Abstract
The expression of the transmembrane protein 25 gene (Tmem25) is strongly influenced by glutamate ionotropic receptor kainate type subunit 4, and its function remains unknown. Here, we showed that TMEM25 was primarily localized to late endosomes in neurons. Electrophysiological experiments suggested that the effects of TMEM25 on neuronal excitability were likely mediated by N-methyl-d-aspartate receptors. TMEM25 affected the expression of the N-methyl-d-aspartate receptor NR2B subunit and interacted with NR2B, and both were colocalized to late endosome compartments. TMEM25 induced acidification changes in lysosome compartments and accelerated the degradation of NR2B. Furthermore, TMEM25 expression was decreased in brain tissues from patients with epilepsy and epileptic mice. TMEM25 overexpression attenuated the behavioral phenotypes of epileptic seizures, whereas TMEM25 downregulation exerted the opposite effect. These results provide some insights into TMEM25 biology in the brain and the functional relationship between TMEM25 and epilepsy.
Keywords: Neuroscience
Keywords: Epilepsy
Introduction
Transmembrane protein 25 (TMEM25) has been identified as a member of the immunoglobulin superfamily, as codons 42–112 of TMEM25 are a C-2 type immunoglobulin domain homologous to titin and nephrin (1). Studies have shown that TMEM25 may be a biomarker of breast and colorectal cancers (2, 3); however, little is currently known about the function of TMEM25 in the brain. According to a preliminary search of the online Allen Mouse Brain Atlas, Tmem25 is expressed at high levels in the CNS, particularly in the CA1 and CA3 regions of the hippocampus. Furthermore, Tmem25 expression is strongly influenced by glutamate ionotropic receptor kainate type subunit 4, which regulates mood, memory, and hippocampal seizure activity (4). Therefore, we hypothesized that TMEM25 may play a specific role in neuronal excitability and neurotransmission.
Epilepsy, a common neurological disease that affects approximately 50 million individuals worldwide, is characterized by recurrent spontaneous seizures (5, 6). Although dozens of antiepileptic drugs (AEDs) have been developed for clinical use, nearly one-third of patients with epilepsy still suffer from intractable seizures (7). Surgical resection of the epileptic focus is suitable for only a small proportion of patients who are resistant to medication (8). An imbalance between neuronal excitation and inhibition that results in hypersynchronous discharge of neurons in the brain is thought to be an important mechanism in epilepsy (6). Therefore, further understanding of the molecular mechanisms that influence this imbalance is necessary for exploring novel antiepileptic targets.
We performed immunofluorescence staining to explore the distribution and subcellular location of TMEM25. We subsequently assessed changes in the electrophysiology of CA1 pyramidal neurons in mice following lentivirus-mediated interference or overexpression of TMEM25 in the hippocampus. TMEM25 altered N-methyl-d-aspartate receptor–mediated (NMDAR–mediated) excitatory currents and influenced the protein level of NR2B without affecting protein levels of NR1 and NR2A. Moreover, we performed coimmunoprecipitation assays to examine the interactions between TMEM25 and the NMDA receptor NR2B subunit. TMEM25 colocalized with NR2B in late endosome/lysosome compartments; thus, we performed a cycloheximide (CHX) degradation assay to investigate the role of TMEM25 in NR2B degradation. To investigate the role of TMEM25 in epilepsy, we examined the expression of TMEM25 in patients with temporal lobe epilepsy (TLE) as well as in pentylenetetrazole (PTZ) kindling and kainic acid–induced (KA–induced) epilepsy mouse models. Furthermore, we detected the behavioral phenotypes in 2 epilepsy mouse models. Our results shed light on the subcellular location and cellular functions of TMEM25 and the mechanisms underlying its regulation of epileptic seizures. Moreover, these findings suggest that TMEM25 may play a critical role in the regulation of neuronal excitability and epileptic seizures by influencing NMDA receptor NR2B subunit degradation.
Results
TMEM25 colocalized with microtubule-associated protein 2 in late endosome/lysosome compartments.
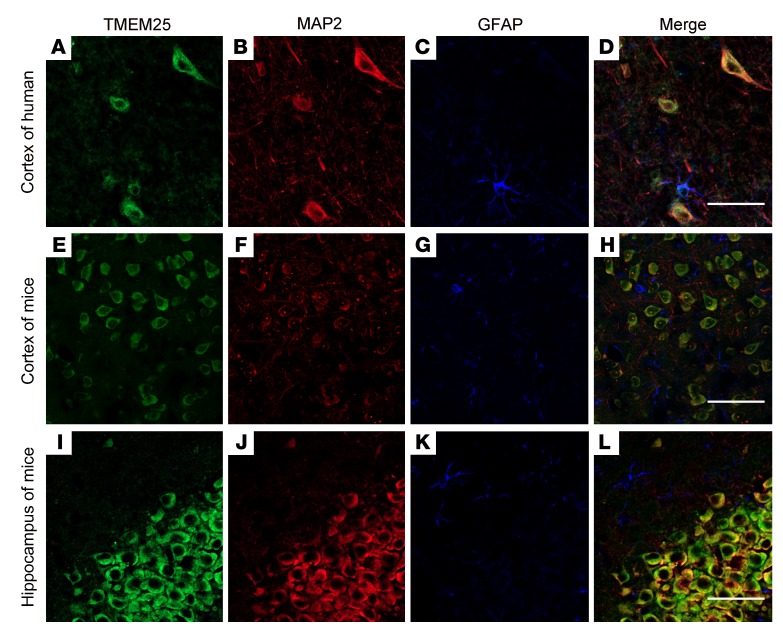
Tmem25 is expressed at high levels in the CNS, and in situ hybridization experiments have shown that it is mainly concentrated within the hippocampus (9). To date, little is known about the precise distribution of the TMEM25 protein in the brain. Here, we obtained similar results indicating that TMEM25 protein is expressed throughout the brain and is particularly concentrated in the pyramidal cell layer of the hippocampal CA1 and CA3 regions. TMEM25 is also expressed at high levels within the hippocampal dentate gyrus region and cerebellum and in scattered neurons in the cerebral cortex (Supplemental Figure 1). To further explore the location of endogenous TMEM25, we performed triple-labeled immunofluorescence staining to determine its cellular location, and the results suggested that TMEM25 colocalized with microtubule-associated protein 2 (MAP2), a marker of dendrites (Figure 1, A–D). The findings from the mouse hippocampus and cortex were consistent with the results obtained in the human temporal cortical tissues (Figure 1, E–L).
Figure 1. Localization of TMEM25 in nonepileptic brain tissues.
(A–D) In temporal cortical tissue from nonepileptic patients, TMEM25 colocalized with MAP2. (E–L) In the cortex and hippocampus from normal mice, TMEM25 colocalized with MAP2. Scale bars: 50 μm.
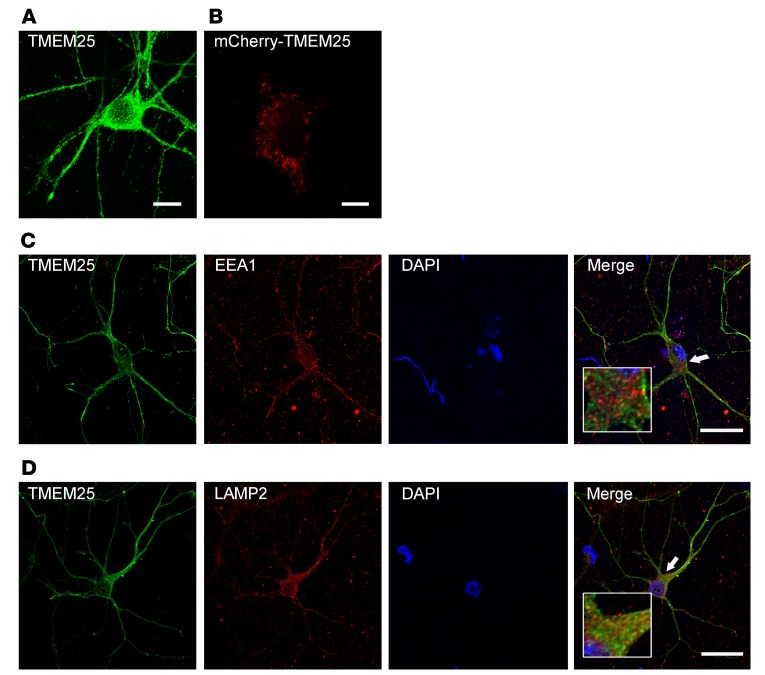
To investigate the subcellular localization of TMEM25, we first assessed its expression in cultured neurons by performing immunofluorescence staining and transfecting human embryonic kidney 293 (HEK293) cells with mCherry-tagged TMEM25. A large pool of TMEM25 was localized in intracellular particles (Figure 2, A and B). Further confirming the identities of these intracellular particles, an abundance of endogenous TMEM25 colocalized with the late endosome and lysosome marker lysosomal-associated membrane protein 2 (LAMP2), but rarely colocalized with the early endosome marker early endosome antigen 1 (EEA1) in cultured primary neurons (Figure 2, C and D).
Figure 2. TMEM25 localizes to late endosome/lysosome compartments.
(A) Neurons were stained with anti-TMEM25 antibodies on DIV 14. Scale bar: 10 μm. (B) HEK293 cells were transfected with mCherry-tagged TMEM25. Scale bar: 10 μm. (C and D) Neurons were stained with anti-TMEM25 and anti-EEA1 or anti-LAMP2 antibodies. Representative images show the colocalization of TMEM25 with LAMP2 (D), whereas TMEM25 and EEA1 did not colocalize (C). Scale bars: 50 μm. High-magnification photomicrographs corresponding to specific cells are shown at the bottom.
TMEM25 is involved in NMDAR-mediated neurotransmission.
