Abstract
Nature exploits cage-like proteins for a variety of biological purposes, from molecular packaging and cargo delivery to catalysis. These cage-like proteins are of immense importance in nanomedicine due to their propensity to self-assemble from simple identical building blocks to highly ordered architecture and the design flexibility afforded by protein engineering. However, delivery of protein nanocages to the renal tubules remains a major challenge because of the glomerular filtration barrier, which effectively excludes conventional size nanocages. Here, we show that DNA-binding protein from starved cells (Dps) — the extremely small archaeal antioxidant nanocage — is able to cross the glomerular filtration barrier and is endocytosed by the renal proximal tubules. Using a model of endotoxemia, we present an example of the way in which proximal tubule–selective Dps nanocages can limit the degree of endotoxin-induced kidney injury. This was accomplished by amplifying the endogenous antioxidant property of Dps with addition of a dinuclear manganese cluster. Dps is the first-in-class protein cage nanoparticle that can be targeted to renal proximal tubules through glomerular filtration. In addition to its therapeutic potential, chemical and genetic engineering of Dps will offer a nanoplatform to advance our understanding of the physiology and pathophysiology of glomerular filtration and tubular endocytosis.
Keywords: Nephrology
Keywords: Diagnostic imaging, Drug therapy, Nanotechnology
Introduction
One of the major challenges in translational research is the development of therapeutics that are precisely targeted to the desired site of action. Precise targeting is also important for minimizing systemic side effects. However, optimizing the delivery of individual prototype drugs is time consuming and sometimes not feasible. The use of cage-like protein nanoparticles holds great promise for overcoming this bottleneck problem (1). First, by harnessing their intrinsic tropism or by tagging a molecular zip code, site specificity can be achieved. Second, these nanoparticles can incorporate a variety of payloads, thus offering the ability to treat various disease processes. Conversely, many disease conditions may share similar pathophysiology (e.g., ischemia-reperfusion injury in various organs). In that case, the same therapeutics can be mounted to different nanoparticles in order to attain tissue specificity. Finally, the internal cavity of nanoparticles can be used as a nanoscale reaction vessel, template for material synthesis, or a place to sequester cytotoxic molecules, providing layers of potential applications.
In the kidney, proximal tubules are particularly susceptible to various nephrotoxic insults, such as endotoxin (2, 3). Therefore, development of proximal tubule–specific nanoparticles is highly desirable. However, the existence of the glomerular filtration barrier poses a major challenge to delivering nanoparticles to the proximal tubules. For example, ferritin, a prototype of very small cage-like protein (diameter 12 nm), is still too large to pass through the glomerular filtration barrier (4–6). To date, only non–protein based synthetic nanoparticles, such as quantum dots, are known to pass the filtration barrier (7–9). These synthetic nanoparticles have no modifiable inner space and present biocompatibility and biodegradability challenges.
In this study, we report that DNA-binding protein from starved cells (Dps), a very small protein-based nanoparticle, passes through the glomeruli and reaches the proximal tubule from the apical side. Dps is a hollow dodecameric protein cage that possesses putative cytoprotective properties in its native form. By adding a dinuclear manganese (Mn) cluster, we demonstrate that Dps can be converted into a powerful antioxidant that mitigates proximal tubular damage in a model of sepsis-induced kidney injury. This is the first-in-class protein cage nanoparticle that can be targeted to renal proximal tubules via glomerular filtration.
Results
Dps passes the glomerular filtration barrier and reaches the proximal tubule.
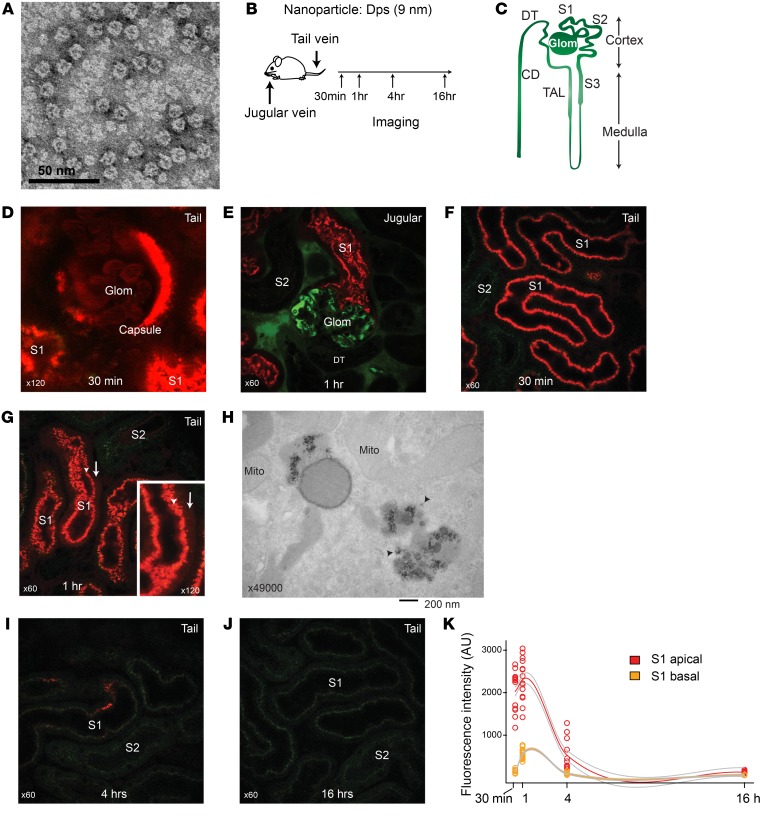
We have previously identified, isolated, and characterized the archaeal antioxidant Dps from hyperthermophilic acidophile Sulfolobus solfataricus (10–12). The Dps protein self assembles into a hollow dodecameric structure with tetrahedral symmetry, giving rise to one of the smallest nature-derived cages. The outer shell diameter is 9 nm, and the interior diameter is 5 nm (Figure 1A). The cut-off diameter of glomerular filtration for a spheroidal molecule is thought to be 6 to 7 nm based on studies of synthetic nanoparticles, though deviations from this general size dependency exist (7–9, 13–16). Using Texas red–conjugated Dps and intravital microscopy, we tested to determine whether Dps can cross the glomerular filtration barrier (Figure 1, B and C). We found that Dps was readily filtered and reabsorbed by the proximal tubules (Figure 1, D and E). Within 30 minutes of systemic administration, Dps localized to the S1 segment of proximal tubules, predominantly in the endosome/lysosome compartments (Figure 1F). In parallel, Dps decreased from the vasculature during that time frame. After 1 hour, a diffuse cytoplasmic Dps signal became apparent, suggesting that some Dps escaped from the endosome/lysosome compartments (Figure 1, G and H). Four hours after injection, the overall Dps signal in the proximal tubules was reduced. This occurred without a concomitant increase of Dps signal in the bloodstream, suggesting that Dps was metabolized in the proximal tubules, although transcytosis and recycling of Dps back into circulation cannot be fully ruled out (Figure 1, I–K, and Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI127511DS1). In addition, there was minimal Dps signal in the lumen of distal tubules, indicating that filtered Dps was effectively endocytosed by the upstream proximal tubules (Supplemental Figure 1B). The residual Dps was concentrated in the inner medulla and appeared in the final urine (Supplemental Figure 1, A and C–J). No difference was found in Dps signal kinetics between jugular vein and tail vein injections (Supplemental Figure 1K).
Figure 1. Dps is filtered through glomeruli and endocytosed by proximal tubules.
