes processed and presented by β cells, thereby providing the first HLA-I peptidome catalog of human β cells (84). Finally, our group used EndoC-βH1 cells to develop models and identify markers of human β cell dedifferentiation (85, 86). All of the above examples depended on experiments that would have been difficult to perform with human islets because of limitations in cell purity or cell number and that would have produced different results in rodent β cell lines because of species differences. Interestingly, human β cell lines have now been validated by the pharmaceutical industry as a screening model to identify novel drug target candidates (87); to determine the effects of fatty acid esters of hydroxylated fatty acids on glucose-stimulated insulin secretion (88); to study the mechanism of secretagogin release by human β cells (89); and to further test a gastrointestinal peptide–based (GIP-based) dual incretin receptor agonist (90).
Figure 1. Distribution of human β cell lines worldwide.
Notably, at the time we published EndoC-βH1, three additional human insulin-releasing cell lines (1.1B4, 1.4E7, and 1.1E7) were generated by electrofusion of human pancreatic β cells with the immortal human PANC-1 cell line that was established from a pancreatic carcinoma of ductal origin (91). These human insulin-releasing cell lines expressed the expected set of human β cell markers and were used to dissect human β cell function and survival (92–95). However, their insulin content was extremely low, around 4 ng per million cells, which is less than a thousandth of the insulin content measured in primary human β cells and 150-fold less than in EndoC-βH1 cells.
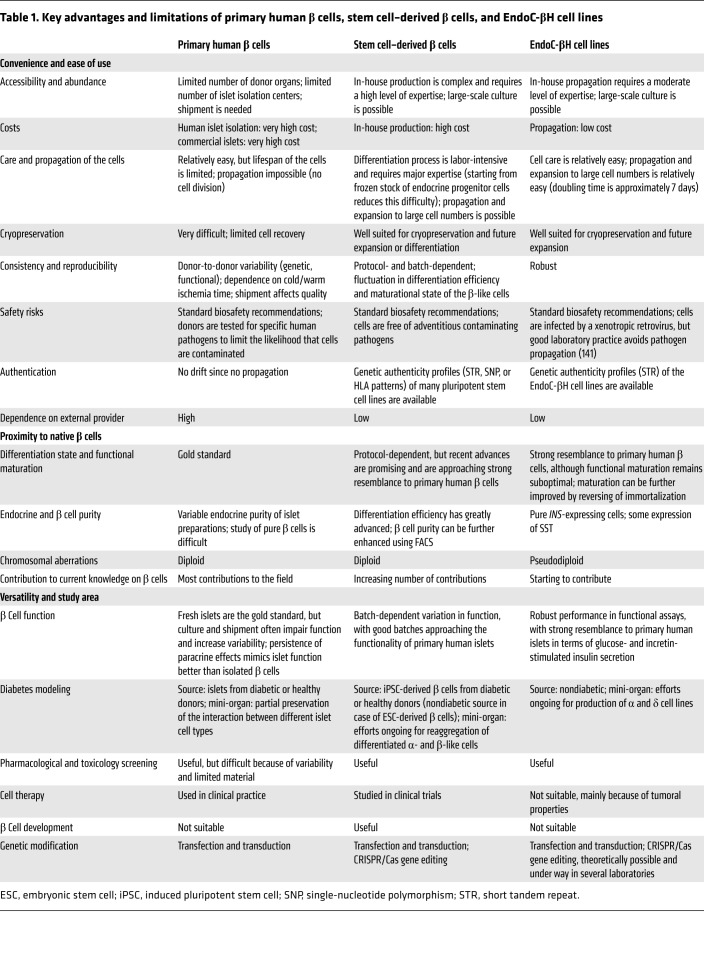
EndoC-βH cells have been used recurrently for multi-omic profiling (68, 70, 96, 97). As a pure human β cell population, they can be used to validate the expression of genes of interest. Surprisingly, a number of proteins detected in primary human β cells by immunostainings are not detected at the transcript level in EndoC-βH cells or primary human β cells. For example, TLR4 (98) and the immunoregulatory antimicrobial peptide CRAMP (99) are not expressed in EndoC-βH cells. Notably, their mRNAs are also absent or at the lowest limit of detection in bulk transcriptional analyses of human islets (27, 100) and undetectable by single-cell transcriptomics (29). Such data contrast sharply with the immunostainings on human islet sections, which show very strong signals for TLR4 and CRAMP in β cells (98, 99). Another example is TGF-β–induced (βig-h3, or TGF-βI), a protein for which immunostainings have been shown in human β cells (101), but again for which the coding mRNA is not found in human β cell lines or human β cells at the single-cell level (29). βig-h3 mRNA was, however, detected in human islets (15), which suggests that it may be expressed by non-β islet cells. A final example concerns CFTR, a chloride channel mutated in patients with cystic fibrosis (102), which is detected in islet preparations (100). However, we know that duct cells, which always contaminate islet preparations (103), express high levels of CFTR (29). While multiple studies report CFTR expression and expand on its function in rodent β cells (104, 105), more recent work debates an intrinsic role of CFTR in β cells (106). We did not detect CFTR mRNA in any of the human β cells, and single-cell RNA-Seq on human islet preparations also showed that β cells barely express CFTR, whereas duct cells express CFTR at high levels (29). Overall, the above-described discrepancies may be due to insufficient antibody validation (107, 108) or to other hitherto unknown reasons that may be related to islet or pancreatic tissue preparation. Importantly, the above examples illustrate the complexity of working on primary human pancreata and islet preparations. In the study of human β cells, EndoC-βH cells can offer a robust and reproducible system that is particularly useful for screening before moving on to primary cells or for dissecting mechanistic aspects of experimental findings. The key advantages and limitations of EndoC-βH cells as compared with primary human β cells and stem cell–derived β cells are shown in Table 1.
Table 1. Key advantages and limitations of primary human β cells, stem cell–derived β cells, and EndoC-βH cell lines.
Stability of human β cell lines
EndoC-βH cells are pseudodiploid, which means they have 46–48 chromosomes per cell and their karyotype is very stable over multiple passages (68). We have, for example, performed comparative genomic hybridization arrays between passages 42 and 103 for ECN90 cells and found almost no genomic changes at the subchromosomal level. Moreover, the overall β cell identity in the different lines proved to be very stable, as we found no significant changes in terms of cell growth, gene expression, insulin content, or insulin secretion over long-term passaging (our unpublished observations). Inherent β cell traits are likely involved in this stability: for example, they express only low levels of lactate dehydrogenase A and produce little lactate, which limits the risk of acidification-induced DNA damage (109). Experimental settings may also be important for phenotypical robustness and may provide selection pressure at multiple levels.
