Abstract
Modulation of chromatin templates in response to cellular cues, including DNA damage, rely heavily on the post-translation modification of histones. Numerous types of histone modifications including phosphorylation, methylation, acetylation and ubiquitylation occur on specific histone residues in response to DNA damage. These histone marks regulate both the structure and function of chromatin, allowing for the transition between chromatin states that function in undamaged condition to those that occur in the presence of DNA damage. Histone modifications play well-recognized roles in sensing, processing and repairing damaged DNA to ensure the integrity of genetic information and cellular homeostasis. This review highlights our current understanding of histone modifications as they relate to DNA damage responses and their involvement in genome maintenance, including the potential targeting of histone modification regulators in cancer, a disease that exhibits both epigenetic dysregulation and intrinsic DNA damage.
Keywords: chromatin, DNA damage, DNA double-strand break repair, histones, modifications, genome integrity, transcription
Introduction
The 6 × 109 base pairs that make up the diploid human genome must be organized into the volume of a single nucleus in cells, while also being dynamic enough to provide access to the genetic information that ensures proper cell function. To achieve this monumental task, nuclear DNA of eukaryotes is bound, arranged and compacted by a nucleoprotein complex called chromatin. The basic unit of chromatin is the nucleosome (Kornberg, 1974), which consists of histone proteins and ~147 bp of DNA (Margueron and Reinberg, 2010). The nucleosome contains two copies each of four core histones, H2A, H2B, H3 and H4, with the DNA wrapped nearly two times around the histone octamer. Higher eukaryotes also contain linker histone H1 proteins, which bind to the entry/exit sites of DNA on the nucleosome to regulate higher-order chromatin structure. In addition to nucleosomes, non-histone proteins also bind to DNA and histone proteins to regulate DNA-templated processes including transcription, replication and DNA repair. To accommodate these various genome functions, chromatin exists in different states throughout the genome, in different cells, and across the cell cycle.
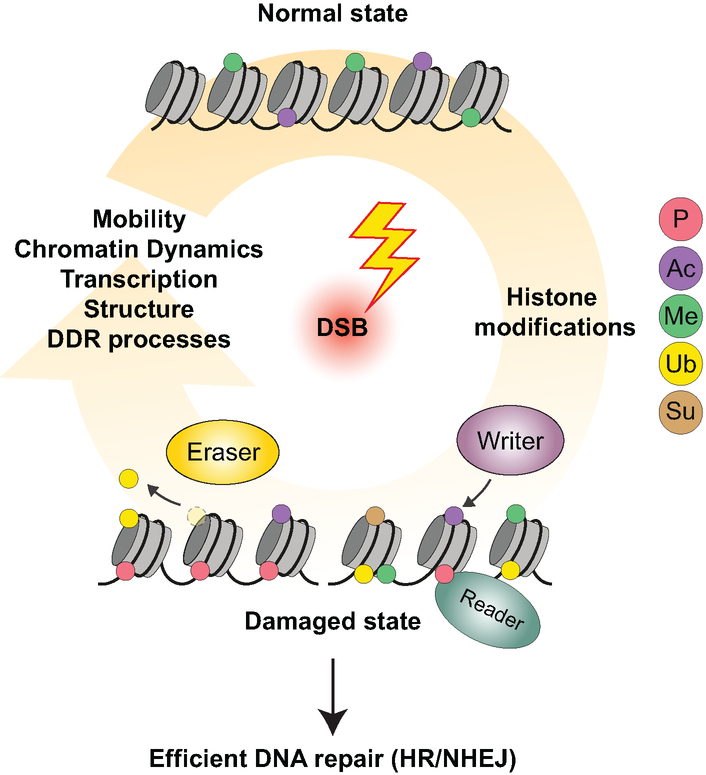
Chromatin states are regulated by several different mechanisms including chromatin remodeling activities that can reposition nucleosomes by sliding, evicting or depositing these proteins in the genome. In addition to the 4 core histones and linker H1, additional histone proteins exist called histone variants. These variants resemble canonical histones but play distinct, specialized roles in cells (Buschbeck and Hake, 2017). For example, histone H2A has several histone variants including H2AX, H2AZ and macroH2A. Finally, histones are highly modified by a host of post-translational modifications (PTMs) including phosphorylation, acetylation, methylation, ubiquitylation and sumoylation. (Kouzarides, 2007; Campos and Reinberg, 2009; Suganuma and Workman, 2011; Miller and Jackson, 2012). These modifications occur on distinct amino acid residues of histones and are dynamic, being catalyzed by “writer” enzymes and removed by “eraser” enzymes, which allows these marks to be reversible (Figure 1).
Figure 1.
Histone modifications pathways. A. Writer and eraser enzymes regulate the addition and removal of histone marks respectively. Reader proteins bind to specific histone PTMs. B. List of reader domains involved in DNA damage signaling and repair.
Histone PTMs can impact chromatin dynamics in several ways. PTMs can influence the non-covalent interactions between DNA and histones. The basic charge of histones is due to the high proportion of positively charged lysine and arginine residues that are found in all histone proteins. Negatively charged DNA interacts electrostatically to regions of the histone that contain these amino acids. Modifications of either lysines or arginines have the potential to disrupt DNA/protein and protein/protein interactions by changing the charge of the modified amino acid (Kouzarides, 2007; Shahbazian and Grunstein, 2007). A prime example of this concept is illustrated by acetylation of histone H4 on lysine 16, which disrupts H4 tail and nucleosome interactions to promote chromatin relaxation (Shogren-Knaak et al., 2006). Histone PTMs can also function as high affinity binding sites for proteins that contain “reader” domains that recognize specific histone marks (Figure 1) (Arrowsmith et al., 2012; Musselman et al., 2012). These histone PTM reader domains are found in many chromatin interacting proteins and exist for many PTMs including phosphorylation, methylation and acetylation. For example, twin-BRCT domains can bind phosphorylated histones, chromodomains recognize methylated lysines and bromodomains interact specifically with acetylated lysine residues (Kouzarides, 2007; Shahbazian and Grunstein, 2007; Ruthenburg et al., 2007). Collectively, these chromatin mechanisms operate to dynamically regulate chromatin compaction, DNA accessibility and protein interactions with chromatin to ensure proper chromatin states and therefore genome access and function.
An important cellular pathway that utilizes chromatin-based mechanisms is DNA damage signaling and repair. Cells exhibit high levels of DNA damage that are constantly threatening the integrity of the genome (Jackson and Bartek, 2009; Ciccia and Elledge, 2010). Sources of DNA damage can originate both endogenously and exogenously. For example, replication errors, reactive oxygen species damage, transcriptional processes, alterations in normal protein levels and mis-expression of enzymes that act on DNA, can all induce DNA damage intrinsically (Tubbs and Nussenzweig, 2017; Xia et al., 2019). Extrinsic factors that damage DNA include ultraviolet radiation from sunlight, carcinogens, and many cancer therapies such as radiation. Regardless of the source, damaged DNA must be detected and signalled to coordinate repair with the cell cycle and other DNA-templated processes that might be in conflict with repair, such as transcription and replication. If the DNA lesions are not repaired, genome instability can occur resulting in mutations that drive cell dysfunction and human diseases including cancer (Negrini, Gorgoulis, and Halazonetis, 2010).
Eukaryotic cells respond to DNA damage by activating a suite of pathways termed the DNA damage response (DDR) (Jackson and Bartek, 2009; Ciccia and Elledge, 2010). The DDR is represented by a large network of cellular pathways that detect lesions within DNA including single and double-strand breaks, DNA mismatches, damaged or inappropriate bases. After detection, the DDR signals the presence of the lesion by activating the DDR, for example through PTMs that activate the requisite cellular response to the lesion leading to the faithful repair and restoration of the genome at the lesion (Jackson and Bartek, 2009; Ciccia and Elledge, 2010). The importance of the DDR is highlighted by the fact that mutations in DDR pathways are commonly found in many different human diseases, such as cancer, neurodegenerative disorders and immune deficiencies (Jackson and Bartek, 2009; Negrini, Gorgoulis, and Halazonetis, 2010; Lord and Ashworth, 2012). Eukaryotic cells contain several different DNA repair pathways that engage and repair a wide variety of DNA lesions that can occur across the genome. For example, UV-induced DNA lesions or other bulky lesions are repaired by nucleotide excision repair (NER) (Marteijn et al., 2014), while DNA mismatch repair (MMR) is used to correct base-base mismatches or insertion/deletion loops generated during DNA replication (Jiricny, 2006). DNA double-strand breaks (DSB) represent a particularly dangerous form of DNA damage since this type of lesion affects both DNA strands. A DSB physically separates the two DNA molecules and exposes free ends, which can be degraded by nucleases or joined to inappropriate DNA molecules. For these reasons, DSBs can result in multiple types of mutations including deletions, insertions and chromosomal translocations if not repaired faithfully. DSBs can also result in large chromosomal losses and cell death. Due to these potentially catastrophic events, DSBs can be highly cytotoxic and dangerous to cells as they can induce genome instability. However, this characteristic of DSBs is unleashed to kill cancer cells through the induction of DSBs by cancer therapeutic drugs and treatments including topoisomerase and PARP inhibitors, as well as radiation. DSBs also represent an important DNA lesion that is programmed to occur in cells to promote genetic diversity (ex. meiotic recombination and class switch recombination) and extrinsically as a cancer therapy and more recently as a DNA substrate for genome editing including CRISPR/Cas9. These examples highlight the importance of DSB signaling and repair, events that are likely to be highly regulated by chromatin and its modifications given that they occur within the chromatin environment.
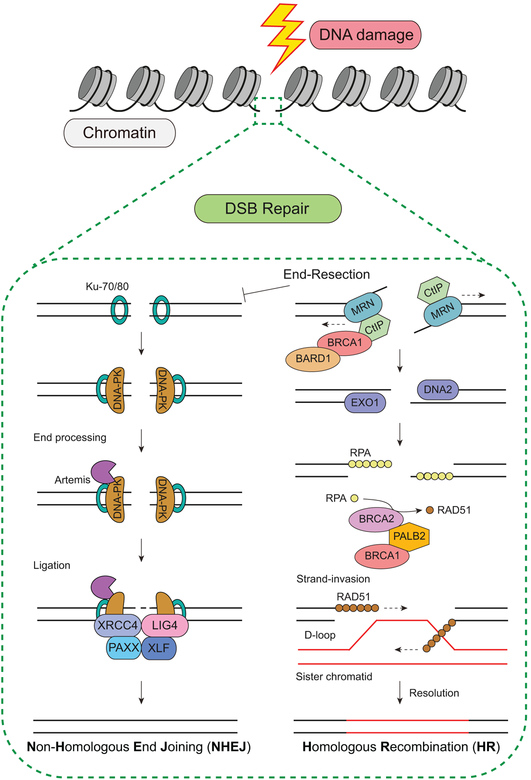
In mammalian cells, DSBs are repaired by two main pathways: homologous recombination (HR) and non-homologous end-joining (NHEJ) (Huertas, 2010). HR involves the use of a template, which allows for the accurate repair of a DSB. NHEJ, on the other hand, does not use a template and involves the ligation of the two free DNA ends to repair the break. While HR is highly accurate due to the use of a template, NHEJ can result in mutations due to processing of the breaks prior to ligation. Interestingly, NHEJ is the predominant repair pathway in mammalian cells and although it is active in other cell cycle phases, NHEJ is the dominant pathway in the G1 phase of the cell cycle due to the lack of a sister chromatid template (Beucher et al., 2009; Lieber, 2010). Unlike NHEJ, HR is restricted to S and G2 phases of the cell cycle when a template is available for repair. How repair pathway utilization is regulated in DSB repair is an important question since both HR and NHEJ can be active at the same time (Chapman, Taylor, and Boulton, 2012). DSB repair does not occur on naked DNA but rather takes place within chromatinized DNA substrates in cells. For these reasons, in addition to threatening the integrity of the genome, DSBs may also impact the epigenome and its function in cells. Thus, a deep mechanistic understanding of these pathways and how they impact genome and epigenome stability, both in normal cellular homeostasis and in disease, will require knowledge of how these repair and signaling pathways operate within chromatin. As is true with transcription for different genes, DSBs occurring in different genomic locations may require unique processes to signal and repair breaks. For example, repair in euchromatin versus heterochromatin or ribosomal DNA versus intron/exon DNA sequences may be unique. Topological states may also regulate DSB repair signalling and repair, as well as cell-type specific repair given that cells have unique functions and sets of proteins and RNA. As chromatin is highly dynamic and involved in cell identity and genome compartmentalizing and function, DSB repair and the DDR may similarly be regulated by these processes, including through chromatin mechanisms.
Histone PTMs, in addition to being involved in transcription, are important contributors to DDR functions, including DSB repair (Miller and Jackson, 2012; Lukas, Lukas, and Bartek, 2011; Gong and Miller, 2013; Jackson and Durocher, 2013; Schwertman, Bekker-Jensen, and Mailand, 2016; Gong, Chiu, and Miller, 2016). Key histone modifications that have been reported to participate in DSB repair are indicated in Figure 3. Histone PTMs can be dynamically regulated by DNA damage. For example, one of the best-characterized histone PTMs involved in the DDR is the phosphorylation of the histone variant H2AX. Upon DNA damage, this histone variant is phosphorylated on Ser139 (called γH2AX) (Rogakou et al., 1998) by the DDR related PIKK kinases (ATM, ATR and DNA-PK). γH2AX is bound by the BRCT phospho-reader domains of MDC1 to promote DDR signaling and recruitment of repair proteins (Stucki et al., 2005). 53BP1 represents another example of a histone modification reader protein that is involved in DSB repair and determine the choice between NHEJ and HR repair pathways (Panier and Boulton, 2014; Zimmermann and de Lange, 2014). 53BP1 binds to chromatin surrounding a DSB lesion through its bivalent interactions with two histone PTMs, H4K20me2 and H2AK15ub (Botuyan et al., 2006; Fradet-Turcotte et al., 2013; Huyen et al., 2004). These examples illustrate well-characterized scenarios where DNA damage-induced histone PTMs provide binding platforms for DDR factors to interact with chromatin at DNA lesions to promote the DDR. Histone PTMs can also modulate chromatin structure to facilitate repair (Polo and Jackson, 2011). While several chromatin-based DDR pathways involving histone PTMs have been described, a comprehensive understanding for how DNA damage impacts chromatin in cells and how chromatin is utilized to repair DSBs across the genome to promote repair and function of the damaged loci remains incomplete.
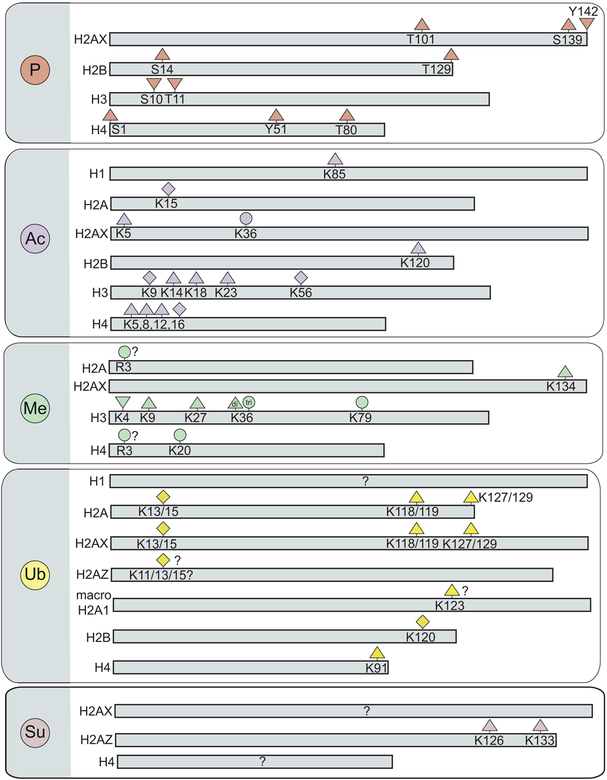
Figure 3.
Histone post-translational modifications involved in the DDR. Reported changes in histone modification levels after DNA damage are indicated with the following symbols; increased (△), decreased (▽), both increased and decreased (◇), and unchanged (○).
The 2018 Albert Laskar Award for outstanding discovery in basic biomedical research were given to two pioneers in chromatin biology, David Allis and Michael Grunstein (Allis, 2018; Williams, 2018). Their fundamental studies on chromatin and its regulation by histone post-translational modifications regulate gene expression paved the way for analyzing these pathways in other DNA-templated reactions, including the DDR. It seems timely then to review here our current understanding of histone PTMs and how they are involved in promoting the repair of DNA lesions in cells. Here, we focus on DNA double-strand breaks in mammalian cells, aiming to provide a framework for understanding the types of histone PTMs and their mechanisms that promote DNA damage signaling and repair of DSBs, pathways that are vital for maintaining the integrity of the genome. We will also highlight the importance of these pathways in understanding the interplay between the DDR and chromatin in human diseases and the potential for epigenetic and DNA damaging drugs to be used as therapeutic agents (Jackson and Bartek, 2009; Lord and Ashworth, 2012; Gong, Chiu, and Miller, 2016; Luijsterburg and van Attikum, 2011; Jeggo, Pearl, and Carr, 2016).
Histone modifications and DNA double-strand break repair: 53BP1 and BRCA1
In DSB repair, the DDR factors 53BP1 and BRCA1 antagonize each other to help regulate the pathway choice between NHEJ and HR respectively (Chapman, Taylor, and Boulton, 2012; Bunting et al., 2010). This is physiologically relevant given that BRCA1-deficient cells are defective in HR repair and mice lacking BRCA1 are embryonically lethal. Remarkably, 53BP1 loss rescues this lethality and HR defects, findings that have been attributed to 53BP1 blocking HR in the absence of BRCA1, including at the level of DNA end resection (Bunting et al., 2010; Cao et al., 2009). These genetic findings raise the question regarding the fine-tuned regulation of both NHEJ and HR pathway that coordinate the repair process to maximize genome maintenance, without interference from two active pathways that can act on the same DSB substrate (Figure 2). Several studies have established that histone modifications play essential roles in regulating DSB repair pathway choice regulation by 53BP1 and BRCA1. For these reasons, we begin with this pathway to provide a well-established example for how multiple histone modifications function to regulate DSB repair pathway.
Figure 2.
DNA double-strand break repair in mammalian cells. DNA is organized into chromatin and following DNA damage, DSBs are repaired by two main pathways, NHEJ and HR.
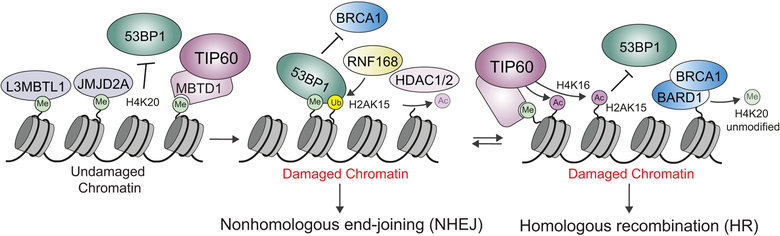
53BP1 is a large, 1972 amino acid mediator protein that interacts with DDR proteins and histone modifications to regulate several cellular pathways including V(D)J and class switch recombination, as well as DSB repair. 53BP1 acts as a bivalent histone PTM reader, binding H4K20me2 with its tudor reader domain and damage-induced RNF168-mediated H2AK15-Ub, with a ubiquitin binding region termed UDR, at DSBs (Botuyan et al., 2006; Doil et al., 2009; Fradet-Turcotte et al., 2013). In undamaged regions, two methylation binding proteins, L3MBTL1 and JMJD2A, bind H4K20me2 presumably to inhibit 53BP1 interactions with chromatin in the absence of DNA damage (Figure 4) (Panier and Boulton, 2014). Another histone methylation binding protein, MBTD1, which is a subunit of TIP60/NuA4 acetyltransferase complex, also competes with 53BP1 dynamics by competing for H4K20me2 binding (Jacquet et al., 2016). Upon DNA damage, H4K20me2 becomes available for binding by 53BP1. H2AK15 becomes ubiquitylated and is also bound by 53BP1. This dual engagement of two histone marks likely increases the specificity and binding of 53BP1 to chromatin at DNA damage sites. As mentioned previously, 53BP1 antagonizes BRCA1 to promote NHEJ. In the case where HR repair is favored, there are additional PTM-mediated pathways that similarly antagonize 53BP1 to promote BRCA1 binding and HR repair. In this pathway, the histone acetyltransferase (HAT) TIP60 acetylates two different histones at H4K16 and H2AK15, after DNA damage. The acetylation of H4K16 provides a steric obstruction to 53BP1 for its binding to the adjacent methylation of H4K20 (Tang et al., 2013). The acetylation of H2A on lysine 15 directly blocks ubiquitylation by modifying the same residue (Jacquet et al., 2016). There are additional eraser enzymes that are also involved in this pathway. For example, HDAC1 and HDAC2 are recruited to DNA damage sites where they deacetylate H4K16 and promote NHEJ (Miller et al., 2010). Similarly, other enzymes that remove these marks are likely to be able to impact the pathway choice for DSB repair between NHEJ and HR (Figure 4). Interestingly, a recent study identified a novel histone interaction domain within BARD1, the heterodimer partner for BRCA1 E3 ligase complex (Nakamura et al., 2019). Thus, H4K20me2 will itself block BRCA1-BARD1 interactions with chromatin. This suggests that unmodified H4K20 is required for BRCA1-BARD1 interactions with chromatin and HR repair. As H4K20 is unmodified in post-replicative cells, these data provide a compelling mechanism for how HR mediated by BRCA1 can be advantageously promoted once the sister-chromatid is available for templated-HR repair. Taken together, these histone PTM-based mechanisms of DSB repair pathway choice by 53BP1 and BRCA1 provide a prime example for the necessity and complexity of remodeling chromatin from an undamaged to a damaged state. Although there are numerous other examples, this system highlights many themes that are observed with other reader, writer and eraser histone PTM pathways involved in the DDR. Next we will systematically summarize the involvement of histone PTMs, including phosphorylation, acetylation, methylation, ubiquitylation and sumoylation that have been reported to be involved in the DDR and in particular DSB repair (Table 1).