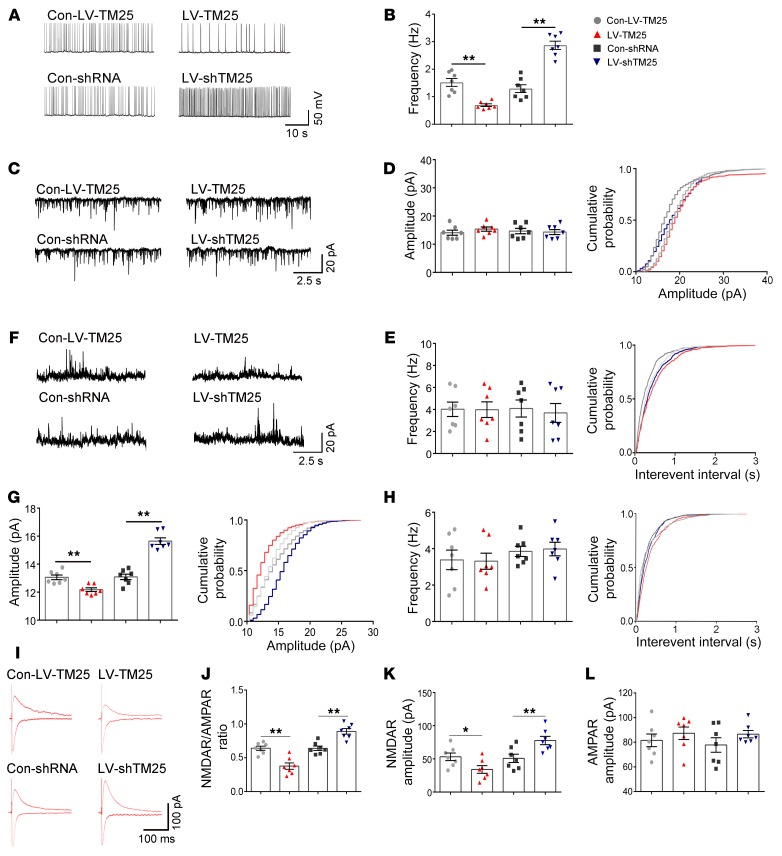
We performed whole-cell patch-clamp electrophysiology in CA1 pyramidal neurons in mouse hippocampal slices 14 days after lentivirus vector (LV) injection to evaluate the role of TMEM25 in neural activity. We recorded spontaneous action potentials (sAPs) from pyramidal neurons treated with Con-LV-TM25, LV-TM25, Con-shRNA, and LV-shTM25 to measure neuronal excitability. In the LV-TM25–treated group, the frequency of action potentials (APs) from pyramidal neurons was significantly lower than the frequency of APs in the Con-LV-TM25–treated group, while the AP frequency in the LV-shTM25–treated group was significantly higher than that in the Con-shRNA–treated group (Figure 3, A and B).
Figure 3. Effect of TMEM25 on the excitability of pyramidal neurons in the hippocampal CA1 region.
(A and B) Representative traces of sAPs and a summary of the sAP frequency among the groups. (C–E) Representative traces of AMPAR-mediated mEPSCs recorded from the examined groups and a summary of the frequency and amplitude of AMPAR-mediated mEPSCs. (F–H) Representative traces of NMDAR-mediated mEPSCs recorded from the examined groups and a summary of the frequency and amplitude of NMDAR-mediated mEPSCs. (I–L) Representative traces of NMDAR- and AMPAR-mediated EPSCs and a summary of the NMDAR/AMPAR ratio and NMDAR- and AMPAR-mediated EPSC amplitude. Data are presented as means ± SEM, n = 7 per group. *P < 0.05; **P < 0.01, by Kruskal-Wallis test followed by Dunn’s test (B), or 1-way ANOVA followed by LSD t test (D, E, G, H, J, K, and L).
We recorded miniature excitatory postsynaptic currents (mEPSCs) and miniature inhibitory postsynaptic currents (mIPSCs) following lentivirus infection to determine whether TMEM25 affected the imbalance in excitatory and inhibitory transmission. Both α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors (AMPARs) and NMDARs mediate the glutamatergic neurotransmission essential for the excitability of neurons; thus, we recorded AMPAR- and NMDAR-mediated mEPSCs. We observed an increase in the amplitudes of the NMDAR-mediated mEPSCs in the hippocampal slices from the TMEM25 knockdown mice and a decrease in the slices from the LV-TM25–treated group compared with those of their corresponding control groups, while no significant differences were observed in the frequency of NMDAR-mediated mEPSCs (Figure 3, F–H). No significant differences in the frequency or amplitude of AMPAR-mediated mEPSCs and mIPSCs were observed among the examined groups (Figure 3, C–E, and Supplemental Figure 2, A–E). The paired-pulse ratio (PPR) is widely considered a measure of the presynaptic release probability (10). No significant differences in the PPR were identified among the examined groups (Supplemental Figure 2, F and G). Furthermore, we measured the NMDAR/AMPAR ratio of the excitatory postsynaptic currents (EPSCs) to further confirm whether the TMEM25 levels in the examined groups affected excitatory transmission. We observed increases in the NMDAR/AMPAR ratio and in the average amplitude of the NMDAR-mediated EPSCs in the hippocampal slices from the TMEM25 knockdown mice and decreases in the LV-TM25–treated group. However, compared with the Con-LV-TM25 and Con-shRNA treatments, the LV-TM25 and LV-shTM25 treatments failed to alter the average amplitude of the AMPAR-mediated EPSCs (Figure 3, I–L). Thus, consistent with the NMDAR-mediated mEPSC amplitude increases, the effect of TMEM25 on neuronal excitability was likely a result of altered NMDAR-mediated currents.
TMEM25 modulates NR2B-mediated but not NR2A-mediated currents.
We recorded evoked EPSCs in the presence of different subtype-selective NMDAR blockers to determine which subunit was responsible for the effects of TMEM25 on NMDAR-mediated currents. The Δ amplitudes of the NMDAR currents in the LV-TM25 group and the LV-shTM25 group before and after Ro 25-6981 (0.5 μM) application were significantly different from those of the respective control groups (Supplemental Figure 3, B and D). By contrast, TMEM25 had no effect on the Δ amplitude of NMDAR currents in different groups before and after 50 nM NVP-AAM077 application (Supplemental Figure 3, A and C). These results imply that TMEM25 likely altered the NMDAR-mediated currents via the NR2B subunit.
Next, we constructed LVs to overexpress or knock down NR2B subunit expression to determine whether this approach may partially rescue the TMEM25-induced changes in neuronal excitability. After performing stereotaxic injections of the viruses (Con-LV-TM25, LV-TM25, LV-TM25+LV-NR2B, Con-shRNA, LV-shTM25, and LV-shTM25+LV-shNR2B) into the hippocampus, we analyzed the NMDAR/AMPAR ratio. Consistent with the results shown in Figure 3J, the NMDAR/AMPAR ratio was decreased in the TMEM25 overexpression group, and coinfection of LV-TM25 with an LV-NR2B that encoded mouse NR2B partially rescued the decrease in the NMDAR/AMPAR ratio (Supplemental Figure 3, E and F). The NMDAR/AMPAR ratio was increased in the TMEM25 downregulation group, and coinfection of LV-shTM25 with an LV-shNR2B partially rescued the increase in the NMDAR/AMPAR ratio (Supplemental Figure 3, E and F). These results further demonstrate the important role of NR2B levels in the regulation of electrophysiological phenotypes by TMEM25.
TMEM25 affects the NR2B expression pattern in the hippocampus.
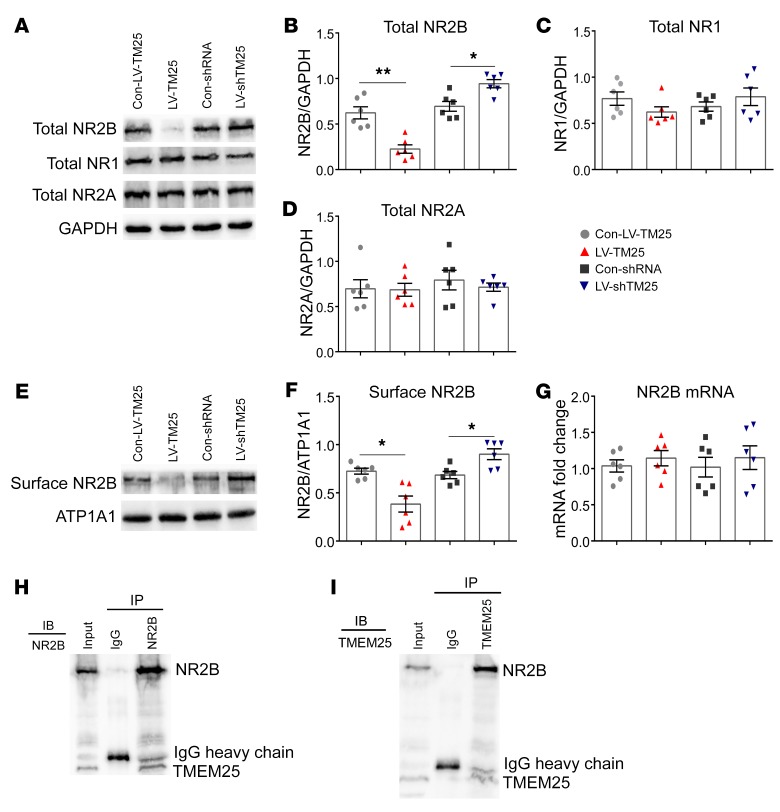
To investigate the mechanism by which TMEM25 affects NMDAR-mediated currents (specifically NR2B-dependent currents), we conducted a series of biochemical experiments to assess the levels of NMDAR in hippocampal tissues following lentiviral intervention. According to the Western blotting analysis, downregulation of TMEM25 resulted in an increase in the total level of NR2B subunits, and the mice treated with LV-TM25 showed a reduced level of total NR2B (Figure 4, A and B). Similarly, a smaller amount of NR2B was observed on the membrane in the LV-TM25–treated group than on that in the Con-LV-TM25–treated group, and a larger amount of NR2B was detected on the membrane in the LV-shTM25–treated group than on that in the Con-shRNA–treated group (Figure 4, E and F). However, no differences in the levels of total NR1 and NR2A proteins in the hippocampus were observed among the examined groups (Figure 4, C and D). Overall, the up- and downregulation of TMEM25 may affect the overall expression level of NR2B subunits and the subsequent membrane expression of NR2B, which may further influence NMDAR-mediated EPSCs. Thus, we speculated that TMEM25 may be involved in the synthesis or degradation of NR2B.
