Abstract
BACKGROUND
While the human fetal immune system defaults to a program of tolerance, there is a concurrent need for protective immunity to meet the antigenic challenges encountered after birth. Activation of T cells in utero is associated with the fetal inflammatory response, with broad implications for the health of the fetus and of the pregnancy. However, the characteristics of the fetal effector T cells that contribute to this process are largely unknown.
METHODS
We analyzed primary human fetal lymphoid and mucosal tissues and performed phenotypic, functional, and transcriptional analysis to identify T cells with proinflammatory potential. The frequency and function of fetal-specific effector T cells was assessed in the cord blood of infants with localized and systemic inflammatory pathologies and compared with that of healthy term controls.
RESULTS
We identified a transcriptionally distinct population of CD4+ T cells characterized by expression of the transcription factor promyelocytic leukemia zinc finger (PLZF). PLZF+CD4+ T cells were specifically enriched in the fetal intestine, possessed an effector memory phenotype, and rapidly produced proinflammatory cytokines. Engagement of the C-type lectin CD161 on these cells inhibited TCR-dependent production of IFN-γ in a fetal-specific manner. IFN-γ–producing PLZF+CD4+ T cells were enriched in the cord blood of infants with gastroschisis, a natural model of chronic inflammation originating from the intestine, as well as in preterm birth, suggesting these cells contribute to fetal systemic immune activation.
CONCLUSION
Our work reveals a fetal-specific program of protective immunity whose dysregulation is associated with fetal and neonatal inflammatory pathologies.
FUNDING
This work was supported by the UCSF Clinical and Translational Science Institute (CTSI) Pilot Award for Basic and Translational Investigators (2014908), UCSF (K12HD072222), the NIAID (K08 AI128007 and 1F31AI136336-01), a National Science Foundation (NSF) Graduate Research Fellowship (1650113 ), and an Academy for Medical Sciences Clinical Lecturer grant (535274).
Keywords: Development, Immunology
Keywords: T cell development
Introduction
In a healthy human pregnancy, fetal immunity is uniquely adapted both to promote tolerance in utero and to subsequently provide protection after birth. Unlike in mice, which lack peripheral TCR-αβ cells in utero (1), in humans, these T cells begin to populate peripheral organs by 11 to 14 weeks of gestation (2, 3). Fetal adaptive tolerance is driven in large part by the predilection of naive CD4+ T cells to differentiate into Tregs (4), yet less is known about the development of protective adaptive immunity. Activation of the human fetal immune system and the ensuing fetal inflammatory response are associated with premature termination of pregnancy and severe neonatal morbidity (5, 6). Activated T cells and reduced suppressive Treg activity are evident during fetal inflammation (7–10), and maternal-reactive fetal Th1 cells are implicated in the pathophysiology of preterm birth (PTB) (11). Animal models of intrauterine inflammation identify the fetal intestine as a potential site for the initiation of immune activation (12–14). The existence of organized intestinal lymphoid structures by the second trimester (15) and the presence of antigens within swallowed amniotic fluid point to an instructive role for the intestinal mucosa in the development of fetal adaptive immunity. However, the identity of the effector T cells that contribute to the initiation of inflammation in the human fetal intestine is not known.
Protective fetal adaptive immunity can develop in response to specific pathogens and vaccines (16, 17), and memory T cells are present in the fetus and infant (18–22), indicating that adaptive memory originates in utero. Innate-like T cells with rapid effector functions, such as γδ T cells, mucosa-associated invariant T (MAIT) cells, and innate-like NKT cells, are also present in fetal tissues (23–25). Promyelocytic leukemia zinc finger (PLZF), a transcriptional regulator that directs the differentiation of innate-like T cells (26, 27), is widely expressed in human immune cells and is commonly associated with expression of CD161, a C-type lectin receptor (28). The human fetal thymus uniquely produces a subset of CD4+ T cells, distinct from NKT cells and MAIT cells, that expresses the transcription factor PLZF (29). However, the role of fetal PLZF+CD4+ T cells in the development of protective immunity and their contribution to perinatal immune dysregulation is not known.
Here, we performed a detailed analysis of human CD4+ T cell phenotype and function in fetal lymphoid and mucosal tissues. We show that Vα7.2–PLZF+TCR-αβ+CD4+ T cells (herein referred to as PLZF+CD4+ T cells) specifically accumulated in the fetal intestine and were absent from the adult. Fetal PLZF+CD4+ T cells represent a transcriptionally unique subset of CD4+ T cells that are distinct from either innate-like, semi-invariant Va7.2+ T cells or PLZF–CD4+ T cells. Consistent with a primarily T effector memory (TEM) phenotype, PLZF+CD4+ T cells rapidly produced TNF-α and IFN-γ in response to both T cell receptor (TCR) activation and cytokine stimulation. Engagement of CD161, coexpressed on the majority of intestinal PLZF+CD4+ T cells, inhibited TCR-mediated activation in a fetal-specific manner. PLZF+CD4+ T cells with enhanced capacity for Th1 cytokine production accumulated in the cord blood of infants with gastroschisis, an abdominal wall defect associated with systemic inflammation originating from intestinal injury (30), and were also increased in the cord blood of preterm infants. Finally, dexamethasone, routinely prescribed to pregnant women in impending preterm labor, inhibited Th1 cytokine production in PLZF+CD4+ T cells, which, unlike their PLZF– counterparts, were resistant to glucocorticoid-induced apoptosis.
We propose a model in which functional maturation of PLZF+CD4+ T cells occurs in a spatially defined manner in the fetal small intestine (SI), where CD161 contributes to the regulation of their effector function and allows for the accumulation of potentially proinflammatory cells within the tightly regulated, tolerogenic fetal environment. Further, the intestinal segregation of PLZF+CD4+ T cells positions them at a critical interface for the initiation of the fetal inflammatory response associated with many fetal and neonatal morbidities.
Results
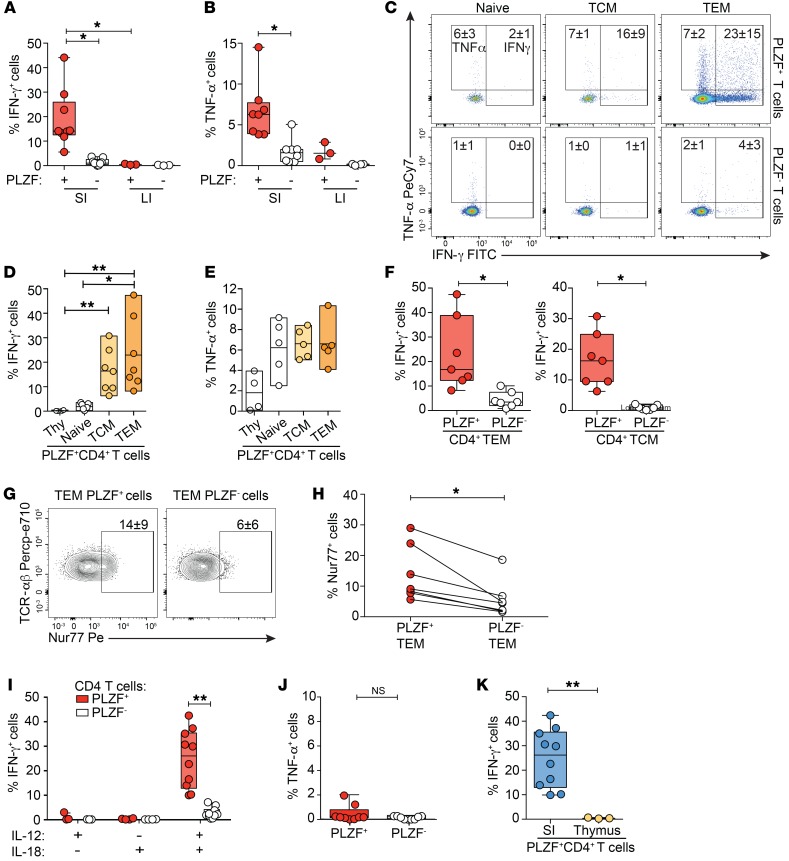
PLZF+CD4+ T cells are highly abundant in the fetal intestine.
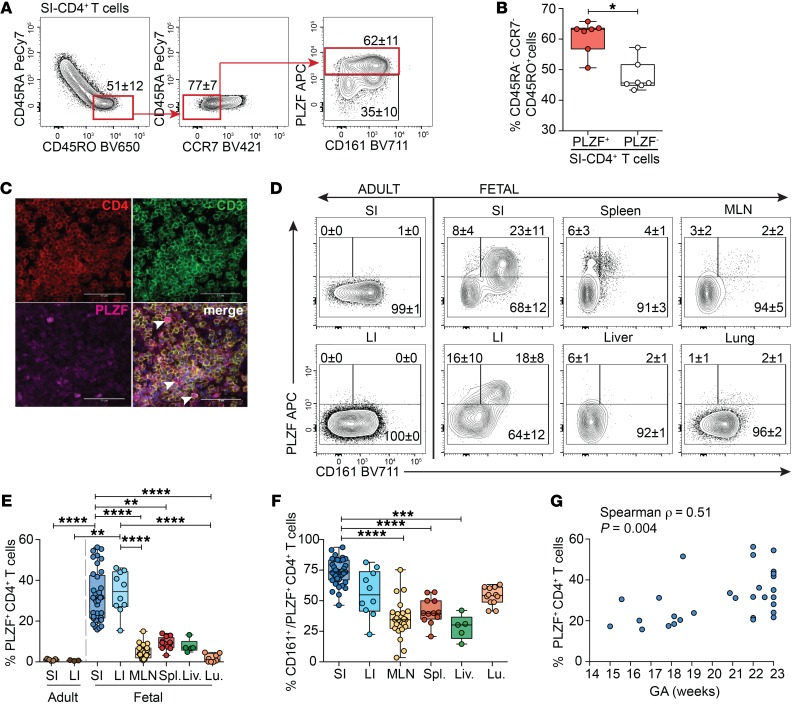
The presence of adaptive memory in the second trimester human fetal intestine (19, 21, 22) led us to examine the composition of these memory T cells. As many human innate cells express PLZF (28), we restricted our analysis to TCR-αβ+Vα7.2–CD4+ T cells (herein referred to as CD4+ T cells), which excluded MAIT cells, γδ T cells, NK cells, and innate lymphoid cells (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI125957DS1). Intestinal memory CD4+ T cells were predominantly of an effector memory phenotype (CD45RO+CD45RA–CCR7–) (TEM); the majority of these expressed PLZF (Figure 1, A and B). Immunofluorescent microscopy confirmed abundant PLZF expression among lamina propria (LP) CD4+ T cells (Figure 1C). In stark contrast to the adult SI or large intestine (LI), the accumulation of PLZF+CD4+ T cells was specific to fetal tissues (Figure 1, D and E). PLZF+CD4+ T cells accounted for approximately 30% and approximately 35% of CD4+ T cells of the fetal SI and LI, respectively, and averaged less than 10% of CD4+ T cells across other fetal tissues. CD161, the human homolog of the murine NK cell receptor NKR-P1, is expressed by many human T cell subsets, including TCR-γδ and TCR-αβ T cells, MAIT cells, and NKT cells, and is associated with adult memory T cells (26, 27, 31, 32). Tissue-specific patterns of CD161 coexpression indicated significantly higher proportions of CD161+ cells among PLZF+CD4+ T cells in the fetal SI compared with those in lymphoid tissue and liver (Figure 1, D and F). Although thymic production of PLZF+CD4+ T cells declines in the third trimester (29), the intestinal accumulation of PLZF+CD4+ T cells was positively associated with advancing gestation and could in part explain the observed variability among samples (Figure 1G).The age-associated and fetal-specific enrichment of PLZF+CD4+ T cells suggest a specific adaptation to the in utero environment.
Figure 1. PLZF+Vα7.2–CD4+TCR-αβ+ T cells are highly abundant in the fetal intestine.
