Current management of Xanthomonas perforans on tomato plants mainly relies on the frequent application of pesticides. However, the lack of effective pesticides and the development of strain tolerance to certain bactericides limit the ability to control outbreaks in production fields. Better knowledge of probable sources of X. perforans inoculum during tomato production is required to refine management strategies. Tomato plants are typically established in the field using transplants. This study aimed to determine if strains from field epidemics were coming from transplant facilities or resulted from local field outbreaks. The overall goal was to identify potential sources of inoculum and subsequently develop strategies to reduce carryover from transplant production to the field. Our results indicate that tomato producers should shift disease management efforts to transplant facilities to reduce disease in the field. Improved transplant health should reduce the likelihood of bacterial spot outbreaks and subsequently reduce pesticide usage in the field.
KEYWORDS: DAPC, PCA, SNP, bacterial spot, cgMLST, ecology, epidemics, genome, seedlings, tracking
ABSTRACT
Outbreaks of bacterial spot on tomato (BST) caused by Xanthomonas perforans are a major concern for sustainable crop production. BST is a common occurrence in tomato transplants grown for field production. We hypothesized that BST outbreaks in commercial fields originate from X. perforans strains inadvertently introduced from commercial transplant facilities. To test this hypothesis, we used a genome-wide single-nucleotide polymorphism (SNP) analysis to characterize X. perforans strains recovered from tomato transplant facilities and fields in commercial production areas. X. perforans strains were isolated from symptomatic transplants prior to roguing at two commercial transplant growers. Then, the same groups of transplants were tracked to commercial fields to recover X. perforans strains from diseased plants prior to harvest. Whole-genome sequencing was carried out on 84 strains isolated from transplant and field plants from Florida and South Carolina. SNPs were called using three reference strains that represented the genetic variation of the sampled strains. Field strains showing genetic similarity to transplant strains had a difference of 2 to 210 SNPs. Transplant and field strains clustered together by grower within each phylogenomic group, consistent with expectations. The range of genetic divergence among strains isolated from field plants was similar to the range obtained from strains on transplants. Using the range of genetic variation observed in transplants, we estimate that 60% to 100% of field strains were an extension of the transplant strain population. Our results stress the importance of BST management to reduce X. perforans movement from transplant to field and to minimize subsequent disease outbreaks.
IMPORTANCE Current management of Xanthomonas perforans on tomato plants mainly relies on the frequent application of pesticides. However, the lack of effective pesticides and the development of strain tolerance to certain bactericides limit the ability to control outbreaks in production fields. Better knowledge of probable sources of X. perforans inoculum during tomato production is required to refine management strategies. Tomato plants are typically established in the field using transplants. This study aimed to determine if strains from field epidemics were coming from transplant facilities or resulted from local field outbreaks. The overall goal was to identify potential sources of inoculum and subsequently develop strategies to reduce carryover from transplant production to the field. Our results indicate that tomato producers should shift disease management efforts to transplant facilities to reduce disease in the field. Improved transplant health should reduce the likelihood of bacterial spot outbreaks and subsequently reduce pesticide usage in the field.
INTRODUCTION
Tomato plants are a widely grown economically important crop throughout the United States. Nationwide, Florida accounts for roughly one-third of fresh market tomato production (1). In addition to fresh market tomato production, Florida is the second largest vegetable transplant producer in the United States. Tomato ranks first, at roughly 50% of the total vegetable transplants produced in Florida. Furthermore, one-third of the transplants produced are used by the transplant growers themselves, whereas half of the transplants are used by local growers in the state and the remaining transplants are shipped out of state (2). Commercial growers produce tomato transplants year round in rudimentary greenhouse-like structures, such as aluminum-trussed bow houses or Quonset-like structures with polyethylene plastic roofs and adjustable sidewalls (2, 3). These greenhouse-like structures lack a temperature-controlled environment, which limits the ability to modify conditions of high temperature and humidity that are optimal for disease outbreaks. Tomato transplants are commonly grown in 200-cell foam or plastic trays at a density ranging from 600 to 1,000 plants per square meter. Furthermore, overhead irrigation is typically used to water and fertilize trays during transplant production, which favors the rapid splash dispersal of pathogens (3). Bacterial spot, caused by the bacterium Xanthomonas perforans, is a major disease occurring in commercial transplant production. Commercial operations typically rogue trays with symptomatic transplants to limit outbreaks. However, the efficacy of roguing can be limited by the rate at which the bacterium can spread and the incubation period. As a result, infected tomato transplants with few or no observable symptoms can be shipped locally or even regionally. In the field, disease outbreaks are common due to Florida’s subtropical climate, including periodic episodes of rain and wind-driven rain which results in dispersal of Xanthomonas (4). Xanthomonas can survive epiphytically on crop residue, weeds, and in the soil for a short period of time. These alternate survival niches are not relevant in initiating disease outbreaks (5). Growers rely heavily on the application of copper and copper-mancozeb mixtures to limit disease outbreaks in transplants and field plants, with limited success due to widespread bacterial resistance against copper products (4).
Molecular epidemiology is a branch of epidemiology that investigates disease outbreaks and the transmission, global movement, and population structure of bacterial pathogens using genomic data (6, 7). Although molecular epidemiology has been extensively used to describe disease epidemics associated with human or animal bacterial pathogens, it has not been as widely applied for plant-pathogenic bacteria (8, 9). Nevertheless, whole-genome single-nucleotide polymorphism (SNP)-based analyses were applied in three pathosystems, Pseudomonas syringae pv. tomato, P. syringae pv. actinidiae, and Xanthomonas campestris pv. musacearum (8, 10, 11). In these studies, SNPs from whole-genome data were successfully applied to detect outbreak sources at a global or continental level (10, 11). For instance, up to 50 SNPs were detected between closely related strains of P. syringae pv. tomato (11). These SNPs were commonly found in strains across the globe (11). Furthermore, outbreaks of X. campestris pv. musacearum were identified in central African countries based on 86 SNPs that defined two lineages causing outbreaks (10). Molecular subtyping tools applied to bacterial pathogens include restriction-fragment length polymorphism, pulsed-field gel electrophoresis (PFGE), multiple-locus variable-number tandem-repeat analysis (VNTR/MLVA), multilocus sequence analysis/typing (MLSA/MLST), virulence gene typing, and DNA fingerprinting patterns (12). Recent studies characterizing bacterial outbreaks in plants focused on using MLSA/MLST or MLVA (13–16). However, these aforementioned tools have significant limitations because they examine only a subset of the bacterial genome of interest and thus restrict molecular epidemiological analyses (12). For instance, virulence- or effector-based typing can be misleading when used to infer ancestry, due to the loss and acquisition of virulence genes (17, 18).
