Summary
The intracellular nucleotide‐binding domain leucine‐rich repeat (NLR) class of immune receptors plays an important role in plant viral defence. Plant NLRs recognize viruses through direct or indirect association of viral proteins, triggering a downstream defence response to prevent viral proliferation and movement within the plant. This review focuses on current knowledge of intracellular perception of viral pathogens, activation of NLRs and the downstream signalling components involved in plant viral defence.
Keywords: Effector‐triggered immunity (ETI), Immune receptors, immune signalling, NLR activation, nucleotide‐binding domain leucine‐rich repeat receptors (NLRs), plant viruses, viral effectors
Introduction
Plants have a two‐layered innate immune system to defend against pathogen infections (Caplan et al., 2008b; Jones and Dangl, 2006; Macho and Zipfel, 2014). The first layer consists of extracellular recognition of pathogen‐associated molecular patterns (PAMPs) such as bacterial flagellin or fungal chitin by cell surface‐localized pattern recognition receptors (PRRs), leading to pattern‐triggered immunity (PTI) (Macho and Zipfel, 2014). Highly adapted pathogens breach PTI by injecting pathogen‐encoded proteins called effectors into plant cells. The second layer of the plant immune system consists of intracellular recognition of pathogen effectors by corresponding nucleotide‐binding leucine‐rich repeat (NLR) receptors, leading to effector‐triggered immunity (ETI) (Caplan et al., 2008b; Jones and Dangl, 2006). Both PTI and ETI culminate in a rapid burst of extracellular reactive oxygen species (ROS), change in ion fluxes, increase in cytosolic calcium, activation of mitogen‐activated protein kinases (MAPKs) and calcium‐dependent protein kinases (CPKs), elevation of phytohormones, and transcriptional and proteome reprogramming (Peng et al., 2018; Yu et al., 2017). Amplitude of these responses during ETI is much higher than in PTI and often leads to induction of hypersensitive response (HR), a type of programmed cell death (PCD), at the site of infection that restricts the pathogen to the sites of localized cell death.
Viruses are intracellular obligate pathogens with RNA or DNA genomes and cause significant crop losses (Anderson et al., 2004). Since viruses encode a minimal number of proteins (approximately 5–10), they hijack host cellular machinery in order to replicate their genome, produce the viral capsid protein for encapsidation of its genome, facilitate cell‐to‐cell movement and for long distance transport within the plant. Most of our understanding of the plant antiviral immune response against virus infections comes from studies involving host‐induced RNA interference (RNAi) and the NLR‐mediated ETI response (Caplan et al., 2008b; Guo et al., 2019; Padmanabhan and Dinesh‐Kumar, 2014). Another major form of antiviral defence in plants is through recessive resistance, in which the plant fails to produce factors required for the virus to complete its life cycle (Wang and Krishnaswamy, 2012). Since plant viruses are intracellular pathogens, it is widely accepted that plants would not invoke PTI responses against viral infection. However, there has been some recent indirect evidence for PTI‐like responses against viral infection (Macho and Lozano‐Duran, 2019; Teixeira et al., 2019). In this review, we primarily discuss NLR‐mediated intracellular perception of viruses by plants, as well as some of the downstream signalling pathways that lead to the innate immune response against viral pathogens.
Intracellular perception of viruses by NLRs
Intracellular NLRs play an important role in both plants and animals for the perception of pathogens and activation of defence (Caplan et al., 2008b; Jones and Dangl, 2006). In plants, canonical NLRs are subdivided based on the presence of a Toll‐Interleukin‐1‐receptor (TIR) homology domain or coiled‐coil (CC) domain at the N‐terminus. Unlike mammalian NLRs, which perceive PAMPs including viral nucleic acid, plant NLRs recognize pathogen effector proteins to activate defence responses that often lead to induction of HR at the site of infection and subsequent containment of the pathogen to the infection site. The first TIR‐NLR cloned was the tobacco N gene, which confers resistance to Tobacco mosaic virus (TMV) (Whitham et al., 1994). Since the cloning of N, at least 15 NLRs that confer resistance to various viruses have been cloned or identified, many of which encode CC‐NLRs (Gouveia et al., 2016; Padmanabhan and Dinesh‐Kumar, 2014).
