Abstract
A population of NK cells expressing the activating receptor NKG2C and the maturation marker CD57 expands in response to human CMV (HCMV) infection. CD3–CD56dimCD57+NKG2C+ NK cells are similar to CD8+ memory T cells with rapid and robust effector function upon restimulation, persistence, and epigenetic remodeling of the IFNG locus. Chronic antigen stimulation drives CD8+ memory T cell proliferation, while also inducing genome-wide epigenetic reprograming and dysfunction. We hypothesized that chronic stimulation could similarly induce epigenetic reprograming and dysfunction in NK cells. Here, we show that chronic stimulation of adaptive NK cells through NKG2C using plate-bound agonistic Abs in combination with IL-15 drove robust proliferation and activation of CD3–CD56dimCD57+NKG2C+ NK cells, while simultaneously inducing high expression of the checkpoint inhibitory receptors LAG-3 and PD-1. Marked induction of checkpoint inhibitory receptors was also observed on the surface of adaptive NK cells cocultured with HCMV-infected endothelial cells. Chronically stimulated adaptive NK cells were dysfunctional when challenged with tumor targets. These cells exhibited a pattern of epigenetic reprograming, with genome-wide alterations in DNA methylation. We believe our study has important implications for cancer immunotherapy and propose that exhausted NK cells could be targeted with inhibitory checkpoint receptor blockade.
Keywords: Immunology
Keywords: NK cells
Introduction
NK cells are innate lymphocytes with direct cytotoxic activity against infected or malignant cells. They also help orchestrate adaptive immune responses through the secretion of inflammatory cytokines such as TNF and IFN-γ. The importance of NK cells in host protection is underscored by reports of individuals with primary NK cell deficiencies who present with severe and recurrent herpesvirus infections including herpes simplex virus (HSV), EBV, varicella zoster virus (VSV), and CMV. Nearly half of patients with an identified NK cell deficiency died prematurely, highlighting the severity of their disease (1).
In both mice and humans, CMV infection is associated with clone-like expansions of NK cells that directly recognize viral antigens. Studies of mouse CMV (MCMV) infection identified a subset of NK cells in C57BL/6 mice expressing the activating receptor Ly49H that specifically recognize the MCMV-encoded glycoprotein m157 (2, 3). The Ly49H-m157 interaction is critical for host control of MCMV infection. In response to MCMV infection, Ly49H+ NK cells undergo expansion, contraction, and persistence phases and confer specific protection against MCMV rechallenge. Thus, these cells are regarded as MCMV-specific memory cells (4). Additionally, animal models of influenza, HSV, and SIV have indicated the potential existence of NK cells with memory attributes (5–7).
Compared with mice, the human NK cell response to CMV appears to be somewhat more complex. Many, but not all, individuals seropositive for human CMV (HCMV) have elevated frequencies of NK cells expressing the lectin-like heterodimeric receptor CD94/NKG2C circulating in their peripheral blood (8). The expansion of NKG2C+ NK cells appears to occur specifically in response to HCMV infection, as no expansions of NKG2C+ NK cells have been observed in studies of patients with recurrent HSV infections (9) or acute EBV infections (10). The expansion of NKG2C+ NK cells is dependent on expression of the HLA-E class Ib molecule on infected cells. HLA-E constitutes the major ligand for CD94/NKG2 NK cell receptors. Both NKG2C and its inhibitory counterpart NKG2A bind HLA-E (11, 12). The affinity of NKG2C and NKG2A for HLA-E is strongly influenced by HLA-E–bound peptides (13). In HCMV-infected cells, HLA-E can also present virus-encoded UL40 peptides and induce NKG2C+ NK cell expansion (14, 15).
NKG2C+ NK cells that expand in response to HCMV infection express lower levels of NKG2A (8), rapidly acquire the maturation marker CD57 (16), and exhibit a stable skewing of the killer cell Ig-like receptor (KIR) profile with a bias for both self-specific inhibitory KIRs and activating KIRs (17). Functionally, NKG2C+ NK cells from HCMV-seropositive individuals exhibit decreased responsiveness to IL-12 and IL-18 stimulation but enhanced Ab-dependent cellular cytotoxicity (ADCC) and a greater capacity to produce IFN-γ (17, 18), which is reflected in epigenetic remodeling of the IFNG locus (18, 19). Interestingly, HCMV-seropositive individuals who are homozygously null for KLRC2 (the gene encoding NKG2C) are asymptomatic and healthy, suggesting that NK cells have redundant pathways for responding to HCMV. Part of this redundancy appears to involve expression of high levels of CD2, which can synergistically enhance signaling following CD16 ligation (20). Because NKG2C+ NK cells exhibit pathogen specificity (8), long-term persistence (21), and control of secondary infections (22), we refer to these cells as “adaptive.”
Beyond NKG2C, subsets of NK cells in many HCMV-seropositive individuals lack the signaling adaptor proteins FcεRγ, SYK, and/or EAT-2 in a seemingly stochastic fashion as well as the transcription factors promyelocytic leukemia zinc finger (PLZF) and HELIOS (18, 23). Lack of FcεRγ, SYK, and EAT-2 correlates, but does not completely overlap, with NKG2C expression. Modulation of signaling molecule expression appears to diversify NK cell function, as cells lacking FcεRγ exhibit enhanced ADCC, and those lacking either FcεRγ or EAT-2 manifest significantly lower degranulation in response to activated, autologous T cells. Importantly, examination of genome-wide DNA methylation in NK cell and CD8+ T cell subsets from HCMV-seropositive individuals revealed that adaptive NK cells have significant alterations compared with canonical NK cell subsets, with widespread hyper- as well as hypomethylation. Furthermore, the methylation profile of adaptive NK cells correlated strongly with that of effector memory CD8+ T cells (18).
T cell exhaustion was first identified during chronic lymphocytic choriomeningitis virus (LCMV) infection in mice as virus-specific CD8+ T cells that are unable to produce cytokines (24). Persistent LCMV infection is associated with CD8+ T cell impairment, which occurs in a hierarchical fashion, in which T cells lose the ability to produce IL-2, followed by the ability to make TNF, and then become unable to produce IFN-γ (25). Inhibitory receptors have a key role in T cell exhaustion, and high expression of multiple inhibitory receptors is a hallmark feature of the exhaustion of T cells in both animal models and in humans (26). Programmed cell death 1 (PD-1) is a major inhibitory receptor involved in T cell exhaustion, and blocking PD-1 during chronic LCMV infection reinvigorates virus-specific CD8+ T cell responses and reduces viral load (27). In addition to PD-1, several other cell-surface inhibitory receptors influence T cell exhaustion. Virus-specific CD8+ T cells responding to chronic viral infection in animal models and in humans can also coexpress lymphocyte activation gene 3 (LAG-3), CD244 (2B4), cytotoxic T lymphocyte–associated 4 (CTLA-4), T cell Ig and mucin domains–containing protein 3 (TIM-3), and other inhibitory receptors (28). The pattern of inhibitory receptor induction and the number of receptors simultaneously expressed reflect the severity of dysfunction (29). Recovery of T cell function can be increased considerably by simultaneous blockade of the PD-1 pathway in combination with LAG-3, CTLA-4, or TIM-3 (28, 30, 31). Tumor-infiltrating CD8+ T cells within solid tumors frequently also display high levels of inhibitory receptors and a diminished capacity to produce effector cytokines such as TNF and IFN-γ (32, 33). These phenotypic and functional attributes are epigenetically imprinted and are driven by chronic antigen exposure and T cell receptor (TCR) stimulation (34–37).
Studies on NK cells collected from patients with cancer have reported diminished cytolytic activity and inflammatory cytokine production compared with NK cells from peritumor regions or from the peripheral blood (38). Additionally, PD-1 was found to be upregulated on NK cells from patients with renal cell carcinoma (39), multiple myeloma (40), and Kaposi sarcoma (41). However, the molecular basis of NK cell exhaustion remains largely unexplored. We hypothesized that hallmarks of T cell exhaustion including checkpoint inhibitory receptor expression, dysfunction, and epigenetic imprinting could be induced in adaptive NK cells by chronic stimulation. This hypothesis was based on the findings that adaptive NK cells expand in vivo in response to a chronic infection (HCMV), and that they have a unique epigenetic signature strikingly similar to that of effector memory CD8+ T cells (18). Our study reveals that chronic stimulation of adaptive NK cells through NKG2C ligation leads to robust proliferation and a concomitant decrease in effector function. These dysfunctional cells expressed high levels of LAG-3 and PD-1 and exhibited whole-genome epigenetic reprograming.
Results
Chronic stimulation drives adaptive NK cell proliferation.
Previous work in murine models has shown that tumor-induced CD8+ T cell dysfunction is predominantly driven by persistent TCR signaling during antigen engagement, and while microenvironmental factors may contribute, they are not sufficient to induce exhaustion (34, 35). Thus, we reasoned that we would be able to set up a simple system to study the effects of chronic activating receptor stimulation on primary NK cells cultured ex vivo and profile NK cell exhaustion. To test this hypothesis, we acquired 2 mAbs developed by R&D Systems. The first Ab (clone 134571; anti-NKG2C) is specific for NKG2C, and the second Ab (clone 131415; anti-NKG2A/C) is specific for both NKG2C and NKG2A, which is structurally similar to NKG2C but inhibits NK cell activation through recruitment of the tyrosine phosphatases SHP1 and SHP2 (42). Both Abs were shown to induce robust NK cell cytotoxicity in redirected lysis assays using Ab-coated P815 cells (43).