(A) Transmission electron microscopy image of S. solfataricus Dps. (B) Overview of intravital imaging protocol. (C) Schematic of renal tubule structure. Glom, glomerulus; S1, S2, and S3, proximal tubule subsegments; TAL, thick ascending loop of Henle; DT, distal tubule; CD, collecting duct. (D) Representative image of glomerulus and S1 proximal tubules 30 minutes after Dps i.v. (Texas red; n = 3 independent experiments). Mice were injected with Dps via tail vein, and the distribution of Dps was determined with intravital 2-photon microscopy. Note that male mice have extension of S1 segment into the Bowman’s capsule in the glomerulus. (E) Dps (red) and 70 kDa dextran (green) were injected via jugular vein. In this experiment, the mouse kidney was freshly dissected 60 minutes after Dps injection and imaged ex vivo in order to determine the distribution of Dps in the deep cortex that is beyond the reach of intravital 2-photon microscopy. (F and G) Intravital time-course imaging under indicated time points. Arrowheads point to the endosome/lysosome layer (S1 apical). Arrows point to cytoplasmic Dps signal beyond the endosome/lysosome layer (S1 basal). (H) Electron microscopy image of S1 proximal tubule 60 minutes after Dps i.v. Arrow points to Dps in the cytoplasm. (I and J) Intravital imaging of the kidney under indicated time points. (K) Quantification of Dps fluorescence intensity for the indicated time points and subcellular regions. Locally weighted regression curve fitting was applied for generating the trajectories and error lines (gray).
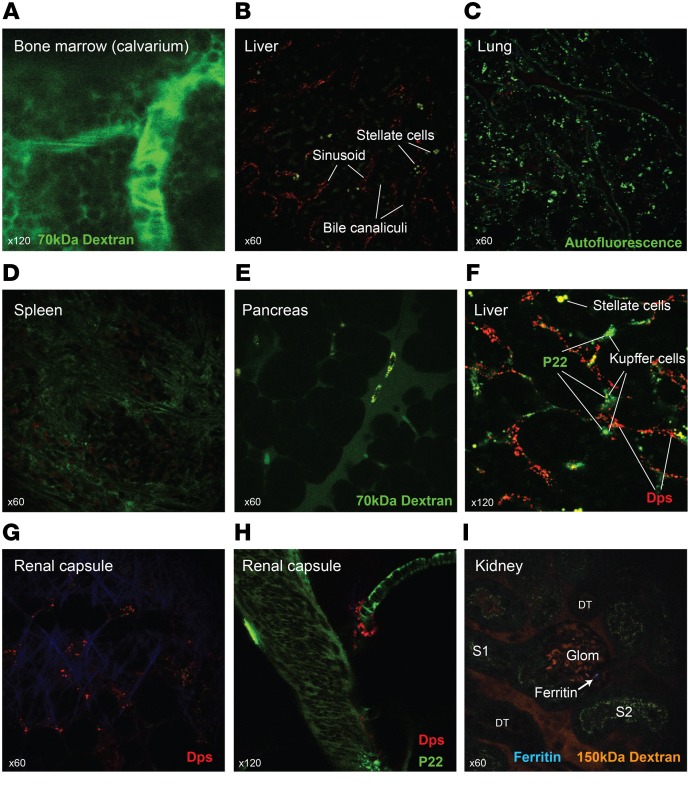
Importantly, other organs showed substantially less Dps signal compared with the kidney. The organs examined included the bone marrow, liver, lung, spleen, pancreas, brain, and heart (Figure 2, A–E, and Supplemental Figure 2, A–F). As is the case for various other nanoparticles, the liver had some accumulation of Dps, albeit much less in quantity compared with that in renal proximal tubules. More specifically, Dps was taken up by the hepatic endothelial cells, not by Kupffer cells (Figure 2B and Supplemental Figure 3, A and B). This was best exemplified when Dps was coadministered with another nanoparticle, P22, resulting in differential distribution of the 2 species in the endothelium and Kupffer cell, respectively (Figure 2F; P22 is a 60 nm icosahedral nanoparticle, ref. 17–19; see Methods regarding the synthesis and characteristics of P22). Distribution of P22 in the kidney was restricted to nonendothelial interstitial cells, as shown in Supplemental Figure 3, C–E. No filtration or tubular uptake of P22 was observed). In contrast to the Dps uptake by hepatic endothelial cells, renal endothelial cells did not accumulate Dps. Additionally, sporadic uptake of Dps by supporting cells around blood vessels was observed in various organs including the ones in the renal capsule (Figure 2, G and H, Supplemental Figure 3F, and Supplemental Video 1).
Figure 2. Distribution of Dps in various organs.
(A) Intravital imaging of calvarium bone marrow niche. No Dps was observed in the bone marrow space. Green 70 kDa dextran was used to identify the structure. (B–E) Ex vivo imaging of liver, lung, spleen, and pancreas, approximately 1 hour after Dps i.v. injection. In some experiments, green 70 kDa dextran was also administered. (F) Dps and P22 (Alexa Fluor 488; green) were coadministered i.v. and imaged 1 hour later. Stellate cells exhibit distinct autofluorescence signal. (G and H) Intravital imaging of the renal capsule 1 hour after Dps alone or Dps and P22 coadministration. (I) Ferritin (Alexa Fluor 405; blue) and 150 kDa TRITC dextran (orange) were administered via jugular vein. Imaging of freshly dissected kidneys revealed occasional ferritin in the glomeruli and interstitium, but not in renal epithelial cells, including S1. n = 3 independent experiments.
In summary, Dps preferentially accumulated in the kidney via glomerular filtration followed by proximal tubular endocytosis. To our knowledge, this is the first example of a filterable cage-like protein nanoparticle. The slightly larger cage-like protein, ferritin, did not pass through the glomeruli, consistent with previous reports (refs. 5, 6, and Figure 2I; see Methods for synthesis of the ferritin nanoparticle).
Modified Dps confers renoprotection in a model of sepsis-induced kidney injury.
We next demonstrated the therapeutic properties of Dps in a model of endotoxemic kidney injury. Systemically administered endotoxin (LPS) was filtered through the glomeruli and caused oxidative damage to proximal tubules during the early phase of sepsis (~4 hours) (20–22). Because Dps has putative catalase-like functions and targets the injury-prone proximal tubules, we hypothesized that Dps has the ability to limit kidney injury by reducing oxidative stress.
To this end, we synthesized 2 types of Dps: unmodified Dps and manganese-bound Dps (MnDps). Unmodified Dps is believed to stabilize DNA through the formation of Dps-DNA complexes in archaea and bacteria (23, 24). In addition, Dps oxidizes ambient iron from Fe2+ to Fe3+ and creates an iron oxide mineral at the ferroxidase center in each Dps subunit. This oxidation and mineralization reaction of iron requires the reduction of H2O2 (10). Therefore, in the presence of tissue oxidative stress, Dps might provide antioxidant activity, depending on the availability of free iron. In contrast, MnDps exerts antioxidant effects similar to those of Mn catalase. The added Mn is tightly bound to the ferroxidase center and not displaced by equimolar amounts of iron (25). Therefore, MnDps could provide stable and powerful antioxidant effects irrespective of free iron availability in vivo.