Despite having cloned many NLRs and their corresponding pathogen effectors, experimental evidence for NLR association (direct or indirect) with effectors has only been demonstrated in a few cases. The tomato Sw‐5b, a CC‐NLR, confers broad‐spectrum resistance to a wide range of agriculturally devastating American‐type tospoviruses, including Tomato spotted wilt virus (TSWV) (Brommonschenkel et al., 2000). In addition to the canonical CC‐NLR domains, Sw‐5b contains an extra N‐terminal Solanaceae domain (SD) (Chen et al., 2016). Sw‐5b recognizes the cell‐to‐cell movement protein, NSm, of TSWV (Hallwass et al., 2014; Peiro et al., 2014; Zhu et al., 2017) (Fig. 1). Interestingly, a highly conserved 21 amino acid peptide region within NSm (NSm21) from American‐type tospoviruses is sufficient to induce Sw‐5b mediated defences (Zhu et al., 2017). The NB‐LRR region of Sw‐5b directly interacts with NSm21 (Zhu et al., 2017). Furthermore, domain swap and mutational analyses indicate that the LRR domain of Sw‐5b is involved in the recognition of NSm21. Therefore, Sw‐5b recognizes a small PAMP‐like peptide within a pathogen‐encoded effector to activate broad‐spectrum resistance, analogous to the mechanism of PRR‐mediated recognition of PAMPs. In addition to the NB‐LRR region, the N‐terminal SD also interacts with NSm21 (Li et al., 2019). Interestingly, the SD domain plays an important role in enhancing the ability of NB‐LRR region to recognize low levels of NSm or NSm21 and to induce robust HR (Li et al., 2019) (Fig. 1). These findings indicate that Sw‐5b uses a novel two‐step recognition mechanism involving the SD and LRR domains to perceive TSWV and to activate a robust defence response.
Figure 1.
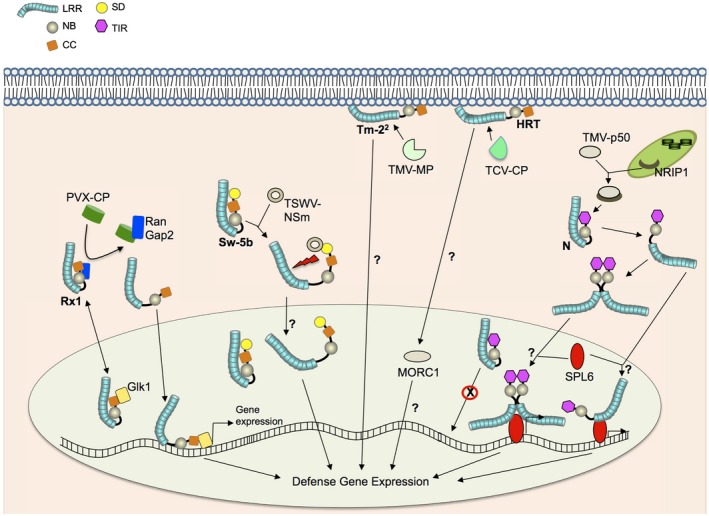
Recognition of viral pathogens by intracellular NLRs. In the absence of infection, the Rx1 CC‐NLR exists in an autoinhibited state stabilized by RanGAP2. Upon recognition of the PVX coat protein (PVX‐CP), Rx1 releases its autoinhibition and enters the nucleus, where it associates with the Golden2‐like transcription factor NbGlk1 to promote the transcription of defence genes. The SD and NB‐LRR domains of the Sw‐5b CC‐NLR interact with the movement protein NSm of TSWV to relieve auto‐inhibition and activate the defence response. Sw‐5b is present in the nucleus in the absence of infection but its role in the nucleus is unknown. It is not known if activated Sw‐5b moves into the nucleus. TM‐22 and HRT are membrane‐associated CC‐NLRs; TM‐22 recognizes the movement protein of tobamoviruses including TMV (TMV‐MP), and HRT recognizes CP of TCV (TCV‐CP). HRT and Rx1 (not shown) interact with MORC1, which has an unknown nuclear function. The nucleocytoplasmic TIR‐NLR N confers resistance to TMV through recognition of the 50 kDa helicase domain of the TMV replicase (TMV‐p50). Recognition of p50 by N requires NRIP1, which relocalizes from the chloroplast to the cytoplasm during infection. Following p50 recognition, N undergoes self‐association; it also associates with the transcription factor SPL6 in the nucleus to promote transcription of defence genes. It is currently unknown which activated form (monomeric or self‐associated N) associates with SPL6 in the nucleus.
N, a TIR‐NLR that confers broad‐spectrum resistance to all strains of tobamoviruses except the Ob strain of TMV, recognizes the 50 kDa helicase domain of the TMV replicase (p50) through its TIR domain (Burch‐Smith et al., 2007; Caplan et al., 2008a); this recognition is mediated by an intermediate protein, N receptor interacting protein 1 (NRIP1) (Caplan et al., 2008a) (Fig. 1). NRIP1 is a sulfurtransferase and is localized to the chloroplast in the absence of infection. Upon TMV infection, NRIP1 relocalizes to the cytoplasm and forms a complex with p50. N then associates with the NRIP1‐p50 complex in the cytoplasm to activate defence (Fig. 1). The LRR domain of N has also been shown to interact with p50 in yeast two‐hybrid and in vitro pull‐down assays (Ueda et al., 2006). Despite the lack of in vivo evidence for an LRR–p50 interaction, it is possible that N uses a two‐step recognition mechanism similar to that of Sw‐5b involving both the TIR and LRR domains to perceive and respond to TMV.