MLSA and MLVA are commonly used tools to characterize bacterial spot of tomato (BST) outbreaks and epidemics caused by X. perforans worldwide (19–28). MLSA-based analysis of X. perforans revealed two subgroups in a global collection of strains (28). Whole-genome sequence analysis of Florida X. perforans populations using core genome MLST (cgMLST) and single nucleotide polymorphisms (SNPs) confirmed MLSA-based phylogenetic observations (18) and revealed a third group of X. perforans in Florida (29). For cgMLST, conserved core genes are used to identify strains of similar genetic background; however, the approach does not consider changes in accessory genomes commonly observed in rapidly evolving bacterial pathogens. These genomic tools are gaining acceptance due to their very high discriminatory power and fine resolution of clonal organisms of low genetic diversity compared to those of traditional tools such as MLST, MLVA, or PFGE (30). SNPs are a better discriminatory tool than MLSA or PFGE for detecting changes in bacterial populations and for tracing epidemiological outbreaks of human-related illnesses (6, 12, 31). For Xanthomonas, mutation rates for X. axonopodis in Rademaker group 9.2 (i.e., X. perforans) were reported at 1.4 × 10−8 per base (32). Also, studies of X. perforans in Florida have shown that recombination contributes to nucleotide variation more than mutation (28, 29).
In this study, we developed a genomic-based approach to study X. perforans strain movement from transplants to field during outbreaks of BST. We collected strains from two commercial tomato growers in central and south Florida, tracking the same batch of transplants to the field for subsequent sampling of plants near the end of the season. We hypothesized that strains collected from transplant facilities are highly similar to field strains as a result of plant movement. We also examined X. perforans strains collected from a BST outbreak occurring in South Carolina fields to determine whether long-distance movement of plant material from transplant facilities results in movement of strains between states. We used these strains to conduct phylogenetic analyses to characterize genetic similarity between strains causing BST outbreaks from transplant facilities and production fields using SNPs and core genes from whole-genome sequencing data.
RESULTS
Bacterial strains and genomes.
A total of 84 strains collected from transplant facilities and fields associated with two commercial grower operations were selected for genetic analysis. We characterized 67 strains from Florida and 17 strains from South Carolina. We obtained a total of 20 strains from grower A; ten strains from transplants and ten strains from field plants were sampled 1 month apart. From grower B, we obtained 47 and 17 strains from locations in Florida and South Carolina. A total of 20 transplant strains were recovered in December 2015 (11 strains) and March 2016 (9 strains), respectively, from grower B transplant facilities. In March 2016, 27 strains were recovered from grower B field plants, which corresponded to transplants grown in December 2015. The strains collected from South Carolina were recovered from diseased field plants that were grown as transplants in grower B facilities in Florida in April 2016 and planted in South Carolina fields. We did not specifically sample these transplants prior to shipping.
Genomes from a total of 103 strains, 84 strains sequenced during this study, were assembled into contigs with an average depth coverage of at least 31.7×. Genome GC content (GC%) ranged from 64.6% to 64.8% across all strains. Genome sizes ranged from 4.6 to 5.2 Mb. See Tables S1 and S2 in the supplemental information for genome size, sequencing coverage, number of genes, GC%, and number of contigs for individual strains. Genome sequences from 19 strains that represent the diversity of X. perforans on tomato plants in Florida in previous surveys were also included in the analyses (18). All strains used in this study are described in Table 1.
TABLE 1.
List of Xanthomonas perforans strains characterized and used in this study
| Grower | Date of collection (mo-yr) | Strain | Location | Source | Reference |
|---|---|---|---|---|---|
| 1991 | Xp91-118 | Floridaa | Field | 4 | |
| 2006 | Xp17-12 | Collier County, FL | Field | 18 | |
| 2010 | Xp2010 | Hendry County, FL | Field | ||
| 2012 | GEV872, GEV893, GEV909, GEV993, GEV940 | Floridab | Transplant | ||
| GEV839, GEV904, GEV915, GEV917, GEV936, GEV968, GEV1001, GEV1026, GEV1044, GEV1054, GEV1063 | Floridab | Field | |||
| A | 9-2015 | GEV2047, GEV2048, GEV2049, GEV2050, GEV2052, GEV2055, GEV2058, GEV2059, GEV2060, GEV2063 | Polk County, FL | Transplant | This study |
| 10-2015 | GEV1989, GEV1991, GEV1992, GEV1993, GEV2004, GEV2009, GEV2010, GEV2011, GEV2013, GEV2015 | Manatee County, FL | Field | ||
| B | 12-2015 | GEV1911, GEV1912, GEV1913, GEV1914, GEV1915, GEV1916, GEV1917, GEV1918, GEV1919, GEV1920, GEV1921 | Collier County, FL | Transplant | |
| 3-2016 | GEV2065, GEV2067, GEV2072, GEV2087, GEV2088, GEV2089, GEV2097, GEV2098, GEV2099 | Collier County, FL | Transplant | ||
| 3-2016 | GEV2108, GEV2109, GEV2110, GEV2111, GEV2112, GEV2113, GEV2114, GEV2115, GEV2116, GEV2117, GEV2118, GEV2119, GEV2120, GEV2121, GEV2122, GEV2123, GEV2124, GEV2125, GEV2126, GEV2127, GEV2128, GEV2129, GEV2130, GEV2132, GEV2133, GEV2134, GEV2135 | Collier County, FL | Field | ||
| 6-2016 | GEV2407, GEV2408 | Hampton County, SC | Field | ||
| GEV2384, GEV2388, GEV2389, GEV2390, GEV2391, GEV2392, GEV2393, GEV2396, GEV2397, GEV2398, GEV2399, GEV2400, GEV2403, GEV2410, GEV2420 | Beaufort County, SC | Field |
County unknown.
Counties for individual strains are listed by Schwartz et al. (18).
SNP statistics based on Xp91-118 genome.
Xp91-118 is a strain collected in Florida in 1991 that has a complete genome sequence and is extensively used as a reference in X. perforans studies. A total of 11,007 polymorphic sites in 102 strains were obtained compared to the reference genome of Xp91-118. The majority of the SNPs (∼83%) were in coding DNA sequences (CDS), while the remaining (∼17%) were in intergenic regions. The SNPs in CDS regions were mostly synonymous substitutions (∼74%), as well as nonsynonymous (∼25%) and nonsense mutations (∼0.2%) causing a disruption of the CDS. The coding and noncoding chromosomal SNPs were utilized in this study. The number of SNPs and types of substitutions, compared to the Xp91-118 reference genome, are summarized in Table S3 for each strain.
SNP analysis based on Xp91-118 genome.