Figure 4.
Summary of histone PTM-mediated 53BP1 and BRCA1 DSB repair pathway choice regulation.
Table 1.
Histone modifications involved in the DDR.
PHOSPHORYLATION
Phosphorylation is one of the most abundant PTMs and is reversibly regulated by kinases that add a phosphate group to mainly Serine, Threonine or Tyrosine amino acids, as well as phosphatases that remove these marks. Many different reader domains have been shown to bind to these phosphorylations on histones, including several that are found in DDR factors including BRCT, BIR and 14-3-3 (Figure 1). Here we report the involvement of histone phosphorylation in DNA damage signaling and repair.
Histone Variant H2AX
We begin our discussion of histone phosphorylation in the DDR with H2AX, as its phosphorylation on S139 to form γH2AX was the first phosphorylation to be described on a histone in response to DSBs. γH2AX is critical for many aspects of the DDR, including the maintenance of genome integrity. This histone modification marks DSBs and is used as a biomarker for DNA damage in cancer cells, making γH2AX the most well-studied histone modification involved in the DDR (Bonner et al., 2008). As mentioned above, H2AX is phosphorylated on S139 by several DDR kinases including ATM (ataxia telangiectasia mutated), ATR (ATM- and Rad3-related) and DNA-PK. The importance of H2AX phosphorylation in DNA repair is validated by the finding that mice deficient for H2AX, as well cells incapable of S139 phosphorylation, are sensitive to DNA damage and display genome instability (Bassing et al., 2002; Celeste et al., 2002). γH2AX also promotes sister chromatid recombination, with a more error-prone single-strand annealing repair pathway being engaged more frequently in its absence (Xie et al., 2004). Thus, H2AX phosphorylation is an important histone mark involved in DNA damage signaling and repair.
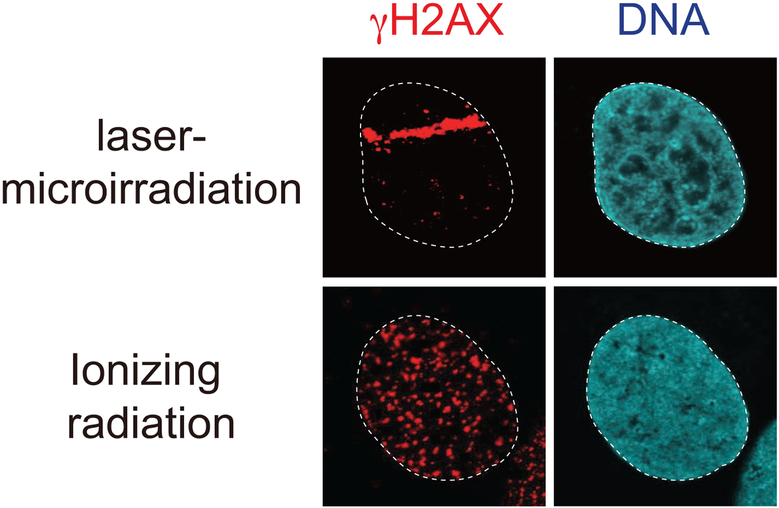
γH2AX marks large, sometimes > 1 Mb of chromatin surrounding a DSB (Rogakou et al., 1998; Rogakou et al., 1999). These large damage-induced γH2AX domains can be observed as foci in response to DNA damage, including IR and laser-microirradiation (Figure 5). γH2AX plays an important role in promoting maximal accumulation of several DDR factors to DSBs including MDC1, 53BP1, BRCA1 and the MRN complex (Miller and Jackson, 2012; Stucki et al., 2005; Celeste et al., 2003; Fernandez-Capetillo, Celeste, and Nussenzweig, 2003; Scully and Xie, 2013; Turinetto and Giachino, 2015{Miller, 2012 #43). MDC1 is a reader of γH2AX, binding selectively to this histone PTM through its twin-BRCT domains (Stucki et al., 2005). MDC1 also promotes the binding of several DDR factors to break sites, which demonstrates how γH2AX provides a recognition signal for a DDR factor that allows for the chromatin binding of this factor to DNA damage sites to trigger the DDR (Coster and Goldberg, 2010). The γH2AX-MDC1 axis also promotes the accumulation of E3 ubiquitin ligases to damage sites, including RNF8 and RNF168 (Jackson and Durocher, 2013; Doil et al., 2009; Scully and Xie, 2013; Huen et al., 2007; Kolas et al., 2007; Mailand et al., 2007; Stewart et al., 2009). Thus, γH2AX is also important for regulating ubiquitylation signaling at DSBs (see ubiquitylation section below).
Figure 5.
DNA damage-induced γH2AX by laser-microirradiation and IR. Human U2OS cancer cells were damaged and analyzed by immunofluorescence using a H2AXS139p antibody. DNA is detected by 4’,6-diamidino-2-phenylindole (DAPI) staining.
γH2AX and its associated readers are antagonized by several phosphatases that can erase S139 phosphorylation on H2AX (Chowdhury et al., 2005; Chowdhury et al., 2008; Douglas et al., 2010; Macurek et al., 2010; Moon et al., 2010; Nakada et al., 2008). As γH2AX is induced at DSBs, the assembled DDR complexes on chromatin must be dismantled following repair to ensure checkpoint inactivation and a restoration of the chromatin. The PP2A phosphatase dephosphorylates γH2AX at damage sites, which is dependent on H2AX. PP2A deficiency results in defective repair and a sustained increase in γH2AX levels, suggesting that dephosphorylation is an important step in post-repair processing of chromatin PTMs associated with break sites, including H2AX. In addition to PP2A, PP4, PP6 and WIP1 phosphatases also dephosphorylate γH2AX (Douglas et al., 2010; Macurek et al., 2010; Moon et al., 2010; Nakada et al., 2008). PP4 was shown to be important in DNA damage checkpoint recovery, pointing to an important role in dephosphorylation of H2AX in resuming the cell cycle following DNA damage (Nakada et al., 2008). The reason that several phosphatases act on γH2AX is unclear, although it could be attributed to the requirement for different phosphatases at unique chromatin environments at DSBs. For example, PP6 interacts with DNA-PK to dephosphorylate γH2AX. This may suggest that this phosphatase is important in erasing DNA-PK mediated γH2AX and not other signals that are independent of DNA-PK. To address this possibility, it would be interesting to determine the regulation of H2AX phosphatases and map them to individual and unique break sites across the genome, for example using the AsiSI-break system (Aymard et al., 2014; Clouaire et al., 2018; Iacovoni et al., 2010).
Although γH2AX is studied extensively in DNA damage response pathways including DNA double-strand break repair, this histone PTM is also involved in several other biological processes including cell cycle, transcription, stem cells and cancer (Bonner et al., 2008; Turinetto and Giachino, 2015). In addition, H2AX contains several other phosphorylation sites, apart from S139 that are regulated by DNA damage. For example, Y141 is phosphorylated by the kinase WSTF and is dephosphorylated by EYA1 (Cook et al., 2009; Xiao et al., 2009). This mechanism has been proposed to act as a repair-apoptosis switch to determine cell fate decisions in response to DNA damage that is mediated by the binding of Fe65-JNK1 complex to Y142p (Cook et al., 2009). H2AXT101 phosphorylation has also been observed to occur in response to DNA damage (Xie et al., 2010). Mutation of this site rendered reconstituted H2AX cells IR-sensitive suggesting that this residue does play a role in regulating H2AX-dependent DDR functions. Much less is known about these phosphorylations, including their potential involvement in H2AX-dependent processes outside of the DDR. While γH2AX is the most common marker used to identify DNA damage, there still remains many mysteries for how this histone variant is regulated to protect the integrity of the genome. It is likely that PTMs in addition to S139 phosphorylation are involved and will require additional studies to decipher their functions. These studies may utilize both mouse and human knockout cells, which are viable and allow for powerful complementation experiments to infer functionality. These capabilities are normally limited when studying histone PTMs due to the infeasibility of making knockout or sole non-modifiable core histones in mammalian cells (Bassing et al., 2002; Celeste et al., 2002; Xie et al., 2004; Xie et al., 2010; Chen et al., 2013; Leung, Emery, and Miller, 2018).
Histone H2B
Several DNA-damage associated phosphorylations have been identified on Histone H2B (Fernandez-Capetillo, Allis, and Nussenzweig, 2004; Lee et al., 2014). In mammalian cells, H2B is phosphorylated on Serine 14 at DSBs (Fernandez-Capetillo, Allis, and Nussenzweig, 2004). The PIKK kinase inhibitor Wortmannin abolished this mark and H2AX was shown to regulate the accumulation of this mark into ionizing radiation-induced foci, although it was not required for its phosphorylation. Little else is known about this DSB-induced histone phosphorylation on H2B including the putative enzymes or readers that act on this mark. H2BT129 is phosphorylated by Tel1 (ATM) and Mec1 (ATR) in yeast in response to DNA damage (Lee et al., 2014). H2BT129p is distributed similarly as γH2AX (H2AS129p in yeast) around large domains surrounding the break site. The involvement of this mark in DSB repair has not yet been deciphered but this mark is promoted by γH2AX, suggesting that these marks may collaborate in the DDR. While yeast H2BT129 is found in a TQ motif, which is the preferred sequence targeted by Tel1 and Mec1 kinases, mammalian H2B does not contain this motif. However, the C-terminus of mammalian H2B contains many phosphorylatable residues including Ser, Thr and Tyr, although there is no evidence that these are phosphorylated in response to DNA damage. Whether C-terminal phosphorylation of H2B occurs and is involved in the DDR in mammalian cells awaits investigation.
Histone H3
Histone H3 phosphorylations on serine 10, threonine 11 and serine 28 occur during mitosis and are involved in chromatin compaction. Several of these phosphorylations have also been reported to be involved in DNA damage pathways (Monaco et al., 2005; Ozawa, 2008; Sharma et al., 2015; Shimada et al., 2008), although these findings have not been observed in all studies (Tjeertes, Miller, and Jackson, 2009). H3S10p levels decrease upon DNA damage induction (Monaco et al., 2005; Ozawa, 2008; Sharma et al., 2015; Tjeertes, Miller, and Jackson, 2009). Aurora-B kinase mediates the phosphorylation of H3 at S10 and upon DNA damage, this kinase activity is inhibited by poly(ADP-ribosylation) by PARP1, which may account for the observed reduction in H3S10p levels (Monaco et al., 2005), perhaps even in G1 cells where H3S10p levels are reduced specifically following DNA damage (Sharma et al., 2015). Other active marks on genes, including H3K9, K14 and K56 acetylations, were also observed to be reduced with chromatin condensation observed, perhaps pointing to a role in transcriptional repression following DNA damage. H3T11 is phosphorylated by the DDR kinase CHK1 and upon DNA damage, the levels of this histone mark are reduced (Shimada et al., 2008). The reduction of this mark correlates with transcriptional repression of genes involved in the cell cycle including cyclin B1 and CDK1. Concomitant loss of H3K9 acetylation by GCN5 was also observed on these genes although a mechanistic explanation for these observations was not established. In another study, the reduction of H3 phosphorylations following DNA damage were proposed to occur through the indirect loss of mitotic cells that occurs due to checkpoint activation from DNA damage (Tjeertes, Miller, and Jackson, 2009). Regardless, the enzymes that regulate these marks, as well as the putative functions for these histone PTMs in the DDR, are currently poorly understood. It is also well documented that H3S10 phosphorylation can affect modifications on other histone residues, including H3K9, making it possible that these changes are acting to regulate proximal modifications to alter chromatin dynamics and reader protein associations in response to DNA damage (Sawicka and Seiser, 2012). Specific DSB repair pathways associated with these histone phosphorylations have also not been determined making it clear that additional studies are needed to clarify the function of H3 phosphorylations in the DDR.
Histone H4
The first H4 phosphorylation site identified to be involved in DSB repair was H4S1 in budding yeast (Cheung et al., 2005). After DSB-induction, H4S1 is phosphorylated by casein kinase II and this mark has been proposed to be involved in NHEJ. Interestingly, this mark was shown to inhibit the NuA4 complex, which is involved in acetylating histones (Utley et al., 2005). These data may also help explain the involvement of histone deacetylation on H4 in NHEJ in human cells (Miller et al., 2010). In support of H4S1 operating in DNA repair in human cells, this mark was found to accumulate at DSBs in human cells (Clouaire et al., 2018). Recently, H4Y51 was shown to be phosphorylated, providing only the second example of histone tyrosine phosphorylation, and the first on H4, that is involved in the DDR. This mark was shown to be catalyzed by the TIE2 receptor and to be involved in NHEJ (Hossain et al., 2016). Interestingly, the proto-oncogene ABL1 was shown to be a reader and transducer of this mark, as binding through its SH2 domain was observed and ABL1 inhibition also reduced NHEJ. Additional studies are required to understand how this pathway mechanistically facilitates NHEJ requires additional studies. Another H4 phosphorylation site in budding yeast was also recently identified as being involved in the DDR. H4T80 is phosphorylated by the kinase Cla4 and this mark was recognized by the histone bivalent histone binding scaffold protein, RTT107 (Millan-Zambrano et al., 2018). Rtt107, through its binding to H4T80p, promotes checkpoint recovery after DNA damage by antagonizing the binding of Rad9 to chromatin. Interestingly, H4T80p has been identified in mass spectrometry experiments in mammalian cells, raising the possibility that similar mechanisms may occur in higher eukaryotes.
It is worth noting that Serine ADP-ribosylation on histone has recently been observed and has been reported to be triggered by DNA damage (Bartlett et al., 2018; Palazzo et al., 2018). These studies propose that serines are the major amino acid acceptor of PAR following DNA damage, although Tyrosine was also observed to be ADP-ribosylated. Linker histone H1.2 is PARylated on Ser188, which is required for H1 removal and ATM activation following DNA damage (Li et al., 2018). As linker histone H1 compacts chromatin, this step may allow for the loosening up of chromatin allowing for activation of the DDR by ATM. Serine residues on core histones, including H3S10, H3S28 and H2BS6, have also been identified as ADP-ribosylation sites following DNA damage. Lysine and glutamate also act as ADP-ribosylation acceptor sites and several of these residues within histones have been shown to be PARylated although their role in the DDR has not yet been examined (Messner and Hottiger, 2011). Given the ability of PARP to antagonize phosphorylation by competing for the same amino acid residue, it will be interesting to investigate this possibility experimentally. In addition, PAR chains could also inhibit neighboring marks on nearby residues. This has been observed for H3K9 acetylation by H3S10-ADPr (Bartlett et al., 2018). These studies provide an interesting example for how histone modifications can be involved in crosstalk with other marks, both on the same or nearby residue, which makes deciphering the impact of these marks on biological outputs including DNA repair challenging.
ACETYLATION
HATs and HDACs
Histone acetylation occurs by the addition of an acetyl group (-COCH3) onto the ε-amino group of lysine residues. In addition to acetylation, lysines can also be modified by methylation, ubiquitylation and sumoylation, which sets up the potential for competition among different histone marks at specific lysines. The writer enzyme for the acetylation of histones are histone acetyltransferases (HATs) while these marks are removed by the eraser enzymes, histone deacetylases (HDACs). HATs and HDACs were first identified for their roles in gene expression regulation (Brownell et al., 1996; Taunton, Hassig, and Schreiber, 1996). Mammalian cells contain at least 17 HATs and 18 HDACs, which fall into distinct classes (Lee and Workman, 2007). HATs can be generally categorized by their HAT domains. Using this classification, there are three major HAT groups in mammals, which include GNAT (GCN5 N-acetyltransferases), MYST (Morf, Ybf2, Sas2, TIP60) and orphan class. HATs are often found in complex with diverse multi-protein complexes. For example, the HATs GCN5 and PCAF can be found in the SAGA complex while TIP60 makes up a large complex that modifies histone H2A and H4. GCN5 and p300 contain bromodomain reader domain and the TIP60 complex contains a bromodomain protein (Lee and Workman, 2007). This highlights the importance of reader domains that bind to specific modifications as part of their regulatory mechanism, including for histone acetylation. These domains play important roles in regulating the binding, activity and specificity of these enzymes.
Mammalian HDACs fall into 4 classes (Yang and Seto, 2008) including Class I HDACs (HDAC 1–3, 8), which are expressed in the nucleus and broadly across cell types. Class III HDACs are the NAD-dependent Sirtuins (SIRT1–7) where SIRT1, 6 and 7 are nuclear, SIRT2 is mainly cytoplasmic and SIRT 3–5 are mitochondrial (Feldman, Dittenhafer-Reed, and Denu, 2012). Class I and III HDACs have been implicated in the DDR and DSB repair so we will focus on these enzymes here (Table I). It is important to emphasize that both HAT and HDAC enzymes also target non-histone substrates, including in the DDR. Given the focus of this review on histone PTMs and DSB repair in mammalian cells, we will limit our discussion to these topics but refer readers to other recent papers and reviews for a more comprehensive overview of protein acetylation, including in the DDR (Choudhary et al., 2009; Choudhary et al., 2014; Elia et al., 2015). For example, proteomic profiling of acetylation after ionizing radiation and UV identified over 16,000 unique acetylation sites in the proteome of the human cancer cell line HeLa. In addition, N-terminal acetylation of proteins, including histones, also occurs but the role of this modification and its associated enzymes are unknown (Aksnes, Ree, and Arnesen, 2019). Thus, acetylation is a diverse modification that not only targets histone proteins but also decorates hundreds if not thousands of other proteins, playing diverse biological roles in addition to genome integrity pathways.
Histone acetylation and the DNA damage response
Smerdon and colleagues first made the link between histone acetylation and DNA damage when they observed that histones were rapidly acetylated after exposure to ultraviolet radiation (U.V.) in human cells (Ramanathan and Smerdon, 1986). Studies of histone acetylation following UV damage provided the data for the authors to propose a framework for chromatin and DNA damage (Smerdon, 1991), later developed into the “access-repair-restore” model (Green and Almouzni, 2002). This model posits that chromatin alterations allow access to DNA damage lesions including repair factors and that following repair, chromatin is reorganized and restored to its original state. This is a very important concept as it denotes why chromatin states are altered after DNA damage. In the case of acetylation, acetylated histones may alter chromatin structure to allow access for repair factors to the lesions. It may also change acetylation state from one DNA-templated process that is occurring, for example transcription, to another state that is more favorable for repair after DNA damage occurs. A large body of work after these initial findings has provided more insights into how these processes take place and the enzymes and histone modifications that are involved. The “access-repair-restore” model is not only applicable for UV damage and repair but also takes place for DSB repair. In mammalian cells, the orchestrated efforts of a host of chromatin modifying enzymes, including HATs and HDACs, as well as chromatin remodeling complexes, histone variants and histone chaperones collectively function to make the appropriate changes in chromatin structure and states to allow DSB repair by HR and NHEJ pathways to take place within the chromatin environment (Soria, Polo, and Almouzni, 2012).
Histone acetylation and DNA double-strand break repair
Acetylation of histone proteins has been shown to play important roles in DSB repair. The chromatin proteins that bind these marks, including bromodomain proteins, are also highly involved in the DDR in mammalian cells (Gong, Chiu, and Miller, 2016; Chiu, Gong, and Miller, 2017; Fujisawa and Filippakopoulos, 2017). For example, over one-third of bromodomain proteins relocalize following DNA damage, including accumulation at DNA breaks (Gong et al., 2015). The specific histone acetylation sites, as well as the writers, erasers and readers of these marks (if known) are summarized in Table 1. Here we discuss these studies in turn, providing the key observations and implications of these histone acetylations in the repair of DSBs.
