Abstract
Long non-coding RNAs (lncRNAs) are emerging as important modulators of cancer progression, among which prostate cancer-associated transcript 1 (PCAT1) has been shown to be an oncogene in several tumors. However, the clinical significance and biological function of PCAT1 in endometrial carcinoma (EC) remain unclear. In this study, we used 89 EC tissues and HEC-1B, Ishikawa, RL95-2 and AN3CA EC cell lines. We found elevated expression levels of PCAT1 in EC tissues and cell lines using reverse transcription qPCR (RT-qPCR). The prognostic value of PCAT1 was determined using Kaplan–Meier survival and Cox regression analysis. The results showed that higher PCAT1 expression was positively correlated with FIGO stage, myometrial invasion, lymph node metastasis, and a shorter overall survival. A series of functional assays showed that the knockdown of PCAT1 by small interfering RNA (siRNA) targeting PCAT1 (siPCAT1) suppressed cell proliferation, migration and invasion, but promoted apoptosis. Western blot analysis further showed that B-cell lymphoma 2 (Bcl-2), vimentin and N-cadherin were downregulated, but E-cadherin and Bcl-2-associated death promoter (Bad) were upregulated in PCAT1-silenced EC cells. Taken together, our results underscore the oncogenic role of PCAT1 in EC and show that PCAT1 may be a potential therapeutic target in EC treatment.
Keywords: Endometrial carcinoma, PCAT1, prognosis, cell proliferation, migration, invasion
INTRODUCTION
Endometrial carcinoma (EC) arises from the lining of the uterus, and it is one of the most frequent gynecological malignancies affecting women around the world [1]. In 2013, the mortality rate of EC was the fourth highest among all malignancies in women in the United States, with 49,500 new cases recorded [2]. Based on the clinicopathological features, EC is mainly divided into two subtypes, endometrioid endometrial carcinomas (EECs) and non-endometrioid endometrial carcinomas (NEECs) for which the prognosis is usually good and poor, respectively [3]. The invasion of the tumor cells into surrounding tissues and spread of cancer to distant organs occur within one year of cancer diagnosis in a minority of patients in advanced stage [4]. Furthermore, the efficacy of surgery, chemotherapy, chemoradiotherapy, and molecular targeted therapies in EC still needs to be improved, which is largely due to poor understanding of the molecular mechanisms underlying EC development. To improve the survival of patients with EC, it is essential to identify candidate genes as well as novel biomarkers for prognosis and targeted therapy.
An emerging evidence indicates that noncoding RNAs (ncRNAs) play critical roles in the modulation of many pathophysiological conditions, including cancer [5]. Among these ncRNAs, the transcripts longer than 200 nucleotides are termed long ncRNAs (lncRNAs). These ncRNAs can operate through various mechanisms and thus impact diverse processes such as cell proliferation, migration, apoptosis, and invasion [6]. Recent studies have demonstrated that aberrant expression of lncRNAs is linked to tumor initiation, progression, and metastasis in human cancer [7]. Due to their function as oncogenes or tumor suppressors, lncRNAs have a role in cancer diagnosis and prognosis [8]. The lncRNA named prostate cancer-associated transcript 1 (PCAT1) was originally discovered in a cohort of prostate tissues and cell lines [9]. This lncRNA is transcribed from a susceptibility locus for prostate cancer at chromosome 8q24 [10]. Prensner et al. [9] identified PCAT1 as a modulator of cell growth in prostate cancer and reported that it is a target of the polycomb repressive complex 2 (PRC2). PCAT1 is upregulated in several cancer types, and its amplification has been linked to an increase in cell proliferation and metastasis in non-small cell lung cancer [11], hepatocellular carcinoma [12], and colorectal cancer [13]. The expression and clinical significance of PCAT1 have also been investigated in gastric cancer [14] and esophageal squamous carcinoma [15], where the overexpression of PCAT1 was associated with advanced cancer stage and poor prognosis. Functional analyses of PCAT1 revealed that the abnormal expression of PCAT1 leads to the upregulation or downregulation of genes involved in mitosis, cell cycle control, and apoptosis among other processes, such as CDKN1A (p21; Cip1/Waf1) [16], BRCA2 [17], c-Myc [18], and FSCN1 [19] genes.
This study was designed to elucidate the clinical significance and biological function of PCAT1 in EC. We analyzed the expression of PCAT1 in EC tissues and cell lines and evaluated whether PCAT1 is a prognostic predictor in EC. Increased knowledge on the biological effects of PCAT1 in EC will lead to better understanding of tumor pathology, enabling the development of novel prognostic biomarkers and targeted therapies.
