Abstract
Background:
Molecular studies have tried to address the unmet need for prognostic biomarkers in prostate cancer (PCa). Some gene expression tests improve upon clinical factors for prediction of outcomes, but additional tools for accurate prediction of tumor aggressiveness are needed.
Methods:
Based on a previously published panel of 23 gene transcripts that distinguished patients with metastatic progression, we constructed a prediction model using independent training and testing datasets. Using the validated mRNAs and Gleason score (GS), we performed model selection in the training set to define a final locked model to classify patients who developed metastatic-lethal events from those who remained recurrence-free. In an independent testing dataset, we compared our locked model to established clinical prognostic factors and utilized Kaplan-Meier curves and ROC analyses to evaluate the model’s performance.
Results:
Thirteen of 23 previously identified gene transcripts that stratified patients with aggressive PCa were validated in the training dataset. These biomarkers plus GS were used to develop a four-gene (CST2, FBLN1, TNFRSF19, ZNF704) transcript (4GT) score that was significantly higher in patients who progressed to metastatic-lethal events compared to those without recurrence in the testing dataset (p-value = 5.7×10−11). The 4GT score provided higher prediction accuracy (AUC = 0.76, 95% CI: 0.69–0.83; pAUC = 0.008) than GS alone (AUC = 0.63, 95% CI: 0.56–0.70; pAUC = 0.002), and it improved risk stratification in subgroups defined by a combination of clinicopathological features (i.e., CAPRA-S).
Conclusions:
Our validated 4GT score has prognostic value for metastatic-lethal progression in men treated for localized PCa and warrants further evaluation for its clinical utility.
Keywords: prostate cancer, prognosis, metastatic-lethal, biomarkers, validation
Introduction
Second to lung cancer, prostate cancer (PCa) is the most common cause of cancer-related mortality in American men. In 2019 there will be 174,650 new cases and 31,620 deaths from the disease.1 PCa is clinically and biologically heterogeneous. Among patients diagnosed with localized tumors who undergo radical prostatectomy (RP), about 30–40% will have adverse clinicopathologic features such as a high Gleason score (GS), positive surgical margins, seminal vesicle involvement, extra capsular extension or a high PSA level that increase risk of metastatic progression.2–4 While only a subset of these men will develop metastatic, life-threatening PCa,5,6 many of these patients will receive secondary therapy such as postoperative radiation and may suffer from treatment-related morbidity. On the other hand, some men without higher risk clinicopathologic features at diagnosis will develop biochemical recurrence (BCR), metastasis and die of PCa. Current clinicopathological factors have limitations in terms of predicting which men are at high risk for metastasis and cancer-related death. Prognostic biomarkers that can accurately predict risk of metastatic-lethal progression are needed to advance personalized medicine.
Over the last decade, molecular studies have tried to address the unmet clinical need for better prognostic biomarkers. Tumor-derived gene expression signatures may have added value to clinical factors for correctly stratifying patients with aggressive PCa,7,8 and three commercially available RNA-based tests have been externally validated.9–16 However, there is room to improve upon these molecular tests by identifying other biomarkers associated with tumor metastatic potential.
Recently, Rubicz et al. identified and validated a panel of 23 prognostic gene transcripts from transcriptome data measured in primary tumors.17 To further investigate the potential clinical utility of these 23 mRNA biomarkers, we evaluated their prognostic performance in a retrospective, nested case-control study dataset.18 We then developed a prognostic four-gene transcript (4GT) score to improve upon GS for predicting metastatic-lethal events. The 4GT score was then validated in a second independent testing dataset.
Materials and Methods
Training dataset
To further evaluate the panel of 23 prognostic mRNAs, we used data from PCa patients identified through the Mayo Clinic (MC) RP Tumor Registry using a nested case-control design as described previously.18 In brief, patients that received RP between 1987 and 2001 were retrospectively classified into outcome groups: 1) no evidence of disease (NED) at ≥7 years of follow-up (n = 213); 2) biochemical recurrence (BCR), defined as two successive PSAs >0.20 ng/ml and no detectable metastasis within 5 years of BCR (n = 213); and, 3) early metastasis defined by a positive bone or CT scan within 5 years after BCR (n = 213). Each metastatic case was matched by birth year, calendar year of PSA increase, and diagnostic Gleason score, to a patient from both the NED and the BCR groups.18 Tumor gene expression data were available from 169 NED patients, 184 BCR patients and 192 metastatic patients.
In comparison to the original study,18 we defined the cases differently and combined all patients who developed metastasis or died of PCa into a metastatic-lethal category, including 20 patients who were classified as BCR in the original study but who developed metastasis during further follow-up. This metastatic-lethal group (n = 212) was compared to NED patients (n = 157) in these analyses. The Affymetrix HuEx 1.0ST array and Single Channel Array Normalization (SCAN) method were used to profile and normalize gene expression data.
