Abstract
Several studies have reported the persistence of HCV RNA in liver and/or peripheral blood mononuclear cells (PBMCs) in spite of undetectable viremia in patients who have achieved sustained virological response (SVR). This event, defined as occult HCV infection, remains controversial and low titers of persistent virus may be underestimated because it has not yet been analyzed by a highly sensitive test such as droplet digital PCR (ddPCR). This method provides an alternate ultra-sensitive detection technique for very low numbers of copies of viral RNA or DNA. The aim of this study was to evaluate the persistence of HCV in HIV-coinfected patients with long-term SVR using ddPCR. For each patient, the presence of HCV RNA in serum and PBMCs at baseline was determined by nested RT-ddPCR. Patients with HCV RNA in PBMCs at baseline were followed until the end of the study. One hundred and twenty-three patients were analyzed for persistence of HCV RNA in serum and PBMCs. Persistence of HCV was not found in serum in any patient. HCV RNA was detected in PBMCs in one patient (0.81%; 95% CI: 0.04–3.94) and resolved spontaneously during follow-up. Persistence of HCV RNA in PBMCs is not a common event in HIV/HCV co-infected patients with long-term SVR evaluated by RT-ddPCR.
Subject terms: Molecular biology, Medical research
Introduction
The objective of chronic HCV infection (CHC) therapy is to achieve sustained virological response (SVR), defined as absence of HCV RNA in serum at twelve or twenty-four weeks after successful treatment1. In the context of HCV infection, therefore, SVR is synonymous with cure, and an HCV RNA determination after SVR is only recommended if the patient has ongoing risk of HCV reinfection1. Several studies however have reported that the persistence of HCV RNA in the liver and/or peripheral blood mononuclear cells (PBMCs) in patients who have achieved SVR can lead to late relapses2–6. This unusual event is recognized as occult HCV infection and defined as the presence of HCV RNA in liver tissue or PBMCs despite undetectable viremia7. This situation remains controversial as several studies have been unable to confirm the presence of HCV RNA in PBMCs or liver tissue in patients with long-term viral clearance8–10. Low titers of persistent virus may be underestimated as this event has not been confirmed with highly sensitive tests with lower limits of detection than reverse transcriptase polymerase chain reaction (RT-PCR) or transcription-mediated amplification (TMA).
Droplet digital PCR (ddPCR) represents an alternative technique, providing ultra-sensitive detection of very low numbers of copies of viral RNA or DNA11–14. In the context of viral infection, ddPCR has been demonstrated to be useful in the measurement of HIV reservoirs15,16. Given that viral persistence in SVR patients remains controversial, the use of ddPCR may be significant for detecting residual HCV RNA in this population. The objective of our study therefore was to use ddPCR to evaluate the persistence of HCV in HIV-coinfected patients with long-term SVR.
Material and Methods
Study populations
HIV-infected patients in follow-up at seven reference hospitals in Andalusia (southern Spain) between January 2015 and January 2018 were included in this prospective longitudinal study. Patients included had to fulfill the following inclusion criteria: (i) treatment-induced SVR, defined as the absence of detectable serum HCV RNA 24 weeks after end of treatment; (ii) at least 12 months of SVR before study inclusion; (iii) annual undetectable serum HCV RNA levels by qRT-PCR (detection limit set at 15–20 IU/mL) after achieving SVR until inclusion in the study; (iv) annual liver stiffness measurements after achieving SVR. Patients with ongoing risk of HCV reinfection were excluded.
Follow-up and variable collection
Using density gradient centrifugation, PBMCs were isolated from whole blood and serum samples collected from all patients (Ficoll-Hypaque). To stabilize RNA, isolated PBMCs and serum samples were treated with RNAlater Solution (Thermo Fisher Scientific, USA) in a 1:1 concentration and cryopreserved at −80 °C.
Nested RT-ddPCR was used to determine the baseline presence of HCV RNA in serum and PBMCs in all patients. Those patients with baseline presence of HCV RNA in PBMCs were followed until the end of the study. Follow-up consisted of an annual determinations of HCV RNA in both PBMCs and serum.
The liver stiffness (LS) records of all patients were collected from achievement of SVR until the end of the study. The cutoff values of LS used to determine liver fibrosis stage in this study were as follows: <6.5 kPa (F0-F1, minimal or absent fibrosis); 6.5–9.4 kPa (F2, significant fibrosis); 9.5–14.5 kPa (F3, advanced fibrosis) and ≥14.6 kPa (F4, cirrhosis)17. Patients whose liver fibrosis increased by at least one stage after achieving SVR were considered to be liver fibrosis progressed. Patients with no identifiable cause of liver fibrosis progression other than HCV infection underwent liver biopsy, and PCR was performed to detect HCV RNA in liver tissue. Patients with HCV RNA present in liver tissue were followed up.
