Whether and how furfural affects genome integrity have not been clarified. Using a Saccharomyces cerevisiae model, we found that furfural exposure leads to in vivo DSBs and elevation in mitotic recombination by orders of magnitude. Gross chromosomal rearrangements and aneuploidy events also occurred at a higher frequency in furfural-treated cells. In a genome-wide analysis, we show that the patterns of mitotic recombination and point mutations differed dramatically in furfural-treated cells and wild-type cells.
KEYWORDS: DNA recombination, furfural, genome sequencing, mutation, yeast
ABSTRACT
Furfural is an important renewable precursor for multiple commercial chemicals and fuels; a main inhibitor existing in cellulosic hydrolysate, which is used for bioethanol fermentation; and a potential carcinogen, as well. Using a genetic system in Saccharomyces cerevisiae that allows detection of crossover events, we observed that the frequency of mitotic recombination was elevated by 1.5- to 40-fold when cells were treated with 0.1 g/liter to 20 g/liter furfural. Analysis of the gene conversion tracts associated with crossover events suggested that most furfural-induced recombination resulted from repair of DNA double-strand breaks (DSBs) that occurred in the G1 phase. Furfural was incapable of breaking DNA directly in vitro but could trigger DSBs in vivo related to reactive oxygen species accumulation. By whole-genome single nucleotide polymorphism (SNP) microarray and sequencing, furfural-induced genomic alterations that range from single base substitutions, loss of heterozygosity, and chromosomal rearrangements to aneuploidy were explored. At the whole-genome level, furfural-induced events were evenly distributed across 16 chromosomes but were enriched in high-GC-content regions. Point mutations, particularly the C-to-T/G-to-A transitions, were significantly elevated in furfural-treated cells compared to wild-type cells. This study provided multiple novel insights into the global effects of furfural on genomic stability.
IMPORTANCE Whether and how furfural affects genome integrity have not been clarified. Using a Saccharomyces cerevisiae model, we found that furfural exposure leads to in vivo DSBs and elevation in mitotic recombination by orders of magnitude. Gross chromosomal rearrangements and aneuploidy events also occurred at a higher frequency in furfural-treated cells. In a genome-wide analysis, we show that the patterns of mitotic recombination and point mutations differed dramatically in furfural-treated cells and wild-type cells.
INTRODUCTION
Furfural is a naturally occurring component of many vegetables and fruits. The formation of furfural during thermal carbohydrate decomposition makes it a ubiquitous compound in numerous processed foods and cigarettes (1). In addition, furfural is an important chemical solvent and a renewable, non-petroleum-based precursor for multiple commercial chemicals and fuels (2, 3). For bioethanol production, furfural (up to more than 11 g/liter) is a main inhibitor in the fermentation medium (especially hydrolysate of cellulosic feedstock) (4, 5). Great efforts have been made to investigate the toxicity of furfural and to develop robust Saccharomyces cerevisiae strains with improved ethanol fermentation performance in the presence of furfural (6–11).
Considering the prevalence of furfural in foods and industrial environments, concerns have been raised about its mutagenicity. Zdzienicka et al. reported the mutagenic activity of furfural in Salmonella enterica serovar Typhimurium (12). Hadi et al. showed that the treatment of λ phage DNA with furfural protected cleavage with certain restriction endonucleases (1). Khan et al. found that furfural had a mutagenic effect on plasmids and resulted in lower transformation efficiency in Escherichia coli (13). It was observed that furfural could induce reactive oxygen species (ROS) accumulation and damage to multiple organelles in yeast cells (14). Also, an increased risk of hepatocellular adenoma and carcinoma was observed in mice given 175 mg of furfural/kg of body weight (15).
As a model organism, the yeast S. cerevisiae has been widely used to determine the genotoxicity of chemicals and environmental pollutants (16–20). In this study, the global effects of furfural on genomic integrity were investigated in a diploid S. cerevisiae strain. Our results showed that furfural exposure leads to elevated frequencies of multiple genetic events, including point mutations, mitotic recombination, and aneuploidy. This work explores the global mutagenicity effects of furfural at the whole-genome level.
RESULTS
Furfural treatment stimulates mitotic homologous recombination in yeast.
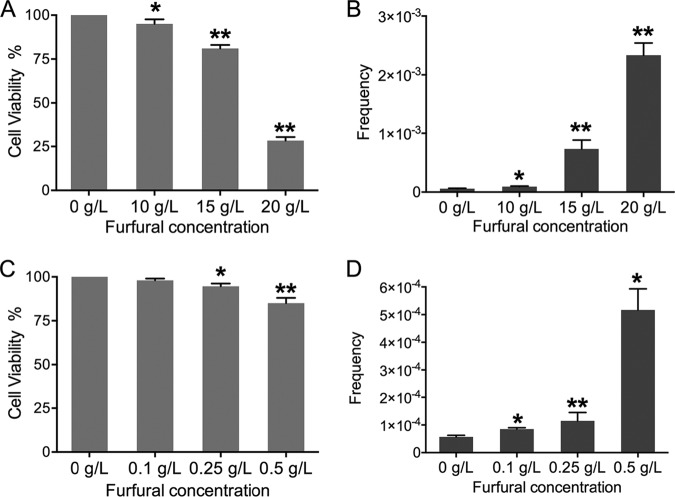
To determine the frequency of furfural-induced mitotic homologous recombination (HR), a yeast colony color-screening assay to identify reciprocal crossover on the right arm of chromosome IV was used. The diploid S. cerevisiae strain JSC25-1 was homozygous for the ade2-1 mutant allele and heterozygous for the ochre-suppressing gene SUP4-o, which was located near the right telomere of YJM789-derived chromosome IV (21, 22). Partial (one copy of SUP4-o) and complete (two copies of SUP4-o) suppression of the ochre mutation of ADE2 in a diploid strain results in pink and white colonies, respectively (21). Crossover events occurring between the centromere and the SUP4-o locus in the first cell cycle after JSC25-1 cells were plated can give rise to red-white sectored colonies (Fig. 1). After acute exposure to 10 g/liter, 15 g/liter, and 20 g/liter furfural for 2 h, about 4%, 19%, and 72% of JSC25-1 cells were killed, respectively (Fig. 2A). The frequency of white-red colonies was elevated by 1.5- to 40-fold compared to that in untreated cells (Fig. 2B) (P < 0.05; t test). In an alternative test, JSC25-1 cells were grown on plates containing 0.1 g/liter, 0.25 g/liter, and 0.5 g/liter furfural. The rates of white-red colonies on furfural-containing plates were 1.5- to 9-fold higher than on normal yeast extract-peptone-dextrose (YPD) plates (P < 0.05; t test) (Fig. 2D), while cell viabilities were almost unaffected (>90%) (Fig. 2C). These results showed that furfural exposure can greatly elevate the frequency of mitotic HR in yeast cells.
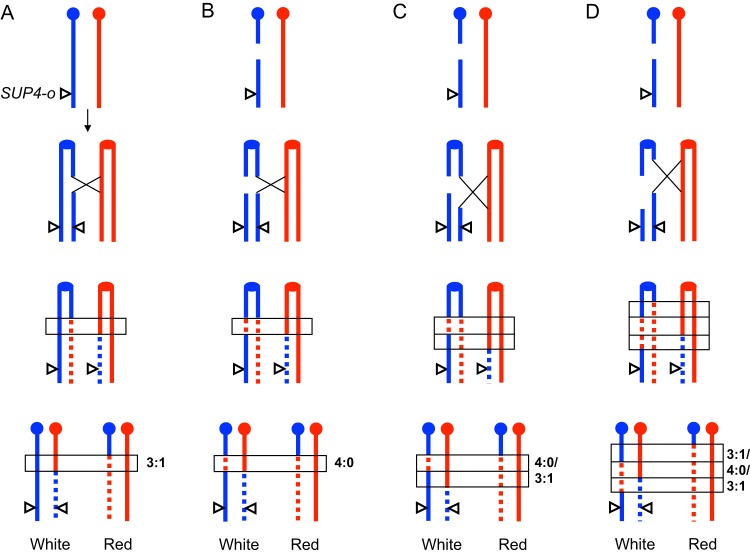
FIG 1.
Crossover events initiated by DSBs. The red and blue lines represent the W303-1A and YJM789 homologs of chromosome IV in strain JSC25-1, respectively. The SUP4-o gene, which suppresses the ochre mutation of ade2-1, was located at the right end of the YJM789-derived homolog. Zero, one, and two SUP4-o (triangles) genes in a diploid yeast strain that is homozygous for ade2-1 led to red, pink, and white colonies, respectively. Reciprocal crossover between CEN4 (solid circles) and SUP4-o generated white-red sectored colonies. (A) Repair of S/G2 phase DSBs usually leads to 3:1 conversion tracts (rectangles) associated with the crossover. (B to D) DSBs in the G1 phase resulted in breaks on both chromatids after chromosome replication. Repair of DSBs at the allelic sites of two chromatids led to 4:0 or hybrid regions within the crossover-associated conversion tracts.