Figure 4. Influence of TMEM25 on NMDAR expression in hippocampal tissues and coimmunoprecipitation of NR2B and TMEM25.
(A–D) Representative images of the expression of total NR2B, total NR1, and total NR2A from the examined groups and quantification of immunoblots. (E and F) Representative images of surface NR2B expression from the examined groups and quantification of immunoblots. (G) Quantitative real-time PCR analysis of the expression of NR2B mRNA. Data are presented as means ± SEM and are representative of at least 3 independent repeats, n = 6 per group. *P < 0.05; **P < 0.01, by 1-way ANOVA followed by LSD t test (B, D and G) or Kruskal-Wallis test followed by Dunn’s test (C and F). (H and I) The interaction between TMEM25 and NR2B as assessed by coimmunoprecipitation.
TMEM25 interacts with NR2B in the hippocampus and colocalizes in late endosome/lysosome compartments.
TMEM25 expression was detected in neurons, and several lines of evidence support the presence of a unique relationship between TMEM25 and NR2B. According to immunofluorescence staining, TMEM25 colocalizes with NR2B in cultured primary hippocampal neurons (Supplemental Figure 4A). We also confirmed this colocalization using HEK293 cells cotransfected with exogenous mCherry-tagged TMEM25 and SEP-NR2B to avoid false-positive staining (Supplemental Figure 4B). Furthermore, endogenous TMEM25 coimmunoprecipitated with NR2B but not with NR2A or AMPAR subunits in the mouse hippocampus (Figure 4, H and I; Supplemental Figure 5, A and B; and Supplemental Figure 6). Notably, a weak interaction was observed between TMEM25 and the NR1 subunit under coimmunoprecipitation conditions (Supplemental Figure 5, C and D), possibly because NR2B must form a heteromeric receptor complex with NR1 (11, 12). We further confirmed these interactions by the colocalization of exogenous mCherry-tagged TMEM25 and SEP-NR2B with LAMP2 in HEK293 cells (Supplemental Figure 4C). Knockdown and overexpression of TMEM25 had no significant effects on the level of the NR2B transcript (Figure 4G). Thus, we hypothesized that the changes in NR2B expression identified in the LV-TM25– and LV-shTM25–treated mice may occur at the protein level. Lysosomes are typically considered the final destination for proteolytic degradation (13). Overall, these results suggest that TMEM25 may play a role in the degradation of NR2B.
TMEM25 participates in the regulation of NR2B degradation.
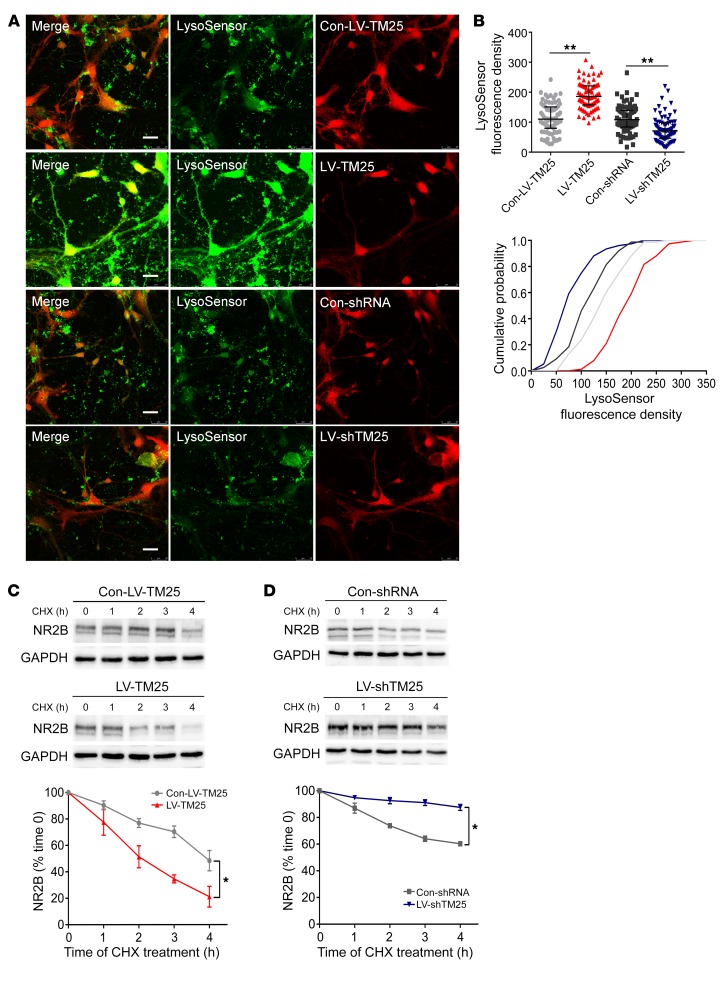
We subsequently investigated the mechanism by which TMEM25 affects the total protein level of NR2B. Considering that the acidic environment is highly important for lysosome functioning (14), we examined whether up- and downregulation of TMEM25 affects lysosomal acidification in primary cultured hippocampal neurons using the pH-sensitive dye LysoSensor Green DND-189. The fluorescence intensity of LysoSensor Green DND-189 within the mCherry-positive area was significantly decreased following downregulation of TMEM25. In contrast, compared with the Con-LV-TM25 treatment, TMEM25 overexpression significantly increased the fluorescence intensity of LysoSensor (Figure 5, A and B). Thus, the expression levels of TMEM25 affect lysosomal acidification.
Figure 5. TMEM25 level alter lysosomal acidification and the rate of NR2B degradation.
(A and B) Representative images of LV-transfected primary cultured hippocampal neurons stained with LysoSensor Green DND-189 and quantification of fluorescence intensity. Scale bar: 25 μm. Data are presented as medians and interquartile ranges and are representative of at least 3 independent repeats, n = 76 sites per group. **P < 0.01, by 1-way ANOVA followed by LSD t test. (C and D) Representative images of the expression of total NR2B in hippocampal neurons transfected with LV and cultured with CHX and quantification of immunoblots. Data are presented as means ± SEM and are representative of at least 3 independent repeats, n = 3 per group. *P < 0.05, by 2-way ANOVA followed by post-hoc test.
Therefore, we hypothesized that changes in lysosomal acidification may modulate NR2B degradation in lysosomes. To investigate whether the TMEM25-induced impairment in the acidification of late endosomes/lysosomes affects endolysosomal function, the cellular turnover rate of NR2B was analyzed in cultured neurons. We performed a CHX-mediated protein degradation assay to address this question. The residual expression of total NR2B in the LV-shTM25–treated neurons increased by 7.82% after 1 hour, 18.86% after 2 hours, 27.12% after 3 hours, and 27.28% after 4 hours. The residual expression of total NR2B in the LV-TM25–treated neurons decreased by 10.03% after 1 hour, 15.06% after 2 hours, 38.22% after 3 hours, and 33.95% after 4 hours, compared with expression of the respective control lentivirus-treated neurons (Figure 5, C and D). The residual expression of the total NR2A, NR1, and AMPAR subunits, however, was not significantly different after the application of different LVs (Supplemental Figures 7–12). In summary, TMEM25 downregulation likely attenuates the degradation rates of total NR2B in neurons in response to CHX treatment, and TMEM25 overexpression accelerates the turnover rate of NR2B.
The TMEM25 expression level is decreased in patients with TLE and in epileptic mice.