Testing dataset
We used an independent study population from the MC Tumor Registry as the testing dataset for validation. This case-cohort study initially sampled 1,010 RP patients treated between 2000 and 2006, and who were at high risk for recurrence based on a preoperative PSA >20 ng/ml, pathological GS ≥8, seminal vesicle involvement or a MC nomogram score of ≥10.10 A random sample of 202 men, together with all remaining men with metastatic progression were drawn from the cohort, resulting in a dataset of 256 men. We combined patients who developed metastasis or died of PCa into a metastatic-lethal category and compared them to patients with NED. The final MC testing dataset included 110 non-recurrent and 76 metastatic-lethal cases.
Statistical Analysis
To evaluate the panel of 23 gene transcripts in the MC training dataset, we calculated the difference between mean expression of each transcript in the non-recurrence and metastatic-lethal groups. For each transcript, we also examined the p-value for the area under the ROC curve (AUC) and the partial area under the ROC curve (pAUC) at 95% specificity using permutation tests, compared to a totally random prediction model. A transcript was considered validated if its expression level between patient groups in the MC training dataset was in the same direction (lower or higher) as that in the earlier study,17 and if the p-value for the AUC, pAUC or the two-sided t-test was significant (p-value <0.05).
We next evaluated the performance of each transcript that validated in the MC training data for its ability to classify men with metastatic-lethal outcomes in combination with GS, which was coded as continuous (i.e., 1: GS ≤6; 2: GS = 7; and, 3: GS ≥ 8). For each validated transcript, we compared the AUC and pAUC from a model of the mRNA combined with GS to a model with GS alone. Likelihood ratio tests were used to compare the goodness of fit for base models with GS alone and alternative models fit with GS and an additional transcript.
We next performed stepwise (forward) logistic regression starting with GS in the model using the MC training dataset. At each step, a mRNA was added to the model one at a time and a likelihood ratio test was performed to compare a model including a single additional transcript with the model from the previous step. The transcript with the lowest p-value was added at each step until all remaining transcripts had a p-value >0.05. The coefficients from the final locked model in the training dataset were then used to calculate a gene transcript score by multiplying with the corresponding mRNA levels and the GS for each patient in the MC testing dataset. Importantly, the testing dataset was not evaluated until after the model from the training set was locked.
A receiver operating characteristic (ROC) analysis and discrimination boxplots of non-recurrent vs. metastatic-lethal patients were performed to evaluate the ability of the gene transcript score to distinguish patient groups. The AUC and pAUC were computed for the final model, as well as for GS alone and for the Cancer of the Prostate Risk Assessment-Surgery (CAPRA-S) score.19 We then dichotomized patients in the testing dataset into low and high gene transcript score groups at zero, which represents the median score. Kaplan-Meier analysis was conducted to compare metastatic-free survival between the low and high gene transcript score groups. The p-value for the log-rank test was used to evaluate the significance of the group difference in the score.
We also examined the performance ability of the gene transcript score in patients stratified by CAPRA-S score. Patients with a CAPRA-S score ≤5 were categorized as low-medium risk and patients with a CAPRA-S score ≥6 were categorized as high risk. Boxplots and a t-test between low-medium risk and high-risk patient groups were performed. All analyses were conducted in R (http://cran.r-project.org).
Results
Selected characteristics of the MC training and testing datasets are listed in Table I. For both datasets, the mean age at diagnosis of patients with metastatic-lethal events was similar to that of patients with no recurrence. As expected, men with adverse outcomes were more likely to have a higher CAPRA-S score, a higher GS, regional stage disease, and higher PSA levels at diagnosis.
Table I.
Characteristics of the prostate cancer patients with gene expression data included in the Mayo Clinic training and testing datasets
| Training Dataset |
Testing Dataset |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No recurrence | Metastatic-lethal | No recurrence | Metastatic-lethal | |||||||||||
| n = 157 |
n = 212 |
n = 110 |
n = 76 |
|||||||||||
| No. | % | Mean (SD) | No. | % | Mean (SD) | P-valuea | No. | % | Mean (SD) | No. | % | Mean (SD) | P-valuea | |
| Age at diagnosis (years) | 64.9 (6.6) | 65.4 (6.4) | 0.5 | 64.0 (7.2) | 62.7 (7.8) | 0.3 | ||||||||
| CAPRA-S score | 4.3 (3.0) | 6.6 (3.1) | <0.01 | 5.1 (2.0) | 7.0 (2.3) | <0.01 | ||||||||
| Gleason score | ||||||||||||||
| ≤ 6 | 28 | 17.8 | 6 | 2.8 | <0.01 | 12 | 10.9 | 0 | 0 | <0.01 | ||||
| 7 | 95 | 60.5 | 76 | 35.8 | 58 | 52.7 | 21 | 42.1 | ||||||
| 8–10 | 34 | 21.7 | 130 | 61.3 | 40 | 36.4 | 44 | 57.9 | ||||||
| Pathologic stageb | ||||||||||||||
| Local | 83 | 52.9 | 58 | 27.4 | <0.01 | 61 | 55.5 | 16 | 21.1 | <0.01 | ||||
| Regional | 74 | 47.1 | 154 | 72.6 | 49 | 44.5 | 60 | 78.9 | ||||||
| PSA (ng/mL) at diagnosis | ||||||||||||||
| < 4.0 | 17 | 10.8 | 21 | 9.9 | 0.03 | 5 | 4.5 | 7 | 9.2 | 0.2 | ||||
| 4.0–9.9 | 79 | 50.3 | 80 | 37.8 | 62 | 56.4 | 31 | 40.8 | ||||||
| 10.0–19.9 | 31 | 19.7 | 44 | 20.8 | 25 | 22.7 | 21 | 27.6 | ||||||
| ≥ 20 | 26 | 16.6 | 60 | 28.3 | 18 | 16.4 | 17 | 22.4 | ||||||
Based on a t-test (age), chi-square or Fisher’s exact test (categorical variables).