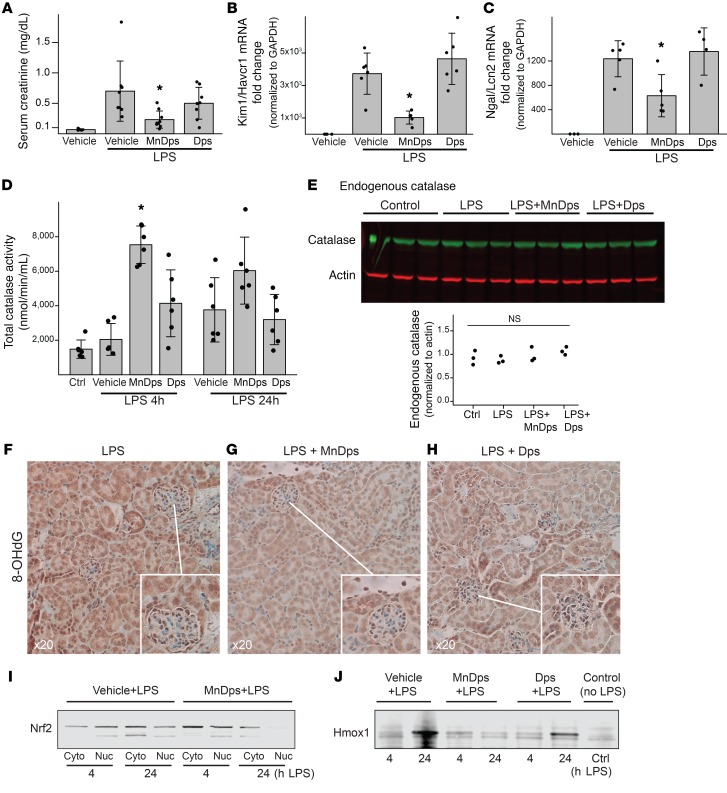
In this model of endotoxemic kidney injury, we found that MnDps treatment significantly improved kidney function as compared with unmodified Dps or vehicle control (Figure 3, A–C, and Supplemental Figure 3, G–I). The therapeutic effect of MnDps was dose dependent (Supplemental Figure 3, J and K). As expected, kidney tissues from mice treated with MnDps had the highest total catalase activity, as measured by an assay that accounts for both endogenous protein catalase and Dps-mediated H2O2 reduction (Figure 3D). Endogenous protein catalase levels were unaffected, suggesting that MnDps-derived catalase activity was the main contributor to the overall reduction of tubular oxidative stress (Figure 3, E–H). Note also that, because of the reduction of oxidative stress, activation of nuclear factor, erythroid 2 like 2 (Nrf2, also known as Nfe2l2), the master regulator of antioxidant responses, and its downstream target Hmox1 (26) were not observed (Figure 3, I and J). There was no difference in the numbers of neutrophils, macrophages, and T and B cells in MnDps-treated mice compared with mice treated with vehicle or unmodified Dps (Supplemental Figure 4). The biodistribution of Dps after systemic endotoxin challenge remained the same as that observed in control animals (Supplemental Figure 3L). Additionally, because endotoxin can increase the permeability of the filtration barrier, we also examined the biodistribution of ferritin after endotoxin challenge. Ferritin remained nonpermeant and was taken up by mobile macrophages (Supplemental Figure 3M). Collectively, these data support a unique role for MnDps as a proximal tubule–specific antioxidant.
Figure 3. MnDps reduces endotoxin-induced renal tissue damage.
(A) Serum creatinine levels 24 hours after 5 mg/kg LPS i.p. with 18 mg/kg unmodified Dps, MnDps, or PBS vehicle i.v. Administration of Dps, MnDps, or vehicle was done via tail vein immediately before LPS i.p. injection. *P < 0.05 vs. LPS+vehicle or LPS+Dps, 1-way ANOVA followed by pairwise t tests with corrections for multiple testing using the Benjamini and Hochberg procedure. n = 8 per condition. Error bars show SD. (B and C) Kidney tissue Kim1/Havcr1 and Ngal/Lcn2 levels under indicated conditions 24 hours after 5 mg/kg LPS i.p., as determined with quantitative PCR. n = 4–5 per condition as depicted with dot plots. *P < 0.05 vs. LPS+vehicle or LPS+Dps. (D) Total catalase activity levels (endogenous and exogenous catalase activities) are shown for indicated conditions. *P < 0.05 vs. LPS 4 hours + vehicle. (E) Endogenous protein catalase levels were determined by Western blot. LPS, 4 hours; catalase, MW, 60 kDa; actin, 42 kDa, n = 3 per condition. (F–H) Staining for 8-OHdG, a marker of oxidative stress/oxidized DNA and RNA, under indicated conditions (LPS 4 hours). Insets provide enlarged views, ×40. n = 3 per condition. (I) Mice were treated under indicated conditions, and kidney tissues were fractionated for cytoplasm and nuclei and analyzed by Western blot for Nrf2 (MW, ~100 kDa). MnDps treatment reduced nuclear translocation of Nrf2 as compared with vehicle control after LPS challenge for indicated durations. (J) Western blot analysis of kidney tissues for Hmox1 under indicated conditions (MW, 32 kDa). Induction of Hmox1 was limited with MnDps treatment.
Characterization of in vivo Dps effects.
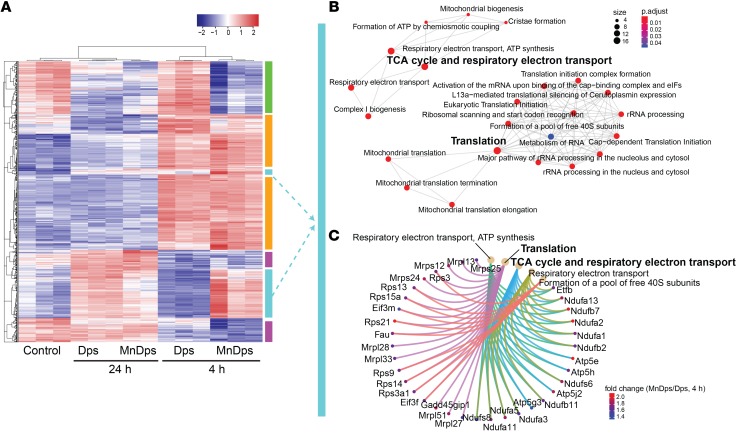
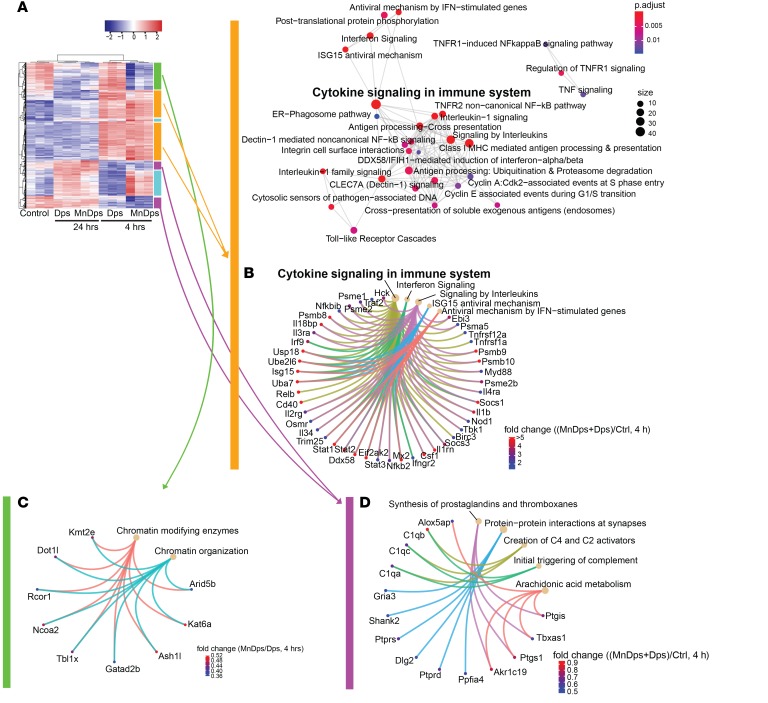
Tolerability and potential adverse effects are major concerns for any type of nanoparticle therapy. Therefore, we next performed global transcriptomics analysis of the kidney from nonseptic mice treated with unmodified Dps, MnDps, or vehicle control. We found that multiple genes were differentially expressed as a result of Dps/MnDps treatment and that these changes were particularly pronounced during the early course of exposure (4 hours; Figure 4A). When comparing unmodified Dps with MnDps, MnDps-treated kidneys had higher gene expression levels in several pathways, including ribosome biogenesis/translation, downregulation of which is crucially involved in sepsis pathology (ref. 27, Figure 4, B and C, and Supplemental Figure 5, C and E). We also noted some activation of innate immune responses in both Dps- and MnDps-treated kidneys (Figure 5, A–D, and Supplemental Figure 5). Whether such responses confer positive immunomodulatory effects or result in low-grade adverse immunogenic reaction remains to be determined. Histopathology analysis revealed no evidence of morphologic damage or infiltration of immune cells up to 24 hours after Dps/MnDps treatment (neutrophils, macrophages, T and B cells; Supplemental Figure 6).
Figure 4. Transcriptomic changes induced by Dps exposure in the kidney.
(A) RNA-Seq analysis of kidney tissues obtained from mice treated with vehicle control, Dps, or MnDps for indicated durations. Heatmap generated from top 1000 genes by ANOVA is shown (n = 3 per condition, 1 MnDps 24-hour sample omitted from the final analysis). The color-coded vertical bars correspond to B and C and Figure 5, in which identical colors are used. (B and C) Pathway enrichment analysis for a set of genes that are upregulated by MnDps treatment, but not by Dps, at 4 hours. The diameter and color of each node indicate gene size and FDR adjusted P values, respectively. (C) Specific genes that contributed to the enriched pathways are depicted. Fold change = MnDps/Dps (4 hours).