The primary outcome of the study was occult HCV infection, defined as the presence of HCV RNA in PBMCs and/or liver tissue, in patients with an undetectable HCV viral load in serum by ddRT-PCR. From the achievement of SVR until the end of follow-up, possible variables related to persistence of HCV RNA in HIV patients were collected, including: (i) HCV infection-related variables: type of HCV therapy, duration of therapy, HCV genotype, baseline HCV viral load, and liver stiffness; (ii) HIV infection-related variables: CD4+ count, HIV RNA, AIDS criteria and use of combination antiretroviral therapy (cART); (iii) demographic and clinical variables: gender, age, alanine aminotransferase (ALT) and aspartate aminotransferase (AST). (iv) risk practices for HCV reinfection.
Nested RT-ddPCR protocol
Viral RNA was extracted from the PBMC samples using the commercial RNeasy plus universal kit (Qiagen, Hilden, Germany). Viral RNA was extracted from serum with the QIAamp minElute virus spin kit (Qiagen, Hilden, Germany). Both extractions were performed using automated QIAcube procedures (QIAgen, Hilden, Germany). Two rounds of PCR were performed for HCV RNA detection, both using primers described previously: sense 5′-CTTCACGCRGAAAGCGYCTA3′ and antisense 5′-CAAGCACCCTATCAGGCAGT-3′ as outer primers, and sense 5′-GCGTTAGTAYGAGTGTYG-3′ and antisense 5′-CRATTCCGGTGTACTCAC-3 as inner primers18. These primers amplify the core region of the HCV genome for four viral genotypes (1, 2, 3 and 4).
The first round of PCR was performed with the iTaq Universal Probes One-Step Kit (Bio-rad, USA). The 20 µL reaction mix contained 10 µL of iTaq universal probes reaction mix (2×), 900 nM of each primer, 0.5 µL of iScript reverse transcriptase and 9.14 µL of template RNA. A Biorad C100 thermal cycler (Biorad; CA, USA) was used, and the cycling conditions were: 50 °C for 10 min, 95 °C for 3 min, and 45 cycles at 95 °C for 20 s, 55 °C for 30 s, and 60 °C for 20 s. The amplified product of 246-bp was then purified with the QIAquick PCR Purification Kit (QIAgen, Hilden, Germany). In the second PCR round, each reaction mix contained 10 µL of ddPCR™ Supermix for Probes (Bio-Rad, USA), 900 nM of each primer, 250 nM of FAM-labeled HCV probe (5′-FAM-CCGCAGACCACTATGGCTC-BHQ1-3′), and 9.35 µL of amplified product from the first round of PCR in a final volume of 20 μL. The length of the second-round PCR amplified product was 89-bp. Nuclease-free water (Sigma-Aldrich Inc., USA) was used as negative control, and both were processed in parallel with cDNA synthesis and analyzed directly by ddPCR. Serum and PBMC samples of patients chronically infected with HCV genotype 1 (serum viral load: 11,909,700 IU/mL), genotype 3 (serum viral load: 4,577,570 IU/mL), genotype 4 (serum viral load: 8,450,166 IU/mL) were used as positive controls, in a 1/200 dilution.
The ddPCR reaction mixes were placed in the Bio-Rad QX200™ Droplet Generator and droplets generated in accordance with the manufacturer’s instructions. The droplets were transferred to a 96-well PCR plate and sealed with Bio-Rad PX1™ PCR Plate Sealer, following the manufacturer’s instructions. Amplification was performed on a Bio-Rad C1000 Touch™ Thermal Cycler at the following cycling temperatures: 95 °C for 10 min, followed by 40 cycles at 94 °C for 30 s and at 55.5 °C for 1 minute and 1 cycle at 98 °C for 10 min, ending with a hold step at 4 °C. After PCR, the plate was immediately read on the Bio-Rad QX200™ Droplet Reader, where the droplets were analyzed.
Use of ddPCR gives an absolute count of genome copies of HCV in PBMCs without the use of standard curves. Fluorescence intensity in every well was analyzed with Bio-Rad QuantaSoft analysis software v. 1.7. After defining the threshold based on fluorescence amplitudes, the droplets were classified as positive or negative. The number of positive and negative droplets was used to calculate the target concentration. This concentration was estimated assuming a Poisson distribution. The QuantaSoft analysis software determined the target concentration by adjusting it to a Poisson algorithm. The formula used by the QuantaSoft software was: [C = -ln(Nneg/N)/Vdroplet], where: C = copies per droplet; Nneg = number of negative droplets; N = total number of droplets; Vdroplet = volume of droplet (0.85nL according to the manufacturer’s instructions).