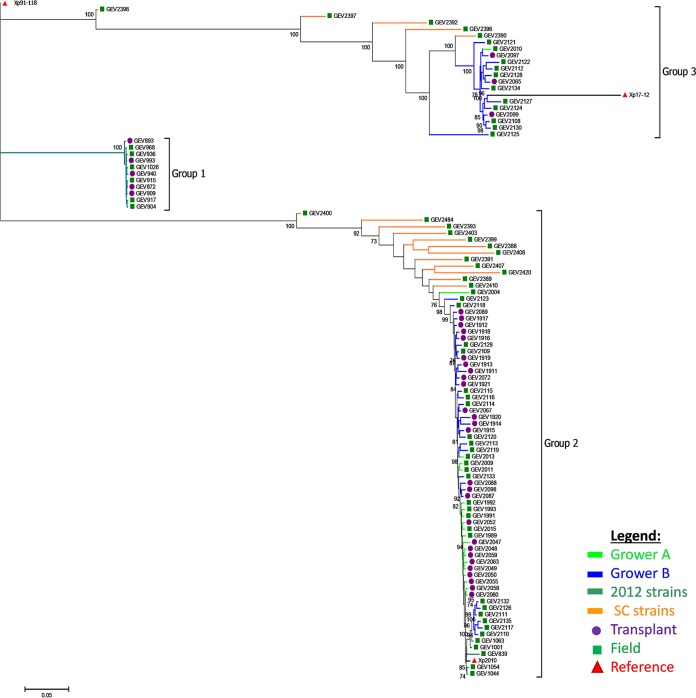
We found two distinct genetic groups among the strains from each grower (Fig. 1). Altogether, the 103 strains utilized in this study formed three distinct clades. Two previously known clades, referred to as groups 1 and 2, contain Xp91-118 and Xp2010 as reference strains, respectively. The third clade containing strain Xp17-12, formerly clustered in group 1 (18), represents a recently described phylogenetic group, group 3 (29). Interestingly, none of the strains sequenced in this study clustered with the previously characterized 2012 group 1 strains (Fig. 1) (18). The 2012 strains were included for comparison, as these strains were collected from the same growers and geographical location in Florida. The majority (>70%) of strains collected from both growers clustered in group 2. There were 18 strains placed in group 3, of which only one strain was from grower A and the remaining strains were from grower B. South Carolina field strains clustered with the Florida strains.
FIG 1.
Maximum likelihood phylogenetic tree of 103 Xanthomonas perforans strains based on 11,007 concatenated SNPs. Tree was rooted with X. perforans 91-118. All strains were isolated from Florida except strains appended to orange-colored branches, which were isolated from South Carolina (SC). 2012 strains were previously examined by Schwartz et al. (18). Green squares, purple circles, and red triangles refer to field, transplant, and reference strains, respectively. Branches are color-coded for the grower location or source of strain. Numbers on branches indicate bootstrap support.
Due to the large genetic distance between and within the clades (see branch lengths in Fig. 1), each group was examined separately for genetic relationships between transplant and field strains. To maximize the resolution between strains within the phylogenomic groups, we used the complete genomes of strains Xp2010 and Xp17-12 as reference strains for groups 2 and 3, respectively, excluding plasmid sequences. We defined transplant populations using discriminant analysis of principal components (DAPC) and assigned field strains to these predefined populations. For group 3, we had only one strain representing grower A (a field strain) and could not define alternative transplant populations. Therefore, we also quantified the level of genetic variation among transplant strains by calculating pairwise genetic distances. We used the maximum pairwise genetic distance as a threshold level for inclusion of field strains within transplant populations.
SNP analysis for group 2 strains based on Xp2010 genome.
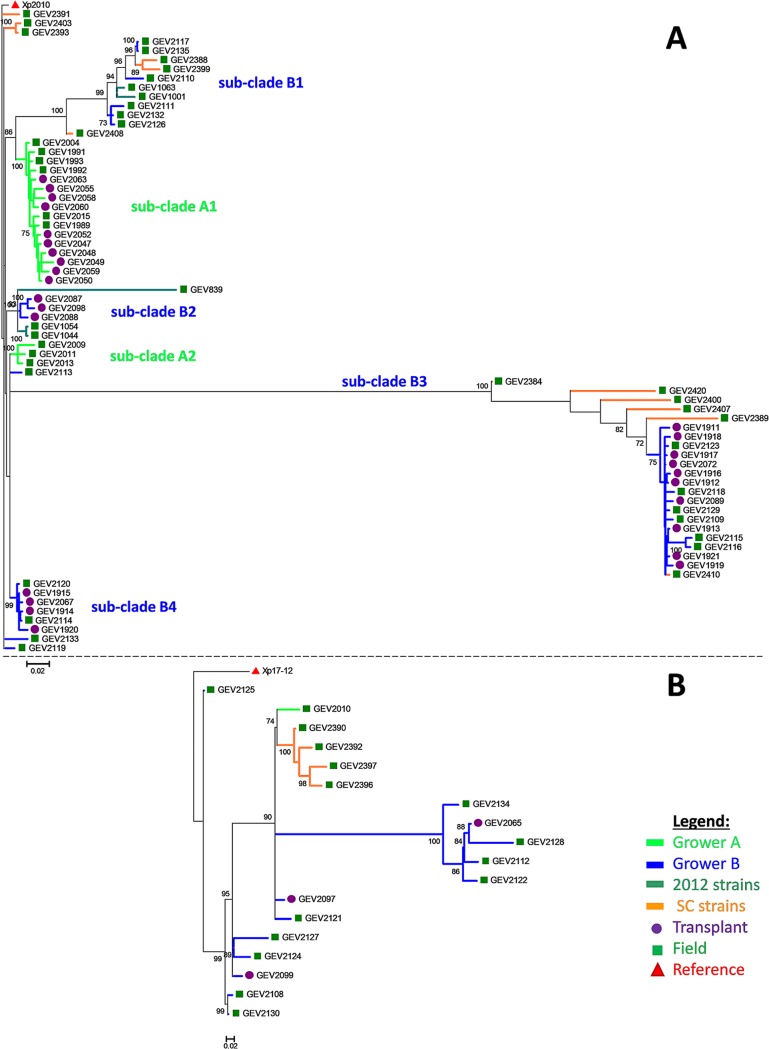
Due to the higher homology of Xp2010 to group 2 strains, we expected higher genetic relatedness and a lower number of SNPs between field and transplant strains due to less genome divergence. A total of 947 SNPs were identified across all strains within group 2 by using reference strain Xp2010. The SNP-based tree for group 2 produced multiple bootstrap-supported subclades (Fig. 2A). Most of the strains from grower A formed a clade (subclade A1), with the exception of three field strains that clustered separately into subclade A2 (Fig. 2A). Grower B strains formed four subclades plus a few strains located outside these clades (Fig. 2A). The following describes the comparisons among field and transplant strains made separately for grower A and grower B.
FIG 2.
Maximum likelihood phylogenetic tree of group 2 (A) and 3 (B) strains of Xanthomonas perforans strains based on 947 and 423 concatenated SNPs using Xp2010 and Xp17-12 as a reference strain. All strains were isolated from Florida except strains appended to orange-colored branches, which were isolated from South Carolina (SC). 2012 strains were previously examined by Schwartz et al. (18). Green squares, purple circles, and red triangles refer to field, transplant, and reference strains, respectively. Trees were rooted with the corresponding reference strain. Branches are color-coded for the grower location. Numbers on branches indicate bootstrap support.