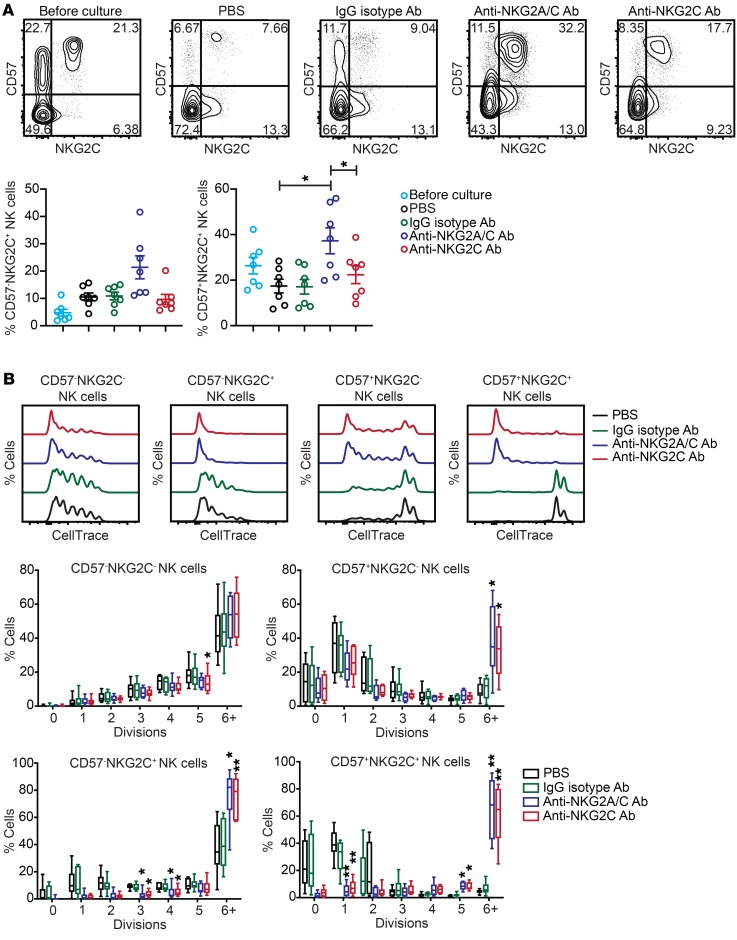
To test whether chronic NKG2C engagement can drive NK cell proliferation, we screened peripheral blood from healthy HCMV-seropositive donors to identify individuals with adaptive NK cell expansions (≥15% of all CD3–CD56dim NK cells having a CD57+NKG2C+ phenotype). PBMCs from these donors were depleted of T and B cells, and the remaining cells (consisting of monocytes and NK cells) were cultured for 7 days with 10 ng/ml IL-15 and either PBS, plate-bound IgG isotype control Ab, plate-bound anti-NKG2A/C Ab, or plate-bound anti-NKG2C Ab. We observed an enrichment of both CD57–NKG2C+ and CD57+NKG2C+ NK cells in anti-NKG2A/C Ab cultures relative to PBS and IgG isotype controls. Interestingly, we found that NKG2C surface expression was significantly lower on NK cells stimulated with the anti-NKG2C Ab relative to those stimulated with the anti-NKG2A/C Ab (Figure 1A). This effect may be due to downmodulation or internalization of NKG2C, a phenomenon that has been previously described upon NK cell stimulation with HLA-E:G peptide complexes (15, 44). For direct measurement of proliferation, we labeled NK cells with CellTrace dye prior to culturing and repeated these culture experiments. We observed significantly more cell division for CD57–NKG2C+, CD57+NKG2C–, and CD57+NKG2C+ NK cells in anti-NKG2A/C and anti-NKG2C Ab cultures relative to PBS and IgG isotype controls (Figure 1B). The increase in proliferation observed for the CD57+NKG2C– subset may be due to low NKG2C expression on these cells or could reflect cells that downmodulated NKG2C after stimulation. Collectively, our data show that IL-15 in combination with NKG2C stimulation is sufficient to drive adaptive NK cell proliferation.
Figure 1. Chronic stimulation through NKG2C expands adaptive NK cells.
CD3/CD19-depleted PBMCs from HCMV-seropositive donors were cultured for 7 days with 10 ng/ml IL-15 and PBS, IgG2b isotype Ab, anti-NKG2A/C Ab, or anti-NKG2C Ab. (A) Representative FACS plots and summary data showing the percentages of NK cell subsets defined by expression of CD57 and NKG2C before and after a 7-day culture (n = 7). Results are from 3 independent experiments. (B) NK cells were labeled with CellTrace dye prior to culturing. Shown are FACS plots of representative donor cells stratified by CD57 and NKG2C expression and summary data (n = 6). Results are from 3 independent experiments. *P ≤ 0.05 by paired t test. P values for multiple group comparisons (A, each group vs. PBS; B, each group vs. IgG2b isotype Ab) were adjusted using the Hommel method.
IL-15 stimulation induces NKG2A expression on the surface of adaptive NK cells.
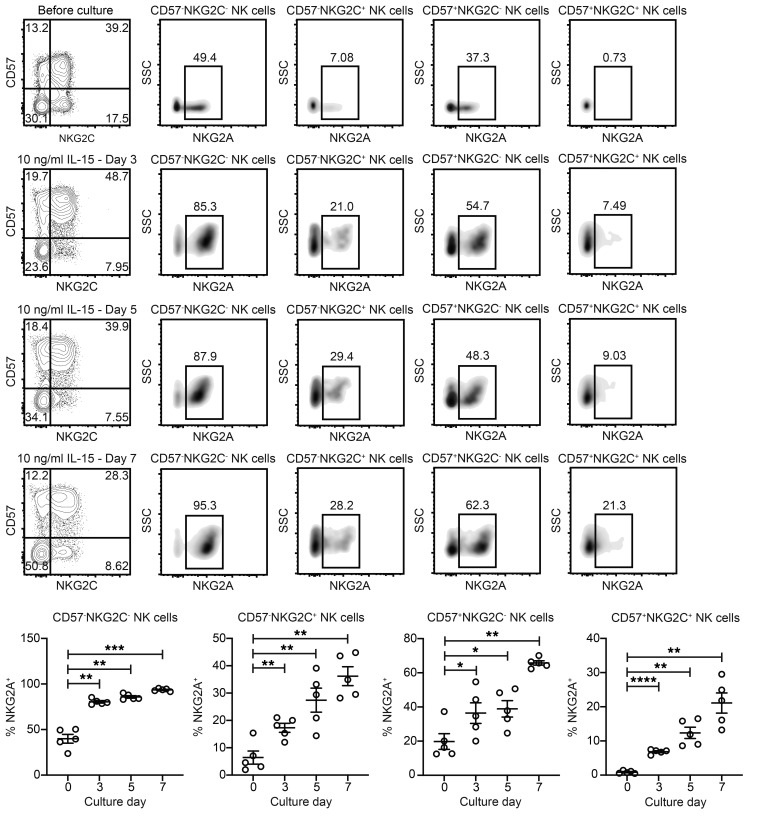
The finding that NKG2C expression was lower on NK cells stimulated with the anti-NKG2C Ab relative to cells stimulated with the anti-NKG2A/C Ab led us to hypothesize that simultaneous engagement of NKG2A might prevent activation-induced NKG2C downmodulation. NKG2A and NKG2C are dichotomously expressed on freshly isolated peripheral blood NK cells from most individuals (45). However, previous studies have shown that common γ-chain cytokines including IL-2 and IL-15 can drive NKG2A upregulation on NK cells and CD8+ T cells (46, 47). To determine whether IL-15 can induce NKG2A expression on adaptive NK cells, we cultured CD3- and CD19-depleted (CD3/CD19-depleted) cells from HCMV-seropositive donors for 7 days with 10 ng/ml IL-15. We found that NKG2A expression increased over time on the surface of all NK cells stimulated with IL-15, including the CD57–NKG2C+ and CD57+NKG2C+ subsets (Figure 2). Similar percentages of NKG2C+ NK cells expressing NKG2A were observed in the anti-NKG2A/C Ab and anti-NKG2C Ab cultures that contained 10 ng/ml IL-15 (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI125916DS1). Additionally, we observed continuous upregulation of NKG2A and a concomitant decrease in NKG2C on sorted NKG2C+ NK cells that were cultured with 10 ng/ml IL-15 (Supplemental Figure 2). Thus, IL-15 stimulation is sufficient to induce NKG2A expression on adaptive NK cells, and engagement of NKG2A may inhibit NKG2C downmodulation in response to stimulation through NKG2C. Although this coexpression explains the differences seen between the anti-NKG2A/C and anti-NKG2C Ab clones, an alternate hypothesis is that differences in affinity between the anti-NKG2A/C and anti-NKG2C Ab clones have an impact on the frequencies of NKG2C+ NK cells after plate-bound stimulation. However, this seems less likely, given the known properties of these Abs (43).
Figure 2. NKG2A is upregulated on the surface of NKG2C– and NKG2C+ NK cells during culture with IL-15.
CD3/CD19-depleted PBMCs from HCMV-seropositive donors were cultured for 7 days with 10 ng/ml IL-15. FACS plots of cells from a representative donor are shown. Summary data (n = 5) show the frequencies of CD3–CD56+ NK cells gated by CD57 and NKG2C that expressed NKG2A before and after culturing. Results are from 2 independent experiments. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.001, by paired t test. P values of multiple comparisons (each culture day vs. day 0) were adjusted using the Hommel method. SSC, side scatter.
Chronically stimulated adaptive NK cells switch from a CD45RA+CD45RO– to a CD45RA–CD45RO+ phenotype.
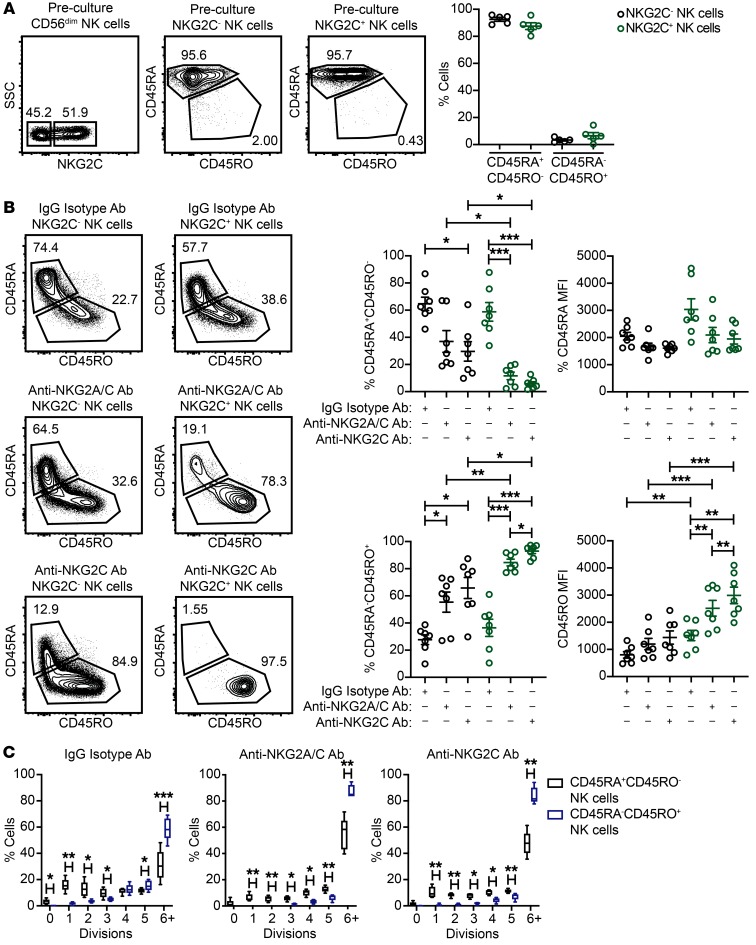
Activation of T cells with mitogens or allogeneic cells for extended periods of time in vitro leads to loss of CD45RA and concomitant gain of CD45RO (48). This switch in isoform expression is a feature of primed T cells with proliferative potential (48), and CD45RO is regarded as a marker for antigen-specific memory T cells. When analyzing freshly isolated NK cells from HCMV-seropositive donors, we observed uniformly high CD45RA expression and low CD45RO expression on the surface of both the NKG2C– and NKG2C+ NK cell subsets (Figure 3A). To determine whether NK cells exhibit a switch in CD45 isoform expression after chronic activation in our system, we analyzed CD45RA and CD45RO expression on NK cells from IgG isotype control, NKG2A/C and NKG2C stimulation cultures. NK cells from IgG isotype control cultures exhibited some skewing to a CD45RA–CD45RO+ phenotype, but generally maintained a CD45RA expression. In contrast, the majority of NK cells from NKG2A/C and NKG2C stimulation cultures switched to a CD45RA–CD45RO+ phenotype. When looking specifically at NKG2C+ NK cells, we found that CD45 isoform switching was evident in the vast majority NKG2C+ NK cells that were chronically stimulated with anti-NKG2A/C or anti-NKG2C Abs. Furthermore, the frequencies of NKG2C+ NK cells expressing CD45RO and the MFI of CD45RO were higher in anti-NKG2C Ab cultures relative to anti-NKG2C/A Ab cultures (Figure 3B). Using CellTrace dye to track proliferation, we found that CD45RA–CD45RO+ NK cells divided at a higher rate than did CD45RA+CD45RO– NK cells in all 3 culture conditions (Figure 3C). Together, our results show that chronic activating receptor stimulation in combination with IL-15 primes NK cells and induces CD45 isoform switching. Furthermore, CD45RA–CD45RO+ NK cells have a high proliferative potential.
