Summary
Rapid alkalinization factor (RALF) genes encode for ubiquitous small peptides that stimulate apoplastic alkalinization through interaction with malectin‐like receptor kinase. RALF peptides may act as negative regulators of plant immune response, inhibiting the formation of the signal receptor complex for immune activation. Recently RALF homologues were identified in different fungal pathogen genomes contributing to host infection ability. Here, FaRALF‐33‐like gene expression was evaluated in strawberry fruits inoculated with Colletotrichum acutatum, Botrytis cinerea, or Penicillium expansum after 24 and 48 h post‐infection. To investigate the role of FaRALF‐33‐like in strawberry susceptibility, transient transformation was used to overexpress it in white unripe fruits and silence it in red ripe fruits. Agroinfiltrated fruits were inoculated with C. acutatum and expression, and histological analysis of infection were performed. Silencing of FaRALF‐33‐like expression in C. acutatum‐inoculated red fruits led to a delay in fruit colonization by the fungal pathogen, and infected tissues showed less penetrated infective hyphae than in wild‐type fruits. In contrast, C. acutatum‐inoculated white unripe fruits overexpressing the FaRALF‐33‐like gene decreased the ontogenic resistance of these fruits, leading to the appearance of disease symptoms and penetrated subcuticular hyphae, normally absent in white unripe fruits. The different response of transfected strawberry fruits to C. acutatum supports the hypothesis that the FaRALF‐33‐like gene plays an important role in the susceptibility of fruits to the fungal pathogen C. acutatum.
Keywords: Colletotrichum acutatum, Fragaria × ananassa, fungi, RALF, ripening
Introduction
Plants are naturally provided with very sophisticated and strong immune mechanisms and the development of diseases is generally dependent on the ‘switch‐off’ state of these mechanisms or the ‘switch‐on’ state of susceptibility mechanisms. Besides suppression or plant immunity evasion, pathogens, in particular biotrophs and hemibiotrophs, require cooperation of the host for the establishment of a compatible interaction. Accommodating the pathogen involves enabling it to establish feeding structures inside the host cell to obtain nutrients. All plant genes that facilitate infection and support compatibility can be considered susceptibility genes. Mutation or silencing of a susceptibility gene can therefore limit the ability of the pathogen to cause disease (van Schie and Takken, 2014). Lately, the possibility of controlling susceptibility genes has emerged as a more promising approach to develop resistant crops than the control of resistance genes. However, knowledge of the susceptibility factors present in plants favouring pathogen invasion and/or growth remains limited.
Small secreted peptides have achieved significant relevance as signalling molecules in plants since they play a key role in the regulation of plant defence and development. Among them, the members of the rapid alkalinization factor (RALF) family were first identified through their ability to trigger a rapid increase in extracellular pH when added to plant cell suspensions (Pearce et al., 2001). RALF peptides have been linked to several physiological and developmental processes, including cell expansion (Haruta et al., 2014), lateral root development (Murphy et al., 2016), root hair growth (Wu et al., 2007), pollen tube elongation (Covey et al., 2010), nodulation in legumes (Djordjevic et al., 2015) and stress (Atkinson et al., 2013). In silico analysis of the Arabidopsis thaliana genome identified 37 genes belonging to the RALF family, named RALF1 to RALF37 (Olsen et al., 2002), and the homologues identified in other species have been named according to their similarities to the members of the A. thaliana family (Campbell and Turner, 2017). The RALF pleiotropic role indicates that these peptides are key players in plant development. Dobón et al. (2015) showed that zfp2, bhlh99, pap2 and At1g66810 Arabidopsis mutants, which are susceptible to Botrytis cinerea and Plectosphaerella cucumerina, share a common transcriptional signature of 77 up‐regulated genes, including RALF. These genes function as positive regulators of disease susceptibility, and their expression is induced during the course of a pathogen infection.
Colletotrichum acutatum, the hemibiotrophic agent of anthracnose disease, causes severe economic losses to strawberry (F. × ananassa) production all over the world. The heaviest economic losses are the result of fruit infections, which can occur on either immature fruits preharvest or mature fruits at harvest or in the postharvest storage stage. In all cases, however, anthracnose symptoms become apparent only during storage or shelf life, when fruit production reaches its highest value. This is because of the ability of Colletotrichum species to develop latent infections on immature fruits, becoming quiescent until fruit ripens (Prusky, 1996). On white fruits, C. acutatum becomes quiescent as melanized appressoria after 24 h of interaction (Guidarelli et al., 2011). Quiescence is a well‐known phenomenon among fruit fungal pathogens. Colletorichum acutatum is one of the most common species causing quiescent infections, developing distinct appressoria which are melanized in the immature state of the fruits (Emmett and Parbery, 1975). Appressoria formation, while enabling penetration of the host, is key for fungus survival under unfavourable conditions (Adikaram et al., 2015). Therefore, the growth arrest as melanized appressoria is a hallmark of the dormant state of the fungus (Guidarelli et al., 2011). Only when fruit ripens to the red stage the pathogen restores its growth and symptoms become apparent within 3 days.
Guidarelli et al. (2011) found that a RALF gene, a homologue to AtRALF33, is up‐regulated after 24 h of C. acutatum inoculation in red ripe strawberry fruits, where the pathogen is active, whereas no difference in the expression of this gene was found in inoculated white unripe fruits.
Here, we aimed to analyse the putative role as a susceptibility gene of FaRALF33‐like gene in anthracnose development in strawberry after C. acutatum inoculation. To test whether RALF up‐regulation also occurs in fungal infections other than Colletotricum, the FaRALF‐33‐like expression in white unripe and red ripe fruits inoculated with C. acutatum, Botrytis cinerea, or Penicillium expansum was analysed at 24 and 48 h post‐inoculation (hpi). Thereafter, Agrobacterium tumefaciens‐mediated transient transformation was used to silence FaRALF‐33‐like gene in red ripe strawberry fruits and, in parallel, to overexpress this gene in white unripe fruits, all inoculated with C. acutatum. Anthracnose symptoms of transfected white and red strawberries inoculated with C. acutatum were monitored following histological analysis of the infected tissues. The results indicate that this gene influences the appearance of disease symptoms in strawberry fruits, suggesting an important role for FaRALF‐33‐like gene in the different susceptibility of these fruits to the pathogen.
Results
FaRALF‐33‐like expression was up‐regulated in red ripe strawberry fruits at 24 h on C. acutatum or B. cinerea inoculation, whereas no difference was observed on P. expansum inoculation
The full‐length cDNA of FaRALF33‐like gene previously found to be up‐regulated in 24 h Colletotrichum acutatum‐inoculated red Fragaria × ananassa fruits was isolated by PCR using primers designed on the FvRALF‐33‐like (GenBank accession number: RRID:XM_011460413.1). FaRALF‐33‐like cDNA is 345 bp long and codes for a protein of 114 amino acids. Peptide sequence alignment with the homologous proteins AtRALF33 and FvRALF‐33‐like highlights typical sequence motifs present in known RALF peptides, with an N‐terminal signal peptide for cell secretion and the proteolytic cleavage site motif RRILA for the pro‐peptide processing by the subtilisin‐like protease and release of mature peptide (Supplementary Fig. S1). According to Campell and Turner (2017) RALF sequence classifications FaRALF33‐like belongs to the clade I subfamily of RALF peptides, since its mature form contains the YISY, YYNC and CRC motifs typical of this clade (Supplementary Fig. S1).