Local stage = pT2, N0/NX, M0; regional stage = pT3/T4 and/or N1, M0.
We first examined the 23 biomarkers in the training dataset, where transcripts from 13 genes (ALDH1A2, CENPE, CST2, DPT, FBLN1, PI15, PLCL2, SELE, SPRY4, SRD5A2, TNFRSF19, TPX2, ZNF704) had the same direction of expression as in the original study 17 and had a significant p-value (<0.05) for the two-sided t-test, AUC test or the pAUC test (Table II). In this training dataset, we next evaluated the ability of the 13 validated mRNAs to improve upon GS for differentiating men with adverse events (Supplemental Table I). The AUC for GS alone in the MC training dataset was 0.72, and addition of each transcript to a model with GS improved all the AUCs (range: 0.73 to 0.76). Likelihood ratio tests for differential expression between the two patient groups were significant (p-value <0.05) for 11 mRNAs. The forward stepwise logistic regression model based on GS and the most robust of the 13 transcripts led to a final locked model that included four transcripts (CST2, FBLN1, TNFRSF19, ZNF704).
Table II.
Performance of twenty-three gene transcripts for classifying metastatic-lethal vs. non-recurrent prostate cancer in the Mayo Clinic training dataset
| Transcript ID | Gene | Mean expression no recurrence | Mean expression metastatic-lethal | Difference in mean expression | P-value t-test | AUC | P-value AUC | pAUC | P-value pAUC |
|---|---|---|---|---|---|---|---|---|---|
| ILMN_1748538 | ALDH1A2 | 0.23 | 0.20 | −0.03 | 3 × 10−4 | 0.61 | < 1.00 × 10−6 | 0.0023 | |
| ILMN_1786125 | CCNA2 | 0.05 | 0.06 | 0.01 | 0.464 | 0.49 | 0.950 | 0.0014 | 0.496 |
| ILMN_1716279 | CENPE | −0.10 | −0.08 | 0.02 | 0.014 | 0.56 | 0.036 | 0.0025 | 0.091 |
| ILMN_1694584 | CLTCL1 | −0.04 | −0.05 | −0.01 | 0.097 | 0.55 | 0.090 | 0.0042 | 0.005 |
| ILMN_1673843 | CST2 | 0.23 | 0.30 | 0.07 | 2 × 10−4 | 0.61 | 1 × 10−3 | 0.0031 | 0.041 |
| ILMN_1708107 | DPT | 0.26 | 0.19 | −0.07 | 1 × 10−4 | 0.62 | < 1.00 × 10−6 | 0.0041 | 4 × 10−3 |
| ILMN_1700541 | FBLN1 | 0.21 | 0.15 | −0.06 | < 1.00 × 10−6 | 0.63 | < 1.00 × 10−6 | 0.0039 | 4 × 10−3 |
| ILMN_1756358 | FBXO36 | 0.03 | 0.02 | −0.01 | 0.273 | 0.52 | 0.586 | 0.0014 | 0.535 |
| ILMN_2406084 | ITGA11 | 0.07 | 0.07 | 0.00 | 0.770 | 0.51 | 0.652 | 0.0017 | 0.339 |
| ILMN_1702738 | KLC3 | −0.02 | −0.04 | −0.02 | 0.341 | 0.52 | 0.485 | 0.0008 | 0.865 |
| ILMN_1661895 | PI15 | 0.51 | 0.49 | −0.03 | 0.491 | 0.54 | 0.141 | 0.0039 | 4 × 10−3 |
| ILMN_1734810 | PJA1 | 0.36 | 0.36 | 0.00 | 0.781 | 0.51 | 0.769 | 0.0008 | 0.846 |
| ILMN_1737025 | PLCL2 | 0.08 | 0.05 | −0.03 | 5.3 × 10−3 | 0.58 | 4 × 10−3 | 0.0026 | 0.076 |
| ILMN_1794490 | PNMAL1 | −0.03 | −0.04 | −0.01 | 0.301 | 0.53 | 0.368 | 0.0025 | 0.114 |
| ILMN_1739393 | SELE | 0.16 | 0.13 | −0.03 | 1.3 × 10−3 | 0.60 | 1 × 10−3 | 0.0033 | 0.032 |
| ILMN_1730295 | SIGLEC8 | −0.04 | −0.05 | −0.01 | 0.550 | 0.50 | 0.813 | 0.0002 | 0.988 |