Figure 3. Chronic stimulation of adaptive NK cells through NKG2C leads to a CD45 isoform switch.
CD3/CD19-depleted PBMCs from HCMV-seropositive donors were cultured for 7 days with 10 ng/ml IL-15 and IgG2b isotype Ab, anti-NKG2A/C Ab, or anti-NKG2C Ab. (A) Representative FACS plots and summary data of the percentages of CD45RA+RO– and CD45RA–CD45RO+ NK cells within the CD3–CD56dimNKG2C– and CD3–CD56dimNKG2C+ subsets prior to culturing (n = 5). (B) Representative FACS plots and summary data on the percentages and MFIs of CD45RA and CD45RO on CD3–CD56+NKG2C– NK cells and CD3–CD56+NKG2C+ NK cells after a 7-day culture (n = 7). Results are from 3 independent experiments. (C) Summary data for CD45RA+CD45RO– and CD45RA–CD45RO+ NK cell proliferation after culturing (n = 5), as measured by CellTrace dye dilution. Results are from 2 independent experiments. *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001, by paired t test. P values for multiple group comparisons (within the NKG2C– NK cell groups, within the NKG2C+ NK cell groups, and between the NKG2C– and NKG2C+ NK cell groups) were adjusted using the Hommel method.
Chronic stimulation induces checkpoint receptor expression on adaptive NK cells and leads to dysfunction.
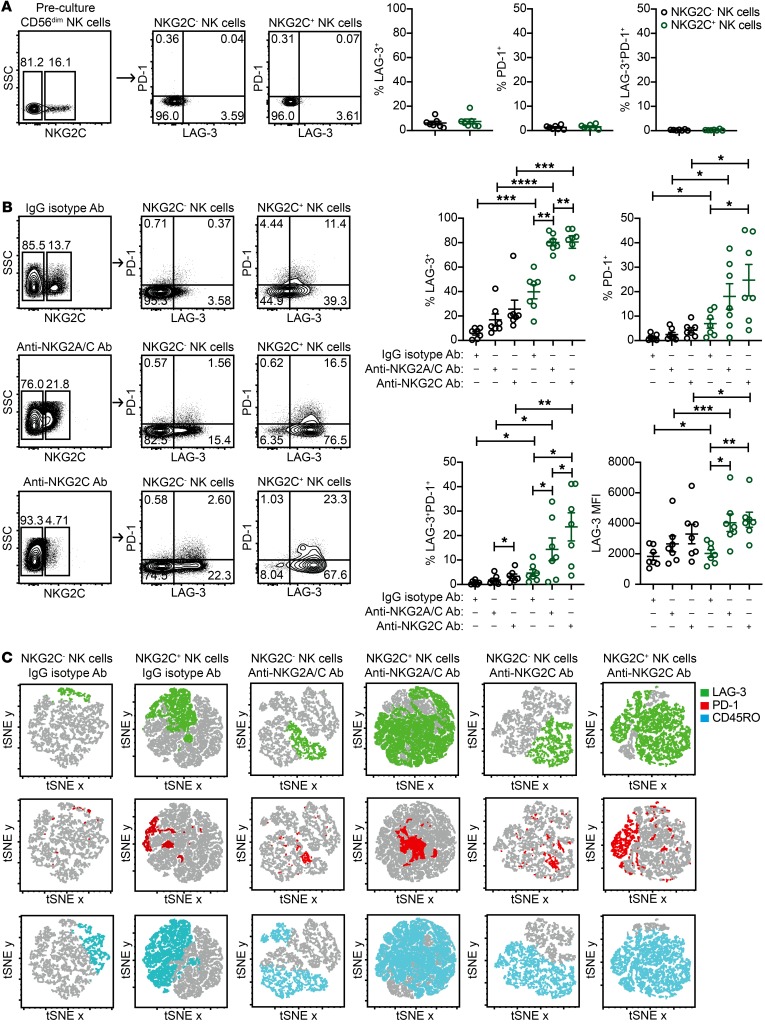
Whether chronic activation through stimulatory receptors is directly involved in the induction of LAG-3 and PD-1 expression on NK cells has not been reported to our knowledge. To address this question, we performed flow cytometry to assess LAG-3 and PD-1 expression on NKG2C– and NKG2C+ NK cells prior to culturing and from control IgG isotype Ab, anti-NKG2A/C Ab, and anti-NKG2C Ab cultures. When analyzing freshly isolated peripheral blood NK cells from HCMV-seropositive donors prior to culturing, we detected no LAG-3 or PD-1 expression on the surface of NKG2C– or NKG2C+ NK cells (Figure 4A). Although LAG-3 and PD-1 expression was moderately upregulated on NKG2C+ NK cells from control IgG isotype Ab cultures, NKG2A/C and NKG2C stimulation induced significantly higher checkpoint inhibitory receptor expression in terms of both frequency and MFI. Of note, the percentage of NKG2C+ NK cells expressing both LAG-3 and PD-1 was higher in anti-NKG2C Ab cultures relative to that observed in anti-NKG2A/C Ab cultures (Figure 4B). A representative example of the full gating scheme for flow cytometric analysis is shown in Supplemental Figure 3. To determine the degree of coexpression of NKG2C, CD45RO, LAG-3, and PD-1 on NK cells from control IgG isotype- and NKG2C-stimulated cultures, we performed t-distributed stochastic neighbor embedding (tSNE) analyses. By this method, we found that all 4 receptors were expressed by the same subset of NK cells (Figure 4C).
Figure 4. Adaptive NK cells chronically stimulated through NKG2C upregulate LAG-3 and PD-1.
CD3/CD19-depleted PBMCs from HCMV-seropositive donors were cultured for 7 days with 10 ng/ml IL-15 and IgG2b isotype Ab, anti-NKG2A/C Ab, or anti-NKG2C Ab. (A) Representative FACS plots and summary data of the percentages of LAG-3 and PD-1 expression on CD3–CD56dimNKG2C– and CD3–CD56dimNKG2C+ NK cell subsets prior to culturing (n = 7). (B) Representative FACS plots and summary data of the percentages and MFIs of LAG-3 and PD-1 on CD3–CD56+NKG2C– NK cells and CD3–CD56+NKG2C+ NK cells after culturing (n = 7). Results are from 3 independent experiments. (C) tSNE images of FACS data. tSNE plots represent a composite of NK cells from 3 donors stimulated with either IgG2b isotype or anti-NKG2A/C Ab. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001, by paired t test. P values for multiple group comparisons in B (within the NKG2C– NK cells groups, within the NKG2C+ NK cells groups, and between the NKG2C- and NKG2C+ NK cell groups) were adjusted using the Hommel method.
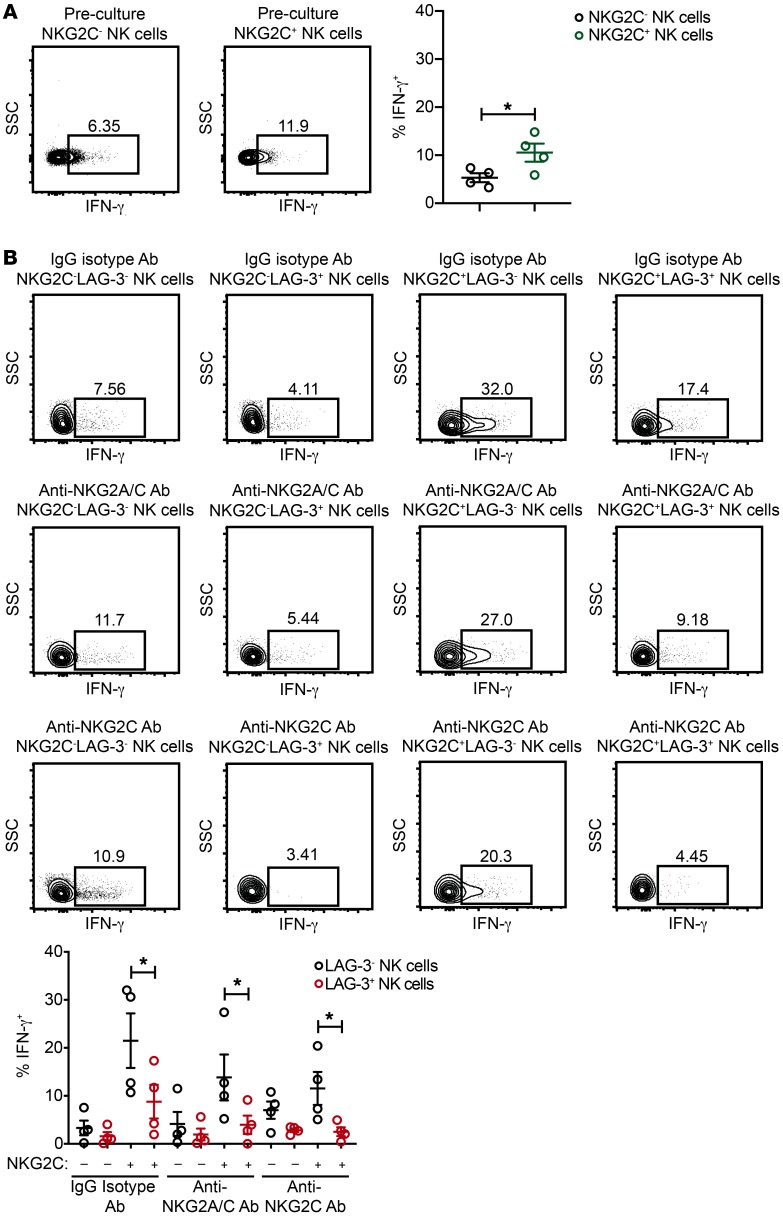
In addition to phenotypic characterization of chronically stimulated adaptive NK cells from HCMV-seropositive individuals, we analyzed intracellular IFN-γ production in NK cell subsets defined by the expression of NKG2C and LAG-3 in response to K562 myeloid leukemia cells. When comparing the NKG2C– and NKG2C+ NK cell subsets from freshly isolated cells before culturing, we observed higher frequencies of IFN-γ production within the NKG2C+ NK cell subset (Figure 5A), consistent with previous studies demonstrating an enhanced capacity for IFN-γ production by adaptive NK cells relative to canonical NK cells in response to K562 targets (49). However, when we compared NKG2C+LAG-3– NK cells with NKG2C+LAG-3+ NK cells from control IgG isotype Ab, anti-NKG2A/C Ab, and anti-NKG2C Ab cultures, we found lower levels of intracellular IFN-γ production by NKG2C+ NK cells that were LAG-3+ relative to those that were LAG-3– (Figure 5B). Of note, we did not observe a statistically significant defect in degranulation (as measured by surface CD107a expression) in NKG2C+LAG-3+ NK cells (Supplemental Figure 4). This finding agrees with those models of T cell exhaustion in which exhausted T cells still retain the ability to degranulate (25). Thus, our data show that adaptive NK cells from HCMV-seropositive donors are “primed” for the induction of inhibitory checkpoint receptors and dysfunction in response to chronic activating receptor engagement.