The FaRALF‐33‐like gene is up‐regulated 24 h post C. acutatum inoculation (hpi) in red ripe strawberries fruits, where the pathogen is active (Guidarelli et al., 2011). No difference in the expression of this gene was found in inoculated white fruits. Moreover, previous histological studies showed that at 16 hpi most conidia germinated on both white unripe and red ripe fruit stages, and only at 24 hpi, C. acutatum arrests its growth as a quiescent melanized appressoria in white strawberries, while it develops intercellular hyphae on red fruits (Guidarelli et al., 2011). In order to find out whether the up‐regulation of FaRALF‐33‐like gene is specifically related to the susceptibility of ripe stages or if it also occurs in white unripe fruits at later times of infection or in fruits inoculated with other fungal pathogens, a gene expression analysis was performed in white and red fruits at 24 and 48 hpi with C. acutatum, B. cinerea or P. expansum. FaRALF‐33‐like gene was quantified by quantitative real‐time reverse transcription‐polymerase chain reaction (qRT‐PCR). Consistently with previous findings, a significant increase in the expression level of FaRALF‐33‐like gene was observed at 24 hpi in red ripe C. acutatum‐inoculated fruits when compared to the mock‐inoculated strawberries whereas no difference was detected in white unripe fruits (Fig. 1). This indicates that the expression of this gene is indeed specific to the red ripe stage of the strawberry fruits interacting with actively growing C. acutatum as subcuticular intercellular hyphae, thereby suggesting a possible involvement of this gene in the susceptibility of the red ripe stage. At 48 hpi C. acutatum did not alter the expression level of FaRALF‐33‐like gene in either red ripe fruits or white unripe fruits (Fig. 1).
Figure 1.
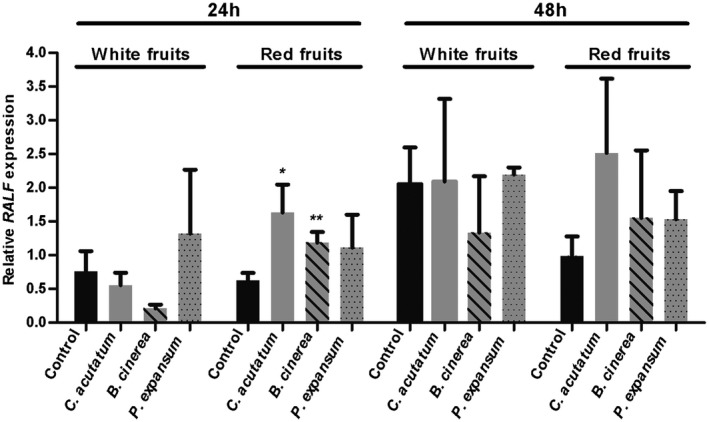
Analysis of gene expression displays an increase in FaRALF‐33‐like gene expression in red ripe strawberry fruits at 24 h after inoculation with Colletotrichum acutatum or Botrytis cinerea, whereas no changes in FaRALF‐33‐like gene expression were observed after inoculation with Penicillium expansum. Expression of FaRALF‐33‐like gene transcripts at 24 h (left) or 48 h (right) after C. acutatum (grey bar), B. cinerea (striped bar) or P. expansum (dotted bar) inoculation in white and red fruits. Expression levels in mock‐inoculated white and red fruits are displayed in black bars. Data were normalized to the transcript level of the housekeeping elongation factor 1α gene. The data are the means and SD of three biological replicates. The asterisks indicate significant difference compared with the control (Student's t‐test: *P < 0.05, **P < 0.01).
When red ripe strawberry fruits were inoculated with B. cinerea, the causal agent of grey mould, FaRALF‐33‐like gene expression increased at 24 hpi, similarly to what was described for C. acutatum‐inoculated fruits, whereas no difference was observed in the white unripe stage at either 24 hpi or 48 hpi (Fig. 1). This may indicate that FaRALF‐33‐like gene participates in the susceptibility of the red ripe fruits against fungal pathogens not only restricted to C. acutatum. In contrast, neither white nor red strawberry fruits increased FaRALF‐33‐like expression at 24 or 48 h after P. expansum inoculation (Fig. 1), the causal agent of blue mould.
Silencing of FaRALF‐33‐like gene in red ripe strawberries did not alter the susceptibility to C. acutatum but decreased the infection process after C. acutatum inoculation
To gain insight in the possible role of FaRALF‐33‐like gene in the different susceptibility to C. acutatum of unripe and ripe strawberry fruits, Agrobacterium transient transformation was used to transiently silence the expression of this gene in red fruits. Fruits were harvested six days post‐agroinfiltration (pai) and inoculated with C. acutatum for 72 h before analysing the fruit response to the anthracnose disease both as visual analysis of symptoms and observation of the fungal infection structure through microscopy analysis. The following conditions were evaluated in order to appreciate the influence of altered FaRALF‐33‐like gene expression in fruit susceptibility: pK7:FaRALF‐33‐like fruits (agroinfiltrated with the silencing vector), pK7:00 (agroinfiltrated with empty vector, as control) and wild‐type non‐agroinfiltrated red strawberries. The silencing FaRALF‐33‐like expression in red fruits is expected to render these fruits more susceptible to C. acutatum infection. The expression of FaRALF‐33‐like gene was significantly induced in both wild‐type and control infiltrated strawberries (pK7:00) at 72 hpi with C. acutatum. The time (72 hpi) was chosen in order to observe the symptoms before sampling for gene expression analysis. Here, RALF gene expression increased by about 5‐ and 4‐fold, respectively. In contrast, in pK7:FaRALF‐33‐like transfected strawberries the induction of FaRALF‐33‐like gene expression was almost abolished (Fig. 2A), indicating that the RNA interference mechanism induced by the expression of RALF self‐complementary hairpin RNA silenced this gene.
Figure 2.
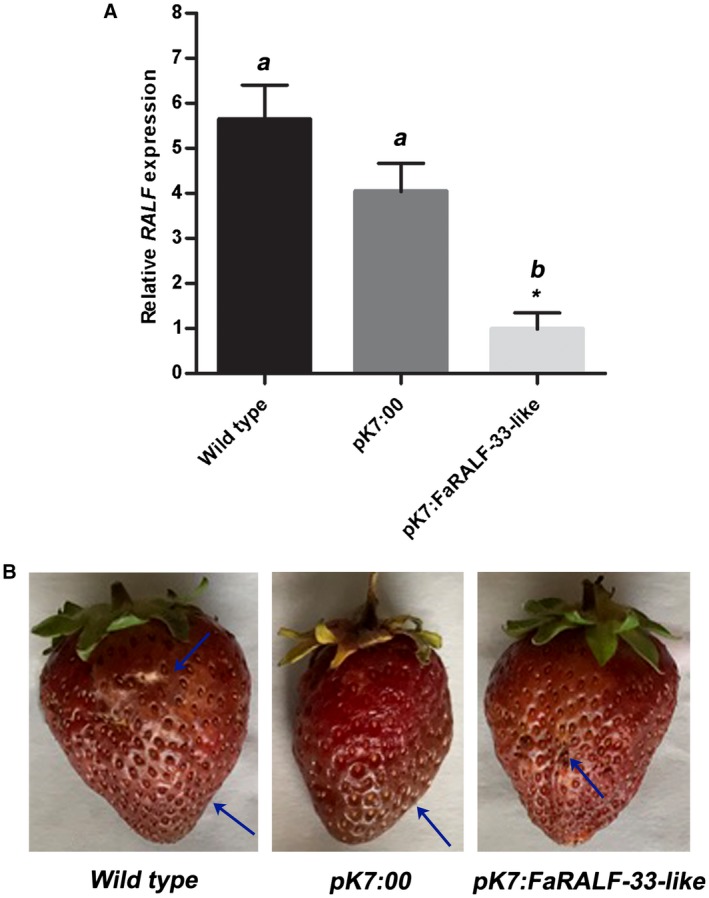
Silencing of FaRALF‐33‐like gene renders red ripe strawberries less susceptible to Colletotrichum acutatum. (A) The transcript levels of FaRALF‐33‐like gene in wild‐type red strawberries were compared with those of mock‐silenced red fruits (pK7:00) and FaRALF‐33‐like‐silenced red fruits (pK7:FaRALF‐33‐like), all inoculated for 72 h with C. acutatum. Amplification of RALF gene untranslated region was normalized to the transcript level of the housekeeping elongation factor 1α gene. The data are the means and SD of three biological replicates. The letters indicate significant difference (Student's t‐test) compared with wild‐type or mock‐treated fruits. The asterisks indicate P < 0.05. (B) Disease symptom analysis in wild‐type infected red fruits (left), mock‐silenced red fruits (pK7:00) (middle) and FaRALF‐33‐like‐silenced red fruits (pK7:FaRALF‐33‐like) (right), all infected for 72 h with C. acutatum. One representative infected red fruit of each condition (wild‐type, pK7:00, pK7:FaRALF‐33‐like) is shown. Arrows indicate anthracnose symptoms.
