Abstract
Paragonimiasis is an important food-borne zoonosis caused by Paragonimus flukes and is endemic to western Japan. However, there have been few epidemiological studies in the Tohoku district of northeastern Japan. In this study, Paragonimus metacercariae (mc) was detected in Geothelphusa dehaani (Japanese freshwater crab or Sawagani) in Iwate Prefecture. Out of the 207 Sawagani collected from 35 localities, 12 individuals from six localities were infected with Paragonimus mc. The mc were identified as P. skrjabini miyazakii based on the sequences of nuclear ribosomal internal transcribed spacer 2 and mitochondrial cytochrome c oxidase I. This is the first report of P. s. miyazakii mc infection in Sawagani in Iwate Prefecture.
Keywords: cox1, Geothelphusa dehaani, nuclear ribosomal internal transcribed spacer 2, Iwate Prefecture, Paragonimus skrjabini miyazakii
Paragonimiasis is one of the severe food-borne parasitic zoonosis caused by Paragonimus flukes, parasitizing the lungs of carnivora and/or omnivorous mammals and humans [2]. In Japan, two Paragonimus species, P. westermani (including the diploid and triploid forms) and P. miyazakii, were recognized as infectious agents in humans [11], and the latter species has been recently referred to as P. skrjabini miyazakii [3, 6]. Both the species require two intermediate hosts; the first is a freshwater snail and the second is a freshwater crab [7]. Human paragonimiasis is caused by consuming the uncooked freshwater crabs infected with metacercariae (mc) [7]. Therefore, many epidemiological studies on Paragonimus infection in crabs [9] have been carried out from southwestern to central Japan where human paragonimiasis is endemic [8]. On the other hand, epidemiological studies in Tohoku district, northeastern Japan have been limited [9], and there has been no report on Paragonimus infection in freshwater crabs in Iwate Prefecture, Japan. This study is the first to report the detection of P. s. miyazakii metacercariae (mc) in the fresh water crabs, Geothelphusa dehaani, in this region.
A total of 207 Japanese freshwater crabs G. dehaani, also called “Sawagani” in Japanese, were collected from mountain streams from 35 localities in Iwate Prefecture from September to October 2016 and from April to September 2017 (Fig. 1, Table 1). The crabs were individually crushed and digested with stirring at 37°C for 15–30 min in 100 ml digestive solution (1,000 ml distilled water, 1.4 ml hydrochloric acid (Wako Pure Chemical Industries, Osaka, Japan), 1.3 g pepsin (proteolytic activity 1:10,000) (MP Biomedicals Inc., Solon, OH, U.S.A.)). The resultant solutions were filtered by a tea strainer for removing shell debris, and the sediments were repeatedly washed with 0.85% NaCl solutions. The resultant sediments were examined for mc under a stereo