(i) Grower A.
Grower A transplant strains were directly compared to strains collected from field plants 1 month after the initial sampling in the transplant facility. Pairwise comparison between grower A transplant strains produced genetic distances ranging from 0.005 to 0.035 (5 to 25 SNPs) (see Table S4). SNP differences ranging from 7 to 54 nucleotides were observed between transplant and field strains. Six of nine (∼60%) grower A field strains had genetic distances ranging from 0.009 to 0.028 (7 to 24 SNPs) that fell within the range of genetic distance found among transplant strains, indicating genetic similarity. Three field strains comprising subclade A2 (GEV2009, GEV2011, and GEV2013) had higher genetic distances, ranging from 0.044 to 0.071 (39 to 55 SNPs), than grower A transplant strains (Fig. 2A). These three strains were also the only grower A field strains assigned to the grower B transplant population based on DAPC (see Table S5). Therefore, there is greater uncertainty as to whether these three strains originated from the transplant population. All other grower A field strains were assigned to the grower A transplant population. The complete genetic pairwise comparison for individual grower A strains are listed in Table S4.
(ii) Grower B.
Grower B strains were distributed across multiple bootstrap-supported subclades, and as a result, transplant strains produced a wider range of genetic distances (from 0.003 to 0.633 [3 to 458 SNPs]) than grower A strains (from 0.005 to 0.035). Although the grower B transplant strains were collected at two different times, December 2015 and March 2016, both samples produced similar ranges of genetic distances (Table S5). The most direct comparison is between transplant strains collected from grower B in December 2015 and strains sampled 3 months later (i.e., March 2016) in the field (Table 1). These strains were distributed across different subclades. The overall pairwise comparison between grower B field-to-transplant strains showed a range from 2 in closely related transplant strains up to 532 SNPs in distantly related strains. Subclades B2, B3, and B4 contained both field and transplant strains. On the other hand, subclade B1 contained six (GEV2110, GEV2111, GEV2117, GEV2126, GEV2132, and GEV2135) and two (GEV2388 and GEV2399) field strains collected from Florida and South Carolina, respectively, that clustered separately from transplant strains. Subclade B1 field strains had genetic distances of 0.074 to 0.167 (68 to 122 SNPs) to transplant strains in subclades B2 and B4 (Table S5). Subclade B2 contained only transplant strains that clustered with 2012 field strains. Field strains in subclades B3 and B4 were similar to transplant strains within their respective clades at genetic distances of 0.004 to 0.171 (4 to 132 SNPs) in B3 and 0.002 to 0.012 (2 to 11 SNPs) in B4 (Fig. 2A). Three field strains each from Florida (GEV2113, GEV2119, and GEV2133) and South Carolina (GEV2391, GEV2393, and GEV2403) were not members of the above clades (Fig. 2A) but were genetically similar to transplant strains in clades B2 and B4. For grower B, pairwise distances between grower B transplant and all field strains showed that 100% of the field strains were within the above-described range and similar to at least a partial number of transplant strains (Fig. 2A and 3B). Likewise, the DAPC analysis assigned all grower B field strains to the grower B transplant population, except for the subclade B2 strains, which were assigned to the population defined by reference strains (Table S5). The reference strain population (i.e., 2012 strains) was genetically similar to the grower B population (see Fig. S1). The complete pairwise comparisons for individual grower B strains are listed in Table S6.
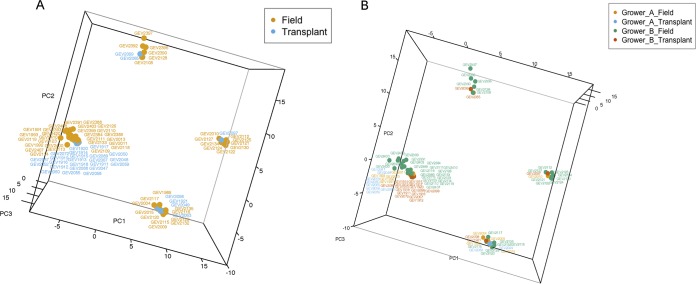
FIG 3.
Principal-component analysis of strains based on cgMLST of 1,356 genes. (A) Strains isolated from field versus transplants. (B) Location of strain isolation.
SNP analysis for group 3 strains based on Xp17-12 genome.
We utilized the whole genome of Xp17-12 as the reference genome for phylogenomic group 3 strains. A total of 380 variant sites were obtained across all group 3 strains, which included 3 transplant and 15 field strains from grower B and one grower A field strain (Fig. 2B). Pairwise distances among transplant strains from grower B ranged from 0.156 to 0.632 (46 to 210 SNPs). All grower B field strains, as well as the grower A strain, were within the range of 0.053 to 0.366 (22 to 92 SNPs). The complete pairwise comparisons for group 3 strains are listed in Table S7.
Clustering based on cgMLST.
To examine genetic similarity based on highly conserved core genes (29), we used principal-component analysis of sequence types of 1,356 core genes to examine clustering of strains. The results were similar to the SNP analyses (Fig. 3A and B). Strains isolated from South Carolina were the most variable, and half of the field strains tightly clustered with strains isolated from transplants (Fig. 3B; Table S5).
DISCUSSION
In this study, we isolated and sequenced X. perforans strains to compare whole-genome SNPs and core genes of transplant and field strains at two different grower sites in order to determine if strains move to the field on contaminated tomato transplants. Our SNP-based analysis combined with strain metadata showed one large population that coexists both on transplants and in the field. This finding suggests that X. perforans strains move to the field in association with transplants. SNP-based analysis using Xp91-118 was useful in showing the genetic divergence among sampled strains; however, none of the strains in this study were associated with Xp91-118 and the 2012 reference population. This indicates a population shift and increased genetic diversity of X. perforans. Recently, Timilsina et al. (29) showed increased diversity in X. perforans brought by recombination. Therefore, utilizing more closely related reference genomes to our strains showed better genetic relationships among strains from each grower. Most of the collected strains clustered within phylogenomic group 2; therefore, we had more resolution within this group to examine genetic relationships between transplant and field strains than within group 3. Overall, more than half of grower A field strains were highly similar to one or more transplant strains. On the other hand, all of the grower B field strains showed similarity to one or more transplant strains.