HCV antigenomic strand determination
PBMC samples positive by ddPCR were also analyzed for a negative-strand HCV determination. PCR was performed using the iTaq Universal Probes One-Step Kit (Bio-rad, USA). The sense and antisense primers used were 5′-AGACTCACTCCCCTGTGAGGAA-3′ and 5′- TGAGTGCACGGTCTACGAGACCTC-3′ respectively. The 20 µL reaction mix contained 10 µL of iTaq universal probes reaction mix (2×), 500 nM of each primer, 0.5 µL of iScript reverse transcriptase and 8.5 µL of template RNA. A thermal cycler Biorad C100 (Biorad; CA, USA) was used and the following thermal cycling conditions applied: 50 °C for 10 min, 95 °C for 3 min and 45 cycles at 95 °C for 20 s, 67 °C for 30 s, and 60 °C for 20 s. The amplified 311 bp product was then purified by QIAquick PCR Purification Kit (QIAgen, Hilden, Germany).
Statistical analysis
Continuous variables were expressed as means (standard deviation) or median and quartiles (Q1-Q3). Categorical variables were expressed as numbers of cases and percentages. Analyses were carried out using the statistical software package SPSS version 18.0 (IBM Corporation, Somers, NY, USA) and GraphPad Prism version 6 (Mac OS X version; GraphPad Software, San Diego, CA, USA).
Ethical statement
This study was designed and performed according to the Helsinki Declaration. All patients signed an informed consent form. The CEIC (Clinical Trial and Ethical Committee) of the Hospital Universitario Reina Sofía de Córdoba, the coordinating Hospital, approved the study protocol.
Results
Study population
One hundred and twenty-three HIV-infected patients with treatment-induced SVR were included in the study. The median time from SVR was 51 months (IQR: 27–76). The distribution of patients by time from SVR was: 12–24 months, n = 27 (21.9%); 25–36 months, n = 15 (12.2%); 37–48 months, n = 14 (11.4%), 49–60 months, n = 14 (11.4%) and more than 60 months, n = 53 (43.1%). The main patient characteristics are summarized in Table 1.
Table 1.
Main baseline patient characteristics.
| Clinical characteristics | Condition | N = 123 |
|---|---|---|
| Age (years), median (IQR) | — | 51 (48–55) |
| Sex, n (%) | Male | 102 (82.9) |
| Female | 21 (17.1) | |
| HIV viral load, n (%) | Undetectable | 119 (96.7) |
| Detectable | 4 (3.3) | |
| CD4 count at inclusion study (cell/mL), median (IQR) | — | 560 (396–726.5) |
| Use of ART, n (%) | Yes | 122 (99.2) |
| No | 1 (0.8) | |
| HCV genotype, n (%) | 1 | 59 (48) |
| 2 | 4 (3.2) | |
| 3 | 54 (43.9) | |
| 4 | 6 (4.9) | |
| Type of HCV therapya | Peg-IFN + RBV | 102 (82.9) |
| DAAb + Peg-IFN + RBV | 20 (16.3) | |
| IFN-free | 1 (0.8) | |
| Liver stiffness (kPa) at inclusion in study, median (IQR) | — | 6 (4.7–7.7) |
| Liver stiffness (kPa) at SVR, median (IQR) | — | 7.2 (5.4–11.7) |
| Time from SVR (months), median (IQR) | — | 51 (27–76) |
Interquertile range (IQR); n (number of cases); human immunodeficiency virus (HIV); antiretroviral therapy (cART); hepatitis C virus (HCV); pegylated interferon (Peg-IFN); ribavirin (RBV); direct-acting antiviral (DAA).
aHCV therapy which induced SVR;
bdaclatasvir (n = 7); telaprevir or boceprevir (n = 13).