| ILMN_2086105 | SPRY4 | 0.20 | 0.24 | 0.04 | 4.2 × 10−3 | 0.57 | 0.013 | 0.0038 | 0.012 |
| ILMN_1788895 | SRD5A2 | −0.12 | −0.17 | −0.05 | 1 × 10−4 | 0.62 | < 1.00 × 10−6 | 0.0032 | 0.029 |
| ILMN_1704154 | TNFRSF19 | 0.25 | 0.19 | −0.07 | < 1.00 × 10−6 | 0.64 | < 1.00 × 10−6 | 0.0026 | 0.103 |
| ILMN_2089875 | TNFSF4 | 0.04 | 0.05 | 0.01 | 0.487 | 0.52 | 0.411 | 0.0027 | 0.081 |
| ILMN_1796949 | TPX2 | 0.18 | 0.24 | 0.06 | < 1.00 × 10−6 | 0.64 | < 1.00 × 10−6 | 0.0057 | < 1.00 × 10−6 |
| ILMN_1748124 | TSC22D3 | 0.09 | 0.08 | 0.00 | 0.648 | 0.50 | 0.784 | 0.0018 | 0.285 |
| ILMN_1656192 | ZNF704 | 0.30 | 0.36 | 0.06 | < 1.00 × 10−6 | 0.61 | < 1.00 × 10−6 | 0.0053 | < 1.00 × 10−6 |
Validated transcripts (n = 13) with the same direction of expression difference as in the original datasets (Rubicz et al., 2017); gene transcripts with significant p-values are shown in boldface.
The coefficients from this final locked model (Table III) were then applied to the testing dataset to calculate a four-gene transcript (4GT) score, which ranged from −4.09 to 5.55 (recurrence-free: range −4.09 to 5.55; metastatic-lethal: range −0.82 to 4.45). The mean 4GT score differed significantly between patient groups (Figure 1A: mean difference = 1.44; p-value = 5.7×10−11). Based on the logistic regression analysis, each unit increase in the 4GT score was associated with a significantly elevated odds ratio of 2.05 (95% CI = 1.60 2.70; p-value = 6.4×10−8). Stratifying by CAPRA-S score, the 4GT score in the low-medium risk group had a mean of −0.37, and in the high-risk group the mean was 1.26 (Figure 1B: mean difference = 1.63; p-value = 2.3×10−13).
Table III.
Coefficients from the prediction model based on four gene transcripts and Gleason score
| Variable | Coefficient Estimate | P-value | 95% Confidence Interval |
|---|---|---|---|
| Intercept | −3.03 | <0.001 | −4.35, −1.78 |
| Gleason score | 1.36 | <0.001 | 0.95, 1.79 |
| TNFRSF19 | −3.17 | 0.001 | −5.16, −1.32 |
| FBLN1 | −3.26 | 0.004 | −5.54, −1.07 |
| CST2 | 2.05 | 0.006 | 0.63, 3.54 |
| ZNF704 | 2.80 | 0.007 | 0.84, 4.90 |
Figure 1.
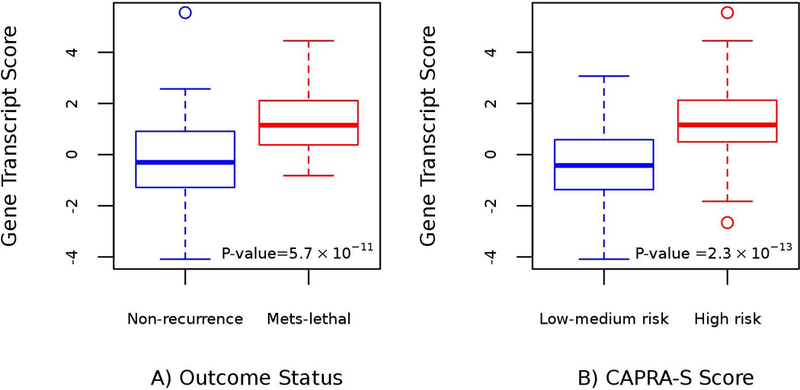
Box plots of the four-gene transcript (4GT) score in the MC testing dataset stratified by: A) prostate cancer outcome status (no recurrence vs. metastatic-lethal progression); and B) CAPRA-S score category (low-medium risk vs. high risk group)
We next evaluated time to metastasis in the low and high 4GT score groups (dichotomized at a score of 0) using Kaplan-Meier analysis (Figure 2). Median follow-up time of all patients was 5.1 years. Twelve of 72 (16.7%) patients in the low 4GT score group developed metastases whereas 64 of 114 (56.1%) patients in the high 4GT score group had metastatic progression. The overall metastasis-free survival was significantly different between the groups (p-value = 3.7×10−7).