microscope. The detected mc were measured in diameter using an optical microscope with a digital camera DP26 and imaging software cellSens (Olympus, Tokyo, Japan). The mc were excysted with tweezers and total DNAs were extracted using High Pure PCR Template Preparation Kit (Roche, Mannheim, Germany). Polymerase chain reaction (PCR) was performed for the nuclear ribosomal internal transcribed spacer 2 (ITS2) and the mitochondrial cytochrome c oxidase subunit 1 (cox1) gene. The primers used were 3S and A28 for ITS2 [1], and JB3 and JB4.5 for cox1 [4]. PCR was performed in a final volume of 50 µl containing 1 µl of template DNA, 0.25 µM of each primer, 1 unit of Tks Gflex DNA polymerase (TaKaRa, Kusatsu, Japan), and 25 µl of manufacturer’s supplied reaction buffer. Thermal cycling conditions were as follows: 1 min at 94°C, followed by 35 cycles at 98°C for 10 sec, 60°C (ITS2) or 55°C (cox1) for 15 sec, and 68°C for 30 sec. PCR amplicons were purified using a NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel, Düren, Germany), and then sequenced in both directions with the same single PCR primers, using a BigDye Terminator v3.1 Cycle Sequence Kit (Applied Biosystems, Foster City, CA, U.S.A.) in an ABI 3500 Genetic Analyzer (Applied Biosystems, Waltham, MA, U.S.A.). The resulting sequences were initially assembled using ATGC ver. 6.0.3 (Genetyx Co., Tokyo, Japan) and aligned using Clustal W [14]. The accession numbers of reference sequence used for homology calculations were as follows: P. s. miyazakii (ITS2: AB713405, AY618742, U96912; cox1: AY618807, AY618809, AY618812-AY618814, AY618816, AY618817, AY618820-AY618823, AY618830, AY618833, U97215), P. westermani (ITS2: AB354214, U96907, U96908, U96909, U96910; cox1: AB354223, AB354225, AY140671, AY140672, AY140676, AY140682, AY140686, AY140695, U97205), P. ohirai (ITS2: U96911; cox1: AF008189, U97214) and P. s. skrjabini (ITS2: U96913, AY618734; cox1: AB325522, AB703455, AY618759-AY618761, AY618783, AY618786, AY618788, AY618789, AY618793-AY618795, AY618798, AY618800, AY618801, AY618805, U97216). A phylogenetic tree based on the cox1 sequences was constructed employing a Maximum Likelihood method [13]. The tree was constructed based on the Hasegawa-Kishino-Yano model [5] with a proportion of invariant sites, which were selected with the maximum likelihood test based on Akaike’s information criteria and evaluated using bootstrap tests with 1000 replications. These analyses were performed using MEGA6 [13].
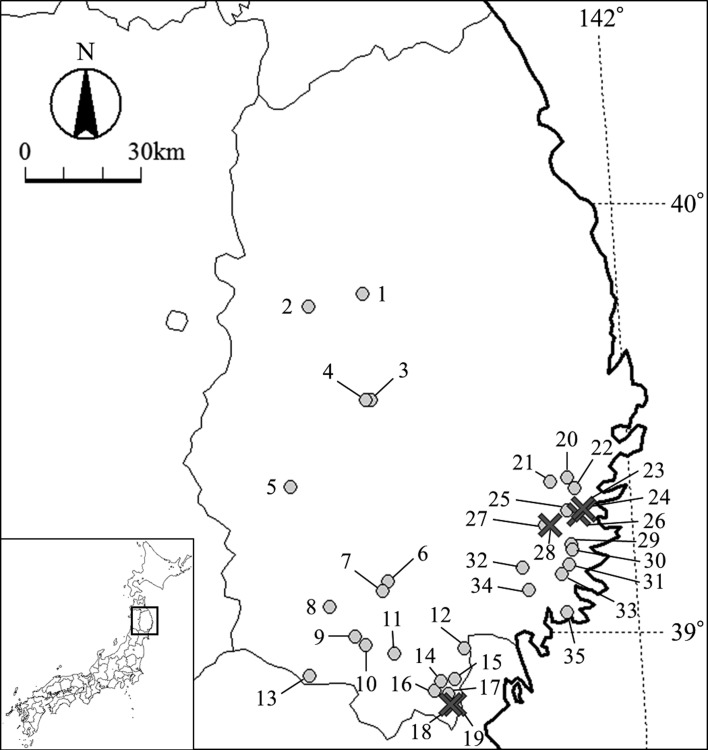
Fig. 1.
Map of Iwate Prefecture showing the sampling localities. The black crosses and gray circles show localities from where Paragonimus skrjabini miyazakii metacercariae infected and non-infected Sawagani, respectively, were collected. The numbers indicate the locality numbers shown in Table 1.