Figure 5. Chronically stimulated adaptive NK cells expressing LAG-3 exhibit impaired IFN-γ production.
CD3/CD19-depleted PBMCs from HCMV-seropositive donors were cultured for 7 days with 10 ng/ml IL-15 and IgG2b isotype Ab, anti-NKG2A/C Ab, or anti-NKG2C Ab. (A) Representative FACS plots and summary data of intracellular IFN-γ levels in preculture CD3–CD56dimNKG2C– and CD3–CD56dimNKG2C+ NK cells cocultured with K562 targets at an E/T ratio of 2:1 (n = 4). Results are from 2 independent experiments. (B) NK cells after culturing were used as effectors in functional assays with K562 targets at an E/T ratio of 2:1. Shown are representative FACS plots and summary data of IFN-γ production by NK cell subsets stratified by NKG2C and LAG-3 expression (n = 4). Results are from 2 independent experiments. *P ≤ 0.05, by paired t test to compare LAG-3– and LAG-3+ NK cells in each condition.
We performed similar experiments comparing cultures containing 1 ng/ml IL-15 versus cultures containing 10 ng/ml IL-15 and 3 days of culture versus 7 days of culture. In combination with anti-NKG2A/C Ab stimulation, we found that 10 ng/ml IL-15 induced significantly higher expression of LAG-3, PD-1, and CD45RO on NKG2C+ NK cells relative to 1 ng/ml IL-15 (Supplemental Figure 5). These data suggest that the strength of cytokine signaling affects the degree to which adaptive NK cells are activated and checkpoint inhibitory receptors are induced (Supplemental Figure 5). Additionally, we set up 7-day culture experiments to test the effects of 10 ng/ml IL-15 in combination with IgG isotype control Ab, anti-NKG2A/C Ab, or anti-NKG2C Ab on NK cells from HCMV-seronegative donors. In contrast to the results observed with NK cells from HCMV-seropositive donors, NKG2C+ NK cells from HCMV-seronegative donors did not preferentially expand in anti-NKG2A/C Ab cultures. We observed a modest upregulation of LAG-3 on NKG2C+ cells after culturing, but no additive effect of NKG2A/C or NKG2C stimulation. Furthermore, NKG2C+ NK cells from HCMV-seronegative donors did not upregulate PD-1 in any culture condition, and we observed no decrease in intracellular IFN-γ production in cultures with anti-NKG2A/C or anti-NKG2C Abs (Supplemental Figure 6). Because most experiments were performed using CD3/CD19-depleted PBMCs, there was the potential that monocyte activation through engagement of Fc receptors could be a confounding factor. To address this issue, we sorted NKG2C– and NKG2C+ NK cells from the peripheral blood of HCMV-seropositive donors and cultured these cells for 7 days with 10 ng/ml IL-15 in combination with 2 different IgG isotype control Abs: anti-NKG2A/C Ab or anti-NKG2C Ab. As with the results obtained from CD3/CD19-depleted PBMC cultures, NKG2C downregulation, CD45 isoform switching, and upregulation of LAG-3 and PD-1 were evident on NKG2C+ NK cells stimulated with IL-15 in combination with agonist Abs (Supplemental Figure 7).
Chronic NKp30 and NKG2D stimulation in combination with IL-15 induces checkpoint inhibitory receptor expression on adaptive but not canonical NK cells.
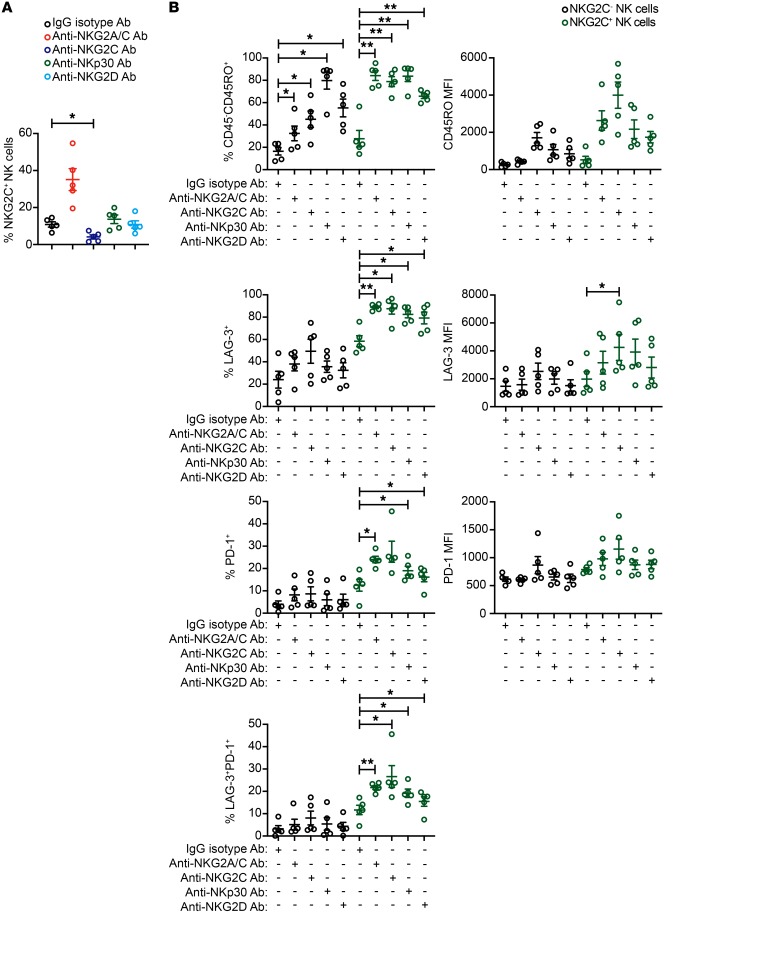
Once engaged, NKG2C propagates an activating signal through its association with the protein tyrosine kinase DNAX activation protein 12 (DAP12) (50). We sought to determine whether NKG2C engagement was unique or whether chronic stimulation through other activating receptors could also drive adaptive NK cell expansion, activation, and checkpoint inhibitory receptor expression. To test this, we prepared additional cultures using CD3/CD19-depleted cells from HCMV-seropositive donors and included plate-bound agonist Abs against NKp30 and NKG2D. NKp30 signals through CD3ξ (51). The signaling cascade downstream of the DAP12 and CD3ξ subunits is similar to the TCR signaling pathway and involves phosphorylation of multiple Src family kinases (52). In humans, NKG2D associates with DAP10 and signals through Vav1, RhoA, Rac1, and PLC-γ2 (53). We found that stimulation with agonist Abs against NKG2C, but not NKp30 or NKG2D, promoted the relative expansion of NKG2C+ NK cells (Figure 6A). Chronic stimulation by all 3 receptors induced higher frequencies of CD45 isoform switching in NKG2C+ NK cells relative to NKG2C– NK cells, with NKG2C engagement showing the strongest effect. Similarly, chronic stimulation individually through each of the 3 receptors induced LAG-3 and PD-1 expression preferentially on adaptive NKG2C+ NK cells (Figure 6B). Thus, our results show that adaptive NKG2C+ NK cells differ from canonical NKG2C– NK cells in their propensity to upregulate checkpoint inhibitory receptors in response to chronic activating receptor stimulation.
Figure 6. Chronic stimulation through NKp30 or NKG2D preferentially induces inhibitory checkpoint receptor expression on adaptive NK cells.
CD3/CD19-depleted PBMCs from HCMV-seropositive donors were cultured for 7 days with 10 ng/ml IL-15 and either IgG2b isotype Ab, anti-NKG2A/C Ab, anti-NKG2C Ab, anti-NKp30 Ab or anti-NKG2D Ab. (A) Summary data of the frequencies of CD3–CD56+NKG2C+ NK cells in each condition. (B) Summary data of the percentages and MFIs of CD45RO, LAG-3, and PD-1 on CD3–CD56+NKG2C– and CD3–CD56+NKG2C+ NK cells in each condition (n = 5). Results are from 2 independent experiments. *P ≤ 0.05 and **P ≤ 0.01, by t test. P values of multiple group comparisons (A, each group vs. IgG isotype Ab; B, within the NKG2C– NK cell groups and within the NKG2C+ NK cell groups) were adjusted using the Hommel method.