Histone H2A and H2AX acetylation
Histone H2A acetylation on K15 plays important roles in modulating 53BP1 and BRCA1 interaction at breaks sites to promote DSB repair (see Figure 4 and associated text). To date, this is the sole acetylation site on H2A that has been shown to play a role in DNA repair in mammalian cells. For the histone variant H2AX, several acetylation sites have been identified and shown to be involved in the DDR (Table 1). While γH2AX plays well-established roles in the DDR, the acetylations on H2AX are less well understood (Miller and Jackson, 2012; Gong and Miller, 2013; Rogakou et al., 1998; Scully and Xie, 2013). The HAT TIP60, in addition to acetylating H2A on lysine 15, also acetylates lysine 5 on H2AX upon DNA damage (Ikura et al., 2007). The eraser of this mark has been shown to be SIRT1, whose depletion results in H2AX K5Ac hyperacetylation and defective DNA damage signaling in response to DSBs (Yamagata and Kitabayashi, 2009). The dynamics of H2AX and its ubiquitylation at break sites also relies on this histone mark. H2AX is also acetylated on lysine 36, which appears to be written by the HATs p300 and CBP as their overexpression results in hyperacetylation of this site (Xie et al., 2010; Jiang, Xu, and Price, 2010). Functionally, H2AXK36Ac, unlike K5Ac, appears to be involved in IR-resistance as H2AX−/− mouse ES cells reconstituted with non-acetylatable H2AXK36R results in sensitivity to IR, unlike H2AXK5R (Xie et al., 2010; Jiang, Xu, and Price, 2010). In the same study, these mutants were analyzed for roles in H2AX-mediated HR. H2AXK36R was found to restore HR defects that are observed in H2AX null ES cells while H2AXK5R stimulated HR. Although the mechanisms whereby these marks are regulated are not fully identified, these marks do play roles in DSB repair and the DDR. For example, no HDAC or reader protein has been reported for H2AX K36Ac. It is unclear if this mark regulates chromatin structure, function or both. Interestingly, H2A also has a lysine at its 36 amino acid. Whether or not this lysine is also acetylated in H2A or if it is uniquely catalyzed on H2AX is unknown. This raises an important point about histones and variants as these proteins contain high homology but do show differences (Buschbeck and Hake, 2017). A complete picture for how histone modifying machinery discriminates between specific histones and their related variants, for example H2AX and H2A, is not well understood. Other acetylation residues on H2A and H2AX have been identified but their putative roles in the DDR and in DSB repair have not yet been established (Xie et al., 2010; Wyrick and Parra, 2009).
Histone H2B
A recent comprehensive screen of histone PTMs at site-specific DSBs in human cancer cells identified H2B acetylation at lysine 120 among many other histone modifications, which changed levels at DNA breaks (Clouaire et al., 2018). Interestingly, H2B is also ubiquitylated on this lysine residue, which was shown to decrease at DSBs, suggesting the presence of a ubiquitin to acetylation switch on H2B following DNA damage. H2B ubiquitylation is known to block higher-order chromatin structure, which may be altered by its loss and subsequent acetylation (Fierz et al., 2011). H2BK120Ac may also provide a binding interface for a reader protein involved in repair. While this study suggested that the SAGA complex, which contains the HATs PCAF and GCN5, may regulate this mark, another study suggested that the HAT CBP can also regulate this histone mark during transcription (Clouaire et al., 2018; Chen et al., 2014). Interestingly, H2B acetylation was also shown to require the histone variant macroH2A and PARP-1, two factors involved in DSB repair (Khurana et al., 2014; Ray Chaudhuri and Nussenzweig, 2017). Other acetylations on H2B have been identified, including K5,12, 15 and 20 {Wyrick, 2009 #212}, although their involvement in the DDR has not been established. Taken together, these studies have identified regulators of H2B K120 acetylation and that this mark is increased at DSB sites. Additional studies are required to identify mechanistically how this histone mark promotes DNA repair and whether or not all these factors participate in this DDR pathway.
Histone H3
Histone H3 contains several acetylation sites that impact DSB repair. In response to DNA damage, acetylations on H3K9Ac, H3K14Ac and H3K56Ac are regulated (Miller et al., 2010; Tjeertes, Miller, and Jackson, 2009; Kim et al., 2009; Das et al., 2009; McCord et al., 2009). H3K14Ac is promoted by the nucleosomal binding protein HMGN1 following treatment with IR (Kim et al., 2009). HMGN1 deficiency led to aberrant ATM signaling, providing a potential link between histone acetylation of H3K14 and DNA damage signaling by the kinase ATM. Analyzing damaged chromatin by isolating γH2AX identified increased levels of H3K9, K14, K18 and K23 acetylations (Lee et al., 2010). This appeared to require γH2AX, as a S139A mutant of γH2AX abolished these DNA damage associated histone acetylations. The HAT GCN5 catalyzed these H3 acetylations, which were involved in the recruitment of the SWI/SNF chromatin remodeling complex to damaged chromatin. The bromodomain-containing protein, BRG1, a component of SWI/SNF complex, reads H3K14Ac through bromodomain-dependent interactions. In support of the importance of this interaction in the DDR, mutation of the bromodomain of BRG1 resulted in an inability to repair DSBs efficiently as well as IR sensitivity (Lee et al., 2010). This study provides an early example of how acetylated chromatin binds a reader protein to promote the recruitment of a complex to damaged chromatin to promote repair of DSBs. Other studies have suggested that p300/CBP-dependent H3K18Ac similarly promotes SWI/SNF chromatin remodeling activity at DSBs to promote DSB repair by NHEJ. Indeed, the activity of SWI/SNF is required for the efficient recruitment of the NHEJ factor KU, and inhibition of this pathway results in defective NHEJ (Ogiwara et al., 2011). The HDAC SIRT7 deacetylates H3K18 at DSBs to promote NHEJ (Vazquez et al., 2016). This function is believed to facilitate the binding of 53BP1 to damage sites, to channel these breaks into the NHEJ pathway while blocking HR repair. Thus, whether or not histone marks are written or erased at damage sites may be regulated by DSB repair pathway choice as well as the chromatin context in which the damage appears.
Finally, deacetylation of the histone mark H3K56Ac has been shown to be reduced at DSBs (Miller et al., 2010; Ogiwara et al., 2011). This observation was determined to require the Class I HDACs, HDAC1 and HDAC2. These deacetylases are themselves recruited to DNA damage sites where they deacetylate H4K16Ac and H3K56Ac to promote NHEJ (Miller et al., 2010). H4K16Ac and H3K56Ac are also deacetylated by the Sirtuins SIRT1 and SIRT6 respectively at DSBs (McCord et al., 2009; O’Hagan, Mohammad, and Baylin, 2008; Toiber et al., 2013). SIRT6 promotes the recruitment of the chromatin remodeler SNF2H and deacetylation of these histone marks may participate in chromatin changes that promote DSB repair. Indeed, cells lacking SIRT6 display defects in the recruitment of many DDR factors including BRCA1, 53BP1 and DNA-PK (McCord et al., 2009; Toiber et al., 2013), suggesting that these chromatin marks and their regulators act early on in the DDR. There is also evidence that Class I HDACs and Sirtuins cooperate to regulate histone acetylation and DSB repair. In neurons, HDAC1 is deacetylated by SIRT1 following DNA damage, which results in the activation of HDAC1, H4K16 deacetylation and repair of DSBs by NHEJ (Dobbin et al., 2013). H4K16 is also acetylated at DSBs and promotes HR repair (see below), highlighting the importance of this histone mark in DSB repair. HDAC1 also deacetylates linker histone H1 following DNA damage (Li et al., 2018). Lysine 85 of linker histone H1 is acetylated by PCAF and erased by HDAC1 after DNA damage. Unlike other histone acetylations, removal of H1K85Ac is thought to decompact chromatin due to a loss in binding of the heterochromatin proteins HP1. How HP1 is targeted to histone H1 acetylation and which aspect of DSB repair this pathway is involved in has yet to be elucidated.
Taken together, these studies highlight the numerous H3 acetylations that are involved in altering chromatin structure at DNA damage sites as well as providing recruitment platforms for DNA repair factors. Several of these studies have identified both local and global changes for these histone marks, suggesting potential additional roles in transcription or other processes that do not occur directly at the break site. Collectively, these activities appear to facilitate DDR activation and DSB repair. These reactions are likely to be regulated by many factors including cell cycle, genome location, cell-type and growth conditions. For example, a recent study using a site-specific DSB systems coupled with ChIP-Seq using histone modification antibodies did not identify changes in H3 acetylation following DNA damage induction (Clouaire et al., 2018). Although there are many potential explanations for the differences between these studies, this work highlights the differences that have been observed when studying histone modifications and DNA damage responses. It will be important to not only catalog the differences in histone PTMs at break sites across the genome, but also as vital is understanding the function of the enzymes that regulate these marks, including the writers, erasers and readers, in the DDR. By identifying the involvement of these proteins in the DDR, additional mechanistic insights can be obtained beyond charting changes in histone modifications, whose levels may not easily reveal their function in DNA damage signaling and repair.
Histone H4
Several acetylation sites on histone H4 are involved in the DDR. Human MOF acetylates H4K16Ac after IR exposure and also interacts with ATM. MOF deficiency results in aberrant ATM activation and DSB repair (Gupta et al., 2005). Employing a site-specific DSB system using I-SceI (site-specific homing endonuclease), analysis of a single DSB in mouse ES cells showed that H4 was hyperacetylated at K5, K8, K12, K16 (Murr et al., 2006). Similar results were also observed using another I-Sce1 system (Ogiwara et al., 2011). In one study, the Trrap-TIP60 complex mediated these acetylations on H4 while another study identified p300 and CBP as the HATs responsible in mediating acetylation. It is possible that both HATs function in a single pathway to promote H4 acetylations. In support of this notion, p300, CBP and TIP60 were all involved in promoting efficient NHEJ. The Trrap-TIP60 complex was shown to be required for efficient HR as well. Interestingly, the use of an HDAC inhibitor was able to restore HR, supporting the importance of H4 acetylation in HR repair. While these studies provide important insights into the role of histone acetylation in DSB repair, it is important to consider that these systems rely on persistent single-locus DSB systems that may exhibit differences in comparison to DSBs formed in other endogenous locations that have not been engineered to contain a restriction enzyme site and reporter construct, which may interfere with the steady-state chromatin environment around this break site. It is likely that generating DSBs through endogenous mechanisms or using exogenous sources such as IR, may trigger different chromatin-related responses that warrant additional studies to compare the results obtained from these different experimental sources of damaging DNA.
Acetylation of H4K16 increases after DNA damage and this site appears to play a critical role in DSB repair (Miller et al., 2010; Gupta et al., 2005; Li et al., 2010). Using laser-induced DNA damage, H4K16Ac levels were observed to have a bi-phasic response, decreasing rapidly after DNA damage induction followed by an acetylation phase resulting in the accumulation of this mark at DNA lesions (Miller et al., 2010). Interestingly, DSB repair in human cells displays a similar behaviour as NHEJ rapidly repairs breaks while a slower phase of repair occurs that is dependent on HR. The deacetylation of H4K16 was determined to be controlled by the histone deacetlyases, HDAC1 and HDAC2 (Miller et al., 2010). These enzymes are highly related and depletion of both HDAC1 and HDAC2 were required to alter histone acetylations regulated by these enzymes, including H4K16Ac (Miller et al., 2010). Loss of HDAC1 and HDAC2 rendered cells deficient for NHEJ, suggesting the importance of this deacetylation step in promoting this DSB repair pathway. In support of this notion, TIP60 was shown to promote the accumulation of the HR factor BRCA1 while inhibiting the NHEJ promoting 53BP1 factors recruitment to DSBs (Tang et al., 2013). Acetylation of H4K16 was found to be inhibit the binding of 53BP1 to H4K20me2, which provides a molecular explanation for why deacetylation of H4K16 would promote NHEJ while acetylation would promote HR (Tang et al., 2013). Although a reader protein may regulate this DSB repair pathway choice governed by this mark, other explanations are possible. For example, H4K16Ac regulates higher order chromatin structure and its presence results in unfolded chromatin (Shogren-Knaak et al., 2006). Thus, H4K16Ac may function to increase chromatin accessibility to repair factors to promote HR while its deacetylation may reduce chromatin flexibility and folding. These alterations may preferentially promote the joining of two adjacent DNA ends in NHEJ. Given that several HATs and HDACs have been shown to regulate H4Ac, it is also unclear how these activities are coordinated. It is possible that different enzymes are required at different locations in the genome to promote repair. For example, pre-existing histone marks or chromatin structures may channel specific proteins to act within these states while others may call on different sets of factors. It is also likely that the chromatin structure and function of the genomic location before DNA damage also regulates these processes. Additional studies are needed to test these hypotheses and provide a better understanding of how seemingly similar pathways that target histone acetylations are uniquely required for DSB repair in mammalian cells.
Given that over half of all HATs and HDACs are recruited to DNA damage sites and many acetylation sites are regulated by DNA damage, acetylation signaling plays an important role in promoting DNA repair and the DDR (Table 1) (Gong and Miller, 2013). It is clear that HATs and HDACs function to orchestrate the transition from undamaged to damaged chromatin that accompanies the presence of DNA damage. The regulation of these activities during both the initiation of these responses and following repair are still not completely understood. There are likely to be both local and global acetylation effects associated with DNA damage. There is also the potential issue of pre-existing chromatin states, which may influence these responses. As most of these responses have been studied in cancer cells, this also raises the question of whether or not these responses are universal or are selected for in cancer, given that these cells exhibit high levels of endogenous DNA damage and often mutations in both chromatin and DNA repair pathways. The development of genomic approaches to study these responses, including site-specific DSB systems, ChIP-Seq and other genome-wide techniques will also likely provide important insights in the future for how these pathways are regulated to promote repair. Although acetylations following DNA damage have been identified by antibody and mass spectrometry approaches, additional acetyl-lysine interactomes are needed to identify the reader proteins associated with these histone marks. These studies can provide important information for identifying the pathways and activities that interact with chromatin at break sites to facilitate repair.
METHYLATION
Histone methylation occurs by the addition of a methyl group (-CH3) onto a lysine or arginine amino acid residue (Murray, 1964; Greer and Shi, 2012). Methylation can be added as mono- (me), di- (me2) or tri- (me3) on the ε-amino group of lysine; while arginines can be mono-methylated (me) or di-methylated symmetrically (me2s) or asymmetrically (me2a). Histone methylations are written by histone methyltransferases (HMTs), which donated a methyl group from S-adenosylmethionine to their target residue (Greer and Shi, 2012). HMTs are stratified into three distinct families; SET domain, Dot1-like enzyme and Arginine N-methyltransferase (PRMTs) enzymes. SET-domain and Dot1-like enzymes methylate lysines (KMTs) (Black, Van Rechem, and Whetstine, 2012), while the third family methylates arginines (Yang and Bedford, 2013; Bedford and Clarke, 2009; Blanc and Richard, 2017). Histone demethylases (HDMs) erase methyl groups from lysines or arginines (Dimitrova, Turberfield, and Klose, 2015; Kooistra and Helin, 2012). Lysine demethylases (KDMs) include amine oxidases and jumonji C (JmjC)-domain containing iron-dependent dioxygenases (Shi et al., 2004; Tsukada et al., 2006; Whetstine et al., 2006). Methylations, in particular on lysine residues of histone, play important roles in transcription as well as in the DDR (Kouzarides, 2007; Musselman et al., 2012; Black, Van Rechem, and Whetstine, 2012; Blanc and Richard, 2017; Dimitrova, Turberfield, and Klose, 2015; Kooistra and Helin, 2012). Dysregulated histone methylation also participates in human diseases including cancer, where it is found to be altered (Greer and Shi, 2012; Yang and Bedford, 2013; Chi, Allis, and Wang, 2010; Albert and Helin, 2010). Interestingly, methylation not only occurs on proteins but also on DNA and RNA (Shelton, Reinsborough, and Xhemalce, 2016; Bird, 2002; Klose and Bird, 2006; Paik, Paik, and Kim, 2007; Biggar and Li, 2015). In addition to histone methylation in DSB repair, the modification of both DNA and RNA are also involved in the DDR (Cuozzo et al., 2007; Xiang et al., 2017). Although we focus here on histone methylation, lysine methylations on non-histone proteins also function in various cellular functions, including the DDR (Biggar and Li, 2015; Hamamoto, Saloura, and Nakamura, 2015). p53 is a critical DDR regulator that controls checkpoint activation, cell cycle arrest and apoptosis in response to DNA damage (Reinhardt and Schumacher, 2012; Lakin and Jackson, 1999). p53 methylation is well-studied and provides a primary example of functional lysine methylation on a non-histone protein that is involved in the DDR (West and Gozani, 2011). DNA damage-mediated methylation dynamics have been identified on several lysine residues (Table 1). Here we review how KMTs and KDMs regulate specific histone methylations to modulate DNA damage signaling and repair in response to DSB lesions.
Histone arginine methylation
Histone methylations involved in the DDR are mainly found to occur on lysines and these pathways are better characterized compared to arginine methylation. Nine arginine methyltransferases have been identified in mammalian cells (Bedford and Clarke, 2009) and no systematic study of these enzymes in DSB repair has been performed. H2AR3me2 and H4R3me2 have been identified and linked with the DDR. These methylations appear to be written by PRMT7 (Karkhanis et al., 2012). Working in conjunction with the SWI/SNF chromatin remodeling complex, PRMT7 regulates methylation on H2AR3 and H4R3 to repress the transcription of several DNA repair genes. In the absence of PRMT7, cells exhibit resistance to DNA damage, suggesting that this pathway may involve resistance mechanisms in cells treated with DNA damaging agents. The mechanistic details on how this pathway acts in normal cells to ensure repair and genome instability, and the exact repair pathways that it regulates, has not been studied. Although arginine methylation on non-histone proteins has been shown to be involved in cell cycle regulation and the DDR (Raposo and Piller, 2018), given the potential for arginine methylation to regulate chromatin, additional studies are warranted to examine the involvement of these pathways in the DDR, including DSB repair.
H2AXK134 methylation
Histone H2A and H2B have not yet been shown to have lysine methylations involved in the DDR. The histone H2AX however is dimethylated on lysine134 (H2AXK134me2), which has been implicated in the DDR (Sone et al., 2014). SUV39H2 (KMT1B) was demonstrated to be able to di-methylate H2AXK134 both in vitro and in vivo. SUV39H2 null mouse MEF cells display reduced γH2AX and 53BP1 foci formation after treatment with the topoisomerase inhibitor doxorubicin, which can generate DSBs. Consistent with a role in DSB repair, SUV39H2 deficiency resulted in sensitivity to IR. Mechanistically, H2AXK134me2 appears to play a critical role in the formation of γH2AX in response to DNA damage. This may be at the level of phosphorylation as H2AXK134me2 modified H2AX resulted in an increased phosphorylation of S139 on H2AX by ATM in in vitro kinase assays (Sone et al., 2014). Thus, H2AXK134me2 may act in cis to promote γH2AX signaling by ATM. It is unclear if SUV39H2 is involved directly in DSB repair and whether or not methylation of H2AX occurs at DNA damage sites. Other regulators, including erasers and readers, have not been identified for this histone mark. Future studies are needed to further define the regulation and function of H2AXK134me2 in DSB repair.
H3K4 methylation
Methylation of H3K4me3 is associated with active transcription and DNA DSBs can result in transcriptional repression. (Barski et al., 2007). These observations suggest that active transcription is not entirely compatible with DSB repair and may require tuning the chromatin environment from a state permissive for transcription to another that promotes DNA damage signaling and repair. Interestingly, demethylation of H3K4me3 has been shown to occur at DNA damage sites and is involved in transcriptional repression following DSB formation (Li et al., 2014; Mosammaparast et al., 2013; Gong et al., 2017; Hendriks et al., 2015). There are several KDMs that are able to demethylate H3K4 that have been found to accumulate at DSB sites (Li et al., 2014; Mosammaparast et al., 2013; Gong et al., 2017; Hendriks et al., 2015). For example, KDM5B (JARID1B) demethylates H3K4me2/3 and was found to accumulate at I-SceI-induced DSB sites in a PARP1 and macroH2A1.1 dependent manner (Li et al., 2014). KDM5B impairment results in both HR and NHEJ defects as the NHEJ factor KU and the HR factor BRCA1 do not associate efficiently with damaged DNA in these cells. KDM5A (JARID1A or RBP2), another H3K4me2/3-specific KDM, also associates with DNA damage where it demethylates H3K4me3 (Gong et al., 2017). This KDM5A-dependent demethylation on H3K4me3 was shown to be required for the recruitment of the ZMYND8-NuRD chromatin remodeling complex to DSB sites, where these proteins act to repress transcription (Gong et al., 2015; Gong et al., 2017). PARP also promotes the recruitment of the ZMYND8-NuRD chromatin remodeling complex to damaged chromatin where it participates in repressing transcription in the vicinity of DSBs (Gong et al., 2015; Chou et al., 2010; Polo et al., 2010; Spruijt et al., 2016). KDM5A deficient cells also display a defect in transcriptional DNA damage repression, which was suggested to be due to hypermethylation of H3K4me3, a mark that blocks NuRD binding to chromatin (Gong et al., 2017). Finally, KDM5A loss, as well as ZMYND8-NuRD, results in defective HR repair pointing to a potential link between transcriptional repression by these factors and HR (Gong, Chiu, and Miller, 2016; Gong et al., 2015; Gong et al., 2017; Spruijt et al., 2016; Gong and Miller, 2018; Savitsky et al., 2016). These results may be explained by the finding that breaks associated with active genes are preferentially repaired by HR, which may require tuning of their transcription to allow HR to occur (Aymard et al., 2014).
