Summary
Autophagy is a conserved self‐cleaning and renewal system required for cellular homeostasis and stress tolerance. Autophagic processes are also implicated in the response to ‘non‐self’ such as viral pathogens, yet the functions and mechanisms of autophagy during plant virus infection have only recently started to be revealed. Compelling evidence now indicates that autophagy is an integral part of antiviral immunity in plants. It can promote the hypersensitive cell death response upon incompatible viral infections or mediate the selective elimination of entire particles and individual proteins from compatible viruses in a pathway similar to xenophagy in animals. Several viruses, however, have evolved measures to antagonize xenophagic degradation or utilize autophagy to suppress disease‐associated cell death and other defence pathways like RNA silencing. Here, we highlight the current advances and gaps in our understanding of the complex autophagy–virus interplay and its consequences for host immunity and viral pathogenesis in plants.
Keywords: ATG8, autophagy, innate immunity, plants, selective cargo receptor, virus, xenophagy
Introduction
Autophagy is a major degradation and recycling system required to maintain cellular homeostasis and organismal health during normal development and in response to stress conditions (Boya et al., 2013; Klionsky and Codogno, 2013; Mizushima, 2018). Autophagy has also been implicated in the regulation of programmed cell death (PCD) and its dysfunction is linked to various pathological conditions and diseases (Doherty and Baehrecke, 2018; Levine and Kroemer, 2019). Initially regarded as a largely unspecific (‘bulk’) process for nutrient recycling and energy supply, it is now evident that autophagy engages highly selective mechanisms to eliminate surplus, harmful and/or damaged cytoplasmic constituents ranging from individual proteins to entire organelles (Gatica et al., 2018; Zaffagnini and Martens, 2016). Autophagic targets are sequestered and transported in specialized double‐membrane vesicles, called autophagosomes, that originate from an expanding isolation membrane (or phagophore) and fuse with lysosomal or vacuolar compartments for lytic breakdown of the cargo (Feng et al., 2014; Yu et al., 2018). The complex series of events underlying autophagosome initiation and maturation is dependent on the coordinated action of a conserved set of AUTOPHAGY‐RELATED (ATG) proteins (Mizushima et al., 2011; Rubinsztein et al., 2012). Among these, ATG8/LC3 family proteins have emerged as central players in both autophagosome biogenesis and cargo recruitment (Abdollahzadeh et al., 2017; Slobodkin and Elazar, 2013). They are anchored as conjugates with the membrane lipid phosphatidylethanolamine (PE) in the growing phagophore and facilitate selectivity through interaction with a large repertoire of autophagic receptors and adaptors. ATG8 lipidation is mediated by two ubiquitin‐like conjugation pathways involving the E1‐like ligase ATG7, the E2‐ligase ATG3 and the E3‐like ligase ATG5‐ATG12‐ATG16 complex. Other functional units comprise the ATG1 kinase complex required for autophagy induction, the ATG6/Beclin1‐containing phosphatidylinositol 3‐kinase (PI3K) complex involved in protein recruitment and curvature formation during phagophore nucleation, and ATG9 vesicles essential for lipid delivery during phagophore expansion (Feng et al., 2014; Marshall and Vierstra, 2018; Rubinsztein et al., 2012; Yu et al., 2018).
Given the evolutionary ancient role of autophagy as a selective ‘self‐eating’ mechanism in cellular quality control and stress adaptation, it is not surprising that autophagy has also evolved as a key factor in the defence against ‘non‐self’ such as intracellular pathogens (Gomes and Dikic, 2014; Sharma et al., 2018; Wileman, 2013). In metazoans, it is well established that selective autophagy contributes to immunity against viruses by directly degrading their particles or individual proteins in a process known as xenophagy or virophagy (Choi et al., 2018; Judith et al., 2013; Orvedahl et al., 2011; Shelly et al., 2009). The targeting of viral structures typically involves autophagy receptors that recognize specific ‘eat‐me’ signals like ubiquitination and bind to phagophore‐localized ATG8 through the LC3/ATG8‐interacting region (LIR) (Gatica et al., 2018; Lai and Devenish, 2015). Additional antiviral functions of autophagy are linked to the exposure of viral nucleic acids to immune receptors and the promotion of viral antigen processing, thereby facilitating the activation of innate and adaptive immune responses (Choi et al., 2018; Lee et al., 2007; Paludan et al., 2005). During the long‐lasting evolutionary battle between viral pathogens and their hosts, successful animal viruses have developed various measures to counteract and utilize autophagy to their own advantage. While some viruses exploit autophagosome formation and transport for viral replication and non‐lytic release of virions, others manipulate autophagy pathways to generate metabolites and energy or to prolong cellular lifespan by suppression of host cell death (Dong and Levine 2013, Chiramel et al., 2013; Heaton and Randall 2010; Jackson, 2015).