Evaluation of low-titer persistent HCV in serum and PBMCs
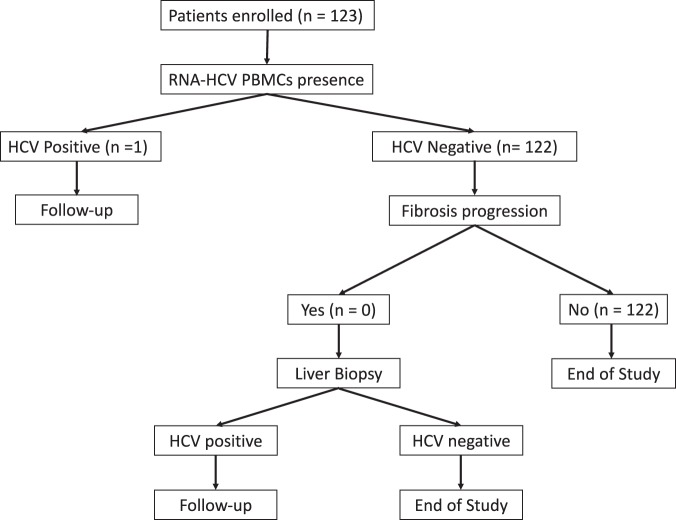
The flow chart of patients in the study is shown in Fig. 1. One hundred and twenty-three patients were evaluated for HCV persistence in serum and HCV RNA was detected in none. The persistence of HCV RNA was then analyzed in PBMCs and was detected in 1 patient (0.81%; 95%CI: 0.04–3.94).
Figure 1.
Flow chart of patients evaluated in the study.
Liver fibrosis progression
The median LS of included patients at the time of SVR was 7.2 kPa (IQR: 5.4–11.7). The median LS value at inclusion was 6 kPa (IQR: 4.7–7.7). None of the patients had criteria for fibrosis progression (increased liver stiffness) between achieving SVR and inclusion in the study and none therefore fulfilled the criterion for liver biopsy. Of the patients included, 14.6% had a liver stiffness score of over 14.5 kPa at achievement of SVR and the percentage of those with liver stiffness values of 14.5 kPa or more decreased to 6.5% at the start of the study.
Follow-up of patient with occult infection
Of all the patients included in the study, only 1 patient fulfilled the criteria for occult HCV infection. This patient was male, infected with genotype 1 HCV, had AIDS criteria in the past, carried IL28B genotype CC, and had SVR for 69 months before inclusion in the study. The patient achieved SVR after a 24-week course of treatment with pegylated interferon plus ribavirin (Peg-IFN/RBV). HCV RNA continued to be present in this patient’s PBMCs until the last study visit, with a decreasing number of HCV-positive droplets between the baseline visit and the end of the study (Table 2 and Fig. S1). CD4+ counts during follow-up increased from 291 cells/mL to 558 cells/mL at the last visit (Table 2). The antigenomic HCV strand was detected at the first follow-up visit (Table 2).
Table 2.
Follow-up of the patient detected with HCV RNA.
| Baseline | First visit | Second visit | Third visit | |||||
|---|---|---|---|---|---|---|---|---|
| Droplets analyzed | Positive droplets | Droplets analyzed | Positive droplets | Droplets analyzed | Positive droplets | Droplets analyzed | Positive droplets | |
| HCV RNA in PBMCs | 12.034 | 12.034 | 11.380 | 11.106 | 14.575 | 4 | 13.726 | 0 |
| HCV RNA in serum | 14.615 | 0 | 14.197 | 0 | 14.353 | 0 | 13.295 | 0 |
| Antigenomic HCV strand | Negative | Positive | Negative | Negative | ||||
| CD4 (cells/mL) | 290 | 406 | 663 | 558 | ||||
| Liver stiffness [kPa (IQR)] | 3.7 (0.7) | 4.6 (1.0) | 5.5 (1.0) | 5.1 (0.8) | ||||
| ALT (U/L) | 19 | 20 | 29 | 24 | ||||
| AST (U/L) | 26 | 24 | 31 | 28 | ||||
| HIV RNA (copies/mL) | <20 | <20 | <20 | <20 | ||||
Abbreviations: peripheral blood mononuclear cells (PBMCs); positive droplets (PD); kilopascal (kPA); alanine aminotransferase (ALT); aspartate aminotransferase (AST).
Discussion
To the best of our knowledge, this is the first study to evaluate the persistence of HCV in PBMCs after achieving long-term SVR using an ultra-sensitive method such as ddPCR. HCV RNA persistence in PBMCs was found in 1 patient who had achieved SVR with sustained clearance of serum HCV RNA for 8 years. Our findings confirm therefore that HCV RNA can persist in PBMCs after long-term SVR, but suggest that this is a rare event. Several studies have described the persistence of HCV RNA in long-term SVR, even after spontaneous resolution of infection19–25. In a cohort of 54 patients with long-term SVR, Hedenstierna et al. detected HCV RNA in PBMC samples from two patients, and the time between SVR and positivity of samples was 5 and 9 years, respectively. These patients were later HCV RNA-negative in a re-analysis 5 and 4 years later, respectively20. Similarly, in a study analyzing whole blood samples from 52 patients with long-term SVR using ultracentrifugation combined with RT-PCR, Lybeck et al. found HCV RNA in 2 patients. These determinations were made 8 and 9 years respectively after SVR. A second determination performed months later was HCV RNA-negative21. Finally, in a longitudinal study conducted by Garcia-Bengoechea et al., 10 SVR patients were followed and showed a gradual decrease in HCV RNA in PBMCs and late disappearance during follow-up, while HCV RNA remained undetectable in serum in all patients22. In our study, the patient was HCV RNA-positive in PBMCs 8 years after achieving SVR, but remained undetectable in serum. The patient remained positive at the first visit after baseline, but presented very few positive droplets at the second. Finally, at the last visit, the patient tested HCV RNA-negative. The antigenomic strand was also detected at the first follow-up visit. In line with the studies mentioned, it may be concluded that after achieving SVR, the probability of finding residual HCV RNA decreases over time.