Figure 2.
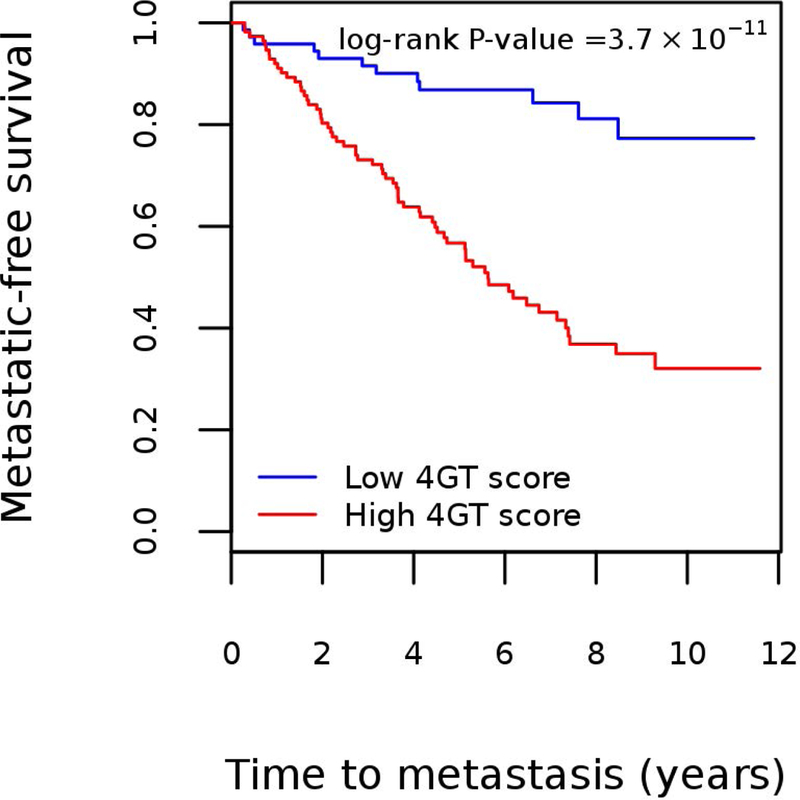
Kaplan-Meier estimates for metastatic-free survival in the MC testing dataset stratified by the four-gene transcript (4GT) score (low vs. high).
In the testing dataset, a ROC curve analysis (Figure 3A) confirmed that the 4GT score had better predictive performance (AUC = 0.76, 95% CI = 0.69 0.83; 95% pAUC = 0.008) than GS alone (AUC = 0.63, 95% CI = 0.56 0.70; pAUC = 0.002) or the CAPRA-S score (AUC = 0.75, 95% CI = 0.68 0.82; 95% pAUC = 0.005). The AUC for the 4GT score model was significantly higher than the AUC for the GS alone model (p-value <0.001), but not the CAPRA-S alone model (p-value = 0.9). Addition of the 4GT score to a model that included CAPRA-S score increased the AUC to 0.77 (95% CI = 0.70 0.83).
Figure 3.
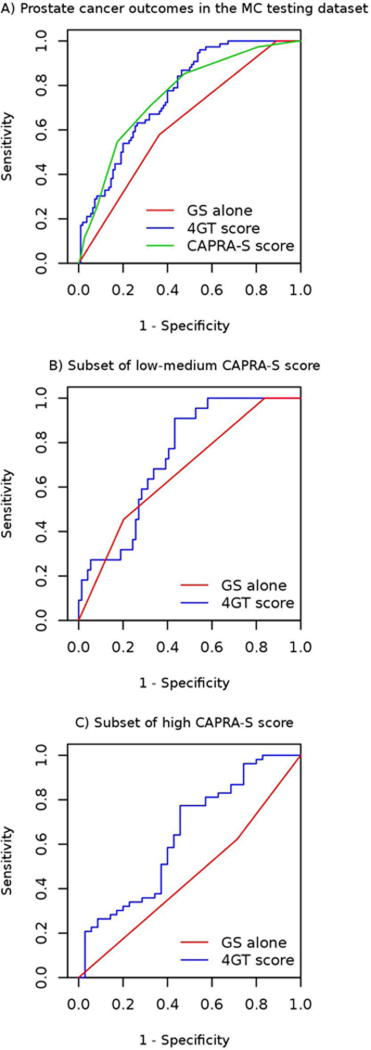
A) Receiver Operating Characteristic curves (ROCs) for predicting metastatic-lethal vs. non-recurrent prostate cancer outcomes in the MC testing dataset The red line shows the prediction performance of Gleason score alone (AUC= 0.63; pAUC= 0.002), the green line shows the prediction performance of CAPRA-S score alone (AUC= 0.75; pAUC= 0.005), and the blue line shows the prediction performance of the four gene transcript (4GT) score based on Gleason score plus four validated transcripts (AUC= 0.76; pAUC= 0.008). B) ROCs for predicting metastatic-lethal vs. non-recurrent prostate cancer outcomes in the subset of low-medium CAPRA-S scores (96 patients): The red line shows the prediction performance of Gleason score alone (AUC= 0.67; pAUC= 0.003), and the blue line shows the prediction performance of the four gene transcript (4GT) score based on Gleason score plus four validated transcripts (AUC= 0.74; pAUC= 0.008). C) ROCs for predicting metastatic-lethal vs. non-recurrent prostate cancer outcomes in the subset of high CAPRA-S scores (88 patients): The red line shows the prediction performance of Gleason score alone (AUC= 0.45; pAUC= 0.001), and the blue line shows the prediction performance of the four gene transcript (4GT) score based on Gleason score plus four validated transcripts (AUC= 0.64; pAUC= 0.004).
