Abstract
The long noncoding RNAs (lncRNAs) have been proven to be involved in the development of alcoholic hepatitis (AH), which has been regarded as a severe form of acute liver injury with a high mortality rate. Through the GEO database, the differentially expressed LINC01093 and intercellular cell adhesion molecule-1 (ICAM-1) were identified in AH. Then, to clarify their specific role and underlying mechanism in AH, we constructed an AH mouse model by using Lieber-Decarli alcoholic feed. It was found that LINC01093 was poorly expressed and ICAM-1 was highly expressed in AH mice. After that, the interactions among LINC01093, ICAM-1, and NF-κB signaling pathway were explored, which verified that LINC01093 could target ICAM-1 and inhibit the NF-κB signaling pathway. Finally, after the hepatocytes were isolated from AH mice, the expression of LINC01093 was up- or downregulated or that of ICAM-1 was silenced to evaluate their effect on cell viability and apoptosis. The corresponding results demonstrated that after overexpression of LINC01093 or silencing of ICAM-1, cell viability was increased and cell apoptosis was reduced in the hepatocytes of AH mice. Moreover, the silencing of LINC01093 was observed to inhibit the viability and promote the apoptosis of hepatocytes of AH mice. Altogether, these results provide evidence that overexpression of LINC01093 could effectively suppress hepatocyte apoptosis and promote proliferation by inhibiting the ICAM-1-mediated NF-κB signaling pathway, thus playing a functional role in AH and hepatic fibrosis.
Keywords: long non-coding RNA LINC01093, ICAM-1, NF-κB signaling pathway, alcoholic hepatitis, hepatic fibrosis, hepatocyte proliferation, apoptosis
Introduction
Alcoholic hepatitis (AH) is considered the severest form of alcoholic liver disease, which is characterized by hepatocellular damage, steatosis, and peri-cellular fibrosis.1 AH generally presents in patients with excessive alcohol use and is known for its rapid onset of jaundice and complications as a result of hepatic insufficiency.2 Because of the limitations in the currently used therapeutic methods, the mortality rate of AH patients remains extremely high.3 Some treatment options, such as corticosteroids and pentoxifylline, have low efficacy and result in the development of side effects; in addition, the short-term mortality of AH might reach 20%–30% because of probable poor characterization in AH patients.4 Previously conducted studies have provided evidence that long noncoding RNAs (lncRNAs) participate in several pathways related to the pathogenesis of alcoholic liver disease.5 Furthermore, the lncRNAs related to liver disease have been regarded as essential biological indicators that are involved in the diagnosis, prognosis, and treatment of disease,6 which we used as a basis when conducting our study on the effect of LINC01093 in AH therapy.
The lncRNAs form a class of transcripts longer than 200 nucleotides that have been found to be dysregulated in several diseases and cancer and are involved in the modulation of several cellular processes, including cell proliferation, apoptosis, migration, and invasion.7, 8, 9 A previous study revealed that lncRNAs were involved in the development of liver diseases, particularly liver cancer.10 Intercellular cell adhesion molecule-1 (ICAM-1), also known as CD54, is a membrane-bound glycoprotein that is mainly expressed in all leukocyte subsets, platelets, fibroblasts, and others.11 ICAM-1 has been identified as a crucial marker for the diagnosis of nonalcoholic steatohepatitis (NASH),12 and previous studies have demonstrated an upregulation in ICAM-1 expression in AH.13, 14 However, little research has been conducted on the regulatory mechanism of ICAM-1 in alcoholic liver disease, and few studies have detailed the high expression of ICAM-1 in alcoholic liver disease. The high expression of ICAM-1 and its solubility have been observed in several inflammatory conditions, chronic diseases, and other malignancies.15 In the present study, the expression profile of AH (GEO: GSE28619) was searched, and based on GEO dataset analysis, it was found that the ICAM-1 gene was highly expressed and LINC01093 was poorly expressed in AH. Furthermore, the correlation analysis for LINC01093 and ICAM-1 (GEO: GSE28619) revealed a negative correlation between LINC01093 and ICAM-1. Therefore, we hypothesized that the interaction between LINC01093 and ICAM-1 may be involved in the development of AH. Moreover, a previously conducted study identified lncRNA as an essential component of the complex modulatory structure that regulates the NF-κB signaling pathway.16 NF-κB was considered a critical transcription factor associated with the upregulation of cytokine or chemokine gene upregulation in alcoholic liver disease.17 The previous study provided the evidence that overactivated nuclear factor κB (NF-κB) could promote liver inflammation and that NF-κB was linked to the induction of B cell lymphoma-2 (Bcl-2), which is an anti-apoptosis gene.18 Our study aimed to explore the effect of LINC01093 on AH by regulating the NF-κB signaling pathway with the involvement of ICAM-1. In addition, this study aimed to explore the cause of the high expression of ICAM-1 in alcoholic liver disease and the key molecules that induced ICAM-1 overexpression. This study can potentially reveal the molecular mechanism of occurrence of alcoholic liver disease and provide a theoretical basis for the prevention and treatment of alcoholic liver disease.
Results
AH Mice Present with Severe Lesions and Increased Apoptosis and Inflammation in Hepatic Tissues
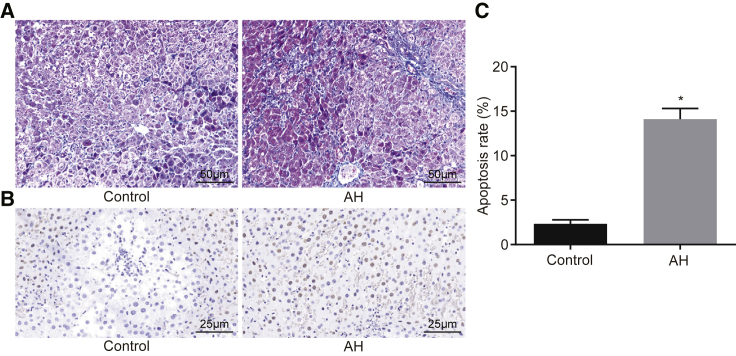
Following the successful establishment of the AH mouse model, Masson staining was employed to observe the pathological changes of hepatic tissues in mice. As shown in Figure 1A, the results revealed that the hepatic lobules of the mice in the control group were clear, and the hepatocytes were arranged in an orderly and radiant manner with a small amount of collagenous fibers in the central vein and the portal area. In contrast with the control group, the mice in the AH group were observed to have severe hepatic fibrosis, with some mice presenting with a disorderly hepatic lobule structure, and the collagen fibers were increased and extended to the perisinusoidal space with an increase of perisinusoidal fibrous tissue.
Figure 1.
Mice with the AH Group Show Serious Hepatic Fibrosis and Increased Apoptosis and Inflammation in Hepatic Tissues
(A) Hepatic tissues of normal mice and AH mice examined by Masson staining (200×). (B) Hepatic tissues of normal mice and AH mice examined by TUNEL staining (400×). (C) Apoptosis rate of hepatic tissues of normal mice and AH mice. *p < 0.05 versus the control group. AH, alcoholic hepatitis.
Cell apoptosis in cells was detected using terminal deoxynucleotidyl transferase (TdT)-mediated dUTP nick-end labeling (TUNEL) staining. As shown in Figures 1B and 1C, the results showed that a few apoptotic cells were occasionally seen in the control group. Compared with the control group, an increase in the number of the apoptotic hepatocytes was found in the AH group and scattered in lobules. The number of apoptotic hepatocytes increased with the aggravation of hepatic inflammation and fibrosis. Compared with the control group, there was a significant increase in the apoptosis index (AI) of the AH group (p < 0.05). ELISA was used to detect the contents of inflammatory factors tumor necrosis factor (TNF) alpha, hyaluronic acid (HA), laminin (LN), and procollagen III (PC III) in the serum of mice in the control and AH groups. The results, as shown in Table 1, revealed that in comparison with the control group, there was significant enhancement in the contents of TNF-α, HA, LN, and PC III in the AH group (p < 0.05). These findings suggested that there was high expression in inflammatory factors in the serum of mice with AH and that the mice with AH exhibited severe lesions in the hepatic tissues, higher AI, and aggravated inflammation.
Table 1.
Contents of TNF-α, HA, LN, and PC III in the Hepatic Tissue of Mice in Control and AH Groups
| Group | TNF-α (ng/L) | HA (μg/L) | LN (μg/L) | PC III (μg/L) |
|---|---|---|---|---|
| Control | 118.43 ± 9.24 | 132.65 ± 10.81 | 50.97 ± 4.66 | 15.58 ± 1.73 |
| AH | 142.36 ± 11.57* | 280.19 ± 13.03* | 89.28 ± 5.32* | 26.35 ± 1.41* |
*p < 0.05 versus the control groups. TNF-α, tumor necrosis factor alpha; HA, hyaluronic acid; LN, laminin; PC III, procollagen III.