(A) Representative flow plots of sequential gating strategy for the identification of intestinal memory T cells indicating the proportion of fetal intestinal effector memory (CD45RO+CD45RA–CCR7–) CD4+TCR-αβ+ T cells that express PLZF and CD161. (B) Frequencies of PLZF+ and PLZF– cells among effector memory CD4+TCR-αβ+ T cells of the fetal SI. (C) Representative imaging of PLZF expression among CD3+CD4+ T cells in tissue sections of the SI (n = 3). Arrowheads indicate triple-positive cells. Scale bar: 50 mm. Original magnification, ×400. (D) Representative flow plots of PLZF and CD161 expression among Vα7.2–CD4+TCR-αβ+ T cells in fetal tissues. (E) Frequencies of PLZF+Vα7.2–CD4+TCR-αβ+ T cells and (F) proportion of CD161+ cells among PLZF+Vα7.2–CD4+TCR-αβ+ T cells in fetal tissues. MLN, mesenteric lymph node; Spl., spleen; Liv., liver; Lu, lung. (G) Association between frequencies of intestinal PLZF+Vα7.2–CD4+TCR-αβ+ T cells and gestational age (GA), Spearman’s rank correlation. Numbers in flow cytometry plots correspond to the mean ± SD of gated populations. Circles represent individual donors. Box plot whiskers span minimum and maximum; lines represent median. **P < 0.01; ***P < 0.001; ****P < 0.0001, Wilcoxon’s matched-pairs signed rank test (B), Kruskal-Wallis paired ANOVA with Dunn’s multiple comparison test (E and F).
The majority of fetal PLZF+CD4+ T cells exhibit a memory phenotype.
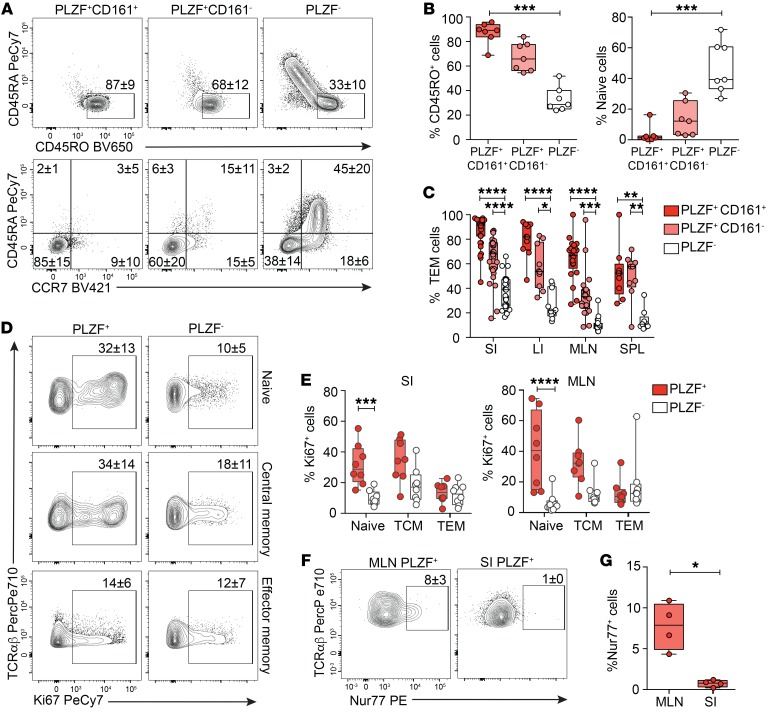
PLZF is the master regulator of the effector phenotype in murine innate-like populations (26, 27), which led us to examine whether human fetal SI PLZF+CD4+ T cells uniformly expressed a TEM phenotype. Compared with SI CD4+ T cells which did not express PLZF, PLZF+CD4+ T cells possessed significantly higher proportions of memory cells and fewer naive cells (Figure 2, A and B). Although the majority of SI PLZF+CD4+ T cells expressed CD45RO and lacked expression of both CD45RA and CCR7 (85% of PLZF+CD161+ and 60% of PLZF+CD161–), a naive phenotype was consistently evident among PLZF+CD4+ T cells. A naive-like CD1d-restricted T cell population with reduced levels of PLZF expression is present in humans, and murine transgenic studies demonstrate that a threshold level of PLZF expression is required for effector conversion (33). Similarly, naive phenotype fetal PLZF+CD4+ T cells expressed lower levels of PLZF compared with those with a memory phenotype (Supplemental Figure 2, A and B). The predominantly TEM phenotype of PLZF+CD161+CD4+ T cells was conserved across lymphoid tissues (Figure 2C). Ex vivo Ki-67 staining of intestinal and mesenteric lymph node (MLN) T cells demonstrated significantly more proliferation among naive PLZF+CD4+ T cells as compared with their PLZF– counterparts, whereas proliferation among memory cells was comparable between the 2 populations (Figure 2, D and E). Similar to adult mucosal memory T cells (34, 35), most fetal intestinal PLZF+CD4+ T cells were CD69+, and a fraction of these (~20%) also expressed CD103, suggestive of a T resident memory (TRM) phenotype (Supplemental Figure 2, C and D). We examined the draining lymph nodes for endogenous expression of Nur77, induction of which is specific to antigen receptor signaling in human T cells (36). Ex vivo Nur77 expression was significantly more frequent among PLZF+CD4+ T cells of the MLN as compared with those of the intestine and was consistently low among PLZF–CD4+ T cells (Figure 2, F and G, and Supplemental Figure 2E). These data indicate that fetal PLZF+CD4+ T cells primarily comprise effector memory phenotype cells as well as a small but highly proliferative fraction of naive T cells. Endogenous Nur77 expression provides evidence of in situ TCR signaling in tissue-draining lymph nodes, which may contribute to the activation of fetal PLZF+CD4+ T cells.
Figure 2. Fetal PLZF+CD4+ T cells exhibit a memory phenotype.
(A) Representative flow plots of CD45RA vs. CD45RO (top) and CD45RA vs. CCR7 (bottom) expression among indicated populations of Vα7.2–CD4+TCR-αβ+ cells in fetal SI. (B) Frequencies of memory (CD45RA–CD45RO+) and naive (CD45RA+, CD45RO–, CCR7+) cells among SI CD4+ T cells. (C) Frequencies of TEM within subsets of Vα7.2–CD4+TCR-αβ+ cells in fetal SI, LI, MLN, and spleen (SPL). (D) Representative flow plots of Ki67 expression among SI naive, central memory, and effector memory Vα7.2–CD4+TCR-αβ+ cells. (E) Frequencies of proliferating cells among paired samples of SI and MLN Vα7.2–CD4+TCR-αβ+ cells. (F–G) (F) Representative flow plots and (G) frequencies of endogenous Nur77 expression among SI and MLN T cells. Circles represent individual donors. Box plot whiskers span minimum and maximum; lines represent median. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, Kruskall-Wallis paired ANOVA with Dunn’s multiple comparison test (B, C, E) and Mann-Whitney U test (G). Numbers in flow cytometry plots represent frequencies of gated populations ± SD.
Fetal PLZF+CD4+ T cells are a transcriptionally distinct population of intestinal T cells.
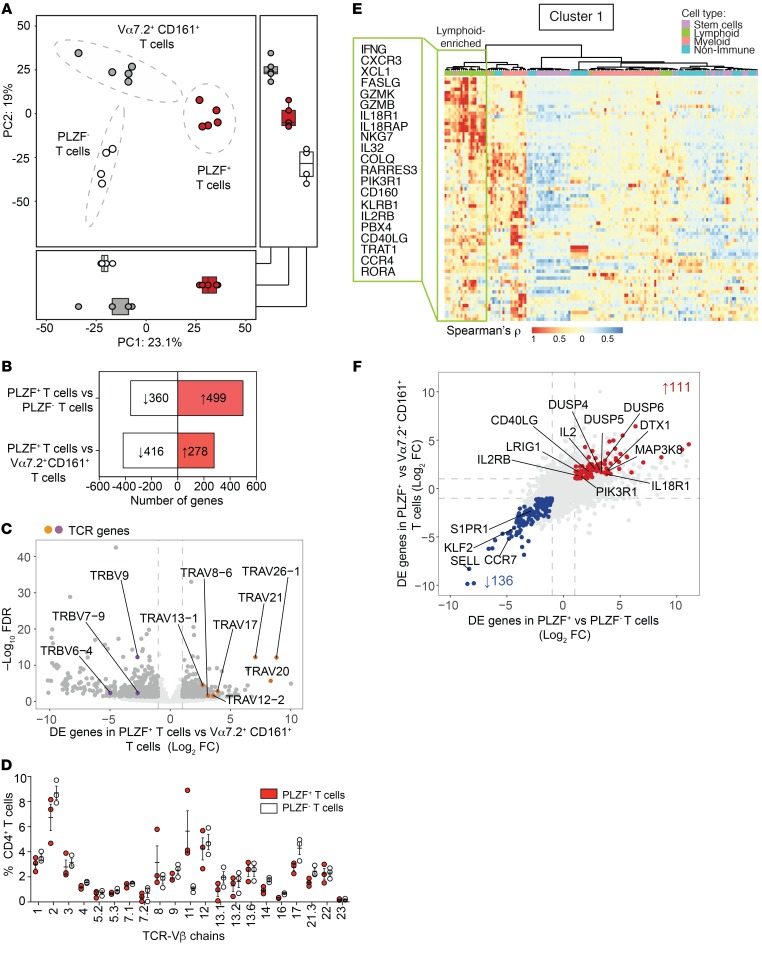
We characterized the global gene expression profile of PLZF+CD4+ T cells to examine their relationship to semiinvariant Vα7.2+CD161+ T cells and conventional memory PLZF–CD4+ T cells of the fetal SI (Supplemental Figure 3A and Supplemental Table 1). We isolated semiinvariant innate T cells by sorting TCR-αβ+Vα7.2+CD161+ cells (Supplemental Figure 3B), which exhibit phenotypic and functional characteristics of innate T cells (24). Of these, only a small fraction (4% ± 2%) were MR1 tetramer–positive MAIT cells (Supplemental Figure 3C) (37). Differential expression of IL-18R and PD1 allowed for the separation of PLZF+ from PLZF– memory CD4+ T cells (Supplemental Figure 3, A and B). The expression of ZBTB16, IL18R, PDCD1, and KLRB1 (the respective genes for PLZF, IL18R, PD1, and CD161) mirrored their protein expression and validated our sorting strategy (Supplemental Figure 3, D and E). Intestinal PLZF+CD4+ T cells were significantly distinct from both Vα7.2+CD161+ T cells and PLZF– memory CD4+ T cells (permutational multivariate ANOVA [PERMANOVA], P < 0.001, r2 = 0.92), reflecting a divergent transcriptional state by principal component analysis (PCA) (Figure 3A).
Figure 3. PLZF+CD4+ T cells are a transcriptionally distinct population of intestinal T cells.
(A) PCA of the top 1000 variable genes identified by RNA-Seq of fetal intestinal PLZF+CD4+ T cells (n = 5), PLZF–CD4+ T cells (n = 4), and Vα7.2+CD161+ T cells (n = 5). Ellipses denote the 95% confidence intervals of population means (PERMANOVA R2 0.92, P < 0.001). Box plots indicate scores for PC1 (bottom) and PC2 (right). (B) Numbers of genes with more than 2-fold (FDR < 0.05) increase (red) or decrease (white) in expression levels between populations. (C) DE genes (>2 fold, FDR < 0.05) in PLZF+CD4+ T cells compared with Vα7.2+CD161+ T cells. DE genes (dark grey), TCR genes, enriched (orange) or diminished (purple), in PLZF+CD4+ T cells are labeled. (D) Frequencies (mean ± SEM) of expression of 21 TCR-Vβ chains by flow cytometry in fetal intestinal PLZF+CD4+ T cells (red) compared with PLZF–CD4+ T cells (white). Circles indicate individual donors. (E) Heatmap shows color-coded relative enrichment of cluster 1 DE genes in PLZF+CD4+ T cells relative to PLZF–CD4+ T cells among indicated cell types identified by correlation analysis using the Human Primary Cell Atlas. Box indicates lymphoid-enriched genes associated with immune activation. (F) Multi-way comparison of DE genes (>2 fold, FDR < 0.05) in PLZF+CD4+ T cells compared with Vα7.2+CD161+ T cells (y axis) and compared with PLZF–CD4+ T cells (x axis) identifies a core signature of genes uniquely enriched (red) and diminished (blue) within intestinal PLZF+CD4+ T cells. Selected genes involved in immune response, immune regulation, and leukocyte migration are labeled.