MATERIALS AND METHODS
Tumor tissues and patient follow-up
A total of 89 patients, whose tumor and adjacent tissues were collected after surgical resection at the First People’s Hospital of Lanzhou city (Gansu, China), were diagnosed with EC by pathology. After resection, the collected tissues were immediately stored at −80°C until RNA extraction. Patients who were not in gestation or lactation and did not receive hormone therapy within 3 months before surgery were included in the study. Patients with adenomyosis, dysfunctional uterine bleeding, polycystic ovary syndrome, or other hormone-dependent disease were excluded. The basic clinical characteristics of patients are summarized in Table 1. After the surgical intervention, the patients were followed up every 2–4 months for 5 consecutive years. The period of time from diagnosis to death was considered as the overall survival of these patients. Each participant signed a written informed consent, and the study was approved by the Ethics Committee of the First People’s Hospital of Lanzhou city (No. jix20170405).
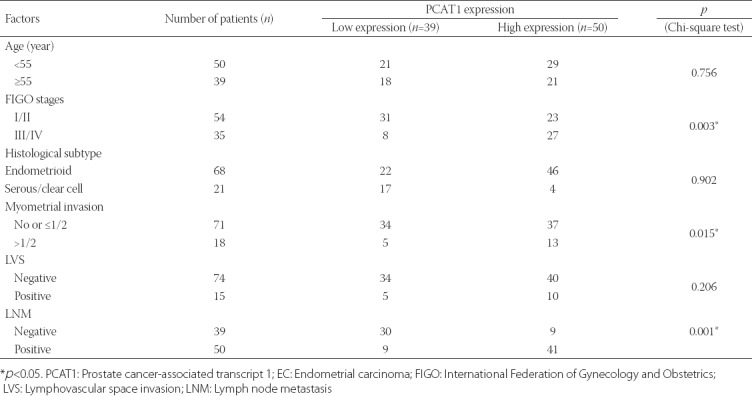
TABLE 1.
Association between PCAT1 expression and clinicopathological characteristics in EC patients (n=89)
Cell culture and transfection
The human EC cell lines HEC-1B, Ishikawa, RL95-2, and AN3CA were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). The normal endometrial stromal cell line ESC was obtained from the Tumor Cell Bank of the Chinese Academy of Medical Sciences (Beijing, China). Ishikawa cells were cultured in the Roswell Park Memorial Institute 1640 (RPMI-1640, Invitrogen, Carlsbad, CA, USA) culture medium and 10% fetal bovine serum (FBS, Gibco, Grand Island, NY). The other four cell lines were cultured in Dulbecco’s modified Eagle’s medium (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with penicillin/streptomycin (100 U/mL) and 10% FBS (Gibco). All cell lines were maintained at 37°C in a humidified atmosphere containing 5% CO2.
The sequence of siRNA targeting PCAT1 (si-PCAT1: 5’-CCTATCTACAGGTCACGTATT-3’) and its corresponding negative control (si-NC: 5’-TTCTCCGAACGTGTCACGT-3’) were synthesized by RiboBio Co. Ltd (Guangzhou, China). For PCAT1 knockdown, HEC-1B or Ishikawa cells were cultured in 6-well plates overnight and transfected with either 50 nM of si-PCAT1 or si-NC using Lipofectamine 2000 Reagent (Invitrogen, Carlsbad, CA, USA), following the manufacturer’s instructions. At 48 hours post-transfection, the cells were used to examine the transfection efficiency and for in vitro experiments.
Reverse transcription quantitative PCR (RT-qPCR)
Total RNA from tissues or cell lines was extracted using TRIzol reagent (Invitrogen). The RNA was reversely transcribed using HiScript first Strand cDNA Synthesis kit (VAzyme Biotech, Nanjing, China). The expression levels of PCAT1 were determined on the CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) using the SYBR-Green PCR Master Mix kit (Takara, Dalian, China) and the 2−ΔΔCt method. The primers were as follows: PCAT1, forward, 5’’-TTGTGGAAGCCCCGCAAGGCCTGAA -3’ and reverse, 5’- TGTGGGGCCTGCACTGGCACTT-3’; β-actin, forward, 5’- TCACCAACTGGGACGACATG-3’ and reverse, 5’- GTCACCGGAGTCCATCACGAT-3’. The β-actin was used as an internal control.
Cell proliferation assay
Cell proliferation was determined with a Cell Counting Kit-8 (CCK-8, Dojindo Laboratories, Kamimashiki-gun, Japan) according to the instructions provided by the manufacturer. Briefly, cells were seeded in 96-well plates (3,000 cells per well) and incubated with 10 µl CCK-8 reagent at indicated time points (0, 24, 48, and 72 hours), followed by continuous culture for 1 hour. The absorbance was measured at 450 nm by a Microplate Reader (Bio-Rad, USA).