LINC01093 Is Downregulated while ICAM-1 Is Upregulated in Hepatic Tissue of AH Mice
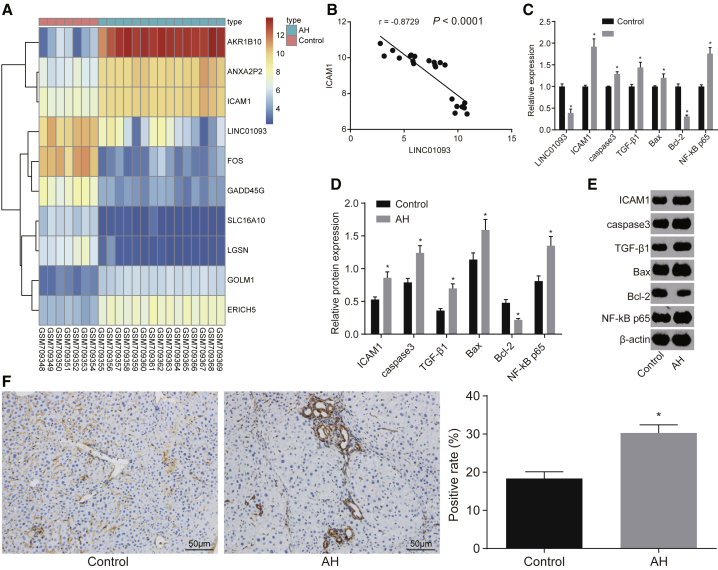
According to microarray data, GEO: GSE28619 consisted of 15 samples of AH patients and 7 samples from healthy individuals.1 R software was used to conduct differential analysis for the gene expression in tissues of these 22 samples, and the results found 108 downregulated genes and 108 upregulated genes, after which the heatmap for the top 10 differentially expressed genes (DEGs) was plotted (Figure 2A). The results found that LINC01093 was poorly expressed in AH, while ICAM-1 was highly expressed in AH. The correlation analysis for LINC01093 and ICAM-1 in GEO: GSE28619 was conducted, and the results in Figure 2B showed that there was a negative correlation between LINC01093 and ICAM-1. Compared with the control group, the expression of LINC01093 in the AH group decreased significantly (p < 0.05). The results in Figures 2C–2E showed that mRNA and protein levels of ICAM-1, caspase-3, transforming growth factor β (TGF-β) 1, Bcl-2-associated X protein (Bax), and p65 subunit of nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB p65) were upregulated significantly in the AH group, while those of Bcl-2 were reduced significantly in contrast to the control group (both p < 0.05). In addition, immunohistochemistry was adopted to detect the positive expression rate of ICAM-1 in hepatic tissue in each group. The results in Figure 2F showed that ICAM-1 was expressed in a few sinusoidal cells and fibroblasts in the control group. Compared with the control group, the immunoreaction of ICAM-1 in the AH group was strongly positive, the positive particles were mainly expressed in the hepatic cell membrane, and some stained hepatic cytoplasm appeared to be positive. The particle density of the ICAM-1-positive reaction of the hepatic tissue in the AH group increased significantly (p < 0.05). These finding indicated that there was a higher positive expression rate of ICAM-1 in the hepatic tissue of mice in the AH group.
Figure 2.
LINC01093 Is Decreased but ICAM-1 Is Increased in Hepatic Tissue of AH Mice
(A) Heatmap of the top 10 DEGs in GEO: GSE28619 (abscissa indicates sample number, and ordinate indicates gene; the upper dendrogram refers to clustering of sample types; the chromatic histogram at the right side refers to gene expression, with each square presenting one gene expression in one sample; and the dendrogram at left side indicates clustering of gene expression). (B) Correlation between LINC01093 and ICAM-1 in GEO: GSE28619. (C) Relative expression of LINC01093 and the mRNA levels of ICAM-1, caspase-3, TGF-β1, Bax, NF-κB p65, and Bcl-2 in hepatic tissue of the normal and AH mice examined by qRT-PCR. (D and E) Relative protein levels of ICAM-1, caspase-3, TGF-β1, Bax, NF-κB p65, and Bcl-2 in hepatic tissue of the normal and AH mice detected by western blot analysis. (F) Expression of ICAM-1 in hepatic tissue of the normal and AH mice examined by immunohistochemistry. *p < 0.05 versus the control group. ICAM-1, intercellular cell adhesion molecule-1; TGF-β1, transforming growth factor β1; NF-κB p65, p65 subunit of nuclear factor κB light-chain enhancer of activated B cells; Bcl-2, B cell lymphoma-2; Bax, Bcl-2-associated X protein; AH, alcoholic hepatitis; DEGs, differentially expressed genes.
LINC01093 Regulates ICAM-1 by Mediating FOXP3
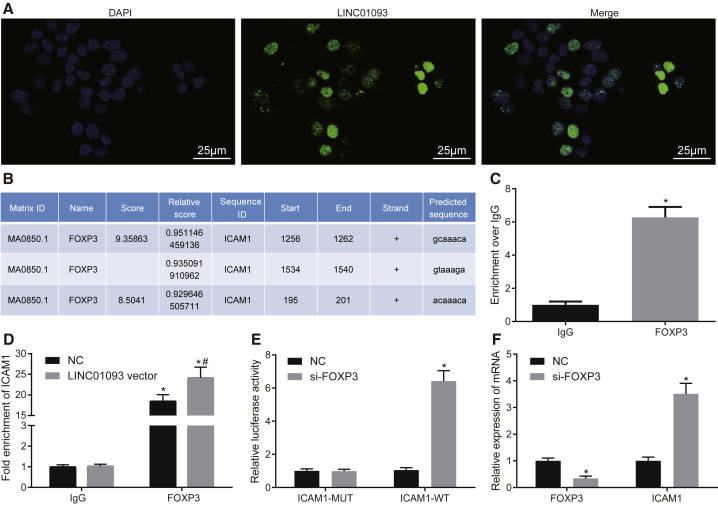
The results from fluorescence in situ hybridization (FISH) assay revealed that LINC01093 was localized in the nucleus of hepatocytes of mice (Figure 3A), suggesting that LINC01093 might interact with transcription factors.19 Following the analysis of the online website PROMO (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3), the results in Figure 3B showed that LINC01093 was bound to transcription factor forkhead box P3 (FOXP3), and the results from the JASPAR website (http://jaspar.genereg.net/) revealed that FOXP3 was bound to ICAM-1 promoter region. RNA binding protein immunoprecipitation (RIP) assay was also conducted, and the results showed that FOXP3 was bound to LINC01093 (Figure 3C). Chromatin immunoprecipitation (ChIP) assay further revealed that FOXP3 was bound to ICAM-1, and the overexpression of LINC01093 promoted the binding of FOXP3 and ICAM-1 (Figure 3D). The results from the dual-luciferase reporter gene assay revealed that the silencing of FOXP3 inhibited the luciferase activity of ICAM-1 wild type (WT) but had no effect on the luciferase activity of ICAM-1 mutant type (MUT) (Figure 3E). In addition, qRT-PCR was applied to examine the effect of the silencing of FOXP3 on expression of FOXP3 and ICAM-1. Compared with the negative control (NC) group, the siRNA against FOXP3 (si-FOXP3) group showed significantly decreased expression of FOXP3 but significantly increased expression of ICAM-1 (Figure 3F). The aforementioned findings suggested that LINC01093 regulated ICAM-1 expression via FOXP3.
Figure 3.
LINC01093 Targets ICAM-1 and Binds to FOXP3
(A) Location of LINC01093 in hepatic tissue of mice examined by FISH assay (400×; scale bar, 25 μm). (B) Binding of the FOXP3 and ICAM-1 promoter region predicted by the JASPAR website (http://jaspar.genereg.net/). (C) Binding of FOXP3 and LINC01093 determined by RIP assay. *p < 0.05 versus the IgG. (D) Binding of FOXP3 and ICAM-1 detected by ChIP assay. *p < 0.05 versus IgG; #p < 0.05 versus the NC group. (E) Luciferase activity of ICAM-1-WT and ICAM-1-Mut after transfection; *p < 0.05 versus the NC group. (F) Expression of FOXP3 and ICAM-1 after FOXP3 silencing examined by qRT-PCR. *p < 0.05 versus the NC group. ICAM-1, intercellular cell adhesion molecule-1; FOXP3, forkhead box P3; ChIP, chromatin immunoprecipitation; IgG, immunoglobulin G; NC, negative control.