Comparison with Vα7.2+CD161+ T cells revealed 278 genes that were differentially enriched in PLZF+CD4+ T cells (>2-fold, FDR < 0.05) (Figure 3B and Supplemental Table 2). In contrast to semiinvariant Va7.2+CD161+ T cells, which preferentially expressed transcripts for TCR-β variable 9 (TRBV9), TRBV7–9, and TRBV6–4, suggestive of an oligoclonal repertoire, PLZF+CD4+ T cells were enriched for multiple TCR-α variable (TRAV) genes and did not display preferential TRBV gene expression (Figure 3C). The diversity of the T cell repertoire of PLZF+CD4+ T cells was supported by similar Vβ use among both PLZF+ and PLZF– memory CD4+ T cells and was comparable to that reported in fetal blood (38), with no single chain accounting for more than 10% of TCR-Vβ families (Figure 3D).
Consistent with a memory phenotype, similarities between SI PLZF+CD4+ T cells and conventional PLZF–CD4+ T cells revealed transcriptional overlap in the expression of genes associated with T cell activation and memory (21) (Supplemental Figure 4A). We identified 499 genes that differentiated these 2 memory populations (>2-fold, FDR < 0.05) (Figure 3B and Supplemental Table 3) and utilized the Human Primary Cell Atlas to identify genes that cluster together by cell type using correlation analysis (39). This approach identified 6 gene clusters, of which clusters 1, 2, and 4 were enriched for transcripts involved in the immune response (Supplemental Figure 4, B and C). Cluster 1 contained significant overlap with lymphoid-associated genes, and clusters 2 and 4 comprised predominantly myeloid-associated genes (Supplemental Figure 4, D and E). Immune activation genes in cluster 1 overlapped with those enriched in TEM cells, NK cells, and γδ T cells and were suggestive of Th1-effector (IFN-G, CXCR3, XCL1) and cytotoxic functions (GZMB, GZMK, FASLG; Figure 3E). Myeloid genes in clusters 2 and 4 were associated with the immune response and regulation of the immune response (Supplemental Figure 4, B–D). The atypical composition of myeloid- and lymphoid-associated genes in the transcriptome of fetal intestinal PLZF+CD4+ T cells suggested shared attributes with both innate and adaptive immune cells.
We identified a core signature of 111 genes that were enriched and 137 genes that were depleted in PLZF+CD4+ T cells compared with both PLZF–CD4+ memory T cells and Va7.2+CD161+ T cells (>2 fold change; FDR < 0.05) (Figure 3F and Supplemental Table 4). The core signature of PLZF+CD4+ T cells contained transcripts involved in T cell activation (IL2, CD40LG, IL18R1, IL2RB, MAP3K8, PIK3R1) as well as T cell regulation (DUSP4, DUSP5, DUSP6, LRIG1, and DTX1). Among the genes specifically depleted in PLZF+CD4+ T cells were those required for lymph node homing and tissue egress (SELL, CCR7, S1PR1, KLF2) (40, 41), supporting the characterization of some of these cells as human TRM cells. In sum, Vα7.2– PLZF+CD4+ TCR-αβ+ cells represent a transcriptionally distinct subset of intestinal T cells with a diverse TCR repertoire and share gene expression profiles with innate immune cells, suggestive of a rapid effector function.
Fetal PLZF+CD4+ T cells possess inflammatory potential.
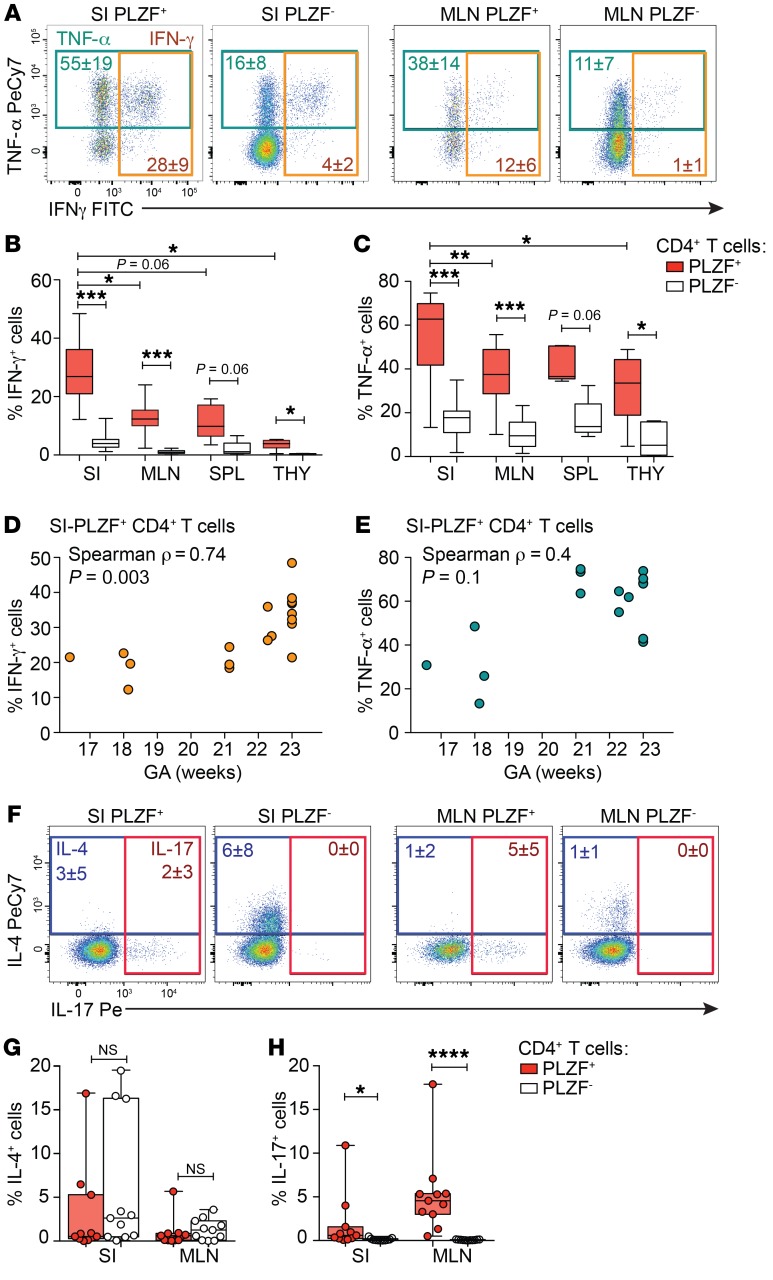
We next examined the effector potential of SI PLZF+CD4+ T cells in response to short-term stimulation with PMA and ionomycin. SI PLZF+CD4+ T cells robustly produced Th1 cytokines: on average 55% produced TNF-α and 28% produced IFN-γ (Figure 4A). Although CD161 is associated with maturation of effector function in human T cells (42), production of TNF-α and IFN-γ was similar among CD161+ and CD161– PLZF+CD4+ T cells (Supplemental Figure 5, A and B). A significantly larger proportion of PLZF+CD4+ T cells produced IFN-γ compared with PLZF–CD4+ T cells, and a larger fraction of intestinal PLZF+CD4+ T cells produced IFN-γ compared with those of the MLN, spleen, or thymus (Figure 4B). TNF-α was abundantly produced by all fetal CD4+ T cells, yet production of TNF-α was significantly more frequent among PLZF+ as compared with PLZF–CD4+ T cells (Figure 4C). The proportion of IFN-γ–producing intestinal PLZF+CD4+ T cells increased with advancing gestation, while the ability to produce TNF-α was not significantly associated with gestational age (GA) (Figure 4, D and E). Among PLZF–CD4+ T cells, neither the ability to produce IFN-γ nor TNF-α correlated with advancing gestation (Supplemental Figure 5, C and D). PLZF has been associated with production of Th2-type cytokines (26, 27), yet production of IL-4 did not differ significantly between PLZF+ and PLZF–CD4+ T cells (Figure 4, F and G) and fetal IL-10–producing CD4+ T cells were nearly undetectable (Supplemental Figure 5E). PLZF is required for the differentiation of human Th17 cells (43), and although low in frequency, IL-17–producing cells were significantly more abundant among PLZF+CD4+ T cells in both the MLN and the intestine (Figure 4, F and H). Consistent with the presence of IL-2 in their core signature (Figure 3F), a greater proportion of SI PLZF+CD4+ T cells produced IL-2 than PLZF‑ CD4+ T cells (Supplemental Figure 5, F and G), and production of IL-8 (CXCL8) was equivalent between these populations (Supplemental Figure 5, H and I). In sum, PLZF+CD4+ T cells are a polyfunctional effector population and a potentially abundant source of Th1 cytokines among fetal CD4+ T cells.
Figure 4. PLZF+CD4+ T cells possess Th-1 effector potential.
(A) Representative flow plots of intracellular TNF-α and IFN-γ staining of indicated populations of SI- and MLN-derived Vα7.2–TCR-αβ+CD4+ T cells following PMA/ionomycin stimulation (n = 18). (B, C) Frequencies of (B) IFN-γ+ and (C) TNF-α+ cells within indicated populations of Vα7.2+TCR-αβ+CD4+ T cells after PMA/ionomycin stimulation within fetal SI (n = 18), MLN (n = 11), spleen (n = 5), and thymus (THY) (n = 6). (D and E) Association between (D) IFN-γ and (E) TNF-α production by SI PLZF+CD4+ T cells in response to PMA/ionomycin and GA, Spearman’s rank correlation. (F–H) (F) Representative flow plots of and frequencies of (G) IL-4 and (H) IL-17 production among indicated populations of SI- and MLN-derived Vα7.2– Va24–TCR-αβ+CD4+ T cells following PMA/ionomycin stimulation. Numbers in flow cytometry plots correspond to the mean ± SD of gated populations. Circles represent individual donors. Box plot whiskers span minimum and maximum, and lines represent median. Kruskal-Wallis paired ANOVA with Dunn’s multiple comparison test (B, C, G, H). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Fetal PLZF+CD4+ T cells produce cytokines in response to both TCR-dependent and TCR-independent activation.
The striking potential of fetal SI PLZF+CD4+ T cells to produce Th1 cytokines led us to investigate the stimulatory signals required to elicit this phenotype. SI PLZF+CD4+ T cells were more responsive to in vitro TCR activation, resulting in a higher proportion of cells producing both IFN-γ and TNF-α compared with PLZF–CD4+ T cells (Figure 5, A and B). In contrast, the proportion of PLZF+CD4+ T cells of the LI capable of IFN-γ production was minimal, suggesting compartmentalization of the effector potential along the intestine. The IFN-γ response of both PLZF+ and PLZF–CD4+ T cells segregated with their maturation state with significantly higher proportions of IFN-γ–producing cells among TEM phenotype cells and minimal IFN-γ production among either intestinal naive cells or mature thymocytes (Figure 5, C and D, and Supplemental Figure 6A). In contrast, TNF-α production did not differ significantly between naive and memory phenotypes for either PLZF+ or PLZF–CD4+ T cells (Figure 5E and Supplemental Figure 6B). SI PLZF+CD4+ T cells with either a TEM or T central memory (TCM) phenotype produced significantly more IFN-γ than their PLZF– counterparts, and a similar trend was observed for TNF-α (Figure 5F and Supplemental Figure 6C), suggesting that the enhanced cytokine production of SI PLZF+CD4+ T cells could not be attributed to the prevalence of a memory phenotype. In vitro activation of fetal T cells induced significantly higher proportions of Nur77-expressing cells in SI PLZF+ TEM cells compared with PLZF– TEM cells (Figure 5, G and H), suggesting a lower TCR signaling threshold.
Figure 5. PLZF+CD4+ T cells produce cytokines in response to both TCR-dependent and TCR-independent activation.