As regards the fruit phenotype, wild‐type and pK7:00 control agroinfiltrated red strawberries at 72 hpi showed typical anthracnose symptoms with dark and sunken lesions on the fruit surface (Fig. 2B). Symptom development was very similar in the pK7:FaRALF‐33‐like‐silenced fruits, indicating that other mechanisms besides RALF gene induction are involved in disease susceptibility to C. acutatum in red ripe strawberries fruits.
The histological analysis of 48 hpi red fruits revealed the presence of penetrated hyphae underneath the fruit surface in wild‐type and mock‐silenced red fruits. Similar penetrated hyphae were apparent also in FaRALF‐33‐like‐silenced red fruits (Fig. 3A, B and C) although here tissue colonization by Colletotrichum hyphae appeared less deep than in control fruits. To evaluate the fungal growth, the expression of the housekeeping gene β‐tubulin, specific to C. acutatum (Brown et al., 2008), was analysed by qRT‐PCR. In FaRALF‐33‐like‐silenced red fruits the fungus grows similarly to wild‐type red fruits but less than in mock‐silenced red fruits (Fig. 3D).
Figure 3.
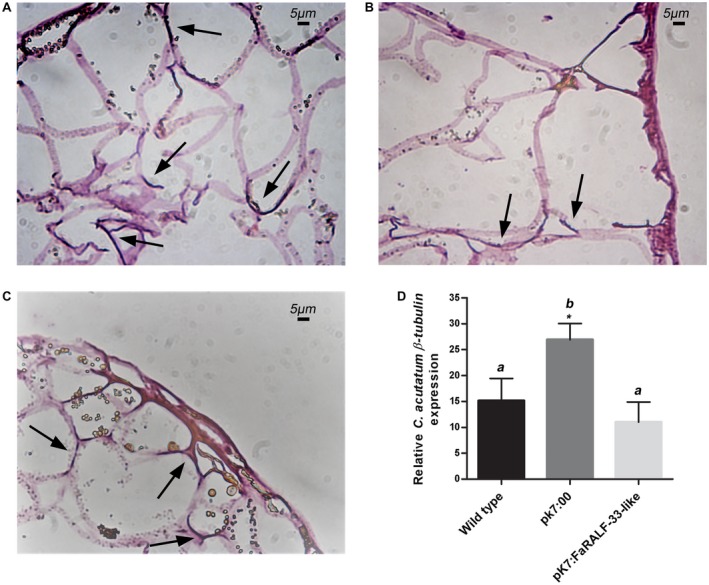
Histological analysis of FaRALF‐33‐like‐silenced 48 h Colletotrichum acutatum‐infected red fruits displays only superficial infection which correlates with C. acutatum similar growth in wild‐type red fruits and in FaRALF‐33‐like‐silenced fruits. Optical microscopy of wild‐type red fruits (A), mock‐silenced red fruits (B) and FaRALF‐33‐like silenced red fruits (C). Tissue slices were stained with haematoxylin and eosin. Inter‐intracellular hyphae are indicated with arrows. Bar: 5 μm. (D) Colletotrichum acutatum β‐tubulin expression in wild‐type red strawberries (black bar) was compared with those of mock‐silenced red fruits (pK7:00, dark grey) and FaRALF‐33‐like‐silenced red fruits (pK7:FaRALF‐33‐like, light grey). The data are the means and SD of three biological replicates. The letters indicate significant difference (Student's t‐test) compared with wild‐type or mock‐treated fruits. The asterisks indicate P < 0.05.
Overexpression of FaRALF‐33‐like gene in white unripe strawberries led to increased susceptibility of fruits to C. acutatum
The overexpression of FaRALF‐33‐like was induced in white unripe strawberries by infiltration for 6 days with Agrobacterium carrying the plasmid 35S:FaRALF‐33‐like. Overexpression in these strawberries was evaluated by qRT‐PCR at 3 dpi (days post‐inoculation) with C. acutatum. As mentioned above, the time (72 h) was chosen in order to observe the symptoms before sampling for gene expression analysis. The expression of FaRALF‐33‐like gene increased 50‐fold with respect to the control white fruit (infected wild‐type or infected 35S:00) (Fig. 4A), indicating that overexpression was efficient and that agroinfiltration itself did not alter FaRALF‐33‐like expression. Seventy‐two hours after C. acutatum inoculation, wild‐type white fruits did not show anthracnose symptoms, and 35S:00 agroinfiltrated control fruits showed light anthracnose symptoms, probably as consequence of agroinfiltration stress (Fig. 4B). On the other hand, symptom development was apparent in 35S:FaRALF‐33‐like white fruits, which were rotten.
Figure 4.
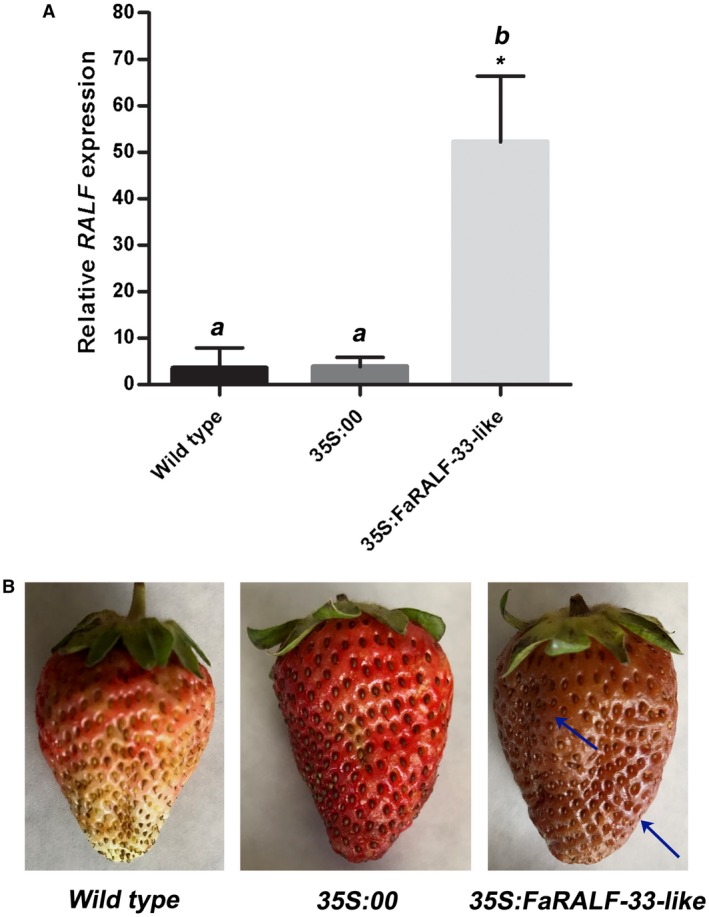
Overexpression of FaRALF‐33‐like gene renders white unripe strawberries more susceptible to Colletotrichum acutatum. (A) The transcript levels of FaRALF‐33‐like gene in wild‐type white strawberries were compared with those of mock‐overexpressing white fruits (35S:00) and FaRALF‐33‐like‐overexpressing white fruits (35S:FaRALF‐33‐like), all inoculated for 72 h with C. acutatum. The FaRALF‐33‐like primers used were mapped in the RALF untranslated region. Data were normalized to the transcript level of the housekeeping elongation factor 1α gene. The data are the means and SD of three biological replicates. The letters indicate significant difference (Student's t‐test) compared with wild‐type or mock‐treated fruits. The asterisks indicate P < 0.05. (B) Disease symptom analysis in wild‐type white fruits (left), mock‐overexpressing white fruits (35S:00) (middle) and FaRALF‐33‐like‐overexpressing white fruits (35S:FaRALF‐33‐like) (right), all infected for 72 h with C. acutatum. One representative infected red fruit of each condition (wild type, 35S:00, 35S:FaRALF‐33‐like) is shown. Arrows indicate anthracnose symptoms.
The histological analysis of 48 h‐infected unripe tissues showed that C. acutatum internal hyphae were distinguishable in the superficial layer of epidermal cells of all the three types of fruits (Fig. 5A, B and C); however, in white FaRALF‐33‐like‐overexpressing fruits, a higher percentage of penetration events, with deeper internal hyphae than in wild‐type and mock‐overexpressing white fruits, were distinguishable. Additionally, the fungus grew more in FaRALF‐33‐like‐overexpressing white fruits compared to wild‐type and mock‐overexpressing fruits (Fig. 5D), suggesting a higher susceptibility of these fruits to C. acutatum.