LINC01093 Increases Bcl-2 Expression while Decreasing the Expression of ICAM-1, Caspase-3, TGF-β1, Bax, and NF-κB p65 in Hepatic Tissues of Mice with AH
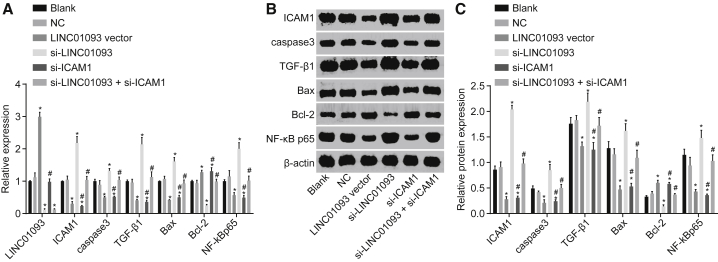
qRT-PCR and western blot analysis were conducted to examine the expression of LINC01093, Bcl-2, ICAM-1, caspase-3, TGF-β1, Bax, and NF-κB p65 in cells following transfection. The results (Figure 4) revealed that there was no significant difference observed between blank and NC groups (p > 0.05). Compared with the blank and NC groups, the expression of LINC01093 and Bcl-2 in the LINC01093 vector group was enhanced significantly (p < 0.05) with a significant decline in mRNA levels of ICAM-1, caspase-3, TGF-β1, Bax, and NF-κB p65 (p < 0.05). In contrast, the expression of LINC01093 and Bcl-2 in the si-LINC01093 group was reduced significantly (p < 0.05), while that of ICAM-1, caspase-3, TGF-β1, Bax, and NF-κB p65 was upregulated (p < 0.05). There was evident downregulation of mRNA levels of ICAM-1, caspase-3, TGF-β1, Bax, and NF-κB p65 in the si-ICAM-1 group (p < 0.05) with an increase of Bcl-2 mRNA expression, while there was no significant difference observed in LINC01093 expression in the si-ICAM-1 group (p > 0.05). The expression of LINC01093 in the si-LINC01093 + si-ICAM-1 group was decreased significantly (p < 0.05), while there was no remarkable difference in the expression of ICAM-1, caspase-3, TGF-β1, Bax, Bcl-2, and NF-κB p65 (p > 0.05). There was no significant difference in the protein levels of ICAM-1, caspase-3, TGF-β1, Bax, Bcl-2, and NF-κB p65 between the NC and the blank groups (p > 0.05). In comparison with the blank and NC groups, the protein level of Bcl-2 in the LINC01093 vector and si-ICAM-1 groups was increased significantly (p < 0.05), while the protein levels of ICAM-1, caspase-3, TGF-β1, Bax, and NF-κB p65 were decreased significantly (p < 0.05). The protein level of Bcl-2 in the si-LINC01093 group was downregulated significantly (p < 0.05) with an increased protein levels of ICAM-1, caspase-3, TGF-β1, Bax, and NF-κB p65 (p < 0.05). There was no significant difference in the expression of Bcl-2, ICAM-1, caspase-3, TGF-β1, Bax, and NF-κB p65 in the si-LINC01093 + si-ICAM-1 group (p > 0.05). These results indicated that overexpressed LINC01093 could result in the downregulation of mRNA and protein levels of ICAM-1, caspase-3, TGF-β1, Bax, and NF-κB p65 with an increase in Bcl-2.
Figure 4.
Upregulated LINC01093 Suppresses ICAM-1, Caspase-3, TGF-β1, Bax, and NF-κB p65 Expression and Increases the Expression of Bcl-2
(A) Relative expression of LINC01093 and the mRNA levels of Bcl-2, ICAM-1, caspase-3, TGF-β1, Bax, and NF-κB p65 in cells in each group examined by qRT-PCR. (B and C) Protein levels of Bcl-2, ICAM-1, caspase-3, TGF-β1, Bax, and NF-κB p65 in cells in each group examined by western blot analysis. *p < 0.05 versus the blank and NC groups; #p < 0.05 versus the si-LINC01093 group. NC, negative control; ICAM-1, intercellular cell adhesion molecule-1; TGF-β1, transforming growth factor β1; NF-κB p65, p65 subunit of nuclear factor κB light-chain enhancer of activated B cells; Bcl-2, B cell lymphoma-2; Bax, Bcl-2-associated X protein.
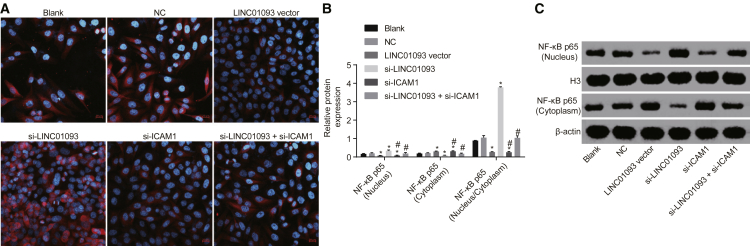
NF-κB p65 Nuclear Translocation Is Inhibited by Overexpression of LINC01093
Immunofluorescence assay was used to detect NF-κB p65 nuclear translocation in each group. As shown in Figure 5, the findings showed that compared with the blank and NC groups, there was a slight NF-κB p65 nuclear translocation in the LINC01093 vector and si-ICAM-1 groups, and there was a decrease in the content of NF-κB p65 in the cytoplasm (p < 0.05) even as it increased in nucleus (p < 0.05). There was no significant difference of nuclear translocation in the si-LINC01093 + si-ICAM-1 group (p > 0.05). The nuclear translocation in the si-LINC01093 group was found to be the most evident (p < 0.05), and the content of NF-κB p65 was increased in cytoplasm (p < 0.05) even as it was decreased in nucleus (p < 0.05). These findings revealed that the overexpression of LINC01093 led to reduction in the nuclear translocation of NF-κB p65.
Figure 5.
NF-κB p65 Nuclear Translocation Is Suppressed by Overexpressed LINC01093
(A) NF-κB p65 nuclear translocation in each group (200×). (B and C) Relative protein levels of NF-κB p65 in nucleus and cytoplasm in each group. *p < 0.05 versus the blank and NC groups; #p < 0.05 versus the si-LINC01093 group. NC, negative control; NF-κB p65, nuclear factor κB light-chain enhancer of activated p65.
Overexpression of LINC01093 Promotes Cell Viability while Inhibiting Cell Apoptosis
The 3-(4, 5-Dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay was used in the evaluation of the cell viability in each group. As shown in Figure 6A, there was no significant difference in cell viability between the blank and the NC groups at each time point (p > 0.05). Compared with the blank and NC groups, there was a significant increase in cell viability in the si-ICAM-1 and LINC01093 vector groups (p < 0.05), while the cell viability in the si-LINC01093 group was inhibited (p < 0.05). There were no evident alterations in the si-LINC01093 + si-ICAM-1 group (p > 0.05). These findings highly indicated that the overexpression of LINC01093 could promote cell viability.
Figure 6.
Cell Viability Is Improved but Cell Apoptosis Is Repressed after Overexpression of LINC01093
(A) Cell viability in each group. (B) Apoptotic cells in each group. (C) Cell apoptosis rate in each group. (D) Cell cycle of apoptotic cells in each group. (E) Cell number in G0/G1, S, and G2 phases. *p < 0.05 versus the blank and NC groups; #p < 0.05 versus the si-LINC01093 group. NC, negative control.
Furthermore, cell cycle and cell apoptosis were assessed in each group using flow cytometry. The results (Table 2; Figures 6B–6E) showed that compared with the blank and NC groups, the si-ICAM-1 group and the LINC01093 vector group showed that the cell proportion was decreased in the G0/G1 phase while it was increased in the S phase, with a significantly decreased cell apoptosis rate (p < 0.05), but the si-LINC01093 group presented with more cells arrested in the G0/G1 phase, fewer cells in the S phase, and a relatively increased apoptosis rate (p < 0.05). There was no statistical significance in the si-LINC01093 + si-ICAM-1 group (p > 0.05). These results identified that cell apoptosis could be inhibited through the overexpression of LINC01093.
Table 2.