Cell apoptosis assay
Cell apoptosis was analyzed using the Annexin V-FITC Apoptosis Detection Kit (BD Biosciences, San Jose, CA, USA). Briefly, transfected cells were treated with trypsin and fixed with 70% ice-cold methanol for 30 minutes at 4°C. After resuspension in binding buffer the cells were stained with 5 µl of Annexin V-FITC and 1 µl of propidium iodide (PI, 50 µg/ml). The percentage of apoptotic cells, including early apoptosis (Annexin V+/PI-) and late apoptosis (Annexin V+/PI+), was analyzed by flow cytometry analysis (BD Biosciences).
Wound healing assay
Wound healing assay was performed to evaluate the migration of EC cells. Briefly, cells were seeded into 6-well plates at a density of 5 × 105 cells per well and cultured for 24 hours to reach 95% confluency. Subsequently, the monolayers were wounded by scraping with a 200 µL tip. Next, the wound images were captured at 0 and 24 hours after scratching. The relative migration distance was calculated according to the following formula: (wound width at 0 h-at 24 h)/wound width at 24 h × 100%.
Cell migration assay
For the invasion assay, transwell chambers (8 µm pores; Corning Inc., Corning, NY, USA) coated with Matrigel (BD Biosciences, San Jose, CA, USA) were used. In brief, transfected cells were placed into the upper chamber in serum-free media and media containing 10% FBS were added into the lower chamber. After 24 hours of incubation, cells that migrated to the lower chamber were fixed with methanol and stained with 0.1% crystal violet. The invasive cells were observed and counted in random five fields under a light microscope.
Western blot analysis
Total proteins from cells were extracted using RIPA buffer (Roche, Basel, Switzerland). After protein quantification with a Bio-Rad protein assay kit (Bio-Rad, Hercules, CA, USA), approximately 30 µg of proteins were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electrotransferred to polyvinylidene fluoride membranes (Thermo Fisher Scientific). The membranes were blocked with 5% skim milk for 2 hours and incubated with primary antibodies against Bcl-2-associated death promoter (Bad), B-cell lymphoma 2 (Bcl-2), E-cadherin, N-cadherin, vimentin, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) overnight at 4°C, followed by incubation with horseradish peroxidase-coupled secondary antibody at room temperature for 1.5 hour. Next, the protein signal was detected using ECL™ Western blotting system (Merck, Darmstadt, Germany) and the intensity of the bands was quantified by densitometry (Quantity One software, Bio-Rad).
Statistical analysis
All statistical analyses were performed using the SPSS Statistics for Windows, Version 17.0. (SPSS Inc., Chicago, IL, USA). Quantitative data were expressed as mean ± standard deviation (SD). The Chi-square test was performed to analyze the relationship between PCAT1 expression levels and clinicopathological parameters. Differences were assessed using Student’s t test (between two groups) and one-way ANOVA and Tukey’s multiple comparison tests (between multiple groups). Overall survival curves were plotted using Kaplan–Meier and the log-rank test. Univariate and multivariate Cox regression analyses were performed to confirm the prognostic predictors affecting overall survival. The p-values <0.05 were considered statistically significant.
RESULTS
PCAT1 is overexpressed in EC tissues and cell lines
To investigate the clinical significance of PCAT1 in EC, the expression of PCAT1 was first determined in a total of 89 pairs of tumor and adjacent tissues using RT-qPCR. As shown in Figure 1A, we found higher expression levels of PCAT1 in tumor tissues than in adjacent tissues. In addition, the expression levels of PCAT1 were significantly elevated in four EC cell lines (HEC-1B, Ishikawa, RL95-2, and AN3CA) compared with the normal endometrial stromal cell line ESC (Figure 1B, p < 0.01, p < 0.001). Together, these data clearly indicate that the PCAT1 gene was overexpressed in EC tissues and cell lines.
FIGURE 1.
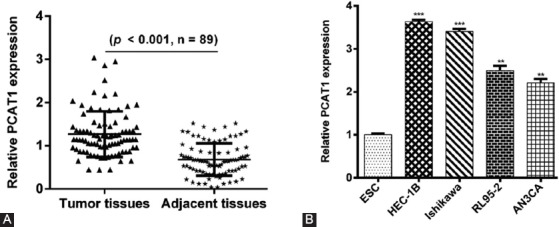
Prostate cancer-associated transcript 1 (PCAT1) was overexpressed in endometrial carcinoma (EC) tissues and cell lines. (A) The relative expression levels of PCAT1 in 89 EC tissues and paired adjacent tissues using reverse transcription quantitative PCR (RT-qPCR) analysis. (B) The relative expression levels of PCAT1 in EC cell lines (HEC-1B, Ishikawa, RL95-2, and AN3CA) and normal endometrial stromal cell line ESC using RT-qPCR analysis. Data are presented as mean ± standard deviation. **p < 0.01, ***p < 0.001.