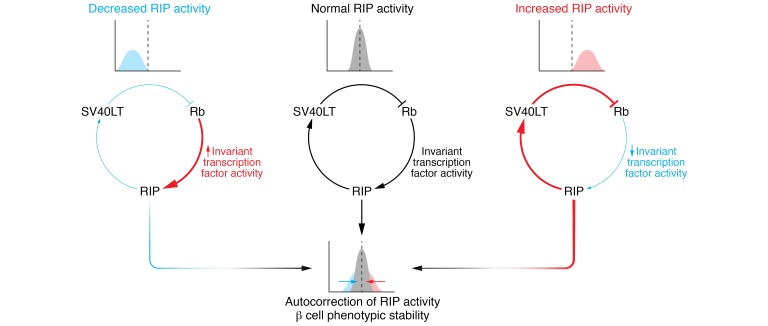
In summary, the stability of EndoC-βH cells comes down to SV40T expression requiring RIP activity and in turn selecting for cells that transcribe INS (Figure 2). Mechanistically, the need to maintain RIP activity drives the selection of cells expressing transcription factors that permit RIP activity. This set of RIP-activating transcription factors probably transactivates β cell genes, among which is INS as well as additional sets of transcription factors, and installs a regulatory module of transcription factors that preserves not only INS expression but also a more general β cell phenotype over time. Suggestive of such a strong selection pressure and the existence of a regulatory module are the remarkably similar expression levels of many key β cell transcription factors across the different human β cell lines. Indeed, at least some of these invariant transcription factors have been found to regulate each other or themselves and are thought to cooperatively transactivate INS and other β cell genes (110–115). The proposed regulatory module in the human β cell lines is thus likely to include these invariant transcription factors. Curiously, despite these invariant transcription factors and, accordingly, invariant RIP activity (as reflected in SV40T expression levels; see below), INS expression shows a much larger variability across the different EndoC-βH cell lines. This variable INS expression may be due to distinct regulation at a region not present in RIP or by transcription factors that do not belong to the regulatory module.
Figure 2. Potential mechanisms underlying the stability of EndoC-βH cell lines and the reversion of their immortalization.
Positive or negative variation in RIP activity is counteracted by its effects on SV40LT expression levels at both an individual and a population level. At the individual cell level, an increase (or decrease) in SV40LT expression decreases (or increases) RIP activity by a mechanism involving the Rb pathway. At the cell population level, decrease in SV40LT expression level in a given cell impairs its viability or proliferation compared with the other cells. The result of these mechanistic and selective forces (red and blue arrows) is to keep RIP activity constant in most cells (narrow bell curve). This maintains stable activity within a putative RIP-regulating module, which in turn ensures long-term phenotypic stability. Persistent subthreshold SV40LT expression, as in experiments aimed to reverse immortalization, triggers cell cycle exit and terminal differentiation. For more details, see main text. Rb, activity of the retinoblastoma protein family; RIP, rat insulin promoter; SV40LT, simian virus 40 large T antigen expression.
Reversibility of immortalization
A notable feature of EndoC-βH cell lines is the reversibility of their immortalized phenotype. Upon excision of both hTERT and SV40LT (70, 71, 116), or knockdown of SV40LT mRNA (21, 117), immortalized human β cells exit the cell cycle and deepen β cell–specific traits. For example, they upregulate the expression of IAPP (encoding islet amyloid polypeptide), INSULIN, SLC2A2 (encoding the glucose transporter GLUT2), and the ABCC8 and KCNJ11 subunits of the ATP-sensitive potassium channel, which are targets of the sulfonylurea drugs, and they downregulate disallowed genes such as LDH-A and MCT-1 (70). Reversibility of immortalization was previously shown in the mouse β cell line BetaTC-Tet (118) by the shutting off of SV40LT, in rat β cell lines upon Cre-(ER)–mediated excision of SV40LT and hTERT encoding retroviral vectors (119), and in lines of other cell types, such as neurons (120) and cardiomyocytes (121). Some other cell lines, however, succumb to oncogene loss. Primate hepatic progenitor cell lines, for example, die after SV40T deletion (122). Cell-specific traits may underlie the ability of β cells to withstand the loss of SV40T and hTERT. One such trait might be that senescence is associated with functional maturation in β cells (116). However, as discussed above, let us consider the experimental settings and especially how RIP activity drives the expression of SV40LT (and hTERT) in the human β cell lines. To support proliferation, immortalized β cells need to maintain SV40LT expression, but high SV40LT expression likely results in dedifferentiation and thus in a reduced activity of the RIP-regulating module. Conversely, decreased SV40LT expression will strengthen the activity of the module and upregulate RIP activity and SV40LT expression. Thus, SV40LT expression is self-corrected, as any variation in its level imparts a counteracting regulation. Mechanistically, SV40LT represses the activity of the retinoblastoma (RB) family and p53 tumor suppressors. Interestingly, RB pathway is decreased, but not completely lost, in mouse insulinomas with a RIP-SV40 early region transgene (123). The unexpected, residual RB pathway may restrict proliferation in one way, but it would also be expected to maintain RIP activity and thus SV40LT expression and proliferation. Indeed, RB knockdown in (SV40LT-expressing) EndoC-βH2 cells quells both their β identity and their RIP activity (117). As noted above, SV40LT expression is almost constant across the different human β cell lines, and even a small decrease (about 2-fold) in SV40LT expression reduces their proliferation and triggers an increase in P21 (CDKN1A), INSULIN, and IAPP expression (117). Thus, human β cell lines need SV40LT expression between a lower and upper threshold to ensure proliferation on one hand and conservation of the β cell phenotype and viability (with residual RB activity) on the other hand. These thresholds for SV40LT expression level imply a narrow range of RIP activity, which might in turn stabilize the activity of the regulatory module and thus promote phenotype robustness. The resulting trade-off is the stabilization of SV40LT expression at the lowest possible level sufficient for proliferation. This leaves the cells poised to withdraw from cycling and to undergo terminal differentiation if SV40LT expression drops below the bottom threshold for a critical length of time. In summary, RIP-mediated control of SV40LT might be instrumental for both the stability of phenotype and the reversibility of the immortalization (Figure 2).