Figure 5. Transcriptomic changes induced by Dps exposure in the kidney.
(A and B) Genes and pathways that are upregulated by both MnDps and Dps treatment at 4 hours are shown. Fold change = (mean of MnDps and Dps)/control (4 hours). (C) Genes and pathways that are downregulated by MnDps treatment, but not by Dps, at 4 hours are shown. Fold change = MnDps/Dps (4 hours). (D) Genes and pathways that are downregulated by both MnDps and Dps at 4 hours. Fold change = (mean of MnDps and Dps)/control (4 hours).
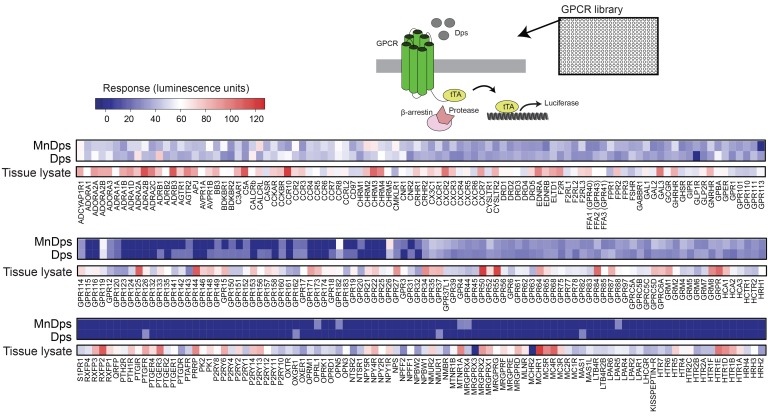
To extend our safety analyses, we next performed a large-scale GPCR screening and examined potential interactions between Dps and GPCRs (Figure 6). GPCRs constitute the largest class of cell-surface receptors, including a diverse range of cytokine/chemokine receptors, and are thus an important class of cell-surface receptors to interrogate (28). Reassuringly, of all 300+ GPCRs assayed, we found no significant activation in any of the receptors by Dps or MnDps. In summary, Dps was well tolerated and had no overt adverse effects.
Figure 6. Dps does not activate GPCRs.
PRESTO-Tango GPCR assay system consists of 315 constructs in individual plasmids fused to a cleavable tTA transcription factor. These plasmids were transfected into 293T cell–derived cell line HTLA, which expresses a β-arrestin2-TEV fusion gene and tTA-induced luciferase reporter. When a specific GPCR is activated, the β-arrestin2-TEV fusion gene is recruited to the membrane-bound GPCR, where it cleaves tTA, allowing for its transport to the nucleus and subsequent activation of the tTA-activated luciferase reporter gene. Using a substrate to quantify luciferase expression, the activation of GPCRs by agonists was measured (median values of quadruplicates; 2 independent experiments). As a reference, activation of various GPCRs induced by kidney tissue lysates is shown in the separate columns.
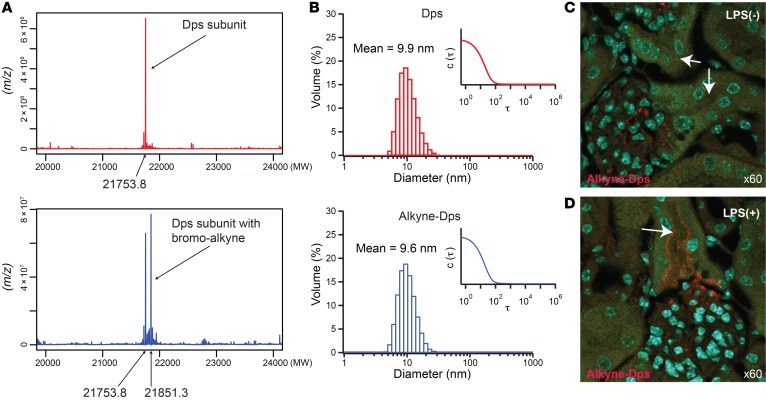
Finally, we present an example of Dps modulation whereby its biodistribution markedly changed. This was done by adding an alkyne moiety to the Cys101 residue (Figure 7A). This modification did not significantly increase the size of Dps (Figure 7B). However, alkyne modification rendered the glomerular filtration barrier much less permeable to Dps, as indicated by the absence of alkyne-Dps signal in the proximal tubule of healthy animals (Figure 7C). Tubular alkyne-Dps signal was detected only in endotoxin-treated mice, indicating that endotoxin disrupted the filtration barrier (Figure 7D). What constitutes the filtration barrier remains controversial (7–9, 13–15, 29–37), and determining the cause of the filtration pattern change of alkyne-Dps is beyond the scope of this study. Notwithstanding, our findings underscore the importance of even minor physicochemical modifications and their effects on the glomerular permeability of nanoparticles in vivo. This also underscores the need for careful experimental determination of glomerular permeability of each nanoparticle regardless of size. In addition to their effects on glomerular permeability, physicochemical alterations can also affect the differential uptake of substances by various tubular segments. This is demonstrated using neutral and anionic low molecular weight dextran as a model molecule (Supplemental Figure 7, A–D). Collectively, these studies illustrate the challenges and promises of designing nanoparticles that target renal proximal tubules.
Figure 7. Modulation of surface characteristics and its effects on biodistribution.
(A) Synthesis of alkyne-Dps. Deconvoluted mass spectrometry data for Dps subunit (MW, 21753.8) and bromo-alkyne–labeled Dps subunit (N-propargyl bromoacetamide; MW, 21851.3) are shown. (B) Volume-averaged hydrodynamic diameter and corresponding correlation function (inset) of Dps and that labeled with N-propargyl bromoacetamide (alkyne-Dps) measured with dynamic light scattering. (C and D) Differential distribution of alkyne-Dps in vivo with and without LPS challenge is shown (red). Arrows point to S1 proximal tubules where alkyne-Dps was detected only in LPS-challenged mice. Alkyne-Dps and LPS were administered i.v. and i.p., respectively, 4 hours before tissue harvest. To visualize alkyne-Dps, Alexa Fluor 555–azide was conjugated to alkyne-Dps using copper(I)-catalyzed azide-alkyne cycloaddition (click chemistry) after tissue harvest. Nuclei were stained blue with DAPI; green shows autofluorescence.
Discussion
In this study, we investigated the in vivo effects of Dps — the smallest class of cage-like protein nanoparticles. We found that Dps is readily filtered and transiently accumulates in the proximal tubules. While Dps did not exert antioxidant effects in its native form in vivo, addition of Mn to the ferroxidase center resulted in reduction of oxidative stress and significant renoprotection in an animal model of sepsis. Even though sepsis is a systemic disease, the use of organ-targeted nanoparticles is essential in order to avoid potential off-target effects. For example, whereas antioxidant therapy is beneficial to damaged renal epithelial cells, unintended delivery of antioxidants to immune cells could negatively affect their ability to fight pathogens.
We noted that the fluorescence signal from Texas red– and Alexa Fluor–conjugated Dps was attenuated after the 3- to 4-hour time points. However, MnDps-mediated antioxidant effects and catalase-like activity were observed up to 24 hours after LPS. Whether this was due to ongoing catalytic activity derived from residual Dps subunits or secondary gene expression changes is unclear (e.g., reduced gene expression of oxidizing enzymes; the fluorophores used are stable in a wide range of pH in vivo, including the kidney [refs. 38, 39], and thus, fluorescence artifacts are less likely).