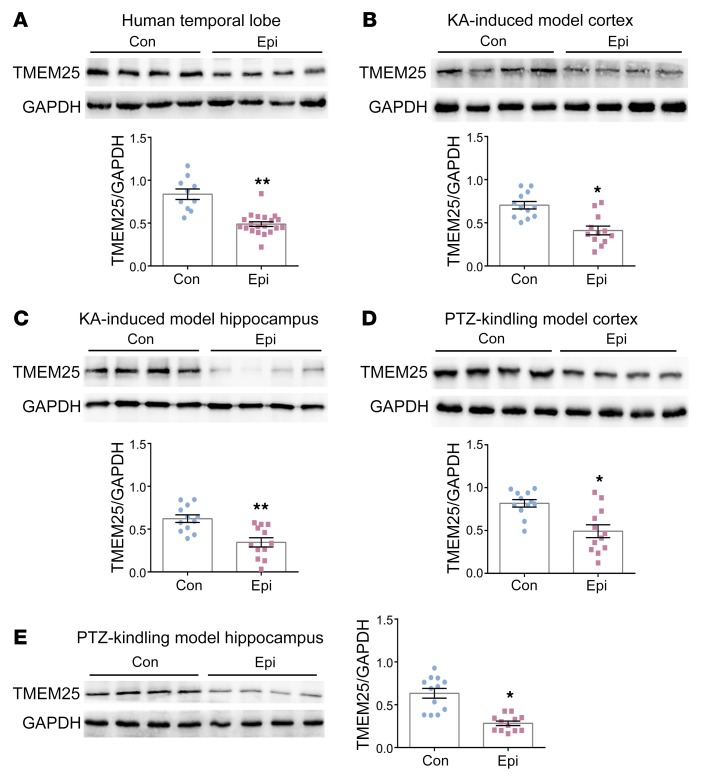
We assessed TMEM25 expression in temporal lobe tissues from patients with TLE and controls to examine the relationship between TMEM25 and epilepsy. The Western blotting results showed decreased levels of TMEM25 in epileptic brain tissues from patients with TLE compared with those in control patients (Figure 6A). We established 2 mouse models to further determine whether the TMEM25 expression level is altered after epileptogenesis. The results suggested that the expression levels of TMEM25 in the cortex and hippocampus derived from KA-induced epileptic mice with spontaneous recurrent seizures (SRSs) or from mice fully kindled by PTZ (15) were reduced compared with those in the controls (Figure 6, B–E). We also used immunofluorescence to detect the expression level of TMEM25 in chronic epileptic tissues. The immunofluorescence results were consistent with the Western blotting results, as the level of TMEM25 was significantly decreased in the cortex and/or hippocampus in patients with TLE (Supplemental Figure 13A) and in KA-induced and PTZ kindling epileptic models (Supplemental Figure 13, B–E). However, no difference in the cellular localization characteristics of TMEM25 was found between epileptic brain tissues and normal brain tissues (Figure 1 and Supplemental Figure 14). In addition, considering the association between TMEM25 and NR2B summarized previously, we also assessed NR2B protein levels by Western blotting in corresponding epileptic brain tissues from animal models and human patients with TLE. The results suggested that NR2B expression is increased in both human and animal epileptic brain tissues compared with that in controls (Supplemental Figure 15).
Figure 6. Expression of TMEM25 in the brain tissues of patients and mice with epilepsy.
(A) Representative images of TMEM25 expression in brain tissues from pharmacoresistant patients with TLE (n = 20) and control individuals with head trauma (n = 10) and quantification of immunoblots. (B–E) Representative images of the expression of TMEM25 in the cortex and hippocampus from chronic epileptic mice (n =12) and normal mice (n = 12) and quantification of immunoblots. Data are presented as means ± SEM and are representative of at least 3 independent repeats. *P < 0.05; **P < 0.01, by Mann-Whitney U test (A and E) or independent Student’s t test (B, C and D).
Effects of TMEM25 on seizure recurrence and epileptic severity.
As previously discussed, TMEM25 is decreased in epileptic tissues. We subsequently performed behavioral tests and analyzed local field potentials (LFPs) to examine whether up- and downregulation of TMEM25 exerted specific effects on seizure activities and epileptiform discharge using the 2 epileptic models. At the end of the behavioral observations, LV-encoded mCherry autofluorescence was detected in the mouse hippocampus, indicating successful lentiviral infection (Supplemental Figure 16A). A significantly lower level of TMEM25 was detected throughout the bilateral hippocampus 6 weeks after LV injection in the mice treated with LV-shTM25 than in the mice treated with Con-shTM25. Furthermore, a significantly higher TMEM25 level was detected in the LV-TM25 group than the Con-LV-TM25 group (Supplemental Figure 16B). Thus, injection of the LVs into the hippocampus effectively altered the endogenous expression of TMEM25.
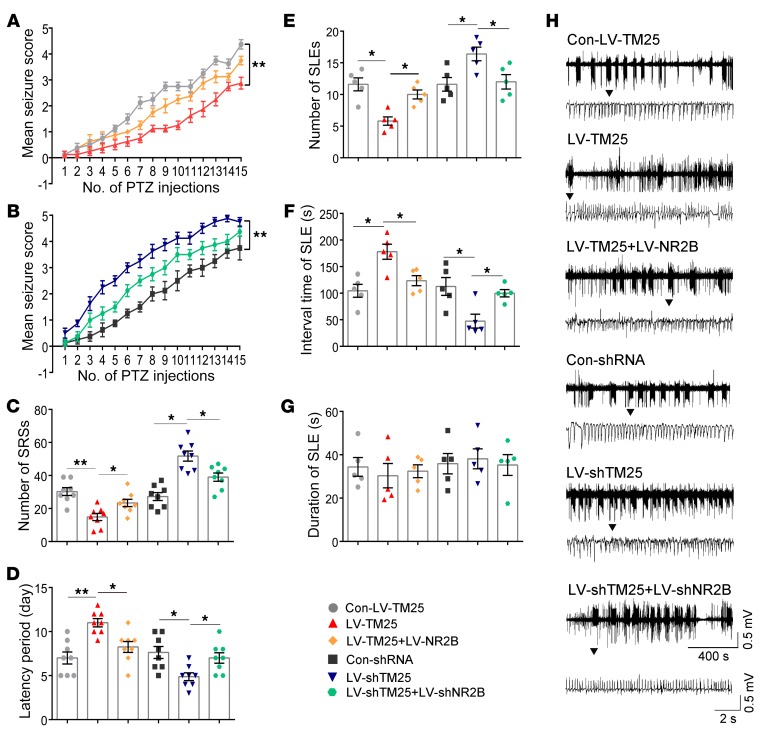
In the PTZ kindling mouse model, we quantified the seizure scores according to Racine’s standard scale during all assessed periods (16). The results suggested that the TMEM25-overexpressing mice had lower seizure scores and that the LV-shTM25–treated mice had higher seizure scores than their respective control groups (Figure 7, A and B). We also assessed whether the SRSs were changed in the mouse KA-induced epilepsy model. According to analyses of the continuous video monitoring system for weeks 1–4 after status epilepticus (SE), fewer SRSs were recorded in the LV-TM25–treated mice than in the Con-LV-TM25–treated mice. The LV-shTM25–treated mice exhibited more SRSs than the Con-shRNA–treated mice (Figure 7C), indicating that the LV-TM25 treatment attenuated the severity of SRSs, whereas the LV-shTM25 treatment aggravated the severity of the SRSs. The latency was significantly decreased in the LV-shTM25 group compared with that in the Con-shRNA group. In contrast, the latency was significantly increased in the LV-TM25 group compared with that in the Con-LV-TM25 group (Figure 7D). During a 30-minute LFP recording, TMEM25 knockdown in the LV-shTM25 group led to seizure-like events (SLEs) that occurred more frequently than in the Con-shRNA controls, and the interval of SLEs in the epileptic mice was significantly decreased. In contrast, TMEM25 overexpression resulted in fewer SLEs in the LV-TM25–treated mice than in the Con-LV-TM25 control mice, and the interval of SLEs in the epileptic mice was significantly increased (Figure 7, E and F). Thus, the expression level of TMEM25 and seizure severity may be correlated.
Figure 7. TMEM25 modulates epileptic seizure activity.
(A and B) Summary of mean seizure scores recorded after i.p. injection of PTZ on alternate days. Data are presented as means ± SEM, n = 8 per group. **P < 0.01, by generalized estimating equation analysis. (C and D) Quantitative analysis of the total number of SRS and latency of SRS in the KA-induced epilepsy model from different groups. Data are presented as means ± SEM, n = 8 per group. *P < 0.05; **P < 0.01, by 1-way ANOVA followed by LSD t test. (E–G) Quantitative analysis of the number, interval time, and duration of SLE of the LFPs from different groups. Data are presented as means ± SEM, n = 5 per group. *P < 0.05, by 1-way ANOVA followed by LSD t test. (H) Representative traces of LFPs from different groups.
As previously stated, TMEM25 probably influenced neuronal excitability by changing the total protein level of NR2B subunits and NMDAR-mediated neurotransmission in neurons. Therefore, exogenously reversing the expression of the NR2B subunit in vivo may rescue the epileptic behaviors and LFP changes mediated by the TMEM25 intervention. Here, NR2B rescue experiments were performed using LVs that carried specific plasmids encoding mouse NR2B or an shRNA directed against NR2B, which were administered via intrahippocampal injection. The results suggested that TMEM25-induced behavioral changes were partially rescued by NR2B (Figure 7).
Discussion
In the present study, we investigated the distribution, subcellular location, and function of TMEM25 in neurons. TMEM25 was mainly expressed in the cytoplasm in neurons and was localized to late endosome/lysosome compartments. Knocking down TMEM25 in pyramidal neurons in the hippocampal CA1 region with lentivirus shRNA induced increases in the amplitude of NMDAR-mediated EPSCs and in the total expression levels of NR2B. In contrast, overexpression of TMEM25 had the opposite effect. TMEM25 was coimmunoprecipitated with NR2B, and these factors colocalized in late endosome/lysosome compartments. TMEM25 participated in the regulation of endolysosomal acidification and NR2B degradation. In addition, we also identified an important role of TMEM25 in epilepsy. TMEM25 expression in the 2 mouse models of epilepsy and in brain tissues from patients with TLE was decreased compared with that in the respective controls. Furthermore, lentiviral-mediated up- or downregulation of TMEM25 in the hippocampus reduced or increased the susceptibility and severity of seizures, respectively. Exogenously manipulating NR2B expression partially rescued the epileptic behavior and electrophysiological changes induced by TMEM25 both in vivo and vitro.
TMEM25 has been reported to be a member of the immunoglobulin superfamily (1), and the various functions of this family include cell adhesion, which plays an important role in synaptic functions as growth factors in the vertebrate nervous system. Our study is the first investigation, to our knowledge, of the biological and electrophysiological roles of TMEM25 in the brain using in vivo and in vitro experiments. We determined that TMEM25 was expressed primarily in neurons rather than astrocytes in the mouse hippocampus and cortex and human brain tissue. Furthermore, TMEM25 displayed cytoplasmic expression and localized to late endosome/lysosome compartments but not early endosomes.