Reversibility of immortalization as an experimental system
The reversibility of immortalization raises interesting questions that can be experimentally addressed. For instance, we don’t know whether proliferating β cell lines escape from aging, or whether they accumulate time-related damage such as genotoxic insults that would contribute to their fate upon SV40LT depletion. Another example is whether reintroduction of SV40LT can awaken proliferation in resting, previously SV40LT-depleted, human β cells. And if so, until what time (duration after SV40LT depletion) and what events (reasons underlying the loss of reversibility) is this capacity retained? Answering such questions may help us understand why adult human β cells are so resistant to immortalization. Oncogene depletion in EndoC-βH cells not only decreases their proliferation but also induces functional maturation, as reflected by increased expression of the transcription factor MAFA, an increase in glucose-stimulated insulin secretion (70), higher mitochondrial activity, and expression of senescence markers (116). This in vitro process resembles the in vivo maturation during postnatal life (124–127) and may thus provide an amenable system to study β cell maturation. The phenotype observed upon reversion of immortalization represents a striking illustration of the mutual exclusion between β cell proliferation and maturation. This mutual exclusion was observed both in natural settings such as during postnatal β cell maturation (127) and weaning (128) and in experimental settings, for example, upon reduced insulin production or by tuning the expression of Myc (129, 130). Its mechanistic details are still not fully understood. Accordingly, how SV40LT depletion enhances INS expression in rodent and human β cell lines (70, 71, 118) remains largely unknown at a molecular level. Again, reversible immortalization may help in elucidating the underlying mechanisms of this fundamental aspect of β cell physiology.
From human β to non-β endocrine cell lines
Human islets also consist of non-β endocrine cells such as α, δ, pancreatic polypeptide (PP, formerly known as γ), and ε cells. These cells interact with each other and with β cells in feedback loops that determine islet function. Cell lines of these non-β endocrine cells could help us reconstruct and dissect the interactions between the different endocrine cells within human pancreatic islets (7, 131). Such cell lines are not available for human and are very limited for rodent. In rodents, few α cell lines have been described and used. An early one, In-R1-G9, was derived from a transplantable hamster insulinoma (132, 133). A second example is represented by the αTC cell line that was derived from a glucagonoma generated in transgenic mice expressing SV40 early region under the control of the glucagon promoter (134, 135). It is thus evident that more lines are needed from both rodents and humans.
Using our experience in generating human β cell lines, we tried to produce human α cell lines. We replaced the RIP that controls oncogene expression with glucagon promoters of different length. This approach failed to generate glucagonomas and α cell lines. There are different possible explanations for this failure, but mainly it is because, compared with the insulin promoter (136, 137), the glucagon promoter has been less studied (138). We may thus lack crucial information on its activity. For example, a recent study compared the specificity of expression of the insulin and glucagon promoters to target β and α cells, respectively, in a lentiviral context. While the insulin promoter is highly specific for β cells (60, 62, 139), the specificity of the tested glucagon promoters for α cells is far lower (139). The lack of specificity in targeting α cells when protocols based on targeted oncogenesis are used is thus an important problem. Moreover, the glucagon promoter is weaker than the insulin promoter (140), and SV40LT expression levels driven by a glucagon promoter may be insufficient to induce immortalization of human pancreatic cells. Third, the generation of functional non-β islet cell lines may depend on more than just strong and specific promoters; it may require that the activity of the promoter harness a regulatory module equivalent to that proposed above in the human β cell lines. We are convinced that more work, effort, and targeted funding are needed to generate functional rodent and human α, δ, PP, and ε cells.
Conclusion
Despite major efforts, the diabetes research community did not have human β cell lines a decade ago. By developing a robust protocol based on targeted oncogenesis, we generated a series of functional human β cell lines from fetal, and recently neonatal, human pancreatic fragments. We have started to dissect the mechanistic reasons for our success in generating stable human β cell lines. Such information may increase our knowledge on primary human β cells. We think that with increased effort and financial support, it will be possible to generate human α and δ cell lines. These are islet cell types of which we know strikingly little in both physiological and pathological conditions such as diabetes. An arsenal of different human endocrine cell lines would enable access to an unlimited supply of pseudoislets that could be used, for example, to dissect inter-endocrine cell-cell interactions under pathophysiological conditions.
Acknowledgments
RS would like to thank two long-term collaborators involved in the generation of human β cell lines, Paul Czernichow (Assistance Publique–Hôpitaux de Paris, Paris, France) and Philippe Ravassard (CNRS, Paris, France). WS is supported by a postdoctoral grant from Agence Nationale de la Recherche (Laboratoire d’Excellence Revive, Investissement d’Avenir; ANR-10-LABX-73). The RS laboratory received funding from Agence Nationale de la Recherche (ANR BromoBeta), the Innovative Medicines Initiative programs IMIDIA, RHAPSODY, and INNODIA, Fondation Bettencourt Schueller, the Laboratoire d’Excellence consortium Revive, and Fondation Francophone pour la Recherche sur le Diabete (FFRD).
Version 1. 09/03/2019
Print issue publication
Footnotes
Conflict of interest: RS is a shareholder in and consultant for Univercell-Biosolutions.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(9):3511–3520.https://doi.org/10.1172/JCI129484.
