Abstract
Bacterial biofilms pose a major threat to public health because they are resistant to most current therapeutics. Conventional antibiotics exhibit limited penetration and weakened activity in the acidic microenvironment of a biofilm. Here, the development of biofilm-responsive nanoantibiotics (rAgNAs) composed of self-assembled silver nanoclusters and pH-sensitive charge reversal ligands, whose bactericidal activity can be selectively boosted in the biofilm microenvironment, is reported. Under neutral physiological conditions, the bactericidal activity of rAgNAs is self-quenched because the toxic silver ions’ release is largely inhibited; however, upon entry into the acidic biofilm microenvironment, the rAgNAs not only exhibit charge reversal to facilitate local accumulation and retention but also disassemble into small silver nanoclusters, thus enabling deep penetration and accelerated silver ions release for dramatically amplified bactericidal activity. The superior antibiofilm activity of rAgNAs is demonstrated both in vitro and in vivo, and the mortality rate of mice with multi-drug-resistant biofilm-induced severe pyomyositis can be significantly reduced by rAgNAs treatment, indicating the immense potential of rAgNAs as highly efficient nanoscale antibacterial agents to combat resistant bacterial biofilm-associated infections.
Short abstract
Smart nanoantibiotics, that can achieve biofilm homing, deep penetration, and locally amplified bactericidal activity via controlled assembly/disassembly, are proposed for efficient biofilm treatments.
Introduction
Most pathogens involved in recent emerging infectious diseases (EIDs) events are bacteria,1,2 and the emergence of drug-resistant bacterial strains is typically represented.3 Although antibiotics are used to combat bacterial EIDs,4 the systemic antibiotic treatment can cause adverse side effects including hepatic and renal toxicity.5,6 Worse still, conventional antibiotics show very limited effectiveness and induce antibiotic resistance of bacteria.7−9 Biofilm formation is considered to be vital to the drug resistance of bacteria,10−12 and the biofilm matrix acts as a natural barrier to drug penetration and activation.13 The biofilm matrix containing exopolymeric substances (EPS) causes diffusion-reaction inhibition for antibiotics.14−16 Moreover, because of the metabolic activity of the bacteria and host immune response, there is an inherent acidic microenvironment (pH values of 4.5–6.5) within the biofilm,17−21 which reduces the activity of most antibiotics.22
Recent advances in nanotechnology have provided promising alternative approaches to combat biofilms.23−26 Nanoparticles with intrinsic antimicrobial activity, including silver nanoparticles,27 zinc oxide nanoparticles,28 and iron oxide nanoparticles,29 are of great potential as nanoscale nanoantibiotics. Among them, silver nanoparticles (AgNPs) have been demonstrated to be effective in a broad spectrum of antibacterial applications and widely used in daily life.30−33 Silver ions leaching from AgNPs play the main role in killing bacteria by interacting with thiol groups of vital enzymes in bacteria.34,35 Unlike traditional antibiotics, the ion-leaching-based bactericidal effect of AgNPs is not suppressed in the acidic microenvironment of the biofilm.36,37 Particularly for extremely small silver nanoclusters, their huge specific surface areas are highly susceptible to oxidative dissolution, facilitating silver ion release for enhanced activity.38−40 However, the limited penetration in the biofilm41−44 and the potential toxicity of released silver ions during the in vivo circulation of AgNPs45 remain knotty problems to be settled for in vivo bactericidal applications.
It is noteworthy that particle size and surface charge are crucial factors in nanoparticle-based drug delivery systems, which determine their blood half-lives, biodistribution, tissue penetration, biological activity, and so on.46−50 Long-circulating nanoparticles are liable to accumulate at the infection site because the host inflammatory response can increase the vascular permeability of the local tissue.51 After then, the negative surface of bacteria can be utilized to enhance the retention of nanoparticles via electrostatic interaction.52,53 Moreover, compared to large nanoparticles of limited biofilm penetration capability,52 small nanoparticles with a size of <20 nm have been reported to penetrate the biofilm with deep layers.53−55 Nevertheless, homing and deep penetration into the biofilm and in situ amplified bactericidal activity are of great importance for the effective antibiofilm application yet are challenging for currently existing nanoantibiotics.