Similar to N, tomato Tm‐22, a CC‐NLR, confers resistance to some strains of tobamoviruses, including TMV. Tm‐22 recognizes the movement protein (MP) from TMV but not p50 (Fig. 1). Furthermore, Tm‐22 associates with MP at the plasma membrane (Chen et al., 2017) and recognition of MP by Tm‐22 may involve an intermediate MP Interacting Protein 1 (NbMIP1) (Du et al., 2013). However, evidence for the existence of Tm‐22, MP and NbMIP1 in a complex is currently lacking.
Unlike N, Sw‐5b, Tm‐22 and other NLRs that recognize virus‐encoded effectors, Vat (Virus aphid transmission), a CC‐NLR from melon, activates defence against aphids, which in turn prevents the transmission of Cucumber mosaic virus (CMV) and several potyviruses (Dogimont et al., 2014). The virus resistance induced by Vat is only observed during aphid transmission. This suggests that activation of Vat is linked to the recognition of effector(s) released by aphids during their initial feeding. Consistent with this, Vat fails to induce a defence response when viruses are introduced mechanically.
Viral NLR activation
As reported for N, the association between NLRs and their corresponding effector is not sufficient to activate NLR and to induce defence (Padmanabhan et al., 2013). N not only associates with p50 from defence‐eliciting TMV‐U1 but also with p50 from non‐defence‐eliciting TMV‐Ob (Padmanabhan et al., 2013). However, only N associated with p50‐U1 will activate the defence response. Furthermore, an ATP‐binding site mutation in N protein that fails to initiate defence signalling can also interact with p50‐U1. Similar to N, SW‐5b is able to associate with non‐eliciting NSm21 mutants, but fails to induce HR (Zhu et al., 2017). Therefore, pathogen recognition and subsequent defence activation are two independent processes in NLR‐mediated defence.
Early studies of Rx1, a CC‐NLR, which recognizes the CP of Potato virus X (PVX), indicated that intramolecular interactions between the CC domain and NB or LRR domains retain NLRs in an auto‐inhibited state (Fig. 1) (Moffett et al., 2002; Rairdan et al., 2008; Slootweg et al., 2013). These interactions are disrupted upon effector association. The exchange of ADP for ATP at the NB domain is then predicted to activate NLRs. Compared to Rx1, the SD and CC domains of Sw‐5b additively retain Sw‐5b in an auto‐inhibited state (Chen et al., 2016). Consistent with the intramolecular domain interaction model, mutations in NB or LRR domains of NLRs that disrupt these interactions have been shown to activate NLRs in the absence of their cognate effectors (Rairdan and Moffett, 2006; Zhu et al., 2017). In Sw‐5b, residue R927 in the LRR domain is critical for auto‐inhibition; association with NSm21 modulates R927 and releases the interaction between the NB and LRR domains (Zhu et al., 2017). Compared to Rx1 and Sw‐5b, intramolecular interactions have not been observed for N (Mestre and Baulcombe, 2006). However, activation of N requires self‐association/dimerization after p50 recognition (Fig. 1), which is dependent on a functional NB domain. Interestingly, a mutation in the TIR domain that fails to induce HR can still undergo self‐association (Mestre and Baulcombe, 2006), suggesting that self‐association is not sufficient to activate downstream defence. Therefore, understanding steps downstream of N self‐association should provide mechanistic insights into how viral NLRs function to induce defence.
In contrast to the Rx1 and Sw‐5b studies described above, a recently reported cryo‐EM structure of ZAR1, a CC‐NLR, suggests that NLR activation may not involve major disruption of intramolecular interaction (Wang et al., 2019b). ZAR1 together with RKS1 kinase recognizes the uridylyltransferase effector AvrAC from Xanthomonas campestris pv. campestris (Wang et al., 2015). AvrAc‐uridylated PBL2 kinase (PBL2UMP) associates with the ZAR1‐RKS1 complex through interaction with RKS1 (Wang et al., 2015). The structure of the ZAR1‐RKS1 complex showed that ADP binding stabilizes the intramolecular interaction within ZAR1 domains and keeps ZAR1 in an inactive monomeric state. The structure of ZAR1‐RKS1‐PBL2UMP indicated that RKS1 binding to PBL2UMP results in a slight conformational change in only the NB domain of ZAR1, indicating that PBL2UMP binding to RKS1 releases bound ADP from ZAR1. However, the ZAR1‐RKS1‐PBL2UMP complex is also monomeric and the conformation of this complex is similar to that of the inactive ZAR1‐RKS1 complex. Therefore, contrary to previously proposed models, activation of ZAR1 does not involve major disruption of intramolecular interactions between N‐terminal domains and the C‐terminal LRR. Furthermore, ADP release is not sufficient to activate ZAR1. ZAR1‐RKS1‐PBL2UMP forms a higher‐order oligomeric complex in the presence of ATP or dATP, indicating that this resistome complex is the active form (Wang et al., 2019a). This higher‐order complex forms a pentameric wheel involving all domains of ZAR1 (Wang et al., 2019a). Based on the structure‐guided mutagenesis experiments, Wang et al. suggest that the funnel‐shaped pentameric complex of ZAR1 associates with the plasma membrane to cause HR (Wang et al., 2019a). Further biochemical and structural analyses of other plant NLRs should shed light on whether the ZAR1 model is universal in the activation of NLRs and defence.