References
- 1.Innes JT, Carey LC. Normal pancreatic dimensions in the adult human. Am J Surg. 1994;167(2):261–263. doi: 10.1016/0002-9610(94)90088-4. [DOI] [PubMed] [Google Scholar]
- 2.Ferrannini E. The stunned beta cell: a brief history. Cell Metab. 2010;11(5):349–352. doi: 10.1016/j.cmet.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 3.Scheuner D, Kaufman RJ. The unfolded protein response: a pathway that links insulin demand with beta-cell failure and diabetes. Endocr Rev. 2008;29(3):317–333. doi: 10.1210/er.2007-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rorsman P, Braun M. Regulation of insulin secretion in human pancreatic islets. Annu Rev Physiol. 2013;75:155–179. doi: 10.1146/annurev-physiol-030212-183754. [DOI] [PubMed] [Google Scholar]
- 5.American Diabetes Association. 2. Classification and diagnosis of diabetes: Standards of Medical Care in Diabetes — 2018. Diabetes Care. 2018;41(suppl 1):S13–S27. doi: 10.2337/dc18-S002. [DOI] [PubMed] [Google Scholar]
- 6.Lacy PE, Kostianovsky M. Method for the isolation of intact islets of Langerhans from the rat pancreas. Diabetes. 1967;16(1):35–39. doi: 10.2337/diab.16.1.35. [DOI] [PubMed] [Google Scholar]
- 7.Caicedo A. Paracrine and autocrine interactions in the human islet: more than meets the eye. Semin Cell Dev Biol. 2013;24(1):11–21. doi: 10.1016/j.semcdb.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scharfmann R, Rachdi L, Ravassard P. Concise review: in search of unlimited sources of functional human pancreatic beta cells. Stem Cells Transl Med. 2013;2(1):61–67. doi: 10.5966/sctm.2012-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu JS, Hebrok M. All mixed up: defining roles for β-cell subtypes in mature islets. Genes Dev. 2017;31(3):228–240. doi: 10.1101/gad.294389.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hart NJ, Powers AC. Use of human islets to understand islet biology and diabetes: progress, challenges and suggestions. Diabetologia. 2019;62(2):212–222. doi: 10.1007/s00125-018-4772-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ravassard P, et al. A genetically engineered human pancreatic β cell line exhibiting glucose-inducible insulin secretion. J Clin Invest. 2011;121(9):3589–3597. doi: 10.1172/JCI58447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weir GC, Bonner-Weir S. Finally! A human pancreatic β cell line. J Clin Invest. 2011;121(9):3395–3397. doi: 10.1172/JCI58899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldman M. Public-private partnerships need honest brokering. Nat Med. 2012;18(3):341. doi: 10.1038/nm0312-341. [DOI] [PubMed] [Google Scholar]
- 14.Kaddis JS, Pugliese A, Atkinson MA. A run on the biobank: what have we learned about type 1 diabetes from the nPOD tissue repository? Curr Opin Endocrinol Diabetes Obes. 2015;22(4):290–295. doi: 10.1097/MED.0000000000000171. [DOI] [PubMed] [Google Scholar]
- 15.Nica AC, et al. Cell-type, allelic, and genetic signatures in the human pancreatic beta cell transcriptome. Genome Res. 2013;23(9):1554–1562. doi: 10.1101/gr.150706.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang YJ, Kaestner KH. Single-Cell RNA-Seq of the pancreatic islets — a promise not yet fulfilled? Cell Metab. 2019;29(3):539–544. doi: 10.1016/j.cmet.2018.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dai C, et al. Stress-impaired transcription factor expression and insulin secretion in transplanted human islets. J Clin Invest. 2016;126(5):1857–1870. doi: 10.1172/JCI83657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen H, et al. PDGF signalling controls age-dependent proliferation in pancreatic β-cells. Nature. 2011;478(7369):349–355. doi: 10.1038/nature10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodyer WR, Gu X, Liu Y, Bottino R, Crabtree GR, Kim SK. Neonatal β cell development in mice and humans is regulated by calcineurin/NFAT. Dev Cell. 2012;23(1):21–34. doi: 10.1016/j.devcel.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang YJ, et al. Single-cell transcriptomics of the human endocrine pancreas. Diabetes. 2016;65(10):3028–3038. doi: 10.2337/db16-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arda HE, et al. Age-dependent pancreatic gene regulation reveals mechanisms governing human β cell function. Cell Metab. 2016;23(5):909–920. doi: 10.1016/j.cmet.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang YJ, et al. Single-cell mass cytometry analysis of the human endocrine pancreas. Cell Metab. 2016;24(4):616–626. doi: 10.1016/j.cmet.2016.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brissova M, et al. α Cell function and gene expression are compromised in type 1 diabetes. Cell Rep. 2018;22(10):2667–2676. doi: 10.1016/j.celrep.2018.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Babon JA, et al. Analysis of self-antigen specificity of islet-infiltrating T cells from human donors with type 1 diabetes. Nat Med. 2016;22(12):1482–1487. doi: 10.1038/nm.4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delong T, et al. Pathogenic CD4 T cells in type 1 diabetes recognize epitopes formed by peptide fusion. Science. 2016;351(6274):711–714. doi: 10.1126/science.aad2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michels AW, et al. Islet-derived CD4 T cells targeting proinsulin in human autoimmune diabetes. Diabetes. 2017;66(3):722–734. doi: 10.2337/db16-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fadista J, et al. Global genomic and transcriptomic analysis of human pancreatic islets reveals novel genes influencing glucose metabolism. Proc Natl Acad Sci U S A. 2014;111(38):13924–13929. doi: 10.1073/pnas.1402665111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Solimena M, et al. Systems biology of the IMIDIA biobank from organ donors and pancreatectomised patients defines a novel transcriptomic signature of islets from individuals with type 2 diabetes. Diabetologia. 2018;61(3):641–657. doi: 10.1007/s00125-017-4500-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Segerstolpe Å, et al. Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metab. 2016;24(4):593–607. doi: 10.1016/j.cmet.2016.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lawlor N, et al. Single-cell transcriptomes identify human islet cell signatures and reveal cell-type-specific expression changes in type 2 diabetes. Genome Res. 2017;27(2):208–222. doi: 10.1101/gr.212720.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ediger BN, et al. LIM domain-binding 1 maintains the terminally differentiated state of pancreatic β cells. J Clin Invest. 2017;127(1):215–229. doi: 10.1172/JCI88016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haliyur R, et al. Human islets expressing HNF1A variant have defective β cell transcriptional regulatory networks. J Clin Invest. 2019;129(1):246–251. doi: 10.1172/JCI121994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tancredi M, et al. Histopathology and ex vivo insulin secretion of pancreatic islets in gestational diabetes: a case report. Islets. 2011;3(5):231–233. doi: 10.4161/isl.3.5.15940. [DOI] [PubMed] [Google Scholar]
