Abstract
Background:
Chronic rhinosinusitis (CRS) is a multifaceted disease with a significant genetic component. The importance of taste receptor signaling has recently been highlighted in CRS; single nucleotide polymorphisms (SNPs) of bitter tastant-responsive G protein-coupled receptors (GPCRs) have been linked with CRS and with altered innate immune responses to multiple bacterially-derived signals.
Objective:
To characterize in CRS the frequency of six SNPs in genes with known bitter tastant signaling function.
Methods:
Genomic DNA was isolated from 74 CRS volunteers in West Virginia and allele frequency was determined and compared to demographically-matched data from the 1000 Genomes database.
Results:
For two SNPs in a gene recently associated with bitterant signaling regulation, RGS21, there were no associations with CRS (although the frequency of the minor allele of RGS21 rs7528947 was seen to increase with increasing Lund-Mackay CT staging score). Two TAS2R bitter taste receptor variants (TAS2R19 rs10772420 and TAS2R38 rs713598), identified in prior CRS genetics studies, were found to have similar associations in this study.
Conclusion:
Unique to our study is the establishment of an association between CRS in this patient population and GNB3 rs5443, a variation in an established G protein component downstream of bitterant receptor signal transduction.
Keywords: bitterant signaling, chronic rhinosinusitis, genetics, innate immune response, single nucleotide polymorphism
Introduction
Chronic rhinosinusitis (CRS) is a persistent inflammatory state of the sinus mucosa with symptoms of nasal obstruction or discharge with or without facial pain and changes in smell present for more than 12 weeks [1]. Prevalence estimates range from 10–12% of the population in USA and Europe [2], resulting in >$770 million healthcare costs in the USA alone [3]. CRS patients have poor quality of life scores, similar to other chronic diseases including CHF and COPD [4]. Evidence suggests a genetic component to CRS [5]. In a large population study, there was a 2.4-fold increased risk of CRS in first-degree relatives [6]; this number is higher in patients with nasal polyps. Furthermore, several well-characterized genetic syndromes are associated with CRS, including cystic fibrosis [7] and primary ciliary dyskinesia [8].
An emerging concept regarding the chronic nature of CRS relates to a potential deficit in sensing and clearing bacterial infections; receptors for bitter and sweet taste have been found in the airway and are thought to play a role in sensing bacteria and regulating innate immune responses [9]. Bitter taste is initiated in the oral cavity through members of the T2R family of G protein-coupled receptors (GPCRs) [10–12]. However, T2R bitter receptors are not limited to taste bud cells; they are also found in solitary chemosensory cells (SCCs) and ciliated epithelial cells of the nasal and sinus cavities, functioning in these locales to “taste” secreted bacterial products such as acyl-homoserine lactones (AHLs) [13]. Gram negative bacteria such as Pseudomonas aeruginosa use AHLs as quorum-sensing molecules [14]. Binding of these bacterial products activates innate immune responses, such as release of antimicrobial peptides and nitric oxide (NO).
Genome-wide association studies (GWAS) have identified several genes potentially associated with CRS [15, 16]. Recent genetic studies have underscored the importance of taste receptor signaling in innate immunity of upper and lower airways, and paranasal sinuses [13]; single nucleotide polymorphisms (SNPs) in taste receptor genes have since been associated with altered bacterial immune response and, thus, with CRS [17, 18]. There are ~25 Type 2 taste receptors (TAS2R “bitterant receptors”), coupled to G protein signaling, that are expressed in multiple tissues. In the ciliated mucosa of the sinuses, these TAS2Rs respond to chemoirritants and bacterially-produced secretions [19]. Some of us also recently showed that an inhibitor of GPCR signaling, Regulator of G protein Signaling-21 (RGS21), opposes TAS2Rmediated bitterant signaling in immortalized airway epithelial cells and, given its expression in sinus mucosa and airway epithelia, may be involved in mucociliary clearance [20, 21]. Here, investigating candidate genes involved in bitterant signaling from among the TAS2R genes and RGS21, we identified three SNPs more common in West Virginia CRS patients, including within GNB3, a downstream component of bitterant signaling previously unassociated with CRS.