Acetylation also regulates KDM5A pathway as ZMYND8 is a bromodomain reader protein and the HAT TIP60 is required for the recruitment of these factors to DNA damage sites. This is consistent with histone modification binding studies with ZMYND8 BRD that identified interactions with H4 acetylation, a known substrate of TIP60. Thus, this pathway represents an acetylation-demethylation switch that acts to transform a transcription-associated chromatin state to a repair-associated state, which appears to be at the cost of transcription as this process is repressed under these conditions. There are likely many additional proteins and factors that regulate these processes and whether or not transcriptional repression is a universal response to DSBs is unclear. Additional studies are needed to further delineate how these pathways are regulated and to define the interplay between transcription and DSB repair.
In addition to H3K4me3 demethylases, LSD1 (KDM1A) demethylates H3K4me1/2 (Shi et al., 2004) and accumulates at DSBs (Mosammaparast et al., 2013). The DDR function of LSD1 appears to be different than other H3K4me2/3 specific KDMs (Li et al., 2014; Gong et al., 2017; Hendriks et al., 2015). Unlike KDM5A and KDM5B that facilitate HR, LSD1 loss increases HR repair (Mosammaparast et al., 2013). How demethylation of chromatin by LSD1 suppresses HR to allow proper DSB repair is unknown. Taken together, these studies highlight H3K4 methylation as a key residue involved in the DDR. Given the importance of H3K4 methylation in transcription, this may explain the importance of methylation on this histone H3 residue in coordinating DNA repair activities. In contrast to KDMs, very little is known about KMTs that methylate H3K4 and their potential role in the DDR. Methylation of H3K4 has been reported at I-SceI-induced DSBs, which required the E3 ligase RNF40 (Nakamura et al., 2011). Given that there are several KMTs that target H3K4 methylation and the examples of demethylase that regulate this mark, it is likely that these writer enzymes either act to promote repair or at the very least to restore chromatin and its modifications after repair so that they can resume functioning in transcription regulation. Additionally, the readers of H3K4 methylation are poorly characterized in the DDR, making it difficult to delineate a clear mechanism for how H3K4 methylation is involved in DSB repair.
H3K9 methylation
Trimethylated H3 lysine 9 is correlated with gene silencing and heterochromatin (Barski et al., 2007; Bannister et al., 2001). Several H3K9-specific KMTs, including SUV39H1 (KMT1A), SETDB1 (KMT1E) and PRDM2 (KMT8A or RIZ1) associate with DNA damage sites where they write H3K9me2/3 at DSB sites (Khurana et al., 2014; Sun et al., 2009; Ayrapetov et al., 2014; Alagoz et al., 2015). H3K9me3 associated with DNA damage may function in several ways. H3K9me3 may serve to stimulate histone acetylation as the HAT TIP60, binds H3K9me3 through its chromodomain, which stimulates its activity (Sun et al., 2009). Increased activity of TIP60 is thought to promote acetylation of chromatin as well as non-histone proteins like ATM that function to facilitate HR repair (Tang et al., 2013; Sun et al., 2009). H3K9me3 at DSBs also provides a binding site for HP1 and its associated proteins, the histone methyltransferases SUV39H1 and KAP1 (Ayrapetov et al., 2014). SUV39H1 writes H3K9me3 so this is a mechanism that can promote spreading and the amplification of this signal along chromatin. TIP60-mediated ATM signaling phosphorylates KAP1 to release this complex, providing a mechanism for fine tuning signaling in the presence of breaks (Ayrapetov et al., 2014).
Methylation on H3K9 and the histone H2A variant macroH2A1 act to form repressive chromatin around DSBs (Khurana et al., 2014). Identified by an RNAi screen to discover chromatin proteins involved in HR, macroH2A1 and KMT PRDM2 accumulate at DNA lesions along with H3K9me2 (Khurana et al., 2014). MacroH2A1 facilitates PRDM2 association with DNA damage, placing this histone variant upstream of this methyl writer. MacroH2A1 or PRDM2 loss led to a defect in HR, including DNA end-resection and damage loading of BRCA1 (Khurana et al., 2014). How this pathway promotes HR remains unclear. The macroH2A histone variant and H3K9 methylation could alter chromatin structure or recruit proteins that promote HR repair. The connection between BRCA1 and H3K9 methylation might be explained by the finding that BARD1 interacts with the H3K9me binding protein HP1 to recruit BRCA1 (Wu et al., 2015). Another H3K9me2/3, KMT SETDB1, is also recruited to DNA damage sites and the loss of SETDB1, similar to PRDM2, results in HR defects (Alagoz et al., 2015). The H3K9me1/2 KMTs, G9a (KMT1C) and GLP (KMT1D), are also linked to the DDR. Upon DSB formation, these enzymes are degraded by the proteosome, which may act to alter histone methylation levels resulting in transcriptional and/or DNA repair effects (Takahashi et al., 2012).
The H3K9 demethylases KDM4B (JMJD2B) and KDM4D (JMJD2D) also localize to DNA damage sites (Young, McDonald, and Hendzel, 2013; Khoury-Haddad et al., 2014). These KDMs also use PARP-dependent mechanisms to associate with damage sites (Young, McDonald, and Hendzel, 2013; Khoury-Haddad et al., 2014). KDM4D is PARylated by PARP1 in response to DNA damage (Khoury-Haddad et al., 2014). How PARylated KDM4D facilitates DSB repair is unclear as this modification may only be involved in damage recruitment, but may also regulate its enzymatic activity or interactions with other proteins. Loss of KDM4D in cells results in defective ATM signaling, including the phosphorylation of H2AX, KAP1 and CHK2. These cells also exhibit impaired RAD51 and 53BP1 damage accrual and diminished HR and NHEJ, which further supports the importance of KDM4D and H3K9 methylation in DSB repair (Khoury-Haddad et al., 2014). These studies have demonstrated that H3K9 methylation pathways are involved in DSB repair. Given the central importance of H3K9 methylation in transcription, the involvement of these pathways in the DDR should be analyzed for their putative involvement with transcriptional responses to DNA damage, including silencing. It is likely that restoration of this mark should also be part of the DDR, which could act to ensure that the correct chromatin state is formed following post-DNA damage repair.
H3K27 methylation
H3K27me3 is associated with repressive chromatin and transcriptional repression, although it has been observed to accumulate at DNA damage sites, including DSBs (O’Hagan, Mohammad, and Baylin, 2008; Chou et al., 2010; O’Hagan et al., 2011). The PRC2 complex associated methylatransferase EZH2 (KMT6), a H3K27me2/3 specific KMT, has been shown to accumulate at DNA damage sites where it methylates H3K27 (O’Hagan, Mohammad, and Baylin, 2008; Chou et al., 2010; O’Hagan et al., 2011; Campbell et al., 2013). Upon DNA damage, EZH2 accumulates at promoters of actively transcribed genes along with other factors associated with silencing of transcription, including SIRT1, DNMT1 and DNMT3B (O’Hagan, Mohammad, and Baylin, 2008; O’Hagan et al., 2011). Like other silencing factors, the DNA damage recruitment of EZH2 is PARP-dependent (Chou et al., 2010; Campbell et al., 2013). Although not tested, PARP-mediated EZH2 recruitment and formation of H3K27me3 at damage sites may function with other repressors to inhibit transcription and promote DNA repair. EZH2-mediated H3K27me3 may also regulate transcriptional responses of DDR genes following DNA damage induction. Whether or not an eraser or reader is also involved with H3K27me3 DDR functions is unknown. However, the expression of a H3K27M mutation, that blocks H3K27 methylation, was found to inhibit NHEJ and alter 53BP1 foci formation through the dysregulation of FANCD2 (Zhang et al., 2018). These findings hint that there are additional functions for H3K27 methylation in the DDR that are yet-to-be identified
H3K36 methylation
The writers, erasers and readers of methylated H3K36 have been demonstrated to be involved in DSB repair. SETD2 (KMT3A) writes H3K36me3 (Wagner and Carpenter, 2012) and is enriched on gene bodies due to its involvement in transcriptional elongation (Barski et al., 2007; Wagner and Carpenter, 2012; Bannister et al., 2005). Unlike most other methylations, this mark does not change levels upon DNA damage but its pre-existing levels have been linked to repairing DSBs associated with actively transcribing chromatin (Aymard et al., 2014; Pfister et al., 2014). SETD2 and H3K36me3 were shown to help repair DSBs proximal to active genes using HR (Aymard et al., 2014). For example, depletion of SETD2 led to defective DNA damage signaling and DNA end-resection, resulting in defective HR (Aymard et al., 2014; Pfister et al., 2014; Carvalho et al., 2014). H3K36me3 is recognized by the methyl reader protein, LEDGF (p75), which encodes a PWWP domain that binds to methylation marks. LEDGF has been shown to interact with the resection-promoting factor CtIP to promote HR at DSBs, functions reliant on SETD2 and H3K36me3 (Daugaard et al., 2012). The importance of H3K36 methylation regulation in DSB repair is further supported by the finding that overexpression of the H3K36me3 demethylase KDM4A/JMJD2A reduces HR (Pfister et al., 2014).
H3K36me2 is also associated with DSB repair. IR-treatment induces H3K36me2 at DSBs, which is required for NHEJ factors to localize to DNA lesions (Fnu et al., 2011). The SET domain containing methyltransferase Metnase (SETMAR) modulates H3K36me2 levels at DNA breaks (Fnu et al., 2011; Lee et al., 2005). Reduction of H3K36me2 can occur either through the loss of Metnase or overexpression of the H3K36me2-specific demethylase KDM2A (JHDM1A), hindering NHEJ-dependent repair of DSBs (Fnu et al., 2011). Degradation of the H3K36 methylation erases KDM2A and KDM4A arises after DNA damage, which may aid to maintain or increase H3K36 methylation at DNA breaks (Cao et al., 2016; Mallette et al., 2012). This increase in H3K36me2 levels around a DSB site is thought to foster the association of the MRN (MRE11-RAD50-NBS1) complex at damage sites to promote DNA repair (Cao et al., 2016). Thus, H3K36 methylation plays an important role in repair of DSBs by NHEJ. These data have revealed the importance of H3K36 methylation regulation by KMTs and KDMs that are involved in DSB repair.
H3K79 methylation
Histone H3K79 methylation is catalyzed by mammalian DOT1L (KMT4) (or yeast Dot1), which is involved in transcription, cell cycle regulation and DSB repair (Farooq et al., 2016; Nguyen and Zhang, 2011). H3K79 methylation by Dot1 promotes the damage recruitment of Rad9, the budding yeast ortholog of 53BP1 (Wysocki et al., 2005), and H3K79 methylation is involved in yeast DNA damage signaling (Huyen et al., 2004; Nguyen and Zhang, 2011). In mammalian cells, DOT1L-mediated H3K79me2 was reported to be bound by 53BP1 in response to DSBs (Huyen et al., 2004; Wakeman et al., 2012), but later studies have determined that the tandem tudor domain of 53BP1 preferentially binds H4K20me2 at DSBs (Panier and Boulton, 2014; Zimmermann and de Lange, 2014; Botuyan et al., 2006). Thus, it remains unclear how DOT1L and H3K79 methylation participates in DSB repair in mammalian cells. In addition, the demethylase and additional reader proteins of this mark are not well studied and may yet constitute additional factors involved in the DDR.
H4K20 methylation
Unlike H3, which contains several methylated lysine residues involved in the DDR, H4 methylation on K20 is the only reported residue to be methylated and involved DNA damage signaling and repair (Jorgensen, Schotta, and Sorensen, 2013). Mono- and di-methylation of H4K20 (H4K20me1/2) provide docking sites for the DDR factor 53BP1 (Botuyan et al., 2006) and as already discussed, the E3 Ub ligase RNF168 ubiquitylates H2A/H2AX on K15 to allow bivalent binding of 53BP1 to these histone marks (Fradet-Turcotte et al., 2013; Mattiroli et al., 2012). These results help to explain why RNF168 deficiency resulted in impaired 53BP1 binding to damage sites (Doil et al., 2009; Stewart et al., 2009). As for methylation of H4K20, several enzymes have been reported to methylate this residue on H4 including PR-SET7 (KMT5A or SETD8) and MMSET (KMT3G, WHSC1 or NSD2), which both localize at DSBs to regulate mono- and di- methylation on H4K20 respectively (Hajdu et al., 2011; Pei et al., 2011; Oda et al., 2010). However, MMSET has been reported to preferentially methylate H3K36, not H4K20 (Nimura et al., 2009) and MMSET KO mouse cells display normal 53BP1 accrual at damage sites (Hartlerode et al., 2012). Thus, while methylation on H4 regulates 53BP1 binding, the detailed mechanisms that regulate H4 methylation in this pathway are still being developed.
In addition to the methyltransferases that catalyze H4K20 methylation, readers and erasers of this mark also regulate 53BP1. The Polycomb protein L3MBTL1 and histone demethylase KDM4A compete with 53BP1 for H4K20me2 binding (Min et al., 2007; Lee et al., 2008). Upon DNA damage, these proteins are removed through distinct mechanisms. L3MBTL1 is evicted by the chaperone VCP (p97) (Acs et al., 2011) and KDM4A is degraded by the proteosome through a RNF8- and RNF168-dependent pathway (Mallette et al., 2012). The removal of these proteins allow for H4K20me2 to be free for binding by 53BP1 recruitment. As previously discussed, acetylation on H4K16 prevents the interaction between 53BP1 and H4K20me2 to promote HR (Tang et al., 2013; Hsiao and Mizzen, 2013). Other H4K20me2/3-specific KMTs (KMT5B/C or Suv4–20h1/2) are also involved in the DDR. Cells lacking these enzymes exhibit high levels of H4K20me1 and display reduced 53BP1 foci upon IR and inefficient DSB repair (Schotta et al., 2008). Thus, many proteins regulate the DDR through H4K20 methylation. It is unclear why so many pathways are required to regulate this specific histone mark and whether or not 53BP1 is the only factor that functions through H4K20 methylation in the DDR. It may be that different genomic locations and/or cell cycle stages regulate these pathways, requiring complex mechanisms to ensure that these repair pathways happen at the right location and in the correct cell cycle stage. This idea is supported by the finding that 53BP1 is inhibited in mitosis and if this inhibition is overridden, telomere fusions occur (Orthwein et al., 2014). This study suggests that telomeres are competent to be fused in mitosis, which necessitates the suppression of 53BP1-mediated NHEJ to maintain genome integrity. Thus, there are many consequences of DNA repair activities throughout the cell cycle and it appears that H4K40 methylation acts as a key histone PTM to regulate these pathways to ensure that repair takes place in a coordinated manner to maximize genome maintenance.
It is interesting to note that the same histone residue can regulate the choice of DSB pathways, as shown by the finding that H3K36me3 is important for HR repair, while H3K36me2 is involved in NHEJ (Aymard et al., 2014; Pfister et al., 2014; Fnu et al., 2011). One explanation that should be explored is that methylation reader proteins recognize these different marks to promote specific branches of DSB repair, i.e. HR and/or NHEJ. For several of the enzymes involved in these pathways, it is unclear how the recruitment of these enzymes to DNA damage sites are regulated. PARP signaling appears to play a prominent role in this aspect of the signaling cascade as the damage recruitment of many KMTs and KDMs, including SUV39H1, EZH2, KDM5A, KDM5B, KDM4B, KDM4D, which are reliant on PARP (Li et al., 2014; Gong et al., 2017; Chou et al., 2010; Ayrapetov et al., 2014; Young, McDonald, and Hendzel, 2013; Khoury-Haddad et al., 2014; Campbell et al., 2013). Whether these enzymes are PARylated or are themselves PAR binding proteins is unknown. These mechanistic details of these pathways should be addressed experimentally to advance our understanding of PARP signaling and histone methylation in the DDR.
UBIQUITYLATION
Ubiquitin is a small and highly conserved protein that exists in most eukaryotic cells (Goldstein et al., 1975). The ubiquitylation process occurs through the addition of ubiquitin onto a lysine amino acid residue in a substrate protein (Pickart, 2001). This process covalently modifies the substrate protein via a cascade of three-enzymatic steps that result in the attachment of ubiquitin to its substrate protein. The ubiquitin activating enzyme (E1) first activates the ubiquitin protein by thioester linkage between the C-terminus of ubiquitin and the E1 cysteine sulfhydryl group in an ATP-dependent manner (Schulman and Harper, 2009). Ubiquitin is then transferred from the E1 to the active site of an ubiquitin conjugating enzyme (E2). The ubiquitin ligase (E3) facilitates the transfer of ubiquitin from the E2 (RING type E3) or E3 (HECT type E3) to the substrate protein by generating an isopeptide bond between the C-terminal glycine of ubiquitin and a lysine residue of the substrate protein (Swatek and Komander, 2016). Ubiquitin can also be added to other ubiquitins to create chains of polyubiquitin (Akutsu, Dikic, and Bremm, 2016). Ubiquitin contains seven lysine residues, which are K6, K11, K27, K29, K33, K48, and K63. These lysine residues can be targeted for ubiquitin linkage, resulting in the generation of various types of ubiquitin chains. K48-linked polyubiquitin chains are associated with proteasomal degradation of the target protein while K63-linked polyubiquitin chains regulate cellular processes including the DNA damage response that are not reliant on degradation (Jackson and Durocher, 2013).
Histone ubiquitylation and the DDR
Histones are substrates for ubiquitylation processes. The ubiquitylation of histone H2A at lysine 119 was first discovered in 1977 and represents one of the most abundantly ubiquitylated proteins in mammalian cells (Hunt and Dayhoff, 1977; Goldknopf and Busch, 1977). RNF2 in the polycomb repressive complex 1 (PRC1) ubiquitylates histone H2A during global transcriptional repression as well as around DNA damage sites (Wang et al., 2004; Li et al., 2006; Gao et al., 2012; Buchwald et al., 2006). Histone H2A and H2B are the major histone targets for ubiquitylation in the DNA damage response. Here, we review our current understanding of histone ubiquitylations that are involved in the DDR.
H2A K13/15 ubiquitylation
H2A K13/15 ubiquitylation has been shown to be an important modification for regulating 53BP1 and BRCA1 in the DDR (Figure 4). 53BP1 directly binds to H2A K15 monoubiquitylation via its ubiquitin dependent recruitment motif (UDR) and H4K20 methylation via its tudor domain (Fradet-Turcotte et al., 2013). Both histone modifications are required for 53BP1 translocation to DNA damage sites. It was originally proposed that RNF8 was initiating H2A ubiquitylation (Huen et al., 2007; Kolas et al., 2007; Mailand et al., 2007). RNF8 is recruited to DNA damage sites via an ATM-mediated phosphorylation cascade. RNF8 contains a forkhead-associated (FHA) domain, which is important for binding to phosphorylated MDC1. RNF8 ubiquitylates histone H2A via K63 linked ubiquitin chains and recruits another E3 ubiquitin ligase RNF168 to DNA damage sites. Interestingly, RNF8 is inactive towards nucleosomal H2A whereas RNF168 monoubiquitylates H2A on K13/15 (Mattiroli et al., 2012). Interestingly, this pathway is also regulated by histone ubiquitylation, which was determined by the finding that RNF8 ubiquitylates linker histone H1 via K63-linked ubiquitin chain (Thorslund et al., 2015). RNF168 then binds these K63-linked chains, allowing its recruitment to damage sites and subsequent mono-ubiquitylation of H2A K13/15. RNF168 regulates DNA repair pathway choice by controlling the recruitment of 53BP1 to DNA damage sites, which inhibits DNA end-resection and promotes NHEJ repair. H2A K13/15 ubiquitylation by RNF168 is also required to protect reversed forks from MRE11-dependent degradation following replication stress (Schmid et al., 2018). This replication stress pathway requires similar factors as DSB repair, suggesting that the DNA ends at reversed forks engage the same pathway for fork protection, including the chromatin marks H2A K13/15ub and presumably histone methylation to target 53BP1 to these structures. 53BP1 selectively binds to mono-H2AK15ub, suggesting that K15 is a key residue for ubiquitylation in this chromatin-recognition cascade by RNF168 and 53BP1 on histone H2A (Fradet-Turcotte et al., 2013).
H2A-K15Ub is not only bound by 53BP1, but also helps to facilitate the recruitment of BRCA1 to damage sites. RNF8 adds ubiquitin chains onto K13/15 site to generate K63 chains, which are important for BRCA1-A complex recruitment (Sobhian et al., 2007). This complex consists of RAP80 (UIMC1), Abraxas (FAM175A), MERIT40 (BABAM1), BRCC36 (BRCC3), BRCC45 (BRE) and BRCA1/BARD1. RAP80 recognizes K63 ubiquitin chains via its ubiquitin-interacting motif (UIM) and recruits this BRCA1-A complex to DNA damage sites. This complex acts to limit DNA end-resection and hyper-active HR (Hendriks et al.; Hu et al., 2011; Coleman and Greenberg, 2011). ZMYM3 is another factor that regulates this pathway and has recently been investigated (Leung et al., 2017). ZMYM3 is a zinc finger protein that binds to chromatin directly, as well as several members of the BRCA1-A, to fine-tune the interactions of this complex with damage sites to promote efficient HR repair (Vilas et al., 2018).