Compared to animals, the knowledge of the roles of autophagy during plant virus infection is still very limited. However, several reports have started to shed light on autophagic mechanisms involved in host immunity and viral pathogenesis, suggesting that plant viruses interact with autophagy in a similar complex and versatile manner as their animal counterparts (Clavel et al., 2017; Hofius et al., 2017; Wu et al., 2019). In this review, we will summarize the recently emerged anti‐ and proviral functions of plant autophagy. In particular, we will highlight the current advances and gaps in our understanding of how viruses are recognized and targeted by autophagy as part of the immune response, and how successful viral pathogens are able to escape, subvert or exploit the autophagic system for enhanced pathogenicity.
Autophagy and Antiviral Immunity: Perception and Degradation
Autophagy has been initially linked to both the restriction and promotion of the antiviral hypersensitive response (HR), a local PCD reaction typically triggered upon nucleotide‐binding leucine‐rich repeat (NLR) immune receptor activation during effector‐triggered immunity (ETI). Silencing of autophagy‐related genes in the NLR‐type N gene‐containing Nicotiana benthamiana plants resulted in enhanced viral levels in HR lesions as well as unrestricted spreading of cell death into non‐infected tissue upon challenge with an avirulent strain of Tobacco mosaic virus (TMV) (Fig. 1) (Liu et al., 2005). Intriguingly, ATG3‐mediated autophagy enhancement through down‐regulation of cytosolic glyceraldehyde‐3‐phosphate dehydrogenase (GAPC) genes stimulated the HR and disease resistance against TMV (Han et al., 2015). These observations added to the emerging view of autophagy acting as mediator of immunity‐related PCD while suppressing stress‐ and disease‐associated (necrotic) cell death in different plant pathosystems (Hofius et al., 2009; Minina et al., 2014; Munch et al., 2014; Ustun et al., 2017). The molecular details underlying the dual role of autophagy during ETI are still unclear. Autophagy induction likely involves salicylic acid (SA) signalling associated with both local NLR protein activation and systemic stress responses (Hofius et al., 2009; Yoshimoto et al., 2009). However, the specific targets of autophagic degradation that impact the pro‐death and pro‐survival decisions inside and outside of HR lesions await to be identified (Ustun et al., 2017).
Figure 1.
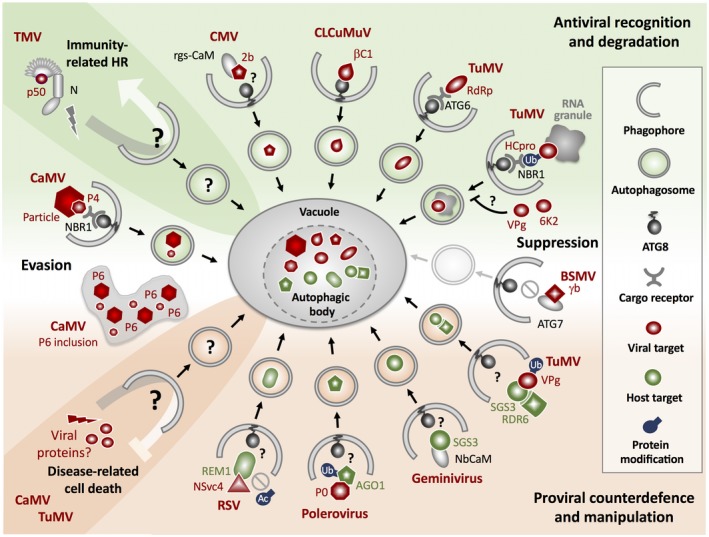
Anti‐ and proviral roles of autophagy in plants. Autophagy has emerged as a central component of antiviral immunity. During infection with an avirulent strain of Tobacco mosaic virus (TMV), autophagy is activated upon N gene‐mediated recognition of the viral p50 protein and contributes to the hypersensitive response (HR). The mechanistic details and autophagic targets underlying the death‐promoting activity of autophagy in effector‐triggered immunity are still unknown. During virulent virus infection, selective autophagy employs the cargo receptor NBR1 to mediate the degradation of non‐assembled capsid protein (P4) and particles of Cauliflower mosaic virus (CaMV). CaMV evades xenophagy by sequestering P4 and particles in autophagy‐resistant inclusions formed by the viral protein P6. NBR1 targets the RNA silencing suppressor HCpro of Turnip mosaic virus (TuMV) in association with potyvirus‐induced RNA granules, but the viral proteins VPg and to a lesser extent 6K2 are able to block NBR1 flux by as yet unknown mechanisms. NBR1 targeting of viral proteins occurs in an ubiquitin‐dependent (TuMV HCpro) or ‐independent manner (CaMV P4). The RNA‐dependent RNA polymerase (RdRp) of TuMV is inhibited via direct interaction with ATG6/Beclin1, which