Materials and Methods
Recruitment
Seventy-four volunteers were seen at an academic tertiary referral center in West Virginia (WV). The study was approved by the Institutional Review Board of WVU’s Office of Research Integrity and Compliance. All volunteers met EPOS criteria [22] for CRS, and informed consent obtained under IRB 1410476782. Age, sex, allergy, asthma, migraine, smoking status, presence of polyps, and previous surgical history were recorded (Table 1). CT scan of the sinuses was available for 66 of the 74 patients. Lund-Mackay CT score was calculated based on the standard radiologic staging of CRS for these patients [23]. Sino-nasal Outcome Test-22 (SNOT22) scores were obtained from all patients; total SNOT22 and the five standard quality of life domain scores were calculated (Table 1) [24]. Patients with cystic fibrosis, primary ciliary dyskinesia, known immunodeficiency, and craniofacial abnormalities were excluded from the study population.
Table 1.
Patient Demographics
| Patient variable | N (74 total) |
Percentage | |
|---|---|---|---|
| Mean age | 55.0 | n/a | |
| Male | 38 | 52% | |
| Female | 36 | 48% | |
| Allergy | 42 | 57% | |
| Asthma | 28 | 38% | |
| Migraine | 8 | 11% | |
| Current smoker | 9 | 11% | |
| History of smoking | 19 | 26% | |
| Polyps | 45 | 61% | |
| Previous Surgery | 29 | 39% | |
|
N (66 total) |
percentage | ||
| Lund-Mackay Score 1−8 | 22 | 33% | |
| Lund-Mackay Score 9−16 | 29 | 44% | |
| Lund-Mackay Score 17−24 | 15 | 23% | |
| average | CI (5%−95%) | ||
| Lund-Mackay Score CT Score | 66 | 11.6 | 10.0−13.3 |
| Score range | average | CI (5%−95%) | |
| SNOT 22 (Total) | 0−110 | 49.0 | 43.5−54.6 |
| Rhinologic Symptoms | 0−30 | 13.0 | 11.4−14.5 |
| Extranasal Rhinologic Symptoms | 0−15 | 7.0 | 6.1−8.0 |
| Ear/Facial Symptoms | 0−25 | 9.7 | 8.4−11.1 |
| Psychological Dysfunction | 0−35 | 17.5 | 15.1−19.8 |
| Sleep Dysfunction | 0−25 | 12.4 | 10.8−14.0 |
Tissue Collection and DNA extraction
Volunteers provided two buccal swabs, which were each placed in a 15-ml polystyrene conical centrifuge tube and stored at −80° C until DNA extraction using QIAamp DNA Mini Kits (exactly as per manufacturer’s protocol). Purity and yield were assessed by 260/280 nm absorbance on a QIAxpert microfluidic spectrophotometer.
Genotyping
Genotyping was performed using TaqMan primer-probe sets (Table 2) and Type-it Fast SNP Probe PCR Kits (Qiagen) exactly as per manufacturer’s protocols. Reactions were run on a Qiagen Rotor-Gene Q in duplicate; no-DNA negative control reactions were also performed for each primer-probe set (e.g., Figure 1 displays representative data obtained).
Table 2.