Nanoparticle delivery to the proximal tubules is a product of the interplay among glomerular filtration, tubular reabsorption, and secretion (40). The use of intravital microscopy allowed high spatial and temporal resolution and was crucial for determining the contribution of these various processes to the final delivery of Dps to the proximal tubules. Microscopic examination further revealed axial differences in proximal tubule endocytosis of nanoparticles. Dps localized to the S1 proximal tubule, but not to S2/S3 subsegments. Importantly, S1 is also the primary site of endotoxin uptake, which in turn causes oxidative damage to downstream S2/S3 subsegments (20). We have previously proposed a model in which S1 acts as the sensor and sink for endotoxin in the filtrate (3). Activated S1 then mediates injury signals to the S2/S3. We speculate that MnDps therapy abrogated the propagation of injury signals by restoring the redox balance in S1 with MnDps catalase activity. Interestingly, S2/S3, but not S1, subsegments were enriched with peroxisomes and endogenous catalase (refs. 20, 41, and Supplemental Figure 7E). The interplay and crosstalk between S1 and S2/S3 subsegments are highly complex and not well understood.
In conclusion, we have shown the in vivo application of the extremely small cage-like protein nanoparticle targeting proximal tubules. The feasibility of translating Dps therapy to human diseases remains to be determined. As a step toward advancing nanoparticle therapy, further characterization and optimization are necessary, including the minimization of potential immunogenicity (42). In this regard, rapid renal clearing of nanoparticles may be advantageous because it limits their contact time with immune cells. In addition to mitigating endotoxin-induced kidney injury, this unique cage-like protein nanoparticle could be applied as a therapy to other forms of renal tubular injury. The self-organizing property of the cage subunit enables effective modulation of their protein characteristics because introduction of a single amino acid mutation in the monomer translates into the entire assembly. Therefore, they can be tailored to improve their effectiveness in renal diseases with widely varying pathology. Moreover, we believe that Dps and other nanoparticles can serve as unique tools to advance our understanding of the physiology of glomerular filtration and tubular endocytosis.
Methods
Preparation of protein cage nanoparticles.
S. solfataricus Dps, mouse heavy-chain ferritin, and virus-like particle derived from bacteriophage P22 were heterologously expressed using E. coli expression system and purified by previously described procedures with some minor modifications (43–46). Briefly, Dps and ferritin were cloned into the pET-30a(+) vector (Novagen). P22 scaffolding protein and coat protein (S39C mutant) were cloned into the first and the second multicloning sites of the pRSFDuet-1 vector. All constructs were transformed into ClearColi BL21(DE3) (Lucigen), which is an engineered E. coli strain that generates nonimmunostimulatory LPS. Transformed cells were grown on Miller’s LB medium at 37°C. Expression of proteins was induced by isopropyl β-d-thiogalactopyranoside when optical density at 600 nm of the culture media reached 0.6. The cells were harvested by centrifugation, and cell pellets were stored at –80°C until subjected to a protein purification process.
To purify protein cage nanoparticles, cell pellets were resuspended in lysis buffer (50 mM MES, 100 mM NaCl, pH 6.5 for Dps, 50 mM HEPES, 100 mM NaCl, pH 7.5 for ferritin, 50 mM sodium phosphate, 100 mM NaCl, pH 7.0 for P22). Lysozyme, DNase, and RNase were supplemented to final concentrations of 50, 60 and 100 μg/mL, respectively. The cell suspensions were incubated at room temperature for 30 minutes and then lysed by sonication. The cell debris was removed by centrifugation. Dps was purified from the supernatants by size-exclusion chromatography (10 × 300 mm Superose 6, GE Healthcare Life Sciences, in 20 mM NaCl, 20 mM Tris, pH 7.5). Eluted fractions corresponding to Dps protein were combined and dialyzed against a Tris buffer (20 mM Tris, 20 mM NaCl, pH 7.5) and then purified further by anion exchange chromatography (Mono Q; GE Healthcare Life Sciences) in a gradient of 20 to 1000 mM NaCl in 20 mM Tris, pH 7.5. Similarly, ferritin was purified by size-exclusion chromatography (50 mM HEPES, 100 mM NaCl, pH 7.5) followed by anion exchange chromatography in a gradient of 20 to 1000 mM NaCl in 20 mM Tris, pH 7.5. P22 was purified by ultracentrifugation of the lysis supernatant at 215,600 g (Piramoon Technologies, F50L-8 × 39 rotor) for 50 minutes through a 35% (wt/vol) sucrose cushion. The resulting protein pellets were resuspended in the lysis buffer and then purified by size-exclusion chromatography (16 × 600 mm Sephacryl S-500; GE Healthcare Life Sciences).
Dps, ferritin, and P22 protein cages were conjugated with Texas red maleimide, Alexa Fluor 405 succinimidyl ester, and Alexa Fluor 488 C5 maleimide (Invitrogen), respectively. In some experiments, Dps protein cages were conjugated with Alexa Fluor 488 C5 maleimide. For Dps, C126S mutant generated by site-directed mutagenesis was used for the dye conjugation. C126S Dps possesses 1 endogenous cysteine (Cys101) adjacent to the ferroxidase site (11). The protein (1.5 mg/mL in Dulbecco’s PBS [DPBS]) was mixed with Texas red maleimide dye in a stoichiometric ratio of 0.5 molar equivalent per subunit and reacted at room temperature for 4 hours with gentle rocking. The reaction was quenched with β-mercaptoethanol (1 mM final concentration), and then the protein was purified by size-exclusion chromatography with DPBS to remove unreacted dye. For conjugation of ferritin, the protein (2.5 mg/mL in 100 mM sodium bicarbonate buffer pH 8.5) was reacted with the Alexa Fluor 405 succinimidyl ester in a concentration of 3 molar equivalents per subunit at room temperature for 5 hours. The reaction was quenched with ethanoleamine (4 mM final concentration), and then the protein was purified by size-exclusion chromatography with DPBS to remove unreacted dye. For conjugation of P22, the protein (1.5 mg/mL in DPBS) was reacted with Alexa Fluor 488 maleimide in a stoichiometric ratio of 0.25 molar equivalents per subunit at room temperature for 4 hours. The reaction was quenched with β-mercaptoethanol (1 mM final concentration). Unreacted dye was removed by 2× ultracentrifugation at 215,600 g for 50 minutes. The protein pellet was resuspended in DPBS. UV-Vis absorbance spectra and fluorescence emission spectra of dye- conjugated protein cages were measured with a UV-Vis spectrometer (Agilent 8453) and a microplate reader (BioTek Synergy H1), respectively. Fluorescence dye per protein subunit ratio of the purified Dps, ferritin, and P22 determined from UV-Vis spectra were 0.38, 0.23, and 0.25, respectively. Dps (C126S) was also decorated with an alkyne group via chemical conjugation at Cys101 with N-propargyl bromoacetamide. N-propargyl bromoacetamide was synthesized as previously described (47). Dps protein (1.5 mg/mL in 100 mM HEPES, 50 mM NaCl, pH 6.5) was reacted with N-propargyl bromoacetamide (3.7 mM dissolved in DMF) at the ratio of 10-fold molar excess per protein subunit for 4 hours at room temperature. The modified protein was purified from the unreacted alkyne molecule by size-exclusion chromatography.
S. solfataricus
Dps exhibits a catalase-like activity when Mn is bound to an active site of the protein subunits (2 Mn per subunit) (11, 25). Mn loading to Dps was performed following an established protocol (25). A stock solution of Mn (0.4 M) was prepared by dissolving MnCl2•4H2O in water. The Mn solution was added to Dps protein (1.5 mg/mL) in a stoichiometry of 4-fold Mn per Dps subunit, and the mixture was incubated at room temperature for 4 hours. Unbound Mn was removed by dialyzing the Dps solution against DPBS 2 times.
Animals.