The precise function of TMEM25 in neurons is entirely unknown. Using a whole-cell patch-clamp technique, we determined that TMEM25 knockdown increased the AP frequency and NMDAR-mediated EPSCs (particularly NR2B-sensitive currents). Interestingly, TMEM25 had no effect on mIPSCs, PPR, or AMPA receptor-mediated EPSCs. Thus, TMEM25 affects excitatory postsynaptic transmission, likely by controlling NMDA receptors. Based on the currently available electrophysiological evidence, we used biochemical research methods to further identify specific changes in the receptor subunits. We determined that downregulation of TMEM25 increased the total and membrane expression levels of the NR2B subunit without affecting the expression of NR1 and NR2A. Therefore, the effects of TMEM25 on neuronal excitability may be attributed to changes in the total expression of NR2B.
NMDA receptors dynamically change in response to neuronal activity, and the regulated insertion and removal of NMDA receptors are essential for altering synaptic transmission in the mammalian CNS (17, 18). NMDARs can selectively undergo clathrin-mediated endocytosis and follow different pathways after internalization, incorporating into early endosomes, translocating to late endosomes for degradation or recycling, and then returning to the surface of synapses (19, 20). Thus, NMDAR receptors are regulated not only by stimulus-dependent gene expression, but also by protein degradation. Because TMEM25 has no significant effect on the transcriptional level of NR2B, we hypothesized that the changes in NR2B expression may occur at the protein level. NR2B subunits can be delivered to late endosomes/lysosomes and subsequently degraded (13). Therefore, we investigated the relationship between TMEM25 and NR2B; TMEM25 interacted with NR2B, and these factors colocalized in late endosomes/lysosomes, but not in early endosomes. Accordingly, we hypothesized that TMEM25 likely regulates NR2B degradation in late endosomes/lysosomes.
Late endosome/lysosome functioning is essential for the health of neurons, and these vesicles create an enclosed environment with an acidic pH system that is typically considered the final destination in proteolytic degradation (21). In most neurons, the pH of late endosomes/lysosomes is as low as 4.5 and is critical for multiple physiological functions, including the release of ligands from internalized receptors and recycling of receptors back to the cell surface (22, 23). The lysosomal pH may influence the activity of enzymes in endomembrane compartments and is expected to exert global effects on proteome clearance, particularly for membrane proteins, which rely on this pathway for their regulation and degradation (21). We used the pH-sensitive dye LysoSensor Green DND-189 to investigate the impact of TMEM25 on the late endosome/lysosome pH. Downregulation of TMEM25 significantly increased the lysosomal pH, and TMEM25 overexpression significantly decreased the lysosomal pH. Moreover, TMEM25 downregulation attenuated the rate of total NR2B degradation in response to CHX treatments in neurons, and TMEM25 overexpression accelerated the rate of NR2B degradation. Thus, TMEM25 likely regulates NR2B expression by changing the acidification of late endosomes/lysosomes where NR2B is degraded. Additional studies exploring the regulatory mechanism by which TMEM25 induces the acidification of late endosomes/lysosomes and the link between TMEM25 and NR2B degradation are necessary to test these possibilities.
NMDA receptor NR2B subunits play an important role in the pathophysiology of several neurological diseases, including epilepsy (24–26). Many in vivo and in vitro studies have suggested that NR2B is closely related to epileptogenesis (27, 28). TLE is the most common form of medically intractable epilepsy (29), and we determined that TMEM25 expression is decreased in tissue samples from patients with TLE. Although the hippocampus is the most widely studied brain region in human and experimental epilepsy (30), we could not perform an equivalent comparison of hippocampal TMEM25 expression between patients with TLE and controls for practical and ethical reasons. However, the animal studies provided direct evidence that TMEM25 expression was lower in the cortex and hippocampus of epileptic mice. In fact, behavioral and molecular events associated with PTZ kindling and KA-induced epilepsy in mice largely reproduce the corresponding events in human TLE (31, 32). Furthermore, using both overexpression and knockdown strategies, we demonstrated that TMEM25 interventions can alleviate or aggravate epileptic seizures, providing in vivo evidence for the involvement of TMEM25 in the development of epilepsy.
Evidence from our electrophysiological and behavioral rescue experiments further demonstrated that TMEM25 involvement in epilepsy is based on regulation of NR2B, which may contribute to the imbalance between excitation and inhibition (6). From this perspective, downregulation of TMEM25 may not directly lead to the occurrence of epilepsy, but may rather reduce the susceptibility to epileptogenesis. In other words, TMEM25, which functions based on the NR2B subunit, is a regulatory or influencing factor in epileptic seizures, and a decrease in TMEM25 levels in the brain is unlikely to directly produce seizures. In addition, according to the behavioral and expression data obtained in our study, a causal relationship between TMEM25 and epileptogenesis remains difficult to distinguish. TMEM25-mediated degradation of NR2B is involved in regulation of the development of epilepsy, and seizure recurrence may further promote abnormal TMEM25 expression.
In summary, for what we believe is the first time, we demonstrated the role and mechanism of TMEM25 in regulating neuronal excitability and NR2B function in vivo and in vitro through behavioral, electrophysiological, and pathological studies and provided potential targets for seizure control and disease modification.
Methods
Primary neuron culture.
To prepare primary hippocampal neurons from early postnatal (P0-P1) C57BL/6 mice, postnatal brain tissue was dissected, and the neurons were obtained by enzymatic digestion with trypsin and mechanical dissociation. We diluted the cell suspension with DMEM supplemented with 20% FBS (both from Gibco, Thermo Fisher Scientific). Neurons were plated at a density of 100,000 cells on poly-l-lysine–coated 35 mm dishes or glass coverslips in 6-well plates for further experimental analyses, and the neurons were incubated in a cell culture incubator at 37°C for 4 hours. Four hours after plating, the cells were maintained in neurobasal medium supplemented with B27, 2-mM l-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin (all obtained from Invitrogen) (33).
Cell culture and transfection.
HEK293 cells (Chinese Academy of Sciences) were maintained in DMEM supplemented with 10% FBS, 100 U/mL penicillin (Invitrogen), and 100 μg/mL streptomycin (Invitrogen) at 37°C in 5% CO2. At 24 hours prior to transfection, the HEK293 cells were seeded onto a 35 mm dish at a density of 1.5 × 104 cells per well in 3 mL of culture medium. Transient transfection of the HEK293 cells was performed using Lipofectamine 2000 (Invitrogen) after 24 hours (34). Before the transfection experiment, the cells were rinsed twice with warm PBS (pH 7.4) and subsequently transfected with 2 μg of pCI-SEP-NR2B (Addgene, plasmid 23998) and mCherry-TM25 (GeneChem) on day in vitro (DIV) 14. Four hours after transfection at 37°C in a 5% CO2 atmosphere, the cells were rinsed with warm PBS and supplied with 3 ml of culture medium containing 10% FBS. After 48 hours of incubation to establish enhanced green and red fluorescent protein expression, the cells were rinsed twice with PBS, fixed, and then stained.
Immunofluorescence staining.
The animals were rapidly euthanized, and the brains were immediately postfixed in 4% paraformaldehyde for 24 hours at 4°C. The brains were immersed in a 30% sucrose PBS solution for 48 hours and subsequently sliced using a freezing microtome (Leica); 16 μm sections were collected on glass slides. The sections were blocked with 10% goat serum (BOSTER Biological Technology) and 0.1% Triton X-100 in PBS for 2 hours. The cultured neurons were fixed on DIV 14 with 4% paraformaldehyde/4% sucrose in PBS for 10 minutes and permeabilized with 0.3% Triton X-100 in PBS for 15 minutes at room temperature. The primary antibodies were diluted in 1% (wt/vol) BSA/PBS, added to the coverslip, and incubated overnight in a humidified chamber at 4°C. The coverslips were washed 3 times with PBS and incubated with the secondary fluorescent antibody diluted in 1% (wt/vol) BSA/PBS at room temperature for 1 hour. The following primary antibodies were used: rabbit polyclonal anti-TMEM25 (Proteintech, catalog 24361-1-AP), mouse monoclonal anti-MAP2 (Zhongshan Golden Bridge, catalog TA337109), chicken polyclonal anti-GFAP (Abcam, catalog ab4674), rabbit polyclonal anti-GFP (Invitrogen, catalog A-11122), mouse monoclonal anti-EEA1 (Proteintech, catalog 66218-1-Ig), and mouse monoclonal anti-LAMP2 (Proteintech, catalog 66301-1-Ig). The following secondary antibodies were used: Alexa Fluor 488-conjugated goat anti-rabbit IgG (Zhongshan Golden Bridge, catalog ZF-0511), DyLight 594-conjugated goat anti-mouse IgG (Zhongshan Golden Bridge, catalog ZF-0513), Alexa Fluor 405-conjugated goat anti-chicken IgG (Abcam, catalog 175674), and AMCA-conjugated goat anti-mouse IgG (Beyotime Biotechnology, catalog A0413). Images were captured using laser-scanning confocal microscopy (Nikon) under an Olympus IX 70 inverted microscope equipped with a Fluoview FVX confocal scan head. The fluorescence intensity was analyzed using Image-Pro Plus software, and colocalization analyses were performed using NIS-viewer (Nikon).
Lentivirus vector construction.
As a result of their high levels of expression, consistent targeting of various cells and efficient mediation of stable gene transfer, LVs were specifically selected for our experiments (35). An shRNA with a targeting sequence 5′-ATCCAACCTTCAGCTCAAT-3′ directed against Tmem25 was carried by an LV, designated LV-shTM25, to reduce hippocampal TMEM25 levels. The LV-transferred plasmids encoding mouse TMEM25 are hereafter designated as LV-TM25. The LVs also contained a separate transcription cassette for the red fluorescent protein mCherry. The control vectors Con-LV-TM25 and Con-shRNA expressed mCherry under the control of the same promoter that was used for LV-TM25 and LV-shTM25, respectively. An shRNA with a targeting sequence 5′-GCTGGTGATAATCCTTCTGAA-3′ directed against NR2B was carried by an LV, designated LV-shNR2B, to knock down the expression of NR2B. The LV-transferred plasmids that encoded mouse NR2B are hereafter designated as LV-NR2B.