- 34.In’t Veld P, et al. Beta-cell replication is increased in donor organs from young patients after prolonged life support. Diabetes. 2010;59(7):1702–1708. doi: 10.2337/db09-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ricordi C, Lacy PE, Finke EH, Olack BJ, Scharp DW. Automated method for isolation of human pancreatic islets. Diabetes. 1988;37(4):413–420. doi: 10.2337/diab.37.4.413. [DOI] [PubMed] [Google Scholar]
- 36.Murdoch TB, McGhee-Wilson D, Shapiro AM, Lakey JR. Methods of human islet culture for transplantation. Cell Transplant. 2004;13(6):605–617. doi: 10.3727/000000004783983602. [DOI] [PubMed] [Google Scholar]
- 37.Nielsen JH, Brunstedt J, Andersson A, Frimodt-Møller C. Preservation of beta cell function in adult human pancreatic islets for several months in vitro. Diabetologia. 1979;16(2):97–100. doi: 10.1007/BF01225457. [DOI] [PubMed] [Google Scholar]
- 38.Poitout V, et al. A call for improved reporting of human islet characteristics in research articles. Diabetologia. 2019;62(2):209–211. doi: 10.1007/s00125-018-4784-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Movahedi B, Gysemans C, Jacobs-Tulleneers-Thevissen D, Mathieu C, Pipeleers D. Pancreatic duct cells in human islet cell preparations are a source of angiogenic cytokines interleukin-8 and vascular endothelial growth factor. Diabetes. 2008;57(8):2128–2136. doi: 10.2337/db07-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lyon J, et al. Research-focused isolation of human islets from donors with and without diabetes at the Alberta Diabetes Institute IsletCore. Endocrinology. 2016;157(2):560–569. doi: 10.1210/en.2015-1562. [DOI] [PubMed] [Google Scholar]
- 41.Gazdar AF, et al. Continuous, clonal, insulin- and somatostatin-secreting cell lines established from a transplantable rat islet cell tumor. Proc Natl Acad Sci U S A. 1980;77(6):3519–3523. doi: 10.1073/pnas.77.6.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Asfari M, Janjic D, Meda P, Li G, Halban P, Wolheim K. Establishement of 2-mercaptoethanol-dependent differentiated insulin secreting cell lines. Endocrinology. 1992;130(1):167–178. doi: 10.1210/endo.130.1.1370150. [DOI] [PubMed] [Google Scholar]
- 43.Santerre RF, et al. Insulin synthesis in a clonal cell line of simian virus 40-transformed hamster pancreatic beta cells. Proc Natl Acad Sci U S A. 1981;78(7):4339–4343. doi: 10.1073/pnas.78.7.4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hanahan D. Heritable formation of pancreatic beta-cell tumours in transgenic mice expressing recombinant insulin/simian virus 40 oncogenes. Nature. 1985;315(6015):115–122. doi: 10.1038/315115a0. [DOI] [PubMed] [Google Scholar]
- 45.Efrat S, Leiser M, Surana M, Tal M, Fusco-Demane D, Fleischer N. Murine insulinoma cell line with normal glucose-regulated insulin secretion. Diabetes. 1993;42(6):901–907. doi: 10.2337/diab.42.6.901. [DOI] [PubMed] [Google Scholar]
- 46.Miyazaki J, et al. Establishment of a pancreatic beta cell line that retains glucose-inducible insulin secretion: special reference to expression of glucose transporter isoforms. Endocrinology. 1990;127(1):126–132. doi: 10.1210/endo-127-1-126. [DOI] [PubMed] [Google Scholar]
- 47.Wang S, Beattie GM, Mally MI, Lopez AD, Hayek A, Levine F. Analysis of a human fetal pancreatic islet cell line. Transplant Proc. 1997;29(4):2219. doi: 10.1016/S0041-1345(97)00306-0. [DOI] [PubMed] [Google Scholar]
- 48.Halvorsen TL, Leibowitz G, Levine F. Telomerase activity is sufficient to allow transformed cells to escape from crisis. Mol Cell Biol. 1999;19(3):1864–1870. doi: 10.1128/MCB.19.3.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de la Tour D, et al. Beta-cell differentiation from a human pancreatic cell line in vitro and in vivo. Mol Endocrinol. 2001;15(3):476–483. doi: 10.1210/mend.15.3.0604. [DOI] [PubMed] [Google Scholar]
- 50.Narushima M, et al. A human beta-cell line for transplantation therapy to control type 1 diabetes. Nat Biotechnol. 2005;23(10):1274–1282. doi: 10.1038/nbt1145. [DOI] [PubMed] [Google Scholar]
- 51.Hohmeier HE, Newgard CB. Islets for all? Nat Biotechnol. 2005;23(10):1231–1232. doi: 10.1038/nbt1005-1231. [DOI] [PubMed] [Google Scholar]
- 52.Miller DG, Adam MA, Miller AD. Gene transfer by retrovirus vectors occurs only in cells that are actively replicating at the time of infection. Mol Cell Biol. 1990;10(8):4239–4242. doi: 10.1128/MCB.10.8.4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mezza T, et al. Insulin resistance alters islet morphology in nondiabetic humans. Diabetes. 2014;63(3):994–1007. doi: 10.2337/db13-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robitaille K, et al. High-throughput functional genomics identifies regulators of primary human beta cell proliferation. J Biol Chem. 2016;291(9):4614–4625. doi: 10.1074/jbc.M115.683912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hwang IH, et al. Tetraspanin-2 promotes glucotoxic apoptosis by regulating the JNK/β-catenin signaling pathway in human pancreatic β cells. FASEB J. 2016;30(9):3107–3116. doi: 10.1096/fj.201600240RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jin J, et al. Detection of differential proteomes of human beta-cells during islet-like differentiation using iTRAQ labeling. J Proteome Res. 2009;8(3):1393–1403. doi: 10.1021/pr800765t. [DOI] [PubMed] [Google Scholar]
- 57.Hahn WC, Weinberg RA. Rules for making human tumor cells. N Engl J Med. 2002;347(20):1593–1603. doi: 10.1056/NEJMra021902. [DOI] [PubMed] [Google Scholar]
- 58.Roy NS, et al. Telomerase immortalization of neuronally restricted progenitor cells derived from the human fetal spinal cord. Nat Biotechnol. 2004;22(3):297–305. doi: 10.1038/nbt944. [DOI] [PubMed] [Google Scholar]
- 59.Sanvito F, et al. TGF-β1 overexpression in murine pancreas induces chronic pancreatitis and together with TNF-α, triggers insulin-dependent diabetes. Biochem Biophys Res Commun. 1995;217(3):1279–1286. doi: 10.1006/bbrc.1995.2906. [DOI] [PubMed] [Google Scholar]