Viral NLR signalling
Activated NLRs use downstream signalling components to initiate immune responses. ENHANCED DISEASE SUSCEPTIBILITY 1 (EDS1) act as a shared, central signalling node for TIR‐NLRs including N‐mediated resistance to TMV (Cui et al., 2015). EDS1 resides in the cytoplasmic and nuclear pools, and interacts dynamically with two regulators of defence, PHYTOALEXIN DEFICIENT 4 (PAD4) and SENESCENCE‐ASSOCIATED GENE 101 (SAG101). While EDS1‐SAG101 complexes are formed exclusively in the nucleus, EDS1‐PAD4 complexes are found in both the cytoplasm and the nucleus, and EDS1 homodimers are strictly cytoplasmic (reviewed in Cui et al. (2015)). Nuclear EDS1 is required for immune receptor‐mediated induction of transcriptional reprogramming and HR (Cui et al., 2015). EDS1 and SAG101 are also required by HRT, a CC‐NLR that recognizes the CP of Turnip crinkle virus (TCV‐CP) (Zhu et al., 2011).
N TIR‐NLR‐mediated resistance to TMV requires NRG1 (N Required Gene 1), a CC‐NLR (Peart et al., 2005). Based on this, it was proposed that NLRs, which recognize pathogens, recruit other NLRs for downstream signalling (Peart et al., 2005). Consistent with this earlier observation, recent studies reported that other pathogen‐recognizing NLRs (called sensor NLRs) also require other NLRs or a network of NLRs (called helper NLRs) for immune signalling (Wu et al., 2017). Sw‐5b and Rx1 require NRC2/3/4 NLRs for the induction of TSWV‐NSm and PVX‐CP‐mediated HR‐PCD, respectively (Li et al., 2019; Wu et al., 2017). Recently, EDS1 has been shown to associate with NRG1 (Qi et al., 2018). Since silencing of NRG1 does not affect N self‐association/dimerization (Peart et al., 2005), the NRG1‐EDS1 complex must function downstream of the dimerization step.
Evidence suggests that some NLRs may function as transcriptional co‐regulators to control transcriptional reprogramming during defence through direct association with transcription factors (Cui et al., 2015; Padmanabhan and Dinesh‐Kumar, 2010, 2014). N associates with the Squamosa Promoter Binding Protein Like 6 (SPL6) in the nucleus only in the presence of defence‐eliciting p50‐U1 but not in the presence of non‐defence‐eliciting p50‐Ob, indicating that N‐SPL6 association is dependent on the N activation step (Fig. 1) (Padmanabhan et al., 2013). However, it is currently unknown if activated monomeric N from the cytoplasm moves to the nucleus and associates with SPL6 or if activated dimeric N from the cytoplasm moves to the nucleus and associates with SPL6 (Fig. 1). Rx1 associates with Ran GTPase‐activating protein 2 (RanGAP2), which is required for defence against PVX (Sacco et al., 2007; Tameling and Baulcombe, 2007) as well as the nucleocytoplasmic partitioning of Rx1 (Tameling et al., 2010) (Fig. 1). Rx1 has been shown to bind DNA following activation with PVX‐CP effector (Fenyk et al., 2015). Furthermore, Rx1 interacts with a Golden2‐like transcription factor, NbGlk1, that binds to DNA when Rx1 is activated by PVX‐CP (Townsend et al., 2018) (Fig. 1). Sw‐5b is also a nucleocytoplasmic protein (De Oliveira et al., 2016) (Fig. 1); however, its role in the nucleus remains unknown.