Figure 5.
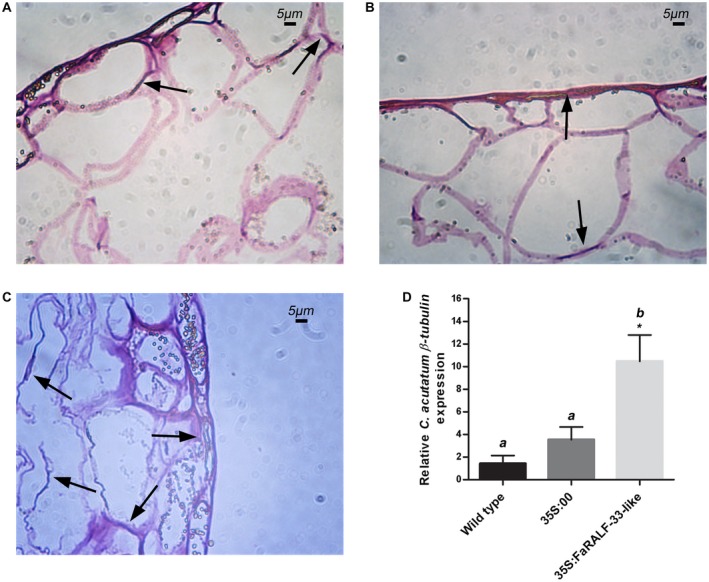
Histological analysis of FaRALF‐33‐like overexpressing 48 h Colletotrichum acutatum‐infected white strawberry fruits shows a high percentage of penetration events which correlates with C. acutatum increased growth in FaRALF‐33‐like‐overexpressing fruits. Optical microscopy of wild‐type white fruits (A), mock‐overexpressing white fruits (B) and FaRALF‐33‐like overexpressing white fruits (C). Tissue slices were stained with haematoxylin and eosin. Inter‐intracellular hyphae are indicated. Bar: 5 μm. (D) Colletotrichum acutatum β‐tubulin expression in wild‐type white strawberries (black bar) was compared with those of mock‐overexpressing white fruits (35S:00, dark grey), and FaRALF‐33‐like‐overexpressing white fruits (35S:FaRALF‐33‐like, light grey). The data are the means and SD of three biological replicates. The letters indicate significant difference (Student's t‐test) compared with wild‐type or mock‐treated fruits. The asterisks indicate P < 0.05.
Silencing or overexpression of FaRALF‐33‐like gene led to a different pattern expression of plant defence genes in strawberry fruits
To gain insight into the different susceptibility to C. acutatum in FaRALF‐33‐like‐silencing or overexpressing fruits, the expression of genes associated with plant defence was evaluated. They consist of a Chitinase gene, a gene encoding for a PR‐10 (pathogenesis‐related protein 10) named Fra a 1E, a polygalacturonase‐inhibiting protein PGIP gene and genes encoding for two transcription factors of the WRKY family, FaWRKY51 and FaWRKY42. These two WRKY genes are homologous to A. thaliana AtWRKY 46 and AtWRKY 33 and here they are named FaWRKY51 and FaWRKY42, respectively, accordingly to the nomenclature proposed by Wei et al. (2016). These genes all have a consolidated function in plant immune response to fungal pathogens (Amil‐Ruiz et al., 2011; De Lorenzo et al., 2001; Jain and Kumar, 2015; Pandey and Somssich, 2009; Sharma et al., 2011) and in our previous microarray analysis were found to be differently regulated by C. acutatum inoculation at 24 hpi in white and red fruits (Guidarelli et al., 2011). In particular, in the microarray analysis, Chitinase and FaWRKY42 were found to be up‐regulated in both white and red fruit upon C. acutatum infection, whereas Fra a E1 and FaWRKY51 were found to be up‐regulated only in white fruits. Here, in FaRALF‐33‐like‐silenced red fruits the levels of transcripts of Chitinase were unaltered with respect to wild‐type fruits, whereas Fra a 1E and FaWRKY42 levels remained similar to the mock‐silenced red fruits control, but lower than the wild‐type, suggesting that agroinfiltration (and not RALF gene silencing) was determining a decrease in expression. On the other hand, PGIP and FaWRKY51 gene expression was decreased (Fig. 6A) with respect to both controls (wild‐type and mock‐silenced fruits), as specific response associated with RALF gene silencing. When FaRALF‐33‐like was overexpressed in white fruits, the expression of Chitinase, Fra a 1E and FaWRKY51 increased compared to wild‐type or mock‐overexpressing white fruits, whereas PGIP and FaWRKY42 did not display any significant differences compared to the mock‐overexpressing control. This could suggest that the white fruits that become more susceptible to C. acutatum by RALF gene overexpression counteract pathogen colonization by increasing the expression of key defence genes.
Figure 6.
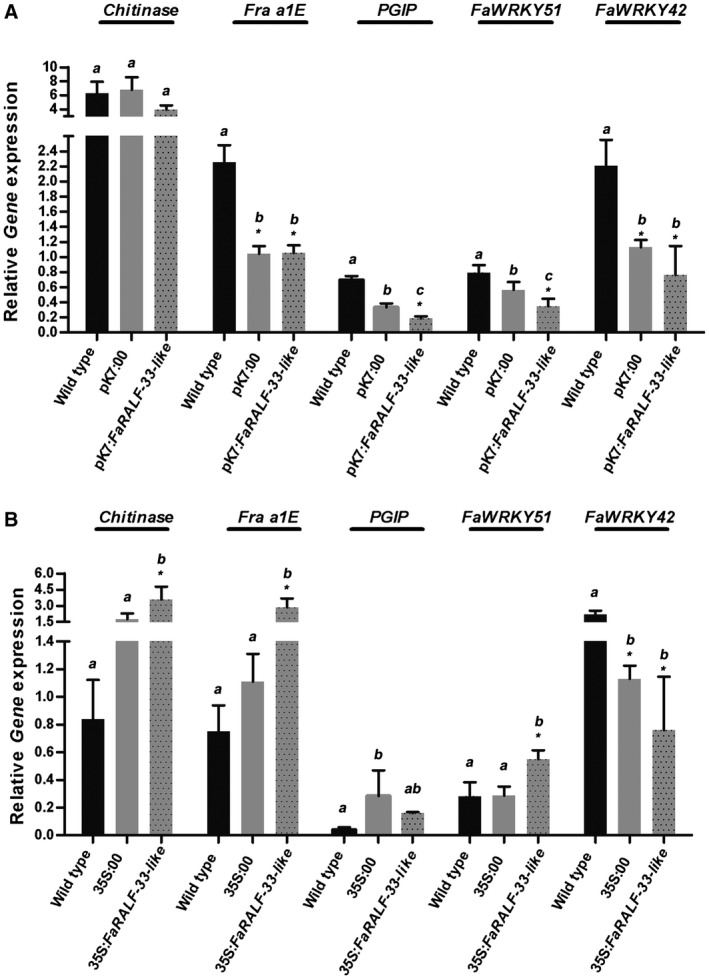
Silencing (A) or overexpression (B) of FaRALF‐33‐like gene triggers a different defence‐related gene expression pattern. (A) The transcript levels of Chitinase, Fra a 1E, PGIP, FaWRKY51 and FaWRKY42 genes in wild‐type red strawberries were compared with those of mock‐silenced red fruits (pK7:00) and FaRALF‐33‐like‐silenced red fruits (pK7:FaRALF‐33‐like), all inoculated for 72 h with Colletotrichum acutatum. (B) The transcript levels of Chitinase, Fra a 1E, PGIP, FaWRKY51 and FaWRKY42 genes in wild‐type white strawberries were compared with those of mock‐overexpressing white fruits (35S:00) and FaRALF‐33‐like‐overexpressing white fruits (pK7:FaRALF‐33‐like), all inoculated for 72 h with C. acutatum. Data were normalized to the transcript level of the housekeeping elongation factor 1α gene. The data are the means and SD of three biological replicates. The letters indicate significant difference (Student's t‐test) compared with wild‐type or mock‐treated fruits. The asterisks indicate P < 0.05.