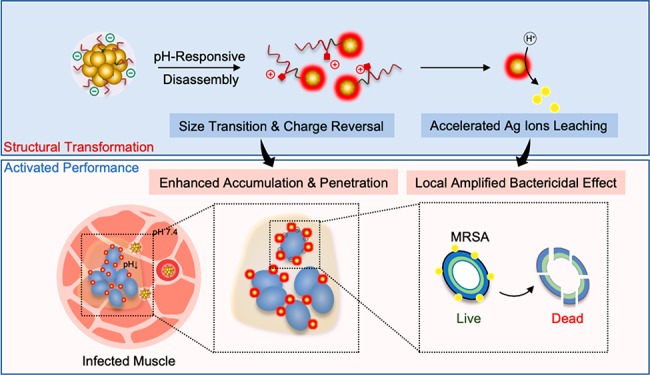
Controllable assembly/disassembly is a reliable way to fine tune the biological activities of nanomedicines.56−61 A series of stimuli-responsive peptide self-assemblies have been developed by Wang’s group for in vivo pathogen-specific detection and therapy.51,62−64 Moreover, our previous works have demonstrated that retaining the stable assembled form but performing in situ disassembly triggered by the disease microenvironment stimulus is highly effective for targeted delivery and retention.65−68 Herein, we hypothesized that the simultaneous biofilm homing and deep penetration and locally amplified bactericidal activity of inorganic nanoparticle-based nanoantibiotics can be achieved via controlled assembly/disassembly strategy using rationally designed biofilm-responsive surface ligands. To demonstrate the proof of concept, we synthesized biofilm-responsive nanoantibiotics (rAgNAs) composed of self-assembled extremely small silver nanoclusters (AgNCs) and the pH-sensitive charge reversal ligand of poly(ethylene glycol)-poly(aminopropyl imidazole-aspartate)-polyalanine (PEG-PSB-PALA). The rAgNAs with high colloid stability and negative charge are supposed to circulate for a long time and gradually accumulate in the biofilm infection site. Once arriving there, the protonation of imidazole groups of PEG-PSB-PALA triggered by the acidic microenvironment of the biofilm22,52,69−71 leads to the increased electrostatic repulsion and consequently induces disassembly. Importantly, the silver ion leaching can be tremendously accelerated from the surface of the disassembled nanoantibiotics in the acidic microenvironment. In contrast to previously reported nanoantibiotics, the herein proposed biofilm-responsive nanoantibiotics can not only achieve enhanced accumulation and penetration in the biofilm but also have a locally activated bactericidal effect (Figure 1). On the basis of the unique properties of as-constructed nanoantibiotics involving acidic biofilm microenvironment-responsive structural transition and bactericidal effect amplification, we successfully demonstrate the highly effective treatments against bacterial biofilm-associated severe pyomyositis.
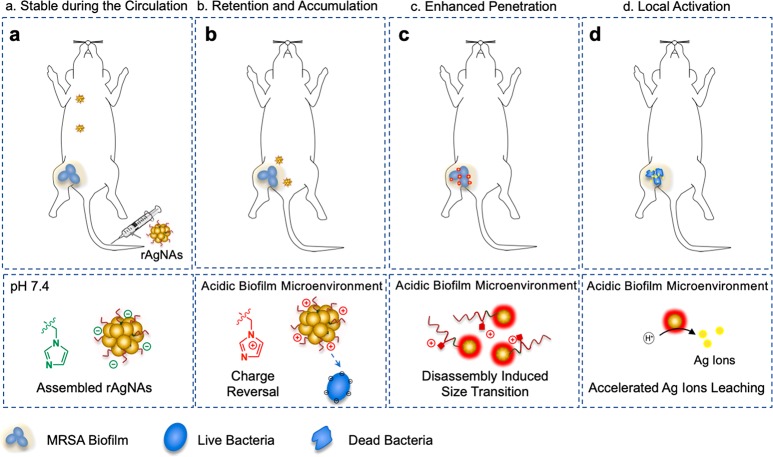
Figure 1.