- 60.Castaing M, Guerci A, Mallet J, Czernichow P, Ravassard P, Scharfmann R. Efficient restricted gene expression in beta cells by lentivirus-mediated gene transfer into pancreatic stem/progenitor cells. Diabetologia. 2005;48(4):709–719. doi: 10.1007/s00125-005-1694-6. [DOI] [PubMed] [Google Scholar]
- 61.Scharfmann R, Xiao X, Heimberg H, Mallet J, Ravassard P. Beta cells within single human islets originate from multiple progenitors. PLoS One. 2008;3(10):e3559. doi: 10.1371/journal.pone.0003559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ravassard P, et al. A new strategy to generate functional insulin-producing cell lines by somatic gene transfer into pancreatic progenitors. PLoS One. 2009;4(3):e4731. doi: 10.1371/journal.pone.0004731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Polak M, Bouchareb-Banaei L, Scharfmann R, Czernichow P. Early pattern of differentiation in the human pancreas. Diabetes. 2000;49(2):225–232. doi: 10.2337/diabetes.49.2.225. [DOI] [PubMed] [Google Scholar]
- 64.Riedel MJ, Asadi A, Wang R, Ao Z, Warnock GL, Kieffer TJ. Immunohistochemical characterisation of cells co-producing insulin and glucagon in the developing human pancreas. Diabetologia. 2012;55(2):372–381. doi: 10.1007/s00125-011-2344-9. [DOI] [PubMed] [Google Scholar]
- 65.Jennings RE, Berry AA, Strutt JP, Gerrard DT, Hanley NA. Human pancreas development. Development. 2015;142(18):3126–3137. doi: 10.1242/dev.120063. [DOI] [PubMed] [Google Scholar]
- 66.Castaing M, Péault B, Basmaciogullari A, Casal I, Czernichow P, Scharfmann R. Blood glucose normalization upon transplantation of human embryonic pancreas into beta-cell-deficient SCID mice. Diabetologia. 2001;44(11):2066–2076. doi: 10.1007/s001250100012. [DOI] [PubMed] [Google Scholar]
- 67.Capito C, et al. Mouse muscle as an ectopic permissive site for human pancreatic development. Diabetes. 2013;62(10):3479–3487. doi: 10.2337/db13-0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lawlor N, et al. Multiomic profiling identifies cis-regulatory networks underlying human pancreatic β cell identity and function. Cell Rep. 2019;26(3):788–801.e6. doi: 10.1016/j.celrep.2018.12.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hastoy B, et al. Electrophysiological properties of human beta-cell lines EndoC-βH1 and -βH2 conform with human beta-cells. Sci Rep. 2018;8(1):16994. doi: 10.1038/s41598-018-34743-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Scharfmann R, et al. Development of a conditionally immortalized human pancreatic β cell line. J Clin Invest. 2014;124(5):2087–2098. doi: 10.1172/JCI72674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Benazra M, et al. A human beta cell line with drug inducible excision of immortalizing transgenes. Mol Metab. 2015;4(12):916–925. doi: 10.1016/j.molmet.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Carlier G, et al. Human Fucci pancreatic beta cell lines: new tools to study beta cell cycle and terminal differentiation. PLoS One. 2014;9(9):e108202. doi: 10.1371/journal.pone.0108202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Culina S, et al. Islet-reactive CD8+ T cell frequencies in the pancreas, but not in blood, distinguish type 1 diabetic patients from healthy donors. Sci Immunol. 2018;3(20):eaao4013. doi: 10.1126/sciimmunol.aao4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Meier JJ, et al. Beta-cell replication is the primary mechanism subserving the postnatal expansion of beta-cell mass in humans. Diabetes. 2008;57(6):1584–1594. doi: 10.2337/db07-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gregg BE, et al. Formation of a human β-cell population within pancreatic islets is set early in life. J Clin Endocrinol Metab. 2012;97(9):3197–3206. doi: 10.1210/jc.2012-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pajalunga D, Puggioni EM, Mazzola A, Leva V, Montecucco A, Crescenzi M. DNA replication is intrinsically hindered in terminally differentiated myotubes. PLoS One. 2010;5(7):e11559. doi: 10.1371/journal.pone.0011559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rieck S, et al. Overexpression of hepatocyte nuclear factor-4α initiates cell cycle entry, but is not sufficient to promote β-cell expansion in human islets. Mol Endocrinol. 2012;26(9):1590–1602. doi: 10.1210/me.2012-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.American Type Culture Collection Standards Development Organization Workgroup ASN-0002. Cell line misidentification: the beginning of the end. Nat Rev Cancer. 2010;10(6):441–448. doi: 10.1038/nrc2852. [DOI] [PubMed] [Google Scholar]
- 79.Yu M, et al. A resource for cell line authentication, annotation and quality control. Nature. 2015;520(7547):307–311. doi: 10.1038/nature14397. [DOI] [PubMed] [Google Scholar]
- 80.Cardozo AK, et al. Cytokines downregulate the sarcoendoplasmic reticulum pump Ca2+ ATPase 2b and deplete endoplasmic reticulum Ca2+, leading to induction of endoplasmic reticulum stress in pancreatic beta-cells. Diabetes. 2005;54(2):452–461. doi: 10.2337/diabetes.54.2.452. [DOI] [PubMed] [Google Scholar]
- 81.Brozzi F, et al. Cytokines induce endoplasmic reticulum stress in human, rat and mouse beta cells via different mechanisms. Diabetologia. 2015;58(10):2307–2316. doi: 10.1007/s00125-015-3669-6. [DOI] [PubMed] [Google Scholar]
- 82.Scoville DW, et al. MLL3 and MLL4 methyltransferases bind to the MAFA and MAFB transcription factors to regulate islet β-cell function. Diabetes. 2015;64(11):3772–3783. doi: 10.2337/db15-0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jian X, Felsenfeld G. Insulin promoter in human pancreatic β cells contacts diabetes susceptibility loci and regulates genes affecting insulin metabolism. Proc Natl Acad Sci U S A. 2018;115(20):E4633–E4641. doi: 10.1073/pnas.1803146115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gonzalez-Duque S, et al. Conventional and neo-antigenic peptides presented by β cells are targeted by circulating naïve CD8+ T cells in type 1 diabetic and healthy donors. Cell Metab. 2018;28(6):946–960.e6. doi: 10.1016/j.cmet.2018.07.007. [DOI] [PubMed] [Google Scholar]