Cell Cycle in Each Group after Transfection
| Group | G0/G1 (%) | S (%) | G2 (%) |
|---|---|---|---|
| Blank | 58.33 ± 3.15 | 31.60 ± 2.10 | 10.07 ± 1.05 |
| NC | 57.26 ± 2.89 | 32.45 ± 1.69 | 10.29 ± 1.20 |
| LINC01093 vector | 31.62 ± 3.07* | 57.63 ± 2.46* | 10.75 ± 1.11* |
| si-LINC01093 | 74.88 ± 2.75* | 13.42 ± 1.73* | 11.70 ± 1.02* |
| si-ICAM-1 | 34.67 ± 3.94*# | 54.60 ± 2.71*# | 10.73 ± 1.96*# |
| si-LINC01093 + si-ICAM-1 | 56.30 ± 1.70# | 31.14 ± 1.31# | 12.56 ± 1.14# |
*p < 0.05 versus the blank and NC groups; #p < 0.05 versus the si-LINC01093 group. NC, negative control; ICAM-1, intercellular cell adhesion molecule-1.
Discussion
AH, like several other types of liver injury, develops as a result of chronic alcohol abuse and presents with jaundice and liver failure in patients who had severe alcohol abuse for decades.20 As the standard-of-care therapy, the efficacy of prednisolone for patients with AH was limited with many side effects, and no other therapies could retain survival benefit consistently.21 Therefore, new targeted therapies are required for patients with AH, because the current therapies are not completely effective. It has been reported that lncRNAs contribute to the development of hepatic disease in particular.9 Our study was conducted with the objective of verifying the effects of LINC01093 and the NF-κB signaling pathway on hepatocyte proliferation and apoptosis in AH and hepatic fibrosis treatment. After a thorough investigation, we found that LINC01093 could promote hepatocyte proliferation, thus attenuating hepatic fibrosis in AH through the inhibition of NF-κB signaling pathway by negatively regulating ICAM-1.
The results from our study revealed that the AH mice presented with reduced LINC01093 and apoptosis factor Bcl-2 while the expression of ICAM-1, TNF-α, and NF-κB p65 increased. A previous study concluded that the overexpression of LINC01093 predicted good prognosis for hepatocellular carcinoma.22 Similarly, overexpressed lncRNA growth arrest-specific transcript 5 could attenuate liver fibrosis by inhibiting the activation and proliferation of hepatic stellate cell.23 The expression of NF-κB target genes like ICAM-1 has been previously assessed, and the findings indicated that the inflammatory response and inhibition of NF-κB via blockade of TNF-α could lead to apoptosis in transformed hepatocytes and suppressed hepatocarcinogenesis.24 A previous report showed that the expression of ICAM-1 could be enhanced by NF-κB and that TNF-α significantly increased the translocation of NF-κB p65 from cytoplasm to nucleus.25 With the evident inactivation of NF-κB, the expression of TGF-β1 and several fibrotic factors, including α-smooth muscle actin (SMA) and type I collagen (COL1), was also reduced, thus indicating that the suppression of NF-κB might contribute to liver fibrosis prevention and treatment.26 Moreover, hepatic fibrosis was featured by excessive accumulation of the extracellular matrix and accompanied by the release of excessive cytokine, especially the entry of monocytes into damaged liver like ICAM-1.27 Several receptors of the TNF superfamily have been found to be involved in AH with higher expression and the pro-fibrogenic cytokine TGF-β1, which was also enhanced significantly in AH.1
Hepatocyte apoptosis and cell death have been identified as critical triggering factors that participated in liver fibrogenesis, and several lncRNAs have been identified to regulate apoptosis.28 The findings from our study demonstrated that LINC01093 could target ICAM-1. We also found that highly expressed LINC01093 could inhibit the NF-κB signaling pathway, thereby inhibiting cell apoptosis. A previous study identified that lncRNAs exerted regulatory effects on many biological processes of the liver, such as the alcoholic liver disease.29 NF-κB transcription factor, a cell survival regulator, participated in cancer development by promoting the major inflammatory pathway activated during liver injury.30 The previous report described that caspase-3 was considered a central executor involved in the mitochondrial-dependent apoptosis pathway and Bax, which also participated in the inactivation of the intrinsic apoptosis pathway.31 A previous report has revealed that the low expression of lncRNA HOX transcript antisense RNA could upregulate Bax and caspase-3 expression and decreased Bcl-2.32 These findings are in line with the results in our study. Moreover, TGF-β was considered a critical activator of fibrosis, and lncRNAs regulated by TGF-β were enriched in superenhancers, suggesting that lncRNAs may exert critical effects on hepatic stellate cell function and fibrosis.33 Thus, we can conclude that the high expression of LINC01093 could suppress hepatocyte apoptosis in AH by negatively regulating the ICAM-1-mediated NF-κB signaling pathway.
In addition, ICAM-1 serves as a crucial marker for the diagnosis of nonalcoholic steatohepatitis (NASH),12 and previous studies have provided evidence that there was high expression in ICAM-1 in AH.13, 14 Upregulated ICAM-1 induced the activation of the NF-κB signaling pathway and promoted hepatic fibrosis in AH. Our study also found that LINC01093 resulted in the upregulation of ICAM-1, after which we detected the levels of ICAM-1 and NF-κB signaling pathway-related factors after ectopic expression or depletion of LINC01093.
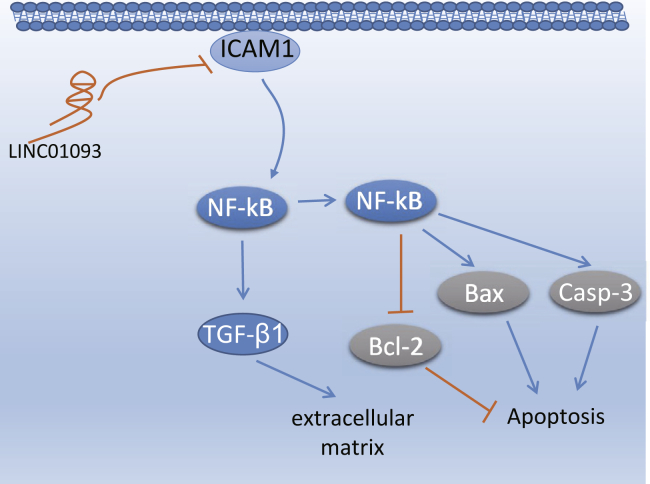
In conclusion, our findings indicated that the overexpression of LINC01093 could inhibit hepatocyte apoptosis in AH and alleviate hepatic fibrosis through inhibition of the NF-κB signaling pathway by targeting ICAM-1, as evidenced by decreased mRNA and protein levels of ICAM-1 and NF-κB p65 (Figure 7). Therefore, the findings from our study provided a new theoretical basis for the treatment of AH.
Figure 7.
Mechanism of the NF-κB Signaling Pathway Mediated by LINC01093
Overexpression of LINC01093 can target ICAM-1 negatively and then inhibit apoptosis and enhance proliferation by suppressing the NF-κB signaling pathway, thereby attenuating AH. NF-κB, nuclear factor κB; ICAM-1, intercellular cell adhesion molecule-1; AH, alcoholic hepatitis.
Materials and Methods
Ethical Statements
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the NIH. The protocol was approved by the Institutional Animal Care and Use Committee of the First Hospital of Jilin University.
Bioinformatics
The GEO database (https://www.ncbi.nlm.nih.gov/geo/) was used to download the AH-related microarray data (GEO: GSE28619) (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE28619) and annotated probe files (Affymetrix Human Genome U133 Plus 2.0 Array [HG-U133_Plus_2]). The Affy package of R software was used for background correction and normalization of microarray data.34 Next, the linear model-empirical Bayes statistics of the Limma package, in combination with t test, was used to screen out differentially expressed microRNA (miRNA) and lncRNA via non-specific filtration,35 with |logFC| > 2 and p < 0.05 as the screening criteria.
Study Subjects
A total of 84 C57BL/6 male mice weighing 20–25 g were purchased from the Department of Laboratory Animal Science, Peking University Health Science Center (Beijing, China) and randomly grouped into control group (n = 42; fed with Lieber-Decarli non-alcoholic feed) and AH group (n = 42; fed with Lieber-Decarli alcoholic feed of the same heat to establish the AH mice models according to previous studies).36, 37, 38 These mice were housed at a specific pathogen-free (SPF) level of animal laboratory at 24°C ± 2°C with a humidity of 40% to ∼70% and artificial day and night circulation lighting (from 8:00 to 20:00) for one month. The criteria for evaluating the mouse models of AH39 were as follows: following the successful establishment of the model, the mice appeared to have unkempt and lusterless hair, diminished activity, slow reaction, and slowly increasing weight. Thirty-six AH mice models were successfully established in the AH group, with a 85.7% model rate, and were then anaesthetized with ether after 24 h. Following the removal of the eyeball, 0.5 mL of blood sample was collected, added into a 2-mL test tube, and placed in the refrigerator at 4°C for 15 min without anticoagulation. After centrifugation at 8,000 rpm for 10 min, the blood serum was collected and stored in liquid nitrogen. The mice livers were removed and fixed with 4% polyoxymethylene. Afterward, the mice were euthanized through ether overdose.