pH-controlled assembly/disassembly strategy for the fabrication of biofilm-responsive nanoantibiotics (rAgNAs) to enhance the treatment of biofilm-associated infections. (a) rAgNAs with high colloid stability and negative charge are supposed to circulate for a long time in vivo. (b) Once in the acidic microenvironment of the biofilm, the rAgNAs go through charge reversal triggered by the protonation of the imidazole groups and tend to accumulate in the biofilm area owing to the electrostatic interaction. (c) Increased electrostatic repulsion consequently leads to disassembly, and a size transition takes place, which is beneficial to the enhanced penetration of the biofilm. (d) Silver ion leaching can be tremendously accelerated from the surface of the disassembled nanoantibiotics in the acidic microenvironment, and thus rAgNAs are able to exhibit locally amplified bactericidal effects.
Results and Discussion
Synthesis and Characterization of the rAgNAs
rAgNAs were constructed by the self-assembly of silver nanoclusters and pH-sensitive charge reversal ligand (PEG-PSB-PALA). The imidazole group of PEG-PSB-PALA is ionized as the pH value decreases to enable the disassembly of rAgNAs in an acidic microenvironment (Figure 2a). Silver nanoclusters were synthesized in an organic solvent72 with a diameter of ∼3.8 nm (Figure 2b), and a high-resolution transmission electron microscope (HRTEM) image (Figure S1a) as well as an X-ray diffraction (XRD) pattern (Figure S1b) demonstrated the face-centered-cubic structure of silver nanoparticles according to powder diffraction standards file no. 04-0783. PEG-PSB-PALA was synthesized through ring-opening polymerization and aminolysis with 1-(3-aminopropyl)imidazole (API) (details in Figures S2 and S3). PEG-PSB-PALA solution had a buffering capability during titration, with a pKa of about 6.5 due to the ionization of the imidazole groups (Figure S4). Spherical silver nanoassemblies (rAgNAs) were subsequently formed via the thin-film dispersion method.73 Infrared spectroscopy (IR) spectra revealed the disappearance of the amino group after self-assembly (Figure S5a), indicating the affinity between the hydrophobic segment of the polymer (polyalanine) and AgNCs.74 Moreover, according to the XRD pattern of rAgNAs, the assembly process did not change the crystalline phase of AgNCs (Figure S5b). As the pH decreased from 7.4 to 5.5, transmission electron microscope (TEM) images revealed that rAgNAs were well dispersed in neutral solution while being disassembled into isolated AgNCs in acidic solution (Figure 2c), accompanied by a dramatically reduced hydrodynamic size from ∼150 to ∼8 nm as observed via the dynamic light scattering (DLS) measurement (Figure 2d) and the solution becoming transparent (Figure 2e and Figure S6a). Besides, a charge switch was also presented from −3.85 ± 0.52 to 28.77 ± 1.00 mV as the pH decreased (Figure 2f). The pH-induced disassembly process of rAgNAs was also monitored by ultraviolet-visible (UV-vis) absorption spectrophotometry,75−78 and the absorption peak was blue-shifted from 425 to 411 nm with a narrower width as the pH decreased (Figure S6b,c). Moreover, rAgNAs remained stable in the 10% serum-containing media without a noticeable hydrodynamic size change for 1 week (Figure S7), indicating the excellent colloidal stability in vivo. Furthermore, by removing the imidazole group from the system, silver nanoassemblies (uAgNAs) composed of AgNCs and poly(ethylene glycol)-poly(β-benzil-ι-aspartate)-polyalanine (PEG-PIB-PALA) were not able to present pH-dependent performance, as evidenced by neither a size transition (Figure S8a,b) nor a peak shift in UV–vis absorption spectra (Figure S8c,d). Interestingly, the silver ions’ leaching of the rAgNAs was tremendously accelerated in an acidic environment as attributed to the pH-responsive disassembly (Figure 2g), indicating the reduced systemic side effects and local activation in the biofilm.
Figure 2.