is proposed to act as cargo receptor for RdRp degradation. Other viral proteins subjected to autophagic destruction include the Cucumber mosaic virus (CMV) silencing suppressor 2b, potentially involving the host protein rgs‐CaM, and the virulence factor βC1 of Cotton leaf curl Multan virus (CLCuMuV) through ATG8 binding. Barley stripe mosaic virus (BSMV) subverts antiviral autophagy with the help of the γb protein, which disrupts ATG7–ATG8 interaction and thus impairs autophagosome formation. Different viruses hijack autophagy mechanisms to enhance viral pathogenicity. The TuMV VPg protein mediates the autophagic degradation of the host RNA silencing component SGS3 and its partner protein RDR6. SGS3 is also targeted during geminivirus infection involving the host protein NbCaM. The poleroviral protein P0 triggers ubiquitination and autophagic breakdown of AGO1, another component of the RNA silencing pathway. The movement protein NSvc4 of Rice stripe virus (RSV) induces the autophagic turnover of the group 1 remorin (REM1) protein through interference with its S‐acetylation, thereby preventing the REM1‐mediated negative regulation of viral cell‐to‐cell movement. Viruses like CaMV and TuMV also benefit from autophagy‐mediated suppression of disease‐associated host cell death, but it is unclear if and how viral proteins are involved in autophagy activation and which host components are targeted to mediate the cytoprotective effects. See also text for further details. Graphical elements are partly adapted from Marshall and Vierstra (2018).
More recent studies focused on unravelling the immune functions of autophagy during virulent virus infections. Autophagy appears to be activated in response to various unrelated DNA and RNA viruses with negative consequences for virus accumulation, suggesting the integration of autophagic mechanisms in basal antiviral defences (Hafren et al., 2017, 2018; Haxim et al., 2017; Li et al., 2018). A seminal example in this regard is given by Cauliflower mosaic virus (CaMV), a double‐stranded DNA pararetrovirus that is targeted by selective autophagy to impair the establishment of infection in Arabidopsis (Fig. 1) (Hafren et al., 2017). Strikingly, the cargo receptor NEIGHBOR OF BRCA1 (NBR1) was found to directly bind to and mediate the vacuolar clearance of the viral capsid protein and particles, thereby establishing xenophagy as antiviral pathway in plants (Hafren and Hofius, 2017; Hafren et al., 2017). NBR1‐dependent processes were further shown to limit infection of Arabidopsis with Turnip mosaic virus (TuMV), a positive‐stranded RNA potyvirus (Fig. 1) (Hafren et al., 2018). However, instead of removing entire particles, selective autophagy targets the viral suppressor of RNA silencing (VSR) HCpro in association with proviral RNA granules (Hafren et al., 2015), suggesting also features of granulophagy as part of the antiviral response (Hafren et al., 2018). Importantly, cargo recognition and binding by NBR1 appeared to be ubiquitin‐independent for the CaMV coat protein/particle but ubiquitin‐dependent for TuMV HCpro, which agrees with different targeting mechanisms of viral proteins and particles by the related animal cargo receptor p62/SQSTM1 (Berryman et al., 2012; Judith et al., 2013; Orvedahl et al., 2010, 2011; Shelly et al., 2009). Previous work in tobacco suggested the involvement of the calmodulin‐like protein rgs‐CaM in the autophagic turnover of potyviral HCpro and unrelated VSRs such as the 2b protein of Cucumber mosaic virus (CMV) (Fig. 1) (Nakahara et al., 2012). rgs‐CaM was proposed to interact with the viral proteins via its affinity to double‐stranded (ds) RNA binding domains, but it is not known if and how rgs‐CaM connects to the autophagy machinery.