FIG 2.
Cell viabilities and frequencies of white-red sectored colonies of yeast strain JSC25-1. (A) Cell viabilities of JSC25-1 cells (OD600, 2 in 1 ml of YPD medium) after furfural treatment for 2 h. (B) Frequencies of white-red colonies formed on YPD plates after furfural treatment. (C) Viabilities of JSC25-1 cells grown on furfural-containing plates. (D) Frequencies of white-red sectored colonies when JSC25-1 cells were grown on furfural-containing plates. Experiments were performed three times. The error bars represent standard deviations. *, P < 0.05, and **, P < 0.01; t test.
Analysis of gene conversion associated with crossover using a chromosome IV-specific single nucleotide polymorphism (SNP) microarray.
In the current models of HR, crossovers are usually initiated by double-strand breaks (DSBs) and are associated with the formation of heteroduplexes (21, 23). Repair of mismatches in the heteroduplex can result in gene conversions (21, 23). Based on the patterns of gene conversation tracts, one can infer in which phase the recombination-initiating DSBs occur (21, 22). As shown in Fig. 1, repair of the DSBs that occur in the S/G2 phases leads to crossover-associated gene conversion tracts (shown within narrow horizontal rectangles) with a 3:1 pattern (three chromatids have W303-1A-derived sequences, and one has YJM789-derived sequences) (Fig. 1A). If DSBs occur in the G1 phase, both chromatids harbor breaks after chromosome duplication. Repairs of breaks at the allelic sites of two chromatids usually produce gene conversion tracts with 4:0 (Fig. 1B) or hybrid (Fig. 1C and D) patterns (21).
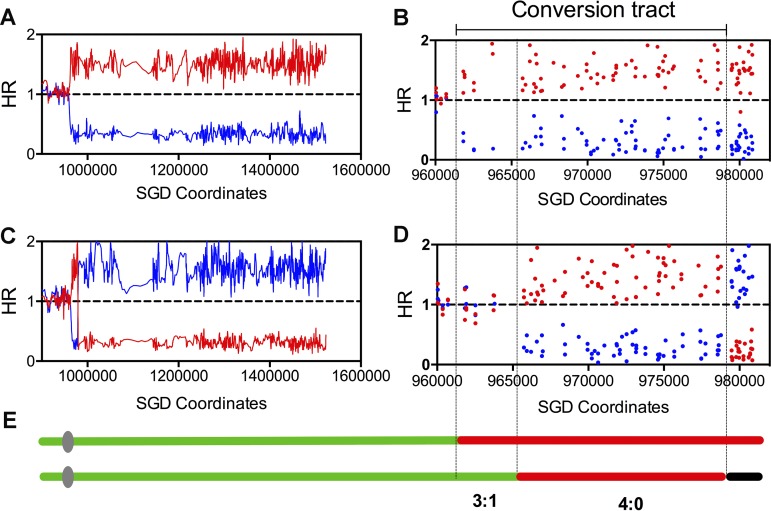
To determine which recombinational DNA lesions were responsible for furfural-induced mitotic recombination, JSC25-1 cells were arrested in the G1 phase and treated with 17 g/liter furfural for 1 h. In this scenario, we observed that ∼30% of the cells remained alive and red-white colonies appeared at a frequency of 0.03%. Using chromosome IV SNP-specific microarrays that could examine 2,300 SNPs on the right arm of chromosome IV (21), the cells purified from both red and white sectors of 19 sectored colonies (QLS1 to -19) were analyzed. Genomic DNA of sectored colonies that are heterozygous for SNPs at a given position hybridizes equally well to probes specific for W303-1A- and YJM789-derived SNPs. The changes of the hybridization ratio indicate copy number variations of either one or both homologs (24). An example of the analysis of sectored colony QLS8 is shown in Fig. 3. The red and blue lines and points represent the normalized hybridization ratios of the examined genomic DNA to oligonucleotides that were specific to SNPs from W303-1A and YJM789, respectively. In Fig. 3A, a transition from heterozygous to homozygous for W303-1A-derived SNPs was observed at around 960 kb in the red sector. From this transition site to the right telomere, the SNPs were homozygous for the W303-1A-derived homolog. At higher resolution (Fig. 3B), we found that the transition occurred between SNPs with chromosomal coordinates 960,710 and 961,833. In the white sector, there were two transitions: one from heterozygous SNPs to homozygous for W303-1A-specific SNPs was between Saccharomyces Genome Database (SGD) coordinates 964,391 and 965,740, and the other transition was between coordinates 978,763 and 979,477 (Fig. 3C and D). Within the crossover-associated gene conversion tract (from 964,391 to 979,477), we observed a 3:1/4:0 hybrid pattern (Fig. 3E). As shown in Fig. 1, such a pattern indicates that the initial DSB occurred in the G1 phase on the W303-1A-derived chromosome. The broken homolog was replicated to generate two broken chromatids and then repaired by HR during S/G2. The length of the gene conversion tract for this crossover was 17.8 kb (averaging the minimal length [the distance between the first and last homozygous SNPs within the tract] and the maximal length [the distance between heterozygous SNPs flanking the conversion region]).
FIG 3.
Mapping a selected crossover event initiated by DSB occurring in the G1 phase by chromosome IV-specific SNP microarray. The hybridization ratio is shown on the y axis, and the values on the x axis are chromosome coordinates. The hybridization ratio values 0, 1, and 1.5 represent 0, 1, and 2 copies of W303-1A-derived (red points or lines) or YJM789-derived (blue points or lines) homologs. (A) Low-resolution depiction of reciprocal crossover in the red part of the selected colony, QLS8. (B) High-resolution depiction of the same event shown in panel A. (C) Low-resolution depiction of reciprocal crossover in the white part of the selected colony, QLS8. (D) High-resolution depiction of the same event shown in panel C. (E) The regions between the lines show 4:0/3:1 conversion tracts associated with the crossover event. The green, red, and black segments represent heterozygous SNPs, homozygous for W303-1A-specific SNPs, and homozygous for YJM789-specific SNPs, respectively.
The coordinates for all the transitions of these 19 selected crossover events are shown in Data Set S1 in the supplemental material and are schematically depicted in Data Set S2 in the supplemental material. The patterns of the conversion tracts associated with these crossovers were as follows: (i) 2 simple 3:1 conversions, (ii) 1 simple 4:0 conversion, (iii) 11 3:1/4:0, 4:0/3:1, or 3:1/4:0/3:1 hybrid conversions, and (iv) 5 other complex hybrid conversions. The complex hybrid conversion tract with several transitions might reflect “patchy” repair of mismatches within long heteroduplexes or template switching between homologs (21, 22). In summary, most (17 out of 19; 89%) crossovers were interpreted as being initiated by furfural-induced DSBs in the G1 phase. Previously, St. Charles and Petes analyzed 139 spontaneous reciprocal crossovers on the right arm of chromosome IV in JSC25-1 using the same SNP microarray system used in this study (24). Most (121/139) of the crossovers were associated with a contiguous gene conversion event: 29 simple 3:1 conversions, 7 simple 4:0 conversions, 46 simple 4:0/3:1 or 3:1/4:0/3:1 hybrid conversions, and 39 tracts with more complicated patterns of conversion (21). In comparison to untreated JSC25-1 cells, furfural-treated cells showed a higher ratio (89% versus 66%) of G1-associated crossovers (Fisher exact test; P < 0.01). Also, we found that the median length of the crossover-associated conversion tracts (21 kb; 95% confidence interval [CI], 14.9 to 28.7 kb) in furfural-treated cells was longer than that (10.6 kb; 95% CI, 8.2 to 13.6 kb) in the wild-type JSC25-1 isolates (P < 0.01; Mann-Whitney test). Because HR occurs only in S/G2, the broken ends generated in the G1 phase have a longer time for 5′-to-3′ resection prior to HR repair and result in longer gene conversion tracts (21). The higher ratio of G1-associated DSBs in furfural-treated isolates may partly explain the longer conversion tracts that are associated with furfural-induced crossovers versus spontaneous crossovers.
Detection of furfural-induced DSBs by CHEF and Southern blot analyses.