Many deubiquitylase eraser enzymes (DUBs) regulate H2A ubiquitylation in the DDR. siRNA screening for DNA damage-dependent ubiquitylation revealed that OTUB1 regulates K63 ubiquitin levels in the DNA damage site (Nakada et al., 2010). Depletion of OTUB1 leads to persistent 53BP1 after DNA damage. OTUB1 directly inhibits the E2 enzyme UBC13, independently of its catalytic activity. This E2 suppression by OTUB1 inhibits RNF168 mediated K63 polyubiquitylation in the DNA damage site. USP3 deubiquitylates H2A K13/15 in response to UV induced DNA damage (Sharma et al., 2014). Ectopic expression of USP3 inhibits K13/K15 monoubiquitylation and 53BP1, BRCA1 foci formation. One of the components of BRCA1-A complex, BRCC36, specifically cleaves K63 polyubiquitin chains on H2A (Feng, Wang, and Chen, 2010). Individual DUB overexpression screens have implicated that USP44 counteracts RNF8/RNF168-mediated 53BP1 recruitment (Mosbech et al., 2013). USP44 recruitment to DNA damage sites is dependent on RNF168. Once localized, this DUB inhibits RNF8/RNF168 ubiquitylation. Depletion of USP51 also leads to an increase in 53BP1 foci formation, suggesting a role in deubiquitylating H2AK13/15 (Wang et al., 2016). Genetic screens have also revealed that USP26 and USP37 are involved in RNF8/RNF168 mediated HR pathways (Typas et al., 2015). A20 is a non-canonical DUB that inhibits H2A ubiquitylation by directly binding to RNF168 and further inhibiting the interaction between RNF168 and H2A, independent of its catalytic activity (Yang et al., 2018). USP11 and DUB3 also deubiquitylate RNF8/RNF168 mediated H2AX mono-ubiquitylation (Delgado-Diaz et al., 2014; Yu et al., 2016). Thus, there are numerous enzymes that act on H2A ubiquitylation in the DDR. Why there are so many enzymes and how these enzymes themselves are regulated by DNA damage remains an open question.
H2A K127/129 ubiquitylation
H2A K127/129 ubiquitylation has been shown to be an important modification to promote DNA end resection. BRCA1 forms a heterodimer with BARD1 via its N-terminal RING domain, which functions as an E3 ligase for ubiquitin (Lorick et al., 1999; Wu et al., 1996). This complex ubiquitylates K127 on H2A (Kalb et al., 2014). This mark is proposed to regulate repositioning of 53BP1 to overcome the chromatin bound barrier created by this factor binding to chromatin to promote DNA end-resection through the actions of the chromatin remodeler SMARCAD1 (Densham et al., 2016). SMARCAD1 binds BRCA1/BARD1-mediated H2AK127/129Ub to allow its association with damaged chromatin. Once localized to damage sites, SMARCAD1 can act to remodel chromatin to promote HR. This pathway is antagonized by USP48, which removes H2AK127/129Ub to limit DNA end-resection (Uckelmann et al., 2018). Interestingly, in the absence of USP48, single-stranded annealing is promoted, which is likely due to the increased resection that occurs if BRCA1 is not properly constrained by USP48.
H2A K119 ubiquitylation
H2A K119 ubiquitylation is vital for regulating transcription following DNA damage (Shanbhag et al., 2010). H2AK119ub is regulated by the polycomb repressive complexes 1 (PRC1) (Wang et al., 2004). The polycomb repressive complex is multi-protein complex that contains several subunits including the E3 ligases RING1B and BMI1, which have distinct functions (Gao et al., 2012). The PRC1 ubiqutylates H2AK119 while another polycomb complex, PRC2, methylates H3K27me3. Both complexes function to regulate transcription and chromatin (Schwartz and Pirrotta, 2013). Members of both the PRC1 and PRC2 complexes, including RING1B, BMI1 and EZH2, have been shown to localize to DNA damage sites (Campbell et al., 2013; Ginjala et al., 2011; Ismail et al., 2010; Sanchez et al., 2016; Ui, Nagaura, and Yasui, 2015). Along with the PRC1 and PRC2 complexes, the PBAF chromatin remodeling complex promotes transcriptional repression after DNA damage through the regulation of H2AK119ub (Sanchez et al., 2016; Ui, Nagaura, and Yasui, 2015; Kakarougkas et al., 2014). PBAF is also shown to be required for NHEJ, suggesting the importance of this PBAF-dependent pathway for DNA repair of DSBs. The H2AK119ub complex FBXL10-RNF68-RNF2 is also involved in transcriptional repression and is recruited to DNA damage sites as well as promoting H2AK119ub (Rona et al., 2018). Of note, deficiencies in this complex result in defects in H2AZ loading and HR repair, suggesting additional pathways that rely on H2AK119ub in the DDR. Thus, several chromatin changes including H2A ubiquitylation act to change local chromatin structure and transcriptional processes in response to DNA damage. Interestingly, these changes are reversible as H2AK119ub mediated transcriptional silencing is regulated by USP16, which erases H2AK119ub (Shanbhag et al., 2010). Depletion of USP16 increases H2A ubiquitylation and restores the otherwise repressed transcription at DNA damage sites. The DUB BAP1 is also recruited to DNA damage sites and is phosphorylated by ATM (Ismail et al., 2014; Yu et al., 2014). BAP1 is activated by ASXL1 and deubiquitylates mono-ubiquitylation of H2AK119 (Daou et al., 2015; Sahtoe et al., 2016). BAP1 deficient cells exhibit defects in HR, suggesting that dysregulated removal of H2AK119Ub impacts DSB repair. H2AX is also ubiquitylated on K119 and has been suggested to be involved in the DDR (Pan et al., 2011; Wu et al., 2011). However, several studies have observed that H2AX, which cannot be ubiquitylated on this residue, rescues several H2AX-dependent DDR functions (Xie et al., 2010; Leung et al., 2014). How these pathways are coordinated to regulate H2AK119 ubiquitylation, as well as other pathways that impinge upon transcription, following DNA damage remains an active question.
PARP is required for transcriptional repression (Chou et al., 2010) as well as regulating several complexes that promote transcriptional repression following DNA damage. These complexes include ZMYND8 and FBXL10-RNF68-RNF2, which require PARP activity to localize to damage sites (Gong et al., 2015; Rona et al., 2018). Transcriptional repression involves several kinases including ATM and DNA-PK (Pankotai et al., 2012), as well PARP, and these pathways have been linked to both NHEJ and HR. Given the involvement of so many pathways and factors, additional studies are needed to organize known observations into a model. This can provide the framework for understanding local transcriptional responses to DNA damage and how the DDR functions within this context to regulate DSB repair while maintaining other DNA-templated processes including transcription.
H2B K120 ubiquitylation
The function of H2BK120 ubiquitylation in the DNA damage response remains to be mechanistically deciphered. This histone mark has been implicated in DNA damage checkpoint activation. RNF20/RNF40 are recruited to DNA damage sites in an ATM-dependent manner where they then monoubiquitylate K120 on H2B (Nakamura et al., 2011; Moyal et al., 2011). RNF20/RNF40 mediated H2BK120 monoubiquitylation promotes DNA repair protein (XRCC4, KU80) recruitment to DNA damage sites and is required for both HR and NHEJ repair (Nakamura et al., 2011; Moyal et al., 2011). RNF20/RNF40 also regulates γH2AX levels, suggesting a link between H2B ubiquitylation and H2AX phosphorylation. The precise mechanisms involved in regulating γH2AX levels, as well as which HR-promoting factors H2BK120Ub regulates, requires additional studies. The SAGA DUB module involved in H2BK120 deubiquitylation contains USP22, which is mainly involved in H2BK120 deubiquitylation. Depletion of the SAGA complex protein impaired γH2AX foci formation in an ATM-dependent manner and SAGA deficiency resulted in defective HR and NHEJ repair (Ramachandran et al., 2016). Ubiquitylation of H2BK120 also has been shown to regulate H3K79 methylation as H2BK120 ubiquitylation is critical for efficient methylation of H3K79. Recent studies had elucidated molecular mechanisms of this crosstalk-mediated activation. Biochemical and cryo-EM structure analysis reveals that DOT1L binds to ubiquitylated H2BK120 and directly promotes DOT1L catalytic activity, which may have implications for chromatin-based DDR mechanisms involving these histone marks (McGinty et al., 2008; McGinty et al., 2009; Chatterjee et al., 2010; Worden et al., 2019).
Other histone ubiquitylations
The use of Ubiquitin-Activated Interaction Traps (UBAITs) revealed that H2A.Z is a substrate for RNF168 (O’Connor et al., 2015). H2A.Z promotes loading of Ku70/80 to DNA damage sites (Xu et al., 2012). However, the role of RNF168-mediated H2A.Z ubiquitylation in the DDR is unknown. Tandem affinity purification assays using four UBA domains enriched the ubiquitylated protein in BRCA1/BARD1 overexpression cells. These assays identified macroH2A1 as a substrate of BRCA1/BARD1. BRCA1/BARD1 ubiquitylates K123 on macroH2A1, which is involved in cellular senescence (Kim et al., 2017). H4K91 is ubiquitylated by BBAP E3 ligase (Yan et al., 2009). Disruption of H4K91 ubiquitylation resulted in impaired H4K20 methylation, leading to delayed 53BP1 recruitment to DNA damage sites. Interestingly, H4 is also neddylated by RNF111, which is bound by RNF168 to promote its accumulation and activity at DNA damage sites (Ma et al., 2013). Neddylation is a ubiquitin-like protein that shares 80% homology with ubiquitin and is also tethered to lysine residues. Another ubiquitin-like modification, UFMylation, has also been shown to regulate the DDR. The ubiquitin-fold modifier1 (UFM1) is conjugated to lysine 31 of histone H4 by the E3 ligase UFL1 after DNA damage. UFMylated H4 promotes the recruitment of Suv39h1 histone methyltransferase, which recruits the histone acetyltransferase TIP60 to activate ATM (Qin et al., 2019). MRE11, which is a component of the MRN complex, is also UFMylated (Wang et al., 2019), suggesting UFMylation modification as an important PTM involved in DSB repair signaling by ATM (Qin et al., 2019; Wang et al., 2019). Taken together, beyond H2A and H2B, several other ubiquitylation and ubiquitin-like modifications occur on other histones to regulate the DDR.
SUMOYLATION
SUMO (small ubiquitin-related modifier) is related to ubiquitin and is similarly attached by isopeptide linkages to lysine residues. Like ubiquitin, SUMO is catalyzed by writer E3 ligases and erased by peptidases. SUMO accumulates at DNA damage sites in mammalian cells and the SUMO E3 ligases PIAS1 and PIAS4 are important in promoting DSB repair by regulating the accrual of the DNA repair factors BRCA1 and 53BP1 at DNA damage sites (Jackson and Durocher, 2013; Galanty et al., 2009; Morris et al., 2009). The role of histone sumoylation in the DDR is poorly understood. Core histone H4, as well as H2AX, has been shown to be sumoylated in mammalian cells (Chen et al., 2013; Shiio and Eisenman, 2003). H2AX is preferentially sumoylated by SUMO1, although the E3 ligase that writes this mark, as well as its modification site(s), have not yet been identified (Chen et al., 2013). Another histone H2A variant, H2AZ, has been shown to be sumoylated in both human and yeast cells (Fukuto et al., 2018; Kalocsay, Hiller, and Jentsch, 2009). In yeast, H2AZ was shown to be sumoylated in response to a persistent DSB, promoting nuclear periphery localization and DNA end-resection. SUMOylation was mapped to lysines 126 and 133 of H2AZ (Kalocsay, Hiller, and Jentsch, 2009). Mutant H2AZ in these two lysine residues did not display DNA damage sensitivity or DNA damage localization defects but did have defective relocalization of the DSB to the nuclear periphery compared to wild-type yeast. H2AZ is also sumoylated in mammalian cells by PIAS4 in a DNA damage-dependent manner (Fukuto et al., 2018). Depletion of PIAS4 blocked the histone exchange of H2AZ-2 at damage sites. Whether or not this was due directly to H2AZ sumoylation is unknown. In other studies, H2AZ has been shown to regulate DSB repair, including by promoting BRCA1 and NHEJ factors at break sites (Xu et al., 2012). Given that PIAS4 also regulates BRCA1 accrual at DNA lesions (Galanty et al., 2009), it is tempting to speculate that sumoylation of histones, including H2AZ, may regulate this step of DSB repair although additional studies are required. In addition, SUMO reader or eraser proteins that act on histones have not yet been reported to be involved in the DDR. It is likely that additional factors are involved in the activation and deactivation (i.e. reversibility) of DSB repair pathways involving sumoylation, including through modified histones.
Discussion
Modification of histones is important for the signaling and repair of DSBs. Numerous writers, erasers and readers of histone modifications are recruited to DNA damage sites where they act to modify chromatin to facilitate chromatin-based DDR activities (Table 1, Figure 6). Histone PTMs can regulate chromatin structure and function through diverse mechanisms. For example, alterations in the chromatin landscape surrounding a DSB may alter the repair of that break by channelling the lesion into a particular pathway via a histone code for repair (reviewed in (Clouaire and Legube, 2019). Although we have deciphered many factors and their assoicated histone PTM involved in chromatin-based mechanisms of DSB repair that we have reviewed here (Table 1), many questions still remain.
Figure 6.
Summary of histone PTM-mediated DSB repair.
Many chromatin modifiers are recruited to DNA damage and display increased associations with DNA lesions. It is still unclear how the normal chromatin state participates in DNA damage signaling and repair in addition to those activities that are modulated within this chromatin environment following DSB induction (Figure 6). Do histone modifications work alone or in concert with other PTMs to elicit specific instructions for a particular repair pathway? The ability for multiple histone modification to occur on the same amino acid residue and the demonstrated crosstalk that takes place between different histone marks make answering this question challenging. Regions on the nucleosome structure also coordinate histone PTMs. For example, the nucleosome acidic patch interacts with multiple E3 Ub ligases to facilitate their targeting of chromatin, including RING1/BMI1, RNF168 and BRCA1 (reviewed in (Agarwal and Miller, 2016). How chromatin proteins interface with histones themselves to allow selective targeting and binding of histone PTMs remains an area of active research. In addition, once the damage chromatin state is created, what functions does this state participate in for repair and how do these vary between the different repair processes, genome loci and cell types? DSBs have been shown to be mobile following their creation, perhaps to promote homology search during HR (Marnef and Legube, 2017). As we have reviewed, chromatin dynamics are altered around break sites as is transcription and chromatin structure. How these changes in chromatin behavior mechanistically interfaces with DDR processes including DNA end-resection, homology search and repair are all exciting questions that the field are working to address (Figure 6). Finally, very little is known about how the normal chromatin state is restored following repair and how defects in this process affect genome and epigenome integrity. This process may be particularly important when considering how DSBs affect transcription of both active and inactive genes. Given the importance of chromatin in transcription and cell identity, it is clear that unscheduled changes in chromatin that may occur after DNA damage, including by PTMs, could result in aberrant gene regulation. Given the prevalence of genome instability and aberrant transcription in cancer, the connection between these two processes that will likely involve histone PTMs warrant further investigation.
Mutations or altered expressions/activities of epigenetic modifiers are critical events in cancer development (Chi, Allis, and Wang, 2010; Dawson, 2017; Dawson and Kouzarides, 2012) and chromatin alterations occurring in diseases including cancer are known to influence DNA repair. This has been especially well-established in cancer treatments as DNA damaging agents are used as frontline therapies to treat cancer and chromatin-based mechanisms have been identified as mediators of these responses. A study that exemplifies this concept showed that cancer cells resistant to DNA damaging agents including cisplatin can be reversed to a drug-sensitive state through the reprogramming of chromatin by histone deacetylase inhibitors (Sharma et al., 2010). HDAC, HAT and bromodomain proteins are potential therapeutic anti-cancer targets (Gong, Chiu, and Miller, 2016; Barbieri, Cannizzaro, and Dawson, 2013; Di Cerbo and Schneider, 2013; Groselj et al., 2013) and histone methylation and DDR pathways, are areas of active research for their potential as therapeutic targets for human diseases including cancer (Lord and Ashworth, 2012; Dawson, 2017; Dawson and Kouzarides, 2012; O’Connor, 2015; Hojfeldt, Agger, and Helin, 2013; Pfister and Ashworth, 2017; Rodriguez and Miller, 2014). Dissecting these pathways mechanistically is warranted to provide mechanistic insights for the connections between histone PTMs and DDR, which may be involved in transcription, DNA repair and diseases including cancer. For example, PARP inhibitors are used and being developed to treat HR-deficient cancers including breast and ovarian (Lin and Kraus, 2017). As PARP regulates many DDR and chromatin pathways, how PARP inhibitors impact these pathways and whether or not they are involved in its drug mechanism in cancer is unclear. We propose that the distinction between DDR factors and epigenetic regulators are being tested by these studies showing that many factors are involved in multiple pathways including the DDR and chromatin. For example, histone modification pathways identified in cancer may alter transcription, which may consequently also impact DNA repair. If mutations in histone modification pathways reduce DNA repair, these tumors may exhibit genome instability and be susceptible to treatment with PARP inhibitors. Given the interplay between DNA repair and chromatin pathways, understanding chromatin dynamics, including histone PTMs, and DNA repair is fundamental for understanding genome maintenance. However, the interconnected functions of chromatin and the DDR make it likely that these pathways participate collectively in human diseases including cancer. The involvement of these pathways in disease further motivates research efforts to provide novel insights into how chromatin and the DDR function together to ensure proper genome function, as well as how genome integrity and epigenome pathways can be targeted for therapeutic interventions to treat diseases including cancer.
Acknowledgements
We thank Ramhari Kumbhar, Jullian Perren and Anusha Sriraman for critical reading of the manuscript. We apologize to colleagues whose work could not be included due to space limitations and the focused nature of this review.
Funding
The K.M.M. laboratory is supported by the NIH National Cancer Institute (R01CA198279 and RO1CA201268) and the American Cancer Society (RSG-16-042-01-DMC).
Footnotes
Disclosure statement
The authors declare no conflict of interest.