Fabrication and characterization of pH-responsive nanoantibiotics (rAgNAs). (a) Schematic illustration of the design and construction of rAgNAs as well as the mechanism explanation. (b) TEM image of silver nanoclusters (AgNCs). (c) TEM images of rAgNAs at pH 7.4 and 5.5. (d) DLS size measurement of rAgNAs at pH 7.4 and 5.5. (e) Transmittance of rAgNAs in PBS with different pH values. The concentration of rAgNAs was 500 μg mL–1. The photographs (inset) presented the appearance of rAgNAs under different pH conditions. (f) Surface charge of rAgNAs along with changes in the pH value (n = 3). (g) Cumulative silver ion release amount at pH 7.4 and 5.5 (n = 3). The concentration for the releasing study of assemblies was 1 mg mL–1. *P < 0.05, **P < 0.01, and ****P < 0.0001.
Bactericidal Effect of rAgNAs in Vitro
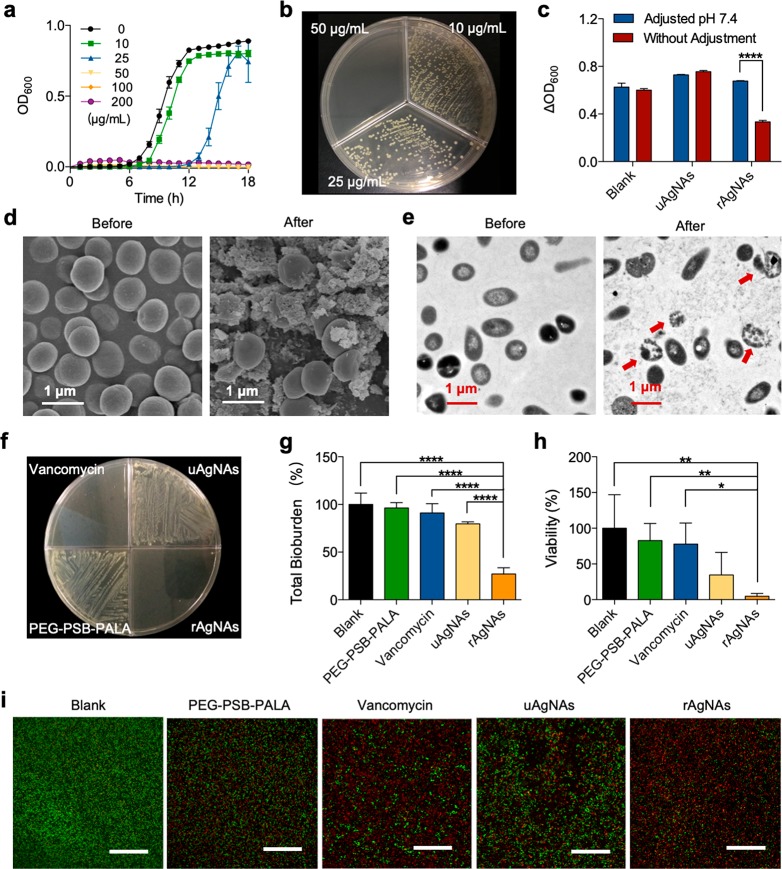
Drug-resistant bacteria has posed a huge threat to public health.79 Here the methicillin-resistant Staphylococcus aureus (MRSA) isolated from clinical samples was used as the typical pathogen. The pH-responsive bactericidal effect of rAgNAs was explored on both planktonic bacteria and a bacterial biofilm. For planktonic bacteria, the growth curve of bacteria that was treated with rAgNAs over 18 h was recorded (Figure 3a), and the visible growth of bacteria was completely inhibited by rAgNAs treatment with a minimal inhibitory concentration (MIC) of ∼50 μg mL–1. Then, a bacterial suspension that had been treated for 18 h was placed on an agar plate to incubate for another 24 h. From the results of bacterial colonies formed in agar plates (Figure 3b), the minimal bactericidal concentration (MBC) of rAgNAs was ∼50 μg mL–1. The observed MBC was identical to the MIC, indicating minimal tolerance (MBC/MIC) and a bactericidal effect of rAgNAs.80 Moreover, under the acidic condition, the MIC of rAgNAs is significantly reduced, showing an enhanced antibacterial effect (Figure S9). It has been reported that a large amount of organic acid is secreted during bacterial metabolism, which naturally decreases the extracellular pH and influences the activity of rAgNAs.81 To further investigate the pH-responsive bactericidal activity, a coculture of bacteria with rAgNAs for 12 h in the medium that maintained a neutral pH by adjusting the pH every 2 h was used (Figure S10). As shown in Figure 3c, in contrast to uAgNAs, the activity of rAgNAs can be significantly reduced in the medium maintained at pH 7.4, demonstrating the selective amplified activity of rAgNAs in an acidic microenvironment. According to the morphology of planktonic bacteria that were cocultured with rAgNAs for 6 h, membrane blebbing and structural damage to bacteria were observed via scanning electron microscopy (SEM) (Figure 3d) and TEM images (Figure 3e), implying that the bactericidal effect of rAgNAs to planktonic bacteria was caused by silver ion leaching to destroy the integrity of the membrane.82 Notably, the in vitro bactericidal effect of rAgNAs on planktonic bacteria was comparable to that of vancomycin (Figure 3f).
Figure 3.