Autophagy was also shown to target the geminiviral VSR and virulence determinant βC1 from Cotton leaf curl Multan virus (CLCuMuV), but through direct binding by ATG8f instead of recruiting a specific cargo receptor (Fig. 1) (Haxim et al., 2017). Disruption of the βC1–ATG8f interaction as well as autophagy deficiency mediated by silencing of ATG5 and ATG7 resulted in increased accumulation of viral DNA, whereas activation of autophagy upon GAPC down‐regulation enhanced disease resistance in N. benthamiana. The inhibitory effect of autophagy extended also to other geminiviruses, including Tomato yellow leaf curl China virus (TYLCCV) (Haxim et al., 2017). Notably, TYLCCV βC1 was previously shown to be polyubiquitinated by a RING E3 ligase in tobacco, resulting in the turnover of βC1 via the ubiquitin/26S proteasome system (Shen et al., 2016). Future studies might reveal whether ubiquitination is also involved in the autophagic recognition of βC1, and how the different degradation routes intersect and are coordinated during geminivirus infection.
The RNA‐dependent RNA polymerase (RdRp) of TuMV was recently identified as another target of selective autophagic degradation (Li et al., 2018). In this case, the core autophagy protein ATG6/Beclin1 was proposed to act as cargo receptor, which suppresses viral polymerase activity upon binding and recruits the viral protein to autophagosomes through specific interaction with ATG8a (Fig. 1) (Li et al., 2018). Consequently, reduced expression of ATG6 and ATG8a increased TuMV accumulation in N. benthamiana and Arabidopsis plants. However, ATG6‐mediated suppression of virus replication was still evident in the absence of ATG8a binding and RdRp degradation, suggesting the involvement of autophagy‐independent processes in this antiviral response that also affected other RNA viruses (Li et al., 2018).
Autophagy and Viral Pathogenesis: Evasion and Manipulation
Besides being activated as part of the antiviral immune response, autophagy may be frequently induced as a by‐product of virus infection to cope with associated stress conditions. Since the viral life cycle strictly depends on living host cells, a functional cytoprotective autophagy pathway should be beneficial for virus propagation and transmission. Indeed, autophagy‐deficient mutants displayed reduced lifespan and were almost devoid of detectable amounts of CaMV and TuMV at late stages of infection (Hafren et al., 2017, 2018). Hence, any viral strategy that interferes with overall autophagy to prevent degradation by selective xenophagy encounters a potential trade‐off in plant fitness. The CaMV P6 protein has been proposed to block autophagy through activation of the TARGET OF RAPAMYCIN (TOR) (Zvereva et al., 2016), but the virus‐induced enhancement of autophagic flux seems to be rather maintained during infection (Hafren et al., 2017). Instead, CaMV escapes xenophagic targeting by the protective functions of autophagy‐resistant viral inclusions. These are mainly formed by P6 and sequester non‐assembled coat protein for particle assembly and storage (Fig. 1) (Hafren et al., 2017). In contrast to CaMV, TuMV is able to specifically interrupt NBR1 flux, and thus HCpro degradation, by the functions of distinct viral proteins (i.e., VPg and 6K2) (Fig. 1), yet the mechanistic details and host targets remain to be resolved (Hafren et al., 2018). Autophagy is also suppressed during infection with Barley stripe mosaic virus (BSMV), another positive‐sense RNA virus. The BSMV γb protein was found to mediate this effect through direct binding to ATG7, thereby disrupting its autophagy‐stimulating interaction with ATG8 (Fig. 1) (Yang et al., 2018). It will be important to decipher if and how BSMV is able to maintain a certain level of autophagy for plant survival while interfering with the core autophagy machinery.