All T cell populations gated on live Vα7.2–CD4+TCR-αβ+ cells. (A and B) Frequencies of (A) IFN-γ+ and (B) TNF-α+ cells within indicated populations of CD4+ T cells of the SI and LI after stimulation with anti-CD3+ anti-CD28. (C–F) (C) Representative flow cytometry plots and frequencies of (D) IFN-γ+ and (E) TNF-α+ cells within indicated populations of SI CD4+ T cells after stimulation with anti-CD3+ anti-CD28 for 24 hours. (F) Frequencies of IFN-γ+ cells within indicated populations of intestinal TEM (left) and TCM (right) CD4+ T cells after stimulation with anti-CD3+ anti-CD28 for 24 hours. (G) Representative flow plots of intracellular Nur77 staining in intestinal CD4+ TEM cells after 4 hours of anti-CD3+ anti-CD28 stimulation. Mean frequency ± SD of gated populations (n = 7). (H) Frequencies of induced Nur77 expression within indicated populations of intestinal CD4+ TEM cells after 4 hours of anti-CD3+ anti-CD28 stimulation. Lines join paired samples. (I and J) Frequencies of (I) IFN-γ+ cells and (J) TNF-α+ among intestinal PLZF+ and PLZF–CD4+ T cells in response to cytokine stimulation with IL-12 plus IL-18 for 24 hours. (K) Frequencies of IFN-γ+ cells within PLZF+CD4+ T cells of the SI and thymus in response to cytokine stimulation with IL-12 plus IL-18 for 24 hours. Numbers in flow cytometry plots correspond to the mean ± SD of gated populations. Box plot whiskers span minimum and maximum; lines represent median. Circles represent individual donors. Kruskal-Wallis paired ANOVA with Dunn’s multiple comparison test (A, B, D, E, I), Mann-Whitney U test (K), and Wilcoxon’s matched-pairs signed rank test (F, H, J). *P < 0.05, **P < 0.01.
Transcriptional overlap with innate-like T cells and myeloid cells (Figure 3E and Supplemental Figure 4, D and E) prompted us to assess the response of SI PLZF+CD4+ T cells to TCR-independent activation. SI PLZF+CD4+ T cells produced IFN-γ, but not TNF-α, in response to the combination of IL-12 and IL-18 (Figure 5, I and J, and Supplemental Figure 6D). Unlike classical innate-like T cells, which acquire effector function during thymic development (44), thymic PLZF+CD4+ T cells did not produce detectable IFN-γ in response to these cytokines (Figure 5K). The ability to respond to cytokines alone was significantly greater among PLZF+CD4+ TEM cells than PLZF–CD4+ TEM cells (Supplemental Figure 6E). Our data reveal that SI PLZF+ TEM cells are poised to produce IFN-γ in response to either TCR- or cytokine-mediated activation and indicate an association between effector memory phenotype and function.
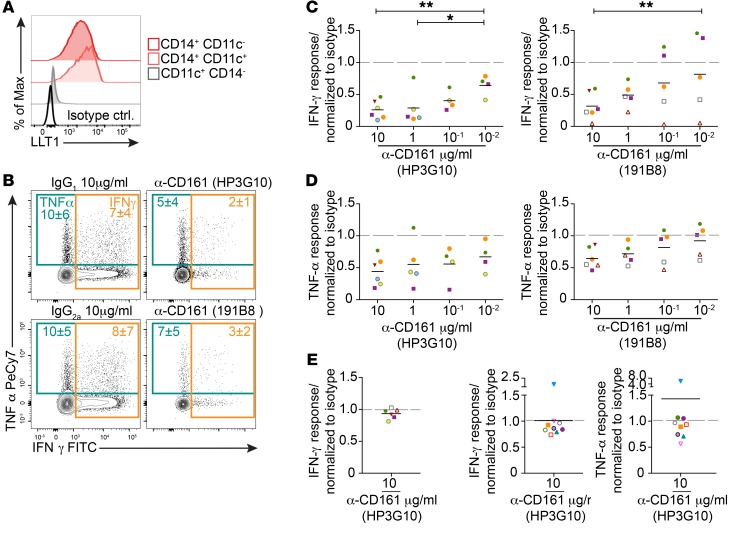
CD161 inhibits cytokine production in SI PLZF+CD4+ T cells.
The dual activation potential of SI PLZF+ T cells led us to hypothesize that their effector function would be tightly regulated. CD161 is expressed by the majority of SI PLZF+CD4+ T cells (Figure 1, D and F) and has been ascribed conflicting roles in either the activation or inhibition of human immune cells (45–47). LLT1, the natural ligand for CD161, is broadly distributed in human tissues, with the highest levels of expression in immune-privileged sites (48, 49). LLT1 was expressed on the majority of intestinal CD14+ antigen-presenting cells (APCs), but not CD11c+CD14– DCs (Figure 6A and Supplemental Figure 7, A and B). Intestinal CD14+ APCs were CD68+, CD163+, CD209+, and CD103– and expressed intermediate levels of HLA-DR, consistent with a tissue-resident macrophage phenotype (50) (Supplemental Figure 7C). Segregation of LLT1 expression to intestinal macrophages, which in adult intestine exhibit antiinflammatory properties (51), led us to explore the role of CD161 in the regulation of fetal SI PLZF+CD4+ T cells. Crosslinking of CD161 on SI PLZF+CD4+ T cells during TCR activation resulted in a dose-dependent inhibition of Th1 cytokines, although this effect was only significant for IFN-γ (Figure 6, B–D). Two different clones of monoclonal antibodies to CD161 were equally effective at a high dose (10 μg/mL), and clone HP3G10 inhibited cytokine production at a lower dose (10–1 μg/mL). Engagement of CD161 had no effect on SI PLZF+CD4+ T cell IFN-γ production in response to cytokine stimulation alone (Figure 6E and Supplemental Figure 7D), which prompted us to examine the effect of CD161 engagement on TCR signaling. However, ligation of CD161 did not appreciably dampen induction of Nur77 in response to TCR activation (Supplemental Figure 7E). Consistent with previous reports (52), engagement of CD161 on adult CD4+ T cells did not significantly affect cytokine production (Figure 6F and Supplemental Figure 7F). Collectively, our results indicate that ligation of CD161 results in fetal-specific inhibition of cytokine production in response to TCR activation.
Figure 6. CD161 inhibits cytokine production in SI PLZF+CD4+ T cells.
(A) Representative histograms of LLT1 expression among intestinal lineage– HLA-DR+CD14+ APCs (n = 10). (B) Representative flow plots of intracellular cytokine staining in PLZF+CD4+ T cells after stimulation with anti-CD3+ anti-CD28 in the presence of 2 different clones of crosslinked anti-CD161 antibodies or isotype controls (10 mg/ml). (C and D) Relative proportion of (C) IFN-γ and (D) TNF-α production by SI PLZF+CD4+ T cells normalized to isotype response after stimulation with anti-CD3+ anti-CD28 in the presence of crosslinked anti-CD161 antibodies. (E) Relative proportions of IFN-γ production by SI PLZF+CD4+ T cells normalized to isotype response after stimulation with IL-12+ IL-18 in the presence of crosslinked anti-CD161 antibody. (F) Relative proportion of IFN-γ (left panel) and TNF-α (right panel) production by adult CD161+CD4+ T cells normalized to isotype response after stimulation with anti-CD3+ anti-CD28 in the presence of crosslinked anti-CD161 antibodies. Lines represent mean. Each color represents an individual donor. Numbers in flow cytometry plots represent mean frequency ± SD of the gated populations. LME modeling in comparison with lowest dose (C and D). *P < 0.05; **P < 0.01.
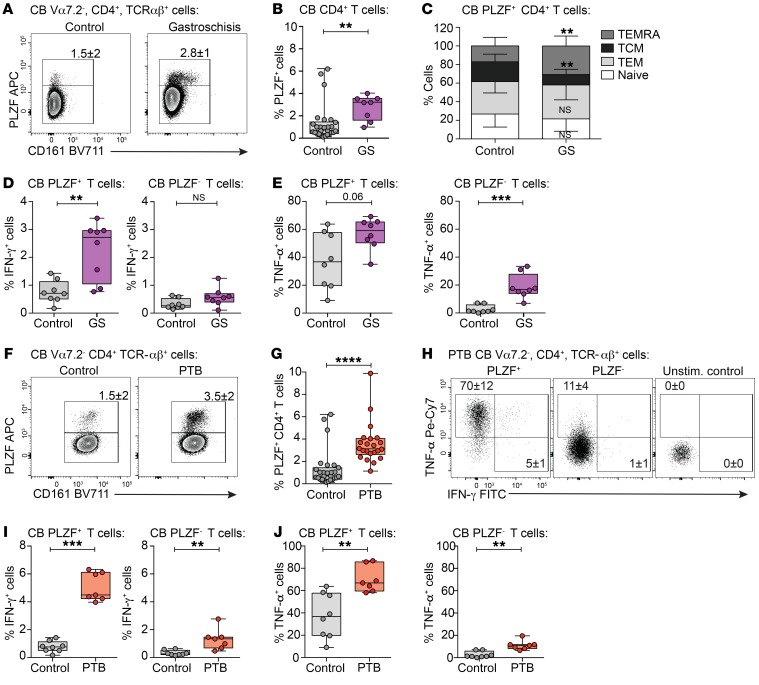
IFN-γ–producing fetal PLZF+CD4+ T cells are enriched in the umbilical cord blood of infants with systemic inflammation.
We next asked whether we could detect effector PLZF+CD4+ T cells in the circulation of infants with gastroschisis, a congenital abdominal wall defect associated with systemic fetal inflammation thought to originate from the intestine (30) (Supplemental Table 5). While present in low numbers in healthy term infants, PLZF+CD4+ T cells were enriched in the cord blood of infants with gastroschisis (Figure 7, A and B). The composition of circulating PLZF+CD4+ T cells reflected that of the fetal intestine, with significantly more effector and memory cells in contrast to the primarily naive phenotype of PLZF–CD4+ T cells (Supplemental Figure 8, A–C). Consistent with an increase in the relative frequency of terminally differentiated effector memory (TEMRA) cells (Figure 7C), a subset of TEM cells that reexpress CD45RA after antigenic stimulation, PLZF+CD4+ T cells from gastroschisis samples exhibited higher proportions of IFN-γ–producing cells compared with healthy controls (Figure 7D and Supplemental Figure 8D). An increase in the proportion of TNF-α–producing cells among both PLZF+ and PLZF–CD4+ T cells was also observed (Figure 7E and Supplemental Figure 8D). The Th1 effector potential of cord blood PLZF+ T cells was consistently greater than that of their PLZF– counterparts in both healthy controls and infants with gastroschisis (Supplemental Figure 8, E and F).
Figure 7. IFN-γ–producing fetal PLZF+CD4+ T cells are enriched in the cord blood of infants with systemic inflammation.
(A) Representative flow plots of PLZF and CD161 expression and (B) frequencies of PLZF+ cells in Vα7.2– TCR-αβ+CD4+ T cells in the cord blood (CB) of infants with gastroschisis (GS) and healthy controls. (C) Frequencies of naive, TCM, TEM, and TEMRA cells among cord blood PLZF+CD4+ T cells in infants with gastroschisis (n = 8) compared with healthy controls (n = 23). (D and E) Frequencies of (D) IFN-γ+ and (E) TNF-α+ cells within PLZF+ (left) and PLZF– (right) populations of cord blood Vα7.2–TCR-αβ+CD4+ T cells in healthy controls compared with infants with gastroschisis after PMA/ionomycin stimulation. (F) Representative flow plots of PLZF and CD161 expression and (G) frequencies of cord blood PLZF+CD4+ T cells in healthy controls and PTB samples. (H) Representative flow plots of TNF-α and IFN-γ production among indicated populations of cord blood Vα7.2–TCR-αβ+CD4+ T cells in PTB samples after PMA/ionomycin stimulation. (I and J) Frequencies of (I) IFN-γ+ and (J) TNF-α+ cells within PLZF+ (left) and PLZF– (right) populations of cord blood Vα7.2–TCR-αβ+CD4+ T cells in healthy controls compared with PTB samples after PMA/ionomycin stimulation. Circles represent individual donors. Bar graphs represent mean with SD. Box plot whiskers span minimum and maximum; lines represent median. Mann-Whitney U test (B–E, G, I, J). **P < 0.01; ***P < 0.001; ****P < 0.0001.