References
- Kornberg RD. (1974).Chromatin structure: a repeating unit of histones and DNA. Science, 184, 868–71 [DOI] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. (2010).Chromatin structure and the inheritance of epigenetic information. Nature reviews Genetics, 11, 285–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschbeck M, Hake SB. (2017).Variants of core histones and their roles in cell fate decisions, development and cancer. Nature reviews Molecular cell biology, 18, 299–314 [DOI] [PubMed] [Google Scholar]
- Kouzarides T (2007).Chromatin modifications and their function. Cell, 128, 693–705 [DOI] [PubMed] [Google Scholar]
- Campos EI, Reinberg D. (2009).Histones: annotating chromatin. Annual review of genetics, 43, 559–99 [DOI] [PubMed] [Google Scholar]
- Suganuma T, Workman JL. (2011).Signals and combinatorial functions of histone modifications. Annual review of biochemistry, 80, 473–99 [DOI] [PubMed] [Google Scholar]
- Miller KM, Jackson SP. (2012).Histone marks: repairing DNA breaks within the context of chromatin. Biochemical Society transactions, 40, 370–6 [DOI] [PubMed] [Google Scholar]
- Shahbazian MD, Grunstein M. (2007).Functions of site-specific histone acetylation and deacetylation. Annual review of biochemistry, 76, 75–100 [DOI] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. (2006).Histone H4–K16 acetylation controls chromatin structure and protein interactions. Science, 311, 844–7 [DOI] [PubMed] [Google Scholar]
- Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. (2012).Epigenetic protein families: a new frontier for drug discovery. Nature reviews Drug discovery, 11, 384–400 [DOI] [PubMed] [Google Scholar]
- Musselman CA, Lalonde ME, Cote J, Kutateladze TG. (2012).Perceiving the epigenetic landscape through histone readers. Nature structural & molecular biology, 19, 1218–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Li H, Patel DJ, Allis CD. (2007).Multivalent engagement of chromatin modifications by linked binding modules. Nature reviews Molecular cell biology, 8, 983–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SP, Bartek J. (2009).The DNA-damage response in human biology and disease. Nature, 461, 1071–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. (2010).The DNA damage response: making it safe to play with knives. Molecular cell, 40, 179–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubbs A, Nussenzweig A. (2017).Endogenous DNA Damage as a Source of Genomic Instability in Cancer. Cell, 168, 644–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Chiu LY, Nehring RB, Bravo Nunez MA, Mei Q, Perez M, Zhai Y, Fitzgerald DM, Pribis JP, Wang Y, Hu CW, Powell RT, LaBonte SA, Jalali A, Matadamas Guzman ML, Lentzsch AM, Szafran AT, Joshi MC, Richters M, Gibson JL, Frisch RL, Hastings PJ, Bates D, Queitsch C, Hilsenbeck SG, Coarfa C, Hu JC, Siegele DA, Scott KL, Liang H, Mancini MA, Herman C, Miller KM, Rosenberg SM. (2019).Bacteria-to-Human Protein Networks Reveal Origins of Endogenous DNA Damage. Cell, 176, 127–43 e24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini S, Gorgoulis VG, Halazonetis TD. (2010).Genomic instability--an evolving hallmark of cancer. Nature reviews Molecular cell biology, 11, 220–8 [DOI] [PubMed] [Google Scholar]
- Lord CJ, Ashworth A. (2012).The DNA damage response and cancer therapy. Nature, 481, 287–94 [DOI] [PubMed] [Google Scholar]
- Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JH. (2014).Understanding nucleotide excision repair and its roles in cancer and ageing. Nature reviews Molecular cell biology, 15, 465–81 [DOI] [PubMed] [Google Scholar]
- Jiricny J (2006).The multifaceted mismatch-repair system. Nature reviews Molecular cell biology, 7, 335–46 [DOI] [PubMed] [Google Scholar]
- Huertas P (2010).DNA resection in eukaryotes: deciding how to fix the break. Nature structural & molecular biology, 17, 11–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beucher A, Birraux J, Tchouandong L, Barton O, Shibata A, Conrad S, Goodarzi AA, Krempler A, Jeggo PA, Lobrich M. (2009).ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. The EMBO journal, 28, 3413–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR. (2010).The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annual review of biochemistry, 79, 181–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JR, Taylor MR, Boulton SJ. (2012).Playing the end game: DNA double-strand break repair pathway choice. Molecular cell, 47, 497–510 [DOI] [PubMed] [Google Scholar]
- Lukas J, Lukas C, Bartek J. (2011).More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nature cell biology, 13, 1161–9 [DOI] [PubMed] [Google Scholar]
- Gong F, Miller KM. (2013).Mammalian DNA repair: HATs and HDACs make their mark through histone acetylation. Mutation research, 750, 23–30 [DOI] [PubMed] [Google Scholar]
- Jackson SP, Durocher D. (2013).Regulation of DNA damage responses by ubiquitin and SUMO. Molecular cell, 49, 795–807 [DOI] [PubMed] [Google Scholar]
- Schwertman P, Bekker-Jensen S, Mailand N. (2016).Regulation of DNA double-strand break repair by ubiquitin and ubiquitin-like modifiers. Nature reviews Molecular cell biology, 17, 379–94 [DOI] [PubMed] [Google Scholar]
- Gong F, Chiu LY, Miller KM. (2016).Acetylation Reader Proteins: Linking Acetylation Signaling to Genome Maintenance and Cancer. PLoS genetics, 12, e1006272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. (1998).DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. The Journal of biological chemistry, 273, 5858–68 [DOI] [PubMed] [Google Scholar]
- Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. (2005).MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell, 123, 1213–26 [DOI] [PubMed] [Google Scholar]
- Panier S, Boulton SJ. (2014).Double-strand break repair: 53BP1 comes into focus. Nature reviews Molecular cell biology, 15, 7–18 [DOI] [PubMed] [Google Scholar]
- Zimmermann M, de Lange T. (2014).53BP1: pro choice in DNA repair. Trends in cell biology, 24, 108–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, Mer G. (2006).Structural basis for the methylation state-specific recognition of histone H4–K20 by 53BP1 and Crb2 in DNA repair. Cell, 127, 1361–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradet-Turcotte A, Canny MD, Escribano-Diaz C, Orthwein A, Leung CC, Huang H, Landry MC, Kitevski-LeBlanc J, Noordermeer SM, Sicheri F, Durocher D. (2013).53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature, 499, 50–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyen Y, Zgheib O, Ditullio RA Jr., Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD. (2004).Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature, 432, 406–11 [DOI] [PubMed] [Google Scholar]
- Polo SE, Jackson SP. (2011).Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes & development, 25, 409–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allis CD. (2018).Pursuing the Secrets of Histone Proteins: An Amazing Journey with a Remarkable Supporting Cast. Cell, 175, 18–21 [DOI] [PubMed] [Google Scholar]
- Williams CL. (2018).Michael Grunstein and David Allis receive the 2018 Lasker Basic Medical Research Award. J Clin Invest, 128, 4201–03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luijsterburg MS, van Attikum H. (2011).Chromatin and the DNA damage response: the cancer connection. Molecular oncology, 5, 349–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeggo PA, Pearl LH, Carr AM. (2016).DNA repair, genome stability and cancer: a historical perspective. Nature reviews Cancer, 16, 35–42 [DOI] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, Xu X, Deng CX, Finkel T, Nussenzweig M, Stark JM, Nussenzweig A. (2010).53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell, 141, 243–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Xu X, Bunting SF, Liu J, Wang RH, Cao LL, Wu JJ, Peng TN, Chen J, Nussenzweig A, Deng CX, Finkel T. (2009).A selective requirement for 53BP1 in the biological response to genomic instability induced by Brca1 deficiency. Molecular cell, 35, 534–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, Lukas J, Lukas C. (2009).RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell, 136, 435–46 [DOI] [PubMed] [Google Scholar]
- Fradet-Turcotte A, Canny MD, Escribano-Diaz C, Orthwein A, Leung CC, Huang H, Landry MC, Kitevski-Leblanc J, Noordermeer SM, Sicheri F, Durocher D. (2013).53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquet K, Fradet-Turcotte A, Avvakumov N, Lambert JP, Roques C, Pandita RK, Paquet E, Herst P, Gingras AC, Pandita TK, Legube G, Doyon Y, Durocher D, Cote J. (2016).The TIP60 Complex Regulates Bivalent Chromatin Recognition by 53BP1 through Direct H4K20me Binding and H2AK15 Acetylation. Molecular cell, 62, 409–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Cho NW, Cui G, Manion EM, Shanbhag NM, Botuyan MV, Mer G, Greenberg RA. (2013).Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nature structural & molecular biology, 20, 317–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KM, Tjeertes JV, Coates J, Legube G, Polo SE, Britton S, Jackson SP. (2010).Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nature structural & molecular biology, 17, 1144–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Saredi G, Becker JR, Foster BM, Nguyen NV, Beyer TE, Cesa LC, Faull PA, Lukauskas S, Frimurer T, Chapman JR, Bartke T, Groth A. (2019).H4K20me0 recognition by BRCA1-BARD1 directs homologous recombination to sister chromatids. Nature cell biology, 21, 311–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, Pommier Y. (2008).GammaH2AX and cancer. Nature reviews Cancer, 8, 957–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassing CH, Chua KF, Sekiguchi J, Suh H, Whitlow SR, Fleming JC, Monroe BC, Ciccone DN, Yan C, Vlasakova K, Livingston DM, Ferguson DO, Scully R, Alt FW. (2002).Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proceedings of the National Academy of Sciences of the United States of America, 99, 8173–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, Reina-San-Martin B, Coppola V, Meffre E, Difilippantonio MJ, Redon C, Pilch DR, Olaru A, Eckhaus M, Camerini-Otero RD, Tessarollo L, Livak F, Manova K, Bonner WM, Nussenzweig MC, Nussenzweig A. (2002).Genomic instability in mice lacking histone H2AX. Science, 296, 922–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie A, Puget N, Shim I, Odate S, Jarzyna I, Bassing CH, Alt FW, Scully R. (2004).Control of sister chromatid recombination by histone H2AX. Molecular cell, 16, 1017–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Boon C, Redon C, Bonner WM. (1999).Megabase chromatin domains involved in DNA double-strand breaks in vivo. The Journal of cell biology, 146, 905–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A. (2003).Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nature cell biology, 5, 675–9 [DOI] [PubMed] [Google Scholar]
- Fernandez-Capetillo O, Celeste A, Nussenzweig A. (2003).Focusing on foci: H2AX and the recruitment of DNA-damage response factors. Cell cycle, 2, 426–7 [PubMed] [Google Scholar]
- Scully R, Xie A. (2013).Double strand break repair functions of histone H2 AX. Mutation research, 750, 5–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turinetto V, Giachino C. (2015).Multiple facets of histone variant H2AX: a DNA double-strand-break marker with several biological functions. Nucleic acids research, 43, 2489–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coster G, Goldberg M. (2010).The cellular response to DNA damage: a focus on MDC1 and its interacting proteins. Nucleus, 1, 166–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. (2007).RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell, 131, 901–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, Pelletier L, Jackson SP, Durocher D. (2007).Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science, 318, 1637–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. (2007).RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell, 131, 887–900 [DOI] [PubMed] [Google Scholar]
- Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, Oldreive C, Wildenhain J, Tagliaferro A, Pelletier L, Taubenheim N, Durandy A, Byrd PJ, Stankovic T, Taylor AM, Durocher D. (2009).The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell, 136, 420–34 [DOI] [PubMed] [Google Scholar]
- Chowdhury D, Keogh MC, Ishii H, Peterson CL, Buratowski S, Lieberman J. (2005).gamma-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Molecular cell, 20, 801–9 [DOI] [PubMed] [Google Scholar]
- Chowdhury D, Xu X, Zhong X, Ahmed F, Zhong J, Liao J, Dykxhoorn DM, Weinstock DM, Pfeifer GP, Lieberman J. (2008).A PP4-phosphatase complex dephosphorylates gamma-H2AX generated during DNA replication. Molecular cell, 31, 33–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas P, Zhong J, Ye R, Moorhead GB, Xu X, Lees-Miller SP. (2010).Protein phosphatase 6 interacts with the DNA-dependent protein kinase catalytic subunit and dephosphorylates gamma-H2AX. Molecular and cellular biology, 30, 1368–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macurek L, Lindqvist A, Voets O, Kool J, Vos HR, Medema RH. (2010).Wip1 phosphatase is associated with chromatin and dephosphorylates gammaH2AX to promote checkpoint inhibition. Oncogene, 29, 2281–91 [DOI] [PubMed] [Google Scholar]
- Moon SH, Lin L, Zhang X, Nguyen TA, Darlington Y, Waldman AS, Lu X, Donehower LA. (2010).Wild-type p53-induced phosphatase 1 dephosphorylates histone variant gamma-H2AX and suppresses DNA double strand break repair. The Journal of biological chemistry, 285, 12935–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada S, Chen GI, Gingras AC, Durocher D. (2008).PP4 is a gamma H2AX phosphatase required for recovery from the DNA damage checkpoint. EMBO reports, 9, 1019–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aymard F, Bugler B, Schmidt CK, Guillou E, Caron P, Briois S, Iacovoni JS, Daburon V, Miller KM, Jackson SP, Legube G. (2014).Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nature structural & molecular biology, 21, 366–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouaire T, Rocher V, Lashgari A, Arnould C, Aguirrebengoa M, Biernacka A, Skrzypczak M, Aymard F, Fongang B, Dojer N, Iacovoni JS, Rowicka M, Ginalski K, Cote J, Legube G. (2018).Comprehensive Mapping of Histone Modifications at DNA Double-Strand Breaks Deciphers Repair Pathway Chromatin Signatures. Molecular cell, 72, 250–62 e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacovoni JS, Caron P, Lassadi I, Nicolas E, Massip L, Trouche D, Legube G. (2010).High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. The EMBO journal, 29, 1446–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook PJ, Ju BG, Telese F, Wang X, Glass CK, Rosenfeld MG. (2009).Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature, 458, 591–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao A, Li H, Shechter D, Ahn SH, Fabrizio LA, Erdjument-Bromage H, Ishibe-Murakami S, Wang B, Tempst P, Hofmann K, Patel DJ, Elledge SJ, Allis CD. (2009).WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature, 457, 57–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie A, Odate S, Chandramouly G, Scully R. (2010).H2AX post-translational modifications in the ionizing radiation response and homologous recombination. Cell cycle, 9, 3602–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WT, Alpert A, Leiter C, Gong F, Jackson SP, Miller KM. (2013).Systematic identification of functional residues in mammalian histone H2AX. Molecular and cellular biology, 33, 111–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung JWC, Emery LE, Miller KM. (2018).CRISPR/Cas9 Gene Editing of Human Histone H2A Variant H2AX and MacroH2A. Methods Mol Biol, 1832, 255–69 [DOI] [PubMed] [Google Scholar]
- Fernandez-Capetillo O, Allis CD, Nussenzweig A. (2004).Phosphorylation of histone H2B at DNA double-strand breaks. J Exp Med, 199, 1671–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CS, Lee K, Legube G, Haber JE. (2014).Dynamics of yeast histone H2A and H2B phosphorylation in response to a double-strand break. Nature structural & molecular biology, 21, 103–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaco L, Kolthur-Seetharam U, Loury R, Murcia JM, de Murcia G, Sassone-Corsi P. (2005).Inhibition of Aurora-B kinase activity by poly(ADP-ribosyl)ation in response to DNA damage. Proceedings of the National Academy of Sciences of the United States of America, 102, 14244–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa K (2008).Reduction of phosphorylated histone H3 serine 10 and serine 28 cell cycle marker intensities after DNA damage. Cytometry A, 73, 517–27 [DOI] [PubMed] [Google Scholar]
- Sharma AK, Bhattacharya S, Khan SA, Khade B, Gupta S. (2015).Dynamic alteration in H3 serine 10 phosphorylation is G1-phase specific during ionization radiation induced DNA damage response in human cells. Mutation research, 773, 83–91 [DOI] [PubMed] [Google Scholar]
- Shimada M, Niida H, Zineldeen DH, Tagami H, Tanaka M, Saito H, Nakanishi M. (2008).Chk1 is a histone H3 threonine 11 kinase that regulates DNA damage-induced transcriptional repression. Cell, 132, 221–32 [DOI] [PubMed] [Google Scholar]
- Tjeertes JV, Miller KM, Jackson SP. (2009).Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. The EMBO journal, 28, 1878–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicka A, Seiser C. (2012).Histone H3 phosphorylation - a versatile chromatin modification for different occasions. Biochimie, 94, 2193–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung WL, Turner FB, Krishnamoorthy T, Wolner B, Ahn SH, Foley M, Dorsey JA, Peterson CL, Berger SL, Allis CD. (2005).Phosphorylation of histone H4 serine 1 during DNA damage requires casein kinase II in S. cerevisiae. Curr Biol, 15, 656–60 [DOI] [PubMed] [Google Scholar]
- Utley RT, Lacoste N, Jobin-Robitaille O, Allard S, Cote J. (2005).Regulation of NuA4 histone acetyltransferase activity in transcription and DNA repair by phosphorylation of histone H4. Molecular and cellular biology, 25, 8179–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MB, Shifat R, Johnson DG, Bedford MT, Gabrusiewicz KR, Cortes-Santiago N, Luo X, Lu Z, Ezhilarasan R, Sulman EP, Jiang H, Li SS, Lang FF, Tyler J, Hung MC, Fueyo J, Gomez-Manzano C. (2016).TIE2-mediated tyrosine phosphorylation of H4 regulates DNA damage response by recruiting ABL1. Sci Adv, 2, e1501290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millan-Zambrano G, Santos-Rosa H, Puddu F, Robson SC, Jackson SP, Kouzarides T. (2018).Phosphorylation of Histone H4T80 Triggers DNA Damage Checkpoint Recovery. Molecular cell, 72, 625–35 e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett E, Bonfiglio JJ, Prokhorova E, Colby T, Zobel F, Ahel I, Matic I. (2018).Interplay of Histone Marks with Serine ADP-Ribosylation. Cell reports, 24, 3488–502 e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo L, Leidecker O, Prokhorova E, Dauben H, Matic I, Ahel I. (2018).Serine is the major residue for ADP-ribosylation upon DNA damage. eLife, 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Li Y, Tang M, Peng B, Lu X, Yang Q, Zhu Q, Hou T, Li M, Liu C, Wang L, Xu X, Zhao Y, Wang H, Yang Y, Zhu WG. (2018).Destabilization of linker histone H1.2 is essential for ATM activation and DNA damage repair. Cell Res, 28, 756–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messner S, Hottiger MO. (2011).Histone ADP-ribosylation in DNA repair, replication and transcription. Trends in cell biology, 21, 534–42 [DOI] [PubMed] [Google Scholar]
- Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. (1996).Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell, 84, 843–51 [DOI] [PubMed] [Google Scholar]
- Taunton J, Hassig CA, Schreiber SL. (1996).A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science, 272, 408–11 [DOI] [PubMed] [Google Scholar]
- Lee KK, Workman JL. (2007).Histone acetyltransferase complexes: one size doesn’t fit all. Nature reviews Molecular cell biology, 8, 284–95 [DOI] [PubMed] [Google Scholar]
- Yang XJ, Seto E. (2008).The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nature reviews Molecular cell biology, 9, 206–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman JL, Dittenhafer-Reed KE, Denu JM. (2012).Sirtuin catalysis and regulation. The Journal of biological chemistry, 287, 42419–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. (2009).Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science, 325, 834–40 [DOI] [PubMed] [Google Scholar]
- Choudhary C, Weinert BT, Nishida Y, Verdin E, Mann M. (2014).The growing landscape of lysine acetylation links metabolism and cell signalling. Nature reviews Molecular cell biology, 15, 536–50 [DOI] [PubMed] [Google Scholar]
- Elia AE, Boardman AP, Wang DC, Huttlin EL, Everley RA, Dephoure N, Zhou C, Koren I, Gygi SP, Elledge SJ. (2015).Quantitative Proteomic Atlas of Ubiquitination and Acetylation in the DNA Damage Response. Molecular cell, 59, 867–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksnes H, Ree R, Arnesen T. (2019).Co-translational, Post-translational, and Non-catalytic Roles of N-Terminal Acetyltransferases. Molecular cell [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan B, Smerdon MJ. (1986).Changes in nuclear protein acetylation in u.v.-damaged human cells. Carcinogenesis, 7, 1087–94 [DOI] [PubMed] [Google Scholar]
- Smerdon MJ. (1991).DNA repair and the role of chromatin structure. Current opinion in cell biology, 3, 422–8 [DOI] [PubMed] [Google Scholar]
- Green CM, Almouzni G. (2002).When repair meets chromatin. First in series on chromatin dynamics. EMBO reports, 3, 28–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria G, Polo SE, Almouzni G. (2012).Prime, repair, restore: the active role of chromatin in the DNA damage response. Molecular cell, 46, 722–34 [DOI] [PubMed] [Google Scholar]
- Chiu LY, Gong F, Miller KM. (2017).Bromodomain proteins: repairing DNA damage within chromatin. Philos Trans R Soc Lond B Biol Sci, 372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa T, Filippakopoulos P. (2017).Functions of bromodomain-containing proteins and their roles in homeostasis and cancer. Nature reviews Molecular cell biology, 18, 246–62 [DOI] [PubMed] [Google Scholar]