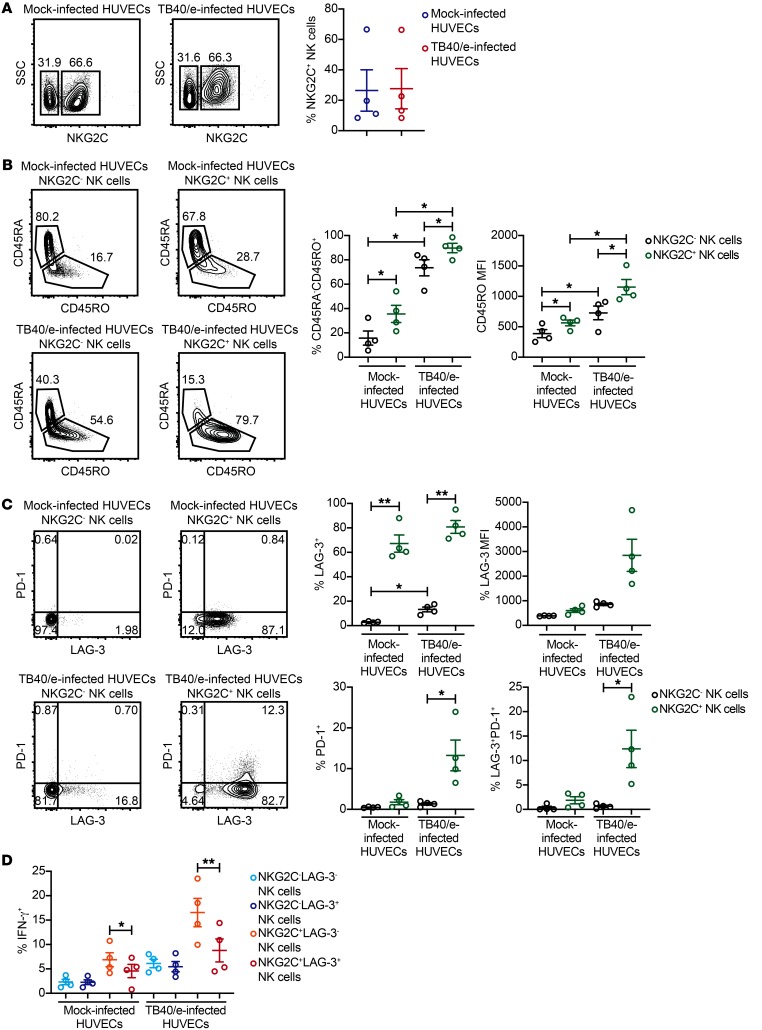
Adaptive NK cells cocultured with HCMV-infected HUVECs exhibit checkpoint inhibitory receptor upregulation and dysfunction.
Since all previous experiments were performed with plate-bound agonist Abs, we sought to determine whether a more physiologic stimulation could also drive upregulation of checkpoint inhibitory receptors and dysfunction. To this end, we infected HUVECs with a version of the TB40/e strain of HCMV that is engineered to express GFP. TB40/e is a highly ednotheliotropic and macrophage-tropic strain of the virus (54). CD3/CD19-depleted PBMCs from HCMV-seropositive donors were cocultured for 7 days with TB40/e-infected HUVECs or mock-infected HUVECs along with 10 ng/ml IL-15. We observed similar frequencies of NKG2C+ NK cells in mock-infected and TB40/e-infected HUVEC cocultures (Figure 7A). However, NKG2C+ NK cells from TB40/e-infected HUVEC cocultures were stimulated, as indicated by CD45 isoform switching (Figure 7B), and exhibited a marked upregulation of both LAG-3 and PD-1 (Figure 7C). Additionally, when NK cells from each coculture were used as effectors against K562 targets, we observed a significant impairment in IFN-γ production by NKG2C+LAG3+ NK cells relative to NKG2C+LAG3– NK cells from TB40/e-infected HUVEC cocultures (Figure 7D). We also performed these experiments using NK cells from HCMV-seronegative donors. Similar to what we observed with NK cells from HCMV-seronegative donors in plate-bound Ab experiments, we observed moderate upregulation of LAG-3 on the surface of NKG2C+ NK cells but no significant induction of PD-1. Furthermore, no difference in IFN-γ production was observed between NKG2C+LAG3+ and NKG2C+LAG3– NK cells from TB40/e-infected HUVEC cocultures (Supplemental Figure 8). Together, our data show that interaction with HCMV-infected cells can drive checkpoint inhibitory receptor upregulation and dysfunction in adaptive NK cells.
Figure 7. Adaptive NK cells cocultured with HCMV-infected HUVECs upregulate LAG-3 and PD-1 and exhibit impaired IFN-γ production.
HUVECs were infected with a GFP-expressing TB40/e clinical strain of HCMV at a MOI of 0.5 or spun down in parallel without virus (mock-infected) and used for 7-day coculture experiments with CD3/CD19-depleted PBMCs from HCMV-seropositive donors (n = 4). Cultures contained 10 ng/ml IL-15. (A) Representative FACS plots and summary data of the percentages of CD3–CD56+NKG2C+ cells from mock-infected and TB40/e-infected HUVEC cocultures. (B) Representative FACS plots and summary data of the percentages of CD3–CD56+NKG2C– and CD3–CD56+NKG2C+ NK cells expressing CD45RO and CD45RO MFI on each cell subset (n = 4). (C) Representative FACS plots and summary data of LAG-3 and PD-1 expression on CD3–CD56+NKG2C– and CD3–CD56+NKG2C+ NK cells from mock- and TB40/e-infected HUVEC cocultures (n = 4). (D) Summary data of intracellular IFN-γ in CD3–CD56+ NK cells gated by NKG2C and LAG-3 expression from mock-infected and TB40/e-infected HUVEC cocultures stimulated with K562 cells at an E/T ratio of 2:1 (n = 4). Results are from 2 independent experiments. *P ≤ 0.05 and **P ≤ 0.01, by paired t test. P values of multiple group comparisons (A, 4 pairwise comparisons as shown; B, same comparisons as in A) were adjusted using the Hommel method.
Chronic NKG2C stimulation drives a program of epigenetic remodeling similar to that of exhausted CD8+ T cells.
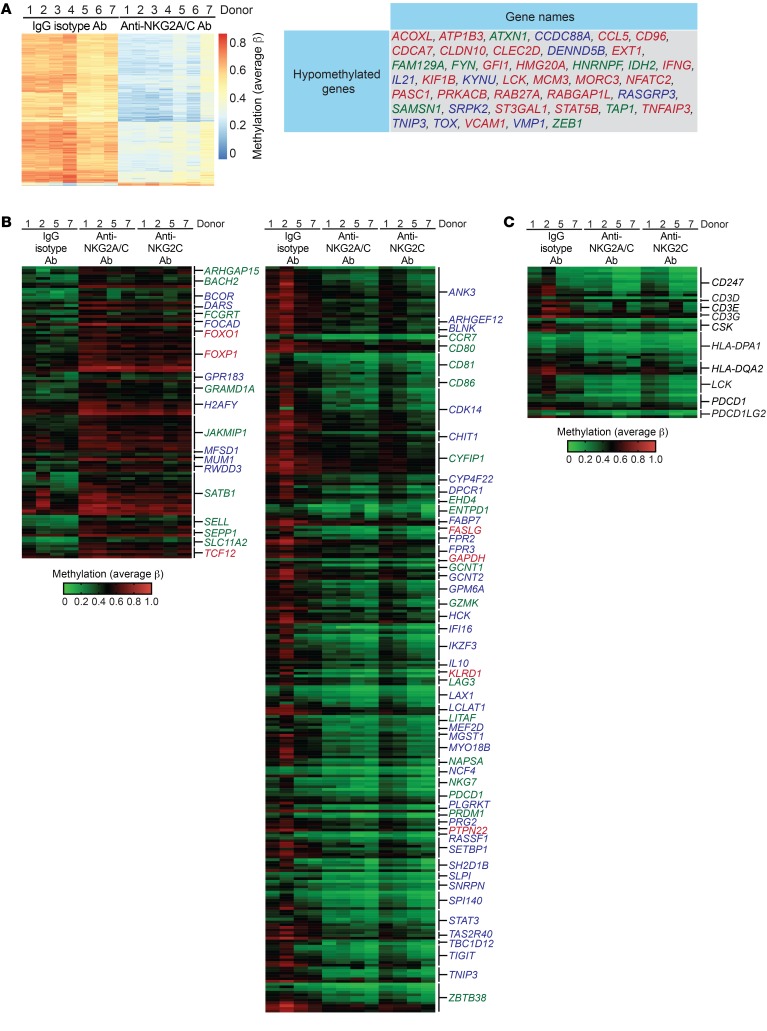
Several studies in mice have provided evidence for widespread epigenetic and transcriptional alterations in exhausted T cells (34–37). We sought to determine whether similar epigenetic changes were also evident in chronically stimulated adaptive NK cells. To this end, we performed whole-genome DNA methylation arrays on NK cells from HCMV-seropositive donors that were cultured for 7 days with control IgG isotype Ab, anti-NKG2C/A Ab, or anti-NKG2C Ab. We identified a strong signature of promoter-associated DNA hypomethylation in chronically stimulated cells. Many of these hypomethylated promoter regions matched to genes shown by either ATAC-Seq or RNA-Seq to be upregulated in exhausted mouse CD8+ T cells (34, 37). This included genes encoding signaling molecules (CCDC88A, FYN, LCK, PRKACB, SAMSN1), cytokines (CCL5, IFNG, IL21), transcription factors (GFI1, NFATC2, STAT5B, TOX, ZEB1), cell-cycle regulators (CDCA7, MCM3), inhibitors of NF-κB activity (TNFAIP3, TNIP3), GTP-binding proteins of the Ras superfamily (RAB27A, RABGAP1L, RASGRP3), and mediators of RNA splicing (HNRNPF, SRPK2) (Figure 8A). We also analyzed methylation levels within additional promoter regions associated with genes with altered expression in exhausted mouse CD8+ T cells that exhibited more variability between donors but still yielded a consistent pattern. Importantly, we observed hypomethylation within the promoter regions of LAG3, PDCD1, and TIGIT in NK cells stimulated with anti-NKG2A/C Ab or anti-NKG2C Ab relative to the IgG isotype control condition. Thus, the observed induction of checkpoint inhibitory receptors on NKG2C+ NK cells from HCMV-seropositive donors after chronic stimulation is epigenetically regulated. Several additional promoter regions that were hypermethylated after chronic NKG2C stimulation are notable and include FOXO1, FOXP1, SATB1, and TCF12, all of which encode transcriptional regulators. Promoter regions for several other genes encoding transcription factors (IKZF3, MEF2D, STAT3, ZBTB38) were hypomethylated after chronic NKG2C stimulation (Figure 8B). Additionally, we looked specifically at promoter regions for genes defined as belonging to the PD-1 signaling pathway as determined by gene set enrichment analysis (GSEA). Promoter DNA methylation varied among donors, but general hypomethylation was observed within this gene set (Figure 8C). Together, these data show that chronic NKG2C stimulation induces epigenetic reprograming toward an imprint indicative of cytotoxic lymphocyte exhaustion.
Figure 8. Chronically stimulated adaptive NK cells exhibit a whole-genome DNA methylation profile indicative of exhaustion.
CD3/CD19-depleted PBMCs from HCMV-seropositive donors were cultured for 7 days with 10 ng/ml IL-15 and either IgG2b isotype Ab, anti-NKG2A/C Ab, or anti-NKG2C Ab. The whole-genome DNA methylation profiles of NK cells from these cultures were analyzed using Illumina Infinium MethylationEPIC BeadChips in 2 independent experiments. (A) Heatmap of annotated genomic sites that exhibited consistent differential methylation across NK cells from 7 donors from IgG2b isotype Ab and anti-NKG2A/C Ab cultures and a list of genes within this data set that match genes shown to be differentially expressed or epigenetically remodeled as a result of CD8+ T cell exhaustion. Genes in blue text were shown to be remodeled in exhausted CD8+ T cells as determined by assay for transposase-accessible chromatin with sequencing (ATAC-Seq) (37). Genes in red text were found to be induced in tumor-specific CD8+ T cells as determined by RNA-Seq (34). Genes in green text were identified in both studies. (B) Heatmap of gene-associated promoter regions that exhibited differential methylation between NK cells cultured with IgG2b isotype Ab, anti-NKG2A/C Ab, or anti-NKG2C Ab and were also reported to be differentially regulated in exhausted CD8+ T cells. (C) Heatmap of gene-associated promoter regions matching to the PD-1 signaling pathway as determined by GSEA.