The pAUC (at 95% specificity) for our model (0.008) in the testing dataset was higher compared to Gleason alone (pAUC = 0.002) or CAPRA-S alone (pAUC = 0.005). This indicates that the 4GT score has a higher probability of correctly classifying men at high risk for metastatic progression, while maintaining a high probability of correctly classifying truly low-risk patients.
To further assess the performance and clinical utility of the 4GT we next stratified the testing dataset by CAPRA-S score, which uses available clinicopathological data from RP to define prognostic risk groups (e.g., low-medium vs. high). Results from these analyses indicate that the 4GT score can improve upon CAPRA-S for more accurate classification of patients within risk strata. For men with low-medium CAPRA-S scores (Figure 3B; 96 patients), the 4GT score (AUC = 0.74, 95% CI = 0.64 0.84; pAUC = 0.008) outperformed GS (AUC = 0.67, 95% CI = 0.56 0.78; pAUC = 0.003). Of the 96 patients in the low-medium risk group, CAPRA-S misclassified 22 (23%) who had metastatic progression, and the majority of these (64%) were correctly classified as high-risk by the 4GT score. In patients with high CAPRA-S scores (Figure 3C; 88 patients), the 4GT score performed much better (AUC = 0.64, 95% CI = 0.52 0.76; pAUC = 0.004) than GS alone (AUC = 0.45, 95% CI = 0.35 0.55; pAUC = 0.001). Of these 88 patients, 35 (40%) remained recurrence-free and may have been misclassified as high-risk by the CAPRA-S score; the 4GT score correctly classified (26%) of these patients as low-risk.
Discussion
In this study, we evaluated a panel of 23 gene transcripts previously validated to predict metastatic-lethal events in men treated for localized PCa,17 and constructed a prediction model in a training dataset based on GS and validated transcripts from four genes (CST2, FBLN1, TNFRSF19, ZNF704). We tested our prediction model on an independent validation dataset in which the 4GT score stratified patients by outcome (recurrence-free vs. metastatic-lethal, p-value = 5.7×10−11) and demonstrated an AUC = 0.76 (95% CI: 0.69 0.83) and a pAUC = 0.008 (at 95% specificity) for prediction of metastatic-lethal progression. The 4GT score provided additional prognostic information within CAPRA-S defined risk strata (i.e., low-medium or high), allowing for more accurate classification of patients.
Our work builds on the three gene expression tests currently available in clinical practice. The Prolaris test measures 46 transcripts, was found to predict BCR after RP,20 and can stratify patients at low risk (i.e., CAPRA-S score ≤2). The test improved prediction accuracy of the CAPRA-S score, which increased from a c-index of 0.73 to 0.77 with the addition of the Polaris panel. The 17 mRNAs in the Oncotype DX test were identified using tumor tissue from low-intermediate risk patients.14 The test was an independent predictor of BCR (HR = 2.7), metastases (HR = 3.8) and adverse pathology (OR = 3.3) in low-intermediate risk patients.16 The AUC for the Oncotype DX genomic score (AUC = 0.68) improved when it was combined with the CAPRA-S score (AUC = 0.73). The Decipher test 9 includes 22 gene transcripts, was developed using a case-control dataset, and was designed to predict metastasis after a rising PSA (AUC = 0.75). Cooperberg et al. 11 found that the Decipher test combined with the CAPRA-S score predicted PCa mortality in high-risk men (c-index = 0.78), all of whom had adverse pathology documented at radical prostatectomy. In the MC population that we used as a testing dataset, the Decipher test was previously applied to predict early metastasis (i.e., within 5 years after RP), and outperformed clinical models with an AUC of 0.79.10 Because these tests were developed using different populations and methodologies, with heterogeneity in the clinicopathologic characteristics of patients and outcomes evaluated, it is difficult to directly compare results across studies.