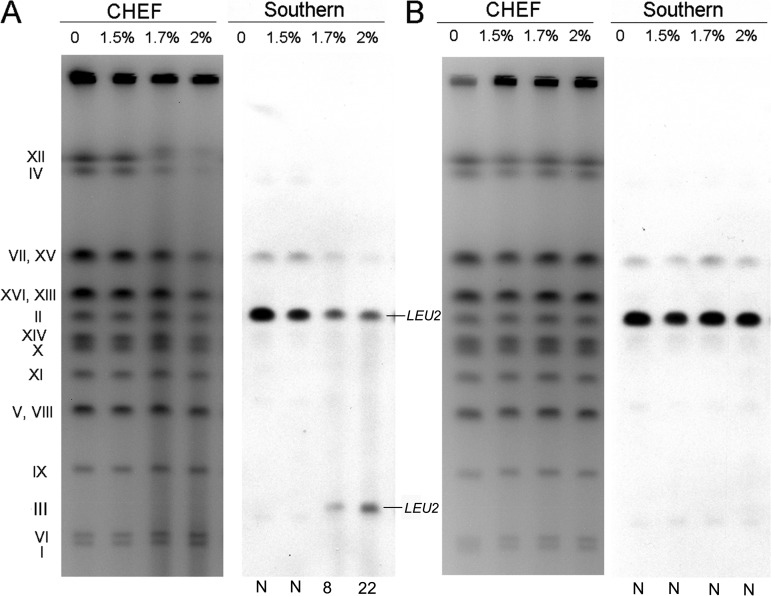
To confirm whether furfural treatment can generate DSBs, a haploid S. cerevisiae strain, Yyy123, with a circularized chromosome III was used for contour-clamped homogeneous electric field (CHEF) and Southern blot analyses. The strain has an insertion of LEU2 on both linear chromosome II and circular chromosome III. A full-size linear yeast chromosome can be separated by CHEF assay, whereas a circular chromosome will stick in the sample well (25). If the circular chromosome III has a break, it can migrate into the gel and be detected by Southern blotting (25). For our experiments, Yyy123 cells were synchronized in the G1 phase, treated with furfural (15 g/liter, 17 g/liter, and 20 g/liter) for 1 h, and embedded in low-melting-point gel for CHEF analysis. We observed that chromosomes were significantly fragmented (Fig. 4A, left) after furfural treatment, and a band representing linear chromosome III was detected using a probe targeted at the gene LEU2 (Fig. 4A, right). Based on the ratio of LEU2-specific hybridization for chromosome II and the linear chromosome III (Fig. 4A), it was calculated that treatment with 17 g/liter and 20 g/liter furfural resulted in 8 and 22 DSBs per genome (the detailed calculation method is described in reference 25). In an in vitro experiment, the chromosomal DNA extracted from Yyy123 cells was exposed to furfural (15 g/liter, 17 g/liter, and 20 g/liter) for 1 h. The treated chromosomes remained intact, and no linear chromosome III was detected (Fig. 4B). These observations suggested that furfural is incapable of cleaving DNA directly but can induce in vivo DSBs depending on endogenous biochemical processes.
FIG 4.
Detection of furfural-induced chromosome breaks in a yeast strain with circular chromosome III. The haploid strain Yyy123 used in the experiment had a circular derivative of chromosome III and a LEU2 insertion on both chromosome III and chromosome II. (A) In an in vivo experiment, yeast cells were treated with furfural (0 to 2%) and prepared for CHEF gel (left) and Southern blot (right) analyses using a LEU2-specific probe described in Materials and Methods. (B) In an in vitro experiment, cell-free DNA prepared in plugs was treated with furfural and then examined by gel electrophoresis as for panel A. The calculated numbers of DSBs per genome are shown below the gel. “N” indicates that DSBs were not detectable.
Furfural-induced ROS were responsible for most mitotic recombination events.
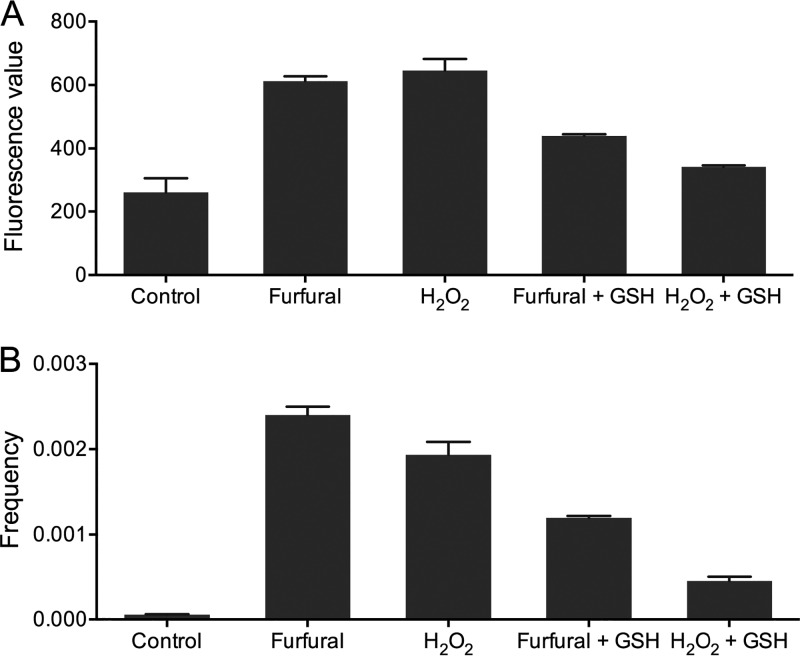
Using 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA) dye, it was determined that furfural-treated (20 g/liter) JSC25-1 cells accumulated 2-fold more ROS than untreated cells (Fig. 5A). To assess whether increased intracellular ROS contributed to elevated mitotic recombination, JSC25-1 cells were treated with 0.05% H2O2, which led to ROS accumulation similar to that in cells treated with 20 g/liter furfural (Fig. 5A). It was observed that treatment with 0.05% H2O2 resulted in 30-fold elevation in the rate of red-white sectored colonies compared to untreated cells (Fig. 5B). In the presence of 5 mM glutathione, ROS accumulations were decreased by 28% and 46%, respectively, in furfural- and H2O2-treated cells (Fig. 5A). Accordingly, the rates of white-red colonies of furfural- and H2O2-treated cells were reduced by about 2- and 4-fold, respectively (Fig. 5B). These results indicate that most furfural-induced mitotic recombination events were strongly associated with accumulation of intracellular ROS.
FIG 5.
Determination of ROS and mitotic-recombination frequencies in yeast cells treated with furfural and H2O2. (A) Detection of ROS accumulation with a fluorescence spectrophotometer in wild-type cells and cells treated with furfural or H2O2. Addition of 5 mM glutathione (GSH) could effectively reduce the generation of ROS during furfural (17 g/liter) and H2O2 (0.05%) treatment. (B) Exposure to furfural or H2O2 increased the frequency of mitotic recombination (measured by the rates of red-white sectored colonies) by 2 orders of magnitude. The presence of GSH (5 mM) in the treatment reaction mixture decreased the frequency of mitotic recombination in furfural- and H2O2-treated cells. The error bars represent standard deviations of the mean from three experiments. *, P < 0.05, and **, P < 0.01; t test.
Whole-genome mapping of mitotic recombination in JSC25-1-derived isolates.
To investigate furfural-induced mitotic HR at the whole-genome level, a custom whole-genome SNP microarray (24, 26) that allowed us to detect 15,000 SNPs between the W303-1A and YJM789 sequences was used. As mentioned in Materials and Methods, 21 JSC25-1-derived isolates that underwent furfural treatment 18 times were analyzed by the custom SNP microarray. Among the 21 isolates, we identified 154 genomic alterations, including 109 internal loss-of-heterozygosity (LOH) events (gene conversions unassociated with crossovers), 41 terminal LOH events, and 4 duplications or deletions (DUP/DEL). The chromosomal coordinates of all the genetic events are shown in Data Set S3 in the supplemental material. According to the patterns (which homolog was duplicated or lost and whether the event was an interstitial or terminal event), we divided these genetic events into 15 classes (see Data Set S4 in the supplemental material).