Masson Staining
Tissue samples obtained from mice in each group were fixed in 4% formaldehyde for 16–18 h, dehydrated, embedded with paraffin, and cut into 5-μm sections. Subsequently, the sections were stained with Weigert hematoxylin staining solution (G1142, Beijing Solarbio Science & Technology, Beijing, China) for 5 min. Once the excess floating color was removed, the sections were rewashed with distilled water for 5 min until the color change to blue was obtained, after which they were added to Masson complex staining solution until the sections were covered and stained for 5–10 min. After the removal of the excess Masson complex staining solution, the sections were immersed into 2% glacial acetic acid solution for 30 s, differentiated with 1% phosphomolybdate water solution for 3 min, and stained with aniline blue or light green water solution for 5 min. Afterward, the sections were immersed into 0.2% glacial acetic acid solution again for 15 s, dehydrated with 95% alcohol and anhydrous alcohol, cleared with dimethylbenzene, and sealed with neutral balsam. Lastly, the pathological changes of hepatic tissues in mice were observed under the light microscope (DSX100, Olympus, Tokyo, Japan).
TUNEL Staining
The TUNEL Kit (C1086, Beyotime Institute of Biotechnology, Shanghai, China) was used to stain the hepatic tissues of mice in the control and AH groups to observe the apoptosis of hepatocytes. The paraffin-embedded sections were dewaxed with xylene, rehydrated with gradient alcohol, and immersed into 3% H2O2 solution at room temperature for 10 min. Next, the sections were washed with PBS for 5 min, added to 50 μL of protease K solution (20 μg/mL, P6556, Sigma-Aldrich Chemical, St. Louis, MO, USA), and hydrolyzed for 20 min at room temperature for the removal of the tissue proteins, after which the sections were added to citrate and repaired by antigen for 30 min). Next, the sections were added to 50 μL of TdT enzyme reaction solution, placed in the wet box, and reacted at 37°C for 1 h, avoiding exposure to light, with the TdT enzyme-free reaction solution used as the NC. After that, the sections were added to 50 μL of peroxidase-labeled anti-digoxigenin and reacted in a wet box at 37°C for 30 min, avoiding exposure to light. The sections were then developed by DAPI solution (C1002, Beyotime) at room temperature for 10 min, sealed with neutral balsam, and observed under a fluorescence microscope (BX53, Olympus). The cells with yellow-stained cytoplasm and nucleus pyknosis were considered positive apoptotic cells. Finally, ten visual fields were randomly selected, and the apoptosis index (AI) was presented as the ratio of positive cells to total cells.40
Immunohistochemistry
The tissues were fixed with 10% formaldehyde, embedded in paraffin, and cut into 4-μm serial sections. Next, the tissue sections were placed in an incubator at 60°C for 1 h. After being dried, the sections were dewaxed with three-cylinder dimethylbenzene (10 min per cylinder) and dehydrated in gradient alcohol with concentrations of 95%, 80%, and 75%, each for 1 min, after which incubation was carried out with 3% H2O2 solution (84885, Sigma-Aldrich Chemical) at 37°C for 30 min. After that, the sections were placed into the citric acid buffer, boiled at 95°C for 20 min, and cooled to room temperature. The sections were blocked with goat serum at room temperature for 10 min, incubated with the primary antibody rabbit anti-human antibody to ICAM-1 (1:200, ab53013, Abcam, Cambridge, MA, USA) at 4°C overnight. Next, the sections were incubated with secondary antibody horseradish peroxidase (HRP)-labeled goat anti-rabbit antibody (DF7852, Shanghai Yaoyun Biotechnology, Shanghai, China) at room temperature for 30 min. The sections were then developed with diaminobenzidine (ab64238, Abcam), counter-stained with hematoxylin, and mounted, with PBS used as a NC. Next, five high-power fields were randomly selected for each section, and 100 mouse hepatocytes were counted for each visual field. The positive cells were regarded as those that were brown. Five high-power fields were randomly selected from each section for the analysis of the positive rate.
ELISA
The serum stored in liquid nitrogen was selected to prepare standard samples in accordance with the instructions of the ELISA kits (PH029RAT, Phygene, Shanghai, China), after which the TNF-α, serum leptin, HA, LN, and PC III were determined. After application of sample, the cells were incubated at 37°C for 60 min. Then, the cells in each well were added to 100 μL of enzyme-labeled antibody, with the plate washed five times with PBS/Tween (PBST) (pH 7.4) solution, followed by coloring with chromogenic substrate, avoiding exposure to light for 10 min. The optical density (OD) value of each well was measured at a 450 nm wavelength. The concentrations of TNF-α, serum leptin, HA, LN, and PC III in the cell supernatant were calculated according to the regression equation of the standard curve. The experiment was conducted in triplicate to obtain the average value.
Cell Treatment
The hepatic tissues that had been extracted from 36 mice with AH were cut into 1 mm3 tissue blocks and detached in a water bath at 37°C for 80 min with 3 mL of enzyme (containing 0.2% type II collagenase and 1% polyvinylpyrrolidone [PVP]), followed by redetachment with 2 mL of enzyme in a water bath at 37°C for 15 min. Next, the tissues were added to 3 mL of M199 culture medium (31100035, Gibco, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS) (16000-044, Gibco) and centrifuged at 1,000 rpm for 10 min with the supernatant discarded. The cells were then resuspended with 3 mL of M199 culture medium containing 10% FBS and cultured in an incubator with 5% CO2 at 37°C. The cell growth was observed under a microscope, and the culture medium was replaced every 2 days. Once cell confluence reached 80%, the cells were rinsed with 0.25% trypsin solution and observed under a microscope. Afterward, M199 culture medium containing 10% FBS was used to terminate detachment once most cells were round. A pipette was used to triturate the cells, and the cell suspension was obtained and reincubated in an incubator with 5% CO2 at 37°C. The culture medium was replaced every 3 days.
The small interfering RNA (siRNA) sequences of LINC01093, FOXP3, and ICAM-1 genes were designed, synthesized, and then directly cloned into pSES_HUS shuttle plasmid after annealing to obtain pSES_HUS_siLINC01093, si-FOXP3, and si-ICAM-1, which were linearized by P Rouge I and homologously recombined with pAdEasy_1 skeleton plasmid in BJ5183 bacteria to construct the si-LINC01093, si-FOXP3, and si-ICAM-1 plasmids. The cells were then assigned to 1 of 6 groups: blank group (cells without transfection), NC group (cells transfected with empty plasmids), LINC01093 vector group (cells transfected with LINC01093 vector plasmids), si-LINC01093 vector (cells transfected with si-LINC01093 plasmids), si-ICAM-1 group (cells transfected with si-ICAM-1 plasmids), and si-LINC01093 + si-ICAM-1 group (cells co-transfected with si-LINC01093 and si-ICAM-1 plasmids). The hepatocytes were seeded into 6-well plates with a density of 3 × 105 cells in each well. Upon reaching 90% confluence, the cells were transfected by using Lipofectamine 2000 kits (11809-019 and 11668-500, Invitrogen, Carlsbad, CA, USA). The serum-free Opti-MEM medium (250 μL) was used to dilute 4 μg of target plasmids and 10 μL of Lipofectamine 2000, respectively; it was then mixed well and placed at room temperature for 5 min. After 20 min, the mixture was added to the culture wells and incubated with 5% CO2 at 37°C. The preceding plasmids were all synthesized by Shanghai Sangon Biotechnology (Shanghai, China). After transfection, the cells were starved for 24 h for follow-up experiments. This experiment was conducted three times.
RIP Assay
The RIP kit (Millipore, Bedford, MA, USA) was applied to detect the binding of LINC01093 and FOXP3. The cells were washed by pre-cooled PBS, after which the supernatant was discarded. Next, the cells were lysed by RIP assay lysis buffer (P0013B, Beyotime) of equal volume in an ice bath for 5 min, and centrifuged at 12,000 × g for 10 min at 4°C, with supernatant obtained. One part of the cell extract was used as input, and the other was co-precipitated with antibodies FOXP3 (ab450, 2 μg/mL of cells, Abcam) and immunoglobulin G (IgG) (ab172730, 1:100, Abcam), which was used as NC. A total of 50 μL of magnetic beads from each co-precipitation reaction system were washed, resuspended in 100 μL of RIP wash buffer, and incubated with 5 μg antibodies for binding. The magnetic bead-antibody complex was then washed, resuspended in 900 μL of RIP wash buffer, and incubated with 100 μL of cell extract at 4°C overnight. The samples were placed on a magnetic base to collect the magnetic bead-antibody complex, after which the samples and the input were detached by protease K to extract RNA, which was used for qRT-PCR.