The differences in tomato transplant operations of both growers may contribute to the differences observed. Grower A produces tomato transplants for short periods throughout the year, typically before the tomato season begins in early fall and in early spring, whereas grower B has continuous transplant operations throughout the year. Grower B produces transplants year round to cater to the different seasons for local and out-of-state production that typically do not overlap. The continuous presence of plants in a transplant facility can introduce more variation and give rise to new mutant strains in the populations. Furthermore, another intriguing observation in grower B transplant operations is the proximity of their greenhouses to the field production site. The presence of transplants year round and the large field production sites are probable factors for the subtle diversity we see across grower B strains, indicating multiple introductions of transplant strains into the field. Grower B transplant strains collected at the same time formed multiple genetically diverged clades, indicating that the transplant population is not homogeneous at a given time point. However, some of the transplant strains sampled in the second collection (March 2016) were similar to transplant strains collected during the first collection (December 2015). Strains collected from South Carolina showed high genetic diversity and overall less genetic similarity to transplant strains than strains collected in Florida fields, indicating that local conditions may also affect which strains cause disease in the field. Nevertheless, we still found that half of the South Carolina strains are similar to a considerable number of transplant strains in Florida. The other distant half might have been introduced from neighboring fields or been due to sampling bias in the transplant facilities. The wide range of SNPs between field and transplant strains varied from 7 to 24 and 2 to 132 for growers A and B, respectively. Therefore, we suggest that strains having up to 132 SNPs were still considered genetically similar to transplant strains. However, this threshold is dependent on the sampled transplant population and should not be used as a cutoff value in future epidemiological studies.
Several factors, such as mutation rates, sequencing error, and sampling bias, can affect our analysis. The X. perforans mutation rate is estimated to be 1.4 × 10−8 per base (32). On the other hand, recombination had a three times higher effect in shaping diversity than mutation events (32). In Florida, X. perforans populations are more prone to recombination events than to point mutations (28, 29). Therefore, the likelihood of spontaneous mutations occurring from the time our strains were introduced from transplants to the field is very low. Mutations introduced due to sequencing errors can also affect the analysis. However, using stringent parameters and statistics to call SNPs using a reference genome reduced the calling of erroneous bases due to homopolymers and insertions or deletions.
Even though SNP-based analysis is an excellent tool to detect genetic variation, identifying the exact source of a disease outbreak remains a difficult task (33). These complex situations are common in medical bacterial epidemics, where showing a connection to a single progenitor strain during an outbreak is often difficult (33). The widespread use and reliance of the PFGE technique created a general guideline for evaluating strain relatedness and relationships; however, with the advent of whole-genome sequencing (WGS) data, these guidelines no longer apply in such context (34). Due to the higher resolving power of WGS, thresholds and cutoff values became harder to determine. Therefore, strain interrelatedness thresholds became organism dependent and on a case-by-case basis (30). Clinical studies of population outbreaks typically include related and unrelated isolates combined with metadata for establishing similarity benchmarks (30). In human epidemics of diseases, the association of strains causing an outbreak with a parental strain was successfully demonstrated by whole-genome SNP analysis (30, 33–35). Although SNP-based analyses are more accurate than other techniques, the reliance on a reference genome has its disadvantages. For instance, queried strains should have the same genomic regions present in the reference genome (6). Also, highly divergent genomic regions are excluded from the comparison, and some queried reads might not be mapped to the reference. Other drawbacks exist for SNP-based analyses, such as the selection of sequencing platform, the need for high computational power for analysis, the cost of sequencing, and the interpretation of complex data (12). In this study, X. perforans strains were not divergent and showed a 99% pairwise average nucleotide identity (P. Abrahamian, unpublished data; 28). On the other hand, major advantages of using reference-based genomes are that they are computationally less time consuming and do not have high computing requirements in comparison to reference-free de novo approaches (6). In addition, de novo assembly-based SNP calling is prone to errors and less accurate due to errors in genome assembly, bias for the consensus from combined reads, and contig joining (36).
We have seen that strains can easily move once established inside a transplant house, that movement within the house occurs rapidly, and that strains spreading through transplants do not immediately cause disease symptoms (P. Abrahamian and G. E. Vallad, unpublished data). To evade severe disease outbreaks during transplant production, growers continuously monitor and remove diseased plants. Management procedures are not standardized across growers, which results in various amounts of rogued infected plants. Disease management practices limited the number of infected transplants that we could sample in this study from both grower sites. Therefore, our limited sample of transplant strains may underestimate the genetic variation contained within each facility. Nevertheless, our data and observations suggest that tomato transplants are a major source of field inoculum. We sampled transplants and field plants only once during the growing season, but outbreaks may have occurred between sampling times during transplant production. As a result, strains from outbreaks occurring after our sampling could have been omitted in the analysis. On the other hand, field strains that are not genetically close to transplant strains might be present due to introduction of external inoculum into the field. External inoculum might cause additional outbreaks after a period of conducive environmental conditions and when sampled sites are in close proximity to neighboring infected tomato fields or transplants. Other external factors might have caused biases in this study, such as the handling of plants, sampling time between outbreaks, horizontal gene transfer, and strain recombination (37). Transplants and field plants are physically handled multiple times during standard operating procedures, which risks the introduction and movement of strains (4).
The introduction of strains to the field is a serious issue that poses a continuous risk for sustainable tomato production. Based on the results in this study, we demonstrated that a high number of strains in the field are similar to strains recovered from the same batch of transplants produced in transplant facilities. This suggests that the primary source of disease outbreaks in the field originates from symptomatic and nonsymptomatic tomato transplants. This study also reinforces the classic concept that the movement of plant material results in long-distance pathogen dispersal, in our case, the movement of strains from transplant houses to fields throughout the state and potentially into other states. This raises a challenge for sustainable vegetable production, in which exotic or local strains are under selection to overcome plant resistance breeding efforts. At the ecological level, X. perforans populations are primarily shaped by the strains that contaminate transplants during production. This results in strains circulating from the transplant house to the open field. Maintaining healthy transplants and excluding X. perforans is crucial to reduce field outbreaks of BST. While transplant growers typically manage BST through repeated pesticide applications, such strategies are not absolutely effective in preventing disease outbreaks (P. Abrahamian and G. E. Vallad, unpublished data). Therefore, alternative BST management strategies are also needed in combination with pesticide applications to reduce disease incidence. Furthermore, BST management in transplant houses is extremely difficult due to the inability of currently used greenhouse-like structures to mitigate conducive environmental factors combined with the use of overhead irrigation. Improved growing conditions would limit disease outbreaks, producing healthy tomato transplants that require fewer pesticide inputs in the field, further reducing pesticide residues in the environment and on fruit during production.
MATERIALS AND METHODS
Sample processing.
Tomato leaf samples with symptoms of bacterial leaf spot were sampled during the 2015 and 2016 growing seasons. Samples were collected from two major transplant and field operations in central (Polk County and Manatee County) and south (Collier County) Florida designated in this study as growers A and B, respectively. The distance between transplant and field operations was approximately 48 km and less than 8 km for growers A and B, respectively. Typical commercial lots consist of approximately 30,000 to 50,000 tomato transplants that require 28 to 35 days from sowing to reach a size suitable for transplanting in the field. During production, transplants were screened and sampled for BST symptoms prior to roguing of diseased plants and shipping to the field site. The remaining transplants were transplanted into the field. Fields are typically planted for one season and are fallowed for the remainder of the year. A tomato field typically takes 90 to 120 days until full development. Production tomato fields were subsequently surveyed and sampled for BST following a zigzag pattern.