Discussion
Although the characteristics of T cell functional exhaustion have been explored in depth, much less is known on this topic with regard to NK cells. Early evidence for NK cell exhaustion came from an analysis of pediatric transplantation patients with posttransplantation lymphoproliferative disorder (PTLD), a life-threatening complication of organ transplantation caused by EBV infection and chronic immunosuppression. NK cells from these patients showed significant upregulation of PD-1 and dysfunction with regard to IFN-γ production. Marginal decreases in degranulation were also reported. Ex vivo culture of NK cells from these patients with PD-1–blocking Abs restored IFN-γ production but not degranulation (55). A subset of CD3–CD56dimCD16+ NK cells expressing PD-1 and exhibiting functional impairment has also been reported in patients with Kaposi sarcoma (41). One recent study in mice demonstrated a contribution of NK cells to effective immunotherapy mediated by PD-1 blockade. PD-1 expression was observed on the majority of tumor-infiltrating NK cells, but PD-1+ cells were not dysfunctional as determined ex vivo by functional responsiveness to engagement of activating receptors (56). However, it is unclear whether the NK cells being tested ex vivo in that study had engaged tumor cells and experienced prior, chronic activation in vivo. Although it is clear that NK cells in tumor microenvironments frequently express checkpoint inhibitory receptors, the mechanistic underpinnings for NK cell exhaustion have not been determined. Given the myriad transcriptional similarities between NK cells and CD8+ T cells (57), we reasoned that both cell types follow a similar path toward exhaustion.
Studies comparing acute and chronic viral infections have shown that differentiation of naive antigen-specific CD8+ T cells toward either a functional memory or a dysfunctional exhaustive state is epigenetically controlled and rapid (approximately 5 to 8 days after infection) (58). Likewise, tumor-specific CD8+ T cells display a dysfunctional state corresponding to genome-wide transcriptional changes early after tumorigenesis and antigen encounter (34). CD8+ T cell epigenetic reprograming and dysfunction appear to be driven predominantly by continuous high-affinity TCR engagement (36). Given these findings, we hypothesized that NK cell exhaustion could be induced using primary cells chronically stimulated ex vivo. We chose to study adaptive CD3–CD56dimNKG2C+ NK cells from HCMV-seropositive individuals for 3 reasons. First, this subset is primed in vivo through NKG2C-specific engagement with HLA-E in response to a chronic viral infection (14, 15). Second, by using an agonist Ab against NKG2C to stimulate cells ex vivo, we could study the effects of continuous activation through the same receptor that primed activation and expansion in vivo. Third, our previous work showed that adaptive NK cells have a unique, whole-genome epigenetic signature approximating that of effector memory CD8+ T cells (18).
In this study, we define characteristics associated with chronic stimulation and NK cell exhaustion. We found that chronic receptor stimulation in combination with IL-15 induced LAG-3 and PD-1 expression on the surface of adaptive NK cells. PD-1 was almost exclusively expressed on a subset of LAG-3+ NK cells and that LAG-3 expression on NKG2C+ NK cells began to increase earlier than did PD-1 expression, suggesting that LAG-3 is turned on early during the exhaustive program and that PD-1 may be induced during terminal NK cell exhaustion. We also observed CD45 isoform switching from CD45RA to CD45RO on dysfunctional NK cells after chronic stimulation through activating receptors. NK cells expanding ex vivo in the presence of irradiated tumor cells have been shown to undergo switching from CD45RA+CD45RO– to CD45RA–CD45RO+ (59). It is possible that activation-induced isoform switching occurs in vivo at sites of viral infection or malignancy, but only those cells that revert back to a CD45RA+CD45RO– phenotype can efficiently re-enter the peripheral blood. Although CD45RO expression is virtually absent on peripheral blood NK cells isolated from healthy individuals, distinct subsets of CD45RO+ cells have been identified in the blood and bone marrow of individuals with various types of hematologic malignancies (60).
We found that high concentrations (10 ng/ml) of IL-15 induced downmodulation of NKG2C on adaptive NK cells and that this effect was potentiated by NKG2C engagement with a high-affinity Ab against NKG2C. These results align with those of previous studies, in which incubation with HLA-G–derived leader peptides was reported to induce NKG2C downmodulation and internalization (15, 44). Intriguingly, we observed a lower frequency of NKG2C downmodulation on adaptive NK cells that were stimulated with IL-15 in combination with the Ab clone that engages both NKG2C and NKG2A relative to the Ab clone that engages only NKG2C. Although adaptive NK cells that were stimulated with the anti-NKG2A/C Ab showed upregulation of LAG-3 and PD-1 along with lower IFN-γ production, the magnitude of exhaustion was less than that observed with anti-NKG2C Ab stimulation. These results suggest that signaling through inhibitory receptors may play a role in preventing activating receptor downmodulation and mitigating exhaustion during chronic stimulation. These findings have important implications for the translation of adaptive NK cell to use in the clinic and support a common theme in immunology in which balanced activation and inhibition may have advantages over unopposed activation alone. Indeed, previous work by Colonna and colleagues aimed at determining the function of NKG2A during viral infections in vivo demonstrated that NKG2A plays an important role in limiting excessive activation, preventing apoptosis, and preserving CD8+ T cell responses during viral infection (61).
In addition to phenotypically and functionally characterizing adaptive NK cell exhaustion, we show that chronic stimulation drives epigenetic reprograming, as evidenced by genome-wide changes in DNA methylation. Many of the same genes that have been previously identified as epigenetically remodeled in exhausted CD8+ T cells exhibited alterations in DNA methylation in our study of chronically stimulated adaptive NK cells. Several genes encoding transcription factors and epigenetic regulators (FOXO1, FOXP1, SATB1, TCF12) that have been linked to T cell exhaustion (62–67) exhibited promoter hypermethylation in response to chronic stimulation. In contrast, several genes encoding transcription factors and epigenetic regulators (IKZF3, NFATC, TOX, ZBTB38) that are induced in exhausted T cells (68–71) exhibited promoter hypomethylation in response to chronic stimulation.
The results presented here show that NK cells and CD8+ T cells share a molecular program of exhaustion in response to chronic activation. This has implications in immunotherapy, in which the goal is to reverse immune exhaustion and rejuvenate cellular cytotoxic responses against tumors. Our data suggest that NK cell exhaustion is not a transient state. Instead, NK cell exhaustion is epigenetically imprinted with a large number of genomic regions displaying differential methylation patterns. Given the epigenetic similarities between NK cell and CD8+ T cell exhaustion, it is likely that the therapeutic benefit of PD-1 and LAG-3 blockade is due to a reversal of exhaustion of both CD8+ T cells and NK cells responding to tumors through a similar molecular mechanism. It will be of considerable interest to compare the epigenetic landscape of tumor-infiltrating NK cells and CD8+ T cells from patients with solid tumors for in vivo confirmation of these results. It will also be of interest to determine whether dysfunctional NK cell subsets that arise in response to chronic HIV and HCV infections (72, 73) in humans also exhibit hallmarks of cytotoxic lymphocyte exhaustion.
Methods
Healthy blood donors.
Peripheral blood products were obtained from the Memorial Blood Bank (Minneapolis, Minnesota, USA). All donors were prescreened for HCMV serostatus, and all blood products were deidentified.
Primary cell isolation and tissue culture.
PBMCs were isolated by Ficoll-Paque (GE Healthcare) density gradient centrifugation. T and B cells were depleted using anti-CD3 and anti-CD19 magnetic beads (STEMCELL Technologies) according to the manufacturer’s protocol. Tissue culture plates (24-well; Corning) were prepared by adding 10 μg/mL mouse IgG2b isotype Ab (BD Biosciences), anti-IgG2a isotype Ab (20102; R&D Systems), anti-NKG2A/C Ab (131415; R&D Systems), anti-NKG2C Ab (134571; R&D Systems), anti-NKp30 Ab (210845; R&D Systems), or anti-NKG2D Ab (149810; R&D Systems) in 0.5 mL PBS. Plates were incubated at 4°C overnight and then washed with PBS to remove unbound Ab. CD3/ CD19-depleted PBMCs were cultured in the prepared plates at a concentration of 0.5 × 106 cells per well in 1 mL B0 media (DMEM plus Ham’s F-12 medium, 2:1, supplemented with 10% heat-inactivated human AB sera, 1% penicillin-streptomycin, 25 μM β-mercaptoethanol, 20 μg/mL ascorbic acid, and 0.05 μg/mL sodium selenite). IL-15 (1 ng/ml or 10 ng/ml; National Cancer Institute) was added at the beginning of the culture. Cells were then cultured for 7 days at 37°C. For experiments designed to track NK cell proliferation, CD3/CD19-depleted PBMCs were labeled with the CellTrace Violet Cell proliferation Kit (Invitrogen) according to the manufacturer’s instructions.
Flow cytometric analysis.
For flow cytometry, staining was performed with fluorochrome-conjugated Abs against the surface epitopes CD3 (OKT4; BioLegend), CD56 (HDCD56; Biolegend), CD57 (NK-1; BD Biosciences), NKG2C (134591; R&D Systems), NKG2A (Z199; Beckman Coulter) LAG-3 (11C3C65; BioLegend), PD-1 (EH12.2H7; BioLegend), CD107a (H4A3; BioLegend), CD45RA (HI100; BioLegend), and CD45RO (UCHL1; BioLegend). Intracellular cytokine staining was performed using a fluorochrome-conjugated Ab against IFN-γ (B27; BD Biosciences). Cells were surface stained with a dead cell stain (Invitrogen, Thermo Fisher Scientific) in FACS buffer (PBS supplemented with 2% FBS and 2 mM EDTA) and fixed in 2% formaldehyde. Flow cytometric data were acquired on an LSR II instrument (BD Biosciences) and analyzed with FlowJo software, version 10 (Tree Star). Cell sorting was performed using a FACS Aria instrument (BD Biosciences).
NK cell coculture with mock- and TB40/e-infected HUVECs.