ChIP Assay
Cells in each group were fixed with formaldehyde for 10 min to produce DNA-protein crosslinks. The cells were then crushed to break the chromatin into fragments using an ultrasonic crusher with 10 s for ultrasound each time and 10 s for the interval (for 15 cycles). Next, the cells were centrifuged at 12,000 × g for 10 min at 4°C to collect supernatant, which was then divided into two tubes and separately added to the IgG (ab172730, 1:100, Abcam) in normal mice with NC antibody and the target protein-specific antibody FOXP3 (ab450, 2 μg/mL of cells, Abcam) for incubation at 4°C overnight. The DNA-protein complex was precipitated by protein agarose or Sepharose and centrifuged at 12,000 × g for 5 min, with the supernatant discarded. The nonspecific complex was washed and de-crosslinked at 65°C overnight, followed by phenol or chloroform extraction and purification to recover DNA fragments.
Dual-Luciferase Reporter Gene Assay
The full length of the promoter region of the ICAM-1 gene was cloned and amplified (forward: 5′-CAGTCGGGCACAGGACAC-3′, reverse: 5′-GTG GAAAACTGAACAGCAGC-3′). The primary hepatocytes of mice in the control group were seeded in 24-well plates and cultured for 24 h. The ICAM-1 dual-luciferase reporter gene vector (Pgl3-BASIC-ICAM-1-3′ UTR) was constructed artificially, and the correct sequences of the luciferase reporter plasmids of wild type (WT) and mutant type (MUT) were co-transfected with si-FOXP3 into hepatocytes of mice. Following a 48 h transfection, the luciferase activity was tested by using a dual-luciferase reporter gene detection system (Dual-Luciferase Reporter Assay System, E1910, Promega, Madison, WI, USA). Each 10 μL cell was added to 50 μL of firefly luciferase working solution to determine the firefly luciferase activity and added to 50 μL of Renilla luciferase to determinate the Renilla luciferase activity. The relative luciferase activity presented as the ratio of luciferase activity of firefly to the Renilla luciferase activity. The experiment was repeated three times.
qRT-PCR
Total RNA was extracted from the hepatic tissues of mice in each group by using a TRIzol one-step method according to the instructions of the TRIzol Kit (15596-026, Invitrogen, Gaithersburg, MD, USA). The nucleic acid protein detector (BioPhotometer D30, Eppendorf, Hamburg, Germany) was used to determine the OD value and RNA concentration at 260 and 280 nm wavelengths (OD260 and OD280). The high purity of the RNA was determined by a ratio of OD260 to OD280 between 1.8 and 2. RNA was reversely transcribed into cDNA according to the instructions of the reverse transcription kit (K1621, Fermentas, Waltham, MA, USA). The primers of RNA LINC01093, ICAM-1, caspase-3, TGF-β1, Bax, Bcl-2, NF-κB p65, and β-actin (Table 3) were designed and synthesized by Shanghai Genechem (Shanghai, China). The mRNA expression of each gene was measured by using a fluorescent qPCR kit (Takara, Dalian, Liaoning, China). Real-time fluorescence qPCR (ABI 7500, ABI, Foster City, CA, USA) was used for determination. With β-actin used as an internal reference, the relative expression of the target genes was calculated using the 2−ΔΔCt method through the following formula: ΔΔCt = ΔCt (average value of target gene in the experimental group − average value of house-keeping gene in the experimental group) − ΔCt (average value of target gene in the control group − average value of house-keeping gene in the control group). Each experiment was conducted in triplicate. This method can also be used to measure the mRNA levels of cells.
Table 3.
Prime Sequences for qRT-PCR
| Gene | Sequences |
|---|---|
| LINC01093 | forward: 5′-CTCTTGCAAACCATGGGCAC-3′ |
| reverse: 5′-CATCTCCCAGTCGGGTTTCC-3′ | |
| ICAM-1 | forward: 5′-GTGATGCTCAGGTATCCATCCA-3′ |
| reverse: 5′-CACAGTTCTCAAAGCACAGCG-3′ | |
| Caspase-3 | forward: 5′-TGTCATCTCGCTCTGGTACG-3′ |
| reverse: 5′-AAATGACCCCTTCATCACCA-3′ | |
| TGF-β1 | forward: 5′-CACCATCCATGACATGAACC-3′ |
| reverse: 5′-TCATGTTGGACAACTGCTCC-3′ | |
| Bax | forward: 5′-AGACAGGGGCCTTTTTGCTAC-3′ |
| reverse: 5′-AATTCGCCGGAGACACTCG-3′ | |
| Bcl-2 | forward: 5′-GAGGAGCTCTTCAGGGACGG-3′ |
| reverse: 5′-GGTGCCGGTGCAGGTACTCA-3′ | |
| NF-κB p65 | forward: 5′-CTGATGTGCATCGG CAAG-3′ |
| reverse: 5′-TGCTGGGAAGGTGTAGGG-3′ | |
| β-actin | forward: 5′-AGCCATGTACGTAGCCATCC-3′ |
| reverse: 5′-TTTGATGTCACGCACGATTT-3′ | |
| FOXP3 | forward: 5′-GGCCCTTCTCCAGGACAGA-3′ |
| reverse: 5′-GCTGATCATGGCTGGGTTGT-3′ |
ICAM-1, intercellular cell adhesion molecule-1; TGF-β1, transforming growth factor β1; Bax, Bcl-2-associated X protein; Bcl-2, B cell lymphoma-2; NF-κB p65, nuclear factor κB light-chain enhancer of activated p65; FOXP3, forkhead box P3.
Western Blot Analysis
A total of 50 mg of hepatic tissues were obtained and added to 0.6 mL of protein lysis buffer (R0010, Beijing Solabio Life Sciences, Beijing, China) to collect the protein. Then, the protein concentration was determined according to the bicinchoninic acid (BCA) protein quantitative kit (23225, Pierce, Rockford, IL, USA), and the concentration was adjusted to 1 μg/μL with deionized water. Next, the protein was added to the sample well (20 μg each well) and electrophoretic separation was performed using 10% SDS-PAGE (P1200, Beijing Solabio). Afterward, the protein samples were transferred to a polyvinylidene fluoride (PVDF) membrane (HVLP04700, Millipore) using the semi-dry method and stained with Ponceau S (P0012, Beijing Solabio). Subsequently, the membrane was washed twice with Tris-buffered saline with Tween 20 (TBST) and blocked with 5% skimmed milk powder at room temperature for 2 h. Then, the membrane was incubated with diluted primary antibodies rabbit antibodies to ICAM-1 (1:1,000, ab179707), caspase-3 (1:500, ab13847), TGF-β1 (1:500, ab92486), Bax (1:2,000, ab32503), Bcl-2 (1:20,000, ab182858), NF-κB p65 (1:1,000, ab16502), β-actin (1:1,000, ab8227), and histone H3 (1:1,000, ab1791) overnight at 4°C. Following this, the membrane was incubated with secondary antibody HRP-labeled goat anti-rabbit antibody to IgG (1:2,000, ab6721) at room temperature for 2 h. All antibodies were purchased from Abcam. The membrane was photographed by using a gel imaging instrument (Gel Doc XR, Bio-Rad, Hercules, CA, USA) and developed by electrochemiluminescence (ECL). The β-actin was used as the internal reference, and the relative expression of the protein was presented as the ratio of the gray value of the target band to the internal reference band. This method can also be used to determine the protein levels of cells. The experiment was conducted in triplicate.
Immunofluorescence Assay
The sub-cultured hepatocytes of mice in each group were obtained and cultured in 6-well plates. Upon reaching 60% to 70% confluence, the cells were centrifuged. The collected cells were suspended in the protein inhibitor (T9128, Sigma-Aldrich Chemical) consisting of 10 μg/mL of leupeptin, aprotinin, and pepstatin. The nuclear protein and cytoplasmic protein extraction kits were used for the extraction of nuclear protein on the ice. The cells (1.0 mL) were then lysed with 1.5 mL of cytoplasmic protein lysis buffer I and then continually lysed with cytoplasmic protein lysis buffer II, after which they were transferred to the centrifuge tube. The supernatant was the cytoplasmic protein solution, which could be used immediately. Afterward, the cells were washed with the cytoplasmic protein lysis buffer I, added to the nuclear lysis buffer for resuspension, and placed in the centrifuge tube at 4°C for 30 min; the nuclear protein was obtained. The cells were then fixed in 2.5% glutaraldehyde for 10 min and permeabilized with 0.1% Triton X-100 for 5 min. Next, the cells were incubated with specific rabbit anti-human antibody to NF-κB p65 (F-6: sc-8008; Santa Cruz Biotechnology, Santa Cruz, CA, USA) at 37°C overnight. Then, the cells were incubated with a secondary antibody, 1 × 1011 infection-forming units (ifu)/mL of fluorescein isothiocyanate (FITC)-labeled antibody (1:1,000, ab6717, Abcam), for 1 h and mounted by a drop of anti-fluorescence quenching sealing solution (Beyotime). Lastly, the cells were observed and photographed under a microscope photograph (200×) under the same conditions. The experiment was conducted in triplicate.