- Gong F, Chiu LY, Cox B, Aymard F, Clouaire T, Leung JW, Cammarata M, Perez M, Agarwal P, Brodbelt JS, Legube G, Miller KM. (2015).Screen identifies bromodomain protein ZMYND8 in chromatin recognition of transcription-associated DNA damage that promotes homologous recombination. Genes & development, 29, 197–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikura T, Tashiro S, Kakino A, Shima H, Jacob N, Amunugama R, Yoder K, Izumi S, Kuraoka I, Tanaka K, Kimura H, Ikura M, Nishikubo S, Ito T, Muto A, Miyagawa K, Takeda S, Fishel R, Igarashi K, Kamiya K. (2007).DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Molecular and cellular biology, 27, 7028–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata K, Kitabayashi I. (2009).Sirt1 physically interacts with Tip60 and negatively regulates Tip60-mediated acetylation of H2AX. Biochemical and biophysical research communications, 390, 1355–60 [DOI] [PubMed] [Google Scholar]
- Jiang X, Xu Y, Price BD. (2010).Acetylation of H2AX on lysine 36 plays a key role in the DNA double-strand break repair pathway. FEBS letters, 584, 2926–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrick JJ, Parra MA. (2009).The role of histone H2A and H2B post-translational modifications in transcription: a genomic perspective. Biochimica et biophysica acta, 1789, 37–44 [DOI] [PubMed] [Google Scholar]
- Fierz B, Chatterjee C, McGinty RK, Bar-Dagan M, Raleigh DP, Muir TW. (2011).Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nature chemical biology, 7, 113–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Ruiz PD, Novikov L, Casill AD, Park JW, Gamble MJ. (2014).MacroH2A1.1 and PARP-1 cooperate to regulate transcription by promoting CBP-mediated H2 B acetylation. Nature structural & molecular biology, 21, 981–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana S, Kruhlak MJ, Kim J, Tran AD, Liu J, Nyswaner K, Shi L, Jailwala P, Sung MH, Hakim O, Oberdoerffer P. (2014).A macrohistone variant links dynamic chromatin compaction to BRCA1-dependent genome maintenance. Cell reports, 8, 1049–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray Chaudhuri A, Nussenzweig A. (2017).The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nature reviews Molecular cell biology, 18, 610–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YC, Gerlitz G, Furusawa T, Catez F, Nussenzweig A, Oh KS, Kraemer KH, Shiloh Y, Bustin M. (2009).Activation of ATM depends on chromatin interactions occurring before induction of DNA damage. Nature cell biology, 11, 92–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das C, Lucia MS, Hansen KC, Tyler JK. (2009).CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature, 459, 113–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCord RA, Michishita E, Hong T, Berber E, Boxer LD, Kusumoto R, Guan S, Shi X, Gozani O, Burlingame AL, Bohr VA, Chua KF. (2009).SIRT6 stabilizes DNA-dependent protein kinase at chromatin for DNA double-strand break repair. Aging, 1, 109–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Park JH, Kim SJ, Kwon SJ, Kwon J. (2010).A cooperative activation loop among SWI/SNF, gamma-H2AX and H3 acetylation for DNA double-strand break repair. The EMBO journal, 29, 1434–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiwara H, Ui A, Otsuka A, Satoh H, Yokomi I, Nakajima S, Yasui A, Yokota J, Kohno T. (2011).Histone acetylation by CBP and p300 at double-strand break sites facilitates SWI/SNF chromatin remodeling and the recruitment of non-homologous end joining factors. Oncogene, 30, 2135–46 [DOI] [PubMed] [Google Scholar]
- Vazquez BN, Thackray JK, Simonet NG, Kane-Goldsmith N, Martinez-Redondo P, Nguyen T, Bunting S, Vaquero A, Tischfield JA, Serrano L. (2016).SIRT7 promotes genome integrity and modulates non-homologous end joining DNA repair. The EMBO journal, 35, 1488–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hagan HM, Mohammad HP, Baylin SB. (2008).Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS genetics, 4, e1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toiber D, Erdel F, Bouazoune K, Silberman DM, Zhong L, Mulligan P, Sebastian C, Cosentino C, Martinez-Pastor B, Giacosa S, D’Urso A, Naar AM, Kingston R, Rippe K, Mostoslavsky R. (2013).SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Molecular cell, 51, 454–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbin MM, Madabhushi R, Pan L, Chen Y, Kim D, Gao J, Ahanonu B, Pao PC, Qiu Y, Zhao Y, Tsai LH. (2013).SIRT1 collaborates with ATM and HDAC1 to maintain genomic stability in neurons. Nat Neurosci, 16, 1008–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li Z, Dong L, Tang M, Zhang P, Zhang C, Cao Z, Zhu Q, Chen Y, Wang H, Wang T, Lv D, Wang L, Zhao Y, Yang Y, Wang H, Zhang H, Roeder RG, Zhu WG. (2018).Histone H1 acetylation at lysine 85 regulates chromatin condensation and genome stability upon DNA damage. Nucleic acids research, 46, 7716–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Sharma GG, Young CS, Agarwal M, Smith ER, Paull TT, Lucchesi JC, Khanna KK, Ludwig T, Pandita TK. (2005).Involvement of human MOF in ATM function. Molecular and cellular biology, 25, 5292–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, Herceg Z. (2006).Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nature cell biology, 8, 91–9 [DOI] [PubMed] [Google Scholar]
- Li X, Corsa CA, Pan PW, Wu L, Ferguson D, Yu X, Min J, Dou Y. (2010).MOF and H4 K16 acetylation play important roles in DNA damage repair by modulating recruitment of DNA damage repair protein Mdc1. Molecular and cellular biology, 30, 5335–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray K (1964).The Occurrence of Epsilon-N-Methyl Lysine in Histones. Biochemistry, 3, 10–5 [DOI] [PubMed] [Google Scholar]
- Greer EL, Shi Y. (2012).Histone methylation: a dynamic mark in health, disease and inheritance. Nature reviews Genetics, 13, 343–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black JC, Van Rechem C, Whetstine JR. (2012).Histone lysine methylation dynamics: establishment, regulation, and biological impact. Molecular cell, 48, 491–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Bedford MT. (2013).Protein arginine methyltransferases and cancer. Nature reviews Cancer, 13, 37–50 [DOI] [PubMed] [Google Scholar]
- Bedford MT, Clarke SG. (2009).Protein arginine methylation in mammals: who, what, and why. Molecular cell, 33, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc RS, Richard S. (2017).Arginine Methylation: The Coming of Age. Molecular cell, 65, 8–24 [DOI] [PubMed] [Google Scholar]
- Dimitrova E, Turberfield AH, Klose RJ. (2015).Histone demethylases in chromatin biology and beyond. EMBO reports, 16, 1620–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooistra SM, Helin K. (2012).Molecular mechanisms and potential functions of histone demethylases. Nature reviews Molecular cell biology, 13, 297–311 [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. (2004).Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell, 119, 941–53 [DOI] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. (2006).Histone demethylation by a family of JmjC domain-containing proteins. Nature, 439, 811–6 [DOI] [PubMed] [Google Scholar]
- Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, Shi Y. (2006).Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell, 125, 467–81 [DOI] [PubMed] [Google Scholar]
- Chi P, Allis CD, Wang GG. (2010).Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nature reviews Cancer, 10, 457–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert M, Helin K. (2010).Histone methyltransferases in cancer. Seminars in cell & developmental biology, 21, 209–20 [DOI] [PubMed] [Google Scholar]
- Shelton SB, Reinsborough C, Xhemalce B. (2016).Who Watches the Watchmen: Roles of RNA Modifications in the RNA Interference Pathway. PLoS genetics, 12, e1006139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A (2002).DNA methylation patterns and epigenetic memory. Genes & development, 16, 6–21 [DOI] [PubMed] [Google Scholar]
- Klose RJ, Bird AP. (2006).Genomic DNA methylation: the mark and its mediators. Trends in biochemical sciences, 31, 89–97 [DOI] [PubMed] [Google Scholar]
- Paik WK, Paik DC, Kim S. (2007).Historical review: the field of protein methylation. Trends in biochemical sciences, 32, 146–52 [DOI] [PubMed] [Google Scholar]
- Biggar KK, Li SS. (2015).Non-histone protein methylation as a regulator of cellular signalling and function. Nature reviews Molecular cell biology, 16, 5–17 [DOI] [PubMed] [Google Scholar]
- Cuozzo C, Porcellini A, Angrisano T, Morano A, Lee B, Di Pardo A, Messina S, Iuliano R, Fusco A, Santillo MR, Muller MT, Chiariotti L, Gottesman ME, Avvedimento EV. (2007).DNA damage, homology-directed repair, and DNA methylation. PLoS genetics, 3, e110. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Xiang Y, Laurent B, Hsu CH, Nachtergaele S, Lu Z, Sheng W, Xu C, Chen H, Ouyang J, Wang S, Ling D, Hsu PH, Zou L, Jambhekar A, He C, Shi Y. (2017).RNA m6A methylation regulates the ultraviolet-induced DNA damage response. Nature, 543, 573–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamoto R, Saloura V, Nakamura Y. (2015).Critical roles of non-histone protein lysine methylation in human tumorigenesis. Nature reviews Cancer, 15, 110–24 [DOI] [PubMed] [Google Scholar]
- Reinhardt HC, Schumacher B. (2012).The p53 network: cellular and systemic DNA damage responses in aging and cancer. Trends in genetics : TIG, 28, 128–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakin ND, Jackson SP. (1999).Regulation of p53 in response to DNA damage. Oncogene, 18, 7644–55 [DOI] [PubMed] [Google Scholar]
- West LE, Gozani O. (2011).Regulation of p53 function by lysine methylation. Epigenomics, 3, 361–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karkhanis V, Wang L, Tae S, Hu YJ, Imbalzano AN, Sif S. (2012).Protein arginine methyltransferase 7 regulates cellular response to DNA damage by methylating promoter histones H2A and H4 of the polymerase delta catalytic subunit gene, POLD1. The Journal of biological chemistry, 287, 29801–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo AE, Piller SC. (2018).Protein arginine methylation: an emerging regulator of the cell cycle. Cell Div, 13, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sone K, Piao L, Nakakido M, Ueda K, Jenuwein T, Nakamura Y, Hamamoto R. (2014).Critical role of lysine 134 methylation on histone H2AX for gamma-H2AX production and DNA repair. Nature communications, 5, 5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. (2007).High-resolution profiling of histone methylations in the human genome. Cell, 129, 823–37 [DOI] [PubMed] [Google Scholar]
- Li X, Liu L, Yang S, Song N, Zhou X, Gao J, Yu N, Shan L, Wang Q, Liang J, Xuan C, Wang Y, Shang Y, Shi L. (2014).Histone demethylase KDM5B is a key regulator of genome stability. Proceedings of the National Academy of Sciences of the United States of America, 111, 7096–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosammaparast N, Kim H, Laurent B, Zhao Y, Lim HJ, Majid MC, Dango S, Luo Y, Hempel K, Sowa ME, Gygi SP, Steen H, Harper JW, Yankner B, Shi Y. (2013).The histone demethylase LSD1/KDM1A promotes the DNA damage response. The Journal of cell biology, 203, 457–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong F, Clouaire T, Aguirrebengoa M, Legube G, Miller KM. (2017).Histone demethylase KDM5A regulates the ZMYND8-NuRD chromatin remodeler to promote DNA repair. The Journal of cell biology [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks IA, Treffers LW, Verlaan-de Vries M, Olsen JV, Vertegaal AC. (2015).SUMO-2 Orchestrates Chromatin Modifiers in Response to DNA Damage. Cell reports, 10, 1778–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou DM, Adamson B, Dephoure NE, Tan X, Nottke AC, Hurov KE, Gygi SP, Colaiacovo MP, Elledge SJ. (2010).A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proceedings of the National Academy of Sciences of the United States of America, 107, 18475–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo SE, Kaidi A, Baskcomb L, Galanty Y, Jackson SP. (2010).Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. The EMBO journal, 29, 3130–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruijt CG, Luijsterburg MS, Menafra R, Lindeboom RG, Jansen PW, Edupuganti RR, Baltissen MP, Wiegant WW, Voelker-Albert MC, Matarese F, Mensinga A, Poser I, Vos HR, Stunnenberg HG, van Attikum H, Vermeulen M. (2016).ZMYND8 Co-localizes with NuRD on Target Genes and Regulates Poly(ADP-Ribose)-Dependent Recruitment of GATAD2A/NuRD to Sites of DNA Damage. Cell reports, 17, 783–98 [DOI] [PubMed] [Google Scholar]
- Gong F, Miller KM. (2018).Double duty: ZMYND8 in the DNA damage response and cancer. Cell cycle, 17, 414–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitsky P, Krojer T, Fujisawa T, Lambert JP, Picaud S, Wang CY, Shanle EK, Krajewski K, Friedrichsen H, Kanapin A, Goding C, Schapira M, Samsonova A, Strahl BD, Gingras AC, Filippakopoulos P. (2016).Multivalent Histone and DNA Engagement by a PHD/BRD/PWWP Triple Reader Cassette Recruits ZMYND8 to K14ac-Rich Chromatin. Cell reports, 17, 2724–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Kato A, Kobayashi J, Yanagihara H, Sakamoto S, Oliveira DV, Shimada M, Tauchi H, Suzuki H, Tashiro S, Zou L, Komatsu K. (2011).Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Molecular cell, 41, 515–28 [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. (2001).Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature, 410, 120–4 [DOI] [PubMed] [Google Scholar]
- Sun Y, Jiang X, Xu Y, Ayrapetov MK, Moreau LA, Whetstine JR, Price BD. (2009).Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nature cell biology, 11, 1376–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayrapetov MK, Gursoy-Yuzugullu O, Xu C, Xu Y, Price BD. (2014).DNA double-strand breaks promote methylation of histone H3 on lysine 9 and transient formation of repressive chromatin. Proceedings of the National Academy of Sciences of the United States of America, 111, 9169–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alagoz M, Katsuki Y, Ogiwara H, Ogi T, Shibata A, Kakarougkas A, Jeggo P. (2015).SETDB1, HP1 and SUV39 promote repositioning of 53BP1 to extend resection during homologous recombination in G2 cells. Nucleic acids research, 43, 7931–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Nishikawa H, Fukuda T, Vittal V, Asano M, Miyoshi Y, Klevit RE, Ohta T. (2015).Interaction of BARD1 and HP1 Is Required for BRCA1 Retention at Sites of DNA Damage. Cancer research, 75, 1311–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A, Imai Y, Yamakoshi K, Kuninaka S, Ohtani N, Yoshimoto S, Hori S, Tachibana M, Anderton E, Takeuchi T, Shinkai Y, Peters G, Saya H, Hara E. (2012).DNA damage signaling triggers degradation of histone methyltransferases through APC/C(Cdh1) in senescent cells. Molecular cell, 45, 123–31 [DOI] [PubMed] [Google Scholar]
- Young LC, McDonald DW, Hendzel MJ. (2013).Kdm4b histone demethylase is a DNA damage response protein and confers a survival advantage following gamma-irradiation. The Journal of biological chemistry, 288, 21376–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury-Haddad H, Guttmann-Raviv N, Ipenberg I, Huggins D, Jeyasekharan AD, Ayoub N. (2014).PARP1-dependent recruitment of KDM4D histone demethylase to DNA damage sites promotes double-strand break repair. Proceedings of the National Academy of Sciences of the United States of America, 111, E728–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hagan HM, Wang W, Sen S, Destefano Shields C, Lee SS, Zhang YW, Clements EG, Cai Y, Van Neste L, Easwaran H, Casero RA, Sears CL, Baylin SB. (2011).Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands. Cancer cell, 20, 606–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S, Ismail IH, Young LC, Poirier GG, Hendzel MJ. (2013).Polycomb repressive complex 2 contributes to DNA double-strand break repair. Cell Cycle, 12, 2675–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chang JF, Sun J, Chen L, Yang XM, Tang HY, Jing YY, Kang X, He ZM, Wu JY, Wei HM, Wang DL, Xu RG, Zhu RB, Shen Y, Zeng SY, Wang C, Liu KN, Zhang Y, Mao ZY, Jiang CZ, Sun FL. (2018).Histone H3K27 methylation modulates the dynamics of FANCD2 on chromatin to facilitate NHEJ and genome stability. J Cell Sci, 131 [DOI] [PubMed] [Google Scholar]
- Wagner EJ, Carpenter PB. (2012).Understanding the language of Lys36 methylation at histone H3. Nature reviews Molecular cell biology, 13, 115–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Schneider R, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. (2005).Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. The Journal of biological chemistry, 280, 17732–6 [DOI] [PubMed] [Google Scholar]
- Pfister SX, Ahrabi S, Zalmas LP, Sarkar S, Aymard F, Bachrati CZ, Helleday T, Legube G, La Thangue NB, Porter AC, Humphrey TC. (2014).SETD2-Dependent Histone H3K36 Trimethylation Is Required for Homologous Recombination Repair and Genome Stability. Cell reports, 7, 2006–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho S, Vitor AC, Sridhara SC, Martins FB, Raposo AC, Desterro JM, Ferreira J, de Almeida SF. (2014).SETD2 is required for DNA double-strand break repair and activation of the p53-mediated checkpoint. eLife, 3, e02482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugaard M, Baude A, Fugger K, Povlsen LK, Beck H, Sorensen CS, Petersen NH, Sorensen PH, Lukas C, Bartek J, Lukas J, Rohde M, Jaattela M. (2012).LEDGF (p75) promotes DNA-end resection and homologous recombination. Nature structural & molecular biology, 19, 803–10 [DOI] [PubMed] [Google Scholar]
- Fnu S, Williamson EA, De Haro LP, Brenneman M, Wray J, Shaheen M, Radhakrishnan K, Lee SH, Nickoloff JA, Hromas R. (2011).Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proceedings of the National Academy of Sciences of the United States of America, 108, 540–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Oshige M, Durant ST, Rasila KK, Williamson EA, Ramsey H, Kwan L, Nickoloff JA, Hromas R. (2005).The SET domain protein Metnase mediates foreign DNA integration and links integration to nonhomologous end-joining repair. Proceedings of the National Academy of Sciences of the United States of America, 102, 18075–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao LL, Wei F, Du Y, Song B, Wang D, Shen C, Lu X, Cao Z, Yang Q, Gao Y, Wang L, Zhao Y, Wang H, Yang Y, Zhu WG. (2016).ATM-mediated KDM2A phosphorylation is required for the DNA damage repair. Oncogene, 35, 402. [DOI] [PubMed] [Google Scholar]
- Mallette FA, Mattiroli F, Cui G, Young LC, Hendzel MJ, Mer G, Sixma TK, Richard S. (2012).RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. The EMBO journal, 31, 1865–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooq Z, Banday S, Pandita TK, Altaf M. (2016).The many faces of histone H3K79 methylation. Mutation research Reviews in mutation research, 768, 46–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AT, Zhang Y. (2011).The diverse functions of Dot1 and H3K79 methylation. Genes & development, 25, 1345–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocki R, Javaheri A, Allard S, Sha F, Cote J, Kron SJ. (2005).Role of Dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Molecular and cellular biology, 25, 8430–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeman TP, Wang Q, Feng J, Wang XF. (2012).Bat3 facilitates H3K79 dimethylation by DOT1L and promotes DNA damage-induced 53BP1 foci at G1/G2 cell-cycle phases. The EMBO journal, 31, 2169–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen S, Schotta G, Sorensen CS. (2013).Histone H4 lysine 20 methylation: key player in epigenetic regulation of genomic integrity. Nucleic acids research, 41, 2797–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattiroli F, Vissers JH, van Dijk WJ, Ikpa P, Citterio E, Vermeulen W, Marteijn JA, Sixma TK. (2012).RNF168 ubiquitinates K13–15 on H2A/H2AX to drive DNA damage signaling. Cell, 150, 1182–95 [DOI] [PubMed] [Google Scholar]
- Hajdu I, Ciccia A, Lewis SM, Elledge SJ. (2011).Wolf-Hirschhorn syndrome candidate 1 is involved in the cellular response to DNA damage. Proceedings of the National Academy of Sciences of the United States of America, 108, 13130–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei H, Zhang L, Luo K, Qin Y, Chesi M, Fei F, Bergsagel PL, Wang L, You Z, Lou Z. (2011).MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature, 470, 124–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Hubner MR, Beck DB, Vermeulen M, Hurwitz J, Spector DL, Reinberg D. (2010).Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage. Molecular cell, 40, 364–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimura K, Ura K, Shiratori H, Ikawa M, Okabe M, Schwartz RJ, Kaneda Y. (2009).A histone H3 lysine 36 trimethyltransferase links Nkx2–5 to Wolf-Hirschhorn syndrome. Nature, 460, 287–91 [DOI] [PubMed] [Google Scholar]
- Hartlerode AJ, Guan Y, Rajendran A, Ura K, Schotta G, Xie A, Shah JV, Scully R. (2012).Impact of histone H4 lysine 20 methylation on 53BP1 responses to chromosomal double strand breaks. PloS one, 7, e49211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J, Allali-Hassani A, Nady N, Qi C, Ouyang H, Liu Y, MacKenzie F, Vedadi M, Arrowsmith CH. (2007).L3MBTL1 recognition of mono- and dimethylated histones. Nature structural & molecular biology, 14, 1229–30 [DOI] [PubMed] [Google Scholar]
- Lee J, Thompson JR, Botuyan MV, Mer G. (2008).Distinct binding modes specify the recognition of methylated histones H3K4 and H4K20 by JMJD2A-tudor. Nature structural & molecular biology, 15, 109–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acs K, Luijsterburg MS, Ackermann L, Salomons FA, Hoppe T, Dantuma NP. (2011).The AAA-ATPase VCP/p97 promotes 53BP1 recruitment by removing L3MBTL1 from DNA double-strand breaks. Nature structural & molecular biology, 18, 1345–50 [DOI] [PubMed] [Google Scholar]
- Hsiao KY, Mizzen CA. (2013).Histone H4 deacetylation facilitates 53BP1 DNA damage signaling and double-strand break repair. Journal of molecular cell biology, 5, 157–65 [DOI] [PubMed] [Google Scholar]
- Schotta G, Sengupta R, Kubicek S, Malin S, Kauer M, Callen E, Celeste A, Pagani M, Opravil S, De La Rosa-Velazquez IA, Espejo A, Bedford MT, Nussenzweig A, Busslinger M, Jenuwein T. (2008).A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes & development, 22, 2048–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orthwein A, Fradet-Turcotte A, Noordermeer SM, Canny MD, Brun CM, Strecker J, Escribano-Diaz C, Durocher D. (2014).Mitosis inhibits DNA double-strand break repair to guard against telomere fusions. Science, 344, 189–93 [DOI] [PubMed] [Google Scholar]