Higher PCAT1 expression is correlated with aggressive clinicopathological features and poor prognosis in EC
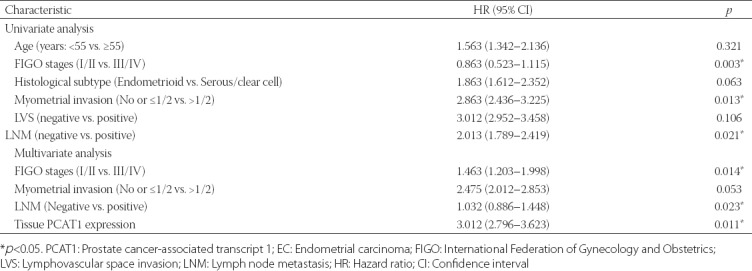
We further evaluated the clinical significance of PCAT1 in EC prognosis. According to the median expression of PCAT1 in EC tissues, patients with EC were divided into high expressing (higher than the median value, n = 50) and low expressing (lower than the median value, n = 39). The Chi-square test showed that high PCAT1 expression was significantly correlated with the International Federation of Gynecology and Obstetrics (FIGO) stages, myometrial invasion, and lymph node metastasis [LNM] (all p < 0.05, Table 1). Kaplan–Meier survival analysis showed that patients with higher PCAT1 expression had poor overall survival rates compared with those with lower PCAT1 expression (Figure 2). Furthermore, univariate and multivariate Cox regression analysis was employed to analyze the effect of PCAT1 expression and other clinical features on overall survival in patients. As shown in Table 2, FIGO stages (hazard ratio [HR] = 1.463, 95% confidence interval [CI] = 1.203–1.998, p = 0.014), LNM (HR = 1.032, 95% CI = 0.886–1.448, p = 0.023), and PCAT1 expression (HR = 3.012, 95% CI = 2.796–3.623, p = 0.011) were independent prognostic factors for overall survival in EC. Collectively, these data show that PCAT1 may be an important prognostic factor for the clinical outcomes in patients with EC.
FIGURE 2.

Kaplan–Meier analysis of prostate cancer-associated transcript 1 (PCAT1) expression and overall survival of patients with endometrial carcinoma [EC] (log-rank p = 0.0032). EC patients with high PCAT1 expression had shorter overall survival than patients with low PCAT1 expression.
TABLE 2.
Univariate and multivariate analysis of prognostic factors affecting overall survival in EC patients
PCAT1 knockdown significantly suppresses proliferation and promotes apoptosis in EC cells
To better understand the biological function of PCAT1 in EC, we performed loss-of-function assays in EC cells. The expression of PCAT1 was markedly higher in HEC-1B and Ishikawa cells than in RL95-2 and AN3CA, so we used HEC-1B and Ishikawa cell lines for the transfection with si-PCAT1 to construct PCAT1 knockdown cell lines. As shown in Figure 3A, the expression level of PCAT1 in HEC-1B and Ishikawa cells transfected with si-PCAT1 was significantly lower than that in HEC-1B and Ishikawa cells transfected with scrambled si-NC, suggesting that the downregulation of PCAT1 in EC cells was achieved by si-PCAT1 transfection (p < 0.001). The results from CCK-8 assay showed that PCAT1 knockdown caused significant inhibition of cell proliferation in HEC-1B and Ishikawa cells (Figure 3B, p < 0.001). Furthermore, the early apoptotic (HEC-1B: 7.7% ± 0.5% vs. 20.5% ± 1.3%, p < 0.001; Ishikawa: 1.7% ± 0.1% vs. 7.6% ± 0.5%, p < 0.01) and late apoptotic rate (HEC-1B: 3.0% ± 0.2% vs. 13.0% ± 0.8%, p < 0.001; Ishikawa: 6.5% ± 0.3% vs. 24.9% ± 0.8%, p < 0.001) were remarkably elevated in si-NC group compared to si-PCAT1 group (Figure 3C). These results demonstrated that the transfection with si-PCAT1 suppressed proliferation and induced apoptosis in EC cells.
FIGURE 3.