MTT Assay
After being transfected and cultured for 48 h, the cells in each group were collected and counted. The transfected cells in each group were seeded into the 96-well plate (1 × 104 each well, 8 parallel wells in each group), and the blank control well was set and added to just the culture medium. The cells were then cultured for 24, 48, and 72 h until cells adhered to the well. The cells in each well were then added to 10 μL of MTT (5 mg/mL, ST316, Beyotime) in an incubator at 37°C for 4 h. With the supernatant discarded, the cells in each well were added to 100 μL of DMSO (D5879, Sigma-Aldrich Chemical), followed by oscillation on a table concentrator for 10 min. Subsequently, the OD value was measured at 490 nm by a microplate reader (MK3, Thermo Scientific, Pittsburgh, PA, USA). The experiment was conducted three times to obtain the average value. The cell viability curve was drawn with the time point as abscissa and OD as ordinate.
Flow Cytometry
After being transfected and cultured for 48 h, the cells were detached with 0.25% trypsin once the culture medium was discarded. The collected cells were centrifuged at 4°C for 5 min at 1,000 rpm, with the supernatant discarded. The cells were centrifuged at 1,000 rpm for 5 min, with the supernatant discarded again; added to 70% pre-cooled ethanol; and fixed at 4°C overnight. Next, the cells were centrifuged at 1,000 rpm for 5 min after being washed with the balanced salt solution PBS, incubated with 10 μL of RNase enzyme at 37°C for 5 min, and stained with 1% propidium iodide (PI; 40710ES03, Shanghai qcbio Science & Technologies, Shanghai, China) for 30 min, avoiding exposure to light. The flow cytometer (FACSCalibur, BD, Franklin Lakes, NJ, USA) was used to test the cell cycle at the excitation wavelength of 488 nm. The experiment was conducted in triplicate.
After a 48 h transfection, the cells were detached with EDTA-free trypsin and collected. Afterward, centrifugation was carried out at 4°C for 5 min at 1,000 rpm, with the supernatant discarded. Then, the cells were centrifuged at 1,000 rpm for 5 min with the supernatant discarded again. The annexin V-FITC and PI apoptosis detection kit (CA1020, Beijing Solarbio) was used to detect cell apoptosis, and cells were washed with binding buffer. The mixture was prepared with annexin V-FITC and binding buffer diluted at a ratio of 1:40; then, the cells were resuspended, oscillated, and mixed at room temperature for 30 min, followed by the addition of the mixture (annexin V-FITC and binding buffer diluted at a ratio of 1:40) cultured at room temperature for 15 min. The apoptosis was detected by flow cytometry. The experiment was conducted in triplicate.
Statistical Analysis
Statistical analyses were conducted by using SPSS 21.0 (IBM, Armonk, NY, USA), Measurement data were expressed as mean ± SD. The t test was used for the comparison between two groups, while the one-factor ANOVA was used for the comparisons among multiple groups. p < 0.05 was considered statistically significant.
Author Contributions
X.S., X.J., and Q.A. designed the study. Y.T., B.Y., T.L., Y.C., and Y.S. collated the data, carried out data analyses, and produced the initial draft of the manuscript. X.S. and X.J. contributed to drafting the manuscript. All authors participated in the revised manuscript and have read and approved the final submitted manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We acknowledge the helpful comments on this paper received from our reviewers. This study was supported by the National Natural Science Foundation of China (81800537 and 81800557), China Postdoctoral Science Foundation (2018M631886), Chinese Foundation for Hepatitis Prevention and Control——TianQing Liver Disease Research Fund Subject (TQGB20190020) and the Science and Technology Development Project of Jilin Province (20170520001JH).
Contributor Information
Ying Shi, Email: shiy707@jlu.edu.cn.
Qing Ai, Email: aiqingjdyy@163.com.
References
- 1.Affò S., Dominguez M., Lozano J.J., Sancho-Bru P., Rodrigo-Torres D., Morales-Ibanez O., Moreno M., Millán C., Loaeza-del-Castillo A., Altamirano J. Transcriptome analysis identifies TNF superfamily receptors as potential therapeutic targets in alcoholic hepatitis. Gut. 2013;62:452–460. doi: 10.1136/gutjnl-2011-301146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mandrekar P., Bataller R., Tsukamoto H., Gao B. Alcoholic hepatitis: Translational approaches to develop targeted therapies. Hepatology. 2016;64:1343–1355. doi: 10.1002/hep.28530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jinjuvadia R., Liangpunsakul S., Translational Research and Evolving Alcoholic Hepatitis Treatment Consortium Trends in Alcoholic Hepatitis-related Hospitalizations, Financial Burden, and Mortality in the United States. J. Clin. Gastroenterol. 2015;49:506–511. doi: 10.1097/MCG.0000000000000161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altamirano J., Miquel R., Katoonizadeh A., Abraldes J.G., Duarte-Rojo A., Louvet A., Augustin S., Mookerjee R.P., Michelena J., Smyrk T.C. A histologic scoring system for prognosis of patients with alcoholic hepatitis. Gastroenterology. 2014;146:1231–1239. doi: 10.1053/j.gastro.2014.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Z., Ross R.A., Zhao S., Tu W., Liangpunsakul S., Wang L. LncRNA AK054921 and AK128652 are potential serum biomarkers and predictors of patient survival with alcoholic cirrhosis. Hepatol. Commun. 2017;1:513–523. doi: 10.1002/hep4.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takahashi K., Yan I., Haga H., Patel T. Long noncoding RNA in liver diseases. Hepatology. 2014;60:744–753. doi: 10.1002/hep.27043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan J.H., Yang F., Wang F., Ma J.Z., Guo Y.J., Tao Q.F., Liu F., Pan W., Wang T.T., Zhou C.C. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 8.Li X., Wu Z., Fu X., Han W. Long Noncoding RNAs: Insights from Biological Features and Functions to Diseases. Med. Res. Rev. 2013;33:517–553. doi: 10.1002/med.21254. [DOI] [PubMed] [Google Scholar]
- 9.Zhang K., Han X., Zhang Z., Zheng L., Hu Z., Yao Q., Cui H., Shu G., Si M., Li C. The liver-enriched lnc-LFAR1 promotes liver fibrosis by activating TGFβ and Notch pathways. Nat. Commun. 2017;8:144. doi: 10.1038/s41467-017-00204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He Y., Wu Y.T., Huang C., Meng X.M., Ma T.T., Wu B.M., Xu F.Y., Zhang L., Lv X.W., Li J. Inhibitory effects of long noncoding RNA MEG3 on hepatic stellate cells activation and liver fibrogenesis. Biochim. Biophys. Acta. 2014;1842:2204–2215. doi: 10.1016/j.bbadis.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 11.Ramos T.N., Bullard D.C., Barnum S.R. ICAM-1: isoforms and phenotypes. J. Immunol. 2014;192:4469–4474. doi: 10.4049/jimmunol.1400135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ito S., Yukawa T., Uetake S., Yamauchi M. Serum intercellular adhesion molecule-1 in patients with nonalcoholic steatohepatitis: comparison with alcoholic hepatitis. Alcohol. Clin. Exp. Res. 2007;31(1, Suppl):S83–S87. doi: 10.1111/j.1530-0277.2006.00292.x. [DOI] [PubMed] [Google Scholar]
- 13.Spahr L., Rubbia-Brandt L., Pugin J., Giostra E., Frossard J.L., Borisch B., Hadengue A. Rapid changes in alcoholic hepatitis histology under steroids: correlation with soluble intercellular adhesion molecule-1 in hepatic venous blood. J. Hepatol. 2001;35:582–589. doi: 10.1016/s0168-8278(01)00190-8. [DOI] [PubMed] [Google Scholar]