- Goldstein G, Scheid M, Hammerling U, Schlesinger DH, Niall HD, Boyse EA. (1975).Isolation of a polypeptide that has lymphocyte-differentiating properties and is probably represented universally in living cells. Proceedings of the National Academy of Sciences of the United States of America, 72, 11–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM. (2001).Mechanisms underlying ubiquitination. Annual review of biochemistry, 70, 503–33 [DOI] [PubMed] [Google Scholar]
- Schulman BA, Harper JW. (2009).Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nature reviews Molecular cell biology, 10, 319–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swatek KN, Komander D. (2016).Ubiquitin modifications. Cell Res, 26, 399–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akutsu M, Dikic I, Bremm A. (2016).Ubiquitin chain diversity at a glance. J Cell Sci, 129, 875–80 [DOI] [PubMed] [Google Scholar]
- Hunt LT, Dayhoff MO. (1977).Amino-terminal sequence identity of ubiquitin and the nonhistone component of nuclear protein A24. Biochemical and biophysical research communications, 74, 650–5 [DOI] [PubMed] [Google Scholar]
- Goldknopf IL, Busch H. (1977).Isopeptide linkage between nonhistone and histone 2A polypeptides of chromosomal conjugate-protein A24. Proceedings of the National Academy of Sciences of the United States of America, 74, 864–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jones RS. (2004).Hierarchical recruitment of polycomb group silencing complexes. Molecular cell, 14, 637–46 [DOI] [PubMed] [Google Scholar]
- Li Z, Cao R, Wang M, Myers MP, Zhang Y, Xu RM. (2006).Structure of a Bmi-1-Ring1B polycomb group ubiquitin ligase complex. The Journal of biological chemistry, 281, 20643–9 [DOI] [PubMed] [Google Scholar]
- Gao Z, Zhang J, Bonasio R, Strino F, Sawai A, Parisi F, Kluger Y, Reinberg D. (2012).PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Molecular cell, 45, 344–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwald G, van der Stoop P, Weichenrieder O, Perrakis A, van Lohuizen M, Sixma TK. (2006).Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1b. The EMBO journal, 25, 2465–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorslund T, Ripplinger A, Hoffmann S, Wild T, Uckelmann M, Villumsen B, Narita T, Sixma TK, Choudhary C, Bekker-Jensen S, Mailand N. (2015).Histone H1 couples initiation and amplification of ubiquitin signalling after DNA damage. Nature, 527, 389–93 [DOI] [PubMed] [Google Scholar]
- Schmid JA, Berti M, Walser F, Raso MC, Schmid F, Krietsch J, Stoy H, Zwicky K, Ursich S, Freire R, Lopes M, Penengo L. (2018).Histone Ubiquitination by the DNA Damage Response Is Required for Efficient DNA Replication in Unperturbed S Phase. Molecular cell, 71, 897–910 e8 [DOI] [PubMed] [Google Scholar]
- Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, Livingston DM, Greenberg RA. (2007).RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science, 316, 1198–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Scully R, Sobhian B, Xie A, Shestakova E, Livingston DM. (2011).RAP80-directed tuning of BRCA1 homologous recombination function at ionizing radiation-induced nuclear foci. Genes & development, 25, 685–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman KA, Greenberg RA. (2011).The BRCA1-RAP80 complex regulates DNA repair mechanism utilization by restricting end resection. The Journal of biological chemistry, 286, 13669–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung JW, Makharashvili N, Agarwal P, Chiu LY, Pourpre R, Cammarata MB, Cannon JR, Sherker A, Durocher D, Brodbelt JS, Paull TT, Miller KM. (2017).ZMYM3 regulates BRCA1 localization at damaged chromatin to promote DNA repair. Genes & development, 31, 260–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilas CK, Emery LE, Denchi EL, Miller KM. (2018).Caught with One’s Zinc Fingers in the Genome Integrity Cookie Jar. Trends in genetics : TIG, 34, 313–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada S, Tai I, Panier S, Al-Hakim A, Iemura S, Juang YC, O’Donnell L, Kumakubo A, Munro M, Sicheri F, Gingras AC, Natsume T, Suda T, Durocher D. (2010).Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature, 466, 941–6 [DOI] [PubMed] [Google Scholar]
- Sharma N, Zhu Q, Wani G, He J, Wang QE, Wani AA. (2014).USP3 counteracts RNF168 via deubiquitinating H2A and gammaH2AX at lysine 13 and 15. Cell cycle, 13, 106–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Wang J, Chen J. (2010).The Lys63-specific deubiquitinating enzyme BRCC36 is regulated by two scaffold proteins localizing in different subcellular compartments. The Journal of biological chemistry, 285, 30982–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosbech A, Lukas C, Bekker-Jensen S, Mailand N. (2013).The deubiquitylating enzyme USP44 counteracts the DNA double-strand break response mediated by the RNF8 and RNF168 ubiquitin ligases. The Journal of biological chemistry, 288, 16579–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zhang H, Liu J, Cheruiyot A, Lee JH, Ordog T, Lou Z, You Z, Zhang Z. (2016).USP51 deubiquitylates H2AK13,15ub and regulates DNA damage response. Genes & development, 30, 946–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Typas D, Luijsterburg MS, Wiegant WW, Diakatou M, Helfricht A, Thijssen PE, van den Broek B, Mullenders LH, van Attikum H. (2015).The de-ubiquitylating enzymes USP26 and USP37 regulate homologous recombination by counteracting RAP80. Nucleic acids research, 43, 6919–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Zang W, Tang Z, Ji Y, Xu R, Yang Y, Luo A, Hu B, Zhang Z, Liu Z, Zheng X. (2018).A20/TNFAIP3 Regulates the DNA Damage Response and Mediates Tumor Cell Resistance to DNA-Damaging Therapy. Cancer research, 78, 1069–82 [DOI] [PubMed] [Google Scholar]
- Delgado-Diaz MR, Martin Y, Berg A, Freire R, Smits VA. (2014).Dub3 controls DNA damage signalling by direct deubiquitination of H2AX. Molecular oncology, 8, 884–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Liu K, Mao Z, Luo J, Gu W, Zhao W. (2016).USP11 Is a Negative Regulator to gammaH2AX Ubiquitylation by RNF8/RNF168. The Journal of biological chemistry, 291, 959–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. (1999).RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proceedings of the National Academy of Sciences of the United States of America, 96, 11364–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LC, Wang ZW, Tsan JT, Spillman MA, Phung A, Xu XL, Yang MC, Hwang LY, Bowcock AM, Baer R. (1996).Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nature genetics, 14, 430–40 [DOI] [PubMed] [Google Scholar]
- Kalb R, Mallery DL, Larkin C, Huang JT, Hiom K. (2014).BRCA1 is a histone-H2A-specific ubiquitin ligase. Cell reports, 8, 999–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Densham RM, Garvin AJ, Stone HR, Strachan J, Baldock RA, Daza-Martin M, Fletcher A, Blair-Reid S, Beesley J, Johal B, Pearl LH, Neely R, Keep NH, Watts FZ, Morris JR. (2016).Human BRCA1-BARD1 ubiquitin ligase activity counteracts chromatin barriers to DNA resection. Nature structural & molecular biology, 23, 647–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uckelmann M, Densham RM, Baas R, Winterwerp HHK, Fish A, Sixma TK, Morris JR. (2018).USP48 restrains resection by site-specific cleavage of the BRCA1 ubiquitin mark from H2A. Nature communications, 9, 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. (2010).ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell, 141, 970–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. (2004).Role of histone H2A ubiquitination in Polycomb silencing. Nature, 431, 873–8 [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V. (2013).A new world of Polycombs: unexpected partnerships and emerging functions. Nature reviews Genetics, 14, 853–64 [DOI] [PubMed] [Google Scholar]
- Ginjala V, Nacerddine K, Kulkarni A, Oza J, Hill SJ, Yao M, Citterio E, van Lohuizen M, Ganesan S. (2011).BMI1 is recruited to DNA breaks and contributes to DNA damage-induced H2A ubiquitination and repair. Molecular and cellular biology, 31, 1972–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail IH, Andrin C, McDonald D, Hendzel MJ. (2010).BMI1-mediated histone ubiquitylation promotes DNA double-strand break repair. The Journal of cell biology, 191, 45–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez A, De Vivo A, Uprety N, Kim J, Stevens SM Jr., Kee Y (2016).BMI1-UBR5 axis regulates transcriptional repression at damaged chromatin. Proceedings of the National Academy of Sciences of the United States of America, 113, 11243–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ui A, Nagaura Y, Yasui A. (2015).Transcriptional elongation factor ENL phosphorylated by ATM recruits polycomb and switches off transcription for DSB repair. Molecular cell, 58, 468–82 [DOI] [PubMed] [Google Scholar]
- Kakarougkas A, Ismail A, Chambers AL, Riballo E, Herbert AD, Kunzel J, Lobrich M, Jeggo PA, Downs JA. (2014).Requirement for PBAF in transcriptional repression and repair at DNA breaks in actively transcribed regions of chromatin. Molecular cell, 55, 723–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rona G, Roberti D, Yin Y, Pagan JK, Homer H, Sassani E, Zeke A, Busino L, Rothenberg E, Pagano M. (2018).PARP1-dependent recruitment of the FBXL10-RNF68-RNF2 ubiquitin ligase to sites of DNA damage controls H2A.Z loading. eLife, 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail IH, Davidson R, Gagne JP, Xu ZZ, Poirier GG, Hendzel MJ. (2014).Germline mutations in BAP1 impair its function in DNA double-strand break repair. Cancer research, 74, 4282–94 [DOI] [PubMed] [Google Scholar]
- Yu H, Pak H, Hammond-Martel I, Ghram M, Rodrigue A, Daou S, Barbour H, Corbeil L, Hebert J, Drobetsky E, Masson JY, Di Noia JM, Affar el B. (2014).Tumor suppressor and deubiquitinase BAP1 promotes DNA double-strand break repair. Proceedings of the National Academy of Sciences of the United States of America, 111, 285–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daou S, Hammond-Martel I, Mashtalir N, Barbour H, Gagnon J, Iannantuono NV, Nkwe NS, Motorina A, Pak H, Yu H, Wurtele H, Milot E, Mallette FA, Carbone M, Affar el B. (2015).The BAP1/ASXL2 Histone H2A Deubiquitinase Complex Regulates Cell Proliferation and Is Disrupted in Cancer. The Journal of biological chemistry, 290, 28643–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahtoe DD, van Dijk WJ, Ekkebus R, Ovaa H, Sixma TK. (2016).BAP1/ASXL1 recruitment and activation for H2A deubiquitination. Nature communications, 7, 10292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan MR, Peng G, Hung WC, Lin SY. (2011).Monoubiquitination of H2AX protein regulates DNA damage response signaling. The Journal of biological chemistry, 286, 28599–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CY, Kang HY, Yang WL, Wu J, Jeong YS, Wang J, Chan CH, Lee SW, Zhang X, Lamothe B, Campos AD, Darnay BG, Lin HK. (2011).Critical role of monoubiquitination of histone H2AX protein in histone H2AX phosphorylation and DNA damage response. The Journal of biological chemistry, 286, 30806–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung JW, Agarwal P, Canny MD, Gong F, Robison AD, Finkelstein IJ, Durocher D, Miller KM. (2014).Nucleosome acidic patch promotes RNF168- and RING1B/BMI1-dependent H2AX and H2A ubiquitination and DNA damage signaling. PLoS genetics, 10, e1004178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankotai T, Bonhomme C, Chen D, Soutoglou E. (2012).DNAPKcs-dependent arrest of RNA polymerase II transcription in the presence of DNA breaks. Nature structural & molecular biology, 19, 276–82 [DOI] [PubMed] [Google Scholar]
- Moyal L, Lerenthal Y, Gana-Weisz M, Mass G, So S, Wang SY, Eppink B, Chung YM, Shalev G, Shema E, Shkedy D, Smorodinsky NI, van Vliet N, Kuster B, Mann M, Ciechanover A, Dahm-Daphi J, Kanaar R, Hu MC, Chen DJ, Oren M, Shiloh Y. (2011).Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Molecular cell, 41, 529–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S, Haddad D, Li C, Le MX, Ling AK, So CC, Nepal RM, Gommerman JL, Yu K, Ketela T, Moffat J, Martin A. (2016).The SAGA Deubiquitination Module Promotes DNA Repair and Class Switch Recombination through ATM and DNAPK-Mediated gammaH2AX Formation. Cell reports, 15, 1554–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. (2008).Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature, 453, 812–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinty RK, Kohn M, Chatterjee C, Chiang KP, Pratt MR, Muir TW. (2009).Structure-activity analysis of semisynthetic nucleosomes: mechanistic insights into the stimulation of Dot1L by ubiquitylated histone H2B. ACS Chem Biol, 4, 958–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee C, McGinty RK, Fierz B, Muir TW. (2010).Disulfide-directed histone ubiquitylation reveals plasticity in hDot1L activation. Nature chemical biology, 6, 267–9 [DOI] [PubMed] [Google Scholar]
- Worden EJ, Hoffmann NA, Hicks CW, Wolberger C. (2019).Mechanism of Cross-talk between H2B Ubiquitination and H3 Methylation by Dot1L. Cell, 176, 1490–501 e12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor HF, Lyon N, Leung JW, Agarwal P, Swaim CD, Miller KM, Huibregtse JM. (2015).Ubiquitin-Activated Interaction Traps (UBAITs) identify E3 ligase binding partners. EMBO reports, 16, 1699–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Ayrapetov MK, Xu C, Gursoy-Yuzugullu O, Hu Y, Price BD. (2012).Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Molecular cell, 48, 723–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BJ, Chan DW, Jung SY, Chen Y, Qin J, Wang Y. (2017).The Histone Variant MacroH2A1 Is a BRCA1 Ubiquitin Ligase Substrate. Cell reports, 19, 1758–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q, Dutt S, Xu R, Graves K, Juszczynski P, Manis JP, Shipp MA. (2009).BBAP monoubiquitylates histone H4 at lysine 91 and selectively modulates the DNA damage response. Molecular cell, 36, 110–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T, Chen Y, Zhang F, Yang CY, Wang S, Yu X. (2013).RNF111-dependent neddylation activates DNA damage-induced ubiquitination. Molecular cell, 49, 897–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin B, Yu J, Nowsheen S, Wang M, Tu X, Liu T, Li H, Wang L, Lou Z. (2019).UFL1 promotes histone H4 ufmylation and ATM activation. Nature communications, 10, 1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gong Y, Peng B, Shi R, Fan D, Zhao H, Zhu M, Zhang H, Lou Z, Zhou J, Zhu WG, Cong YS, Xu X. (2019).MRE11 UFMylation promotes ATM activation. Nucleic acids research [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanty Y, Belotserkovskaya R, Coates J, Polo S, Miller KM, Jackson SP. (2009).Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature, 462, 935–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JR, Boutell C, Keppler M, Densham R, Weekes D, Alamshah A, Butler L, Galanty Y, Pangon L, Kiuchi T, Ng T, Solomon E. (2009).The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature, 462, 886–90 [DOI] [PubMed] [Google Scholar]
- Shiio Y, Eisenman RN. (2003).Histone sumoylation is associated with transcriptional repression. Proceedings of the National Academy of Sciences of the United States of America, 100, 13225–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuto A, Ikura M, Ikura T, Sun J, Horikoshi Y, Shima H, Igarashi K, Kusakabe M, Harata M, Horikoshi N, Kurumizaka H, Kiuchi Y, Tashiro S. (2018).SUMO modification system facilitates the exchange of histone variant H2A.Z-2 at DNA damage sites. Nucleus, 9, 87–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalocsay M, Hiller NJ, Jentsch S. (2009).Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Molecular cell, 33, 335–43 [DOI] [PubMed] [Google Scholar]
- Clouaire T, Legube G. (2019).A Snapshot on the Cis Chromatin Response to DNA Double-Strand Breaks. Trends in genetics : TIG [DOI] [PubMed] [Google Scholar]
- Agarwal P, Miller KM. (2016).The nucleosome: orchestrating DNA damage signaling and repair within chromatin. Biochemistry and cell biology = Biochimie et biologie cellulaire, 94, 381–95 [DOI] [PubMed] [Google Scholar]
- Marnef A, Legube G. (2017).Organizing DNA repair in the nucleus: DSBs hit the road. Current opinion in cell biology, 46, 1–8 [DOI] [PubMed] [Google Scholar]
- Dawson MA. (2017).The cancer epigenome: Concepts, challenges, and therapeutic opportunities. Science, 355, 1147–52 [DOI] [PubMed] [Google Scholar]
- Dawson MA, Kouzarides T. (2012).Cancer epigenetics: from mechanism to therapy. Cell, 150, 12–27 [DOI] [PubMed] [Google Scholar]
- Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, Wong KK, Brandstetter K, Wittner B, Ramaswamy S, Classon M, Settleman J. (2010).A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell, 141, 69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri I, Cannizzaro E, Dawson MA. (2013).Bromodomains as therapeutic targets in cancer. Briefings in functional genomics [DOI] [PubMed] [Google Scholar]
- Di Cerbo V, Schneider R. (2013).Cancers with wrong HATs: the impact of acetylation. Briefings in functional genomics [DOI] [PubMed] [Google Scholar]
- Groselj B, Sharma NL, Hamdy FC, Kerr M, Kiltie AE. (2013).Histone deacetylase inhibitors as radiosensitisers: effects on DNA damage signalling and repair. British journal of cancer, 108, 748–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor MJ. (2015).Targeting the DNA Damage Response in Cancer. Molecular cell, 60, 547–60 [DOI] [PubMed] [Google Scholar]
- Hojfeldt JW, Agger K, Helin K. (2013).Histone lysine demethylases as targets for anticancer therapy. Nature reviews Drug discovery, 12, 917–30 [DOI] [PubMed] [Google Scholar]
- Pfister SX, Ashworth A. (2017).Marked for death: targeting epigenetic changes in cancer. Nature reviews Drug discovery, 16, 241–63 [DOI] [PubMed] [Google Scholar]
- Rodriguez R, Miller KM. (2014).Unravelling the genomic targets of small molecules using high-throughput sequencing. Nature reviews Genetics, 15, 783–96 [DOI] [PubMed] [Google Scholar]
- Lin KY, Kraus WL. (2017).PARP Inhibitors for Cancer Therapy. Cell, 169, 183. [DOI] [PubMed] [Google Scholar]
- Stiff T, O’Driscoll M, Rief N, Iwabuchi K, Lobrich M, Jeggo PA. (2004).ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer research, 64, 2390–6 [DOI] [PubMed] [Google Scholar]
- Ward IM, Chen J. (2001).Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. The Journal of biological chemistry, 276, 47759–62 [DOI] [PubMed] [Google Scholar]
- Downs JA, Lowndes NF, Jackson SP. (2000).A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature, 408, 1001–4 [DOI] [PubMed] [Google Scholar]
- Lee JH, Kang BH, Jang H, Kim TW, Choi J, Kwak S, Han J, Cho EJ, Youn HD. (2015).AKT phosphorylates H3-threonine 45 to facilitate termination of gene transcription in response to DNA damage. Nucleic acids research, 43, 4505–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch T, Florens L, Macdonald WH, Swanson SK, Glaser RL, Yates JR 3rd, Abmayr SM, Washburn MP, Workman JL. (2004).Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science, 306, 2084–7 [DOI] [PubMed] [Google Scholar]
- Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, Chang HY, Bohr VA, Ried T, Gozani O, Chua KF. (2008).SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature, 452, 492–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo R, Chen J, Mitchell DL, Johnson DG. (2011).GCN5 and E2F1 stimulate nucleotide excision repair by promoting H3K9 acetylation at sites of damage. Nucleic acids research, 39, 1390–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan MR, Smerdon MJ. (2014).Histone H3 lysine 14 (H3K14) acetylation facilitates DNA repair in a positioned nucleosome by stabilizing the binding of the chromatin Remodeler RSC (Remodels Structure of Chromatin). The Journal of biological chemistry, 289, 8353–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S, Parthun MR. (2002).Histone H3 and the histone acetyltransferase Hat1p contribute to DNA double-strand break repair. Molecular and cellular biology, 22, 8353–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto H, Hawke D, Kobayashi R, Verreault A. (2005).A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature, 436, 294–8 [DOI] [PubMed] [Google Scholar]
- Barman HK, Takami Y, Ono T, Nishijima H, Sanematsu F, Shibahara K, Nakayama T. (2006).Histone acetyltransferase 1 is dispensable for replication-coupled chromatin assembly but contributes to recover DNA damages created following replication blockage in vertebrate cells. Biochemical and biophysical research communications, 345, 1547–57 [DOI] [PubMed] [Google Scholar]
- Gupta DK, Patra AT, Zhu L, Gupta AP, Bozdech Z. (2016).DNA damage regulation and its role in drug-related phenotypes in the malaria parasites. Sci Rep, 6, 23603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. (2000).Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell, 102, 463–73 [DOI] [PubMed] [Google Scholar]
- Faucher D, Wellinger RJ. (2010).Methylated H3K4, a transcription-associated histone modification, is involved in the DNA damage response pathway. PLoS genetics, 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldeyron C, Soria G, Roche D, Cook AJ, Almouzni G. (2011).HP1alpha recruitment to DNA damage by p150CAF-1 promotes homologous recombination repair. The Journal of cell biology, 193, 81–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luijsterburg MS, Dinant C, Lans H, Stap J, Wiernasz E, Lagerwerf S, Warmerdam DO, Lindh M, Brink MC, Dobrucki JW, Aten JA, Fousteri MI, Jansen G, Dantuma NP, Vermeulen W, Mullenders LH, Houtsmuller AB, Verschure PJ, van Driel R. (2009).Heterochromatin protein 1 is recruited to various types of DNA damage. The Journal of cell biology, 185, 577–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sustackova G, Kozubek S, Stixova L, Legartova S, Matula P, Orlova D, Bartova E. (2012).Acetylation-dependent nuclear arrangement and recruitment of BMI1 protein to UV-damaged chromatin. J Cell Physiol, 227, 1838–50 [DOI] [PubMed] [Google Scholar]
- Horn V, Uckelmann M, Zhang H, Eerland J, Aarsman I, le Paige UB, Davidovich C, Sixma TK, van Ingen H. (2019).Structural basis of specific H2A K13/K15 ubiquitination by RNF168. Nature communications, 10, 1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JB, Qi YK, He QQ, Ai HS, Liu SL, Wang JX, Zheng JS, Liu L, Tian C. (2018).Chemically synthesized histone H2A Lys13 di-ubiquitination promotes binding of 53BP1 to nucleosomes. Cell Res, 28, 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. (2003).Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes & development, 17, 2648–63 [DOI] [PMC free article] [PubMed] [Google Scholar]