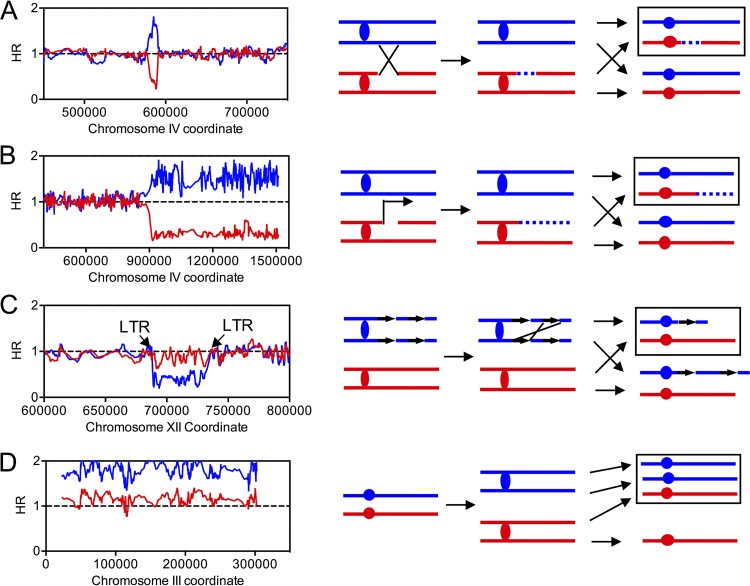
The largest category of furfural-induced genomic alteration is gene conversion events that are unassociated with crossovers in which the DNA sequence of one homolog was replaced by that of the other homolog (see classes b1 and b2 in Data Set S4). An example of such an event is shown in Fig. 6A. The red and blue lines indicate the normalized hybridization ratios of sample DNA to W303-1A- and YJM789-specific SNPs, respectively. If a chromosomal region was heterozygous, the values of the red and blue lines were about 1. A hybridization ratio of ∼1.5 or ∼0.3 indicates DNA duplication or loss, respectively. In this event, we observed that the region between bp 576244 and bp 590872 was homozygous for YJM789-derived SNPs. We interpret this as showing that a DSB occurred on the W303-1A-derived homolog and was repaired by HR, using the YJM789 homolog as a template (Fig. 6A). Most (107 out of 109) gene conversions showed contiguous conversions tracts, like the example shown in Fig. 6A. The median size of these conversion tracts was 9.9 kb (95% CI, 7.4 kb to 12.6 kb). Fig. 6B shows a terminal LOH event in which sequences from the W303-1A homolog were lost, whereas sequences from the YJM789 homolog were duplicated (see class a4 in Data Set S4); the affected regions were extended from the breakpoint to the telomere. Such a terminal LOH event might be a consequence of reciprocal crossover (Fig. 1) or break-induced replication (BIR) (Fig. 6B).
FIG 6.
Examples of LOH events and chromosomal alterations detected by SNP microarrays. The normalized HRs of W303-1A-derived and YJM789-derived SNPs are depicted in red and blue, respectively. (Right) The horizontal lines represent chromosomes or sister chromatids. The boxed regions represent the daughter cell analyzed by microarrays. (A) A gene conversion event on chromosome IV in the isolate QLF18. (B) A terminal LOH event detected on chromosome IV in the isolate QLF18. In addition to the reciprocal crossover depicted in Fig. 1, BIR would also lead to a terminal LOH event. (C) An interstitial deletion that might be caused by unequal crossing over occurred on chromosome XII in isolate QLF17. The breakpoints of this deletion contain two LTR repetitive elements. (D) Analysis of a trisomy that occurred on chromosome III in isolate QLF18.
In addition to LOH events, we detected 4 chromosomal rearrangements, including 2 internal deletions, 1 terminal deletion, and 1 terminal duplication (see Data Sets S3 and S4). For these events, the hybridization ratio of one homolog was decreased (deletions) or elevated (duplications), with the other unchanged. As shown in Fig. 6C, one internal deletion occurred between kb 688 and kb 732 on chromosome XII, flanked by two long terminal repeat (LTR) elements. Such a deletion can result from unequal crossing over between sister chromatids (Fig. 6C). The other internal deletion was located between two fragile sites (FS) (FS1, at approximately kb 149, involves two directed Ty1 elements, and FS2, at approximately kb 175, involves two inverted Ty1 elements) on W303-1A-derived chromosome III (see Data Set S3). The terminal deletion (kb 0 to kb 35 of W303-1A-derived chromosome II) and duplication (kb 0 to kb 177 of W303-1A-derived chromosome XIII) events occurred in the same isolate, QLF12 (see Data Set S3). Such “paired events” may result from repairing a break on chromosome II by the BIR pathway, using chromosome XIII as a template. Among the 21 analyzed isolates, we also identified 7 aneuploidy events: 3 were trisomy (chromosomes III, IV, and XVI), and 4 were uniparental disomy (chromosomes V, VII, X, and XV) (Fig. 6D; see Data Set S3 in the supplemental material). We calculated the frequency of gross chromosomal rearrangements and aneuploidy events as 1.1 × 10−3, which was about 5-fold higher than that observed in the wild-type cells (27).
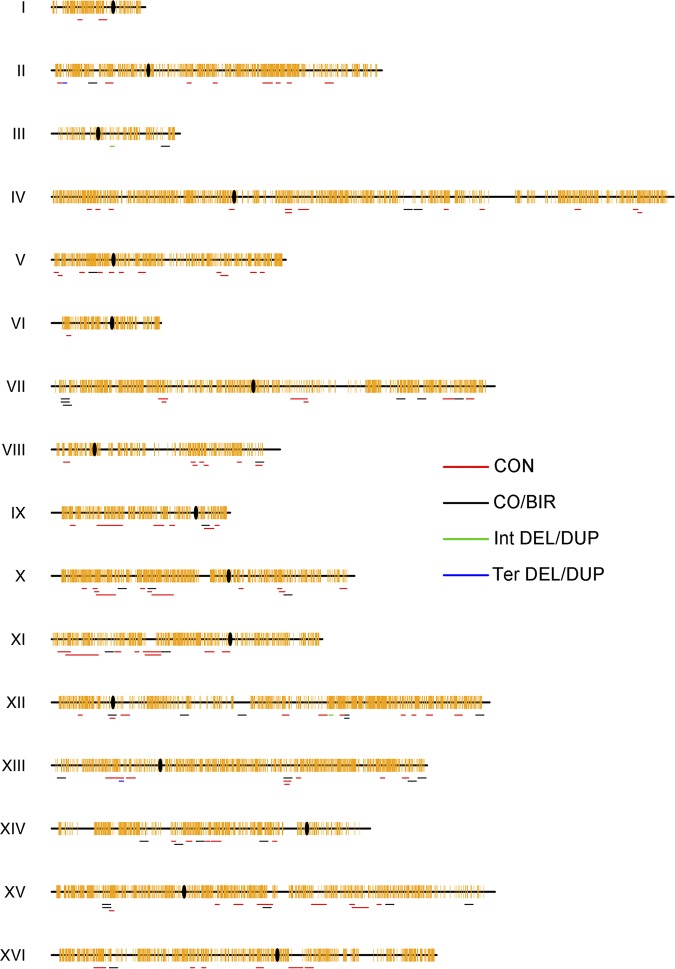
We found that the 154 detected genomic alterations were almost evenly distributed on the 16 chromosomes (Fig. 7). The frequency of genomic alterations was about 154/25/18/21, or 0.02 events per genome per cell division. The numbers of events on each chromosome were positively associated with their sizes (P < 0.001; Pearson correlation analysis). Using the chi-square test-based association analysis described in our previous study (26), we evaluated whether certain chromosomal elements/motifs were enriched within “windows” that might contain the breakpoints of furfural-induced recombination events (Table 1). For internal LOH events, the region between the heterozygous sites of the leftmost and rightmost transitions was defined as a “window” for analysis. To define windows for terminal LOH events, we used 20-kb windows that were 10 kb to each side of the first homozygous SNP. The coordinates of the windows for all LOH events are provided in Data Set S3. As shown in Table 1, our results showed that high-GC regions (the GC content was higher than 42% within 1-kb sliding windows, with 200-bp steps across the yeast genome) were enriched near the breakpoints of the 150 LOH events. In contrast, low-GC (<36%) regions, tRNAs, and LTRs were significantly less represented within the windows (Table 1).
FIG 7.
Distribution of LOH and duplication/deletion events across the yeast genome. CO/BIR, crossover/break-induced replication; CON, gene conversion; TER DEL/DUP, terminal deletions/duplications; INT DEL/DUP, interstitial duplications/deletions. Centromeres are shown as black ovals, and SNPs are shown as yellow vertical lines.
TABLE 1.
Analysis of associations of chromosomal elements with breakpoints of unselected LOH events in furfural-treated cellsa
| Chromosome element | Predicted no. of elements included in microarrays | No. observed |
No. expected |
P valueb | ||
|---|---|---|---|---|---|---|
| Within tracts | Outside tracts | Within tracts | Outside tracts | |||
| tRNA genes | 274 | 36 | 5,718 | 60 | 5,694 | 0.002 |
| Autonomously replicating sequence elements | 317 | 51 | 6,606 | 69 | 6,588 | 0.029 |
| snRNA and snoRNA genes | 83 | 19 | 1,724 | 18 | 1,725 | 0.830 |
| Transposable elements | 47 | 8 | 979 | 10 | 977 | 0.481 |
| Solo LTRs | 274 | 23 | 5,731 | 60 | 5,694 | 0.000 |
| Centromeres | 16 | 3 | 333 | 3 | 333 | 0.793 |
| Palindromic sequences | 570 | 125 | 11,845 | 124 | 11,846 | 0.947 |
| G4 sequences | 544 | 115 | 11,309 | 119 | 11,305 | 0.740 |
| Highly transcribed genes | 330 | 75 | 6,855 | 72 | 6,858 | 0.717 |
| Weakly transcribed genes | 312 | 56 | 6,496 | 68 | 6,484 | 0.143 |
| Binding sites for Rrm3p | 112 | 19 | 2,333 | 24 | 2,328 | 0.271 |
| Regions with high levels of phosphorylated histone H2AX | 631 | 135 | 13,116 | 138 | 13,113 | 0.826 |
| Replication-termination sequences | 71 | 17 | 1,474 | 15 | 1,476 | 0.697 |
| Tandemly repeated sequences | 1,125 | 277 | 23,348 | 245 | 23,380 | 0.042 |
| Regions with high GC content (>42%) | 538 | 215 | 11,083 | 117 | 11,181 | 0.000 |
| Regions with low GC content (<36%) | 407 | 49 | 8,498 | 89 | 8,458 | 0.000 |
| Meiotic hot spots | 136 | 37 | 2,819 | 30 | 2,826 | 0.175 |
The table shows the expected numbers of elements within and outside LOH regions compared to the observed numbers of elements within and outside the regions (see Data Set S3). The chromosomal coordinates of chromosomal elements were provided by Song et al. (38).