Gene variant minor allele frequency (MAF) data obtained from West Virginia CRS clinic patients (N = 74) and public databases with demographically matched cohorts.
| Gene | dbSNP ID | Minor Allele | TaqMan Probe | European MAF* | MAF in CRS probands‡ |
|---|---|---|---|---|---|
| TAS2R38 | rs713598 | C | C___8876467_10 | 0.423 |
0.568 p < 0.001 |
| GNB3 | rs5443 | T | C___2184734_10 | 0.367 (HapMap#) |
0.493 p = 0.014 |
| TAS2R19 | rs10772420 | A | C___1317426_10 | 0.505 |
0.601 p = 0.024 |
| TAS2R20 | rs12226920 | T | C___1326611_10 | 0.383 | 0.458 p = 0.062 |
| RGS21 | rs7528947 | G | C__30007846_20 | 0.516 | 0.547 p = 0.461 |
| RGS21 | rs1175152 | A | C_____68684_10 | 0.433 | 0.439 p = 0.875 |
Minor allele frequency reported on N = 1006 Europeans via 1000 Genomes (#except as otherwise noted: i.e., 116 Europeans from HapMap); MAF comparisons between West Virginia probands and European SNP databases was previously established in Kaski, SW, et al. (2019) J. Opioid Manag. [in press; vol. 15, issue 1].
p-value denotes significance of difference between MAFs derived by comparison between CRS probands and European MAF using binomial test (null hypothesis is “no difference between CRS probands and Europeans”; alternative is “there is difference between CRS probands and Europeans”
Fig. 1.
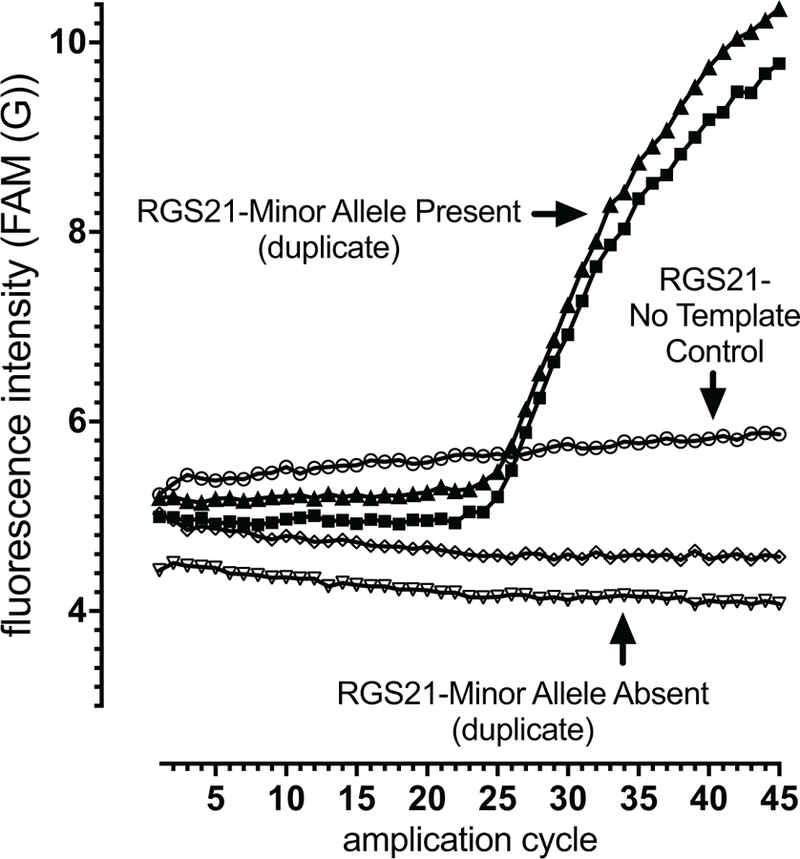
Sample real-time quantitative polymerase-chain reaction (qPCR) results for the detection of the absence or presence of RGS21 SNP rs7528947 (minor allele G) within WV CRS patient genomic DNA using TaqMan primer-probe set C__30007846_20. Examples for probands with the minor allele (closed symbols) and lacking the minor allele (open triangle and open diamond) are shown; a separate qPCR reaction lacking input genomic DNA is also illustrated (open circles).