A GHKL (gyrase, heat‐shock protein 90, histidine kinase, MutL)‐type ATPase, Microchidia 1 [MORC1; formerly called Compromised Recognition of TCV (CRT1)], that interacts with many CC‐NLRs, including viral NLRs such as HRT, Rx1 and RCY1 (which confers resistance to Cucumber mosaic virus), has also been shown to have a nuclear role (Fig. 1) (Kang et al., 2008, 2010, 2012). The interaction between NLRs and MORC1 is disrupted when NLRs are activated (Kang et al., 2008, 2010). Recent evidence suggests that MORC1 binds nucleic acid and possesses endonuclease activity in vitro and activation of immune receptors increases MORC1 levels in the nucleus (Kang et al., 2012). Considering MORC family members participate in transcriptional gene silencing (TGS) and interact with the components of RNA‐dependent DNA methylation (RDDM) and chromatin remodelling complex, it has been proposed that MORC family members may regulate transcriptional reprogramming during immunity (see review in Koch et al. (2017)). Many questions remain with MORC function. How does endosome‐localized MORC1 interact with plasma membrane‐ and nucleocytoplasmic‐localized NLRs? What processes are involved in the disruption of MORC1 interaction with NLRs post‐activation? What processes enhance accumulation of MORC in the nucleus during defence? What is the role of MORC‐mediated chromatin remodelling during immunity?
Conclusions and Perspectives
In this review, we have highlighted some of the recent advances in understanding viral recognition, signalling intermediates, and the mechanisms of NLR regulation and activation. We have also discussed emerging evidence for the possible involvement of cell‐surface receptors in the perception of viruses or viral nucleic acids. However, further studies are needed to establish the existence of “viral PTI” in plants. It has been 25 years since the first plant NLRs were cloned. Despite this, our understanding of how NLRs recognize pathogens and activate defence is far from complete. The recent reporting of the first plant NLR structure has provided insights into the intramolecular interaction and nucleotide‐binding state of ZAR1 (Wang et al., 2019b). To understand whether the model of ZAR1 activation and HR induction (Wang et al., 2019a) is universal to all NLRs will require further biochemical and structural analyses of other NLRs. It will be interesting to see how nucleocytoplasmic NLRs such as N and Rx1, which interact with transcription factors, are activated following effector recognition compared to plasma membrane‐localized ZAR1. Furthermore, structural comparison between TIR‐NLRs and CC‐NLRs as additional structural information becomes available for plant NLRs should provide insights into how these two classes of NLRs function. Further structural studies will also provide information on how NLRs are recognizing effectors. Do effectors form a complex with their cognate NLRs? Or, as in the case of PBL2UMP, do NLRs recognize effector‐modified ligands? Considering CP and MP do not possess any known enzymatic activity and helicase activity of TMV‐p50 is not required to activate N, how would these effectors modify and recruit a decoy like PBL2? Based on the ZAR1 resistome structure, Wang et al. (2019a) suggested that helper CC‐NLRs like NRG1 and NRC4 might form higher‐order pentamer structures like ZAR1 to induce HR during N‐ or Rx1‐mediated immunity, respectively. Considering NRC4 and NRG1 are cytoplasmic proteins, it is unclear how these proteins would be recruited to the plasma membrane to execute cell death. Regardless, the knowledge we have gained over the last few years has provided the framework for future studies to understand the precise molecular and biochemical function of NLRs in antiviral immunity and immunity in general.
Acknowledgements
We could not cite much of the primary literature because of the limited space. Innate immunity work in the S. P. Dinesh‐Kumar laboratory is supported by National Science Foundation grants IOS‐1354434 and IOS‐1339185 and National Institute of Health grant GM097587.
References
- Anderson, P.K. , Cunningham, A.A. , Patel, N.G. , Morales, F.J. , Epstein, P.R. and Daszak, P. (2004) Emerging infectious diseases of plants: pathogen pollution, climate change and agrotechnology drivers. Trends Ecol. Evol. 19, 535–544. [DOI] [PubMed] [Google Scholar]