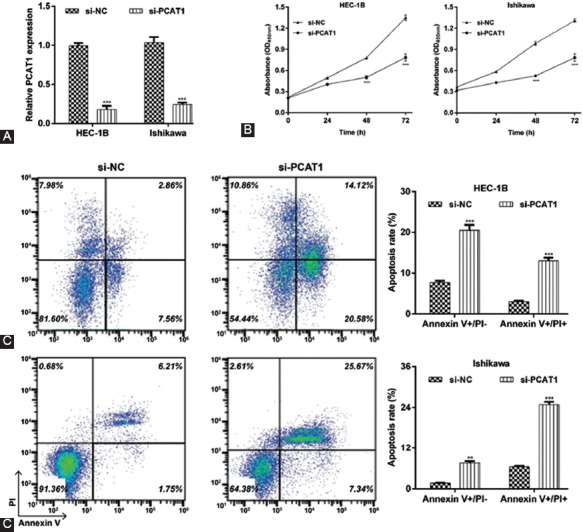
Effects of prostate cancer-associated transcript 1 (PCAT1) knockdown on cell proliferation and apoptosis in endometrial carcinoma (EC) cells. HEC-1B or Ishikawa cells were transfected with si-PCAT1 or si-NC, respectively. (A) The expression of PCAT1 was determined in transfected HEC-1B or Ishikawa cells using reverse transcription quantitative PCR (RT-qPCR) analysis. (B) Cell proliferation was measured by CCK-8 assay in transfected HEC-1B or Ishikawa cells. (C) Cell apoptosis was measured by flow cytometry analysis with Annexin V-FITC/PI double staining. Representative images of apoptotic cells are shown in the left panel. The right panel presents the statistical analysis of early and late apoptotic rate. Data are presented as mean ± standard deviation. **p < 0.01, ***p < 0.001, compared with si-NC group. si-PCAT1: Small interfering (siRNA) targeting PCAT1; si-NC: Negative control.
PCAT1 knockdown significantly suppresses cell migration and invasion in EC cells
We assessed the effects of PCAT1 knockdown on EC cell migration and invasion using a wound healing assay and transwell assay, respectively. As shown in Figure 4A, wound healing assay indicated that the PCAT1 knockdown significantly inhibited relative cell migration distance from 54.9% ± 0.7% to 27.9% ± 0.5% in HEC-1B cells (p < 0.001) and from 30.8% ± 0.6% to 12.2% ± 0.7% in Ishikawa cells (p < 0.01). The transwell assay (Figure 4B) also confirmed that invasive cells were significantly decreased in si-PCAT1 group compared with si-NC group in HEC-1B (53.0 ± 4.4 vs. 10.7 ± 2.1, p < 0.001) and Ishikawa cells (81.0 ± 2.0 vs. 28.3 ± 3.1, p < 0.001). These data indicate that the inhibition of PCAT1 can effectively inhibit the migration and invasion abilities of EC cells.
FIGURE 4.
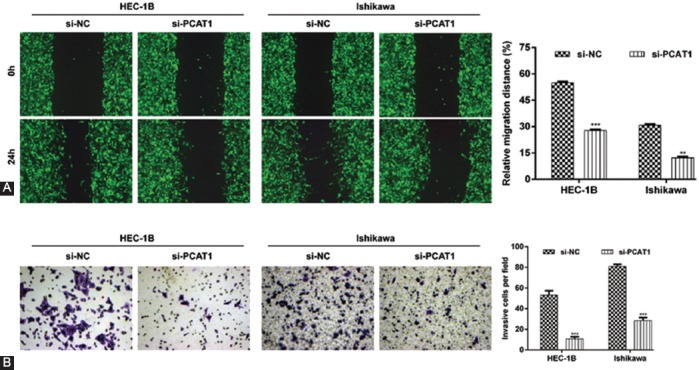
Effects of prostate cancer-associated transcript 1 (PCAT1) knockdown on cell migration and invasion in endometrial carcinoma (EC) cells. HEC-1B or Ishikawa cells were transfected with si-PCAT1 or si-NC, respectively. (A) Representative images (left panel) and quantification (right panel) of wound healing assay in transfected HEC-1B or Ishikawa cell migration. (B) Images (left panel) and quantification (right panel) of transwell assay of transfected HEC-1B or Ishikawa cells. Magnification ×200; data are presented as mean ± standard deviation. **p < 0.01, ***p < 0.001, compared with si-NC group. si-PCAT1: Small interfering (siRNA) targeting PCAT1; si-NC: Negative control.
PCAT1 knockdown notably affects gene expression associated with apoptosis, migration, and invasion in EC cells
We further investigated the molecular mechanisms of PCAT1 in EC cells. Using western blot analysis, we found that the PCAT1 knockdown significantly upregulated the expression of Bad and E-cadherin, but downregulated the expression of Bcl-2, N-cadherin, and vimentin in HEC-1B cells (Figure 5). Therefore, PCAT1 may promote the proliferation, migration, and invasion of EC cells by suppressing pro-apoptotic proteins and activating the expression of epithelial-mesenchymal transition (EMT)-related proteins.
FIGURE 5.