Compared to the tests described above, the 4GT score demonstrated similar or slightly better prognostic performance for distinguishing low-risk vs. high-risk patients for subsequent metastatic-lethal events. Furthermore, the 4GT score may offer some advantages over the other tests. Given that it is based on expression levels of only four mRNAs, the 4GT test is straightforward to calculate and should be less expensive to perform than current tests that cost $3,400 to $4,180. A low-cost prognostic biomarker test could become more widely used as a clinical tool for patient management, especially among patients with limited or no health insurance and in developing countries where PCa diagnoses are predicted to rise. In addition, future work based on prostate biopsy samples may provide further support for molecular tests such as the 4GT score that measure a limited number of biomarkers given the limited quantity of tumor tissue available from needle biopsy specimens. A biopsy-based prognostic test to risk stratify patients prior to treatment could potentially guide therapy and improve patient outcomes. Lastly, in terms of clinical utility, the 4GT score in the testing dataset was shown to have added value to the standard clinicopathologic factors available for RP patients used to calculate the CAPRA-S score, providing more accurate classification of patients in both the low-medium risk and the high-risk CAPRA-S subgroups.
The four genes represented in the 4GT score have previously been associated with biological processes involved in prostate or other cancers. Notably, the tumor necrosis factor receptor superfamily member 19 (TNFRSF19) transcript is also included in the Decipher test.9 In a study of gastric cancer TNFRSF19 was down-regulated in tumor compared to adjacent benign tissue and lower expression levels were associated with worse outcomes.21 This is consistent with our finding of lower gene expression levels in men with metastatic progression.
The basement membrane component fibulin 1 (FBLN1) is suggested to function both as a tumor suppressor and an oncogene. Its role as a tumor suppressor is supported by the fact that FBLN1 was shown to inhibit migration and invasion.22 Downregulation of FBLN1 in bladder cancer was associated with high-grade cancer and lower recurrence-free survival;23 FBLN1 has also been observed to be down-regulated in tumors compared to benign prostate tissue,24 particularly in tumors with overexpression of ERG.25 These observations are supported by our results where lower FBLN1 expression was associated with metastatic-lethal events.
Salivary cystatin family member 2 (CST2) was first suggested to be involved in cancer based its higher expression in breast cancer compared to normal tissues.26 More relevant to our findings are two recent studies that identified nine-gene prognostic signatures for gastric cancer, with CST2 present in both. 27,28 We found increased CST2 expression in patients at high risk of metastatic-lethal progression.
Little is known about zinc-finger protein 704 (ZNF704), although one study suggested that it plays a role in malignant transformation.29 Data from The Cancer Genome Atlas showed lower expression in tumors compared to benign prostate samples, but we found higher ZNF704 expression in patients with metastatic progression. Further work will be needed to understand the role this gene may play in aggressive PCa.
In summary, we have used independent training and testing datasets to develop and validate a 4GT score that is predictive of metastatic-lethal progression in men treated for localized PCa. The 4GT score demonstrated added value for improving prognostic classification of patients defined as low-medium risk or high-risk CAPRA-S scores. The four mRNAs in this score have biological plausibility as prognostic biomarkers and are complimentary to GS for predicting adverse events. Based on these results, the 4GT score warrants further evaluation as a promising clinical tool to improve patient outcomes.
Supplementary Material
Acknowledgements
This work was supported by grants from the National Cancer Institute (K05 CA175147 to JLS, and the Pacific Northwest Prostate Cancer SPORE P50 CA097186), with additional support provided by the Fred Hutchinson Cancer Research Center (P30 CA015704), and the Intramural Research Programs of the National Human Genome Research Institute and the National Institute of Environmental Health Sciences. Access to the Mayo Clinic gene expression datasets was made possible through a collaborative agreement with GenomeDx Biosciences. The authors thank all the men who participated in these studies.