Discussion
Fruits undergo a transcriptional reprogramming as they ripen. This different expression profile induces substantial metabolic and structural changes, prompting increased susceptibility to many fungal infections (Gapper et al., 2014; Guidarelli et al., 2014; Sanchez‐Sevilla et al., 2017). Colletotrichum spp. are pathogens associated with quiescent infections leading to ripe rots during storage and transport of fruits. In order to reduce fruit losses due to postharvest rotting it is important to elucidate the mechanisms underlying the ontogenic variation of fruit susceptibility during ripening.
During a microarray analysis of 24 h C. acutatum‐inoculated unripe and ripe strawberry fruits a gene encoding for a RALF peptide was found to be up‐regulated only in red ripe susceptible stages (Guidarelli et al., 2011). In different plant species, RALF peptides are encoded by a gene family of different size (37 in A. thaliana, 33 in Malus domestica, 4 in Vitis vinifera and 9 in Fragaria vesca). Depending on the sequence similarity and recognition of typical distinctive motifs in primary sequences, RALF peptides have been divided into four main clades (Campbell and Turner, 2017). The Fragaria × ananassa gene up‐regulated in red fruit on C. acutatum interaction shows high similarity with AtRALF33 and F. vesca FvRALF‐33‐like (GenBank accession number : RRID:XM_011460413.1). Given the presence of a number of aminoacidic hallmark motifs and of the typical RRILA aminoacidic recognition sequence for subtilisin‐like peptidase processing of the mature peptide, RALF33 homologues were classified as clade I members. Subtilisin processing was shown to be necessary for RALF peptide activity (Srivastava et al., 2009). Similarly, the aminoacidic motif YISY found at 5–10 amino acid position in RALF mature peptide was shown to be crucial for binding to RALF receptor and downstream signalling (Pearce et al., 2010).
We have shown that silencing FaRALF‐33‐like expression in 72 h C. acutatum‐inoculated red fruits (Fig. 2) can lead to a delay in fruit colonization by the fungal pathogen, appreciable by histological analysis of the infected tissues showing less penetrated infective hyphae than in wild‐type fruit (Fig. 3 and Supplementary Fig. S4) and a decrease in the expression of the plant defence genes PGIP and FaWRKY51 (Fig. 6A). In contrast, 72 h C. acutatum‐inoculated white unripe fruits overexpressing the FaRALF‐33‐like gene decreased the ontogenic resistance of these fruits, leading to the appearance of disease symptoms (Fig. 4) and deep penetrated subcuticular hyphae (Fig. 5 and Supplementary Fig. S7), which are generally absent in white unripe fruits. The different response of agroinfiltrated strawberry fruits to C. acutatum supports the hypothesis that FaRALF‐33‐like plays an important role in the susceptibility of fruits against the fungal pathogen C. acutatum.
Ambient pH is a key player in fungi growth and development (Penalva and Arst, 2004). Intriguingly, fungal pathogens have the ability to adjust the extracellular pH as a way to facilitate infection by regulating a plethora of virulence factors (Alkan et al., 2013; Jennings, 1989). Secretion of organic acids leads to acidification, a feature of necrotrophic pathogens (Cessna et al., 2000). In contrast, hemibiotrophs induce extracellular alkalinization during the early biotrophic stages of infection (Prusky and Yakoby, 2003). Several fruit‐infecting species, including Colletotrichum spp. (Alkan et al., 2013; Prusky et al., 2001) and the root‐infecting fungus Fusarium oxysporum, have been shown to induce host alkalinization by releasing ammonia. In order to alkalinize the environment surrounding the plant tissue, through ammonia secretion, fungi must develop a significant hyphal biomass (Fernandes et al., 2017). However, due to the low number of hyphae in the early stages of infection, fungal pathogens have adapted pH regulatory mechanisms present in the hosts. Fusarium oxysporum displays a functional homologue of the plant RALF peptides and uses it to enhance its own infectious potential by mimicking plant RALF‐induced alkalinization (Masachis et al., 2016). A plausible explanation for the susceptibility role of RALF against C. acutatum in red ripe strawberry fruits is the fact that its expression favours fungal pathogenicity by increasing the extracellular pH in the first stages of strawberry fruit colonization, after breaking the quiescence state. Consistently, we observed that C. acutatum‐inoculated red ripe fruits showed an increased expression of FaRALF‐33‐like compared to the mock‐inoculated fruits at 24 hpi (Fig. 1). In contrast, the expression level of 48 h C. acutatum‐inoculated red ripe fruits is similar to the mock‐inoculated fruits. Colletotrichum acutatum, which is probably not able to secrete ammonia at early times post‐infection, would induce RALF expression in susceptible red ripe fruits favouring its pathogenicity. Thus, our results reinforce the hypothesis that fungi not only encode homologues of RALF in their genome (Masachis et al., 2016), but may also hijack the host RALF pathway by prompting RALF expression in plants, as previously shown by Dobón et al. (2015). Further research is needed to elucidate the way in which C. acutatum could induce RALF expression in the host. However, silencing RALF in red ripe fruits while decreasing anthracnose symptoms could not prevent disease onset. Many other pathways, different from RALF signalling, may be involved in red ripe strawberry fruit susceptibility.
On the other hand, in white unripe fruits that do not display an increase in RALF expression after C. acutatum inoculation (Fig. 1), RALF overexpression seems to stimulate fungal growth in a stage when it is normally quiescent and render fruits susceptible to infection (Fig. 4).
FERONIA was identified as the RALF receptor in plants (Haruta et al., 2014). Interestingly, fungal RALF seems to signal through FERONIA as well (Masachis et al., 2016). Besides its role in plant growth and development, signalling through FERONIA has also been implicated in negative regulation of plant defence (Masachis et al., 2016). Feronia knockout mutants express increased levels of several immunity response marker genes (Masachis et al., 2016), and are more resistant to some bacterial and fungal pathogens (Kessler et al., 2010; Masachis et al., 2016). Thus, the enhanced susceptibility to C. acutatum found in white unripe fruits transfected with RALF may also be explained by the increased signalling through FERONIA leading to a decreased defence response against pathogens. This occurs despite the increase in transcript levels of Chitinase, Fra a 1E and Fa WRKY51, displayed by 35S:FaRALF‐33‐like white fruits (Fig. 6B), which could be due to the immune response of white fruit counteracting infection.
Whilst C. acutatum prompts alkalinization to induce extensive gene expression and colonization (Prusky and Yakoby, 2003), P. expansum and B. cinerea are known to induce tissue acidification to cope effectively with the host environment (Manteau et al, 2003; Prusky et al., 2004). Consistently, we observed no difference in RALF expression in P. expansum‐inoculated red or white fruits at either 24 or 48 hpi (Fig. 1). Unexpectedly, B. cinerea‐inoculated red fruits displayed an increased expression of RALF in red ripe fruits at 24 hpi (Fig. 1), similar to what we observed in C. acutatum‐inoculated fruits.
Fungal pathogens have evolved multiple ways to cope efficiently with host defences. Early findings showed that in filamentous fungi the external pH is sensed and transmitted intracellularly by phenylalanine ammonia‐lyase (PAL) leading to the proteolytic cleavage of the transcription factor PacC (Peñalva and Arst, 2002). The active form of PacC regulates pH‐responsive gene expression, including those with similar functions but differential pH‐expression patterns (Tilburn et al., 1995). Despite the need for an acidic environment, B. cinerea may activate the FERONIA pathway through RALF and consequently inhibit immune responses, rendering fruits more susceptible to colonization and infection.
Despite being classified as acidifiers and alkalinizers, depending on their ability to either decrease or increase the pH of the surrounding host tissue, fungal pathogens regulate the environmental pH depending on carbon availability (Bi et al., 2016). Carbon deprivation induces extracellular accumulation of ammonia and alkalinization, whereas carbon excess triggers acidification by releasing gluconic acid (Bi et al., 2016). Therefore, it is also possible that B. cinerea in that stage of infection and host niche (24 hpi, red ripe strawberry fruits) (Fig. 1) senses low levels of carbon, thus increasing the tissue pH by inducing RALF expression.