Male mouse strain C57BL/6J was obtained from the Jackson Laboratory. The endothelial-specific CreERT2 transgenic mice were generated by crossbreeding Tie2-CreER mice with Rosa26-TD-Tomato transgenic mice, as we described previously (48). Tamoxifen was injected i.p. into Tie2CreER/TD-Tomato mice once per day for 3 days at 50 μg/g body weight (T5648; MilliporeSigma) dissolved in 250 μL sunflower oil. Imaging was performed 1 week after tamoxifen induction. Lysozyme-EGFP transgenic mice, in which myeloid cells are marked with EGFP, were from our established colony (49). All mice were 8 to 12 weeks of age and weighed 20 to 30 g. Munich Wistar Frömter rats were from our established colony. All animals were given water and food ad libitum throughout the study. Mice were subjected to a single-dose, 5 mg/kg LPS i.p. injection (E. coli serotype, 0111:B4, MilliporeSigma). For therapeutic experiments, mice were injected with Dps at indicated doses (9 mg/kg, 18 mg/kg [~2 nmol Dps/30 g mouse] or 27 mg/kg) via tail vein immediately before LPS i.p. Control animals received an equal volume of PBS vehicle. Dps was freshly synthesized before each set of experiments and used within 2 weeks. Stability of Dps in PBS was confirmed using native gel Western blot analysis (Supplemental Figure 7F). For intravital imaging with Dps, ferritin, or P22, we titrated the dose of each nanoparticle according to their dye concentrations (4 nmol dye/mouse per each type of nanoparticles). The average numbers of dyes conjugated to each cage were as follows: 4.5/Dps cage, 5.4/ferritin cage, and 104.8/P22 cage. Neutral 3 kDa dextran (Texas red) and anionic 3 kDa dextran (Cascade Blue) were from Invitrogen, and 4 mg/kg body weight of each type of dextran was administered i.v.
Intravital 2-photon imaging and ex vivo tissue imaging.
Live animal imaging was performed using an Olympus FV1000-MPE confocal/multiphoton microscope equipped with a Spectra Physics MaiTai Deep See laser and gallium arsenide 12-bit detectors. The system is mounted on an Olympus Ix81 inverted microscope stand with a Nikon 20× and 60× NA 1.2 water-immersion objective. The laser was tuned to 810-nm excitation and used for all studies. Animals were placed on the stage with the exposed intact kidney, liver, or calvarium placed in a coverslip-bottomed cell culture dish bathed in isotonic saline, as previously described (50). Other organs were primarily imaged ex vivo in this study. The rectal temperature was monitored using a thermometer. Two ReptiTherm pads (Zoo Med) and a heated water jacket blanket were used to maintain the temperature at around 36°C. Whole-organ ex vivo imaging was performed using a Leica MZ 12.5 stereomicroscope equipped with a Leica MC 170 HD digital microscope camera. Serum and urine fluorescence signals were measured using a CLARIOStar plate reader instrument.
Immunohistochemistry and click chemistry.
To determine the distribution of alkyne-Dps, tissues were harvested from mice that received alkyne-Dps. Alexa Fluor 555 azide was conjugated to alkyne-Dps after tissue harvest using click chemistry. Tissues were fixed with 4% paraformaldehyde, sectioned (10 μm), and deparaffinized. Tissues were incubated for 30 minutes with 100 mM Tris Base, 1 mM CuSO4, 100 mM ascorbic acid, 100 μM Alexa Fluor 555 azide (Molecular Probes), and 0.2% Triton X-100 at room temperature. Tissues were then washed in PBS with 0.1% Triton X-100 for 30 minutes × 3, counterstained with DAPI for 10 minutes, washed in PBS × 3, and imaged with an Olympus FV1000-MPE confocal/multiphoton microscope. For standard immunohistochemistry, the following primary antibodies were used for immunostaining: 8-hydroxy-2′-deoxyguanosine (8-OHdG) (Abcam, catalog ab10802), F4/80 (clone CI:A3–1; AbD Serotec), Ly6G (clone RB6-8c5, Affymetrix 14-5931), and B220 (catalog BD-550286).
Electron microscopy.
Kidney tissues were fixed with 2% paraformaldehyde and 2% glutaraldehyde in phosphate buffer, followed by osmium tetroxide staining with a uranyl acetate replacement (UAR-EMS, Electron Microscopy Sciences). Specimen processing and transmission electron microscopy imaging were performed at the Indiana University Electron Microscopy Center using Tecnai G2 12 Bio Twin. For peroxisome staining, the alkaline DAB method was applied (51). Alkaline DAB solution consisted of 20 mg DAB (MilliporeSigma, D5905) dissolved in 5 mL of 0.01M Teorell-Stenhagen buffer (0.05M stock; 0.05M H3PO4, 0.057M boric acid, 0.035M citric acid, 0.345M NaOH, pH 10.5). Fixed tissues (50 μm vibrato sections) were incubated in alkaline DAB solution for 30 minutes; H2O2 was then added at a final concentration of 0.15% and incubated for another 1 hour at 37°C, washed in PBS, and subsequently processed for imaging.
GPCR assay.
The PRESTO-Tango constructs, developed by the Bryan Roth lab (52), were obtained from Addgene (catalog 1000000068). We first undertook sequencing verification for all of the 315 constructs and found that 2 constructs (BB3-Tango and DRD3-Tango) had mutations as compared with the original sequences. In the DRD3-Tango construct, there were mutations at position 528 bp (GAG to GGG; Glu to Gly) and at 704 bp (AAG to AGG; Lys to Arg). These mutations were corrected using mutagenesis. For BB3-Tango, there was 1 bp mutation at position 927 bp. The amino acid of this protein was not affected (TTC to TTT; Phe to Phe); thus, no mutagenesis was done. Next, using select plasmids, we verified the kinetics of the PRESTO-Tango, β-arrestin–recruitment assay system (Supplemental Figure 7, G and H). Finally, for the entire GPCR screening, 10,000 HTLA cells (HEK 293–derived cell line containing the stably integrated tTA-dependent luciferase reporter gene and a β-arrestin2-TEV fusion gene) were plated per well in 384-well poly-d-lysine–coated plates (Thermo Fisher) in 30 μL DMEM containing 10% FBS and 1% penicillin/streptomycin. Plasmids were transfected overnight into the cells in quadruplicate at a concentration of 0.01 μg/well in 30 μL DMEM containing polyethylenimine (MilliporeSigma; 45 mL DMEM + 900 μL 1 mg/mL PEI). Dps/MnDps was diluted for 30 μL/well (~2.3 μg Dps/well) in fresh complete DMEM, and 30 μL was added to each well following transfection. Cells were incubated overnight, washed 3× with PBS, and incubated for 15 minutes with 30 μL/well BrightGlo (Promega) luciferase substrate. Luminescence was measured using a CLARIOStar plate reader instrument. Negative controls consisted of transfected cells with no Dps/MnDps. Kidney tissues were lysed in 0.4% N-dodecyl β-d-maltoside, and the detergent was removed using Detergent Removal Spin Columns (Pierce, Thermo Fisher) per the manufacturer’s protocol. Tissue lysates were diluted to 1:100 in DMEM + 10% FBS + 1% penicillin/streptomycin for use in the GPCR assay.
RNA-Seq.
Kidneys were snap-frozen and RNA was extracted using QIAGEN Rneasy Plus Midi Kit with gDNA removal column. RNA quality was determined using Agilent 2100 Bioanalyzer (median RIN value 7.4). Sequencing was done at the Indiana University Center for Medical Genomics Core. Illumina TruSeq Stranded mRNA HT (RS-122-21) was used for library construction. Sequence was performed with 2 × 75 bp paired-end configuration on HiSeq4000 using the HiSeq 3000/4000 PE SBS Kit, and the sequenced data were mapped to the mm10 genome using STAR RNA-Seq aligner. Uniquely mapped sequencing reads were assigned to mm10 refGene genes using featureCounts.
Data analysis.
Data were analyzed for statistical significance and visualization with R software 3.4.3. RNA-Seq data were analyzed using edgeR. P values were adjusted with the FDR method as indicated. The following packages were used for data visualization: loess (Figure 1K), heatmap3 (Figure 4A), reactomePA::emapplot and enrichplot::cnetplot (Figure 4, B and C, and Figure 5, A–D), edgeR::glmLRT and plot (Supplemental Figure 5, A–D), edgeR::plotSmear (Supplemental Figure 5, E and F), and levelplot (Figure 6).
Western blotting.