- 85.Diedisheim M, et al. Modeling human pancreatic beta cell dedifferentiation. Mol Metab. 2018;10:74–86. doi: 10.1016/j.molmet.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Oshima M, et al. Virus-like infection induces human β cell dedifferentiation. JCI Insight. 2018;3(3):97732. doi: 10.1172/jci.insight.97732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tsonkova VG, et al. The EndoC-βH1 cell line is a valid model of human beta cells and applicable for screenings to identify novel drug target candidates. Mol Metab. 2018;8:144–157. doi: 10.1016/j.molmet.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pflimlin E, et al. Acute and repeated treatment with 5-PAHSA or 9-PAHSA isomers does not improve glucose control in mice. Cell Metab. 2018;28(2):217–227.e13. doi: 10.1016/j.cmet.2018.05.028. [DOI] [PubMed] [Google Scholar]
- 89.Hansson SF, et al. Secretagogin is increased in plasma from type 2 diabetes patients and potentially reflects stress and islet dysfunction. PLoS One. 2018;13(4):e0196601. doi: 10.1371/journal.pone.0196601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Coskun T, et al. LY3298176, a novel dual GIP and GLP-1 receptor agonist for the treatment of type 2 diabetes mellitus: from discovery to clinical proof of concept. Mol Metab. 2018;18:3–14. doi: 10.1016/j.molmet.2018.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.McCluskey JT, Hamid M, Guo-Parke H, McClenaghan NH, Gomis R, Flatt PR. Development and functional characterization of insulin-releasing human pancreatic beta cell lines produced by electrofusion. J Biol Chem. 2011;286(25):21982–21992. doi: 10.1074/jbc.M111.226795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Milton NG, Chilumuri A, Rocha-Ferreira E, Nercessian AN, Ashioti M. Kisspeptin prevention of amyloid-β peptide neurotoxicity in vitro. ACS Chem Neurosci. 2012;3(9):706–719. doi: 10.1021/cn300045d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vasu S, McClenaghan NH, McCluskey JT, Flatt PR. Effects of lipotoxicity on a novel insulin-secreting human pancreatic β-cell line, 1.1B4. Biol Chem. 2013;394(7):909–918. doi: 10.1515/hsz-2013-0115. [DOI] [PubMed] [Google Scholar]
- 94.Mellado-Gil JM, et al. The type 2 diabetes-associated HMG20A gene is mandatory for islet beta cell functional maturity. Cell Death Dis. 2018;9(3):279. doi: 10.1038/s41419-018-0272-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mohan S, Moffett RC, Thomas KG, Irwin N, Flatt PR. Vasopressin receptors in islets enhance glucose tolerance, pancreatic beta-cell secretory function, proliferation and survival. Biochimie. 2019;158:191–198. doi: 10.1016/j.biochi.2019.01.008. [DOI] [PubMed] [Google Scholar]
- 96.Morán I, et al. Human β cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab. 2012;16(4):435–448. doi: 10.1016/j.cmet.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Akerman I, et al. Human pancreatic beta cell lncRNAs control cell-specific regulatory networks. Cell Metab. 2017;25(2):400–411. doi: 10.1016/j.cmet.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schulthess FT, et al. CXCL10 impairs beta cell function and viability in diabetes through TLR4 signaling. Cell Metab. 2009;9(2):125–139. doi: 10.1016/j.cmet.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 99.Sun J, et al. Pancreatic β-cells limit autoimmune diabetes via an immunoregulatory antimicrobial peptide expressed under the influence of the gut microbiota. Immunity. 2015;43(2):304–317. doi: 10.1016/j.immuni.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 100.Eizirik DL, et al. The human pancreatic islet transcriptome: expression of candidate genes for type 1 diabetes and the impact of pro-inflammatory cytokines. PLoS Genet. 2012;8(3):e1002552. doi: 10.1371/journal.pgen.1002552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Patry M, et al. βig-h3 represses T-cell activation in type 1 diabetes. Diabetes. 2015;64(12):4212–4219. doi: 10.2337/db15-0638. [DOI] [PubMed] [Google Scholar]
- 102.Konrad K, et al. Cystic fibrosis-related diabetes compared with type 1 and type 2 diabetes in adults. Diabetes Metab Res Rev. 2013;29(7):568–575. doi: 10.1002/dmrr.2429. [DOI] [PubMed] [Google Scholar]
- 103.Schuit F, Moens K, Heimberg H, Pipeleers D. Cellular origin of hexokinase in pancreatic islets. J Biol Chem. 1999;274(46):32803–32809. doi: 10.1074/jbc.274.46.32803. [DOI] [PubMed] [Google Scholar]
- 104.Guo JH, et al. Glucose-induced electrical activities and insulin secretion in pancreatic islet β-cells are modulated by CFTR. Nat Commun. 2014;5:4420. doi: 10.1038/ncomms5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Edlund A, Pedersen MG, Lindqvist A, Wierup N, Flodström-Tullberg M, Eliasson L. CFTR is involved in the regulation of glucagon secretion in human and rodent alpha cells. Sci Rep. 2017;7(1):90. doi: 10.1038/s41598-017-00098-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hart NJ, et al. Cystic fibrosis-related diabetes is caused by islet loss and inflammation. JCI Insight. 2018;3(8):98240. doi: 10.1172/jci.insight.98240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Polakiewicz RD. Antibodies: the solution is validation. Nature. 2015;518(7540):483. doi: 10.1038/518483b. [DOI] [PubMed] [Google Scholar]
- 108.Uhlen M, et al. A proposal for validation of antibodies. Nat Methods. 2016;13(10):823–827. doi: 10.1038/nmeth.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jacobs K, et al. Higher-density culture in human embryonic stem cells results in DNA damage and genome instability. Stem Cell Reports. 2016;6(3):330–341. doi: 10.1016/j.stemcr.2016.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chakrabarti SK, Mirmira RG. Transcription factors direct the development and function of pancreatic beta cells. Trends Endocrinol Metab. 2003;14(2):78–84. doi: 10.1016/S1043-2760(02)00039-5. [DOI] [PubMed] [Google Scholar]
- 111.Servitja JM, Ferrer J. Transcriptional networks controlling pancreatic development and beta cell function. Diabetologia. 2004;47(4):597–613. doi: 10.1007/s00125-004-1368-9. [DOI] [PubMed] [Google Scholar]
- 112.Taylor BL, Liu FF, Sander M. Nkx6.1 is essential for maintaining the functional state of pancreatic beta cells. Cell Rep. 2013;4(6):1262–1275. doi: 10.1016/j.celrep.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jia S, et al. Insm1 cooperates with Neurod1 and Foxa2 to maintain mature pancreatic β-cell function. EMBO J. 2015;34(10):1417–1433. doi: 10.15252/embj.201490819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Prado CL, Pugh-Bernard AE, Elghazi L, Sosa-Pineda B, Sussel L. Ghrelin cells replace insulin-producing beta cells in two mouse models of pancreas development. Proc Natl Acad Sci U S A. 2004;101(9):2924–2929. doi: 10.1073/pnas.0308604100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Keller DM, et al. Characterization of pancreatic transcription factor Pdx-1 binding sites using promoter microarray and serial analysis of chromatin occupancy. J Biol Chem. 2007;282(44):32084–32092. doi: 10.1074/jbc.M700899200. [DOI] [PubMed] [Google Scholar]