The accumulation of circulating effector PLZF+CD4+ T cells during a systemic inflammatory response originating from the intestine led us to examine their involvement in the Th1-mediated immune activation associated with PTB (11). Analysis of published gene expression data sets (53, 54) revealed that expression of ZBTB16, the gene encoding PLZF, was significantly increased in the cord blood of PTB samples compared with term controls in 2 independent cohorts (Supplemental Figure 9A). We examined cord blood samples from PTB (<37 weeks) infants (Supplemental Table 6), which reaffirmed the previously reported increase in TCM cells and the concomitant decrease in naive cells among CD4+ T cells in PTB as compared with term (>37 weeks) controls (Supplemental Figure 9, B and C, and refs. 8, 11). In agreement with the gene expression analysis, the frequency of circulating PLZF+CD4+ T cells was significantly higher in PTB compared with term infants (Figure 7, F and G). The accumulation of PLZF+CD4+ T cells in the cord blood of PTB infants is less likely to be attributed to increased thymic production of these cells earlier in gestation, as the proportion of naive PLZF+CD4+ T cells was significantly lower than in term cord blood, with a concomitant increase in TEM cells (Supplemental Figure 9, B and D). Circulating PLZF+CD4+ T cells in PTB samples displayed a significantly greater capacity for IFN-γ and TNF-α production compared with those of term infants (Figure 7, H–J), and the proportion of PLZF+CD4+ T cells capable of Th1 cytokine production was consistently higher than that of their PLZF– counterparts (Supplemental Figure 9E). In sum, our data indicate that PLZF+CD4+ T cells with increased Th1 effector potential accumulate in the circulation of infants with gastroschisis as well as infants with PTB, suggesting that they may contribute to prenatal immune activation.
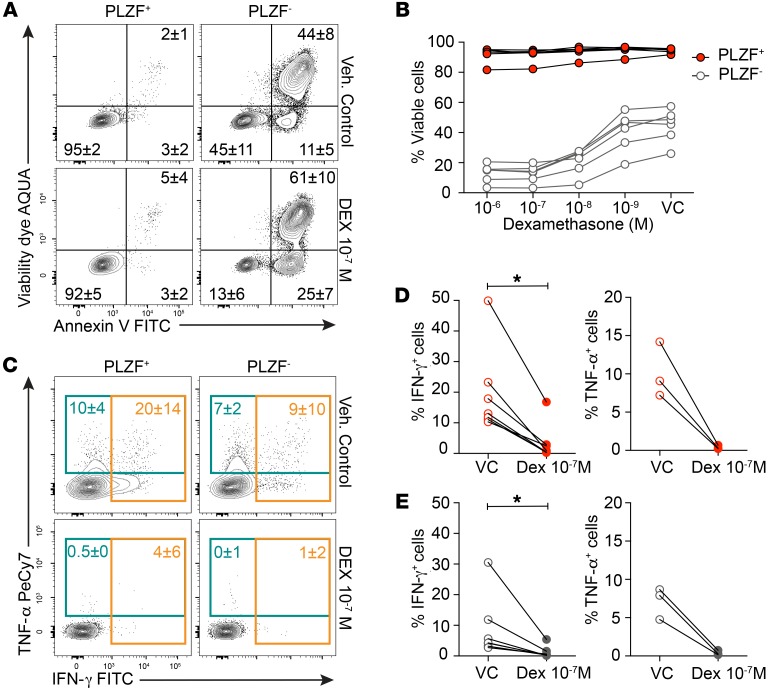
Glucocorticoids exert antiinflammatory effects on fetal PLZF+CD4+ T cells.
Maternal antenatal corticosteroid therapy with betamethasone or dexamethasone is one of the most effective interventions to improve the outcomes of premature infants. Both drugs cross the placenta and have equivalent immunosuppressive function, yet their effect on human fetal T cells is not known. In contrast to PLZF–CD4+ T cells, PLZF+CD4+ T cells were relatively resistant to glucocorticoid-induced apoptosis (Figure 8, A and B). This discordant response was also evident among CD45RA‑ CD4+ T cells and thus could not be attributed to a greater proportion of naive cells among PLZF–CD4+ T cells (Supplemental Figure 10). PLZF+CD4+ T cells are differentially enriched for the antiapoptotic BCL-2 homolog A1 (BCL2A1) (Supplemental Figure 4A), which may contribute to their survival (55). However, dexamethasone significantly inhibited the ability of both SI PLZF+ and PLZF–CD4+ T cells to produce Th1 cytokines (Figure 8, C and D). Thus, despite resistance to apoptosis, PLZF+CD4+ T cells are susceptible to the antiinflammatory effects of glucocorticoids routinely used in antenatal therapy.
Figure 8. SI PLZF+CD4+ T cells are resistant to glucocorticoid-induced apoptosis.
(A and B) (A) Representative flow plots of annexin V and viability dye and (B) frequencies of viable cells among SI PLZF+ and PLZF–CD4+ T cells cultured in the presence of indicated doses of dexamethasone (Dex) or ethanol vehicle control (VC) for 36 hours. (C) Representative flow plots of intracellular IFN-γ and TNF-α staining among SI PLZF+ and PLZF–CD4+ T cells after stimulation with anti-CD3/anti-CD28/anti-CD2 beads in the presence of dexamethasone or vehicle control. (D and E) Frequencies of IFN-γ– (left) and TNF-α –producing (right) (D) SI PLZF+CD4+ T cells and (E) SI PLZF–CD4+ T cells after stimulation with anti-CD3/anti-CD28/anti-CD2 beads in the presence of dexamethasone or vehicle control. Circles represent individual donors. Numbers in flow cytometry plots represent mean frequency ± SD of the gated populations. Wilcoxon’s matched-pairs signed rank test (D and E). *P < 0.05.
Discussion
Perinatal inflammatory pathologies are a primary cause of morbidity and mortality in the fetus and newborn and may affect the developing brain (neurocognitive disorders, cerebral palsy), lung (bronchopulmonary dysplasia), and intestine (necrotizing enterocolitis) (56). Given the contribution of T cells to the fetal inflammatory response, an understanding of fetal effector T cell identity, function, and regulation is essential for the design of effective strategies to treat or prevent these illnesses. Mouse models have provided critical mechanistic insight of neonatal T cell responses in vivo (57), yet extrapolating findings between species is hampered by substantial differences in the timing of immune development (1). This study examines the characteristics of human fetal CD4+ T cells in lymphoid and mucosal tissues and reveals an abundant population of fetal PLZF+Vα7.2–TCR-αβ+CD4+ cells with Th1 effector potential that is not present in the adult intestine. The broad distribution of PLZF+CD4+ T cells in lymphoid organs and the predominant accumulation of effector function in the intestine suggest a role for these cells in fetal immune surveillance and defense.
In utero development occurs under unique conditions that may contribute to the fetal-specific intestinal accumulation of PLZF+CD4+ T cells. Human fetal lymph nodes are enriched for TGF-β and TNF-α (4), cytokines associated with homeostatic proliferation and murine TRM development (58), suggesting a role for cytokine signaling in the expansion and intestinal homing of PLZF+CD4+ T cells. The robust response of fetal T cells to maternal antigens (59) is actively suppressed by fetal Tregs (4, 60), and expression of endogenous Nur77 in MLN-derived PLZF+CD4+ T cells supports the hypothesis that antigen recognition may also inform the development of these cells. Reports of microbial presence in human meconium and amniotic fluid (61–63) point to the growing importance of understanding the development of protective fetal immunity and the effects of bacterial priming that may occur in utero. Intestinal luminal antigens may also direct surface acquisition of LLT1 on fetal macrophages, as LLT1 expression is robustly elicited in human APCs in response to TLR and B cell receptor activation (49, 52, 64). Loss of placental hormones and the sudden increase in oxygen availability at birth, the introduction of an oral diet, and other environmental exposures likely exert different pressures on mucosal immune composition and may explain the absence of PLZF+CD4+ T cells from the adult intestine. The emergence of the neonatal intestinal microbiome may allow for the expansion of new T cell populations that outcompete fetal SI PLZF+CD4+ T cells. Cell-intrinsic properties related to the developmental origin of fetal T cells (65, 66) may additionally contribute to the fetal-specific accumulation of SI PLZF+CD4+ T cells and the inhibitory effect of CD161 engagement. CD161 ligation inhibits human NK cells (45, 64, 67), but exerts costimulatory effects on NKT cells (46) and MAIT cells (47). Perhaps due to conditions unique to the in utero environment, our results demonstrate a fetal-specific inhibitory effect that is the opposite of the primarily costimulatory role for CD161 on adult human innate T cells. CD161 signaling may influence activation downstream of TCR signaling, yet we did not observe an effect on expression of Nur77. Induction of Nur77 in human CD4+ T cells is dependent on PI3K and MEK, as well as Akt and mTOR complex 1 (mTORC1) and mTORC2, and less sensitive to mTORC1 alone and JAK/Stat signaling (36). CD161 engagement results in increased production of ceramide through induction of acid sphingomyelinase (ASM) activity and mediates downstream signaling cascades, including STAT3 and mTOR (68, 69). Thus, the mechanism of CD161 inhibition may involve JAK/Stat and mTORC1 signaling pathways independent of Nur77 induction and will be explored in future studies. The differential enrichment of numerous negative T cell regulators in the gene signature of SI PLZF+CD4+ T cells suggests the existence of additional cell-intrinsic mechanisms of regulation to promote immune homeostasis in utero.
Fetal SI PLZF+CD4+ T cells share functional and transcriptional attributes with both conventional and innate-like T cells. The development of TRM cells is closely coupled to anatomic location (41, 70), suggesting that the intestinal microenvironment may be responsible for the enhanced cytokine production of SI PLZF+CD4+ T cells as well as the preferential accumulation of CD161+ cells. The appearance of Th1 effector cells in the SI precedes that in the LI and supports the regional compartmentalization of the effector response. However, to what extent SI PLZF+CD4+ T cells recirculate to other organs and whether they represent a different population than those in lymphoid tissues is not known. A Th1 effector phenotype is also evident in human fetal γδ T cells, Vα7.2+CD161+ T cells, and NKT cells (23–25), yet unlike Vα7.2+CD161+ T cells and NKT cells, which make up on average approximately 1% of T cells in the fetal intestine (24, 25), fetal PLZF+CD4+ T cells represent an unusually abundant proportion (~30%) of intestinal T cells. The transcriptional overlap with myeloid cells, in conjunction with the ability to respond to cytokines independently of TCR activation, suggests that these cells may serve as a link between fetal innate and adaptive immunity. Expression of PLZF is required for thymic differentiation of NKT cells and is sufficient to confer a memory phenotype and effector function (26, 27). Similarly, acquisition of PLZF expression in MAIT cells coincides with the initiation of functional maturation in the thymus (71). Although PLZF is a master regulator of murine innate-like T cells, its contribution to lineage differentiation in humans appears to be more complex. Comparable to adult naive-like CD1d-restricted T cells (33), fetal naive PLZF+CD4+ T cells express reduced levels of PLZF compared with their memory phenotype counterparts. A correlation between gradients of PLZF expression and acquisition of effector maturation is also evident during human Th17 development (43), although unequivocal proof of this would necessitate lineage-tracing studies that are not feasible in humans. It is possible that the differentiation program of fetal PLZF+CD4+ T cells is imprinted during thymic development; however, functional maturation is not apparent in the thymus. Neither thymus-derived nor naive phenotype PLZF+CD4+ T cells produce significant levels of IFN-γ, which, coupled with the increasing effector potential of SI PLZF+CD4+ T cells with advancing gestation, suggests that additional maturation steps are required after thymic differentiation. A naive T cell phenotype supports lymph node–homing potential, a key step in the initiation of adaptive-like responses, which, together with evidence of antigen-receptor activation in MLN-derived PLZF+CD4+ T cells, could suggest an adaptive-like process in the acquisition of functional maturation. Previous studies implicate MHC class II–dependent selection of PLZF+ T cells during thymic development (29). Determining whether antigen recognition by fetal PLZF+CD4+ T cells in the periphery is restricted to MHC class II and understanding the specific nature of these antigens will help clarify the development of these cells in future studies. In contrast to the oligoclonal repertoire of innate-like T cells, PLZF+CD4+ T cells exhibited variable TCR-α and TCR-β chains, indicative of a polyclonal T cell population. Unlike MAIT cells and γδ T cells, which are hyporesponsive to TCR stimulation (72, 73), fetal SI PLZF+ T cells demonstrated enhanced cytokine production and a lower TCR signaling threshold than conventional PLZF– TEM cells. Thus, although fetal PLZF+CD4+ T cells possess important innate cell attributes, substantial transcriptional, phenotypic, and functional differences set this subset apart from other innate-like T cells.