Kidney tissues were extracted with RIPA buffer with additional 5 mM EDTA, 5 mM EGTA, Halt protease inhibitors (Pierce), phosStop inhibitor (Roche), and benzonase nuclease (EMD Millipore). Total protein levels were determined using a modified Lowry assay (Bio-Rad). Equal amounts of kidney proteins (20 μg) were mixed with NuPAGE LDS sample buffer with 100 mM of DTT and separated by electrophoreses on NuPage 4%–12% Bis-Tris gels and transferred to PVDF membranes. Antibodies used included the following: catalase (Abcam, catalog ab209211), Nrf2 (MilliporeSigma, catalog SAB4501984), Hmox1 (Abcam, catalog ab13243), and actin (Santa Cruz Biotechnology Inc., C-2, catalog sc-8432 AF680). Nuclear fractionation was done using the Subcellular Protein Fractionation Kit for Tissues (Thermo Scientific, catalog87790). Native gel Western blotting was done using blue native gel electrophoresis on 4% to 16% Native PAGE (Novex). Dps nanoparticles were titrated in 2-fold dilutions from 2.8 μg to 0.02 μg before loading on the gel. Samples were overlaid with blue cathode buffer and electrophoresed against cathode buffer at 150 volts for 2 hours. Proteins in the gel were stained using the fast Coomassie G-250 staining protocol (Novex).
Catalase assay.
Total catalase activity was measured using the Cayman Chemical Catalase Assay Kit (catalog 707002) per the manufacturer’s instructions. The assay is based on the reaction with methanol in the presence of an optimal concentration of H2O2, and the formaldehyde produced was measured colorimetrically. Half of a mouse kidney was lysed in 1 mL sample buffer using the Bertin Instruments Minilys Personal Homogenizer at full speed for 45 seconds in Precellys 2 mL Hard Tissue Homogenizing Ceramic Beads CK28. Homogenate was then centrifuged for 5 minutes at 10,000 g (4°C), diluted 1:100, and immediately assayed.
PCR.
RNA extraction from snap-frozen kidneys was performed using TRIzol, and the extracted RNA was reverse-transcribed using High Capacity cDNA Reverse Transcription Kit (Life Technologies). TaqMan gene expression assays used included Havcr1/KIM1 (Mm00506686_m1), Lcn2/NGAL (Mm01324470_ml), Eif2ak2 (Mm01235643_m1), Atf4 (Mm00515325_g1), and glyceraldehyde-3-phosphate dehydrogenase (Mm99999915_g1). Real-time quantitative PCR amplifications were performed for 40 cycles using ViiA 7 Real-Time PCR systems. The ΔΔCt method was used to analyze the relative changes in gene expression.
Data availability.
RNA-Seq data were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE131922).
Statistics.
Data were analyzed for statistical significance and visualization with R software 3.4.3. Error bars show SD. For multiple comparisons, 1-way ANOVA followed by pairwise t tests was performed using the Benjamini and Hochberg method to adjust P values. All analyses were 2-sided, and a P value of less than 0.05 was considered significant.
Study approval.
All animal protocols were approved by the Indiana University Institutional Animal Care Committee and conform to the NIH Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011).
Author contributions
MU, BM, TD, and TH designed and coordinated the study. MU, HKW,SLP, RMS, BM, AZ, TH, ES, JA, and EY performed experiments. SLP and TH performed data analyses. MU, BM, BAM, TD, and TH interpreted data. TH wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Pierre Dagher for his helpful comments and suggestions on the manuscript. RNA-Seq was performed by Xiaoling Xuei, Yunlong Liu, et al. at the Center for Medical Genomics at Indiana University School of Medicine. Electron microscopy was performed by Caroline Miller at the Indiana University Electron Microscopy Center. GPCR plasmid sequencing was done at GenScript. Measurement of serum creatinine concentration was performed by John Moore, Yang Yan, et al. at the University of Alabama at Birmingham/UCSD O’Brien Center Core for Acute Kidney Injury Research (NIH P30DK079337) using isotope dilution liquid chromatography–tandem mass spectrometry. We thank Michelle Martinez for assistance with intravital liver imaging. This work was supported by NIH grant K08-DK113223 and a Paul Teschan research grant (Dialysis Clinics Inc.) to TH. This work was also supported by the Indiana University O’Brien Center grant P30-DK079312.
Version 1. 08/19/2019
Electronic publication
Version 2. 09/03/2019
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(9):3941–3951.https://doi.org/10.1172/JCI127511.
Contributor Information
Masaki Uchida, Email: muchida@indiana.edu.
Bernhard Maier, Email: bfmaier@iupui.edu.
Hitesh Kumar Waghwani, Email: hwaghwan@iu.edu.
Ekaterina Selivanovitch, Email: kselivan@umail.iu.edu.
S. Louise Pay, Email: slpay@iu.edu.
John Avera, Email: itsavera14@gmail.com.
EJun Yun, Email: ejunyun@iu.edu.
Amy Zollman, Email: alzollma@iu.edu.
Trevor Douglas, Email: trevdoug@indiana.edu.
Takashi Hato, Email: thato@iu.edu.
References
- 1.Putri RM, Cornelissen JJ, Koay MS. Self-assembled cage-like protein structures. Chemphyschem. 2015;16(5):911–918. doi: 10.1002/cphc.201402722. [DOI] [PubMed] [Google Scholar]
- 2.Zarjou A, Agarwal A. Sepsis and acute kidney injury. J Am Soc Nephrol. 2011;22(6):999–1006. doi: 10.1681/ASN.2010050484. [DOI] [PubMed] [Google Scholar]
- 3.Hato T, Dagher PC. How the innate immune system senses trouble and causes trouble. Clin J Am Soc Nephrol. 2015;10(8):1459–1469. doi: 10.2215/CJN.04680514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brenner BM, Hostetter TH, Humes HD. Molecular basis of proteinuria of glomerular origin. N Engl J Med. 1978;298(15):826–833. doi: 10.1056/NEJM197804132981507. [DOI] [PubMed] [Google Scholar]
- 5.Bennett KM, et al. MRI of the basement membrane using charged nanoparticles as contrast agents. Magn Reson Med. 2008;60(3):564–574. doi: 10.1002/mrm.21684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Charlton JR, et al. Biocompatibility of ferritin-based nanoparticles as targeted MRI contrast agents. Nanomedicine. 2016;12(6):1735–1745. doi: 10.1016/j.nano.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamaly N, He JC, Ausiello DA, Farokhzad OC. Nanomedicines for renal disease: current status and future applications. Nat Rev Nephrol. 2016;12(12):738–753. doi: 10.1038/nrneph.2016.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du B, Yu M, Zheng J. Transport and interactions of nanoparticles in the kidneys. Nature Reviews Materials. 2018;3(10):358–374. doi: 10.1038/s41578-018-0038-3. [DOI] [Google Scholar]
- 9.Longmire M, Choyke PL, Kobayashi H. Clearance properties of nano-sized particles and molecules as imaging agents: considerations and caveats. Nanomedicine (Lond) 2008;3(5):703–717. doi: 10.2217/17435889.3.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiedenheft B, et al. An archaeal antioxidant: characterization of a Dps-like protein from Sulfolobus solfataricus. Proc Natl Acad Sci U S A. 2005;102(30):10551–10556. doi: 10.1073/pnas.0501497102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gauss GH, Benas P, Wiedenheft B, Young M, Douglas T, Lawrence CM. Structure of the DPS-like protein from Sulfolobus solfataricus reveals a bacterioferritin-like dimetal binding site within a DPS-like dodecameric assembly. Biochemistry. 2006;45(36):10815–10827. doi: 10.1021/bi060782u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maaty WS, et al. Something old, something new, something borrowed; how the thermoacidophilic archaeon Sulfolobus solfataricus responds to oxidative stress. PLoS ONE. 2009;4(9):e6964. doi: 10.1371/journal.pone.0006964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi HS, et al. Renal clearance of quantum dots. Nat Biotechnol. 2007;25(10):1165–1170. doi: 10.1038/nbt1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du B, et al. Glomerular barrier behaves as an atomically precise bandpass filter in a sub-nanometre regime. Nat Nanotechnol. 2017;12(11):1096–1102. doi: 10.1038/nnano.2017.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JW, Chou CL, Knepper MA. Deep sequencing in microdissected renal tubules identifies nephron segment-specific transcriptomes. J Am Soc Nephrol. 2015;26(11):2669–2677. doi: 10.1681/ASN.2014111067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, Masehi-Lano JJ, Chung EJ. Peptide and antibody ligands for renal targeting: nanomedicine strategies for kidney disease. Biomater Sci. 2017;5(8):1450–1459. doi: 10.1039/C7BM00271H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lucon J, et al. Use of the interior cavity of the P22 capsid for site-specific initiation of atom-transfer radical polymerization with high-density cargo loading. Nat Chem. 2012;4(10):781–788. doi: 10.1038/nchem.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jordan PC, et al. Self-assembling biomolecular catalysts for hydrogen production. Nat Chem. 2016;8(2):179–185. doi: 10.1038/nchem.2416. [DOI] [PubMed] [Google Scholar]