In the context of HIV co-infection, effective treatment with cART suppresses HIV replication and restores the total CD4+cell count. Cases have been reported of spontaneous resolution of chronic HCV infection after immune reconstitution based on the use of effective antiretroviral treatment26–28. In the patient identified in our study, the progressive clearance of HCV RNA from PBMCs coincided with a significant progressive increase in CD4+ cells following SVR, which could suggest that HCV RNA clearance from PBMCs could be associated with the timing of progressive immune reconstitution.
The persistence of HCV RNA in PBMCs could be a potential risk for late relapse, defined as viral rebound after attaining SVR. This has been reported in several studies, which included patients treated with Peg-IFN/RBV or IFN-free therapy29–33. In a study including HCV-monoinfected subjects, Desmond et al. found late HCV relapse in 1 out of 147 patients who achieved SVR with Peg-IFN/RBV (0.68%). The relapse was identified 46 weeks after achieving SVR29. Likewise, in a cohort of 129 transplant patients who achieved SVR, Elmasry et al. reported a late HCV relapse rate of 3.8% (n = 5)30. In an SVR registry study including 5,433 patients treated with DAAs in combination or not with Peg-IFN/RBV (NCT01457755), Lawitz et al. identified 6 patients (0.1%) with late HCV relapse between 217 and 429 days after completion of therapy, confirmed by phylogenetic analysis31. Most studies have reported late relapses, even in the first year after achieving SVR29–33. In a study analyzing 262 SVR patients treated with Peg-IFN/RBV, Giannini et al. described two types of patient with positive HCV RNA after SVR: those who were transiently HCV RNA-positive after a long time with SVR (n = 18), and those who experienced a true late relapse (n = 2) in the first year after achieving SVR24. In our study, the median SVR was 51 months, so that we are unable to report possible relapses occurring in the first few months following achievement of SVR. Nonetheless, the patient identified with persistence of HCV RNA did not experience a very late relapse in follow-up, which would be consistent with most studies that have found HCV RNA in long-term SVR patients20–24.
Studies that have found residual HCV RNA did not report liver damage or fibrosis progression20,21,24. Castillo et al. found the antigenomic HCV RNA strand in 15 of 20 SVR patients in a longitudinal study evaluating HCV RNA persistence in paired liver biopsies. Nevertheless, in spite of this evidence of HCV virus replication and persistence, the paired liver biopsies revealed a significant improvement in necroinflammatory activity and liver fibrosis stage after SVR6. In our study, the liver stiffness values of this patient did not progress during follow-up, and the patient did not experience transaminase elevations during the study period, which suggests that HCV viral persistence does not correlate with liver injury or liver fibrosis progression.
Nonetheless, given that patients cured of HCV may be organ or blood donors, the possibility of HCV RNA persistence in patients who have achieved SVR may justify further evaluation with highly sensitive procedures. Similarly, the risk of mother-to-child transmission has been associated with the presence of HCV RNA in PBMCs34,35. This would be critical in women with SVR who hope to become pregnant, because the persistence of HCV RNA in PBMCs could have important implications for vertical transmission.
Our study presents several limitations that should be noted. First, the HCV RNA detected was not sequenced, so that we were unable to accurately define the phylogeny of the virus. In this respect, although the patient did not engage in risky practices for HCV reinfection, the possibility could not be ruled out. Second, measuring HCV RNA in other compartments, such as the liver, was not available in this study. Finally, this study included only HIV/HCV co-infected patients and the results cannot therefore be extrapolated to series of HCV-monoinfected individuals.
In conclusion, HCV RNA persistence in PBMCs is not a common event in HIV/HCV co-infected patients with long-term SVR after evaluation with an ultra-sensitive procedure such as ddPCR. Only one patient had viral persistence and this did not lead to HCV-related clinical complications, such as late relapse or liver fibrosis progression.