In addition to the viral capacities for autophagic evasion and subversion, several plant viruses have evolved measures to co‐opt the autophagy system for degradation of host proteins involved in antiviral defences, particularly in the RNA silencing pathway. For instance, the poleroviral VSR P0 triggers ubiquitination and autophagic degradation of Arabidopsis ARGONAUTE1 (AGO1), the core subunit of the RNA‐induced silencing complex (Fig. 1) (Derrien et al., 2012). Similarly, potyviral VPg interacts with, and mediates the turnover of, the silencing component SUPPRESSOR OF GENE SILENCING3 (SGS3) and its associated partner RNA‐DEPENDENT RNA POLYMERASE6 (RDR6) in N. benthamiana, involving both autophagic and proteasomal degradation pathways (Fig. 1) (Cheng and Wang, 2017). The autophagic breakdown of SGS3 is also induced during geminivirus infection, which seems to be mediated through viral hijacking of the endogenous silencing suppressor NbCaM, the orthologue of tobacco rgs‐CaM (Fig. 1) (Li et al., 2017).
The virus‐induced exploitation of autophagy for enhanced pathogenicity extends also to other defence pathways. In rice and N. benthamiana, Rice stripe virus (RSV) triggers the autophagic destruction of a group 1 remorin (REM1), which is involved in the negative regulation of viral infection through inhibition of cell‐to‐cell transport. The RSV movement protein NSvc4 binds to, and interferes with, the S‐acetylation of REM1, thereby preventing its correct targeting to the plasma membrane and plasmodesmata. Non‐acetylated REM1 accumulates in the endoplasmic reticulum (ER) and is subjected to the autophagy degradation pathway (Fig. 1) (Fu et al., 2018). Whether autophagic turnover of the mislocalized REM1 is triggered by the cellular quality control system and involves selective cargo receptors awaits further investigation.
Conclusions and Future Perspectives
The past few years have witnessed major advances in our understanding of the interplay between autophagy and viruses in plants. It is now well established that this ancient catabolic pathway is a key factor in antiviral immunity by mediating the xenophagic elimination of viral proteins and particles in a comparable fashion as in animals. Nonetheless, the mechanisms underlying the specific recognition and targeting of the viral material are far from being understood. While autophagic receptors such as NBR1 and ‘eat‐me’ signals like ubiquitination have been identified in some cases, there is also support for the involvement of ubiquitin‐independent receptor binding and direct recruitment of viral substrates through ATG8 interaction. Further research is needed to unravel the inventory of plant xenophagy receptors and to identify the ubiquitination signatures and/or alternative post‐translational modifications involved in selectivity and cargo recognition. In this context, it will be important to reveal whether the xenophagy pathway has developed specificity towards distinct isoforms of the expanded ATG8 protein family (Kellner et al., 2017), either through interaction with receptors or directly with the cargo. Furthermore, it remains largely elusive to what extent the autophagic degradation of host proteins regulates and shapes the immune response against viruses, and whether individual autophagy‐related proteins have evolved non‐canonical antiviral functions, as might be the case for ATG6/Beclin1.
Similar to the situation in metazoans, the co‐evolution of viruses with their plant hosts has developed diverse viral capacities to counteract and modulate autophagy for the benefit of infection. Many, if not all, plant viruses share the need of autophagy‐mediated protection from host cell death for survival and epidemiological success, but often face the challenge to simultaneously escape autophagic destruction. Hence, it is still largely unclear how viruses are able to fine‐tune the induction and suppression of distinct autophagy mechanisms and pathways, and thus to achieve a delicate balance of concurrent pro‐ and antiviral autophagic activities for successful pathogenesis. Other important questions relate to the molecular details of virus‐induced autophagy activation and the as yet unknown role of plant autophagy proteins and membranes in viral replication, which is well established in the animal system. Collectively, the anticipated research efforts in the upcoming years will substantially add to our limited knowledge on the mechanisms and functions of autophagy in plant immunity and viral pathogenesis, and thus may help to identify new targets and strategies for improving resistance against economically relevant viruses in crop plants.
Acknowledgements
We wish to apologize to those colleagues whose work could not be cited due to space limitations. Research in the laboratory is supported by SLU, grants from the Knut‐and‐Alice Wallenberg and Carl Tryggers Foundations (to D.H.), as well as the Swedish Research Councils VR (to D.H.) and FORMAS (to D.H. and A.H.)