Table 1. Number of collected crabs and detected metacercariae in each locality.
| No. in Fig. 1 |
Locality | No. of collected crabs (No. of positives) |
Code of metacercariae (Total No. of metacercariae) |
||
|---|---|---|---|---|---|
| Name | Latitude (N) | Longitude (E) | |||
| 1 | Takizawa | 39.7888130 | 141.2286940 | 2 (0) | |
| 2 | Morioka | 39.7604520 | 141.0618990 | 3 (0) | |
| 3 | Shiwa | 39.5426320 | 141.2350930 | 7 (0) | |
| 4 | Shiwa | 39.5430830 | 141.2203890 | 2 (0) | |
| 5 | Kitakami | 39.3402410 | 140.9774910 | 4 (0) | |
| 6 | Oshu | 39.1197140 | 141.2550460 | 9 (0) | |
| 7 | Oshu | 39.0972990 | 141.2368660 | 13 (0) | |
| 8 | Oshu | 39.0609210 | 141.0739780 | 6 (0) | |
| 9 | Hiraizumi | 38.9911820 | 141.1459760 | 8 (0) | |
| 10 | Ichinoseki | 38.9719680 | 141.1760270 | 13 (0) | |
| 11 | Ichinoseki | 38.9526270 | 141.2620780 | 12 (0) | |
| 12 | Ichinoseki | 38.9638740 | 141.4719730 | 4 (0) | |
| 13 | Ichinoseki | 38.8989690 | 141.0029290 | 12 (0) | |
| 14 | Ichinoseki | 38.8863490 | 141.3945950 | 4 (0) | |
| 15 | Ichinoseki | 38.8913540 | 141.4371810 | 2 (0) | |
| 16 | Ichinoseki | 38.8661980 | 141.3749440 | 10 (0) | |
| 17 | Ichinoseki | 38.8580380 | 141.4173890 | 9 (0) | |
| 18 | Ichinoseki | 38.8341680 | 141.4207840 | 8 (1) | Gd094mc02 (1) |
| 19 | Ichinoseki | 38.8322430 | 141.4341460 | 6 (1) | Gd085mc01 (1) |
| 20 | Kamaishi | 39.3613240 | 141.8132759 | 1 (0) | |
| 21 | Kamaishi | 39.3513811 | 141.7600270 | 12 (0) | |
| 22 | Kamaishi | 39.3374470 | 141.8330840 | 1 (0) | |
| 23 | Kamaishi | 39.2909711 | 141.8544840 | 13 (4) | Gd179mc01, Gd180mc01, Gd182mc01, Gd186mc01 (4) |
| 24 | Kamaishi | 39.2857761 | 141.8617921 | 2 (1) | Gd175mc01-Gd175mc04 (4) |
| 25 | Kamaishi | 39.2837900 | 141.8072040 | 6 (0) | |
| 26 | Kamaishi | 39.2755640 | 141.8410420 | 5 (4) | Gd140mc01, Gd141mc01-Gd141mc03, Gd142mc01-Gd142mc12, Gd144mc01-Gd144mn04 (20) |
| 27 | Kamaishi | 39.2501310 | 141.7362790 | 2 (0) | |
| 28 | Kamaishi | 39.2495400 | 141.7542680 | 1 (1) | Gd132mc01-Gd132mc09 (9) |
| 29 | Kamaishi | 39.2065830 | 141.8118979 | 4 (0) | |
| 30 | Kamaishi | 39.1943590 | 141.8151961 | 4 (0) | |
| 31 | Ofunato | 39.1582100 | 141.8036351 | 2 (0) | |
| 32 | Ofunato | 39.1516700 | 141.6631762 | 4 (0) | |
| 33 | Ofunato | 39.1357050 | 141.7797031 | 2 (0) | |
| 34 | Ofunato | 39.0997160 | 141.6773439 | 10 (0) | |
| 35 | Ofunato | 39.0470749 | 141.7874241 | 4 (0) | |
| Total | 207 (12) | ||||
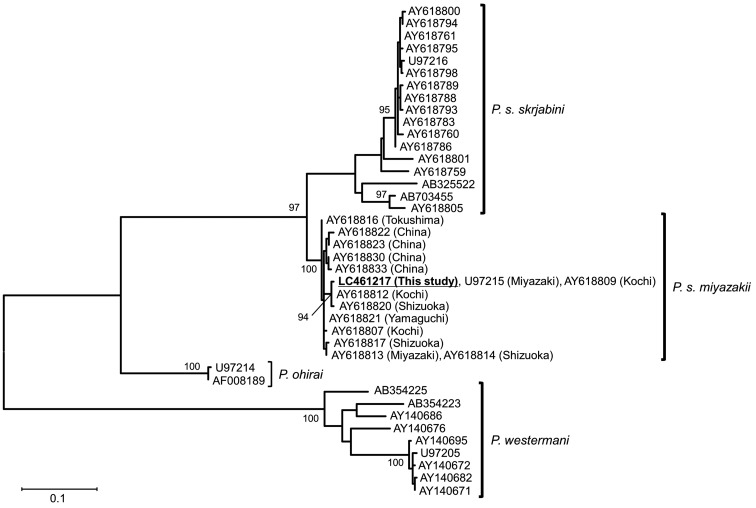