Bacterial strains and isolations.
A total of 77 and 16 strains were recovered from symptomatic leaves in the transplant house and field, respectively, of grower A. A subset of 10 strains each from the transplant house and field were sequenced for this study. Field strains were collected 1 month after the initial collection from the transplant house. For grower B, a total of 11 and 57 strains were recovered from symptomatic leaves in transplant houses and fields during December 2015 and March 2016. A subset of 11 and 27 strains were selected for sequencing from transplant houses and fields, respectively. An additional collection of 42 strains was recovered from transplant houses during March 2016 at grower B, of which nine strains were sequenced. However, no field strains were recovered corresponding to March 2016 transplant strains due to lack of disease at time of sampling. The total numbers of transplant and field strains sequenced from grower B were 20 and 27, respectively. An additional 37 strains were collected from field plants in Beaufort and Hampton Counties in South Carolina during July 2016. Field plants grown in South Carolina originated from the same transplant houses used by grower B. A subset of 17 strains from South Carolina were sequenced and included in this study. In addition to the 84 strains sequenced in this study, 19 previously sequenced strains (18) were included for comparison and as references.
Each bacterial strain was isolated from a single lesion recovered from an individual tomato plant. Briefly, the lesion was excised, surface sterilized, and macerated in sterile distilled water and then streaked on nutrient agar plates amended with cycloheximide (50 mg/ml). Plates were incubated for 48 h at 28°C. Single, yellow mucoid bacterial colonies were recovered from each plate, representative of one lesion, and stored in nutrient broth with 20% glycerol at −80°C. A list and description of all sequenced bacterial strains in this study are listed in Table 1.
Genome sequencing.
Bacterial genome sequencing was carried out according to the Illumina manual (Illumina Inc., San Diego, CA). Briefly, bacterial genomic DNA, from a single colony, was isolated using a Wizard genomic DNA purification kit (Promega, Madison, WI), and libraries were constructed using a Nextera library preparation kit (Illumina Inc., San Diego, CA). Genomes were sequenced in two separate runs. In the first, second, and third runs, 20, 47, and 17 samples, respectively, were multiplexed and pooled into a single lane for sequencing. The genomes were sequenced using the Illumina MiSeq platform at the Interdisciplinary Center for Biotechnology Research, University of Florida. Draft genomes were de novo assembled using CLC Genomics Workbench v.5 (Qiagen, Hilden, Germany) or SPAdes v.3.1.1 assembler with a minimum contig size of 500 bp. The assembled sequences were annotated using the IMG/JGI platform (38).
Calling single-nucleotide variants.
SNPs across all 103 genomes were identified by mapping the raw reads to the complete genome of Xp91-118. A subset of 19 and 70 of the 103 genomes were further mapped against Xp17-12 and Xp2010, respectively, as a reference. Illumina raw reads were converted to Sanger format using FASTQ groomer (version 1.0.4). Paired-end raw sequence reads were aligned against the reference genome using Burrows-Wheeler Aligner (BWA-MEM) (version 1.2.3) using the default parameters. The alignment files, in SAM format, were used to remove duplicate raw reads caused by library construction artifacts in MarkDuplicates (v. 2.7.1.2). To reduce SNP calling errors due to indel misalignments in our read mapping against the reference genome, we used BamLeftAlign (v. 1.1.0.46) using the default settings. SNPs were called using FreeBayes (v. 1.1.0.46). VCF files for each strain were merged into one file using VCFtools. VCF files were filtered with the following parameters: minimum Phred score of 50 (99.999% accuracy), indels were discarded, minimum of 8× read coverage per SNP site. The VCF files containing SNPs for each strain were combined using VCFcombine, part of the VCFlib toolkit. The SNPs were aligned against the complete reference genome (Xp91-118, Xp2010, or Xp17-12) alignment in Geneious v. 10.1.3 (39). Identical bases, gaps and ambiguous bases were manually examined and stripped to create an alignment with high-quality SNPs used for downstream analysis. The effect of SNPs on amino acid changes, distribution, and statistics of SNPs were determined using SnpEff (v. 4.3+T).
Phylogenetic analysis.
The best fit model for the multiple sequence alignment was determined in MEGA7 (40) for SNP outputs from Galaxy. Maximum likelihood trees were constructed using RAxML v8.2.7 in Geneious v. 10.1.3 (39) using the GTR substitution model and GAMMA rate heterogeneity. To evaluate branch support, 1,000 bootstrap samples were used. The following strains were included to improve phylogenetic relationships: Xp91-118 (GenBank identifier [ID] CP019725.1), Xp2010 (JZVJ00000000), Xp17-12 (JZVH00000000), GEV839 (JZVK00000000), GEV872 (JZVL01000000), GEV893 (JZVM00000000), GEV904 (JZVN00000000), GEV909 (JZVO00000000), GEV915 (JZVP00000000), GEV917 (JZVQ00000000), GEV936 (JZVR00000000), GEV940 (JZVS00000000), GEV968 (JZVT00000000), GEV993 (JZVU00000000), GEV1001 (JZVV00000000), GEV1026 (JZVW00000000), GEV1044 (JZVX00000000), GEV1054 (JZVY00000000), and GEV1063 (JZVZ00000000).
Discriminant analysis of principal components.
Group 2 SNPs were imported into R (41) using the vcfR package (42) and converted into a genlight object using the adegenet package (43). Discriminant analysis of principal components was run on transplant and reference strains using three predefined populations representing grower A transplant strains, grower B transplant strains, and reference strains (44). The predict.dapc function was used to map the field strains onto the discriminant functions produced from the transplant and reference strains. The results were compared between analyses performed using three and four principal components.
Core genome MLST.
The core genes from Xp91-118, described in Timilsina et al. (29), were used as a reference to extract corresponding gene sequences from the assembled genomes of the strains used in this study. The single gene sequences were aligned, and allele types were designated to generate a sequence type matrix. Strain clustering was determined by plotting principal components in R using the micropan and ggfortify packages (41, 45). Three-dimensional (3D) visualization was conducted in R using the rgl package.
Accession number(s).
The assembled whole-genome sequences and raw read data of 84 genomes that support the results of this study were deposited in the NCBI database under BioProject numbers PRJNA436012 and PRJNA506994.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Florida Department of Agriculture and Consumer Services through Florida Specialty Crop Block (grant number 00099221), with administrative support from the Florida Specialty Crop Foundation.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01220-19.