The inflammatory potential of fetal PLZF+CD4+ T cells may have broad implications for the health of the fetus during pregnancy and for newborns, particularly those that are born prematurely. The increased frequency of effector IFN-γ+PLZF+CD4+ T cells in the cord blood of infants with gastroschisis as well as infants born preterm supports a role for these cells in the fetal inflammatory response. The insult in gastroschisis is thought to originate from intestinal serosal exposure to amniotic fluid, and it is equally plausible that luminal insults result in the activation of mucosal immune cells. The association of bacteria in intraamniotic infection and PTB is well established (74), suggesting that fetal SI PLZF+CD4+ T cells are ideally situated to encounter antigen or inflammatory modulators in swallowed amniotic fluid. Yet whether immune activation in the intestine contributes to the effector T cell pool in the circulation is unclear. In mice, the vast majority of memory CD8+ T cells within nonlymphoid tissue are TRM, whereas secondary lymphoid organs contain mostly recirculating (TEM and TCM) subsets (75). In humans, defining the relationship between circulating and memory T cells within different anatomic compartments remains a daunting challenge. Our data suggest that PLZF+CD4+ T cells may play a role in fetal immune activation and that disturbances in this population may contribute to diverse inflammatory pathologies that occur during the perinatal period. A recent longitudinal, systems-level immune analysis in newborns revealed that human immune cells reached adult-like phenotypes by 3 months of age, with the notable exception of T cells (76). Whether intestinal PLZF+CD4+ T cells gradually disappear in healthy infants and whether early gut dysbiosis results in the persistence of these T cells will be explored in futures studies.
These data add to our knowledge of the composition and function of the protective arm of the fetal adaptive immune system, in particular to the establishment of tissue-localized responses. The presence of a sizeable fraction of PLZF+CD4+ T cells with Th1 effector potential offers a more nuanced appreciation of human fetal immune development and highlights the unique challenges faced by the developing immune system as it transitions from a program of tolerance to one of protective immunity during the last trimester of pregnancy. We have identified a mechanism of fetal-specific immune suppression through CD161 blockade, which likely works in concert with multiple other regulatory mechanisms to actively promote immune suppression in utero (4, 77, 78) and could be harnessed therapeutically for the prevention or treatment of fetal and neonatal inflammatory pathologies. Beyond the perinatal period, expanding our knowledge of human fetal immune development and function is of utmost importance to our understanding of the early origins of predisposition to adult inflammatory diseases.
Methods
Cell isolation.
PBMCs from adult and cord blood were isolated by Ficoll-Histopaque (Sigma-Aldrich) gradient centrifugation and cryopreserved in freezing medium (90% FBS + 10% DMSO; ATCC) and analyzed in batches. Fetal and adult organs were collected into cold RPMI with 10% FCS, 10 mM HEPES, penicillin, streptomycin, 0.1 mM 2-β-mercaptoethanol, 2 mM l-glutamine, and nonessential amino acids (cgRPMI medium), transported on ice, and processed within 2 hours of collection. The SI and colon were dissected from the mesentery, opened longitudinally, and cut into 1 cm sections. Mucus was removed with 3 washes in 1 mM DTT in PBS for 10 minutes. The epithelial layer was removed with 3 washes in 1 mM EDTA in PBS for 20 minutes. The intestine, MLN, liver, lung, and spleen were minced into smaller pieces and digested with freshly prepared 3 mg/mL collagenase IV (Life Technologies) and 10 mg/mL DNAse (Roche) in cgRPMI for 30 minutes, and dissociated cells were filtered through a 70 μm strainer. Cells were separated in a 20%/40%/80% Percoll density gradient at 400 g for 40 minutes. T cells were recovered at the 40%–80% interface, APCs were recovered at the 20%–40% interface, and all cells were washed twice in cgRPMI. All washes and incubations were performed in a shaking (200 rpm) water bath at 37°C. Thymocytes were isolated by gently pressing small pieces of thymus though a 70 μm strainer. Viability was measured with Trypan Blue (Sigma Aldrich).
Antibodies and flow cytometry.
Intestinal LP T cells were isolated by negative selection using the EasySep Human T Cell Isolation Kit (STEMCELL Technologies). Isolated cells were incubated in 2% FCS in PBS with 1 mM EDTA (staining buffer) with human Fc block (STEMCELL Technologies) and stained with fluorochrome-conjugated antibodies against surface markers. Intracellular protein detection was performed on fixed, permeabilized cells using the Foxp3/Transcription Factor Staining Buffer Set (Tonbo Biosciences). Mouse anti-human mAbs used in this study included the following: TCR-αβ PerCP-e710 (clone IP26, eBioscience, catalog 46-9986-42), Vα7.2 biotin and BV605 (clone 3C10, BioLegend, catalog 351724 and 351720), CD4 APC H7 (clone L200, BD Biosciences — Pharmingen, catalog 560837), CD8α FITC (clone SK1, BD Biosciences, catalog 347313), CD8α PE (clone HIT8a, BD Biosciences, catalog 555635), CD45RA PE (clone HI100, BD Biosciences — Pharmingen, catalog 555489), CCR7 BV421 (clone G043H7, BioLegend, catalog 353208), CD45RO BV650 (clone UCHL1, BioLegend, catalog 304232), CD69 PE (clone FN50, BioLegend, catalog 310906), Vα24Jα18 efluor450 (clone 6B11, Invitrogen, catalog 48-5806-42), CD103-FITC (clone Ber-ACT8, BD Biosciences — Pharmingen, catalog 550259), PLZF-APC (clone 6318100, R&D, catalog IC2944A), CD161-BV711 (clone DX12, BD Biosciences, catalog 563865), IL-18Rα–PE (clone H44, BioLegend, catalog 313808), PD-1 BV605 (clone EH12.2H7, BioLegend, catalog 329924), IFN-γ–FITC (clone 25723.11, BD Biosciences, catalog 340449), IFN-γ–BV650 (clone 4S.B3, BD Biosciences, catalog 563416), TNF-α–PE Cy7 (clone MAB11, BD Biosciences — Pharmingen, catalog 557647), IL-2 (clone 5344.11, BD, catalog 340448), IL-8 BV421 (clone G265-8, BD Biosciences, catalog 563310), IL-10 Pe (clone JES3-19F1, BD Biosciences, catalog 562035), IL-4 PeCy7 (clone 8D4-8, BD Biosciences, catalog 560672), IL-17A Pe (clone eBIO64DEC17, eBioscience, catalog 12-7179-42), CD45 APC (clone HI30, Tonbo, catalog 20-0459), CD14 BV605 (clone M5E2, BD Biosciences — Pharmingen, catalog 564054), CD11c BB515 (clone B-ly6, BD Biosciences — Pharmingen, catalog 564491), HLA-DR APC-R700 (clone G46-6, BD Biosciences, catalog 565128), CD3 BV510 (clone HIT3α, BD Biosciences, catalog 564713), CD19 BV510 (clone SJ25C1, BD Biosciences, catalog 562947), CD56 BV510 (clone NCAM16.2, BD Biosciences, catalog 563041), CD68 PE Cy7 (clone FA-11, BioLegend, catalog 137016), CD163 BV711 (clone GHI/61, BioLegend, catalog 333630), CD209 PerCP Cy 5.5 (clone DCN46, BD Biosciences — Pharmingen, catalog 558263), LLT1 PE (clone 402659 R&D, catalog FAB3480P), and Nur77-PE (clone 12.14, eBioscience, catalog 12-5965-80). TCR-β repertoire profiling was performed by staining with the IOTest Beta Mark TCR V β Repertoire Kit (Beckman Coulter). All cells were stained with Aqua LIVE/DEAD Fixable Dead Cell Stain Kit (Invitrogen) to exclude dead cells from analysis. All data were acquired with an LSR/Fortessa Dual SORP flow cytometer (BD Biosciences) and analyzed with FlowJo V10.0.8 (TreeStar) software.
Immunohistochemistry.
Intestinal segments were fixed in 4% paraformaldehyde for 10 minutes and dehydrated in a sucrose gradient prior to embedding in OCT. Embedded tissues were frozen on dry ice and stored at –80°C, and 10 μm thin cryosections were mounted on frosted charged slides. Histological work, imaging, and image processing were performed by the Gladstone Institutes’ Histology and Light Microscopy Core. Slides were fixed in acetone for 5 minutes at –20°C, rehydrated in PBS for 10 minutes, rinsed in 0.05% PBS-Tween, and permeabilized in 0.1% PBS–Triton X-100 for 15 minutes at room temperature (RT). Slides were blocked using 1% BSA, 0.1% fish skin gelatin, 0.5% Triton X-100,and 0.05% Na-Azide in PBS for 2 hours and stained with purified anti-CD3 (clone Hit3a; BD Biosciences — Pharmingen, catalog 555336) and anti-PLZF (clone NBPI-80894, Novus Biologicals, catalog NBPI-80894) overnight at 4°C, followed by anti-CD4–eFluor 570 (N1UG0, Thermo Fisher, catalog 41-2444-82) at RT for 3 hours. Slides were incubated with secondary antibody goat anti-mouse-DyLight 488 (Thermo Fisher, catalog 35502) and goat anti-rabbit 633 (Thermo Fisher, catalog A-21070) for 1 hour at RT, stained with DAPI (Invitrogen) for 10 minutes at RT, and mounted with Fluormount-G (EMS). Images were captured with a Zeiss Cell Observer Spinning Disk microscope (Carl Zeiss Microscopy) equipped with 405 nm, 488 nm, 561 nm, 633 nm lasers, a prime 95b sCMOS camera (Photometrics), and Zeiss Zen imaging software.
T cell stimulation and CD161 inhibition.
For detection of basal cytokine potential, single cell suspensions from various tissues were cultured directly ex vivo in a 96-well U-bottom plate in cgRPMI and stimulated with 50 ng/ml PMA (Santa Cruz Biotechnology Inc., catalog SC-3576) and 5 μg/ml ionomycin (Sigma-Aldrich) in the presence of Brefeldin A (Sigma-Aldrich, catalog B7651-5MG) for 3 hours at 37°C in 4% O2 to mimic intrauterine hypoxia. For Nur77 induction, T cells were cultured at 500 K/well in 96-well plates and stimulated with plate-bound anti-CD3 (clone HIT3a, BioLegend, catalog 300313) at 1 μg/ml and soluble anti-CD28 (CD28.2) at 2 μg/ml for 4 hours at 37°C in 4% O2. For cytokine production, T cells were cultured at 250 K/well in 96-well plates and stimulated with plate-bound anti-CD3 at 1 μg/ml and soluble anti-CD28 (CD28.2, Invitrogen, catalog MA1-20792) at 2 μg/ml, or cells were activated with 50 μg IL- 12 (PeproTech, catalog 200-12) and IL-18 (R&D Systems, catalog 9124-IL-050) at 50 ng/mL for 16–20 hours at 37°C in 4% O2, and Brefeldin A was added for the last 4 hours. After stimulation, cells were stained for intracellular cytokine production as described above. For the inhibition assays, T cells were first incubated with anti-CD161 biotin mAbs (clone HP3G10, BioLegend, catalog 339932, or clone 191B8, Miltenyi Biotec, catalog 130-092-906) or isotype-biotin controls IgG1 (MOPC-21, BioLegend, catalog 400104) and IgG2a (MOPC-173, BioLegend, catalog 400204) for 15 minutes, washed, and crosslinked with Anti-Biotin Cocktail (STEMCELL Technologies) at 1:50 dilution during stimulation with anti-CD3/anti-CD28 or cytokines as described above. For glucocorticoid-induced apoptosis assays, T cells were incubated in the presence of dexamethasone (Sigma-Aldrich) (10–6–10–9 M) or ethanol vehicle control alone or stimulated with Immunocult Human CD3/CD28/CD2 T cell activator beads (STEMCELL Technologies, catalog 10990) for 36 hours. T cells were then stained for annexin V using the Annexin V Apoptosis Detection Kit FITC (eBioscience, catalog 88-8005-74) according to the manufacturer’s instructions, followed by intracellular staining for PLZF and/or intracellular cytokine production as above.
T cell isolation and RNA extraction for RNA-Seq.
PLZF+ and PLZF–CD4+ TCR-β+ T cells were sorted using FACSAria2 SORP (BD Biosciences). T cells were isolated as described above from the intestine of 5 individual samples. MAIT cells were pooled from the intestine and MLN for RNA-Seq as outlined in Supplemental Figure 3A, and 104 cells were collected per subset for each sample. Postsort purity was determined by flow cytometry following intracellular staining for PLZF as above and was more than 92% for PLZF– T cells and MAIT cells and more than 87% for PLZF+ T cells. RNA was extracted and purified with the Dynabeads mRNA DIRECT Purification Kit (Thermo Fisher Scientific). mRNA libraries were constructed using the Nugen/Nextera XT Library Prep Kit (Illumina), and 15 samples (5 donors, 3 T cell subsets) were sequenced on an Illumina HiSeq 4000 by the Functional Genomics Core Facility at UCSF. The reads from the Illumina HiSeq sequencer in fastq format were verified for quality control using the fastqc software package. Reads were aligned to the human genome (hg38) and read counts aggregated by gene using the Ensembl GRCh38.78 annotation using STAR (79). Differential gene expression analysis was performed with the DESeq2, version 1.16.1, package (80).