HUVECs were obtained from Lonza and propagated in EGM-2 media. Cells were cultured for 3 to 4 passages prior to infection. HUVECs were infected with the TB40/e HCMV strain modified to express GFP (provided by Don J. Diamond, City of Hope, Duarte, California, USA). Briefly, HUVECs were seeded at a concentration of 1.5 × 103 cells/well in a 96-well tissue culture plate (Corning) and cultured at 37°C overnight. The following day, HUVECs were infected with TB40/e at a MOI of 0.5 by a 30-minute spin transduction. Cells were then cultured for 1.5 hours at 37°C, washed twice with prewarmed PBS, and cultured with fresh media overnight at 37°C. TB40/e infection was confirmed by visualization of GFP expression. HUVECs were spun down and cultured in parallel without the addition of TB40/e virus to serve as a mock control for NK cell coculture experiments. The day after infection, CD3/CD19-depleted PBMCs were added at a concentration of 1.5 × 105 cells per well in B0 media containing 10 ng/ml IL-15 (National Cancer Institute). NK cells were harvested for surface staining and function assays after 7 days.
NK cell function assays.
Freshly isolated NK cells or NK cells from 7-day cultures were incubated at an effector-to-target (E/T) ratio of 2:1 with K562 cells. After 4 hours of coincubation, intracellular and surface staining was performed as described above, and NK cells were analyzed on an LSR II instrument (BD Biosciences). K562 cells were purchased from American Type Culture Collection (ATCC) and maintained according to their instructions. For intracellular staining, GolgiStop and GolgiPlug (BD Biosciences) were used to block cytokine secretion, and cells were permeabilized in 0.05% Triton X-100 (MilliporeSigma).
Genomic DNA methylation analysis.
After a 7-day culture with plate-bound anti-NKG2C or IgG isotype Abs, NK cells were lysed and DNA was extracted using the DNeasy Blood & Tissue Kit (QIAGEN). DNA was quantified, bisulfite-converted, and analyzed on Illumina Infinium MethylationEPIC BeadChips at the University of Minnesota Genomics Center according to the manufacturer’s instructions. Raw and processed DNA methylation data have been submitted to the Gene Expression Omnibus (GEO) database curated by the NCBI (GEO GSE131973).
Statistics.
Data are presented as the mean ± SEM. Paired t tests were used for comparisons, maintaining internal pairing within each donor sample. The P values for multiple pairwise comparisons were adjusted using the method of Hommel (74) when comparing more than 2 groups in an experiment. All tests were 2 sided. P values of 0.05 or less were considered statistically significant. Statistical analyses were performed with SAS 9.4 (SAS Institute), and graphs were generated using GraphPad Prism 7 (GraphPad Software). Images of NK cell phenotypes by tSNE were created in FlowJo software, version 10. Average β values from whole-genome DNA methylation arrays were analyzed using GenomeStudio (Illumina) and R package. Genes with adjusted P values of less than 0.5 (rank permutation test) and a diffbeta value higher than 0.3 were selected. A heatmap of the selected genes was plotted with the NMF package.
Study approval.
The present studies involving the use of human peripheral blood products were reviewed and approved by the IRB of the University of Minnesota under protocol 9709M00134. All blood donors provided informed consent prior to donation according to Memorial Blood Bank policies. All blood samples were deidentified prior to arrival in the laboratory.
Author contributions
AM performed experiments, interpreted data, and assisted with preparation of the manuscript. BZ and PD performed experiments. JW performed DNA methylation array analysis. XL performed statisical analyses. JSM and BRB assisted with preparation of the manuscript. FC conceived and designed the study, interpreted data, and wrote the manuscript.
Supplementary Material
Acknowledgments
We thank the University of Minnesota Genomics Center (UMGC) and the University of Minnesota Flow Cytometry Core for their services. We also thank J.P. Houchins at R&D Systems for providing all mAbs used for plate-bound stimulation experiments. This work was supported by NIH grants K99/R00 HL123638 (to FC), P30 CA77598 (to JW), P01 CA111412 (to JSM), P01 CA65493 (to JSM; BRB), R35 CA197292 (to JSM), and R01 HL56067 (to BRB), as well as by a Hematology Research Training Program grant T32 2T32HL007062 (to AM).
Version 1. 06/18/2019
In-Press Preview
Version 2. 08/12/2019
Electronic publication
Version 3. 09/03/2019
Print issue publication
Footnotes
Conflict of interest: FC consults for Fate Therapeutics and has received research funds from this relationship. JSM serves on the Scientific Advisory Board (SAB) and consults for GT BioPharma and Fate Therapeutics. He has received research funds from these relationships. JSM also serves on the SAB for CytoSen and Onkimmune. BRB declares a financial conflict with Tmunity and Kadmon Corporation. He also serves on the SAB for GT Biopharma, Magenta Therapeutics, and Five Prime Therapeutics. He consults for Regeneron and Equillium Inc. None of these companies had a role in funding this research. All conflicts are managed according to institutional policies.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(9):3770–3785.https://doi.org/10.1172/JCI125916.
Contributor Information
Bin Zhang, Email: zhang461@umn.edu.
Xianghua Luo, Email: luox0054@umn.edu.
Jinhua Wang, Email: wangjh@umn.edu.
Bruce R. Blazar, Email: blaza001@umn.edu.
Jeffrey S. Miller, Email: mille011@tc.umn.edu.
Frank Cichocki, Email: cich0040@umn.edu.
References
- 1.Orange JS. Natural killer cell deficiency. J Allergy Clin Immunol. 2013;132(3):515–525. doi: 10.1016/j.jaci.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arase H, Mocarski ES, Campbell AE, Hill AB, Lanier LL. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 2002;296(5571):1323–1326. doi: 10.1126/science.1070884. [DOI] [PubMed] [Google Scholar]
- 3.Smith HR, et al. Recognition of a virus-encoded ligand by a natural killer cell activation receptor. Proc Natl Acad Sci USA. 2002;99(13):8826–8831. doi: 10.1073/pnas.092258599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun JC, Beilke JN, Lanier LL. Adaptive immune features of natural killer cells. Nature. 2009;457(7229):557–561. doi: 10.1038/nature07665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Helden MJ, Zaiss DM, Sijts AJ. CCR2 defines a distinct population of NK cells and mediates their migration during influenza virus infection in mice. PLoS ONE. 2012;7(12):e52027. doi: 10.1371/journal.pone.0052027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abdul-Careem MF, et al. Genital HSV-2 infection induces short-term NK cell memory. PLoS ONE. 2012;7(3):e32821. doi: 10.1371/journal.pone.0032821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reeves RK, et al. Antigen-specific NK cell memory in rhesus macaques. Nat Immunol. 2015;16(9):927–932. doi: 10.1038/ni.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gumá M, Angulo A, Vilches C, Gómez-Lozano N, Malats N, López-Botet M. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood. 2004;104(12):3664–3671. doi: 10.1182/blood-2004-05-2058. [DOI] [PubMed] [Google Scholar]
- 9.Björkström NK, Svensson A, Malmberg KJ, Eriksson K, Ljunggren HG. Characterization of natural killer cell phenotype and function during recurrent human HSV-2 infection. PLoS ONE. 2011;6(11):e27664. doi: 10.1371/journal.pone.0027664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hendricks DW, Balfour HH, Dunmire SK, Schmeling DO, Hogquist KA, Lanier LL. Cutting edge: NKG2C(hi)CD57+ NK cells respond specifically to acute infection with cytomegalovirus and not Epstein-Barr virus. J Immunol. 2014;192(10):4492–4496. doi: 10.4049/jimmunol.1303211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee N, et al. HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proc Natl Acad Sci USA. 1998;95(9):5199–5204. doi: 10.1073/pnas.95.9.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braud VM, et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391(6669):795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 13.Llano M, et al. HLA-E-bound peptides influence recognition by inhibitory and triggering CD94/NKG2 receptors: preferential response to an HLA-G-derived nonamer. Eur J Immunol. 1998;28(9):2854–2863. doi: 10.1002/(SICI)1521-4141(199809)28:09<2854::AID-IMMU2854>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 14.Hammer Q, et al. Peptide-specific recognition of human cytomegalovirus strains controls adaptive natural killer cells. Nat Immunol. 2018;19(5):453–463. doi: 10.1038/s41590-018-0082-6. [DOI] [PubMed] [Google Scholar]
- 15.Rölle A, Meyer M, Calderazzo S, Jäger D, Momburg F. Distinct HLA-E peptide complexes modify antibody-driven effector functions of adaptive NK cells. Cell Rep. 2018;24(8):1967–1976.e4. doi: 10.1016/j.celrep.2018.07.069. [DOI] [PubMed] [Google Scholar]
- 16.Lopez-Vergès S, et al. Expansion of a unique CD57+NKG2Chi natural killer cell subset during acute human cytomegalovirus infection. Proc Natl Acad Sci USA. 2011;108(36):14725–14732. doi: 10.1073/pnas.1110900108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Béziat V, et al. NK cell responses to cytomegalovirus infection lead to stable imprints in the human KIR repertoire and involve activating KIRs. Blood. 2013;121(14):2678–2688. doi: 10.1182/blood-2012-10-459545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schlums H, et al. Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function. Immunity. 2015;42(3):443–456. doi: 10.1016/j.immuni.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luetke-Eversloh M, et al. Human cytomegalovirus drives epigenetic imprinting of the IFNG locus in NKG2Chi natural killer cells. PLoS Pathog. 2014;10(10):e1004441. doi: 10.1371/journal.ppat.1004441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu LL, et al. Critical role of CD2 co-stimulation in adaptive natural killer cell responses revealed in NKG2C-deficient humans. Cell Rep. 2016;15(5):1088–1099. doi: 10.1016/j.celrep.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schlums H, et al. Adaptive NK cells can persist in patients with GATA2 mutation depleted of stem and progenitor cells. Blood. 2017;129(14):1927–1939. doi: 10.1182/blood-2016-08-734236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kheav VD, et al. Favorable impact of natural killer cell reconstitution on chronic graft-versus-host disease and cytomegalovirus reactivation after allogeneic hematopoietic stem cell transplantation. Haematologica. 2014;99(12):1860–1867. doi: 10.3324/haematol.2014.108407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee J, et al. Epigenetic modification and antibody-dependent expansion of memory-like NK cells in human cytomegalovirus-infected individuals. Immunity. 2015;42(3):431–442. doi: 10.1016/j.immuni.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zajac AJ, et al. Viral immune evasion due to persistence of activated T cells without effector function. J Exp Med. 1998;188(12):2205–2213. doi: 10.1084/jem.188.12.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wherry EJ, Blattman JN, Murali-Krishna K, van der Most R, Ahmed R. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J Virol. 2003;77(8):4911–4927. doi: 10.1128/JVI.77.8.4911-4927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Virgin HW, Wherry EJ, Ahmed R. Redefining chronic viral infection. Cell. 2009;138(1):30–50. doi: 10.1016/j.cell.2009.06.036. [DOI] [PubMed] [Google Scholar]