In summary, our results indicate that FaRALF‐33‐like gene expression plays a key role in fruit immune responses to fungal pathogens. Future research is required to elucidate the host mechanisms governing RALF expression in disease and the possible crosstalk between fungi and hosts in inducing the expression of this gene.
Experimental Procedures
Fungal material
Colletotrichum acutatum was isolated from strawberry fruits of different strawberry cultivars showing severe anthracnose symptoms. Monoconidial cultures were identified by morphological analysis and sequencing of ribosomal DNA internal transcribed spacer (ITS) regions. Isolate maya‐3 was grown on potato dextrose agar (Sigma, St. Louis, MO, USA) at 20 °C for 10 days. Conidial suspensions of pathogen were prepared by washing the colonies with 5 mL of sterile distilled water containing 0.05% (v/v) Tween‐80, quantified with a haemocytometer, and diluted to a concentration of about 106 conidia/mL for the infection trials. Infections with B. cinerea B05.10 and with P. expansum (CRIOF collection) were similarly performed.
Plant material
Fragaria × ananassa ‘Alba’ plants, cultivated in pots in a glasshouse, were used for all experiments. Standard growing conditions were maintained at 20 °C with a 16 h photoperiod.
For the analysis of susceptibility, fruits (three replicates of 20 fruits each) at unripe (20 days after flowering) or ripe (30 days after flowering) stage were inoculated with C. acutatum by dipping for 1 min in a suspension of 106 conidia/mL or distilled water (mock control), and stored at 20 °C and 70% relative humidity (RH) and observed every 24 h. After 3 days, the degree of susceptibility of strawberry fruits to the pathogen was scored as the percentage of infected fruits. The fruit surface was then excised with a clean scalpel and immediately frozen in liquid nitrogen and transferred to −80 °C until use. Total RNA was prepared as described by Lopez‐Gomez and Gomez‐Lim (1992) with minor modifications.
qRT‐PCR
For qRT‐PCR experiments, first‐strand cDNA was synthesized from 1 μg of total RNA in a volume of 20 μL with oligo‐d(T)17 and Superscript III (Invitrogen Life Technology, Carlsbad, CA, USA), following the manufacturer’s instructions. The cDNA concentration in the RT mix was quantified using a ND‐1000 UV spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA), and 1 μg of cDNA was used for qRT‐PCR experiments, employing an MX3000 thermal cycler (Stratagene, La Jolla, CA, USA) and Platinum Sybr‐Green Kit (Invitrogen Life Technology), according to the manufacturer’s instructions. The elongation factor 1α gene, having constitutive expression, was used to normalize raw data and to calculate relative transcript levels. The primer sequences used in qRT‐PCR are displayed in Table S1.
Plasmid construction, Agrobacterium transformation and plant transformation
The full‐length cDNA sequence for the FaRALF‐33‐like gene was amplified from the cDNA pool of C. acutatum 24 h inoculated red strawberry fruit using the primers forward 5ʹ‐ATGGCAAAGTCCTCTTCCATT‐3′ and reverse 5ʹ‐TCAACTACGGCAGCGAGTGAT‐3′. The translated peptide sequence was aligned with the A. thaliana RALF cDNAs using ClustalW software (https://www.ebi.ac.uk/Tools/msa/clustalw2/). The pK7GWIWG2(II) RNAi silencing and pK7WG2 overexpression vectors, described in Karimi et al. (2002) [VIB Department of Plant Systems Biology, Ghent University, Belgium (http://gateway.psb.ugent.be)], were used as destination plasmids for inducing silencing or overexpression in agroinfiltrated fruits. For this, the same pENTR/D‐TOPO (Invitrogen Life Technologies) construct containing the full‐length sequence of the FaRALF33‐like cDNA, amplified with forward primer 5ʹ‐CACCATGGCAAAGTCCTCTTCCATT‐3ʹ and reverse primer 5ʹ‐TCAACTACGGCAGCGAGTGAT‐3ʹ, was used in GATEWAY cloning (Invitrogen Life Technologies) (Supplementary Fig. S8). The plasmid constructs were checked by PCR [CaMV 35S promoter forward primer and attB2 reverse primer (5'‐ACCACTTTGTACAAGAAA‐3')], by digestion using restriction enzymes and by DNA sequencing (Supplementary Fig. S9). The resulting plasmids (pK7:FaRALF‐33‐like for silencing, 35S:FaRALF‐33‐like for overexpression) were introduced into A. tumefaciens strain EHA105 using the freeze–thaw shock method (Holsters et al., 1978). The A. tumefaciens EHA105 containing pK7:RALF33 or 35S:RALF33 were grown at 28 °C in Luria–Bertani (LB) medium with appropriate antibiotics. When the culture reached an optical density at 600 nm (OD600) of about 0.8, Agrobacterium cells were harvested and resuspended in a modified MacConkey (MMA) medium [Murashige and Skoog salts, 10 mM 2‐(N‐morpholino)ethanesulphonic acid (MES), pH 5.6, 20 g/L sucrose and 200 μM acetosyringone], according to Spolaore et al. (2001). After 1 h of incubation at 22 °C, the Agrobacterium suspension was injected into fruits still attached to the plant using a sterile syringe. After 6 days fruits were harvested and inoculated with C. acutatum conidia for 3 days, as described above. Tissues from the surface of the whole fruit were collected and RNA was isolated, as previously described. qRT‐PCR was used to evaluate the FaRALF33‐like transcript level, as described above.
FaRALF‐33‐like silencing and overexpression
For silencing, 44 red strawberry fruits at about 27 days after anthesis (3 days before the ‘red’ stage) were agroinfiltrated with pK7:FaRALF‐33‐like for 6 days. They were harvested and inoculated with C. acutatum, as described above. Twenty four of these fruits were collected 48 h after inoculation for histological analysis of the fungal infection, as described by Guidarelli et al. (2011). The other 20 fruits were phenotypically observed after 3 days of inoculation for anthracnose symptom evaluation and used for FaRALF‐33‐like gene expression analysis to confirm gene silencing. For this, RNA was isolated and qRT‐PCR was performed, as described above. Similarly, for overexpression, 44 white fruits at about 17 days after anthesis (3 days before the ‘white’ stage) were agroinfiltrated with plasmid 35S:FaRALF‐33‐like for silencing for 6 days. They were harvested and inoculated with C. acutatum, as described above. The fruits were collected 48 h after inoculation for histological analysis of the fungal infection, as described by Guidarelli et al. (2011). As controls for silencing and overexpression, the same number (44) of white and red fruits were agroinfiltrated with pK7:00 (the empty silencing vector) and 35S:00 (the empty overexpression vector) plasmids or left in the wild‐type condition. Phenotype observation, gene expression and histological analysis were performed as described above.
Statistics
Results were analysed for statistical significance (defined as P < 0.05 and indicated by asterisks in figures) by performing unpaired, two‐sided Student's t‐test with GraphPad Prism 7 Data Analysis Software (GraphPad Software, Inc., La Jolla, CA, USA). Mean and standard deviation (SD) values were calculated from at least three biologically and technically independent experiments.
Supporting information
Fig. S1 FaRALF33‐like protein sequence. (A) Sequence alignment between Fragaria × ananassa FaRALF33‐like named protein, Arabidopsis thaliana RALF33 (Locus AEE83653) and Fragaria vesca subsp. vesca XP_011458715.1 PREDICTED: protein RALF‐like 33. Proteolytic cleavage site (RRILA) for processing by subtilisin‐like serine protease (S1P) (Srivastava et al., 2009) is highlighted by a grey box. The YISY motif, crucial for receptor binding (Pearce et al., 2010), is underlined. The four conserved cysteine residues for disulphide bond formation are indicated by vertical bars. (B) Schematic representation of FaRALF33‐like sequence, signal peptides until residues 25 (dark grey), the variable region until RRILA processing site (light grey) and the mature peptide (black).
Fig. S2 Histological analysis of wild‐type red fruits. Tissue slices were stained with haematoxylin and eosin. Inter‐intracellular hyphae are indicated. Bar: 5 μm.