Statistics.
Groups were compared with Prism, version 6, software (GraphPad). Data were analyzed using Wilcoxon’s test for paired nonparametric data, Mann-Whitney U test for unpaired nonparametric data, and the Kruskal-Wallis with Dunn’s multiple comparison test for comparison of 3 or more groups. Box plot rectangles show first to third quartile, the line shows the median, and the whiskers represent minimum and maximum values, unless otherwise stated. Correlation analysis was measured by Spearman’s correlation coefficient. Bar plots represent the mean and the SEM. P < 0.05 was considered significant. Linear mixed effects (LME) modeling was used to evaluate significance in dose-response inhibition to CD161 antibody crosslinking. Principle component analysis and accompanying confidence intervals were performed using combined functions in the R packages stats, vegan, and ggplot2, and PERMANOVA analysis was used to assess the significance of the Euclidean distances between the groups. Heatmaps were generated using the R packages heatmap and circlize. A coexpression analysis was performed for the differentially expressed (DE) genes using the Human Primary Cell Atlas (39). This atlas contains 713 microarray samples of a wide range of pure cell types and states. The complexity of the data set was reduced to the median gene expression per cell state (n = 157), and Spearman’s correlation coefficients were calculated for each pair of DE genes, revealing clusters of genes that are coexpressed in the same set of cell types. Gene clusters were analyzed for overlap with KEGG and GO biological processes gene sets using the Molecular Signatures Database (MSigDB) (81, 82). No statistical methods were used to predetermine sample size. The experiments were not randomized, and the investigators were not blinded to allocation during experiments and outcome assessment.
Study approval.
Human fetal tissues (15 to 23 weeks GA) were obtained from San Francisco General Hospital (San Francisco, California, USA) from terminations of pregnancy after maternal written informed consent with approval from and under the guidelines of the UCSF Research Protection program. Samples were excluded in the cases of (a) known maternal infection, (b) intrauterine fetal demise, and/or (c) known or suspected chromosomal abnormality. Adult intestinal tissues were obtained after written informed consent from the UCSF Biospecimen Resources Program. Adult SI was obtained from routine resections during Whipple procedures, and adult colon was obtained during hemicolectomies and surgical resections. PBMCs derived from Trima residues from Trima Apheresis collection kits were obtained from healthy donors after written informed consent at the Blood Centers of the Pacific. Approval for healthy, term umbilical cord blood collection was obtained from the University of Texas MD Anderson Cancer Center (Houston, Texas, USA) with written informed consent between January 2017 and June 2018. Eligibility criteria for term infant control samples included healthy infants delivered at more than 37 weeks GA; samples were excluded in the case of known maternal infection, intrauterine fetal demise, and/or known or suspected chromosomal abnormality. Approval for cord blood collections from live infants with gastroschisis (>35 weeks GA) was obtained from the UCSF Research Protection program, and samples were obtained from infants enrolled for a separate study between November 2009 and December 2012 (30). PTB cord blood samples were collected from live preterm infants (<37 weeks GA) at University College Hospital, London (London, United Kingdom) and the Homerton University Hospital (London, United Kingdom) with ethical permission granted by the South Central–Oxford A Research Ethics Committee between June 2017 and June 2018. Samples with known or suspected chromosomal abnormalities were excluded. After written informed consent, cord blood was obtained at the time of birth by sterile cordocentesis. The study was conducted in accordance with the Declaration of Helsinki principles.
Data and software availability.
RNA-Seq data that support the findings of this study were deposited in NCBI’s Sequence Read Archive (SRA BioProject PRJNA438160). Visualization of individual gene expression in RNA-Seq data was performed with DittoSeq, available on GitHub (http://github.com/dtm2451/dittoseq, branch name: master, commit ID: db0304104d29725dbf00ce3604be916ffd0139ed).
Author contributions
JH designed the study and wrote the manuscript. JH and ER performed the research and analyzed the data. SLH, TCM, and TDB provided clinical samples and intellectual expertise and assisted with manuscript development. DA performed bioinformatics analysis. VFM and LRM assisted in performing experiments. All authors discussed the results and edited the manuscript.
Supplementary Material
Acknowledgments
We would like to acknowledge the tissue and blood donors for their participation in this study and Heather Melichar, Michela Frascoli, Dan Bunis, Melissa Ng, Renan Sper, and Adrian Erlebacher for thoughtful critique of this manuscript. UCSF flow core instrumentation assistance was supported by NIH P30 DK063720 and NIH Shared Instrument Grant 1S10OD021822-01. This study was supported by the UCSF Clinical and Translational Science Institute Pilot Award for Basic and Translational Investigators 2014908 (to JH). JH was supported by the National Institute of Child Health and Human Development K12 Career Development Award at UCSF (K12HD072222), and the National Institute of Allergy and Infectious Diseases K08 Mentored Clinical Scientist Development Award (K08 AI128007). ER was supported by National Science Foundation Graduate Research Fellowship 1650113 and a National Institute of Allergy and Infectious Diseases F31 Fellowship (1F31AI136336-01). SH was supported by an Academy for Medical Sciences Clinical Lecturer grant (535274).
Version 1. 05/30/2019
In-Press Preview
Version 2. 07/29/2019
Electronic publication
Version 3. 09/03/2019
Print issue publication
Funding Statement
to Joanna Halkias
K12 Career Development Award to Joanna Halkias
K08 Mentored Clinical Scientist Development Award to Joanna Halkias
Graduate Research Fellowship to Elze Rackaityte
F31 Fellowship to Elze Rackaityte
Starter Grant for Clinical Lecturers to Sara Hillman, grant number 535274
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(9):3562–3577.https://doi.org/10.1172/JCI125957.
Contributor Information
Joanna Halkias, Email: joanna.halkias@ucsf.edu.
Elze Rackaityte, Email: elze.rackaityte@ucsf.edu.
Sara L. Hillman, Email: sara.hillman@ucl.ac.uk.
Dvir Aran, Email: dvir.aran@ucsf.edu.
Ventura F. Mendoza, Email: ventura.mendoza@ucsf.edu.
Lucy R. Marshall, Email: lucy.marshall.17@ucl.ac.uk.
Tippi C. MacKenzie, Email: Tippi.MacKenzie@ucsf.edu.
Trevor D. Burt, Email: trevor.burt@ucsf.edu.
References
- 1.Friedberg SH, Weissman IL. Lymphoid tissue architecture. II. Ontogeny of peripheral T and B cells in mice: evidence against Peyer’s patches as the site of generation of B cells. J Immunol. 1974;113(5):1477–1492. [PubMed] [Google Scholar]
- 2.Orlic D, Lev R. An electron microscopic study of intraepithelial lymphocytes in human fetal small intestine. Lab Invest. 1977;37(6):554–561. [PubMed] [Google Scholar]
- 3.Spencer J, Dillon SB, Isaacson PG, MacDonald TT. T cell subclasses in fetal human ileum. Clin Exp Immunol. 1986;65(3):553–558. [PMC free article] [PubMed] [Google Scholar]
- 4.Mold JE, et al. Maternal alloantigens promote the development of tolerogenic fetal regulatory T cells in utero. Science. 2008;322(5907):1562–1565. doi: 10.1126/science.1164511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gomez R, Romero R, Ghezzi F, Yoon BH, Mazor M, Berry SM. The fetal inflammatory response syndrome. Am J Obstet Gynecol. 1998;179(1):194–202. doi: 10.1016/S0002-9378(98)70272-8. [DOI] [PubMed] [Google Scholar]
- 6.Romero R, et al. A fetal systemic inflammatory response is followed by the spontaneous onset of preterm parturition. Am J Obstet Gynecol. 1998;179(1):186–193. doi: 10.1016/S0002-9378(98)70271-6. [DOI] [PubMed] [Google Scholar]
- 7.Duggan PJ, et al. Intrauterine T-cell activation and increased proinflammatory cytokine concentrations in preterm infants with cerebral lesions. Lancet. 2001;358(9294):1699–1700. doi: 10.1016/S0140-6736(01)06723-X. [DOI] [PubMed] [Google Scholar]
- 8.Luciano AA, Yu H, Jackson LW, Wolfe LA, Bernstein HB. Preterm labor and chorioamnionitis are associated with neonatal T cell activation. PLoS ONE. 2011;6(2):e16698. doi: 10.1371/journal.pone.0016698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crespo M, et al. Neonatal T-cell maturation and homing receptor responses to Toll-like receptor ligands differ from those of adult naive T cells: relationship to prematurity. Pediatr Res. 2012;71(2):136–143. doi: 10.1038/pr.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rueda CM, Wells CB, Gisslen T, Jobe AH, Kallapur SG, Chougnet CA. Effect of chorioamnionitis on regulatory T cells in moderate/late preterm neonates. Hum Immunol. 2015;76(1):65–73. doi: 10.1016/j.humimm.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frascoli M, et al. Alloreactive fetal T cells promote uterine contractility in preterm labor via IFN-γ and TNF-α. Sci Transl Med. 2018;10(438):eaan2263. doi: 10.1126/scitranslmed.aan2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolfs TG, et al. IL-1α mediated chorioamnionitis induces depletion of FoxP3+ cells and ileal inflammation in the ovine fetal gut. PLoS ONE. 2011;6(3):e18355. doi: 10.1371/journal.pone.0018355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolfs TG, et al. Chorioamnionitis-induced fetal gut injury is mediated by direct gut exposure of inflammatory mediators or by lung inflammation. Am J Physiol Gastrointest Liver Physiol. 2014;306(5):G382–G393. doi: 10.1152/ajpgi.00260.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nikiforou M, et al. Intra-amniotic Candida albicans infection induces mucosal injury and inflammation in the ovine fetal intestine. Sci Rep. 2016;6:29806. doi: 10.1038/srep29806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spencer J, MacDonald TT, Finn T, Isaacson PG. The development of gut associated lymphoid tissue in the terminal ileum of fetal human intestine. Clin Exp Immunol. 1986;64(3):536–543. [PMC free article] [PubMed] [Google Scholar]
- 16.Hermann E, et al. Human fetuses are able to mount an adultlike CD8 T-cell response. Blood. 2002;100(6):2153–2158. [PubMed] [Google Scholar]
- 17.Malhotra I, et al. In utero exposure to helminth and mycobacterial antigens generates cytokine responses similar to that observed in adults. J Clin Invest. 1997;99(7):1759–1766. doi: 10.1172/JCI119340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bunders MJ, et al. Memory CD4(+)CCR5(+) T cells are abundantly present in the gut of newborn infants to facilitate mother-to-child transmission of HIV-1. Blood. 2012;120(22):4383–4390. doi: 10.1182/blood-2012-06-437566. [DOI] [PubMed] [Google Scholar]
- 19.Howie D, et al. Extrathymic T cell differentiation in the human intestine early in life. J Immunol. 1998;161(11):5862–5872. [PubMed] [Google Scholar]
- 20.Zhang X, et al. CD4 T cells with effector memory phenotype and function develop in the sterile environment of the fetus. Sci Transl Med. 2014;6(238):238ra72. doi: 10.1126/scitranslmed.3008748. [DOI] [PubMed] [Google Scholar]
- 21.Li N, et al. Memory CD4. Nat Immunol. 2019;20(3):301–312. doi: 10.1038/s41590-018-0294-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schreurs RRCE, et al. Human Fetal TNF-α-Cytokine-Producing CD4. Immunity. 2019;50(2):462–476.e8. doi: 10.1016/j.immuni.2018.12.010. [DOI] [PubMed] [Google Scholar]
- 23.Dimova T, et al. Effector Vγ9Vδ2 T cells dominate the human fetal γδ T-cell repertoire. Proc Natl Acad Sci USA. 2015;112(6):E556–E565. doi: 10.1073/pnas.1412058112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leeansyah E, Loh L, Nixon DF, Sandberg JK. Acquisition of innate-like microbial reactivity in mucosal tissues during human fetal MAIT-cell development. Nat Commun. 2014;5:3143. doi: 10.1038/ncomms4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loh L, Ivarsson MA, Michaëlsson J, Sandberg JK, Nixon DF. Invariant natural killer T cells developing in the human fetus accumulate and mature in the small intestine. Mucosal Immunol. 2014;7(5):1233–1243. doi: 10.1038/mi.2014.13. [DOI] [PubMed] [Google Scholar]