References
- Abdollahzadeh, I. , Schwarten, M. , Gensch, T. , Willbold, D. and Weiergraber, O.H. (2017) The Atg8 family of proteins‐modulating shape and functionality of autophagic membranes. Front. Genet. 8, 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berryman, S. , Brooks, E. , Burman, A. , Hawes, P. , Roberts, R. , Netherton, C. , Monaghan, P. , Whelband, M. , Cottam, E. , Elazar, Z. , Jackson, T. and Wileman, T. (2012) Foot‐and‐mouth disease virus induces autophagosomes during cell entry via a class III phosphatidylinositol 3‐kinase‐independent pathway. J. Virol. 86, 12940–12953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boya, P. , Reggiori, F. and Codogno, P. (2013) Emerging regulation and functions of autophagy. Nat. Cell Biol. 15, 1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, X. and Wang, A. (2017) The potyvirus silencing suppressor protein VPg mediates degradation of SGS3 via ubiquitination and autophagy pathways. J. Virol. 91, e01478–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiramel, A.I. , Brady, N.R. and Bartenschlager, R. (2013) Divergent roles of autophagy in virus infection. Cells, 2, 83–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, Y. , Bowman, J.W. and Jung, J.U. (2018) Autophagy during viral infection – a double‐edged sword. Nat. Rev. Microbiol. 16, 341–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavel, M. , Michaeli, S. and Genschik, P. (2017) Autophagy: a double‐edged sword to fight plant viruses. Trends Plant Sci. 22, 646–648. [DOI] [PubMed] [Google Scholar]
- Derrien, B. , Baumberger, N. , Schepetilnikov, M. , Viotti, C. , De Cillia, J. , Ziegler‐Graff, V. , Isono, E. , Schumacher, K. and Genschik, P. (2012) Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc. Natl. Acad. Sci. USA. 109, 15942–15946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty, J. and Baehrecke, E.H. (2018) Life, death and autophagy. Nat. Cell. Biol. 20, 1110–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, X. and Levine, B. (2013) Autophagy and viruses: adversaries or allies? J. Innate Immun. 5, 480–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, Y. , He, D. , Yao, Z. and Klionsky, D.J. (2014) The machinery of macroautophagy. Cell Res. 24, 24–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, S. , Xu, Y. , Li, C. , Li, Y. , Wu, J. and Zhou, X. (2018) Rice stripe virus Interferes with S‐acylation of remorin and induces its autophagic degradation to facilitate virus infection. Mol. Plant. 11, 269–287. [DOI] [PubMed] [Google Scholar]
- Gatica, D. , Lahiri, V. and Klionsky, D.J. (2018) Cargo recognition and degradation by selective autophagy. Nat. Cell Biol. 20, 233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes, L.C. and Dikic, I. (2014) Autophagy in antimicrobial immunity. Mol. Cell, 54, 224–233. [DOI] [PubMed] [Google Scholar]
- Hafren, A. and Hofius, D. (2017) NBR1‐mediated antiviral xenophagy in plant immunity. Autophagy, 13, 2000–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafren, A. , Lohmus, A. and Makinen, K. (2015) Formation of potato virus A‐induced RNA granules and viral translation are interrelated processes required for optimal virus accumulation. PLoS Pathog. 11, e1005314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafren, A. , Macia, J.L. , Love, A.J. , Milner, J.J. , Drucker, M. and Hofius, D. (2017) Selective autophagy limits cauliflower mosaic virus infection by NBR1‐mediated targeting of viral capsid protein and particles. Proc. Natl. Acad. Sci. USA. 114, E2026–E2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafren, A. , Ustun, S. , Hochmuth, A. , Svenning, S. , Johansen, T. and Hofius, D. (2018) Turnip mosaic virus vounteracts selective autophagy of the viral silencing suppressor HCpro. Plant Physiol. 