- Brommonschenkel, S.H. , Frary, A. and Tanksley, S.D. (2000) The broad‐spectrum tospovirus resistance gene Sw‐5 of tomato is a homolog of the root‐knot nematode resistance gene Mi . Mol. Plant‐Microbe Interact. 13, 1130–1138. [DOI] [PubMed] [Google Scholar]
- Burch‐Smith, T.M. , Schiff, M. , Caplan, J.L. , Tsao, J. , Czymmek, K. and Dinesh‐Kumar, S.P. (2007) A novel role for the TIR domain in association with pathogen‐derived elicitors. PLoS Biol. 5, e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan, J.L. , Mamillapalli, P. , Burch‐Smith, T.M. , Czymmek, K. and Dinesh‐Kumar, S.P. (2008a) Chloroplastic protein NRIP1 mediates innate immune receptor recognition of a viral effector. Cell, 132, 449–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan, J.L. , Padmanabhan, M. and Dinesh‐Kumar, S.P. (2008b) Plant NB‐LRR immune receptors: from recognition to transcriptional reprogramming. Cell Host Microbe, 3, 126–135. [DOI] [PubMed] [Google Scholar]
- Chen, X. , Zhu, M. , Jiang, L. , Zhao, W. , Li, J. , Wu, J. , Li, C. , Bai, B. , Lu, G. , Chen, H. , Moffett, P. and Tao, X. (2016) A multilayered regulatory mechanism for the autoinhibition and activation of a plant CC‐NB‐LRR resistance protein with an extra N‐terminal domain. New Phytol. 212, 161–175. [DOI] [PubMed] [Google Scholar]
- Chen, T. , Liu, D. , Niu, X. , Wang, J. , Qian, L. , Han, L. , Liu, N. , Zhao, J. , Hong, Y. and Liu, Y. (2017) Antiviral resistance protein Tm‐2(2) functions on the plasma membrane. Plant Physiol. 173, 2399–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, H. , Tsuda, K. and Parker, J.E. (2015) Effector‐triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 66, 487–511. [DOI] [PubMed] [Google Scholar]
- De Oliveira, A.S. , Koolhaas, I. , Boiteux, L.S. , Caldararu, O.F. , Petrescu, A.J. , Oliveira Resende, R. and Kormelink, R. (2016) Cell death triggering and effector recognition by Sw‐5 SD‐CNL proteins from resistant and susceptible tomato isolines to Tomato spotted wilt virus . Mol. Plant Pathol. 17, 1442–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogimont, C. , Chovelon, V. , Pauquet, J. , Boualem, A. and Bendahmane, A. (2014) The Vat locus encodes for a CC‐NBS‐LRR protein that confers resistance to Aphis gossypii infestation and A. gossypii‐mediated virus resistance. Plant J. 80, 993–1004. [DOI] [PubMed] [Google Scholar]
- Du, Y. , Zhao, J. , Chen, T. , Liu, Q. , Zhang, H. , Wang, Y. , Hong, Y. , Xiao, F. , Zhang, L. , Shen, Q. and Liu, Y. (2013) Type I J‐domain NbMIP1 proteins are required for both Tobacco mosaic virus infection and plant innate immunity. PLoS Path. 9, e1003659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenyk, S. , Townsend, P.D. , Dixon, C.H. , Spies, G.B. , de San Eustaquio Campillo, A. , Slootweg, E.J. , Westerhof, L.B. , Gawehns, F.K. , Knight, M.R. , Sharples, G.J. , Goverse, A. , Palsson, L.O. , Takken, F.L. and Cann, M.J. (2015) The potato nucleotide‐binding leucine‐rich repeat (NLR) immune receptor Rx1 is a pathogen‐dependent DNA‐deforming protein. J. Biol. Chem. 290, 24945–24960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouveia, B.C. , Calil, I.P. , Machado, J.P. , Santos, A.A. and Fontes, E.P. (2016) Immune receptors and co‐receptors in antiviral innate immunity in plants. Front Microbiol. 7, 2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Z. , Li, Y. and Ding, S.W. (2019) Small RNA‐based antimicrobial immunity. Nat. Rev. Immunol. 19, 31–44. [DOI] [PubMed] [Google Scholar]
- Hallwass, M. , de Oliveira, A.S. , de Campos Dianese, E. , Lohuis, D. , Boiteux, L.S. , Inoue‐Nagata, A.K. , Resende, R.O. and Kormelink, R. (2014) The Tomato spotted wilt virus cell‐to‐cell movement protein (NSM ) triggers a hypersensitive response in Sw‐5‐containing resistant tomato lines and in Nicotiana benthamiana transformed with the functional Sw‐5b resistance gene copy. Mol. Plant Pathol. 15, 871–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kang, H.G. , Kuhl, J.C. , Kachroo, P. and Klessig, D.F. (2008) CRT1, an Arabidopsis ATPase that interacts with diverse resistance proteins and modulates disease resistance to turnip crinkle virus. Cell Host Microbe, 3, 48–57. [DOI] [PubMed] [Google Scholar]