A total of 39 Paragonimus mc were detected in 12 Sawagani from 6 localities (Table 1). The mc were almost circular and their mean diameter (n=25) was 437.5 µm (range 377–485 µm). No stylet was found on their oral suckers. The determined ITS2 sequences (461 bp) were registered to GenBank (LC461214-LC461216) with a single mutation site at the nucleotide position 267. The mutation site was a C in LC461214 (9 mc), a T in LC461215 (3 mc), and heterozygous in LC461216 (27 mc). The homology calculated using the 361 bp ITS2 sequences and ignoring two gap sites was 99.72–100% with P. s. miyazakii, 98.61–100% with P. s. skrjabini, 92.24% with P. ohirai, and 90.86–92.80% with P. westermani. The cox1 sequences (396 bp) were identical among all the mc and registered to GenBank (LC461217). The cox1 sequence homology (340 bp) was 97.65–100% with P. s. miyazakii, 89.71–91.47% with P. s. skrjabini, 86.76–87.05% with P. ohirai, and 80.00–85.29% with P. westermani. In the phylogenetic tree constructed with cox1 (340 bp), the sequence from this study clustered with those of P. s. miyazakii with 100% bootstrap value, and the P. s. miyazakii clade was paraphyletic with the P. s. skrjabini clade (Fig. 2).
Fig. 2.
A maximum likelihood tree constructed based on cox1 seuences of Paragonimus species. The numbersnear the nodes indicate bootstrap values (>90%). The scale bar indicates the number of substitutions per sequence position.
The Sawagani has been reported as the second intermediate host of P. s. miyazakii and diploid P. westermani in Japan [9]. Although the mean diameter of mc obtained in this study was similar to that of both the species [10], the stylet generally observed in P. westermani mc was not observed in this study. The ITS2, which is a useful marker for species discrimination irrespective of the life cycle stages of the lung flukes [1, 12], showed that the present mc were more closely related to P. s. miyazakii and P. s. skrjabini, members of P. skrjabini complex [3], than to P. westermani and P. ohirai. Moreover, the cox1 sequence was completely consistent with those (U97215, AY618809) of P. s. miyazakii previously detected in Japan, belonged to the P. s. miyazakii clade in the phylogenetic tree, and was clearly distinct from the P. s. skrjabini clade with high bootstrap value (Fig. 2). The relationships between morphology and genetic characteristics including cox1 marker among Paragonimus species have been confirmed in previous studies [1, 3]. From these results, the mc in this study were identified as P. s. miyazakii. This is the first report of P. s. miyazakii infection in Sawagani occurring in Iwate Prefecture, Japan and it suggests a possible risk of human paragonimiasis up on consumption of uncooked Sawagani in this region. The first intermediate and definitive hosts of P. s. miyazakii in this region remain unknown; therefore, further studies are needed to elucidate the life cycle of P. s. miyazakii in Iwate Prefecture.
Acknowledgments
We are grateful to the members of the Department of Animal Risk Management, Faculty of Risk and Crisis Management, Chiba Institute of Science.