- 19.Patterson DP, Schwarz B, Waters RS, Gedeon T, Douglas T. Encapsulation of an enzyme cascade within the bacteriophage P22 virus-like particle. ACS Chem Biol. 2014;9(2):359–365. doi: 10.1021/cb4006529. [DOI] [PubMed] [Google Scholar]
- 20.Kalakeche R, et al. Endotoxin uptake by S1 proximal tubular segment causes oxidative stress in the downstream S2 segment. J Am Soc Nephrol. 2011;22(8):1505–1516. doi: 10.1681/ASN.2011020203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hato T, et al. The macrophage mediates the renoprotective effects of endotoxin preconditioning. J Am Soc Nephrol. 2015;26(6):1347–1362. doi: 10.1681/ASN.2014060561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hato T, et al. Endotoxin preconditioning reprograms S1 tubules and macrophages to protect the kidney. J Am Soc Nephrol. 2018;29(1):104–117. doi: 10.1681/ASN.2017060624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeth K. Dps biomineralizing proteins: multifunctional architects of nature. Biochem J. 2012;445(3):297–311. doi: 10.1042/BJ20120514. [DOI] [PubMed] [Google Scholar]
- 24.Janissen R, et al. Global DNA compaction in stationary-phase bacteria does not affect transcription. Cell. 2018;174(5):1188–1199.e14. doi: 10.1016/j.cell.2018.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayden JA, Hendrich MP. EPR spectroscopy and catalase activity of manganese-bound DNA-binding protein from nutrient starved cells. J Biol Inorg Chem. 2010;15(5):729–736. doi: 10.1007/s00775-010-0640-3. [DOI] [PubMed] [Google Scholar]
- 26.Nath KA. Heme oxygenase-1 and acute kidney injury. Curr Opin Nephrol Hypertens. 2014;23(1):17–24. doi: 10.1097/01.mnh.0000437613.88158.d3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hato T, et al. Bacterial sepsis triggers an antiviral response that causes translation shutdown. J Clin Invest. 2019;129(1):296–309. doi: 10.1172/JCI123284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wacker D, Stevens RC, Roth BL. How ligands illuminate GPCR molecular pharmacology. Cell. 2017;170(3):414–427. doi: 10.1016/j.cell.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fissell WH, Miner JH. What Is the Glomerular Ultrafiltration Barrier? J Am Soc Nephrol. 2018;29(9):2262–2264. doi: 10.1681/ASN.2018050490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi CH, Zuckerman JE, Webster P, Davis ME. Targeting kidney mesangium by nanoparticles of defined size. Proc Natl Acad Sci USA. 2011;108(16):6656–6661. doi: 10.1073/pnas.1103573108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams RM, et al. Mesoscale nanoparticles selectively target the renal proximal tubule epithelium. Nano Lett. 2015;15(4):2358–2364. doi: 10.1021/nl504610d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nair AV, Keliher EJ, Core AB, Brown D, Weissleder R. Characterizing the interactions of organic nanoparticles with renal epithelial cells in vivo. ACS Nano. 2015;9(4):3641–3653. doi: 10.1021/acsnano.5b00428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hackel BJ, Sathirachinda A, Gambhir SS. Designed hydrophilic and charge mutations of the fibronectin domain: towards tailored protein biodistribution. Protein Eng Des Sel. 2012;25(10):639–647. doi: 10.1093/protein/gzs036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruggiero A, et al. Paradoxical glomerular filtration of carbon nanotubes. Proc Natl Acad Sci USA. 2010;107(27):12369–12374. doi: 10.1073/pnas.0913667107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alidori S, et al. Targeted fibrillar nanocarbon RNAi treatment of acute kidney injury. Sci Transl Med. 2016;8(331):331ra39. doi: 10.1126/scitranslmed.aac9647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stamatiades EG, et al. Immune monitoring of trans-endothelial transport by kidney-resident macrophages. Cell. 2016;166(4):991–1003. doi: 10.1016/j.cell.2016.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang D, et al. DNA origami nanostructures can exhibit preferential renal uptake and alleviate acute kidney injury. Nat Biomed Eng. 2018;2(11):865–877. doi: 10.1038/s41551-018-0317-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hato T, Winfree S, Dagher PC. Intravital imaging of the kidney. Methods. 2017;128:33–39. doi: 10.1016/j.ymeth.2017.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sandoval RM, et al. Multiple factors influence glomerular albumin permeability in rats. J Am Soc Nephrol. 2012;23(3):447–457. doi: 10.1681/ASN.2011070666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dickson LE, Wagner MC, Sandoval RM, Molitoris BA. The proximal tubule and albuminuria: really! J Am Soc Nephrol. 2014;25(3):443–453. doi: 10.1681/ASN.2013090950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee JW, Chou CL, Knepper MA. Deep sequencing in microdissected renal tubules identifies nephron segment-specific transcriptomes. J Am Soc Nephrol. 2015;26(11):2669–2677. doi: 10.1681/ASN.2014111067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koudelka KJ, Pitek AS, Manchester M, Steinmetz NF. Virus-based nanoparticles as versatile nanomachines. Annu Rev Virol. 2015;2(1):379–401. doi: 10.1146/annurev-virology-100114-055141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Broomell CC, et al. Protein cage nanoparticles as secondary building units for the synthesis of 3-dimensional coordination polymers. Soft Matter. 2010;6(14):3167–3171. doi: 10.1039/c0sm00039f. [DOI] [Google Scholar]
- 44.Uchida M, et al. Targeting of cancer cells with ferrimagnetic ferritin cage nanoparticles. J Am Chem Soc. 2006;128(51):16626–16633. doi: 10.1021/ja0655690. [DOI] [PubMed] [Google Scholar]
- 45.Uchida M, et al. Site-directed coordination chemistry with P22 virus-like particles. Langmuir. 2012;28(4):1998–2006. doi: 10.1021/la203866c. [DOI] [PubMed] [Google Scholar]
- 46.Kang S, Uchida M, O’Neil A, Li R, Prevelige PE, Douglas T. Implementation of p22 viral capsids as nanoplatforms. Biomacromolecules. 2010;11(10):2804–2809. doi: 10.1021/bm100877q. [DOI] [PubMed] [Google Scholar]
- 47.Abedin MJ, Liepold L, Suci P, Young M, Douglas T. Synthesis of a cross-linked branched polymer network in the interior of a protein cage. J Am Chem Soc. 2009;131(12):4346–4354. doi: 10.1021/ja8079862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hato T, et al. Two-photon intravital fluorescence lifetime imaging of the kidney reveals cell-type specific metabolic signatures. J Am Soc Nephrol. 2017;28(8):2420–2430. doi: 10.1681/ASN.2016101153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Faust N, Varas F, Kelly LM, Heck S, Graf T. Insertion of enhanced green fluorescent protein into the lysozyme gene creates mice with green fluorescent granulocytes and macrophages. Blood. 2000;96(2):719–726. [PubMed] [Google Scholar]
- 50.Hato T, Winfree S, Dagher PC. Kidney imaging: intravital microscopy. Methods Mol Biol. 2018;1763:129–136. doi: 10.1007/978-1-4939-7762-8_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bonekamp NA, Islinger M, Lázaro MG, Schrader M. Cytochemical detection of peroxisomes and mitochondria. Methods Mol Biol. 2013;931:467–482. doi: 10.1007/978-1-62703-056-4_24. [DOI] [PubMed] [Google Scholar]
- 52.Kroeze WK, et al. PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nat Struct Mol Biol. 2015;22(5):362–369. doi: 10.1038/nsmb.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq data were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE131922).