Statistical Analyses
Allele frequencies were compared to 1006 Europeans (1000 Genomes Project) [25], given that WV is demographically homogenous [26], i.e., 94% white European ancestry (http://censusviewer.com/state/WV). Given no information for the frequency of the GNB3 SNP rs5443 in 1000 Genomes; its allele frequency was instead compared to 116 Europeans (CEU) from HapMap [27]. Statistical analyses of SNP frequency were performed using Pearson’s chi-square. A p-value < 0.05 was used to infer that the allele frequency from CRS patients is significantly different from demographically matched, public genome data. Subgroup analyses were performed comparing the SNP MAF in patients with (CRSwNP) or without (CRSsNP) nasal polyps, and separately based on Lund-Mackay CT score, using standard Chi-squared statistical tests (Tables 3 and 4).
Table 3.
Gene variant minor allele frequency (MAF) data from CRS patients with and without nasal polyps (n=74).
| Gene | dbSNP ID | Minor Allele | CRSwNP MAF | CRSsNP MAF | p value |
|---|---|---|---|---|---|
| TAS2R38 | rs713598 | C | 0.567 | 0.569 | 0.978 |
| GNB3 | rs5443 | T | 0.522 | 0.448 | 0.380 |
| TAS2R19 | rs10772420 | A | 0.589 | 0.621 | 0.700 |
| TAS2R20 | rs12226920 | T | 0.456 | 0.466 | 0.906 |
| RGS21 | rs7528947 | G | 0.567 | 0.517 | 0.555 |
| RGS21 | rs1175152 | A | 0.389 | 0.517 | 0.125 |
Table 4.
Gene variant minor allele frequency (MAF) data from CRS based on Lund-Mackay (LM) CT score (n=66).
| Gene | dbSNP ID | Minor Allele | LM 1–8 MAF | LM 9–16 MAF | LM 17–24 MAF | p value |
|---|---|---|---|---|---|---|
| TAS2R38 | rs713598 | C | 0.614 | 0.500 | 0.600 | 0.459 |
| GNB3 | rs5443 | T | 0.568 | 0.517 | 0.467 | 0.688 |
| TAS2R19 | rs10772420 | A | 0.591 | 0.603 | 0.600 | 0.992 |
| TAS2R20 | rs12226920 | T | 0.523 | 0.362 | 0.533 | 0.167 |
| RGS21 | rs7528947 | G | 0.386 | 0.603 | 0.700 | 0.017 |
| RGS21 | rs1175152 | A | 0.500 | 0.431 | 0.333 | 0.364 |
Results
Given our recent findings that a negative regulator of bitterant GPCR signaling, RGS21, is expressed in sinus mucosa and airway epithelia [20, 21], we interrogated the status within a CRS patient population of two SNPs within RGS21, as well as four SNPs within other bitterant signaling genes with established or unknown association to CRS (i.e., TAS2R19, TAS2R20, TAS2R38, and GNB3). All chosen SNPs had a minor allele frequency (MAF) greater than 30% so they would be identifiable in this patient population. Buccal swabs from CRS patients (Table 1) were used for genomic DNA isolation and genotyping.
TAS2R19 rs10772420 (Type 2 Taste Receptor 19)
TAS2R19 (previously known as TAS2R48) is a GPCR with bitterant taste receptor activity located on chromosome 12. SNP rs10772420 within TAS2R19 was previously identified as associated with RSV infections in a genome-wide association study (GWAS) [28] and also identified in a GWAS of Canadian patients with sinusitis [15]. The minor allele (A) causes a missense mutation (arginine-299 to cysteine) in the encoded protein and is associated with intense quinine perception [29]. The “minor” allele is actually more prominent in Europeans (MAF of 0.505; Table 2). In the CRS patients in this study, this allele is even more common (MAF 0.601; p=0.024); twenty-three percent of the CRS patients were homozygous (A/A) for the minor allele.