Evaluation of the antibacterial effect of rAgNAs on planktonic bacteria and a bacterial biofilm in vitro. (a) Monitoring of the growth curve of planktonic MRSA cocultured with rAgNAs with different concentrations. (b) Colony formation of planktonic MRSA on the agar after being cocultured with different concentrations of rAgNAs. (c) Evaluation of the relative growth of planktonic MRSA cocultured with rAgNAs or uAgNAs in a medium with or without pH adjustment with a concentration of 100 μg mL–1. ****P < 0.0001. (d, e) Morphologies of planktonic MRSA before and after being cocultured with rAgNAs, characterized via SEM (d) and TEM (e). (f) Colony formation of planktonic MRSA on the agar after being cocultured with a therapeutic agent with a concentration of 200 μg mL–1. (g) Total bioburden of the MRSA biofilm after treatment (n = 4). ****P < 0.0001. (h) Viability of the biofilm bacteria after treatment (n = 4). *P < 0.05 and **P < 0.01. (i) Representative images of Syto green/PI staining of a biofilm to investigate the membrane permeability of bacteria in the biofilm. Scale bar, 50 μm.
To further study the biofilm microenvironment activatable antibacterial performance of rAgNAs, mature biofilms were further constructed80 by seeding MRSA into 96-well plates and incubating at 37 °C for 48 h with gentle shaking. The total bioburden content of the biofilm was determined by crystal violet staining. Figure 3g shows the total biomass content of untreated and various antibacterial agent-treated MRSA biofilms. Among several tested antibacterial agents, rAgNAs of 4-fold MIC-dosage-treated biofilms exhibited a significant reduction (about 72.9%) in the total bioburden whereas uAgNAs or vancomycin caused only an 8–25% reduction, indicating that the MRSA biofilm was more susceptible to the toxicity of rAgNAs. Subsequently, the bacterial viability of the untreated and various antibacterial-agent-treated MRSA biofilms was further explored according to the colony formation on the agar (Figure 3h). rAgNAs-treated biofilm bacteria had the lowest survival of less than 5% of the control at a 4-fold MIC dosage, confirming the results obtained from crystal violet staining. Afterward, Syto green/propidium iodide (PI) staining of the biofilm bacteria was performed to further verify the bactericidal effect of rAgNAs. Syto green is able to stain the live bacteria with an intact membrane showing green fluorescence while PI can enter only the bacteria with damaged membranes representing red fluorescence. According to the typical photographs (Figure 3i) obtained from confocal laser scanning microscopy (CLSM), rAgNAs-treated biofilm bacteria exhibited the highest ratio of red/green fluorescence via quantitative statistics (Figure S11), indicating that rAgNAs could increase the membrane permeability of biofilm bacteria significantly, which was one of the signals of the bactericidal effect. The treatment of MRSA biofilms with vancomycin was not effective owing to the diffusion-reaction inhibition effect83,84 and the reduced activity in the acidic biofilm microenvironment.22,85
Exploration of the Mutual Interaction between Biofilm and rAgNAs
According to the composition of rAgNAs and uAgNAs as well as the silver ions’ release profile, both of them can perform acidic microenvironment accelerated silver ion leaching. However, uAgNAs showed no significant antibiofilm effect according to the above exploration. Thus, apart from the difference in the silver ion leaching rate, the mutual interaction between the biofilm and rAgNAs is hypothetically taken into consideration. By investigation of the z-axis scanning images shown in Figure 4a, rhodamine B isothiocyanate (RITC)-labeled rAgNAs could penetrate and accumulate in the biofilm whereas RITC-labeled uAgNAs were retained a little on the top side of the biofilm. Furthermore, there was an affinity between rAgNAs and biofilm bacteria because RITC-labeled rAgNAs were observed to colocalize with Hoechst 33258-stained bacteria (Figure 4b–d).