- 26.Kovalovsky D, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol. 2008;9(9):1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Savage AK, et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29(3):391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eidson M, et al. Altered development of NKT cells, γδ T cells, CD8 T cells and NK cells in a PLZF deficient patient. PLoS ONE. 2011;6(9):e24441. doi: 10.1371/journal.pone.0024441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee YJ, et al. Generation of PLZF+ CD4+ T cells via MHC class II-dependent thymocyte-thymocyte interaction is a physiological process in humans. J Exp Med. 2010;207(1):237–246. doi: 10.1084/jem.20091519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frascoli M, et al. Heightened Immune Activation in Fetuses with Gastroschisis May Be Blocked by Targeting IL-5. J Immunol. 2016;196(12):4957–4966. doi: 10.4049/jimmunol.1502587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lanier LL, Chang C, Phillips JH. Human NKR-P1A. A disulfide-linked homodimer of the C-type lectin superfamily expressed by a subset of NK and T lymphocytes. J Immunol. 1994;153(6):2417–2428. [PubMed] [Google Scholar]
- 32.Takahashi T, Dejbakhsh-Jones S, Strober S. Expression of CD161 (NKR-P1A) defines subsets of human CD4 and CD8 T cells with different functional activities. J Immunol. 2006;176(1):211–216. doi: 10.4049/jimmunol.176.1.211. [DOI] [PubMed] [Google Scholar]
- 33.Constantinides MG, Picard D, Savage AK, Bendelac A. A naive-like population of human CD1d-restricted T cells expressing intermediate levels of promyelocytic leukemia zinc finger. J Immunol. 2011;187(1):309–315. doi: 10.4049/jimmunol.1100761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sathaliyawala T, et al. Distribution and compartmentalization of human circulating and tissue-resident memory T cell subsets. Immunity. 2013;38(1):187–197. doi: 10.1016/j.immuni.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thome JJ, et al. Spatial map of human T cell compartmentalization and maintenance over decades of life. Cell. 2014;159(4):814–828. doi: 10.1016/j.cell.2014.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ashouri JF, Weiss A. Endogenous Nur77 Is a specific indicator of antigen receptor signaling in human T and B cells. J Immunol. 2017;198(2):657–668. doi: 10.4049/jimmunol.1601301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ben Youssef G, et al. Ontogeny of human mucosal-associated invariant T cells and related T cell subsets. J Exp Med. 2018;215(2):459–479. doi: 10.1084/jem.20171739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rechavi E, et al. Timely and spatially regulated maturation of B and T cell repertoire during human fetal development. Sci Transl Med. 2015;7(276):276ra25. doi: 10.1126/scitranslmed.aaa0072. [DOI] [PubMed] [Google Scholar]
- 39.Mabbott NA, Baillie JK, Brown H, Freeman TC, Hume DA. An expression atlas of human primary cells: inference of gene function from coexpression networks. BMC Genomics. 2013;14:632. doi: 10.1186/1471-2164-14-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skon CN, Lee JY, Anderson KG, Masopust D, Hogquist KA, Jameson SC. Transcriptional downregulation of S1pr1 is required for the establishment of resident memory CD8+ T cells. Nat Immunol. 2013;14(12):1285–1293. doi: 10.1038/ni.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mueller SN, Gebhardt T, Carbone FR, Heath WR. Memory T cell subsets, migration patterns, and tissue residence. Annu Rev Immunol. 2013;31:137–161. doi: 10.1146/annurev-immunol-032712-095954. [DOI] [PubMed] [Google Scholar]
- 42.Fergusson JR, Fleming VM, Klenerman P. CD161-expressing human T cells. Front Immunol. 2011;2:36. doi: 10.3389/fimmu.2011.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh SP, et al. PLZF regulates CCR6 and is critical for the acquisition and maintenance of the Th17 phenotype in human cells. J Immunol. 2015;194(9):4350–4361. doi: 10.4049/jimmunol.1401093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Legoux F, Salou M, Lantz O. Unconventional or preset αβ T cells: evolutionarily conserved tissue-resident T cells recognizing nonpeptidic ligands. Annu Rev Cell Dev Biol. 2017;33:511–535. doi: 10.1146/annurev-cellbio-100616-060725. [DOI] [PubMed] [Google Scholar]
- 45.Aldemir H, et al. Cutting edge: lectin-like transcript 1 is a ligand for the CD161 receptor. J Immunol. 2005;175(12):7791–7795. doi: 10.4049/jimmunol.175.12.7791. [DOI] [PubMed] [Google Scholar]
- 46.Exley M, Porcelli S, Furman M, Garcia J, Balk S. CD161 (NKR-P1A) costimulation of CD1d-dependent activation of human T cells expressing invariant V alpha 24 J alpha Q T cell receptor alpha chains. J Exp Med. 1998;188(5):867–876. doi: 10.1084/jem.188.5.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fergusson JR, et al. CD161 defines a transcriptional and functional phenotype across distinct human T cell lineages. Cell Rep. 2014;9(3):1075–1088. doi: 10.1016/j.celrep.2014.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Llibre A, Garner L, Partridge A, Freeman GJ, Klenerman P, Willberg CB. Expression of lectin-like transcript-1 in human tissues. F1000Res. 2016;5:2929. doi: 10.12688/f1000research.10009.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Llibre A, et al. LLT1 and CD161 expression in human germinal centers promotes B cell activation and CXCR4 downregulation. J Immunol. 2016;196(5):2085–2094. doi: 10.4049/jimmunol.1502462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bain CC, et al. Resident and pro-inflammatory macrophages in the colon represent alternative context-dependent fates of the same Ly6Chi monocyte precursors. Mucosal Immunol. 2013;6(3):498–510. doi: 10.1038/mi.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smythies LE, et al. Human intestinal macrophages display profound inflammatory anergy despite avid phagocytic and bacteriocidal activity. J Clin Invest. 2005;115(1):66–75. doi: 10.1172/JCI19229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rosen DB, et al. Functional consequences of interactions between human NKR-P1A and its ligand LLT1 expressed on activated dendritic cells and B cells. J Immunol. 2008;180(10):6508–6517. doi: 10.4049/jimmunol.180.10.6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee J, et al. Characterization of the fetal blood transcriptome and proteome in maternal anti-fetal rejection: evidence of a distinct and novel type of human fetal systemic inflammatory response. Am J Reprod Immunol. 2013;70(4):265–284. doi: 10.1111/aji.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bukowski R, et al. Onset of human preterm and term birth is related to unique inflammatory transcriptome profiles at the maternal fetal interface. PeerJ. 2017;5:e3685. doi: 10.7717/peerj.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Banuelos J, et al. BCL-2 protects human and mouse Th17 cells from glucocorticoid-induced apoptosis. Allergy. 2016;71(5):640–650. doi: 10.1111/all.12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boyle AK, Rinaldi SF, Norman JE, Stock SJ. Preterm birth: Inflammation, fetal injury and treatment strategies. J Reprod Immunol. 2017;119:62–66. doi: 10.1016/j.jri.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 57.Adkins B, Leclerc C, Marshall-Clarke S. Neonatal adaptive immunity comes of age. Nat Rev Immunol. 2004;4(7):553–564. doi: 10.1038/nri1394. [DOI] [PubMed] [Google Scholar]
- 58.Casey KA, et al. Antigen-independent differentiation and maintenance of effector-like resident memory T cells in tissues. J Immunol. 2012;188(10):4866–4875. doi: 10.4049/jimmunol.1200402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Opiela SJ, Levy RB, Adkins B. Murine neonates develop vigorous in vivo cytotoxic and Th1/Th2 responses upon exposure to low doses of NIMA-like alloantigens. Blood. 2008;112(4):1530–1538. doi: 10.1182/blood-2007-08-106500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Michaëlsson J, Mold JE, McCune JM, Nixon DF. Regulation of T cell responses in the developing human fetus. J Immunol. 2006;176(10):5741–5748. doi: 10.4049/jimmunol.176.10.5741. [DOI] [PubMed] [Google Scholar]
- 61.Ardissone AN, et al. Meconium microbiome analysis identifies bacteria correlated with premature birth. PLoS ONE. 2014;9(3):e90784. doi: 10.1371/journal.pone.0090784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Collado MC, Rautava S, Aakko J, Isolauri E, Salminen S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci Rep. 2016;6:23129. doi: 10.1038/srep23129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chu DM, Ma J, Prince AL, Antony KM, Seferovic MD, Aagaard KM. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat Med. 2017;23(3):314–326. doi: 10.1038/nm.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Germain C, et al. Induction of lectin-like transcript 1 (LLT1) protein cell surface expression by pathogens and interferon-γ contributes to modulate immune responses. J Biol Chem. 2011;286(44):37964–37975. doi: 10.1074/jbc.M111.285312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mold JE, et al. Fetal and adult hematopoietic stem cells give rise to distinct T cell lineages in humans. Science. 2010;330(6011):1695–1699. doi: 10.1126/science.1196509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smith NL, et al. Developmental Origin Governs CD8. Cell. 2018;174(1):117–130.e14. doi: 10.1016/j.cell.2018.05.029. [DOI] [PubMed] [Google Scholar]
- 67.Rosen DB, Bettadapura J, Alsharifi M, Mathew PA, Warren HS, Lanier LL. Cutting edge: lectin-like transcript-1 is a ligand for the inhibitory human NKR-P1A receptor. J Immunol. 2005;175(12):7796–7799. doi: 10.4049/jimmunol.175.12.7796. [DOI] [PubMed] [Google Scholar]
- 68.Pozo D, Valés-Gómez M, Mavaddat N, Williamson SC, Chisholm SE, Reyburn H. CD161 (human NKR-P1A) signaling in NK cells involves the activation of acid sphingomyelinase. J Immunol. 2006;176(4):2397–2406. doi: 10.4049/jimmunol.176.4.2397. [DOI] [PubMed] [Google Scholar]
- 69.Bai A, et al. CD39 and CD161 modulate Th17 responses in Crohn’s disease. J Immunol. 2014;193(7):3366–3377. doi: 10.4049/jimmunol.1400346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thome JJ, Farber DL. Emerging concepts in tissue-resident T cells: lessons from humans. Trends Immunol. 2015;36(7):428–435. doi: 10.1016/j.it.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Koay HF, et al. A three-stage intrathymic development pathway for the mucosal-associated invariant T cell lineage. Nat Immunol. 2016;17(11):1300–1311. doi: 10.1038/ni.3565. [DOI] [PubMed] [Google Scholar]
- 72.Turtle CJ, et al. Innate signals overcome acquired TCR signaling pathway regulation and govern the fate of human CD161(hi) CD8α+ semi-invariant T cells. Blood. 2011;118(10):2752–2762. doi: 10.1182/blood-2011-02-334698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wencker M, et al. Innate-like T cells straddle innate and adaptive immunity by altering antigen-receptor responsiveness. Nat Immunol. 2014;15(1):80–87. doi: 10.1038/ni.2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chu DM, Seferovic M, Pace RM, Aagaard KM. The microbiome in preterm birth. Best Pract Res Clin Obstet Gynaecol. 2018;52:103–113. doi: 10.1016/j.bpobgyn.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 75.Steinert EM, et al. Quantifying memory CD8 T cells reveals regionalization of immunosurveillance. Cell. 2015;161(4):737–749. doi: 10.1016/j.cell.2015.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Olin A, et al. Stereotypic immune system development in newborn children. Cell. 2018;174(5):1277–1292.e14. doi: 10.1016/j.cell.2018.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Elahi S, et al. Immunosuppressive CD71+ erythroid cells compromise neonatal host defence against infection. Nature. 2013;504(7478):158–162. doi: 10.1038/nature12675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McGovern N, et al. Human fetal dendritic cells promote prenatal T-cell immune suppression through arginase-2. Nature. 2017;546(7660):662–666. doi: 10.1038/nature22795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.