Intrahippocampal injections of virus.
Mice were randomly assigned to groups based on the type of virus injected. Briefly, the mice were anesthetized and placed in a stereotaxic apparatus (RWD Life Science). Two microliters of virus particles (0.5-1 × 109 TU/mL) was bilaterally injected into the dorsal hippocampus (anterior/posterior: –2.0 mm, medial/lateral: ±1.5 mm, and dorsal/ventral: –1.5 mm) through a glass pipette (0.2 μL/min) using a glass microsyringe. Following injection, the pipette was maintained in place for an additional 5 minutes to prevent backflow of viral particles through the injection probe.
In vitro electrophysiological analysis.
Fourteen days after virus infection, the mice used for the patch-clamp experiments were deeply anesthetized and subsequently decapitated; coronal slices (300 μm thick) were cut using a vibratome (Leica VT1200S). The slices were cut in an ice-cold solution containing the following components: 60 mM NaCl, 100 mM sucrose, 2.5 mM KCl, 1.25 mM NaH2PO4•2H2O, 20 mM d-glucose, 26 mM NaHCO3, 1 mM CaCl2, and 5 mM MgCl2•6H2O saturated with 95% O2 and 5% CO2. The slices were allowed to recover in modified artificial cerebral spinal fluid (ACSF) containing 125 mM NaCl, 3 mM KCl, 1.25 mM NaH2PO4•2H2O, 15 mM d-glucose, 26 mM NaHCO3, and 2 mM CaCl2 saturated with 95% O2 and 5% CO2 at 32°C for at least 45 minutes before recording. This ACSF composition was used for all in vitro recordings. NVP-AAM077 (50 nM; MedChemExpress) or Ro 25-6981 (0.5 μM; Sigma-Aldrich) was added to the perfusate to block NR2A or NR2B subunits, respectively (36).
Glass pipettes were filled with an internal solution consisting of the following components to measure the sAPs: 60 mM K2SO4, 60 mM N-methyl-d-glucamine, 40 mM HEPES, 4 mM MgCl2•6H2O, 0.5 mM BAPTA, 12 mM phosphocreatine, 2 mM Na2ATP, and 0.2 mM Na3GTP. Spontaneous APs were recorded from neurons at the resting membrane potential using a whole-cell patch clamp in current clamp mode.
Glass microelectrodes were filled with a solution containing 130-mM Cs-methanesulfonate, 10 mM HEPES, 10 mM CsCl, 4 mM NaCl, 1 mM MgCl2•6H2O, 1 mM EGTA, 5 mM N-methyl-d-glucamine, 5 mM MgATP, 0.5 mM Na3GTP, and 12 mM phosphocreatine to measure the NMDAR-mediated mEPSCs. Additionally, 10 μM 6,7-dinitroquinoxaline-2,3(1H,4H)-dione (DNQX), 1-μM tetrodotoxin (TTX), and 100-μM picrotoxin (PTX) were added to the ACSF to record the NMDAR-mediated mEPSCs at a holding potential of +40 mV. The solution in the glass microelectrodes in the experiments designed to measure the AMPAR-mediated mEPSCs was the same as that for NMDAR-mediated mEPSCs: 100 μM PTX, 50 μM 2-amino-5-phosphonovaleric acid, and 1 μM TTX were added to the ACSF to record the AMPAR-mEPSCs when held at –70 mV. Glass microelectrodes were filled with the following solution to record the mIPSCs: 100 mM CsCl, 10 mM HEPES, 1 mM MgCl2•6H2O, 1 mM EGTA, 30 mM N-methyl-d-glucamine, 5 mM MgATP, 12 mM phosphocreatine, and 0.5 mM Na3GTP. Slices were submerged and continuously perfused with ACSF containing 10 μM DNQX, 50 μM 2-amino-5-phosphonovaleric acid, and 1 μM TTX to record the mIPSCs when held at –70 mV.
To record evoked EPSCs, glass microelectrodes were filled with the same solution used to record the mEPSCs. The AMPAR- and NMDAR-mediated synaptic responses were evoked by a bipolar stimulation electrode located approximately 50 μm rostral to the recording electrode in the same layer. In the presence of 100 μM PTX, we obtained stable synaptic responses at a holding potential of –70 mV; the amplitude of these responses represents the AMPAR-specific component. At +40 mV, an amplitude of 50 ms poststimulus was identified as the NMDA-specific response.
For PPR recordings, the holding potential was +40 mV in the presence of 100 μM PTX and 10 μM DNQX. The interval for paired stimulations was set at 50 ms. The PPRs were calculated as the ratio of the second peak amplitude to the first peak amplitude.
The signals were acquired using a MultiClamp 700B amplifier (Axon) and were recorded using pClamp 10 software (Molecular Devices). All electrophysiological signals were filtered at 2 kHz and digitized at 10 kHz.
Western blotting analysis.
The collected brain tissue samples and hippocampal neurons used for total protein collection were homogenized in radioimmune precipitation buffer containing 1% PMSF. The obtained homogenate was incubated on ice for 30 minutes and centrifuged in an ultracentrifuge (Sigma-Aldrich) at 16,000 g for 30 minutes at 4°C, and the supernatant fractions were collected. The membrane fraction was collected using a Minute Plasma Membrane Protein Isolation and Cell Fractionation Kit (Invent Biotechnologies, catalog SM-005) according to the manufacturer’s instructions. Briefly, the tissue was placed in a filter cartridge and homogenized in 200 μL of buffer A using a plastic rod for 1 minute. Then, 300 μL of buffer A was added to the same filter cartridge, and the tube was incubated on ice with constant mixing. The homogenates were centrifuged at 16,000 g for 30 seconds at 4°C. The filter was discarded, and the pellet was vigorously resuspended. The homogenates were centrifuged at 700 g for 1 minute. The supernatant was transferred to a fresh 1.5 mL tube and centrifuged at 4°C for 30 minutes at 16,000 g, and then the pellet was collected. The total and membrane protein concentrations were measured by a BCA protein assay (Beyotime Biotechnology).
Equal amounts of proteins (20 μg) were loaded on 10% SDS-PAGE gels and transferred to polyvinylidene fluoride membranes (Millipore). The membranes were blocked with blocking reagent (Beyotime Biotechnology, catalog P0252) for 15 minutes at room temperature, followed by incubation with primary antibodies overnight at 4°C. The membranes were washed 3 times for 5 minutes per wash with Tris-buffered saline with 0.1% Tween-20, incubated with a peroxidase-conjugated secondary antibody (Proteintech, catalog SA00001-2) for 1 hour at room temperature, and then washed 3 additional times with Tris-buffered saline with 0.1% Tween-20. The blots were imaged and quantified using a Fusion Imaging System. The primary antibodies and dilutions used for the Western blot were as follows: rabbit polyclonal anti-TMEM25 (Proteintech, catalog 24361-1-AP), rabbit polyclonal anti-NR2B (Proteintech, catalog 21920-1-AP), rabbit polyclonal anti-NR2A (Proteintech, catalog 19953-1-AP), rabbit monoclonal anti-NR1 (Abcam, catalog ab109182), rabbit polyclonal anti-GluR1 (Abcam, catalog ab31232), rabbit polyclonal anti-GluR2 (Proteintech, catalog 11994-1-AP), rabbit monoclonal anti-GluR3 (Abcam, catalog ab40845), rabbit polyclonal anti-GluR4 (Proteintech, catalog 23350-1-AP), rabbit polyclonal anti-GAPDH (Proteintech, catalog 10494-1-AP), and rabbit polyclonal anti-ATP1A1 (Proteintech, catalog 14418-1-AP). For quantification, the immunoreactive bands were normalized to a corresponding GAPDH or ATP1A1 reference band.
Coimmunoprecipitation.
The tissues were homogenized in 200 μl of lysis buffer, and the tubes containing the lysates were incubated on ice for 30 minutes. The lysates were centrifuged at 16,000 g for 15 minutes at 4°C, and then the supernatant was collected. Twenty microliters of the supernatant was frozen as a lysis control. Forty microliters of magnetic beads (MedChemExpress, catalog HY-K0202) was resuspended and transferred into a 1.5 mL tube. The beads were washed 3 times with wash buffer, incubated with diluted antibody (5 μg/mL), and then rotated for 2 hours at 4°C. The beads were washed 5 times with wash buffer and then incubated with tissue lysates with rotation for 2 hours at 4°C to allow the Ag to bind to the Protein A/G Magnetic Bead-Ab complex. Then, the Magbead-Ab-Ag complex was collected and washed 5 times using wash buffer. Finally, magnetic separation was performed using 40 μL of elution buffer, and the supernatant was collected. Samples of the final solution were used for denaturing SDS-PAGE. Membranes were blocked with blocking reagent (Beyotime Biotechnology, catalog P0252) for 15 minutes and then incubated overnight at 4°C with rabbit polyclonal anti-TMEM25 (Proteintech, catalog 24361-1-AP), rabbit polyclonal anti-NR2B (Proteintech, catalog 21920-1-AP), rabbit polyclonal anti-NR2A (Proteintech, catalog 19953-1-AP), rabbit monoclonal anti-NR1 (Abcam, catalog ab109182), rabbit polyclonal anti-GluR1 (Abcam, catalog ab31232), rabbit polyclonal anti-GluR2 (Proteintech, catalog 11994-1-AP), rabbit monoclonal anti-GluR3 (Abcam, catalog ab40845), and rabbit polyclonal anti-GluR4 (Proteintech, catalog 23350-1-AP) in Tris-buffered saline with 0.1% Tween-20. After washing, the filters were incubated for 1 hour at room temperature with a peroxidase-conjugated secondary antibody (Proteintech, catalog SA00001-2). The blots were imaged using a Fusion Imaging System.