- 14.Hill D.B., Deaciuc I.V., McClain C.J. Hyperhyaluronanemia in alcoholic hepatitis is associated with increased levels of circulating soluble intercellular adhesion molecule-1. Alcohol. Clin. Exp. Res. 1998;22:1324–1327. [PubMed] [Google Scholar]
- 15.Kotteas E.A., Boulas P., Gkiozos I., Tsagkouli S., Tsoukalas G., Syrigos K.N. The intercellular cell adhesion molecule-1 (icam-1) in lung cancer: implications for disease progression and prognosis. Anticancer Res. 2014;34:4665–4672. [PubMed] [Google Scholar]
- 16.Mao X., Su Z., Mookhtiar A.K. Long non-coding RNA: a versatile regulator of the nuclear factor-κB signalling circuit. Immunology. 2017;150:379–388. doi: 10.1111/imm.12698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bulychev A.A., Dodonova S.O. Effects of cyclosis on chloroplast-cytoplasm interactions revealed with localized lighting in Characean cells at rest and after electrical excitation. Biochim. Biophys. Acta. 2011;1807:1221–1230. doi: 10.1016/j.bbabio.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 18.Zhang B., Roh Y.S., Liang S., Liu C., Naiki M., Masuda K., Seki E. Neurotropin suppresses inflammatory cytokine expression and cell death through suppression of NF-κB and JNK in hepatocytes. PLoS ONE. 2014;9:e114071. doi: 10.1371/journal.pone.0114071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taniue K., Kurimoto A., Takeda Y., Nagashima T., Okada-Hatakeyama M., Katou Y., Shirahige K., Akiyama T. ASBEL-TCF3 complex is required for the tumorigenicity of colorectal cancer cells. Proc. Natl. Acad. Sci. USA. 2016;113:12739–12744. doi: 10.1073/pnas.1605938113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lucey M.R., Mathurin P., Morgan T.R. Alcoholic hepatitis. N. Engl. J. Med. 2009;360:2758–2769. doi: 10.1056/NEJMra0805786. [DOI] [PubMed] [Google Scholar]
- 21.Crabb D.W., Bataller R., Chalasani N.P., Kamath P.S., Lucey M., Mathurin P., McClain C., McCullough A., Mitchell M.C., Morgan T.R., NIAAA Alcoholic Hepatitis Consortia Standard Definitions and Common Data Elements for Clinical Trials in Patients With Alcoholic Hepatitis: Recommendation From the NIAAA Alcoholic Hepatitis Consortia. Gastroenterology. 2016;150:785–790. doi: 10.1053/j.gastro.2016.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dai M., Chen S., Wei X., Zhu X., Lan F., Dai S., Qin X. Diagnosis, prognosis and bioinformatics analysis of lncRNAs in hepatocellular carcinoma. Oncotarget. 2017;8:95799–95809. doi: 10.18632/oncotarget.21329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu F., Zheng J., Mao Y., Dong P., Lu Z., Li G., Guo C., Liu Z., Fan X. Long Non-coding RNA Growth Arrest-specific Transcript 5 (GAS5) Inhibits Liver Fibrogenesis through a Mechanism of Competing Endogenous RNA. J. Biol. Chem. 2015;290:28286–28298. doi: 10.1074/jbc.M115.683813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cho K., Song S.B., Tung N.H., Kim K.E., Kim Y.H. Inhibition of TNF-α-Mediated NF-κB Transcriptional Activity by Dammarane-Type Ginsenosides from Steamed Flower Buds of Panax ginseng in HepG2 and SK-Hep1 Cells. Biomol. Ther. (Seoul) 2014;22:55–61. doi: 10.4062/biomolther.2013.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang Y., Jiang L.L., Maimaitirexiati X.M., Zhang Y., Wu L. Irbesartan attenuates TNF-α-induced ICAM-1, VCAM-1, and E-selectin expression through suppression of NF-κB pathway in HUVECs. Eur. Rev. Med. Pharmacol. Sci. 2015;19:3295–3302. [PubMed] [Google Scholar]
- 26.Shin D.S., Kim K.W., Chung H.Y., Yoon S., Moon J.O. Effect of sinapic acid against dimethylnitrosamine-induced hepatic fibrosis in rats. Arch. Pharm. Res. 2013;36:608–618. doi: 10.1007/s12272-013-0033-6. [DOI] [PubMed] [Google Scholar]
- 27.El-Bassiouni N.E., Mahmoud O.M., El Ahwani E.G., Ibrahim R.A., El Bassiouny A.E. Monocyte adhesion molecules expression in patients with chronic hepatitis C liver disease. Mediterr. J. Hematol. Infect. Dis. 2013;5:e2013054. doi: 10.4084/MJHID.2013.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tu X., Zhang Y., Zheng X., Deng J., Li H., Kang Z., Cao Z., Huang Z., Ding Z., Dong L. TGF-β-induced hepatocyte lincRNA-p21 contributes to liver fibrosis in mice. Sci. Rep. 2017;7:2957. doi: 10.1038/s41598-017-03175-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang S., Wu X., Liu Y., Yuan J., Yang F., Huang J., Meng Q., Zhou C., Liu F., Ma J. Long noncoding RNA H19 inhibits the proliferation of fetal liver cells and the Wnt signaling pathway. FEBS Lett. 2016;590:559–570. doi: 10.1002/1873-3468.12078. [DOI] [PubMed] [Google Scholar]
- 30.Chen Z., Yu C., Zhan L., Pan Y., Chen L., Sun C. LncRNA CRNDE promotes hepatic carcinoma cell proliferation, migration and invasion by suppressing miR-384. Am. J. Cancer Res. 2016;6:2299–2309. [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y., Zou Y., Wang W., Zuo Q., Jiang Z., Sun M., De W., Sun L. Down-regulated long non-coding RNA MEG3 and its effect on promoting apoptosis and suppressing migration of trophoblast cells. J. Cell. Biochem. 2015;116:542–550. doi: 10.1002/jcb.25004. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y., Jiang H., Zhou H., Ying X., Wang Z., Yang Y., Xu W., He X., Li Y. Lentivirus-mediated silencing of HOTAIR lncRNA restores gefitinib sensitivity by activating Bax/Caspase-3 and suppressing TGF-α/EGFR signaling in lung adenocarcinoma. Oncol. Lett. 2018;15:2829–2838. doi: 10.3892/ol.2017.7656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou C., York S.R., Chen J.Y., Pondick J.V., Motola D.L., Chung R.T., Mullen A.C. Long noncoding RNAs expressed in human hepatic stellate cells form networks with extracellular matrix proteins. Genome Med. 2016;8:31. doi: 10.1186/s13073-016-0285-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fujita A., Sato J.R., Rodrigues Lde.O., Ferreira C.E., Sogayar M.C. Evaluating different methods of microarray data normalization. BMC Bioinformatics. 2006;7:469. doi: 10.1186/1471-2105-7-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 36.Lieber C.S., Jones D.P., Decarli L.M. Effects of Prolonged Ethanol Intake: Production of Fatty Liver Despite Adequate Diets. J. Clin. Invest. 1965;44:1009–1021. doi: 10.1172/JCI105200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCullough R.L., McMullen M.R., Poulsen K.L., Kim A., Medof M.E., Nagy L.E. Anaphylatoxin Receptors C3aR and C5aR1 Are Important Factors That Influence the Impact of Ethanol on the Adipose Secretome. Front. Immunol. 2018;9:2133. doi: 10.3389/fimmu.2018.02133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li X., Zhang Y., Jin Q., Xia K.L., Jiang M., Cui B.W., Wu Y.L., Song S.Z., Lian L.H., Nan J.X. Liver kinase B1/AMP-activated protein kinase-mediated regulation by gentiopicroside ameliorates P2X7 receptor-dependent alcoholic hepatosteatosis. Br. J. Pharmacol. 2018;175:1451–1470. doi: 10.1111/bph.14145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lowe P.P., Gyongyosi B., Satishchandran A., Iracheta-Vellve A., Ambade A., Kodys K., Catalano D., Ward D.V., Szabo G. Alcohol-related changes in the intestinal microbiome influence neutrophil infiltration, inflammation and steatosis in early alcoholic hepatitis in mice. PLoS ONE. 2017;12:e0174544. doi: 10.1371/journal.pone.0174544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abiega O., Beccari S., Diaz-Aparicio I., Nadjar A., Layé S., Leyrolle Q., Gómez-Nicola D., Domercq M., Pérez-Samartín A., Sánchez-Zafra V. Neuronal Hyperactivity Disturbs ATP Microgradients, Impairs Microglial Motility, and Reduces Phagocytic Receptor Expression Triggering Apoptosis/Microglial Phagocytosis Uncoupling. PLoS Biol. 2016;14:e1002466. doi: 10.1371/journal.pbio.1002466. [DOI] [PMC free article] [PubMed] [Google Scholar]