176, 649–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, S. , Wang, Y. , Zheng, X. , Jia, Q. , Zhao, J. , Bai, F. , Hong, Y. and Liu, Y. (2015) Cytoplastic glyceraldehyde‐3‐phosphate dehydrogenases interact with ATG3 to negatively regulate autophagy and immunity in Nicotiana benthamiana . Plant Cell, 27, 1316–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haxim, Y. , Ismayil, A. , Jia, Q. , Wang, Y. , Zheng, X. , Chen, T. , Qian, L. , Liu, N. , Wang, Y. , Han, S. , Cheng, J. , Qi, Y. , Hong, Y. and Liu, Y . (2017) Autophagy functions as an antiviral mechanism against geminiviruses in plants. eLife, 6, e23897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton, N.S. and Randall, G. (2010) Dengue virus‐induced autophagy regulates lipid metabolism. Cell Host Microbe, 8, 422–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofius, D. , Schultz‐Larsen, T. , Joensen, J. , Tsitsigiannis, D.I. , Petersen, N.H. , Mattsson, O. , Jørgensen, L.B. , Jones, J.D.G. , Mundy, J. and Petersen, M . (2009) Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell, 137, 773–783. [DOI] [PubMed] [Google Scholar]
- Hofius, D. , Li, L. , Hafren, A. and Coll, N.S. (2017) Autophagy as an emerging arena for plant–pathogen interactions. Curr. Opin. Plant Biol. 38, 117–123. [DOI] [PubMed] [Google Scholar]
- Jackson, W.T. (2015) Viruses and the autophagy pathway. Virology, 479–480, 450–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judith, D. , Mostowy, S. , Bourai, M. , Gangneux, N. , Lelek, M. , Lucas‐Hourani, M. , Cayet, N. , Jacob, Y. , Prévost, M.‐C. , Pierre, P. , Tangy, F. , Zimmer, C. , Vidalain, P.‐O. , Couderc, T. and Lecuit, M . (2013) Species‐specific impact of the autophagy machinery on Chikungunya virus infection. EMBO Rep. 14, 534–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner, R. , De la Concepcion, J.C. , Maqbool, A. , Kamoun, S. and Dagdas, Y.F. (2017) ATG8 expansion: a driver of delective autophagy diversification? Trends Plant Sci. 22, 204–214. [DOI] [PubMed] [Google Scholar]
- Klionsky, D.J. and Codogno, P. (2013) The mechanism and physiological function of macroautophagy. J. Innate Immun. 5, 427–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, S.C. and Devenish, R. (2015) Peering into the 'black box' of pathogen recognition by cellular autophagy systems. Microb. Cell, 2, 322–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H.K. , Lund, J.M. , Ramanathan, B. , Mizushima, N. and Iwasaki, A. (2007) Autophagy‐dependent viral recognition by plasmacytoid dendritic cells. Science, 315, 1398–1401. [DOI] [PubMed] [Google Scholar]
- Levine, B. and Kroemer, G. (2019) Biological functions of autophagy genes: a disease perspective. Cell, 176, 11–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, F. , Zhao, N. , Li, Z. , Xu, X. , Wang, Y. , Yang, X. , Liu, S.‐S. , Wang, A. and Zhou, X . (2017) A calmodulin‐like protein suppresses RNA silencing and promotes geminivirus infection by degrading SGS3 via the autophagy pathway in Nicotiana benthamiana . PLoS Pathog. 13, e1006213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, F. , Zhang, C. , Li, Y. , Wu, G. , Hou, X. , Zhou, X. , and Wang, A . (2018) Beclin1 restricts RNA virus infection in plants through suppression and degradation of the viral polymerase. Nat. Commun. 9, 1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Schiff, M. , Czymmek, K. , Talloczy, Z. , Levine, B. and Dinesh‐Kumar, S.P. (2005) Autophagy regulates programmed cell death during the plant innate immune response. Cell, 121, 567–577. [DOI] [PubMed] [Google Scholar]
- Marshall, R.S. and Vierstra, R.D. (2018) Autophagy: the master of bulk and selective recycling. Annu. Rev. Plant Biol. 69, 173–208. [DOI] [PubMed] [Google Scholar]
- Minina, E.A. , Bozhkov, P.V. and Hofius, D. (2014) Autophagy as initiator or executioner of cell death. Trends Plant Sci. 19, 692–697. [DOI] [PubMed] [Google Scholar]
- Mizushima, N. (2018) A brief history of autophagy from cell biology to physiology and disease. Nat. Cell Biol. 20, 521–527. [DOI] [PubMed] [Google Scholar]
- Mizushima, N. , Yoshimori, T. and Ohsumi, Y. (2011) The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107–132. [DOI] [PubMed] [Google Scholar]