Figure 4.
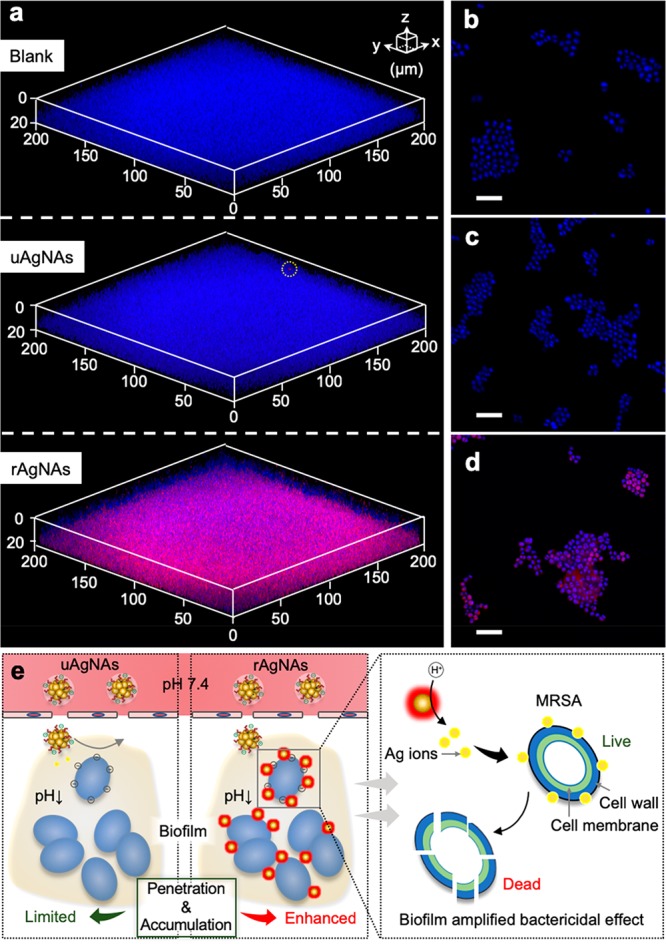
Enhanced accumulation and deep penetration of rAgNAs for the biofilm-amplified bactericidal effect. (a) Penetration of RITC-labeled rAgNAs or uAgNAs into bacterial biofilm as characterized by CLSM. (b–d) Merged fluorescence micrographs of bacteria in the biofilm (blue fluorescence) and PBS (b), RITC-labeled uAgNAs (c), or rAgNAs (d). The scale bar is 5 μm. (e) Schematic illustration of the enhanced accumulation and penetration of rAgNAs to the biofilm via the pH-responsive size transition and charge reversal, which can be attributed to the biofilm amplified bactericidal effect of rAgNAs on the bacteria in the biofilm.