Quantitative real-time PCR analysis of NR2B.
Total RNA was extracted from the hippocampi of the Con-LV-TM25–, LV-TM25–, Con-shRNA–, and LV-shTM25–treated mice using standard protocols. cDNA was subsequently synthetized using HiScript II qRT Super Mix (Vazyme). Real-time PCR was performed using
Ace Q qPCR SYBR Green Master Mix (Vazyme) and an iQ5 Real-Time PCR detection system (Bio-Rad). The PCR primer sequences used to amplify NR2B and GAPDH mRNAs are listed in Supplemental Table 1. The relative mRNA levels were determined according to the average cycle threshold value (Ct) and were normalized to the level of the housekeeping gene Gapdh. Fold changes were quantified using the 2−ΔΔCt method (37).
LysoSensor Green DND-189 staining.
Primary cultured neurons were prepared as previously described and plated onto poly-l-lysine–coated 6-well plates. On DIV 2, the neurons were infected with different lentiviruses that encoded an mCherry reporter. On DIV 14, the neurons were stained with LysoSensor Green DND-189 (1 μM; Yeasen, catalog 40767ES50) for 30 minutes. The neurons were subsequently washed once with neurobasal medium and immediately imaged. The images of the LysoSensor and lentivirus fluorescence were obtained using a ×40 objective lens. All images were uniformly thresholded for the LysoSensor- or mCherry-positive area analysis (34). The fluorescence intensity of LysoSensor in the mCherry-positive areas was analyzed using Image-Pro Plus software.
NR2B degradation assay.
Neurons were cultured as previously described. On DIV 2, neurons were infected with LVs. On DIV 14, primary neurons were treated with CHX (20 μg/mL; MedChemExpress) at 37°C to inhibit further protein synthesis (38). At the indicated time points, total protein was extracted from the cells as previously described, and the total protein expression of the NMDAR and AMPAR subunits was analyzed by Western blotting.
Human samples.
Cortical tissue from the temporal lobes of patients with drug-refractory TLE or brain trauma was surgically obtained from the First Affiliated Hospital of Chongqing Medical University. All patients with TLE had typical epilepsy symptoms and electroencephalographic features, and their seizures persisted despite more than 2 years of medical therapy with 3 or more kinds of AEDs at effective blood drug concentrations. The seizure type was diagnosed based on the proposed classification of epileptic seizures by the International League Against Epilepsy in 2001 (39). Temporal lobe resection was performed in accordance with a conventional standard protocol, namely, the en bloc anterior temporal lobe resection procedure (40). Before surgery, the epileptic lesion was localized by high-resolution magnetic resonance imaging, prolonged video-EEG monitoring, and/or PET. Depth electrodes were embedded in patients whose lesions could not be localized by the previously mentioned techniques. Surgical specimens were subjected to routine histopathological examination. Two neuropathologists independently reviewed all samples. The age- and sex-matched control group consisted of temporal lobe tissue samples obtained from patients who were treated for increased intracranial pressure secondary to traumatic brain injury and had no history of epilepsy, no exposure to AEDs, and no other history of neurological or psychiatric disorders. The 2 neuropathologists also reviewed these cases and confirmed that the histopathological findings of the specimens were normal. The demographic and clinical features of the patients are summarized in Supplemental Table 2.
Mouse seizure models.
For the epilepsy models, specific pathogen–free C57BL/6 male mice (20–25 g) were housed 5 animals per cage in the Experimental Animal Center of Chongqing Medical University. The mice were housed under standard conditions (room temperature, 23 ± 1°C; illumination, 12-hour light/12-hour dark cycle; access to food and water, ad libitum). Two weeks after intrahippocampal injections of viruses into the mice, seizures were induced using the PTZ kindling model or KA model.
In the PTZ kindling model, mice received i.p. injections of 35 mg/kg PTZ (Sigma-Aldrich) every other day for 30 days. Immediately after each injection, the animals were observed for 60 minutes, and the evoked behavioral seizures were evaluated according to Racine’s standard scale (1972; ref. 16).
In the KA model, mice were injected with 1.0 nmol of KA (Sigma-Aldrich) in 50 nl of saline as previously described (41, 42). Two hours after KA injection, nonconvulsive SE was terminated using diazepam. Then, the mice were video recorded for 24 hours to monitor SRSs for 4 weeks. Seizure scores were evaluated on Racine’s standard scale (1972; ref. 16), and only seizures with a score of 4 or 5 were recorded to determine the number of SRSs and the latency period between SE termination and the first SRS (43). Furthermore, intracranial LFP recordings were performed as previously described (41) to assess the effects of the different virus vectors on electrographic seizures at the end of video recording.
Statistics.
For independent samples, the independent Student’s t test (2-tailed) or Mann-Whitney U test was performed to compare 2 groups. Comparisons of more than 2 groups were performed using 1-way ANOVA. Two-way ANOVA was used to detect the differences between 2 or more groups at multiple time points. Generalized estimating equation analysis was used to detect the differences of repeated measurement data between 2 or more groups (SPSS version 17.0). If the data did not meet the assumptions of normality and homogenous variances, the nonparametric Kruskal-Wallis test was used instead. The χ2 test was applied to explore the gender differences between the groups of patients with TLE and the controls. Graphs were prepared by GraphPad Prism 4 software. P < 0.05 was considered statistically significant and was indicated as *P < 0.05, **P < 0.01, or ***P < 0.001 in the summary graphs.
Ethical approval and consent to participate.
The study protocol related to human subjects was performed in accordance with the Declaration of Helsinki and approved by the Ethics Committee of the First Affiliated Hospital of Chongqing Medical University. Written informed consent for the use of tissues in this study was obtained before surgery. All animal studies were approved by the Ethics Committee of the Chongqing Medical University and were conducted in accordance with the principles outlined in the Animal Research: Reporting In Vivo Experiments guidelines.
Author contributions
HQZ, XT, FX, and XFW conceived the project and designed the experiments. HQZ, XT, XL, DMX, YG, YL, YLM, CZC, YY, MY, YY, FL, RJZ, and MQH performed the experiments. HQZ and FX analyzed the data. HQZ, XT, ZFD, FX, and XFW wrote the manuscript. All authors revised and approved the final version of the manuscript. The order of co–first authors in author list was determined by the workload.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (Grant nos. 81671301, 81471319, 81873788, 81622015, and 81571042) and the National Clinical Key Specialty Construction Foundation of China. The authors are sincerely grateful for the support of Beijing Tiantan Hospital, Xuanwu Hospital of Capital University of Medical Sciences, and Daping Hospital of the Third Military Medical University for supplying the brain tissues. In addition, we thank all the patients and their families for participating in this study.
Version 1. 08/19/2019
Electronic publication
Version 2. 09/03/2019
Print issue publication
Funding Statement
This work was supported by grants from the National Natural Science Foundation of China (Grant numbers: 81671301, 81471319, 81873788, 81622015 and 81571042) and the National Clinical Key Specialty Construction Foundation of China. The authors are sincerely grateful for the support of Beijing Tiantan Hospital, Xuanwu Hospital of Capital University of Medical Sciences, and Daping Hospital of the Third Military Medical University for supplying the brain tissues. In addition, we thank all the patients and their families for participating in this study.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(9):3864–3876.https://doi.org/10.1172/JCI122599.
Contributor Information
Haiqing Zhang, Email: cqmuhaiqing@126.com.
Xin Tian, Email: cqmutx@163.com.
Xi Lu, Email: luxi_1988@163.com.
Demei Xu, Email: tiankongfeiyu204@163.com.
Yi Guo, Email: guoyi870720@126.com.
Zhifang Dong, Email: zfdong@aliyun.com.
Yun Li, Email: cqmuyunli@163.com.
Yuanlin Ma, Email: ylinma@126.com.
Chengzhi Chen, Email: chengzhichen@cqmu.edu.cn.
Yong Yang, Email: yangyongdoc@126.com.
Min Yang, Email: yangmin_9535@126.com.
Yi Yang, Email: yangyi_cqm@126.com.
Feng Liu, Email: fengliu_19910405@163.com.
Ruijiao Zhou, Email: ruijiao_zhou@126.com.
Miaoqing He, Email: miaoqing_he@126.com.
Fei Xiao, Email: feixiao_81@163.com.
Xuefeng Wang, Email: cqmuwxf@163.com.