TAS2R38 rs713598 (Type 2 Taste Receptor 38)
TAS2R38, a bitterant receptor key to phenylthiocarbamine perception [30], is expressed in sinonasal ciliated epithelium [31] and upper airways [32]. TAS2R38 is implicated in innate immunity and the response to Pseudomonas [31]. rs713598 is a common missense SNP in TAS2R38 [30], associated with quinine intensity [29] and other taste preferences [33–35]. rs713598 is one of the most common SNPs in the TAS2R38 gene, causing a missense mutation to the encoded GPCR (alanine-49 to proline) [30]. In the CRS patients in this study, this allele is more common (MAF 0.568; p<0.001) than in Europeans of the 1000 Genomes Project (MAF 0.423).
RGS21 rs7528947 (Regulator of G-protein signaling 21)
RGS21 is a Gα GTPase-accelerating protein expressed in lingual epithelium, lung, and gastrointestinal tissues [21]; ablation of Rgs21 in mice blunts bitterant signaling [36]. rs7528947 is a 3’ untranslated region variant of RGS21 with no known disease associations. The rs7528947 MAF in CRS patients is 0.547, similar to the European MAF (0.516; p=0.461). No difference in rs7528947 MAF was observed when stratified between CRS patients with or without nasal polyps (Table 3); however, a significant difference (p = 0.017) was observed across the spectrum of Lund-Mackay CT staging scores (Table 4), with the minor allele appearing more frequently with higher Lund-Mackay scores.
RGS21 intergenic region rs1175152 (Regulator of G-protein signaling 21)
A second RGS21-associated SNP rs1175152 is located in the intergenic region between RGS18 and RGS21 and is also of unknown consequence. To-date, there have been no known studies identifying this marker with any known phenotype or in association with any disease. Its MAF among Europeans of the 1000 Genomes Project is 0.433, similar to the frequency observed in the CRS patients of this study (MAF 0.439; p=0.875).
TAS2R20 rs12226920 (Type 2 Taste Receptor 20)
TAS2R20 (also known as TAS2R49) is bitterant receptor expressed in multiple tissue types [37]. Its missense SNP rs12226920 (histidine-143 to glutamine) was previously associated with CRS, with a biallelic difference of 16% vs controls, suggesting a possible role for variations in this GPCR in CRS pathogenesis [ [15] ]. In the CRS patients of this study, the MAF (0.458) was higher than predicted by 1000 Genomes (European MAF 0.383); however, this difference did not meet our statistical significance threshold (p=0.062).
GNB3 rs5443 (G protein subunit beta 3)
GNB3 encodes a Gβ subunit of the G protein heterotrimer. Its SNP rs5443 has been studied extensively in other contexts and is associated with numerous metabolic-related diseases including obesity [38], coronary artery disease and hypertension [39], as well as the increased efficacy of antidepressants in major depressive disorder [40]. All of these associations are with minor allele (T) carriers. The HapMap European MAF is 0.367, whereas the CRS patients of this study showed an increased MAF of 0.493 (p=0.014).
Discussion
While RGS21 is functionally linked to bitterant GPCR signaling [20, 21, 36], neither RGS21 SNP tested was found in this study to be associated with CRS (although the minor allele of RGS21 SNP rs7528947 was seen more frequently in CRS patients with higher Lund-Mackay CT staging scores). Conversely, GNB3 SNP rs5443, a gene variation in an established component of bitterant GPCR signaling [41] but without a previous association to chronic rhinosinusitis, was found to be more highly prevalent in West Virginia CRS patients, along with two TAS2R variants previously identified as associated with CRS. Gβ subunits of G protein heterotrimers, such as that encoded by GNB3, are known to assemble with E3 ubiquitin ligase complexes responsible for degradation of GRK2, a G protein-coupled receptor kinase [42]. Recent work has shown that GNB3 SNP rs5443, identified more commonly in the CRS patients of this study, is associated with decreased GRK2 ubiquitination [43]. Further studies are therefore required to clarify the role of altered GNB3 function (and potentially GRK2 function) in bitterant signaling, innate immunity activation/regulation, and CRS.