- 27.Barber DL, et al. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439(7077):682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 28.Blackburn SD, et al. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat Immunol. 2009;10(1):29–37. doi: 10.1038/ni.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crawford A, Wherry EJ. The diversity of costimulatory and inhibitory receptor pathways and the regulation of antiviral T cell responses. Curr Opin Immunol. 2009;21(2):179–186. doi: 10.1016/j.coi.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakamoto N, et al. Synergistic reversal of intrahepatic HCV-specific CD8 T cell exhaustion by combined PD-1/CTLA-4 blockade. PLoS Pathog. 2009;5(2):e1000313. doi: 10.1371/journal.ppat.1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin HT, et al. Cooperation of Tim-3 and PD-1 in CD8 T-cell exhaustion during chronic viral infection. Proc Natl Acad Sci USA. 2010;107(33):14733–14738. doi: 10.1073/pnas.1009731107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baitsch L, et al. Exhaustion of tumor-specific CD8+ T cells in metastases from melanoma patients. J Clin Invest. 2011;121(6):2350–2360. doi: 10.1172/JCI46102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gubin MM, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515(7528):577–581. doi: 10.1038/nature13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schietinger A, et al. Tumor-specific T cell dysfunction is a dynamic antigen-driven differentiation program initiated early during tumorigenesis. Immunity. 2016;45(2):389–401. doi: 10.1016/j.immuni.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sen DR, et al. The epigenetic landscape of T cell exhaustion. Science. 2016;354(6316):1165–1169. doi: 10.1126/science.aae0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Philip M, et al. Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature. 2017;545(7655):452–456. doi: 10.1038/nature22367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bengsch B, et al. Epigenomic-guided mass cytometry profiling reveals disease-specific features of exhausted CD8 T cells. Immunity. 2018;48(5):1029–1045.e5. doi: 10.1016/j.immuni.2018.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Platonova S, et al. Profound coordinated alterations of intratumoral NK cell phenotype and function in lung carcinoma. Cancer Res. 2011;71(16):5412–5422. doi: 10.1158/0008-5472.CAN-10-4179. [DOI] [PubMed] [Google Scholar]
- 39.MacFarlane AW, et al. PD-1 expression on peripheral blood cells increases with stage in renal cell carcinoma patients and is rapidly reduced after surgical tumor resection. Cancer Immunol Res. 2014;2(4):320–331. doi: 10.1158/2326-6066.CIR-13-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benson DM, et al. The PD-1/PD-L1 axis modulates the natural killer cell versus multiple myeloma effect: a therapeutic target for CT-011, a novel monoclonal anti-PD-1 antibody. Blood. 2010;116(13):2286–2294. doi: 10.1182/blood-2010-02-271874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beldi-Ferchiou A, et al. PD-1 mediates functional exhaustion of activated NK cells in patients with Kaposi sarcoma. Oncotarget. 2016;7(45):72961–72977. doi: 10.18632/oncotarget.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Le Dréan E, et al. Inhibition of antigen-induced T cell response and antibody-induced NK cell cytotoxicity by NKG2A: association of NKG2A with SHP-1 and SHP-2 protein-tyrosine phosphatases. Eur J Immunol. 1998;28(1):264–276. doi: 10.1002/(SICI)1521-4141(199801)28:01<264::AID-IMMU264>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 43. Downs S, Bonnevier JL, Thomas M, Houchins JP, Ni JHT. Development of antibodies specific for NKG2 family members. R&D Systems Web site. https://www.rndsystems.com/resources/posters/development-antibodies-specific-nkg2-family-members Accessed June 24, 2019.
- 44.Lauterbach N, Wieten L, Popeijus HE, Voorter CE, Tilanus MG. HLA-E regulates NKG2C+ natural killer cell function through presentation of a restricted peptide repertoire. Hum Immunol. 2015;76(8):578–586. doi: 10.1016/j.humimm.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 45.Kusumi M, Yamashita T, Fujii T, Nagamatsu T, Kozuma S, Taketani Y. Expression patterns of lectin-like natural killer receptors, inhibitory CD94/NKG2A, and activating CD94/NKG2C on decidual CD56bright natural killer cells differ from those on peripheral CD56dim natural killer cells. J Reprod Immunol. 2006;70(1-2):33–42. doi: 10.1016/j.jri.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 46.Mori S, Jewett A, Cavalcanti M, Murakami-Mori K, Nakamura S, Bonavida B. Differential regulation of human NK cell-associated gene expression following activation by IL-2, IFN-alpha and PMA/ionomycin. Int J Oncol. 1998;12(5):1165–1170. doi: 10.3892/ijo.12.5.1165. [DOI] [PubMed] [Google Scholar]
- 47.Correia MP, Costa AV, Uhrberg M, Cardoso EM, Arosa FA. IL-15 induces CD8+ T cells to acquire functional NK receptors capable of modulating cytotoxicity and cytokine secretion. Immunobiology. 2011;216(5):604–612. doi: 10.1016/j.imbio.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 48.Akbar AN, Terry L, Timms A, Beverley PC, Janossy G. Loss of CD45R and gain of UCHL1 reactivity is a feature of primed T cells. J Immunol. 1988;140(7):2171–2178. [PubMed] [Google Scholar]
- 49.Foley B, et al. Cytomegalovirus reactivation after allogeneic transplantation promotes a lasting increase in educated NKG2C+ natural killer cells with potent function. Blood. 2012;119(11):2665–2674. doi: 10.1182/blood-2011-10-386995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lanier LL, Corliss B, Wu J, Phillips JH. Association of DAP12 with activating CD94/NKG2C NK cell receptors. Immunity. 1998;8(6):693–701. doi: 10.1016/S1074-7613(00)80574-9. [DOI] [PubMed] [Google Scholar]
- 51.Pende D, et al. Identification and molecular characterization of NKp30, a novel triggering receptor involved in natural cytotoxicity mediated by human natural killer cells. J Exp Med. 1999;190(10):1505–1516. doi: 10.1084/jem.190.10.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol. 2008;9(5):495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Billadeau DD, Upshaw JL, Schoon RA, Dick CJ, Leibson PJ. NKG2D-DAP10 triggers human NK cell-mediated killing via a Syk-independent regulatory pathway. Nat Immunol. 2003;4(6):557–564. doi: 10.1038/ni929. [DOI] [PubMed] [Google Scholar]
- 54.Sinzger C, et al. Cloning and sequencing of a highly productive, endotheliotropic virus strain derived from human cytomegalovirus TB40/E. J Gen Virol. 2008;89(Pt 2):359–368. doi: 10.1099/vir.0.83286-0. [DOI] [PubMed] [Google Scholar]
- 55.Wiesmayr S, et al. Decreased NKp46 and NKG2D and elevated PD-1 are associated with altered NK-cell function in pediatric transplant patients with PTLD. Eur J Immunol. 2012;42(2):541–550. doi: 10.1002/eji.201141832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hsu J, et al. Contribution of NK cells to immunotherapy mediated by PD-1/PD-L1 blockade. J Clin Invest. 2018;128(10):4654–4668. doi: 10.1172/JCI99317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bezman NA, et al. Molecular definition of the identity and activation of natural killer cells. Nat Immunol. 2012;13(10):1000–1009. doi: 10.1038/ni.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Youngblood B, et al. Chronic virus infection enforces demethylation of the locus that encodes PD-1 in antigen-specific CD8(+) T cells. Immunity. 2011;35(3):400–412. doi: 10.1016/j.immuni.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Warren HS, Skipsey LJ. Loss of activation-induced CD45RO with maintenance of CD45RA expression during prolonged culture of T cells and NK cells. Immunology. 1991;74(1):78–85. [PMC free article] [PubMed] [Google Scholar]
- 60.Krzywinska E, et al. Identification of anti-tumor cells carrying natural killer (NK) cell antigens in patients with hematological cancers. EBioMedicine. 2015;2(10):1364–1376. doi: 10.1016/j.ebiom.2015.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rapaport AS, et al. The inhibitory receptor NKG2A sustains virus-specific CD8+ T cells in response to a lethal poxvirus Infection. Immunity. 2015;43(6):1112–1124. doi: 10.1016/j.immuni.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Delpoux A, et al. Continuous activity of Foxo1 is required to prevent anergy and maintain the memory state of CD8+ T cells. J Exp Med. 2018;215(2):575–594. doi: 10.1084/jem.20170697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Feng X, et al. Foxp1 is an essential transcriptional regulator for the generation of quiescent naive T cells during thymocyte development. Blood. 2010;115(3):510–518. doi: 10.1182/blood-2009-07-232694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Feng X, Wang H, Takata H, Day TJ, Willen J, Hu H. Transcription factor Foxp1 exerts essential cell-intrinsic regulation of the quiescence of naive T cells. Nat Immunol. 2011;12(6):544–550. doi: 10.1038/ni.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Duruisseaux M, et al. Epigenetic prediction of response to anti-PD-1 treatment in non-small-cell lung cancer: a multicentre, retrospective analysis. Lancet Respir Med. 2018;6(10):771–781. doi: 10.1016/S2213-2600(18)30284-4. [DOI] [PubMed] [Google Scholar]
- 66.Stephen TL, et al. SATB1 expression governs epigenetic repression of PD-1 in tumor-reactive T cells. Immunity. 2017;46(1):51–64. doi: 10.1016/j.immuni.2016.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.D’Cruz LM, Knell J, Fujimoto JK, Goldrath AW. An essential role for the transcription factor HEB in thymocyte survival, Tcra rearrangement and the development of natural killer T cells. Nat Immunol. 2010;11(3):240–249. doi: 10.1038/ni.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim J, et al. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10(3):345–355. doi: 10.1016/S1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- 69.Martinez GJ, et al. The transcription factor NFAT promotes exhaustion of activated CD8+ T cells. Immunity. 2015;42(2):265–278. doi: 10.1016/j.immuni.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aliahmad P, et al. TOX provides a link between calcineurin activation and CD8 lineage commitment. J Exp Med. 2004;199(8):1089–1099. doi: 10.1084/jem.20040051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pozner A, Hudson NO, Trewhella J, Terooatea TW, Miller SA, Buck-Koehntop BA. The C-terminal zinc fingers of ZBTB38 are novel selective readers of DNA methylation. J Mol Biol. 2018;430(3):258–271. doi: 10.1016/j.jmb.2017.12.014. [DOI] [PubMed] [Google Scholar]
- 72.Mavilio D, et al. Characterization of CD56-/CD16+ natural killer (NK) cells: a highly dysfunctional NK subset expanded in HIV-infected viremic individuals. Proc Natl Acad Sci USA. 2005;102(8):2886–2891. doi: 10.1073/pnas.0409872102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Björkström NK, Ljunggren HG, Sandberg JK. CD56 negative NK cells: origin, function, and role in chronic viral disease. Trends Immunol. 2010;31(11):401–406. doi: 10.1016/j.it.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 74.Hommel G. A stagewise rejective multiple test procedure based on a modified Bonferroni test. Biometrika. 1988;75(2):383–386. doi: 10.1093/biomet/75.2.383. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.