References
- 1.Katoh M, Katoh M. Identification and characterization of human TMEM25 and mouse Tmem25 genes in silico. Oncol Rep. 2004;12(2):429–433. [PubMed] [Google Scholar]
- 2.Hrašovec S, Hauptman N, Glavač D, Jelenc F, Ravnik-Glavač M. TMEM25 is a candidate biomarker methylated and down-regulated in colorectal cancer. Dis Markers. 2013;34(2):93–104. doi: 10.1155/2013/427890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doolan P, et al. TMEM25, REPS2 and Meis 1: favourable prognostic and predictive biomarkers for breast cancer. Tumour Biol. 2009;30(4):200–209. doi: 10.1159/000239795. [DOI] [PubMed] [Google Scholar]
- 4. Lowry ER. The GluK4 kainate receptor subunit regulates memory, mood, and excitotoxic neurodegeneration [dissertation]. New York: Rockefeller University; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ngugi AK, Bottomley C, Kleinschmidt I, Sander JW, Newton CR. Estimation of the burden of active and life-time epilepsy: a meta-analytic approach. Epilepsia. 2010;51(5):883–890. doi: 10.1111/j.1528-1167.2009.02481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morimoto K, Fahnestock M, Racine RJ. Kindling and status epilepticus models of epilepsy: rewiring the brain. Prog Neurobiol. 2004;73(1):1–60. doi: 10.1016/j.pneurobio.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 7.Perucca E, French J, Bialer M. Development of new antiepileptic drugs: challenges, incentives, and recent advances. Lancet Neurol. 2007;6(9):793–804. doi: 10.1016/S1474-4422(07)70215-6. [DOI] [PubMed] [Google Scholar]
- 8.Duncan JS, Sander JW, Sisodiya SM, Walker MC. Adult epilepsy. Lancet. 2006;367(9516):1087–1100. doi: 10.1016/S0140-6736(06)68477-8. [DOI] [PubMed] [Google Scholar]
- 9. [No authors listed]. Allen Brain Atlases and Data. Allen Institute for Brain Science. http://www.brain-map.org Accessed August 7, 2019.
- 10.Zucker RS, Regehr WG. Short-term synaptic plasticity. Annu Rev Physiol. 2002;64:355–405. doi: 10.1146/annurev.physiol.64.092501.114547. [DOI] [PubMed] [Google Scholar]
- 11.Laube B, Hirai H, Sturgess M, Betz H, Kuhse J. Molecular determinants of agonist discrimination by NMDA receptor subunits: analysis of the glutamate binding site on the NR2B subunit. Neuron. 1997;18(3):493–503. doi: 10.1016/S0896-6273(00)81249-0. [DOI] [PubMed] [Google Scholar]
- 12.Ying Z, Babb TL, Mikuni N, Najm I, Drazba J, Bingaman W. Selective coexpression of NMDAR2A/B and NMDAR1 subunit proteins in dysplastic neurons of human epileptic cortex. Exp Neurol. 1999;159(2):409–418. doi: 10.1006/exnr.1999.7188. [DOI] [PubMed] [Google Scholar]
- 13.Scott DB, Michailidis I, Mu Y, Logothetis D, Ehlers MD. Endocytosis and degradative sorting of NMDA receptors by conserved membrane-proximal signals. J Neurosci. 2004;24(32):7096–7109. doi: 10.1523/JNEUROSCI.0780-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diering GH, Numata M. Endosomal pH in neuronal signaling and synaptic transmission: role of Na(+)/H(+) exchanger NHE5. Front Physiol. 2014;4:412. doi: 10.3389/fphys.2013.00412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gelfuso EA, et al. Parawixin2, a novel non-selective GABA uptake inhibitor from Parawixia bistriata spider venom, inhibits pentylenetetrazole-induced chemical kindling in rats. Neurosci Lett. 2013;543:12–16. doi: 10.1016/j.neulet.2013.02.074. [DOI] [PubMed] [Google Scholar]
- 16.Racine RJ. Modification of seizure activity by electrical stimulation. II. Motor seizure. Electroencephalogr Clin Neurophysiol. 1972;32(3):281–294. doi: 10.1016/0013-4694(72)90177-0. [DOI] [PubMed] [Google Scholar]
- 17.Washbourne P, Liu XB, Jones EG, McAllister AK. Cycling of NMDA receptors during trafficking in neurons before synapse formation. J Neurosci. 2004;24(38):8253–8264. doi: 10.1523/JNEUROSCI.2555-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ehlers MD. Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat Neurosci. 2003;6(3):231–242. doi: 10.1038/nn1013. [DOI] [PubMed] [Google Scholar]
- 19.Prybylowski K, Chang K, Sans N, Kan L, Vicini S, Wenthold RJ. The synaptic localization of NR2B-containing NMDA receptors is controlled by interactions with PDZ proteins and AP-2. Neuron. 2005;47(6):845–857. doi: 10.1016/j.neuron.2005.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lavezzari G, McCallum J, Dewey CM, Roche KW. Subunit-specific regulation of NMDA receptor endocytosis. J Neurosci. 2004;24(28):6383–6391. doi: 10.1523/JNEUROSCI.1890-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu YB, Dammer EB, Ren RJ, Wang G. The endosomal-lysosomal system: from acidification and cargo sorting to neurodegeneration. Transl Neurodegener. 2015;4:18. doi: 10.1186/s40035-015-0041-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishida Y, Nayak S, Mindell JA, Grabe M. A model of lysosomal pH regulation. J Gen Physiol. 2013;141(6):705–720. doi: 10.1085/jgp.201210930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pillay CS, Elliott E, Dennison C. Endolysosomal proteolysis and its regulation. Biochem J. 2002;363(pt 3):417–429. doi: 10.1042/0264-6021:3630417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalia LV, Kalia SK, Salter MW. NMDA receptors in clinical neurology: excitatory times ahead. Lancet Neurol. 2008;7(8):742–755. doi: 10.1016/S1474-4422(08)70165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waxman EA, Lynch DR. N-methyl-D-aspartate receptor subtypes: multiple roles in excitotoxicity and neurological disease. Neuroscientist. 2005;11(1):37–49. doi: 10.1177/1073858404269012. [DOI] [PubMed] [Google Scholar]
- 26.Mathern GW, et al. Hippocampal N-methyl-D-aspartate receptor subunit mRNA levels in temporal lobe epilepsy patients. Ann Neurol. 1999;46(3):343–358. doi: 10.1002/1531-8249(199909)46:3<343::AID-ANA10>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 27.Möddel G, et al. The NMDA receptor NR2B subunit contributes to epileptogenesis in human cortical dysplasia. Brain Res. 2005;1046(1–2):10–23. doi: 10.1016/j.brainres.2005.03.042. [DOI] [PubMed] [Google Scholar]
- 28.Di Maio R, Mastroberardino PG, Hu X, Montero L, Greenamyre JT. Pilocapine alters NMDA receptor expression and function in hippocampal neurons: NADPH oxidase and ERK1/2 mechanisms. Neurobiol Dis. 2011;42(3):482–495. doi: 10.1016/j.nbd.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 29.Téllez-Zenteno JF, Hernández-Ronquillo L. A review of the epidemiology of temporal lobe epilepsy. Epilepsy Res Treat. 2012;2012:630853. doi: 10.1155/2012/630853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thom M. Review: Hippocampal sclerosis in epilepsy: a neuropathology review. Neuropathol Appl Neurobiol. 2014;40(5):520–543. doi: 10.1111/nan.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ben-Ari Y. Cell death and synaptic reorganizations produced by seizures. Epilepsia. 2001;42(suppl 3):5–7. doi: 10.1046/j.1528-1157.2001.042suppl.3005.x. [DOI] [PubMed] [Google Scholar]
- 32.Wilczynski GM, et al. Important role of matrix metalloproteinase 9 in epileptogenesis. J Cell Biol. 2008;180(5):1021–1035. doi: 10.1083/jcb.200708213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beaudoin GM, et al. Culturing pyramidal neurons from the early postnatal mouse hippocampus and cortex. Nat Protoc. 2012;7(9):1741–1754. doi: 10.1038/nprot.2012.099. [DOI] [PubMed] [Google Scholar]
- 34.Klein ZA, et al. Loss of TMEM106B ameliorates lysosomal and frontotemporal dementia-related phenotypes in progranulin-deficient mice. Neuron. 2017;95(2):281–296.e6. doi: 10.1016/j.neuron.2017.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bai L, et al. Effect of lentivirus-mediated gene silencing, targeting toll-like receptor 2, on corneal allograft transplantation in rats. Mol Immunol. 2017;91:97–104. doi: 10.1016/j.molimm.2017.08.022. [DOI] [PubMed] [Google Scholar]
- 36.Haseneder R, et al. The xenon-mediated antagonism against the NMDA receptor is non-selective for receptors containing either NR2A or NR2B subunits in the mouse amygdala. Eur J Pharmacol. 2009;619(1–3):33–37. doi: 10.1016/j.ejphar.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 37.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 38.Maier W, et al. LRP1 is critical for the surface distribution and internalization of the NR2B NMDA receptor subtype. Mol Neurodegener. 2013;8:25. doi: 10.1186/1750-1326-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Engel J, International League Against Epilepsy (ILAE) A proposed diagnostic scheme for people with epileptic seizures and with epilepsy: report of the ILAE Task Force on Classification and Terminology. Epilepsia. 2001;42(6):796–803. doi: 10.1046/j.1528-1157.2001.10401.x. [DOI] [PubMed] [Google Scholar]
- 40.Falconer MA, Taylor DC. Surgical treatment of drug-resistant epilepsy due to mesial temporal sclerosis. Etiology and significance. Arch Neurol. 1968;19(4):353–361. doi: 10.1001/archneur.1968.00480040019001. [DOI] [PubMed] [Google Scholar]
- 41.Sada N, Lee S, Katsu T, Otsuki T, Inoue T. Epilepsy treatment. Targeting LDH enzymes with a stiripentol analog to treat epilepsy. Science. 2015;347(6228):1362–1367. doi: 10.1126/science.aaa1299. [DOI] [PubMed] [Google Scholar]
- 42.Riban V, Bouilleret V, Pham-Lê BT, Fritschy JM, Marescaux C, Depaulis A. Evolution of hippocampal epileptic activity during the development of hippocampal sclerosis in a mouse model of temporal lobe epilepsy. Neuroscience. 2002;112(1):101–111. doi: 10.1016/S0306-4522(02)00064-7. [DOI] [PubMed] [Google Scholar]
- 43.Pitkänen A. Therapeutic approaches to epileptogenesis—hope on the horizon. Epilepsia. 2010;51(suppl 3):2–17. doi: 10.1111/j.1528-1167.2010.02602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.