The two sets of data were compared using chi-square tests in Excel, and the resulting P values (for the differences between expected and observed values) are shown. The values in boldface were significant (P < 0.05) after correcting for multiple comparisons.
Whole-genome sequencing to identify small DNA changes.
To find genomic alterations (single base substitutions and small insertions/deletions [indels]) that could not be detected by SNP microarrays, 7 JSC25-1-derived isolates (QLF1, QLF2, QLF5, QLF8, QLF11, QLF18, and QLF19) were also analyzed by Illumina high-throughput sequencing (HTS). All genetic events (48 gene conversions, 13 terminal LOH events, 1 trisomy, 1 internal deletion, and 1 uniparental disomy [UPD] [see Data Set S3]) detected by SNP microarray in these 7 isolates were confirmed by HTS. Because HTS could detect 3-fold more SNPs than the whole-genome SNP microarray (26), we identified 17 new interstitial gene conversion events with a median tract size of 1.7 kb (see Table S1 in the supplemental material). One new gene conversion located on chromosome V in isolate QLF11 is shown in Fig. S1 in the supplemental material. However, HTS and the SNP microarray had the same capability to detect terminal LOH events, chromosomal rearrangements, and aneuploidy events.
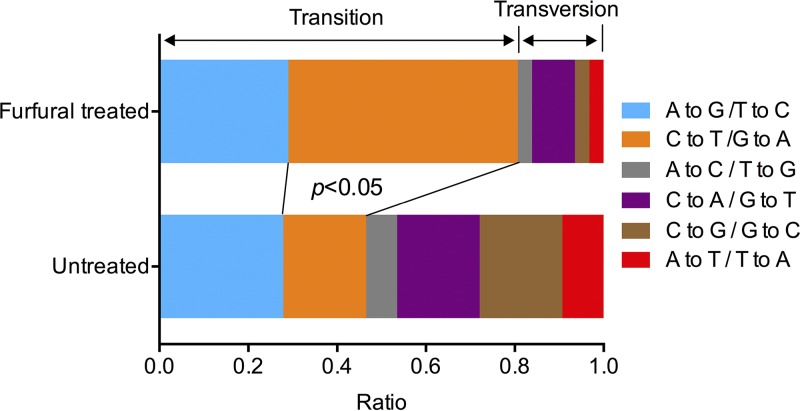
Among the 7 sequenced QLF isolates, we observed 33 point mutations; 31 were single nucleotide variations (SNVs) and 2 were small indels (see Table S2 in the supplemental material). Five point mutations were randomly selected, and all were confirmed by Sanger sequencing (see Table S2). Based on the genome size (24 Mb) and the numbers of sequenced isolates (7 isolates), subcultured generations (18 generations), and cell divisions per generation (25 divisions), we calculated the rates of SNVs and small indels as 4.1 × 10−10 (31/24,000,000/7/18/25) and 2.6 × 10−11 (2/24,000,000/7/18/25) per base per cell division. Among the 20 control isolates (not treated with furfural), we detected 43 SNVs and 3 indels. The rates of spontaneous SNVs (1.8 × 10−10) and indels (1.25 × 10−11) observed in this study were very similar to those reported in previous studies. Our analysis suggested that furfural exposure increased the rate of SNVs by 2.3-fold in JSC25-1-derived isolates. Interestingly, we found that most (25 out of 31) of the observed SNVs among the furfural-treated isolates fell into the category of base transitions (A-to-G/T-to-C, and C-to-T/G-to-A) (Fig. 8), resulting in significantly higher ratios of transition and transversion than of spontaneous SNVs (transition-transversion, 1:1.5) (Fig. 8) (P < 0.05; Fisher exact test). In particular, the ratio of C-to-T/G-to-A transitions was elevated to a greater extent than those of other mutations in furfural-treated cells compared to untreated cells (Fig. 8) (P < 0.05; Fisher exact test).
FIG 8.
Spectra of single base mutations in furfural-treated and untreated isolates derived from JSC25-1. A higher ratio of C-to-T/G-to-A transition was observed in furfural-treated cells than in untreated cells (Fisher exact test; P < 0.05).
DISCUSSION
To improve our understanding of the genetic toxicity of furfural, this study explored whether and how it affects genome stability using a yeast model. Our main findings include the following: (i) furfural exposure can elevate mitotic recombination by 2 orders of magnitude; (ii) most furfural-induced mitotic recombinations resulted from repairing DSBs, which was dependent on ROS accumulation; (iii) genetic events were enriched at GC-rich regions; and (iv) furfural exposure resulted in an increased mutation rate and modified mutation spectra compared to those in wild-type cells. Below, we discuss the implications of our results.
Among the DNA lesions induced by chemicals, DSBs are recognized as the most toxic (22, 23). HR in which DNA lesions are healed using an intact homolog as a template is a revolutionary conserved pathway to repair DSBs in organisms (23). Compared to meiotic HR, mitotic HR occurred at a much lower frequency under normal conditions (21). In the wild-type diploid S. cerevisiae, the frequency of mitotic HR is about 10−4 events per genome per cell division (28). Multiple endogenous and exogenous factors have been proved to greatly stimulate mitotic HR in yeast cells (29). Using a diploid yeast strain, JSC25-1, we showed that furfural (0.1 to 20 g/liter) treatment resulted in a significant elevation (1.5- to 40-fold) in the rate of reciprocal crossover that occurred on the right arm of chromosome IV (Fig. 2). To reveal the nature of furfural-induced DNA lesions that initiated HR, G1-synchronized JSC25-1 cells were treated with furfural to select crossover events. The LOH patterns of the crossover-associated gene conversion tracts indicated that most crossover events were initiated by DSBs generated in the G1 phase. This conclusion was further confirmed by the CHEF gel-based detection of linear chromosome III in the haploid strain Yyy123 (Fig. 4A). However, it is unlikely that furfural can break DNA directly (Fig. 4B). How does furfural exposure result in in vivo DSBs? In yeast, the main pathway to detoxify furfural converts furfural to its corresponding alcohols (30). Growth of yeast in furfural-containing medium would drain the cells of the reductive cofactors NADH and NADPH, contributing to intracellular ROS accumulation (31, 32). ROS have been demonstrated to be a potential inducer of DSBs and genomic instability (33, 34). Scavenging of furfural-induced ROS by glutathione in furfural-treated cells led to 90% reduction of mitotic recombination (Fig. 5). These observations enable us to conclude that furfural-induced in vivo DSBs are largely dependent on cellular metabolism rather than direct interaction between furfural and DNA. Because ROS accumulation can be caused by environmental stimuli and different inhibitors existing in cellulosic feedstocks, we assume that oxidative DNA damage and genomic instability may also occur when yeast cells encounter other stressors. Further analysis of whether other inhibitors result in similar or diverse patterns of genomic instability could shed new light on how the yeast genome evolves under industrial conditions.