Acknowledgement
We thank the American Academy of Otolaryngology—Head and Neck Surgery Foundation (AAO-HNSF) Annual Meeting in Chicago, IL, September 10–13, 2017 for accepting an oral presentation by one of us (B.L.A.) of an interim report on this study.
Funding Sources
Research was supported in part by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number 5U54GM104942-03 to the West Virginia Clinical & Translational Science Institute (to B.L.A. and D.P.S.). Funding support was also provided by the WVU E.J. Van Liere Endowed Medicine Professorship (to D.P.S). Neither of these sources participated in the preparation of data or the manuscript.
Footnotes
Statement of Ethics
The study protocol has been approved by the Institutional Review Board of WVU’s Office of Research Integrity and Compliance. Subjects of this study have given their written informed consent as obtained under WVU IRB 1410476782.
Disclosure Statement
The authors have no conflicts of interest to declare.
References
- 1.Fokkens WJ, Lund VJ, Mullol J, et al. (2012) EPOS 2012: European position paper on rhinosinusitis and nasal polyps 2012. A summary for otorhinolaryngologists. Rhinology 50:1–12 [DOI] [PubMed] [Google Scholar]
- 2.DeConde AS, Soler ZM (2016) Chronic rhinosinusitis: Epidemiology and burden of disease. Am J Rhinol Allergy 30:134–9 [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharyya N (2011) Incremental health care utilization and expenditures for chronic rhinosinusitis in the United States. Ann Otol Rhinol Laryngol 120:423–7 [DOI] [PubMed] [Google Scholar]
- 4.Gliklich RE, Metson R (1995) The health impact of chronic sinusitis in patients seeking otolaryngologic care. Otolaryngol Head Neck Surg 113:104–9 [DOI] [PubMed] [Google Scholar]
- 5.Yoo F, Suh JD (2017) What is the evidence for genetics in chronic rhinosinusitis? Curr Opin Otolaryngol Head Neck Surg 25:54–63 [DOI] [PubMed] [Google Scholar]
- 6.Oakley GM, Curtin K, Orb Q, Schaefer C, Orlandi RR, Alt JA (2015) Familial risk of chronic rhinosinusitis with and without nasal polyposis: genetics or environment. Int Forum Allergy Rhinol 5:276–82 [DOI] [PubMed] [Google Scholar]
- 7.Hamilos DL Chronic Rhinosinusitis in Patients with Cystic Fibrosis. J allergy Clin Immunol Pract 4:605–12 [DOI] [PubMed] [Google Scholar]
- 8.Noone PG, Leigh MW, Sannuti A, Minnix SL, Carson JL, Hazucha M, Zariwala MA, Knowles MR (2004) Primary ciliary dyskinesia: diagnostic and phenotypic features. Am J Respir Crit Care Med 169:459–67 [DOI] [PubMed] [Google Scholar]
- 9.Lee RJ, Cohen NA (2014) Bitter and sweet taste receptors in the respiratory epithelium in health and disease. J Mol Med (Berl) 92:1235–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandrashekar J, Mueller KL, Hoon MA, Adler E, Feng L, Guo W, Zuker CS, Ryba NJ (2000) T2Rs function as bitter taste receptors. Cell 100:703–11 [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Hoon MA, Chandrashekar J, Mueller KL, Cook B, Wu D, Zuker CS, Ryba NJP (2003) Coding of sweet, bitter, and umami tastes: different receptor cells sharing similar signaling pathways. Cell 112:293–301 [DOI] [PubMed] [Google Scholar]
- 12.Chandrashekar J, Hoon MA, Ryba NJP, Zuker CS (2006) The receptors and cells for mammalian taste. Nature 444:288–94 [DOI] [PubMed] [Google Scholar]