REFERENCES
- 1.Blair D., Agatsuma T., Watanobe T., Okamoto M., Ito A.1997. Geographical genetic structure within the human lung fluke, Paragonimus westermani, detected from DNA sequences. Parasitology 115: 411–417. doi: 10.1017/S0031182097001534 [DOI] [PubMed] [Google Scholar]
- 2.Blair D., Xu Z. B., Agatsuma T.1999. Paragonimiasis and the genus Paragonimus. Adv. Parasitol. 42: 113–222. doi: 10.1016/S0065-308X(08)60149-9 [DOI] [PubMed] [Google Scholar]
- 3.Blair D., Chang Z., Chen M., Cui A., Wu B., Agatsuma T., Iwagami M., Corlis D., Fu C., Zhan X.2005. Paragonimus skrjabini Chen, 1959 (Digenea: Paragonimidae) and related species in eastern Asia: a combined molecular and morphological approach to identification and taxonomy. Syst. Parasitol. 60: 1–21. doi: 10.1007/s11230-004-1378-5 [DOI] [PubMed] [Google Scholar]
- 4.Bowles J., Blair D., McManus D. P.1995. A molecular phylogeny of the human schistosomes. Mol. Phylogenet. Evol. 4: 103–109. doi: 10.1006/mpev.1995.1011 [DOI] [PubMed] [Google Scholar]
- 5.Hasegawa M., Kishino H., Yano T.1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22: 160–174. doi: 10.1007/BF02101694 [DOI] [PubMed] [Google Scholar]
- 6.Irie T., Yamaguchi Y., Doanh P. N., Guo Z. H., Habe S., Horii Y., Nonaka N.2017. Infection with Paragonimus westermani of boar-hunting dogs in Western Japan maintained via artificial feeding with wild boar meat by hunters. J. Vet. Med. Sci. 79: 1419–1425. doi: 10.1292/jvms.17-0149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawashima K.2003. Biology of lung flukes. pp. 165–182. In: Progress of Medical Parasitology in Japan (Otsuru, M., Kamegai, S. and Hayashi, S. eds.), Meguro Parasitological Museum, Tokyo. [Google Scholar]
- 8.Nagayasu E., Yoshida A., Hombu A., Horii Y., Maruyama H.2015. Paragonimiasis in Japan: a twelve-year retrospective case review (2001–2012). Intern. Med. 54: 179–186. doi: 10.2169/internalmedicine.54.1733 [DOI] [PubMed] [Google Scholar]
- 9.Nishida H., Shibahara T.2003. Epidemiology of paragonimiasis. pp. 201–217. In: Progress of Medical Parasitology in Japan (Otsuru, M., Kamegai, S. and Hayashi, S. eds.), Meguro Parasitological Museum, Tokyo. [Google Scholar]
- 10.Nishida H., Shibahara T., Torii M., Okamoto K., Sakai M., Gyoten J.1987. Studies on the lung flukes, the genus Paragonimus (Trematoda: Troglotrematidae), found from the freshwater crab, Geothelphusa dehaani in northern part of Kyoto Prefecture, Japan II. Geographical distribution of the lung flukes in the districts Yosa, Miyazu, Kasa and Maizuru. Nihon Seibutsu Chiri Gakkai Kaiho 42: 15–21(in Japanese with English abstract). [Google Scholar]
- 11.Otsuji Y.2003. Paragonimiasis. pp. 183–200. In: Progress of Medical Parasitology in Japan (Otsuru, M., Kamegai, S. and Hayashi, S. eds.), Meguro Parasitological Museum, Tokyo. [Google Scholar]
- 12.Sugiyama H., Morishima Y., Kameoka Y., Kawanaka M.2002. Polymerase chain reaction (PCR)-based molecular discrimination between Paragonimus westermani and P. miyazakii at the metacercarial stage. Mol. Cell. Probes 16: 231–236. doi: 10.1006/mcpr.2002.0417 [DOI] [PubMed] [Google Scholar]
- 13.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S.2013. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. doi: 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson J. D., Higgins D. G., Gibson T. J.1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. doi: 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]