Supplementary information
Acknowledgements
This work was supported by the Fundación Progreso y Salud, Consejería de Salud de la Junta de Andalucía (grants for health research projects: refs. PI0036/2010; PI0187/2013), the Ministerio de Sanidad (RD12/0017/0012) integrated in the Plan Nacional de I + D + I, and cofinanced by the ISCIII-Subdirección General de Evaluación and the Fondo Europeo de Desarrollo Regional (FEDER), the Fundación para la Investigación en Salud (FIS) del Instituto Carlos III (PI15/01017), and the Red de Investigación en SIDA de España ISCIII-RETIC (Grant Number: RD16/0025/0034). A. Rivero Juárez and M. Frías are the recipient of a Miguel Servet and Sara Borrell postdoctoral perfection grants by the Ministerio de Ciencia, Innovación y Universidades of Spain (CP18/00111) and (CD18/00091), respectively.
Author Contributions
Conceived and designed the study: M.F., A.R.J., A.C. and A.R. Funding: A.C., A.R. Collected data: All authors. Analyzed the results: M.F., A.R.J., A.C. and A.R. Wrote the manuscript: M.F., A.R.J., A.C. and A.R. Critical review of the manuscript: All authors.
Competing Interests
We declare no competing interests with the present work. Authors or their institutions did not receive a payment or services from a third party at any time or for any aspect of the submitted work (data monitoring board, study design, manuscript preparation, statistical analysis, etc.).
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-48966-9.
References
- 1.Recommendations for Testing, Managing, and Treating Hepatitis C of the American Association for The Study of Liver Diseases (AASLD) and the Infectious Diseases Society of America (IDSA). Available at, http://www.hcvguidelines.org (last accessed August 2th 2019)
- 2.Carreno V. Occult hepatitis C virus infection: a new form of hepatitis C. World J Gastroenterol. 2006;12:6922–6925. doi: 10.3748/wjg.v12.i43.6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castillo I, et al. Hepatitis C virus replicates in peripheral blood mononuclear cells of patients with occult hepatitis C virus infection. Gut. 2005;254:682–685. doi: 10.1136/gut.2004.057281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pham Tram N.Q., King Dawn, MacParland Sonya A., McGrath Jerry S., Reddy S. Bharati, Bursey Ford R., Michalak Tomasz I. Hepatitis C Virus Replicates in the Same Immune Cell Subsets in Chronic Hepatitis C and Occult Infection. Gastroenterology. 2008;134(3):812–822. doi: 10.1053/j.gastro.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 5.Pham, T. N. & Michalak, T. I. Occult hepatitis C virus infection and its relevance in clinical practice. J Clin Exp Hepatol. Dec, 1(3), 185–9 (2011). [DOI] [PMC free article] [PubMed]
- 6.Castillo I, et al. Hepatitis C virus replicates in the liver of patients who have a sustained response to antiviral treatment. Clin Infect Dis. 2006;43:1277–1283. doi: 10.1086/508198. [DOI] [PubMed] [Google Scholar]
- 7.Welker MW, Zeuzem S. Occult hepatitis C: how convincing are the current data? Hepatology. 2009;49:665–675. doi: 10.1002/hep.22706. [DOI] [PubMed] [Google Scholar]
- 8.Maylin S, et al. Eradication of hepatitis C virus in patients successfully treated for chronic hepatitis C. Gastroenterology. 2008;135:821–829. doi: 10.1053/j.gastro.2008.05.044. [DOI] [PubMed] [Google Scholar]
- 9.Martinot-Peignoux M, et al. Sustained virological response is associated with eradication of hepatitis C virus and decrease in anti-HCV titer in patients treated for chronic hepatitis C [Abstract] J Hepatol. 2008;48(S2):S302. [Google Scholar]
- 10.Bernardin F, et al. Clearance of hepatitis C virus RNA from the peripheral blood mononuclear cells of blood donors who spontaneously or therapeutically control their plasma viremia. Hepatology. 2008;47:1446–1452. doi: 10.1002/hep.22184. [DOI] [PubMed] [Google Scholar]