REFERENCES
- 1.NASS. 2016. Vegetables 2015 summary, census of agriculture, United States summary and state data. United States Department of Agriculture, National Agricultural Statistics Service, Washington, DC. [Google Scholar]
- 2.Vavrina CS, Summerhill W. 1992. Florida vegetable transplant producers survey, 1989–1990. Horttechnology 2:480–483. doi: 10.21273/HORTTECH.2.4.480. [DOI] [Google Scholar]
- 3.McAvoy G, Ozores-Hampton M. 2018. Commercial transplant production in Florida. Florida vegetable production handbook. https://edis.ifas.ufl.edu/cv104. Accessed 9 July 2019.
- 4.Potnis N, Timilsina S, Strayer A, Shantharaj D, Barak JD, Paret ML, Vallad GE, Jones JB. 2015. Bacterial spot of tomato and pepper: diverse Xanthomonas species with a wide variety of virulence factors posing a worldwide challenge. Mol Plant Pathol 16:907–920. doi: 10.1111/mpp.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones J, Pohronezny K, Stall R, Jones J. 1986. Survival of Xanthomonas campestris pv. vesicatoria in Florida on tomato crop residue, weeds, seeds, and volunteer tomato plants. Phytopathology 76:430–434. doi: 10.1094/Phyto-76-430. [DOI] [Google Scholar]
- 6.Deng X, den Bakker HC, Hendriksen RS. 2016. Genomic epidemiology: whole-genome-sequencing–powered surveillance and outbreak investigation of foodborne bacterial pathogens. Annu Rev Food Sci Technol 7:353–374. doi: 10.1146/annurev-food-041715-033259. [DOI] [PubMed] [Google Scholar]
- 7.Didelot X, Bowden R, Wilson DJ, Peto TEA, Crook DW. 2012. Transforming clinical microbiology with bacterial genome sequencing. Nat Rev Genet 13:601–612. doi: 10.1038/nrg3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baltrus DA, McCann HC, Guttman DS. 2017. Evolution, genomics and epidemiology of Pseudomonas syringae. Mol Plant Pathol 18:152–168. doi: 10.1111/mpp.12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bentley SD, Parkhill J. 2015. Genomic perspectives on the evolution and spread of bacterial pathogens. Proc Biol Sci 282:20150488. doi: 10.1098/rspb.2015.0488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wasukira A, Tayebwa J, Thwaites R, Paszkiewicz K, Aritua V, Kubiriba J, Smith J, Grant M, Studholme DJ. 2012. Genome-wide sequencing reveals two major sub-lineages in the genetically monomorphic pathogen Xanthomonas campestris pathovar musacearum. Genes (Basel) 3:361–377. doi: 10.3390/genes3030361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai R, Lewis J, Yan S, Liu H, Clarke CR, Campanile F, Almeida NF, Studholme DJ, Lindeberg M, Schneider D, Zaccardelli M, Setubal JC, Morales-Lizcano NP, Bernal A, Coaker G, Baker C, Bender CL, Leman S, Vinatzer BA. 2011. The plant pathogen Pseudomonas syringae pv. tomato is genetically monomorphic and under strong selection to evade tomato immunity. PLoS Pathog 7:e1002130. doi: 10.1371/journal.ppat.1002130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quainoo S, Coolen JPM, van Hijum S, Huynen MA, Melchers WJG, van Schaik W, Wertheim H. 2017. Whole-genome sequencing of bacterial pathogens: the future of nosocomial outbreak analysis. Clin Microbiol Rev 30:1015–1063. doi: 10.1128/CMR.00016-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bühlmann A, Dreo T, Rezzonico F, Pothier JF, Smits THM, Ravnikar M, Frey JE, Duffy B. 2014. Phylogeography and population structure of the biologically invasive phytopathogen Erwinia amylovora inferred using minisatellites. Environ Microbiol 16:2112–2125. doi: 10.1111/1462-2920.12289. [DOI] [PubMed] [Google Scholar]
- 14.Newberry EA, Babu B, Roberts PD, Dufault NS, Goss EM, Jones JB, Paret ML. 2018. Molecular epidemiology of Pseudomonas syringae pv. syringae causing bacterial leaf spot of watermelon and squash in Florida. Plant Dis 102:511–518. doi: 10.1094/PDIS-07-17-1002-RE. [DOI] [PubMed] [Google Scholar]
- 15.Pruvost O, Magne M, Boyer K, Leduc A, Tourterel C, Drevet C, Ravigné V, Gagnevin L, Guérin F, Chiroleu F, Koebnik R, Verdier V, Vernière C. 2014. A MLVA genotyping scheme for global surveillance of the citrus pathogen Xanthomonas citri pv. citri suggests a worldwide geographical expansion of a single genetic lineage. PLoS One 9:e98129. doi: 10.1371/journal.pone.0098129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ravelomanantsoa S, Vernière C, Rieux A, Costet L, Chiroleu F, Arribat S, Cellier G, Pruvost O, Poussier S, Robène I, Guérin F, Prior P. 2017. Molecular epidemiology of bacterial wilt in the Madagascar highlands caused by Andean (phylotype IIb-1) and African (phylotype III) brown rot strains of the Ralstonia solanacearum species complex. Front Plant Sci 8:2258. doi: 10.3389/fpls.2017.02258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barak JD, Vancheva T, Lefeuvre P, Jones JB, Timilsina S, Minsavage GV, Vallad GE, Koebnik R. 2016. Whole-genome sequences of Xanthomonas euvesicatoria strains clarify taxonomy and reveal a stepwise erosion of type 3 effectors. Front Plant Sci 7:1805. doi: 10.3389/fpls.2016.01805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwartz AR, Potnis N, Timilsina S, Wilson M, Patane J, Martins J, Minsavage GV, Dahlbeck D, Akhunova A, Almeida N, Vallad GE, Barak JD, White FF, Miller SA, Ritchie D, Goss E, Bart RS, Setubal JC, Jones JB, Staskawicz BJ. 2015. Phylogenomics of Xanthomonas field strains infecting pepper and tomato reveals diversity in effector repertoires and identifies determinants of host specificity. Front Microbiol 6:535. doi: 10.3389/fmicb.2015.00535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abdurahman A, Griffin D, Elphinstone J, Struik PC, Schulz S, Schulte-Geldermann E, Sharma K. 2017. Molecular characterization of Ralstonia solanacearum strains from Ethiopia and tracing potential source of bacterial wilt disease outbreak in seed potatoes. Plant Pathol 66:826–834. doi: 10.1111/ppa.12661. [DOI] [Google Scholar]
- 20.Albuquerque P, Caridade CMR, Rodrigues AS, Marcal ARS, Cruz J, Cruz L, Santos CL, Mendes MV, Tavares F. 2012. Evolutionary and experimental assessment of novel markers for detection of Xanthomonas euvesicatoria in plant samples. PLoS One 7:e37836. doi: 10.1371/journal.pone.0037836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Almeida NF, Yan S, Cai R, Clarke CR, Morris CE, Schaad NW, Schuenzel EL, Lacy GH, Sun X, Jones JB, Castillo JA, Bull CT, Leman S, Guttman DS, Setubal JC, Vinatzer BA. 2010. PAMDB, a multilocus sequence typing and analysis database and website for plant-associated microbes. Phytopathology 100:208–215. doi: 10.1094/PHYTO-100-3-0208. [DOI] [PubMed] [Google Scholar]