- Kang, H.G. , Oh, C.S. , Sato, M. , Katagiri, F. , Glazebrook, J. , Takahashi, H. , Kachroo, P. , Martin, G.B. and Klessig, D.F. (2010) Endosome‐associated CRT1 functions early in resistance gene‐mediated defense signaling in Arabidopsis and tobacco. Plant Cell, 22, 918–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, H.G. , Hyong, W.C. , von Einem, S. , Manosalva, P. , Ehlers, K. , Liu, P.P. , Buxa, S.V. , Moreau, M. , Mang, H.G. , Kachroo, P. , Kogel, K.H. and Klessig, D.F. (2012) CRT1 is a nuclear‐translocated MORC endonuclease that participates in multiple levels of plant immunity. Nat. Commun. 3, 1297. [DOI] [PubMed] [Google Scholar]
- Koch, A. , Kang, H.G. , Steinbrenner, J. , Dempsey, D.A. , Klessig, D.F. and Kogel, K.H. (2017) MORC proteins: novel players in plant and animal health. Front. Plant Sci. 8, 1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Huang, H. , Zhu, M. , Huang, S. , Zhang, W. , Dinesh‐Kumar, S.P. and Tao, X. (2019) A plant immune receptor adopts a two‐step recognition mechanism to enhance viral effector perception. Mol. Plant, 12, 248–262. [DOI] [PubMed] [Google Scholar]
- Macho, A.P. and Lozano‐Duran, R. (2019) Molecular dialogues between viruses and receptor‐like kinases in plants. Mol. Plant Pathol. 10.1111/mpp.12812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho, A.P. and Zipfel, C. (2014) Plant PRRs and the activation of innate immune signaling. Mol. Cell, 54, 263–272. [DOI] [PubMed] [Google Scholar]
- Mestre, P. and Baulcombe, D.C. (2006) Elicitor‐mediated oligomerization of the tobacco N disease resistance protein. Plant Cell, 18, 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffett, P. , Farnham, G. , Peart, J. and Baulcombe, D.C. (2002) Interaction between domains of a plant NBS‐LRR protein in disease resistance‐related cell death. EMBO J. 21, 4511–4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan, M.S. and Dinesh‐Kumar, S.P. (2010) All hands on deck – the role of chloroplasts, endoplasmic reticulum, and the nucleus in driving plant innate immunity. Mol. Plant‐Microbe Interact. 23, 1368–1380. [DOI] [PubMed] [Google Scholar]
- Padmanabhan, M.S. and Dinesh‐Kumar, S.P. (2014) The conformational and subcellular compartmental dance of plant NLRs during viral recognition and defense signaling. Curr. Opin. Microbiol. 20, 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan, M.S. , Ma, S. , Burch‐Smith, T.M. , Czymmek, K. , Huijser, P. and Dinesh‐Kumar, S.P. (2013) Novel positive regulatory role for the SPL6 transcription factor in the N TIR‐NB‐LRR receptor‐mediated plant innate immunity. PLoS Pathog. 9, e1003235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peart, J.R. , Mestre, P. , Lu, R. , Malcuit, I. and Baulcombe, D.C. (2005) NRG1, a CC‐NB‐LRR protein, together with N, a TIR‐NB‐LRR protein, mediates resistance against tobacco mosaic virus. Curr. Biol. 15, 968–973. [DOI] [PubMed] [Google Scholar]
- Peiro, A. , Canizares, M.C. , Rubio, L. , Lopez, C. , Moriones, E. , Aramburu, J. and Sanchez‐Navarro, J. (2014) The movement protein (NSm) of Tomato spotted wilt virus is the avirulence determinant in the tomato Sw‐5 gene‐based resistance. Mol. Plant Pathol. 15, 802–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, Y. , van Wersch, R. and Zhang, Y. (2018) Convergent and divergent signaling in PAMP‐triggered immunity and effector‐triggered immunity. Mol. Plant‐Microbe Interact. 31, 403–409. [DOI] [PubMed] [Google Scholar]
- Qi, T. , Seong, K. , Thomazella, D.P.T. , Kim, J.R. , Pham, J. , Seo, E. , Cho, M.-J. , Schultink, A. and Staskawicz, B.J. (2018) NRG1 functions downstream of EDS1 to regulate TIR-NLR-mediated plant immunity in Nicotiana benthamiana . Proc Natl. Acad. Sci., 115, E10979–E10987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rairdan, G.J. and Moffett, P. (2006) Distinct domains in the ARC region of the potato resistance protein Rx mediate LRR binding and inhibition of activation. Plant Cell, 18, 2082–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rairdan, G.J. , Collier, S.M. , Sacco, M.A. , Baldwin, T.T. , Boettrich, T. and Moffett, P. (2008) The coiled‐coil and nucleotide binding domains of the potato Rx disease resistance protein function in pathogen recognition and signaling. Plant Cell, 20, 739–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco, M.A. , Mansoor, S. and Moffett, P. (2007) A RanGAP protein physically interacts with the NB‐LRR protein Rx, and is required for Rx‐mediated viral resistance. Plant J. 52, 82–93. [DOI] [PubMed] [Google Scholar]