- 13.Tizzano M, Gulbransen BD, Vandenbeuch A, Clapp TR, Herman JP, Sibhatu HM, Churchill MEA, Silver WL, Kinnamon SC, Finger TE (2010) Nasal chemosensory cells use bitter taste signaling to detect irritants and bacterial signals. Proc Natl Acad Sci U S A 107:3210–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pearson JP, Passador L, Iglewski BH, Greenberg EP (1995) A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 92:1490–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mfuna Endam L, Filali-Mouhim A, Boisvert P, Boulet L-P, Bossé Y, Desrosiers M (2014) Genetic variations in taste receptors are associated with chronic rhinosinusitis: a replication study. Int Forum Allergy Rhinol 4:200–6 [DOI] [PubMed] [Google Scholar]
- 16.Tournas A, Mfuna L, Bossé Y, Filali-Mouhim A, Grenier J-P, Desrosiers M (2010) A pooling-based genome-wide association study implicates the p73 gene in chronic rhinosinusitis. J Otolaryngol Head Neck Surg 39:188–95 [PubMed] [Google Scholar]
- 17.Freund JR, Lee RJ (2018) Taste receptors in the upper airway. World J Otorhinolaryngol - head neck Surg 4:67–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Workman AD, Palmer JN, Adappa ND, Cohen NA (2015) The Role of Bitter and Sweet Taste Receptors in Upper Airway Immunity. Curr Allergy Asthma Rep 15:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Douglas JE, Cohen NA (2017) Taste Receptors Mediate Sinonasal Immunity and Respiratory Disease. Int J Mol Sci doi: 10.3390/ijms18020437 [DOI] [PMC free article] [PubMed]
- 20.Kimple AJ, Garland AL, Cohen SP, Setola V, Willard FS, Zielinski T, Lowery RG, Tarran R, Siderovski DP (2014) RGS21, a regulator of taste and mucociliary clearance? Laryngoscope 124:E56–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cohen SP, Buckley BK, Kosloff M, et al. (2012) Regulator of G-protein signaling-21 (RGS21) is an inhibitor of bitter gustatory signaling found in lingual and airway epithelia. J Biol Chem 287:41706–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fokkens WJ, Lund VJ, Mullol J, et al. (2012) European Position Paper on Rhinosinusitis and Nasal Polyps 2012. Rhinol Suppl 23:3 p preceding table of contents, 1–298 [PubMed]
- 23.Lund VJ, Mackay IS (1993) Staging in rhinosinusitus. Rhinology 31:183–184 [PubMed] [Google Scholar]
- 24.DeConde AS, Mace JC, Bodner T, Hwang PH, Rudmik L, Soler ZM, Smith TL (2014) SNOT-22 quality of life domains differentially predict treatment modality selection in chronic rhinosinusitis. Int Forum Allergy Rhinol 4:972–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.1000 Genomes Project Consortium, Auton A, Brooks LD, et al. (2015) A global reference for human genetic variation. Nature 526:68–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gall BJ, Wilson A, Schroer AB, Gross JD, Stoilov P, Setola V, Watkins CM, Siderovski DP (2016) Genetic variations in GPSM3 associated with protection from rheumatoid arthritis affect its transcript abundance. Genes Immun 17:139–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thorisson G, Smith A (2005) The international HapMap project web site. Genome Res doi: 10.1101/gr.4413105. [DOI] [PMC free article] [PubMed]
- 28.Salas A, Pardo-Seco J, Cebey-López M, et al. (2017) Whole Exome Sequencing reveals new candidate genes in host genomic susceptibility to Respiratory Syncytial Virus Disease. Sci Rep 7:15888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reed DR, Zhu G, Breslin PAS, Duke FF, Henders AK, Campbell MJ, Montgomery GW, Medland SE, Martin NG, Wright MJ (2010) The perception of quinine taste intensity is associated with common genetic variants in a bitter receptor cluster on chromosome 12. Hum Mol Genet 19:4278–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim U, Jorgenson E, Coon H, Leppert M, Risch N, Drayna D (2003) Positional cloning of the human quantitative trait locus underlying taste sensitivity to phenylthiocarbamide. Science 299:1221–5 [DOI] [PubMed] [Google Scholar]