- Munch, D. , Rodriguez, E. , Bressendorff, S. , Park, O.K. , Hofius, D. and Petersen, M. (2014) Autophagy deficiency leads to accumulation of ubiquitinated proteins, ER stress, and cell death in Arabidopsis. Autophagy, 10, 1579–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara, K.S. , Masuta, C. , Yamada, S. , Shimura, H. , Kashihara, Y. , Wada, T.S. , Meguro, A. , Goto, K. , Tadamura, K. , Sueda, K. , Sekiguchi, T. , Shao, J. , Itchoda, N. , Matsumura, T. , Igarashi, M. , Ito, K. , Carthew, R.W and Uyeda, I. (2012) Tobacco calmodulin‐like protein provides secondary defense by binding to and directing degradation of virus RNA silencing suppressors. Proc. Natl. Acad. Sci. USA. 109, 10113–10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvedahl, A. , MacPherson, S. , Sumpter, R. Jr , Talloczy, Z. , Zou, Z. and Levine, B. (2010) Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe, 7, 115–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvedahl, A. , Sumpter, R. Jr , Xiao, G. , Ng, A. , Zou, Z. , Tang, Y. , Narimatsu, M. , Gilpin, C. , Sun, Q. , Roth, M. , Forst, C.V. , Wrana, J.L. , Zhang, Y.E. , Luby‐Phelps, K. , Xavier, R.J. , Xie, Y. and Levine, B . (2011) Image‐based genome‐wide siRNA screen identifies selective autophagy factors. Nature, 480, 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paludan, C. , Schmid, D. , Landthaler, M. , Vockerodt, M. , Kube, D. , Tuschl, T. and Münz, C. (2005) Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science, 307, 593–596. [DOI] [PubMed] [Google Scholar]
- Rubinsztein, D.C. , Shpilka, T. and Elazar, Z. (2012) Mechanisms of autophagosome biogenesis. Curr. Biol. 22, R29–34. [DOI] [PubMed] [Google Scholar]
- Sharma, V. , Verma, S. , Seranova, E. , Sarkar, S. and Kumar, D. (2018) Selective autophagy and xenophagy in infection and disease. Front. Cell Dev. Biol. 6, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelly, S. , Lukinova, N. , Bambina, S. , Berman, A. and Cherry, S. (2009) Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity, 30, 588–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Q. , Hu, T. , Bao, M. , Cao, L. , Zhang, H. , Song, F. , Xie, Q. and Zhou, X. (2016) Tobacco RING E3 Ligase NtRFP1 mediates ubiquitination and proteasomal degradation of a geminivirus‐encoded βC1. Mol. Plant, 9, 911–925. [DOI] [PubMed] [Google Scholar]
- Slobodkin, M.R. and Elazar, Z. (2013) The Atg8 family: multifunctional ubiquitin‐like key regulators of autophagy. Essays Biochem. 55, 51–64. [DOI] [PubMed] [Google Scholar]
- Ustun, S. , Hafren, A. and Hofius, D. (2017) Autophagy as a mediator of life and death in plants. Curr. Opin. Plant Biol. 40, 122–130. [DOI] [PubMed] [Google Scholar]
- Wileman, T. (2013) Autophagy as a defence against intracellular pathogens. Essays Biochem. 55, 153–163. [DOI] [PubMed] [Google Scholar]
- Wu, X. , Valli, A. , Garcia, J.A. , Zhou, X. and Cheng, X. (2019) The tug‐of‐war between plants and viruses: great progress and many remaining questions. Viruses, 11, E203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, M. , Zhang, Y. , Xie, X. , Yue, N. , Li, J. , Wang, X.B. , Han, C. , Yu, J. , Liu, Y. and Li, D. (2018) Barley stripe mosaic virus yb protein subverts autophagy to promote viral infection by disrupting the ATG7‐ATG8 Interaction. Plant Cell, 30, 1582–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto, K. , Jikumaru, Y. , Kamiya, Y. , Kusano, M. , Consonni, C. , Panstruga, R. , Ohsumi, Y. and Shirasu, K. (2009) Autophagy negatively regulates cell death by controlling NPR1‐dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis . Plant Cell, 21, 2914–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, L. , Chen, Y. and Tooze, S.A. (2018) Autophagy pathway: cellular and molecular mechanisms. Autophagy, 14, 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffagnini, G. and Martens, S. (2016) Mechanisms of selective autophagy. J. Mol. Biol. 428, 1714–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zvereva, A.S. , Golyaev, V. , Turco, S. , Gubaeva, E.G. , Rajeswaran, R. , Schepetilnikov, M.V. , Srour, O. , Ryabova, L.A. , Boller, T. and Pooggin, M.M. (2016) Viral protein suppresses oxidative burst and salicylic acid‐dependent autophagy and facilitates bacterial growth on virus‐infected plants. New Phytol. 211, 1020–1034. [DOI] [PubMed] [Google Scholar]