- Slootweg, E.J. , Spiridon, L.N. , Roosien, J. , Butterbach, P. , Pomp, R. , Westerhof, L. , Wilbers, R. , Bakker, E. , Bakker, J. , Petrescu, A.J. , Smant, G. and Goverse, A. (2013) Structural determinants at the interface of the ARC2 and leucine‐rich repeat domains control the activation of the plant immune receptors Rx1 and Gpa2. Plant Physiol. 162, 1510–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameling, W.I. and Baulcombe, D.C. (2007) Physical association of the NB‐LRR resistance protein Rx with a Ran GTPase‐activating protein is required for extreme resistance to Potato virus X . Plant Cell, 19, 1682–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameling, W.I. , Nooijen, C. , Ludwig, N. , Boter, M. , Slootweg, E. , Goverse, A. , Shirasu, K. and Joosten, M.H. (2010) RanGAP2 mediates nucleocytoplasmic partitioning of the NB‐LRR immune receptor Rx in the Solanaceae, thereby dictating Rx function. Plant Cell, 22, 4176–4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira, R.M. , Ferreira, M.A. , Raimundo, G.A.S. , Loriato, V.A. , Reis, P.A.B. and Fontes, E.P.B. (2019) Virus perception at the cell surface: revisiting the roles of receptor‐like kinases as viral pattern recognition receptors. Mol. Plant Pathol. 10.1111/mpp.12816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend, P.D. , Dixon, C.H. , Slootweg, E.J. , Sukarta, O.C.A. , Yang, A.W.H. , Hughes, T.R. , Sharples, G.J. , Palsson, L.O. , Takken, F.L.W. , Goverse, A. and Cann, M.J. (2018) The intracellular immune receptor Rx1 regulates the DNA‐binding activity of a Golden2‐like transcription factor. J. Biol. Chem. 293, 3218–3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda, H. , Yamaguchi, Y. and Sano, H. (2006) Direct interaction between the Tobacco mosaic virus helicase domain and the ATP‐bound resistance protein, N factor during the hypersensitive response in tobacco plants. Plant Mol. Biol. 61, 31–45. [DOI] [PubMed] [Google Scholar]
- Wang, A. and Krishnaswamy, S. (2012) Eukaryotic translation initiation factor 4E‐mediated recessive resistance to plant viruses and its utility in crop improvement. Mol. Plant Pathol. 13, 795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G. , Roux, B. , Feng, F. , Guy, E. , Li, L. , Li, N. , Zhang, X. , Lautier, M. , Jardinaud, M.F. , Chabannes, M. , Arlat, M. , Chen, S. , He, C. , Noel, L.D. and Zhou, J.M. (2015) The decoy substrate of a pathogen effector and a pseudokinase specify pathogen‐induced modified‐self recognition and immunity in plants. Cell Host Microbe, 18, 285–295. [DOI] [PubMed] [Google Scholar]
- Wang, J. , Hu, M. , Wang, J. , Qi, J. , Han, Z. , Wang, G. , Qi, Y. , Wang, H.W. , Zhou, J.M. and Chai, J. (2019a) Reconstitution and structure of a plant NLR resistosome conferring immunity. Science, 364, eaav5870. [DOI] [PubMed] [Google Scholar]
- Wang, J. , Wang, J. , Hu, M. , Wu, S. , Qi, J. , Wang, G. , Han, Z. , Qi, Y. , Gao, N. , Wang, H.W. , Zhou, J.M. and Chai, J. (2019b) Ligand‐triggered allosteric ADP release primes a plant NLR complex. Science, 364, eaav5868. [DOI] [PubMed] [Google Scholar]
- Whitham, S. , Dineshkumar, S.P. , Choi, D. , Hehl, R. , Corr, C. and Baker, B. (1994) The product of the Tobacco mosaic virus resistance gene N – Similarity to toll and the interleukin‐1 receptor. Cell, 78, 1101–1115. [DOI] [PubMed] [Google Scholar]
- Wu, C.H. , Abd‐El‐Haliem, A. , Bozkurt, T.O. , Belhaj, K. , Terauchi, R. , Vossen, J.H. and Kamoun, S. (2017) NLR network mediates immunity to diverse plant pathogens. Proc. Natl. Acad. Sci. USA. 114, 8113–8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, X. , Feng, B. , He, P. and Shan, L. (2017) From chaos to harmony: responses and signaling upon microbial pattern recognition. Annu. Rev. Phytopathol. 55, 109–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, S. , Jeong, R.D. , Venugopal, S.C. , Lapchyk, L. , Navarre, D. , Kachroo, A. and Kachroo, P. (2011) SAG101 forms a ternary complex with EDS1 and PAD4 and is required for resistance signaling against turnip crinkle virus. PLoS Pathog. 7, e1002318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, M. , Jiang, L. , Bai, B. , Zhao, W. , Chen, X. , Li, J. , Liu, Y. , Chen, Z. , Wang, B. , Wang, C. , Wu, Q. , Shen, Q. , Dinesh‐Kumar, S.P. and Tao, X. (2017) The intracellular immune receptor Sw‐5b confers broad‐spectrum resistance to tospoviruses through recognition of a conserved 21‐amino acid viral effector epitope. Plant Cell, 29, 2214–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]