At the whole-genome level, furfural-induced genomic alterations were revealed by a custom whole-genome microarray (see Data Sets S3 and S4). The frequency of genomic alterations in furfural-treated isolates was about 10-fold higher than that observed in a wild-type diploid strain (∼2 × 10−3 events per genome per cell division) (28). Internal and terminal LOH represent most of the outcomes of repairing furfural-induced DNA lesions, accounting for up to 90% of all detected genetic events. The total size of the LOH regions among the 21 isolates was 17.5 Mb. On average, for individual subcultured colonies, about 7% of the genomic region had LOH. In addition, furfural-induced gross chromosomal rearrangements and aneuploidy events influenced the copy numbers of hundreds of genes among certain isolates. In wild-type yeast, mitotic recombination events are not randomly distributed across the genome (21). It was found that the replication fork is slow moving at certain genomic sites, including centromeres, tRNA genes, Ty elements, DNA replication terminations, G4 motifs, highly transcribed genes, and pause sites for Rrm3p (35–37). Longer stalling of the replication fork increases the risk of replication fork collapse and DSBs in the S phase (38). In addition, certain chromosomal elements, such as two inverted Ty elements located at approximately kb 950 on chromosome IV, are likely to induce DSBs in the G1 phase (21). In this study, association analysis of chromosomal elements and furfural-induced breakpoints showed that only one type of element, a high-GC (>42%) region, was significantly enriched near breakpoints (Table 1). This analysis indicated that furfural-induced DSBs that initiated mitotic recombination occur more frequently in GC-rich regions. In cells, repair of oxidized bases through the base excision repair pathway generates apurinic/apyrimidinic (AP) sites that can be removed by backbone cleavage, end processing, DNA synthesis, and ligation (39, 40). Two recent studies showed, through analysis of the distributions of AP sites across the genome, that oxidative damage accumulates preferentially in GC-rich regions (39, 40). Because incision of close AP sites would give rise to DSBs (25), repair of oxidized bases within GC-rich regions may be a factor that contributes to mitotic recombination in furfural-treated cells.
We then asked whether and how furfural exposure affects point mutations. By whole-genome sequencing of 7 furfural-treated isolates and 20 untreated isolates of JSC25-1, we found that furfural treatment resulted in a mild increase in the point mutation rate. Additionally, the spectra of SNVs are very different between furfural-treated cells and untreated cells (Fig. 8). In the untreated JSC25-1 isolates, like other aerobic organisms, C-to-T/G-to-A transition is the most prominent mutation and accounts for ∼30% of SNVs (27). This substitution was increased by about 2-fold in furfural-treated cells (P < 0.05; Fisher exact test) (Fig. 8). Multiple pieces of evidence show that the transition from C to T/G to A is also the most abundant genetic change that resulted from oxidative DNA damage (41, 42). It was also suggested that oxidized cytosines contribute significantly to the C-to-T transition (41, 42). Oxidation of cytosine can give rise to 5,6-dihydroxy-5,6-dihydrocytosine, which can break down further to form 5-hydroxycytosine, 5-hydroxyuracil, and 5,6-dihydroxy-5,6-dihydrouracil (42). Deamination of the 4-amino group of cytosine causes misincorporation of A opposite C, resulting in C-to-T mutation after DNA replication (42). Taken together, it is likely that furfural-induced ROS not only stimulate mitotic recombination, but also trigger cytosine oxidation and contribute to C-to-T transition.
Among industrial yeast strains, ploidy changes, large-scale deletions/amplification, LOH, and small DNA changes are ubiquitous and contribute to adaptability under certain conditions (43–46). Given the recombinogenic effects of furfural revealed in this study, it is likely that furfural not only acts as a stressor in fermentation medium, but is also a factor that drives phenotypic variations. In the laboratory, directed evolution (iterative rounds of incubation of strains in furfural-containing medium) has proven to be efficient in improving the tolerance of furfural in yeast (4). Our previous study showed that whole-genome duplication (from diploid to triploid or tetraploid) in yeast is advantageous for the tolerance of vanillin (another common inhibitor in cellulosic feedstock), but not of furfural (47). The improved resistance to furfural should be attributed to other genetic events. On the other hand, it should be noted that the prolonged exposure of yeast cells to furfural (particularly in distilleries that use cell cycling) may quickly lead to loss of selected desirable traits (such as the yield of ethanol) within the yeast population.
In summary, this study characterizes the effects of furfural on a number of different types of changes throughout the yeast genome. Our work also provides a paradigm for exploring the global effects of other industrial or environmental chemicals on genomic stability at the whole-genome level.
MATERIALS AND METHODS
Strains and medium.
S. cerevisiae strain JSC25-1 (MATα::HYGr/MATa LEU/leu2-3,112 HIS/his3-11,15 ura3/ura3-1 GAL2/gal2 ade2-1/ade2-1 TRP/trp101 CAN1/can1-100::NATr NAT RAD5/RAD5 IV1510386::KANr-can1-100/IVI1510386::SUP4-o) was constructed by mating two haploid strains, JSC12 (W303-1A background) and JSC21 (YJM789 background) (21). MATα in JSC25-1 was deleted to allow the synchronization of cells in the G1 phase. Strain Yyy123 (MATa lys2::Alu-DIR-LEU2-lys2DEL5′ ura3-DEL leu2-3,112 his7-2 trp1-289 ade5-1 circular chromosome III) was constructed in the Petes laboratory at Duke University. Strain JSC24-2 (MATα::NATr/MATa ura3/ura3-1 ade2-1/ade2-1 SUP4-o/can1-100) was used as a control for SNP microarray analysis (21). The YPD medium contained 20 g/liter dextrose, 20 g/liter peptone, and 10 g/liter yeast extract.
Yeast colony color-screening assay.
Strain JSC25-1 was plated on YPD plates and incubated at 30°C for 72 h. Cells collected from the plates were resuspended in 1 ml of YPD medium (optical density at 600 nm [OD600] = 2) containing 10 to 20 g/liter furfural. After incubation at 30°C for 2 h, the cells were washed with double-distilled H2O (ddH2O) and plated on YPD plates to form colonies at 30°C. Alternatively, JSC25-1 cells from the plates were plated on YPD solid medium containing 0.1 to 0.5 g/liter furfural. The plates were incubated at 30°C for 4 days. The ranges of furfural concentrations (0.1 to 20 g/liter) were close to those existing in various bioethanol fermentation media (4, 5). Also, as described above, exposure to furfural at these concentrations for 2 h significantly stimulated mitotic recombination in our yeast model.
Cell synchronization in the G1 phase and furfural treatment.
JSC25-1 cells were cultured in 7 ml of YPD to the logarithmic phase (OD600, ∼0.2). Cells were collected by centrifugation (6,000 rpm for 5 min) and resuspended in 2 ml of YPD containing 100 ng/ml alpha-factor (Sigma-Aldrich, St. Louis, MO, USA). The cells were incubated at 30°C for 2 h to allow >90% of the cells to be synchronized at the G1 phase. Because we found that G1-synchronized cells were less tolerant of furfural, a lower concentration (17 g/liter) of furfural was added to the cell culture. Culture was continued at 30°C for 1 h. This treatment procedure led to cell viability loss similar to that observed in asynchronous yeast cells treated with 20 g/liter furfural for 2 h. The cells were then collected by centrifugation (6,000 rpm for 5 min) and washed twice with sterile water before plating on YPD plates or plug preparation for CHEF experiments.
Subculture of JSC25-1-derived isolates to accumulate genomic alterations.
To begin the subculture process, JSC25-1 cells were plated on a YPD plate to form colonies. Twenty-one colonies (named QLF1 to QLF21) were picked and independently suspended in 0.5 ml YPD containing 20 g/liter furfural. After exposure to furfural for 2 h at 30°C, the treated cells were washed and plated on YPD plates. From each plate, only one colony was picked for the next round of furfural treatment. This procedure was repeated 18 times to accumulate genomic alterations. The isolates from the 18th subculture were stored in a freezer at −80°C. To determine the rates of spontaneous point mutations in JSC25-1, 20 isolates were subcultured 20 times in a way that was similar to that used for the furfural-treated isolates. The only difference was that no furfural exposure was applied to these 20 control isolates.
CHEF gel analysis.
Yeast chromosomal DNA samples for CHEF analysis were prepared as described in a study by Argueso et al. (48). Briefly, yeast cells were embedded in plugs containing 1 mg/ml zymolyase. The plugs were incubated in a 1-ml Tris-EDTA (TE) solution at 37°C for 14 h. Proteinase K solution (6 mg/ml) was then added, and the incubation was continued at 50°C for 16 h. The plugs were washed in 10 ml of 1× TE buffer for 3 days before electrophoresis. CHEF analysis was carried out using a Bio-Rad CHEF Mapper system under the following conditions: 0.5× Tris-borate-EDTA (TBE), 5 V/cm, and 60 to 120 s switch time ramp for 48 h. The gels were stained with 0.5 mg/ml ethidium bromide for 0.5 h and destained with 400 ml sterile water for 0.5 h.
Southern blot analysis.
After separation by CHEF gel electrophoresis, chromosomal DNA was transferred to nylon membranes (49). Hybridization probes specific for the LEU2 gene were prepared by PCR amplification (primers CTGTGGAGGAAACCATCAAG and TCAATGGCCTTACCTTCTTC) of genomic DNA extracted from strain Yyy123 using the PCR DIG probe synthesis kit (Roche, Basel, Switzerland) (34). Details of the hybridization conditions were provided previously (50).
SNP microarray analysis.