Effects of prostate cancer-associated transcript 1 (PCAT1) knockdown on the downstream molecules associated with apoptosis, migration, and invasion in endometrial carcinoma (EC) cells. HEC-1B cells were transfected with si-PCAT1 or si-NC, respectively. Representative images (left panel) and quantification (right panel) of western blotting analysis of transfected HEC-1B cells; data are presented as mean ± standard deviation. ***p < 0.001, compared with si-NC group. si-PCAT1: Small interfering (siRNA) targeting PCAT1; si-NC: Negative control.
DISCUSSION
In the current study, we first showed that the expression of PCAT1 was upregulated in EC tissues and cell lines compared with paired adjacent tissues and normal endometrial stromal cell line ESC. Based on the Chi-square test, PCAT1 expression was significantly correlated with the FIGO stage, myometrial invasion, and LNM. Moreover, elevated levels of PCAT1 were associated with the poor overall survival of patients with EC and were identified as an independent prognostic factor in multivariate analysis. Similarly, Shi et al. [15] discovered that PCAT1 is a strong predictor of advanced malignant status and poor prognosis in 130 patients diagnosed with esophageal squamous carcinoma. PCAT1 was also considered as a negative prognostic factor in patients with hepatocellular carcinoma [20], gastric cancer [14], and colorectal cancer [10]. In this study, the expression profile and clinical significance of PCAT1 suggest that this lncRNA may play an oncogenic role in EC. Recent studies have revealed that PCAT1 promotes cell growth, migration, and survival in many types of cancers [21,22], which further support our hypothesis that PCAT1 has tumorigenic activities. Furthermore, our loss-of-function analysis showed that the knockdown of PCAT1 decreased the proliferative, migratory, and invasive behavior of EC cells. In addition, the depletion of PCAT1 in EC cells induced early and late apoptosis.
Apoptosis is a critically important biological process that leads to programmed cell death through triggering a series of apoptotic events [23]. Bcl-2 is a crucial component of the mitochondrial apoptotic pathway, which prevents the release of cytochrome c from mitochondria [24]. Bad, a member of the Bcl-2 family, is also a regulator of apoptosis that acts on the mitochondrial membrane and exerts its death-promoting effect through heterodimerization with Bcl2 and Bcl-xl [25]. Our western blot analysis revealed that the knockdown of PCAT1 increased Bad and decreased Bcl-2 protein levels. This indicates that the apoptotic mitochondrial pathway was stimulated by inhibiting the apoptosis inhibitor Bcl2 and inducing the apoptosis promoter Bad, leading to cell death and the inhibition of EC cells proliferation.
In this study, we also found that the lack of PCAT1 expression attenuates the migration and invasion of EC cells. The EMT, a critical developmental process that allows the transdifferentiation of polarized, immotile epithelial cells into motile mesenchymal cells, is frequently activated during cancer migration and invasion [26,27]. The EMT consists of a series of events that include the loss of epithelial features and acquisition of mesenchymal properties through the disruption of E-cadherin and N-cadherin protein homeostasis, and the alteration of cytoskeleton arrangement through increased vimentin synthesis [28]. Cadherins are calcium-dependent cell adhesion molecules that have a principal role in maintaining the epithelial structure in diverse types of tissues [28]. E-cadherin deficiency has been found to be associated with tumor differentiation and deep myometrial invasion in EC [29]. N-cadherin is an adhesion molecule that can direct the EMT and enhance the invasive ability of cells, as demonstrated in various types of cancers, including EC [30]. In our study, higher levels of E-cadherin protein and lower levels of N-cadherin and vimentin proteins were observed in si-PCAT1-transfected HEC-1B cells compared with cells transfected with si-NC. We suggest that the inhibition of EMT process is responsible for decreased migration and invasion of EC cells with PCAT1 knockdown. Besides, our Chi-square test results revealed that the high PCAT1 expression was significantly associated with the FIGO stage, myometrial invasion, and LNM. These results indicate that the dysregulation of EMT markers, especially E-cadherin, is implicated in poor prognosis of EC patients who have high levels of PCAT1.
CONCLUSION
In summary, the present study shows that PCAT1 is an oncogene and a prognostic biomarker in EC. Notably, the mechanistic analysis revealed that the PCAT1 knockdown inhibited malignant progression and induced apoptosis through regulating a series of proteins including Bcl-2, Bad, and EMT markers (E-cadherin, N-cadherin, and vimentin). The results of this study may enhance our understanding of the initiation and development of EC and ultimately facilitate the exploration of lncRNA-directed prognosis and therapy for this disease.
ACKNOWLEDGMENTS
This work was supported by the Project of Lanzhou Science and Technology (Grant No. 2017-4-16).
DECLARATION OF INTERESTS
The authors declare no conflict of interests.