Fig. S3 Histological analysis of mock‐silenced red fruits. Tissue slices were stained with haematoxylin and eosin. Inter‐intracellular hyphae are indicated. Bar: 5 μm.
Fig. S4 Histological analysis of FaRALF‐33‐like‐silenced fruits. Tissue slices were stained with haematoxylin and eosin. Inter‐intracellular hyphae are indicated. Bar: 5 μm.
Fig. S5 Histological analysis of wild‐type white fruits. Tissue slices were stained with haematoxylin and eosin. Inter‐intracellular hyphae are indicated. Bar: 5 μm.
Fig. S6 Histological analysis of mock‐overexpressing white fruits. Tissue slices were stained with haematoxylin and eosin. Inter‐intracellular hyphae are indicated. Bar: 5 μm.
Fig. S7 Histological analysis of FaRALF‐33‐like‐overexpressing fruits. Tissue slices were stained with haematoxylin and eosin. Inter‐intracellular hyphae are indicated. Bar: 5 μm.
Fig. S8 (A) pK7GWIWG2(II):RALF silencing construct, referred to in the text as pk7:FaRALF‐33‐like. (B) pK7WG2:RALF overexpressing construct, referred to in the text as 35S:FaRALF‐33‐like. The size of the attB and attR sites, and RALF_CDS and ccdB_CDS are indicated on the right. Primers used for cloning are indicated in violet and the sequences displayed on the right.
Fig. S9 PCR displaying proper plasmid construction. (A) pK7GWIWG(II):RALF, referred to as pK7:FaRALF‐33‐like in the text. (B) pK7WG2:RALF, referred to as 35S:FaRALF‐33‐like in the text.
Table S1 Oligonucleotide primers used for qRT‐PCR.
Acknowledgements
MCM is a member of the scientific career of the National Council of Scientific Research (CONICET, Argentina) and thanks EMBO for the fellowship granted. We thank the whole technical team of Professor Pession for kind help with histological analysis.
References
- Adikaram, N. , Karunanayake, C. , Sinniah, G. , Vithanage, I.K. and Yakandawala, D. (2015) Fungal quiescence in fruit: an attempt to avoid toxic substances? J. Mycopathol. Res. 53, 1–7. [Google Scholar]
- Alkan, N. , Espeso, E.A. and Prusky, D. (2013) Virulence regulation of phytopathogenic fungi by pH. Antioxid. Redox Signal. 19, 1012–1025. [DOI] [PubMed] [Google Scholar]
- Amil‐Ruiz, F. , Blanco‐Portales, R. , Munoz‐Blanco, J. and Caballero, J.L. (2011) The strawberry plant defense mechanism: a molecular review. Plant Cell Physiol. 52, 1873–1903. [DOI] [PubMed] [Google Scholar]
- Atkinson, N.J. , Lilley, C.J. and Urwin, P.E. (2013) Identification of genes involved in the response of Arabidopsis to simultaneous biotic and abiotic stresses. Plant Physiol. 162, 2028–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi, F. , Barad, S. , Ment, D. , Luria, N. , Dubey, A. , Casado, V. , Glam, N. , Minguez, J.D. , Espeso, E.A. , Fluhr, R. and Prusky, D. (2016) Carbon regulation of environmental pH by secreted small molecules that modulate pathogenicity in phytopathogenic fungi. Mol. Plant Pathol. 17, 1178–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, S.H. , Yarden, O. , Gollop, N. , Chen, S. , Zveibil, A. , Belausov, E. and Freeman, S. (2008) Differential protein expression in Colletotrichum acutatum: changes associated with reactive oxygen species and nitrogen starvation implicated in pathogenicity on strawberry. Mol. Plant Pathol. 9, 171–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, L. and Turner, S.R. (2017) A comprehensive analysis of RALF proteins in green plants suggests there are two distinct functional groups. Front. Plant Sci. 8, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cessna, S.G. , Sears, V.E. , Dickman, M.B. and Low, P.S. (2000) Oxalic acid, a pathogenicity factor for Sclerotinia sclerotiorum, suppresses the oxidative burst of the host plant. Plant Cell, 12, 2191–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covey, P.A. , Subbaiah, C.C. , Parsons, R.L. , Pearce, G. , Lay, F.T. , Anderson, M.A. , Ryan, C.A. and Bedinger, P.A. (2010) A pollen‐specific RALF from tomato that regulates pollen tube elongation. Plant Physiol. 153, 703–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lorenzo, G. , D'Ovidio, R. and Cervone, F. (2001) The role of polygalacturonase‐inhibiting proteins (PGIPs) in defense against pathogenic fungi. Annu. Rev. Phytopathol. 39, 313–335. [DOI] [PubMed] [Google Scholar]
- Djordjevic, M.A. , Mohd‐Radzman, N.A. and Imin, N. (2015) Small‐peptide signals that control root nodule number, development, and symbiosis. J. Exp. Bot. 66, 5171–5181. [DOI] [PubMed] [Google Scholar]
- Dobón, A. , Canet, J.V. , García‐Andrade, J. , Angulo, C. , Neumetzler, L. , Persson, S. and Vera, P. (2015) Novel disease susceptibility factors for fungal necrotrophic pathogens in Arabidopsis . PLoS Pathog. 11, e1004800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmett, R.W. and Parbery, D.G. (1975) Appressoria. Annu. Rev. Phytopathol. 13, 147–167. [Google Scholar]
- Fernandes, T.R. , Segorbe, D. , Prusky, D. and Di Pietro, A. (2017) How alkalinization drives fungal pathogenicity. PLoS Pathog. 13, e1006621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gapper, N.E. , Giovannoni, J.J. and Watkins, C.B. (2014) Understanding development and ripening of fruit crops in an 'omics' era. Hortic. Res. 1, 14034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidarelli, M. , Carbone, F. , Mourgues, F. , Perrotta, G. , Rosati, C. , Bertolini, P. and Baraldi, E. (2011) Colletotrichum acutatum interactions with unripe and ripe strawberry fruits and differential responses at histological and transcriptional levels. Plant Pathol. 60, 685–697. [Google Scholar]
- Guidarelli, M. , Zubini, P. , Nanni, V. , Rasori, A. , Bonghi, C. , Bertolini, P. and Baraldi, E. (2014) Gene expression analysis of peach fruit at different growth stages and with different susceptibility to Monilinia laxa . Eur. J. Plant Pathol. 140, 503–513. [Google Scholar]
- Haruta, M. , Sabat, G. , Stecker, K. , Minkoff, B.B. and Sussman, M.R. (2014) A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science, 343, 408–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsters, M. , de Waele, D. , Depicker, A. , Messens, E. , Van Montagu, M. and Schell, J. (1978) Transfection and transformation of Agrobacterium tumefaciens . Mol. Gen. Genet. 163, 181–187. [DOI] [PubMed] [Google Scholar]
- Jain, S. and Kumar, A. (2015) The pathogenesis related class 10 proteins in plant defense against biotic and abiotic stresses. Adv. Plants Agric. Res. 3, 00077. [Google Scholar]
- Jennings, D.H. (1989) Some perspectives on nitrogen and phosphorus metabolism in fungi In: Boddy L., Machant R. and Read D.J. (Eds.) Nitrogen, Phosphorus and Sulphur Utilization by Fungi. Cambridge: Cambridge University Press, pp. 1–31 [Google Scholar]
- Karimi, M. , Inzé, D. and Depicker, A. (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195 [DOI] [PubMed] [Google Scholar]
- Kessler, S.A. , Shimosato‐Asano, H. , Keinath, N.F. , Wuest, S.E. , Ingram, G. , Panstruga, R. and Grossniklaus, U. (2010) Conserved molecular components for pollen tube reception and fungal invasion. Science, 330, 968–971. [DOI] [PubMed] [Google Scholar]
- Lopez‐Gomez, R. and Gomez‐Lim, M. (1992) A method for extracting intact RNA from fruits rich in polysaccharides using ripe mango mesocarp. HortScience, 27, 440–442. [Google Scholar]