Two kinds of SNP microarrays, a chromosome IV-specific array (21) and a whole-gnome SNP array (24), were used for LOH analysis in this study. Yeast genomic DNA was extracted using the commercial EZNA yeast DNA kit (Omega, Doraville, GA, USA) and sheared into fragments with an average size of about 500 bp by sonication. Genomic DNA from the JSC25-1-derived isolates was labeled with Cy5-dUTP, and control DNA from the fully heterozygous strain JSC24-2 was labeled with Cy3-dUTP. Competitive hybridization of the two DNA samples was performed on the SNP microarrays at 62°C. The ratio of hybridization of the two differentially labeled samples was examined using a GenePix scanner and GenePix Pro6.1 software. Ratios of hybridization for each oligonucleotide were normalized to the Cy5/Cy3 ratio of all of the oligonucleotides on the microarray. The principles distinguishing homozygous and heterozygous SNPs have been described by St. Charles and Petes (24).
Whole-genome sequencing analysis.
Whole-genome sequencing of yeast strains was performed on an Illumina HiSeq 2500 sequencer using a 2- by 150-bp paired-end indexing protocol. Mapping of the reads onto a reference genome and calling of point mutations were performed as described in our previous study (26). Briefly, sequencing reads were aligned with the genome of yeast strain S288c (https://www.yeastgenome.org/) with BWA software (51). Single base substitutions and small insertions/deletions were detected using Samtools (52) and VarScan (53) software.
Detection of intracellular ROS.
Yeast cells were precultured on YPD plates for 2 days at 30°C. The cells (∼3 × 106) from patches on YPD plates were collected and resuspended in 2 ml of YPD, 2 ml of YPD containing furfural, or 2 ml of YPD containing 0.05% H2O2. The cells were incubated at 30°C for 1 h, and DCFH-DA was added to a final concentration of 10 μM (14). Following 1 h of incubation, the cells were washed three times with 1× phosphate-buffered saline (PBS) (10 mM phosphate, 138 mM NaCl, and 2.7 mM KCl, pH 7.4) three times and resuspended in 1 ml of 1× PBS. The fluorescence of each sample was measured with a fluorescence spectrophotometer (RF-6000PC; Shimadzu) at the longest excitation wavelength (488 nm) and the maximum emission wavelength (525 nm) (14). Raw data were extracted with LabSolutions RF software. The mean values of five replicates were used for statistical analysis.
Accession number(s).
The accession numbers for the microarrays used in this study are GSE104879 (whole-genome microarrays of subcultured strains) and GSE104880 (chromosome IV-specific microarrays of sectored colonies). The clear read data have been deposited in the SRA database (accession number PRJNA416056).
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Natural Science Foundation of Zhejiang Province (LY18C060002), the National Natural Science Foundation of China (31800055), the Fundamental Research Funds for the Central Universities, and the Postdoctoral Research Foundation of China (2018M630666).
We thank Tom Petes, who is supported by NIH grants GM24110, GM52319, and R35GM118020, for kindly providing yeast strains and technological support in the SNP microarray experiments.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01237-19.
REFERENCES
- 1.Hadi SM, Shahabuddin Rehman A. 1989. Specificity of the interaction of furfural with DNA. Mutat Res 225:101–106. doi: 10.1016/0165-7992(89)90125-5. [DOI] [PubMed] [Google Scholar]
- 2.Lange JP, van der Heide E, van Buijtenen J, Price R. 2012. Furfural—a promising platform for lignocellulosic biofuels. ChemSusChem 5:150–166. doi: 10.1002/cssc.201100648. [DOI] [PubMed] [Google Scholar]
- 3.Mariscal R, Maireles-Torres P, Ojeda M, Sádaba I, Granados ML. 2016. Furfural: a renewable and versatile platform molecule for the synthesis of chemicals and fuels. Energy Environ Sci 9:1144–1189. doi: 10.1039/C5EE02666K. [DOI] [Google Scholar]
- 4.Heer D, Sauer U. 2008. Identification of furfural as a key toxin in lignocellulosic hydrolysates and evolution of a tolerant yeast strain. Microb Biotechnol 1:497–506. doi: 10.1111/j.1751-7915.2008.00050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Almeida JRM, Modig T, Petersson A, Hähn‐Hägerdal B, Lidén G, Gorwa‐Grauslund MF. 2007. Increased tolerance and conversion of inhibitors in lignocellulosic hydrolysates by Saccharomyces cerevisiae. J Chem Technol Biotechnol 82:340–349. doi: 10.1002/jctb.1676. [DOI] [Google Scholar]
- 6.Song H-S, Jeon J-M, Kim H-J, Bhatia SK, Sathiyanarayanan G, Kim J, Won Hong J, Gi Hong Y, Young Choi K, Kim Y-G, Kim W, Yang Y-H. 2017. Increase in furfural tolerance by combinatorial overexpression of NAD salvage pathway enzymes in engineered isobutanol-producing E. coli. Bioresour Technol 245:1430–1435. doi: 10.1016/j.biortech.2017.05.197. [DOI] [PubMed] [Google Scholar]
- 7.Palmqvist E, Almeida JS, Hahn-Hagerdal B. 1999. Influence of furfural on anaerobic glycolytic kinetics of Saccharomyces cerevisiae in batch culture. Biotechnol Bioeng 62:447–454. doi:. [DOI] [PubMed] [Google Scholar]
- 8.Lopes da Silva T, Santo R, Reis A, Passarinho PC. 2017. Effect of furfural on Saccharomyces carlsbergensis growth, physiology and ethanol production. Appl Biochem Biotechnol 182:708–720. doi: 10.1007/s12010-016-2356-5. [DOI] [PubMed] [Google Scholar]
- 9.Hasunuma T, Ismail KS, Nambu Y, Kondo A. 2014. Co-expression of TAL1 and ADH1 in recombinant xylose-fermenting Saccharomyces cerevisiae improves ethanol production from lignocellulosic hydrolysates in the presence of furfural. J Biosci Bioeng 117:165–169. doi: 10.1016/j.jbiosc.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 10.Lin FM, Qiao B, Yuan YJ. 2009. Comparative proteomic analysis of tolerance and adaptation of ethanologenic Saccharomyces cerevisiae to furfural, a lignocellulosic inhibitory compound. Appl Environ Microbiol 75:3765–3776. doi: 10.1128/AEM.02594-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan FR, Dai LC, Wu B, Qin H, Shui ZX, Wang JL, Zhu QL, Hu QC, Ruan ZY, He MX. 2015. Improving furfural tolerance of Zymomonas mobilis by rewiring a sigma factor RpoD protein. Appl Microbiol Biotechnol 99:5363–5371. doi: 10.1007/s00253-015-6577-2. [DOI] [PubMed] [Google Scholar]
- 12.Zdzienicka M, Tudek B, Zieleńska M, Szymczyk T. 1978. Mutagenic activity of furfural in Salmonella typhimurium TA100. Mutat Res 58:205–209. doi: 10.1016/0165-1218(78)90010-1. [DOI] [PubMed] [Google Scholar]
- 13.Khan QA, Shamsi FA, Hadi SM. 1995. Mutagenicity of furfural in plasmid DNA. Cancer Lett 89:95–99. doi: 10.1016/0304-3835(94)03654-2. [DOI] [PubMed] [Google Scholar]
- 14.Allen SA, Clark W, McCaffery JM, Cai Z, Lanctot A, Slininger PJ, Liu ZL, Gorsich SW. 2010. Furfural induces reactive oxygen species accumulation and cellular damage in Saccharomyces cerevisiae. Biotechnol Biofuels 3:2. doi: 10.1186/1754-6834-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lake B, Edwards A, Price R, Phillips B, Renwick A, Beamand J, Adams T. 2001. Lack of effect of furfural on unscheduled DNA synthesis in the in vivo rat and mouse hepatocyte DNA repair assays and in precision-cut human liver slices. Food Chem Toxicol 39:999–1011. doi: 10.1016/S0278-6915(01)00050-3. [DOI] [PubMed] [Google Scholar]
- 16.Calabrese EJ. 2017. Hormesis commonly observed in the assessment of aneuploidy in yeast. Environ Pollut 225:713–728. doi: 10.1016/j.envpol.2017.03.020. [DOI] [PubMed] [Google Scholar]
- 17.Eki T. 2018. Yeast-based genotoxicity tests for assessing DNA alterations and DNA stress responses: a 40-year overview. Appl Microbiol Biotechnol 102:2493–2507. doi: 10.1007/s00253-018-8783-1. [DOI] [PubMed] [Google Scholar]
- 18.Suzuki H, Sakabe T, Hirose Y, Eki T. 2017. Development and evaluation of yeast-based GFP and luciferase reporter assays for chemical-induced genotoxicity and oxidative damage. Appl Microbiol Biotechnol 101:659–671. doi: 10.1007/s00253-016-7911-z. [DOI] [PubMed] [Google Scholar]