- 11.Arvia Rosaria, Sollai Mauro, Pierucci Federica, Urso Carmelo, Massi Daniela, Zakrzewska Krystyna. Droplet digital PCR (ddPCR) vs quantitative real-time PCR (qPCR) approach for detection and quantification of Merkel cell polyomavirus (MCPyV) DNA in formalin fixed paraffin embedded (FFPE) cutaneous biopsies. Journal of Virological Methods. 2017;246:15–20. doi: 10.1016/j.jviromet.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 12.Sze Marc A., Abbasi Meysam, Hogg James C., Sin Don D. A Comparison between Droplet Digital and Quantitative PCR in the Analysis of Bacterial 16S Load in Lung Tissue Samples from Control and COPD GOLD 2. PLoS ONE. 2014;9(10):e110351. doi: 10.1371/journal.pone.0110351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uchiyama, Y. et al. Ultra–sensitive droplet digital PCR for detecting a low–prevalence somatic GNAQ mutation in Sturge–Weber syndrome. Sci Rep. 6, 22985. Published online 2016 Mar 9, 10.1038/srep22985 (2016). [DOI] [PMC free article] [PubMed]
- 14.Jones M, et al. Low copy target detection by Droplet Digital PCR through application of a novel open access bioinformatic pipeline, ‘definetherain’. J Virol Methods. 2014;202:46–53. doi: 10.1016/j.jviromet.2014.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trypsteen, W., Kiselinova, M., Vandekerckhove, L., De Spiegelaere, W. Diagnostic utility of droplet digital PCR for HIV reservoir quantification. J Virus Erad. Jul, 2(3), 162–169 (2016). [PMC free article] [PubMed]
- 16.Ruelle J, Yfantis V, Duquenne A, Goubau P. Validation of an ultrasensitive digital droplet PCR assay for HIV-2 plasma RNA quantification. J Int AIDS Soc. 2014;17:19675. doi: 10.7448/IAS.17.4.19675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vergara S, et al. The use of transient elastometry for assessing liver fibrosis in patients with HIV and hepatitis C virus coinfection. Clin Infect Dis. 2007;45(8):969–974. doi: 10.1086/521857. [DOI] [PubMed] [Google Scholar]
- 18.Rivero-Juarez A., Caruz A., Real L. M., Martinez-Dueñas L., Marquez F. J., Frias M., Recio E., Gordon A., Pineda J. A., Rivero A., Camacho A. Longitudinal evaluation of hepatitis C viral persistence in HIV-infected patients with spontaneous hepatitis C clearance. European Journal of Clinical Microbiology & Infectious Diseases. 2015;34(11):2171–2175. doi: 10.1007/s10096-015-2463-1. [DOI] [PubMed] [Google Scholar]
- 19.Radkowski Marek, Gallegos-Orozco Juan F., Jablonska Joanna, Colby Thomas V., Walewska-Zielecka Bozena, Kubicka Joanna, Wilkinson Jeffrey, Adair Debra, Rakela Jorge, Laskus Tomasz. Persistence of hepatitis C virus in patients successfully treated for chronic hepatitis C. Hepatology. 2005;41(1):106–114. doi: 10.1002/hep.20518. [DOI] [PubMed] [Google Scholar]
- 20.Hedenstierna M., Weiland O., Brass A., Bankwitz D., Behrendt P., Uhnoo I., Aleman S., Cardell K., Fryden A., Norkrans G., Eilard A., Glaumann H., Pietschmann T., Sällberg M., Brenndörfer E. D. Long-term follow-up of successful hepatitis C virus therapy: waning immune responses and disappearance of liver disease are consistent with cure. Alimentary Pharmacology & Therapeutics. 2015;41(6):532–543. doi: 10.1111/apt.13096. [DOI] [PubMed] [Google Scholar]
- 21.Lybeck, C. et al. Long-term follow-up after cure from chronic hepatitis C virus infection shows occult hepatitis and a risk of hepatocellular carcinoma in noncirrhotic patients. Eur J Gastroenterol Hepatol. Nov 19 (2018). [DOI] [PMC free article] [PubMed]
- 22.Garcia-Bengoechea Manuel, Basaras Miren, Barrio Jesus, Arrese Elisabet, Montalvo Inmaculada I., Arenas Juan I., Cisterna Ramon. Late disappearance of hepatitis C virus RNA from peripheral blood mononuclear cells in patients with chronic hepatitis C in sustained response after alpha-interferon therapy. The American Journal of Gastroenterology. 1999;94(7):1902–1905. doi: 10.1111/j.1572-0241.1999.01227.x. [DOI] [PubMed] [Google Scholar]
- 23.Veerapu, N. S., Raghuraman, S., Liang, T.J., Heller, T. & Rehermann, B. Sporadic reappearance of minute amounts of hepatitis C virus RNA after successful therapy stimulates cellular immune responses. Gastroenterology. Feb, 140(2), 676–685.e1 (2011). [DOI] [PMC free article] [PubMed]