- 31.Lee RJ, Cohen NA (2015) Role of the bitter taste receptor T2R38 in upper respiratory infection and chronic rhinosinusitis. Curr Opin Allergy Clin Immunol 15:14–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shah AS, Ben-Shahar Y, Moninger TO, Kline JN, Welsh MJ (2009) Motile cilia of human airway epithelia are chemosensory. Science 325:1131–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chamoun E, Hutchinson JM, Krystia O, et al. (2018) Single Nucleotide Polymorphisms in Taste Receptor Genes Are Associated with Snacking Patterns of Preschool-Aged Children in the Guelph Family Health Study: A Pilot Study. Nutrients doi: 10.3390/nu10020153 [DOI] [PMC free article] [PubMed]
- 34.Pawellek I, Grote V, Rzehak P, Xhonneux A, Verduci E, Stolarczyk A, Closa-Monasterolo R, Reischl E, Koletzko B, European Childhood Obesity Trial Study Group (2016) Association of TAS2R38 variants with sweet food intake in children aged 1–6 years. Appetite 107:126–134 [DOI] [PubMed] [Google Scholar]
- 35.Deshaware S, Singhal R (2017) Genetic variation in bitter taste receptor gene TAS2 R38, PROP taster status and their association with body mass index and food preferences in Indian population. Gene 627:363–368 [DOI] [PubMed] [Google Scholar]
- 36.Schroer AB, Gross JD, Kaski SW, Wix K, Siderovski DP, Vandenbeuch A, Setola V (2018) Development of Full Sweet, Umami, and Bitter Taste Responsiveness Requires Regulator of G protein Signaling-21 (RGS21). Chem Senses 43:367–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jaggupilli A, Singh N, Upadhyaya J, Sikarwar AS, Arakawa M, Dakshinamurti S, Bhullar RP, Duan K, Chelikani P (2017) Analysis of the expression of human bitter taste receptors in extraoral tissues. Mol Cell Biochem 426:137–147 [DOI] [PubMed] [Google Scholar]
- 38.Siffert W, Forster P, Jöckel KH, et al. (1999) Worldwide ethnic distribution of the G protein beta3 subunit 825T allele and its association with obesity in Caucasian, Chinese, and Black African individuals. J Am Soc Nephrol 10:1921–30 [DOI] [PubMed] [Google Scholar]
- 39.Siffert W (2003) G hypertension. Curr Hypertens Rep 5:47–53 [DOI] [PubMed] [Google Scholar]
- 40.Hu Q, Zhang S-Y, Liu F, et al. (2015) Influence of GNB3 C825T polymorphism on the efficacy of antidepressants in the treatment of major depressive disorder: A meta-analysis. J Affect Disord 172:103–9 [DOI] [PubMed] [Google Scholar]
- 41.Huang L, Shanker YG, Dubauskaite J, Zheng JZ, Yan W, Rosenzweig S, Spielman AI, Max M, Margolskee RF (1999) Ggamma13 colocalizes with gustducin in taste receptor cells and mediates IP3 responses to bitter denatonium. Nat Neurosci 2:1055–62 [DOI] [PubMed] [Google Scholar]
- 42.Zha Z, Han X, Smith MD, et al. (2015) A Non-Canonical Function of Gβ as a Subunit of E3 Ligase in Targeting GRK2 Ubiquitylation. Mol Cell 58:794–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zha Z, Han X-R, Smith MD, Lei Q-Y, Guan K-L, Xiong Y (2016) Hypertension-associated C825T polymorphism impairs the function of Gβ3 to target GRK2 ubiquitination. Cell Discov 2:16005. [DOI] [PMC free article] [PubMed] [Google Scholar]