- 116.Helman A, et al. p16(Ink4a)-induced senescence of pancreatic beta cells enhances insulin secretion. Nat Med. 2016;22(4):412–420. doi: 10.1038/nm.4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Albagli O, et al. New α- SIN γ-retrovectors for safe transduction specfic transgene expression in pancreatic β cell lines. BMC Biotechnol. 2019;19(1):35. doi: 10.1186/s12896-019-0531-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fleischer N, et al. Functional analysis of a conditionally transformed pancreatic beta-cell line. Diabetes. 1998;47(9):1419–1425. doi: 10.2337/diabetes.47.9.1419. [DOI] [PubMed] [Google Scholar]
- 119.Wu HL, et al. Reversible immortalization of rat pancreatic β cells with a novel immortalizing and tamoxifen-mediated self-recombination tricistronic vector. J Biotechnol. 2011;151(3):231–241. doi: 10.1016/j.jbiotec.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 120.Eves EM, Kwon J, Downen M, Tucker MS, Wainer BH, Rosner MR. Conditional immortalization of neuronal cells from postmitotic cultures and adult CNS. Brain Res. 1994;656(2):396–404. doi: 10.1016/0006-8993(94)91484-2. [DOI] [PubMed] [Google Scholar]
- 121.Zhang Y, Nuglozeh E, Touré F, Schmidt AM, Vunjak-Novakovic G. Controllable expansion of primary cardiomyocytes by reversible immortalization. Hum Gene Ther. 2009;20(12):1687–1696. doi: 10.1089/hum.2009.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Delgado JP, et al. Long-term controlled immortalization of a primate hepatic progenitor cell line after Simian virus 40 T-Antigen gene transfer. Oncogene. 2005;24(4):541–551. doi: 10.1038/sj.onc.1208089. [DOI] [PubMed] [Google Scholar]
- 123.Casanovas O, Hager JH, Chun MG, Hanahan D. Incomplete inhibition of the Rb tumor suppressor pathway in the context of inactivated p53 is sufficient for pancreatic islet tumorigenesis. Oncogene. 2005;24(44):6597–6604. doi: 10.1038/sj.onc.1208823. [DOI] [PubMed] [Google Scholar]
- 124.Nishimura W, et al. A switch from MafB to MafA expression accompanies differentiation to pancreatic beta-cells. Dev Biol. 2006;293(2):526–539. doi: 10.1016/j.ydbio.2006.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Asplund K, Westman S, Hellerström C. Glucose stimulation of insulin secretion from the isolated pancreas of foetal and newborn rats. Diabetologia. 1969;5(4):260–262. doi: 10.1007/BF01212095. [DOI] [PubMed] [Google Scholar]
- 126.Blum B, Hrvatin S, Schuetz C, Bonal C, Rezania A, Melton DA. Functional beta-cell maturation is marked by an increased glucose threshold and by expression of urocortin 3. Nat Biotechnol. 2012;30(3):261–264. doi: 10.1038/nbt.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Stolovich-Rain M, et al. Weaning triggers a maturation step of pancreatic β cells. Dev Cell. 2015;32(5):535–545. doi: 10.1016/j.devcel.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 128.Jacovetti C, Matkovich SJ, Rodriguez-Trejo A, Guay C, Regazzi R. Postnatal β-cell maturation is associated with islet-specific microRNA changes induced by nutrient shifts at weaning. Nat Commun. 2015;6:8084. doi: 10.1038/ncomms9084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Szabat M, et al. Reduced insulin production relieves endoplasmic reticulum stress and induces β cell proliferation. Cell Metab. 2016;23(1):179–193. doi: 10.1016/j.cmet.2015.10.016. [DOI] [PubMed] [Google Scholar]
- 130.Puri S, et al. Replication confers β cell immaturity. Nat Commun. 2018;9(1):485. doi: 10.1038/s41467-018-02939-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.van der Meulen T, et al. Urocortin3 mediates somatostatin-dependent negative feedback control of insulin secretion. Nat Med. 2015;21(7):769–776. doi: 10.1038/nm.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Takaki R, et al. Isolation of glucagon-secreting cell lines by cloning insulinoma cells. In Vitro Cell Dev Biol. 1986;22(3 pt 1):120–126. doi: 10.1007/BF02623498. [DOI] [PubMed] [Google Scholar]
- 133.Drucker DJ, Philippe J, Mojsov S. Proglucagon gene expression and posttranslational processing in a hamster islet cell line. Endocrinology. 1988;123(4):1861–1867. doi: 10.1210/endo-123-4-1861. [DOI] [PubMed] [Google Scholar]
- 134.Efrat S, Teitelman G, Anwar M, Ruggerio D, Hanahan D. Glucagon gene regulatory region directs oncoprotein expression to neurons and pancreatic alpha cells. Neuron. 1988;1(7):605–613. doi: 10.1016/0896-6273(88)90110-9. [DOI] [PubMed] [Google Scholar]
- 135.Powers AC, Efrat S, Mojsov S, Spector D, Habener JF, Hanahan D. Proglucagon processing similar to normal islets in pancreatic alpha-like cell line derived from transgenic mouse tumor. Diabetes. 1990;39(4):406–414. doi: 10.2337/diab.39.4.406. [DOI] [PubMed] [Google Scholar]
- 136.Melloul D, Marshak S, Cerasi E. Regulation of insulin gene transcription. Diabetologia. 2002;45(3):309–326. doi: 10.1007/s00125-001-0728-y. [DOI] [PubMed] [Google Scholar]
- 137.Xu Z, Lefevre GM, Felsenfeld G. Chromatin structure, epigenetic mechanisms and long-range interactions in the human insulin locus. Diabetes Obes Metab. 2012;14(suppl 3):1–11. doi: 10.1111/j.1463-1326.2012.01645.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gosmain Y, Cheyssac C, Heddad Masson M, Dibner C, Philippe J. Glucagon gene expression in the endocrine pancreas: the role of the transcription factor Pax6 in α-cell differentiation, glucagon biosynthesis and secretion. Diabetes Obes Metab. 2011;13(suppl 1):31–38. doi: 10.1111/j.1463-1326.2011.01445.x. [DOI] [PubMed] [Google Scholar]
- 139.Pauerstein PT, Park KM, Peiris HS, Wang J, Kim SK. Research resource: genetic labeling of human islet alpha cells. Mol Endocrinol. 2016;30(2):248–253. doi: 10.1210/me.2015-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ishihara H, Maechler P, Gjinovci A, Herrera PL, Wollheim CB. Islet beta-cell secretion determines glucagon release from neighbouring alpha-cells. Nat Cell Biol. 2003;5(4):330–335. doi: 10.1038/ncb951. [DOI] [PubMed] [Google Scholar]
- 141.Kirkegaard JS, et al. Xenotropic retrovirus Bxv1 in human pancreatic β cell lines. J Clin Invest. 2016;126(3):1109–1113. doi: 10.1172/JCI83573. [DOI] [PMC free article] [PubMed] [Google Scholar]