- 24.Giannin, E. G., Basso, M., Savarino, V. & Picciotto, A. Sustained virological response to pegylated interferon and ribavirin is maintained during long-term follow-up of chronic hepatitis C patients. Aliment PharmacolTher. Feb 15, 31(4): 502–8 (2010). [DOI] [PubMed]
- 25.Chen Annie Y., Hoare Matthew, Shankar Arun N., Allison Michael, Alexander Graeme J. M., Michalak Tomasz I. Persistence of Hepatitis C Virus Traces after Spontaneous Resolution of Hepatitis C. PLOS ONE. 2015;10(10):e0140312. doi: 10.1371/journal.pone.0140312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vispo Eugenia, Barreiro Pablo, Plaza Zulema, Fernández-Montero Jose Vicente, Labarga Pablo, de Mendoza Carmen, Sierra-Enguita Rocío, Treviño Ana, Lopez Mariola, Soriano Vicente. Spontaneous hepatitis C virus clearance in HIV patients with chronic hepatitis C bearing IL28B-CC alleles using antiretroviral therapy. AIDS. 2014;28(10):1473–1478. doi: 10.1097/QAD.0000000000000275. [DOI] [PubMed] [Google Scholar]
- 27.Frias Mario, Rivero-Juarez Antonio, Tellez Francisco, Perez-Perez Monserrat, Camacho Angela, Machuca Isabel, Lorenzo-Moncada Sandra, Lopez-Lopez Pedro, Rivero Antonio. Spontaneous clearance of chronic hepatitis C is rare in HIV-infected patients after effective use of combination antiretroviral therapy. PLOS ONE. 2017;12(5):e0177141. doi: 10.1371/journal.pone.0177141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soares J, Santos JV, Sarmento A, Costa-Pereira A. Spontaneous Viral Clearance in Sixteen HIV-Infected Patients with Chronic Hepatitis C. Intervirology. 2018;61(2):64–71. doi: 10.1159/000490056. [DOI] [PubMed] [Google Scholar]
- 29.Desmond C. P., Roberts S. K., Dudley F., Mitchell J., Day C., Nguyen S., Pianko S. Sustained virological response rates and durability of the response to interferon-based therapies in hepatitis C patients treated in the clinical setting. Journal of Viral Hepatitis. 2006;13(5):311–315. doi: 10.1111/j.1365-2893.2005.00685.x. [DOI] [PubMed] [Google Scholar]
- 30.Elmasry Sandra, Wadhwa Sanya, Bang Bo-Ram, Cook Linda, Chopra Shefali, Kanel Gary, Kim Brian, Harper Tammy, Feng Zongdi, Jerome Keith R., Kahn Jeffrey A., Saito Takeshi. Detection of Occult Hepatitis C Virus Infection in Patients Who Achieved a Sustained Virologic Response to Direct-Acting Antiviral Agents for Recurrent Infection After Liver Transplantation. Gastroenterology. 2017;152(3):550-553.e8. doi: 10.1053/j.gastro.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lawitz E.J., Ruane P., Stedman C., Foster G., Hyland R.H., Coogan S., Moody S., Dvory-Sobol H., Knox S.J., Brainard D.M., Abergel A., Agarwal K., Younes Z., Schwabe C. Long-Term Follow-Up of Patients with Chronic HCV Infection following Treatment with Direct Acting Antiviral Regimens: Maintenance of SVR, Persistence of Resistance Mutations and Clinical Outcomes. Journal of Hepatology. 2016;64(2):S612–S613. doi: 10.1016/S0168-8278(16)01134-X. [DOI] [Google Scholar]
- 32.McHutchison J. Hepatic HCV RNA before and after treatment with interferon alone or combined with ribavirin. Hepatology. 2002;35(3):688–693. doi: 10.1053/jhep.2002.31870. [DOI] [PubMed] [Google Scholar]
- 33.Sarrazin Christoph, Isakov Vasily, Svarovskaia Evguenia S., Hedskog Charlotte, Martin Ross, Chodavarapu Krishna, Brainard Diana M., Miller Michael D., Mo Hongmei, Molina Jean-Michel, Sulkowski Mark S. Late Relapse Versus Hepatitis C Virus Reinfection in Patients With Sustained Virologic Response After Sofosbuvir-Based Therapies. Clinical Infectious Diseases. 2016;64(1):44–52. doi: 10.1093/cid/ciw676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baré, P. Hepatitis C virus and peripheral blood mononuclear cell reservoirs. World J Hepatol. Oct 31, 1(1), 67–71 (2009). [DOI] [PMC free article] [PubMed]
- 35.Azzari C, et al. Vertical transmission of HCV is related to maternal peripheral blood mononuclear cell infection. Blood. 2000;96:2045–2048. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.