Footnotes
Conflict of Interest
Dr. Elai Davicioni is employed by GenomeDx Biosciences.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2019. CA Cancer J Clin 2019;69:7–34. [DOI] [PubMed] [Google Scholar]
- 2.Hull GW, Rabbani F, Abbas F, Wheeler TM, Kattan MW, Scardino PT. Cancer control with radical prostatectomy alone in 1,000 consecutive patients. J Urol 2002;167(2 Pt 1):528–534. [DOI] [PubMed] [Google Scholar]
- 3.Patel AR, Stephenson AJ. Radiation therapy for prostate cancer after prostatectomy: adjuvant or salvage? Nat Rev Urol 2011;8(7):385–392. [DOI] [PubMed] [Google Scholar]
- 4.Mishra MV, Champ CE, Den RB, et al. Postprostatectomy radiation therapy: an evidence-based review. Future Oncol 2011;7(12):1429–1440. [DOI] [PubMed] [Google Scholar]
- 5.Eggener SE, Scardino PT, Walsh PC, et al. Predicting 15-year prostate cancer specific mortality after radical prostatectomy. J Urol 2011;185:869–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freedland SJ, Humphreys EB, Mangold LA, et al. Risk of prostate cancer-specific mortality following biochemical recurrence after radical prostatectomy. JAMA 2005;294(4):433–439. [DOI] [PubMed] [Google Scholar]
- 7.Bostrom PJ, Bjartell AS, Catto JWF, et al. Genomic predictors of outcome in prostate cancer. Eur Urol 2015;68:1033–1044. [DOI] [PubMed] [Google Scholar]
- 8.Ross AE, D’Amico AV, Freedland SJ. Which, when and why? Rational use of tissue-based molecular testing in localized prostate cancer. Prostate Cancer and Prostatic Diseases 2016;19:1–6. [DOI] [PubMed] [Google Scholar]
- 9.Erho N, Crisan A, Vergara IA, et al. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS One 2013;8(6):e66855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karnes RJ, Bergstralh EJ, Davicioni E, et al. Validation of a genomic classifier that predicts metastasis following radical prostatectomy in an at risk patient population. J Urol 2013;190(6):2047–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooperberg MR, Davicioni E, Crisan A, Jenkins RB, Ghadessi M, Karnes RJ. Combined value of validated clinical and genomic risk stratification tools for predicting prostate cancer mortality in a high-risk prostatectomy cohort. Eur Urol 2015;67(2):326–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cuzick J, Swanson GP, Fisher G, et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol 2011;12(3):245–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cuzick J, Berney DM, Fisher G, et al. Prognostic value of a cell cycle progression signature for prostate cancer death in a conservatively managed needle biopsy cohort. Br J Cancer 2012;106(6):1095–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klein EA, Cooperberg MR, Magi-Galluzzi C, et al. A 17-gene assay to predict prostate cancer aggressiveness in the context of Gleason grade heterogeneity, tumor multifocality, and biopsy undersampling. Eur Urol 2014;66(3):550–560. [DOI] [PubMed] [Google Scholar]
- 15.Cullen KJ, Yee D, Rosen N. Insulinlike growth factors in human malignancy. Cancer Invest 1991;9(4):443–454. [DOI] [PubMed] [Google Scholar]
- 16.Cullen J, Rosner IL, Brand TC, et al. A biopsy-based 17-gene genomic prostate score predicts recurrence after radical prostatectomy and adverse surgical pathology in a racially diverse population of men with clinically low- and intermediate-risk prostate cancer. Eur Urol 2015;68(1):123–131. [DOI] [PubMed] [Google Scholar]
- 17.Rubicz R, Zhao S, Wright JL, et al. Gene expression panel predicts metastatic‐lethal prostate cancer outcomes in men diagnosed with clinically localized prostate cancer. Mol Oncol 2017;11:140–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakagawa T, Kollmeyer TM, Morlan BW, et al. A tissue biomarker panel predicting systemic progression after PSA recurrence post-definitive prostate cancer therapy. PLoS One 2008;3(5):e2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooperberg MR, Hilton JF, Carroll PR. The CAPRA-S score. Cancer 2011;117:5039–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Punnen S, Freedland SJ, Presti JC, et al. Multi-institutional validation of the CAPRA-S score to predict disease recurrence and mortality after radical prostatectomy. Eur Urol 2014;65(6):1171–1177. [DOI] [PubMed] [Google Scholar]
- 21.Wilhelm F, Boger C, Kruger S, Behrens HM, Rocken C. Troy is expressed in human stomach mucosa and a novel putative prognostic marker of intestinal type gastric cancer. Oncotarget 2017;8(31):50557–50569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kotula E, Berthault N, Agrario C, et al. DNA-PKcs plays role in cancer metastasis through regulation of secreted proteins involved in migration and invasion. Cell Cycle 2015;14(12):1961–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao W, Wang J, Li H, et al. Fibulin-1 is epigenetically down-regulated and related with bladder cancer recurrence. BMC Cancer 2014;14:677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wlazlinski A, Engers R, Hoffmann MJ, et al. Downregulation of several fibulin genes in prostate cancer. Prostate 2007;67(16):1770–1780. [DOI] [PubMed] [Google Scholar]
- 25.Schulz WA, Ingenwerth M, Djuidje CE, Hader C, Rahnenfuhrer J, Engers R. Changes in cortical cytoskeletal and extracellular matrix gene expression in prostate cancer are related to oncogenic ERG deregulation. BMC Cancer 2010;10:505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Egland KA, Vincent JJ, Strausberg R, Lee B, Pastan I. Discovery of the breast cancer gene BASE using a molecular approach to enrich for genes encoding membrane and secreted proteins. Proc Natl Acad Sci USA 2003;100(3):1099–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu X, Wu J, Zhang D, et al. Identification of potential key genes associated with the pathogenesis and prognosis of gastric cancer based on integrated bioinformatics analysis. Frontiers in Genetics 2018;9:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z, Chen G, Wang Q, Lu W, Xu M. Identification and validation of a prognostic 9-genes expression signature for gastric cancer. Oncotarget 2017;8(43):73826–73836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen HQ, Zhao J, Li Y, et al. Gene expression network regulated by DNA methylation and microRNA during microcystin-leucine arginine induced malignant transformation in human hepatocyte L02 cells. Toxicology Letters 2018;289:42–53. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