- 19.Fasullo M, Freedland J, St John N, Cera C, Egner P, Hartog M, Ding X. 2017. An in vitro system for measuring genotoxicity mediated by human CYP3A4 in Saccharomyces cerevisiae. Environ Mol Mutagen 58:217–227. doi: 10.1002/em.22093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lan J, Rahman SM, Gou N, Jiang T, Plewa MJ, Alshawabkeh A, Gu AZ. 2018. Genotoxicity assessment of drinking water disinfection byproducts by DNA damage and repair pathway profiling analysis. Environ Sci Technol 52:6565–6575. doi: 10.1021/acs.est.7b06389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Symington LS, Rothstein R, Lisby M. 2014. Mechanisms and regulation of mitotic recombination in Saccharomyces cerevisiae. Genetics 198:795–835. doi: 10.1534/genetics.114.166140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sung P. 2018. Introduction to the thematic minireview series: DNA double-strand break repair and pathway choice. J Biol Chem 293:10500–10501. doi: 10.1074/jbc.TM118.003212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeggo PA, Pearl LH, Carr AM. 2016. DNA repair, genome stability and cancer: a historical perspective. Nat Rev Cancer 16:35–42. doi: 10.1038/nrc.2015.4. [DOI] [PubMed] [Google Scholar]
- 24.St. Charles J, Petes TD. 2013. High-resolution mapping of spontaneous mitotic recombination hotspots on the 1.1 Mb arm of yeast chromosome IV. PLoS Genet 9:e1003434. doi: 10.1371/journal.pgen.1003434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma W, Resnick MA, Gordenin DA. 2008. Apn1 and Apn2 endonucleases prevent accumulation of repair-associated DNA breaks in budding yeast as revealed by direct chromosomal analysis. Nucleic Acids Res 36:1836–1846. doi: 10.1093/nar/gkm1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng D-Q, Zhang K, Wu X-C, Mieczkowski PA, Petes TD. 2016. Global analysis of genomic instability caused by DNA replication stress in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 113:E8114–E8121. doi: 10.1073/pnas.1618129113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu YO, Siegal ML, Hall DW, Petrov DA. 2014. Precise estimates of mutation rate and spectrum in yeast. Proc Natl Acad Sci U S A 111:E2310–E2318. doi: 10.1073/pnas.1323011111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Connell K, Jinks-Robertson S, Petes TD. 2015. Elevated genome-wide instability in yeast mutants lacking RNase H activity. Genetics 201:963–975. doi: 10.1534/genetics.115.182725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yin Y, Dominska M, Yim E, Petes TD. 2017. High-resolution mapping of heteroduplex DNA formed during UV-induced and spontaneous mitotic recombination events in yeast. Elife 6:e28069. doi: 10.7554/eLife.28069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang H, Li Q, Kuang X, Xiao D, Han X, Hu X, Li X, Ma M. 2018. Functions of aldehyde reductases from Saccharomyces cerevisiae in detoxification of aldehyde inhibitors and their biotechnological applications. Appl Microbiol Biotechnol 24:10439–10456. doi: 10.1007/s00253-018-9425-3. [DOI] [PubMed] [Google Scholar]
- 31.Ask M, Bettiga M, Mapelli V, Olsson L. 2013. The influence of HMF and furfural on redox-balance and energy-state of xylose-utilizing Saccharomyces cerevisiae. Biotechnol Biofuels 6:22. doi: 10.1186/1754-6834-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li K, Xia J, Mehmood MA, Zhao X-Q, Liu C-G, Bai F-W. 2019. Extracellular redox potential regulation improves yeast tolerance to furfural. Chem Eng Sci 196:54–63. doi: 10.1016/j.ces.2018.11.059. [DOI] [Google Scholar]
- 33.Hayashi M, Umezu K. 2017. Homologous recombination is required for recovery from oxidative DNA damage. Genes Genet Syst 92:73–80. doi: 10.1266/ggs.16-00066. [DOI] [PubMed] [Google Scholar]
- 34.Zhang K, Zheng DQ, Sui Y, Qi L, Petes TD. 2019. Genome-wide analysis of genomic alterations induced by oxidative DNA damage in yeast. Nucleic Acids Res 47:3521–3535. doi: 10.1093/nar/gkz027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fachinetti D, Bermejo R, Cocito A, Minardi S, Katou Y, Kanoh Y, Shirahige K, Azvolinsky A, Zakian VA, Foiani M. 2010. Replication termination at eukaryotic chromosomes is mediated by Top2 and occurs at genomic loci containing pausing elements. Mol Cell 39:595–605. doi: 10.1016/j.molcel.2010.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Azvolinsky A, Giresi PG, Lieb JD, Zakian VA. 2009. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol Cell 34:722–734. doi: 10.1016/j.molcel.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Capra JA, Paeschke K, Singh M, Zakian VA. 2010. G-quadruplex DNA sequences are evolutionarily conserved and associated with distinct genomic features in Saccharomyces cerevisiae. PLoS Comput Biol 6:e1000861. doi: 10.1371/journal.pcbi.1000861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song W, Dominska M, Greenwell PW, Petes TD. 2014. Genome-wide high-resolution mapping of chromosome fragile sites in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 111:E2210–E2218. doi: 10.1073/pnas.1406847111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morris LP, Conley AB, Degtyareva N, Jordan IK, Doetsch PW. 2017. Genome‐wide map of Apn1 binding sites under oxidative stress in Saccharomyces cerevisiae. Yeast 34:447–458. doi: 10.1002/yea.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poetsch AR, Boulton SJ, Luscombe NM. 2018. Genomic landscape of oxidative DNA damage and repair reveals regioselective protection from mutagenesis. Genome Biol 19:215. doi: 10.1186/s13059-018-1582-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Degtyareva NP, Heyburn L, Sterling J, Resnick MA, Gordenin DA, Doetsch PW. 2013. Oxidative stress-induced mutagenesis in single-strand DNA occurs primarily at cytosines and is DNA polymerase zeta-dependent only for adenines and guanines. Nucleic Acids Res 41:8995–9005. doi: 10.1093/nar/gkt671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kreutzer DA, Essigmann JM. 1998. Oxidized, deaminated cytosines are a source of C→T transitions in vivo. Proc Natl Acad Sci U S A 95:3578–3582. doi: 10.1073/pnas.95.7.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang K, Di YN, Qi L, Sui Y, Wang TY, Fan L, Lv ZM, Wu XC, Wang PM, Zheng DQ. 2018. Genetic characterization and modification of a bioethanol-producing yeast strain. Appl Microbiol Biotechnol 102:2213–2223. doi: 10.1007/s00253-017-8727-1. [DOI] [PubMed] [Google Scholar]
- 44.Zhang K, Zhang LJ, Fang YH, Jin XN, Qi L, Wu XC, Zheng DQ. 2016. Genomic structural variation contributes to phenotypic change of industrial bioethanol yeast Saccharomyces cerevisiae. FEMS Yeast Res 16:fov118. doi: 10.1093/femsyr/fov118. [DOI] [PubMed] [Google Scholar]
- 45.Zhang K, Tong M, Gao K, Di Y, Wang P, Zhang C, Wu X, Zheng D. 2015. Genomic reconstruction to improve bioethanol and ergosterol production of industrial yeast Saccharomyces cerevisiae. J Ind Microbiol Biotechnol 42:207–218. doi: 10.1007/s10295-014-1556-7. [DOI] [PubMed] [Google Scholar]
- 46.Zheng DQ, Wang PM, Chen J, Zhang K, Liu TZ, Wu XC, Li YD, Zhao YH. 2012. Genome sequencing and genetic breeding of a bioethanol Saccharomyces cerevisiae strain YJS329. BMC Genomics 13:479. doi: 10.1186/1471-2164-13-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang K, Fang Y-H, Gao K-H, Sui Y, Zheng D-Q, Wu X-C. 2017. Effects of genome duplication on phenotypes and industrial applications of Saccharomyces cerevisiae strains. Appl Microbiol Biotechnol 101:5405–5414. doi: 10.1007/s00253-017-8284-7. [DOI] [PubMed] [Google Scholar]
- 48.Argueso JL, Westmoreland J, Mieczkowski PA, Gawel M, Petes TD, Resnick MA. 2008. Double-strand breaks associated with repetitive DNA can reshape the genome. Proc Natl Acad Sci U S A 105:11845–11850. doi: 10.1073/pnas.0804529105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCulley JL, Petes TD. 2010. Chromosome rearrangements and aneuploidy in yeast strains lacking both Tel1p and Mec1p reflect deficiencies in two different mechanisms. Proc Natl Acad Sci U S A 107:11465–11470. doi: 10.1073/pnas.1006281107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao Y, Strope PK, Kozmin SG, McCusker JH, Dietrich FS, Kokoska RJ, Petes TD. 2014. Structures of naturally evolved CUP1 tandem arrays in yeast indicate that these arrays are generated by unequal nonhomologous recombination. G3 (Bethesda) 4:2259–2269. doi: 10.1534/g3.114.012922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. 2012. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.