- 22.Araújo ER, Costa JR, Ferreira M, Quezado-Duval AM. 2017. Widespread distribution of Xanthomonas perforans and limited presence of X. gardneri in Brazil. Plant Pathol 66:159–168. doi: 10.1111/ppa.12543. [DOI] [Google Scholar]
- 23.Constantin EC, Cleenwerck I, Maes M, Baeyen S, Van Malderghem C, De Vos P, Cottyn B. 2016. Genetic characterization of strains named as Xanthomonas axonopodis pv. dieffenbachiae leads to a taxonomic revision of the X. axonopodis species complex. Plant Pathol 65:792–806. doi: 10.1111/ppa.12461. [DOI] [Google Scholar]
- 24.Hamza AA, Robène-Soustrade I, Jouen E, Gagnevin L, Lefeuvre P, Chiroleu F, Pruvost O. 2010. Genetic and pathological diversity among Xanthomonas strains responsible for bacterial spot on tomato and pepper in the Southwest Indian Ocean region. Plant Dis 94:993–999. doi: 10.1094/PDIS-94-8-0993. [DOI] [PubMed] [Google Scholar]
- 25.Kebede M, Timilsina S, Ayalew A, Admassu B, Potnis N, Minsavage GV, Goss EM, Hong JC, Strayer A, Paret M, Jones JB, Vallad GE. 2014. Molecular characterization of Xanthomonas strains responsible for bacterial spot of tomato in Ethiopia. Eur J Plant Pathol 140:677–688. doi: 10.1007/s10658-014-0497-3. [DOI] [Google Scholar]
- 26.Osdaghi E, Taghavi SM, Hamzehzarghani H, Fazliarab A, Lamichhane JR. 2017. Monitoring the occurrence of tomato bacterial spot and range of the causal agent Xanthomonas perforans in Iran. Plant Pathol 66:990–1002. doi: 10.1111/ppa.12642. [DOI] [Google Scholar]
- 27.Roach R, Mann R, Gambley CG, Shivas RG, Rodoni B. 2018. Identification of Xanthomonas species associated with bacterial leaf spot of tomato, capsicum and chilli crops in eastern Australia. Eur J Plant Pathol 150:595–608. doi: 10.1007/s10658-017-1303-9. [DOI] [Google Scholar]
- 28.Timilsina S, Jibrin MO, Potnis N, Minsavage GV, Kebede M, Schwartz A, Bart R, Staskawicz B, Boyer C, Vallad GE, Pruvost O, Jones JB, Goss EM. 2015. Multilocus sequence analysis of xanthomonads causing bacterial spot of tomato and pepper plants reveals strains generated by recombination among species and recent global spread of Xanthomonas gardneri. Appl Environ Microbiol 81:1520–1529. doi: 10.1128/AEM.03000-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Timilsina S, Pereira-Martin JA, Minsavage GV, Iruegas Bocardo F, Abrahamian P, Potnis N, Kolaczkowski B, Vallad GE, Goss EM, Jones J. 2019. Multiple recombination events drive the current genetic structure of Xanthomonas perforans in Florida. Front Microbiol 10:448. doi: 10.3389/fmicb.2019.00448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schürch AC, Arredondo-Alonso S, Willems RJL, Goering RV. 2018. Whole genome sequencing options for bacterial strain typing and epidemiologic analysis based on single nucleotide polymorphism versus gene-by-gene–based approaches. Clin Microbiol Infect 24:350–354. doi: 10.1016/j.cmi.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 31.Aritua V, Harrison J, Sapp M, Buruchara R, Smith J, Studholme DJ. 2015. Genome sequencing reveals a new lineage associated with lablab bean and genetic exchange between Xanthomonas axonopodis pv. phaseoli and Xanthomonas fuscans subsp. fuscans. Front Microbiol 6:1080. doi: 10.3389/fmicb.2015.01080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mhedbi-Hajri N, Hajri A, Boureau T, Darrasse A, Durand K, Brin C, Fischer-Le Saux M, Manceau C, Poussier S, Pruvost O, Lemaire C, Jacques M-A. 2013. Evolutionary history of the plant pathogenic bacterium Xanthomonas axonopodis. PLoS One 8:e58474. doi: 10.1371/journal.pone.0058474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schürch AC, Siezen RJ. 2010. Genomic tracing of epidemics and disease outbreaks. Microb Biotechnol 3:628–633. doi: 10.1111/j.1751-7915.2010.00224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maiden MCJ, van Rensburg MJJ, Bray JE, Earle SG, Ford SA, Jolley KA, McCarthy ND. 2013. MLST revisited: the gene-by-gene approach to bacterial genomics. Nat Rev Microbiol 11:728–736. doi: 10.1038/nrmicro3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Croucher NJ, Finkelstein JA, Pelton SI, Mitchell PK, Lee GM, Parkhill J, Bentley SD, Hanage WP, Lipsitch M. 2013. Population genomics of post-vaccine changes in pneumococcal epidemiology. Nat Genet 45:656–663. doi: 10.1038/ng.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olson ND, Lund SP, Colman RE, Foster JT, Sahl JW, Schupp JM, Keim P, Morrow JB, Salit ML, Zook JM. 2015. Best practices for evaluating single nucleotide variant calling methods for microbial genomics. Front Genet 6:235. doi: 10.3389/fgene.2015.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Canteros BI, Minsavage GV, Jones JB, Stall RE. 1995. Diversity of plasmids in Xanthomonas campestris pv. vesicatoria. Phytopathology 85:1482–1486. doi: 10.1094/Phyto-85-1482. [DOI] [Google Scholar]
- 38.Markowitz VM, Chen IMA, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Jacob B, Huang J, Williams P, Huntemann M, Anderson I, Mavromatis K, Ivanova NN, Kyrpides NC. 2012. IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res 40:D115–D122. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.R Core Team. 2016. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 42.Knaus BJ, Grunwald NJ. 2017. vcfR: a package to manipulate and visualize variant call format data in R. Mol Ecol Resour 17:44–53. doi: 10.1111/1755-0998.12549. [DOI] [PubMed] [Google Scholar]
- 43.Jombart T, Ahmed I. 2011. adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27:3070–3071. doi: 10.1093/bioinformatics/btr521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jombart T, Devillard S, Balloux F. 2010. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11:94. doi: 10.1186/1471-2156-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Snipen L, Liland KH. 2015. micropan: an R-package for microbial pan-genomics. BMC Bioinformatics 16:79. doi: 10.1186/s12859-015-0517-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.