REFERENCES
- 1.Gibson WJ, Hoivik EA, Halle MK, Taylor-Weiner A, Cherniack AD, Berg A, et al. The genomic landscape and evolution of endometrial carcinoma progression and abdominopelvic metastasis. Nat Genet. 2016;48:848–55. doi: 10.1038/ng.3602. http://doi:10.1038/ng.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsujiura M, Mazack V, Sudol M, Kaspar HG, Nash J, Carey DJ, et al. Yes-associated protein (YAP) modulates oncogenic features and radiation sensitivity in endometrial cancer. PLoS One. 2014;9:e100974. doi: 10.1371/journal.pone.0100974. http://doi:10.1371/journal.pone.0100974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.García-Sanz P, Triviño JC, Mota A, Pérez López M, Colás E, Rojo-Sebastián A, et al. Chromatin remodelling and DNA repair genes are frequently mutated in endometrioid endometrial carcinoma. Int J Cancer. 2017;140:1551–63. doi: 10.1002/ijc.30573. http://doi:10.1002/ijc.30573. [DOI] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research. Network K, andoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. http://doi:10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–63. doi: 10.1016/j.ccell.2016.03.010. http://doi:10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martens-Uzunova ES, Böttcher R, Croce CM, Jenster G, Visakorpi T, Calin GA, et al. Long noncoding RNA in prostate, bladder, and kidney cancer. Eur Urol. 2014;65:1140–51. doi: 10.1016/j.eururo.2013.12.003. http://doi:10.1016/j.eururo.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Bartonicek N, Maag JL, Dinger ME. Long noncoding RNAs in cancer:Mechanisms of action and technological advancements. Mol Cancer. 2016;15:43. doi: 10.1186/s12943-016-0530-6. https://doi.org/10.1186/s12943-016-0530-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malek E, Jagannathan S, Driscoll JJ. Correlation of long non-coding RNA expression with metastasis, drug resistance and clinical outcome in cancer. Oncotarget. 2014;5:8027–38. doi: 10.18632/oncotarget.2469. http://doi:10.18632/oncotarget.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29:742–9. doi: 10.1038/nbt.1914. http://doi:10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ge X, Chen Y, Liao X, Liu D, Li F, Ruan H, et al. Overexpression of long noncoding RNA PCAT-1 is a novel biomarker of poor prognosis in patients with colorectal cancer. Med Oncol. 2013;30:588. doi: 10.1007/s12032-013-0588-6. http://doi:10.1007/s12032-013-0588-6. [DOI] [PubMed] [Google Scholar]
- 11.Zhao B, Hou X, Zhan H. Long non-coding RNA PCAT-1 over-expression promotes proliferation and metastasis in non-small cell lung cancer cells. Int J Clin Exp Med. 2015;8:18482–7. [PMC free article] [PubMed] [Google Scholar]
- 12.Wen J, Xu J, Sun Q, Xing C, Yin W. Upregulation of long non coding RNA PCAT-1 contributes to cell proliferation, migration and apoptosis in hepatocellular carcinoma. Mol Med Rep. 2016;13:4481–6. doi: 10.3892/mmr.2016.5075. http://doi:10.3892/mmr.2016.5075. [DOI] [PubMed] [Google Scholar]
- 13.Qiao L, Liu X, Tang Y, Zhao Z, Zhang J, Feng Y, et al. Down regulation of the long non-coding RNA PCAT-1 induced growth arrest and apoptosis of colorectal cancer cells. Life Sci. 2017;188:37–44. doi: 10.1016/j.lfs.2017.08.024. http://doi:10.1016/j.lfs.2017.08.024. [DOI] [PubMed] [Google Scholar]
- 14.Cui WC, Wu YF, Qu HM. Up-regulation of long non-coding RNA PCAT-1 correlates with tumor progression and poor prognosis in gastric cancer. Eur Rev Med Pharmacol Sci. 2017;21:3021–7. [PubMed] [Google Scholar]
- 15.Shi WH, Wu QQ, Li SQ, Yang TX, Liu ZH, Tong YS, et al. Upregulation of the long noncoding RNA PCAT-1 correlates with advanced clinical stage and poor prognosis in esophageal squamous carcinoma. Tumour Biol. 2015;36:2501–7. doi: 10.1007/s13277-014-2863-3. http://doi:10.1007/s13277-014-2863-3. [DOI] [PubMed] [Google Scholar]
- 16.Huang J, Deng G, Liu T, Chen W, Zhou Y. Long noncoding RNA PCAT-1 acts as an oncogene in osteosarcoma by reducing p21 levels. Biochem Biophys Res Commun. 2018;495:2622–9. doi: 10.1016/j.bbrc.2017.12.157. http://doi:10.1016/j.bbrc.2017.12.157. [DOI] [PubMed] [Google Scholar]