- Manteau, S. , Abouna, S. , Lambert, B. and Legendre, L. (2003) Differential regulation by ambient pH of putative virulence factor secretion by the phytopathogenic fungus Botrytis cinerea . FEMS Microbiol. Ecol. 43, 359–366. [DOI] [PubMed] [Google Scholar]
- Masachis, S. , Segorbe, D. , Turra, D. , Leon‐Ruiz, M. , Furst, U. , El Ghalid, M. , Leonard, G. , Lopez‐Berges, M.S. , Richards, T.A. , Felix, G. and Di Pietro, A. (2016) A fungal pathogen secretes plant alkalinizing peptides to increase infection. Nat. Microbiol. 1, 16043. [DOI] [PubMed] [Google Scholar]
- Murphy, E. , Vu, L.D. , Van den Broeck, L. , Lin, Z. , Ramakrishna, P. , van de Cotte, B. , Gaudinier, A. , Goh, T. , Slane, D. , Beeckman, T. , Inze, D. , Brady, S.M. , Fukaki, H. and De Smet, I. (2016) RALFL34 regulates formative cell divisions in Arabidopsis pericycle during lateral root initiation. J. Exp. Bot. 67, 4863–4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, A.N. , Mundy, J. and Skriver, K. (2002) Peptomics, identification of novel cationic Arabidopsis peptides with conserved sequence motifs. Silico. Biol. 2, 441–451. [PubMed] [Google Scholar]
- Pandey, S.P. and Somssich, I.E. (2009) The role of WRKY transcription factors in plant immunity. Plant Physiol. 150, 1648–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce, G. , Moura, D.S. , Stratmann, J. and Ryan, C.A. (2001) RALF, a 5‐kDa ubiquitous polypeptide in plants, arrests root growth and development. Proc. Natl. Acad. Sci. USA. 98, 12843–12847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce, G. , Yamaguchi, Y. , Munske, G. and Ryan, C.A. (2010) Structure‐activity studies of RALF, rapid alkalinization factor, reveal an essential–YISY–motif. Peptides, 31, 1973–1977. [DOI] [PubMed] [Google Scholar]
- Peñalva, M. A. and Arst, H. N. (2002) Regulation of gene expression by ambient pH in filamentous fungi and yeasts. Microbiol. Mol. Biol. Rev. 66, 426–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penalva, M.A. and Arst, H.N., Jr . (2004) Recent advances in the characterization of ambient pH regulation of gene expression in filamentous fungi and yeasts. Annu. Rev. Microbiol. 58, 425–451. [DOI] [PubMed] [Google Scholar]
- Prusky, D. (1996) Pathogen quiescence in postharvest diseases. Annu. Rev. Phytopathol. 34, 413–434. [DOI] [PubMed] [Google Scholar]
- Prusky, D. and Yakoby, N. (2003) Pathogenic fungi: leading or led by ambient pH? Mol. Plant Pathol. 4, 509–516. [DOI] [PubMed] [Google Scholar]
- Prusky, D. , Mcevoy, J.L. , Leverentz, B. and Conway, W.S. (2001) Local modulation of host pH by Colletotrichum species as a mechanism to increase virulence. Mol. Plant‐Microbe Interact. 14, 1105–1113. [DOI] [PubMed] [Google Scholar]
- Prusky, D. , Mcevoy, J.L. , Saftner, R. , Conway, W.S. and Jones, R. (2004) Relationship between host acidification and virulence of Penicillium spp. on apple and citrus fruit. Phytopathology, 94, 44–51. [DOI] [PubMed] [Google Scholar]
- Sanchez‐Sevilla, J.F. , Vallarino, J.G. , Osorio, S. , Bombarely, A. , Pose, D. , Merchante, C. , Botella, M.A. , Amaya, I. and Valpuesta, V. (2017) Gene expression atlas of fruit ripening and transcriptome assembly from RNA‐seq data in octoploid strawberry (Fragaria × ananassa). Sci. Rep. 7, 13737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schie, C.C. and Takken, F.L. (2014) Susceptibility genes 101: how to be a good host. Annu. Rev. Phytopathol. 52, 551–581. [DOI] [PubMed] [Google Scholar]
- Sharma, N. , Sharma, K.P. , Gaur, R.K. and Gupta, V.K. (2011) Role of chitinase in plant defense. Asian J. Biochem. 6, 29–37. [Google Scholar]
- Spolaore, S. , Trainotti, L. and Casadoro, G. (2001) A simple protocol for transient gene expression in ripe fleshy fruit mediated by Agrobacterium. J. Exp. Bot. 52, 845–850. [DOI] [PubMed] [Google Scholar]
- Srivastava, R. , Liu, J.X. , Guo, H. , Yin, Y. and Howell, S.H. (2009) Regulation and processing of a plant peptide hormone, AtRALF23, in Arabidopsis. Plant J. 59, 930–939. [DOI] [PubMed] [Google Scholar]
- Tilburn, J. , Sarkar, S. , Widdick, D.A. , Espeso, E.A. , Orejas, M. , Mungroo, J. and Arst, H.N. Jr . (1995) The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid‐and alkaline‐expressed genes by ambient pH. EMBO J. 14, 779–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, W. , Hu, Y. , Han, Y.T. , Zhang, K. , Zhao, F.L. and Feng, J.Y. (2016) The WRKY transcription factors in the diploid woodland strawberry Fragaria vesca: identification and expression analysis under biotic and abiotic stresses. Plant Physiol. Biochem. 105, 129–144. [DOI] [PubMed] [Google Scholar]
- Wu, J. , Kurten, E.L. , Monshausen, G. , Hummel, G.M. , Gilroy, S. and Baldwin, I.T. (2007) NaRALF, a peptide signal essential for the regulation of root hair tip apoplastic pH in Nicotiana attenuata, is required for root hair development and plant growth in native soils. Plant J. 52, 877–890. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 FaRALF33‐like protein sequence. (A) Sequence alignment between Fragaria × ananassa FaRALF33‐like named protein, Arabidopsis thaliana RALF33 (Locus AEE83653) and Fragaria vesca subsp. vesca XP_011458715.1 PREDICTED: protein RALF‐like 33. Proteolytic cleavage site (RRILA) for processing by subtilisin‐like serine protease (S1P) (Srivastava et al., 2009) is highlighted by a grey box. The YISY motif, crucial for receptor binding (Pearce et al., 2010), is underlined. The four conserved cysteine residues for disulphide bond formation are indicated by vertical bars. (B) Schematic representation of FaRALF33‐like sequence, signal peptides until residues 25 (dark grey), the variable region until RRILA processing site (light grey) and the mature peptide (black).
Fig. S2 Histological analysis of wild‐type red fruits. Tissue slices were stained with haematoxylin and eosin. Inter‐intracellular hyphae are indicated. Bar: 5 μm.
Fig. S3 Histological analysis of mock‐silenced red fruits. Tissue slices were stained with haematoxylin and eosin. Inter‐intracellular hyphae are indicated. Bar: 5 μm.
Fig. S4 Histological analysis of FaRALF‐33‐like‐silenced fruits. Tissue slices were stained with haematoxylin and eosin. Inter‐intracellular hyphae are indicated. Bar: 5 μm.
Fig. S5 Histological analysis of wild‐type white fruits. Tissue slices were stained with haematoxylin and eosin. Inter‐intracellular hyphae are indicated. Bar: 5 μm.
Fig. S6 Histological analysis of mock‐overexpressing white fruits. Tissue slices were stained with haematoxylin and eosin. Inter‐intracellular hyphae are indicated. Bar: 5 μm.
Fig. S7 Histological analysis of FaRALF‐33‐like‐overexpressing fruits. Tissue slices were stained with haematoxylin and eosin. Inter‐intracellular hyphae are indicated. Bar: 5 μm.
Fig. S8 (A) pK7GWIWG2(II):RALF silencing construct, referred to in the text as pk7:FaRALF‐33‐like. (B) pK7WG2:RALF overexpressing construct, referred to in the text as 35S:FaRALF‐33‐like. The size of the attB and attR sites, and RALF_CDS and ccdB_CDS are indicated on the right. Primers used for cloning are indicated in violet and the sequences displayed on the right.
Fig. S9 PCR displaying proper plasmid construction. (A) pK7GWIWG(II):RALF, referred to as pK7:FaRALF‐33‐like in the text. (B) pK7WG2:RALF, referred to as 35S:FaRALF‐33‐like in the text.
Table S1 Oligonucleotide primers used for qRT‐PCR.