- 17.Prensner JR, Chen W, Iyer MK, Cao Q, Ma T, Han S, et al. PCAT-1, a long noncoding RNA, regulates BRCA2 and controls homologous recombination in cancer. Cancer Res. 2014;74:1651–60. doi: 10.1158/0008-5472.CAN-13-3159. http://doi:10.1158/0008-5472.CAN-13-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prensner JR, Chen W, Han S, Iyer MK, Cao Q, Kothari V, et al. The long non-coding RNA PCAT-1 promotes prostate cancer cell proliferation through cMyc. Neoplasia. 2014;16:900–8. doi: 10.1016/j.neo.2014.09.001. http://doi:10.1016/j.neo.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu W, Chang J, Du X, Hou J. Long non-coding RNA PCAT-1 contributes to tumorigenesis by regulating FSCN1 via miR-145-5p in prostate cancer. Biomed Pharmacother. 2017;95:1112–8. doi: 10.1016/j.biopha.2017.09.019. http://doi:10.1016/j.biopha.2017.09.019. [DOI] [PubMed] [Google Scholar]
- 20.Yan TH, Yang H, Jiang JH, Lu SW, Peng CX, Que HX, et al. Prognostic significance of long non-coding RNA PCAT-1 expression in human hepatocellular carcinoma. Int J Clin Exp Pathol. 2015;8:4126–31. [PMC free article] [PubMed] [Google Scholar]
- 21.Liu L, Liu Y, Zhuang C, Xu W, Fu X, Lv Z, et al. Inducing cell growth arrest and apoptosis by silencing long non-coding RNA PCAT-1 in human bladder cancer. Tumour Biol. 2015;36:7685–9. doi: 10.1007/s13277-015-3490-3. http://doi:10.1007/s13277-015-3490-3. [DOI] [PubMed] [Google Scholar]
- 22.Zhen Q, Gao LN, Wang RF, Chu WW, Zhang YX, Zhao XJ, et al. LncRNA PCAT-1 promotes tumour growth and chemoresistance of oesophageal cancer to cisplatin. Cell Biochem Funct. 2018;36:27–33. doi: 10.1002/cbf.3314. http://doi:10.1002/cbf.3314. [DOI] [PubMed] [Google Scholar]
- 23.Hassan M, Watari H, Almaaty AA, Ohba Y, Sakuragi N. Apoptosis and molecular targeting therapy in cancer. Biomed Res Int. 2014;2014:150845. doi: 10.1155/2014/150845. http://doi:10.1155/2014/150845. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Radha G, Raghavan SC. BCL2:A promising cancer therapeutic target. Biochim Biophys Acta Rev Cancer. 2017;1868:309–14. doi: 10.1016/j.bbcan.2017.06.004. http://doi:10.1016/j.bbcan.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 25.Siddiqui WA, Ahad A, Ahsan H. The mystery of BCL2 family:Bcl-2 proteins and apoptosis:An update. Arch Toxicol. 2015;89:289–317. doi: 10.1007/s00204-014-1448-7. http://doi:10.1007/s00204-014-1448-7. [DOI] [PubMed] [Google Scholar]
- 26.Yang J, Weinberg RA. Epithelial-mesenchymal transition:At the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–29. doi: 10.1016/j.devcel.2008.05.009. http://doi:10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 27.Diepenbruck M, Christofori G. Epithelial-mesenchymal transition (EMT) and metastasis:Yes, no, maybe? Curr Opin Cell Biol. 2016;43:7–13. doi: 10.1016/j.ceb.2016.06.002. http://doi:10.1016/j.ceb.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 28.Wang CY, Duran GE, Francisco EB, Juric D, Witten D, Tibshrani R, et al. Abstract 1526:Epithelial mesenchymal transition (EMT) is associated with non-transporter resistance to taxanes in human ovarian cancer cell. Cancer Res. 2010;70(Suppl 8):1526. http://doi:10.1158/1538-7445.AM10-1526. [Google Scholar]
- 29.Onder S, Taskin OC, Sen F, Topuz S, Kucucuk S, Sozen H, et al. High expression of SALL4 and fascin, and loss of E-cadherin expression in undifferentiated/dedifferentiated carcinomas of the endometrium:An immunohistochemical and clinicopathologic study. Medicine (Baltimore) 2017;96:e6248. doi: 10.1097/MD.0000000000006248. http://doi:10.1097/MD.0000000000006248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakajima S, Doi R, Toyoda E, Tsuji S, Wada M, Koizumi M, et al. N-cadherin expression and epithelial-mesenchymal transition in pancreatic carcinoma. Clin Cancer Res. 2004;10:4125–33. doi: 10.1158/1078-0432.CCR-0578-03. http://doi:10.1158/1078-0432.CCR-0578-03. [DOI] [PubMed] [Google Scholar]