Therefore, the superiority of rAgNAs for antibiofilms can be attributed to the (i) acidic microenvironment of the biofilm actuated size transition and charge reversal for enhanced accumulation and penetration in the biofilm and (ii) dual accelerated silver ion leaching by structural disassembly and an acidic microenvironment for the locally amplified bactericidal effect (Figure 4e). Initially, because of the pH-sensitive size transition, rAgNAs could arrive at the deep side of the biofilm. In the meantime, charge reversal occurs and the positive disassembled rAgNAs anchor on the negative surface of bacteria via electrostatic interaction. Subsequently, the reduced size of rAgNAs after disassembly and the acidic microenvironment in the biofilm accelerate silver ion leaching synergistically. Finally, the biofilm amplified bactericidal effect causes the eradication of the biofilm. In contrast, lacking in pH sensitivity, uAgNAs showed limited penetration and accumulation in the biofilm and failed to activate themselves for the antibiofilm.
Exploration of the Antibiofilm Effect of rAgNAs in Vivo
Staphylococcus aureus is a pathogenic bacterium for many diseases, and pyomyositis is one of them. Pyomyositis refers to a bacterial infection of the muscles and most often is caused by Staphylococcus aureus. Moreover, pyomyositis is a deep-seated biofilm-associated infection.86,87 Without effective therapy in time, the progression of pyomyositis can lead to limb dysfunction, a systemic inflammatory response, and even death. Although antibiotics combined with abscess excision is the clinically adopted strategy for controlling the progression of pyomyositis, it is necessary to develop a novel effective bactericidal agent to avoid superinfection during surgery. Considering the potential antibiofilm effect of rAgNAs with enhanced penetration and accumulation as well as the locally amplified bactericidal effect in vitro, a pyomyositis mice model was developed for further in vivo bactericidal effect exploration (Figure 5a). The mice were treated with cyclophosphamide (CP) for neutropenia, and then MRSA was injected intramuscularly into the rear thigh (Figure S12a). The increased creatine kinase level implied the successful establishment of the pyomyositis model (Figure S12b). The biodistribution of rAgNAs in the model mice was first explored. After the intravenous injection of rAgNAs, the amount of silver in the infected muscle significantly increased 8 h postinjection and remained stable for 24 h postinjection (Figure S13a). Moreover, from the ex vivo fluorescence images of the model mice after treatment with RITC-labeled rAgNAs, the high fluorescence intensity in the infected thighs demonstrated the efficient accumulation of rAgNAs in the infection site (Figure S13a). The successful biofilm-targeted delivery of rAgNAs was attributed to their excellent colloidal stability, which enabled a long-term circulation in vivo before gradually accumulating in the biofilm.49,51
Figure 5.
Treatment of multi-drug-resistant biofilm-induced severe pyomyositis using rAgNAs. (a) Schematic illustration of rAgNAs’ potential therapy in biofilm-induced severe pyomyositis. Without effective therapy in time, severe pyomyositis could lead to limb dysfunction and even a systemic inflammatory response, which might finally cause death. However, bacterial-biofilm-activatable nanoantibiotic rAgNAs that could be actuated by the acidic microenvironment and go through enhanced penetration and accumulation as well as local amplified bactericidal effect are able to control the progression. (b) Time line and strategy for the therapeutic process. (c) Percent survival curve after different treatments on the mice with severe pyomyositis (n = 15). (d) Clinical scores of mice during therapy (n = 15). (e) Ultrasound images of the infected thighs of the mice with severe pyomyositis during therapy. (f) Gram staining of the sections from infected thighs. The scale bar is 100 μm. (g) Number of bacteria in thighs in the severe pyomyositis mice model after therapy. **P < 0.01. (h) Levels of serum TNF-α of the mice with severe pyomyositis on day 10 after treatment (n = 5). Mice in the negative control were treated only with CP. *P < 0.05.
The time line and the therapeutic process are illustrated in Figure 5b. As shown in Figure 5c, rAgNAs could reduce the mortality rate significantly after 10 days of treatment, and the survival rates of the control group and of the PEG-PSB-PALA-, vancomycin-, and uAgNAs-treated groups were 22.88, 56.10, 42.42, and 54.17%, respectively. Although vancomycin is considered to be the “last resort” against resistant pathogens, the limitation in the effective delivery and activation in the biofilm suppresses its efficacy in vivo. During 10 days of treatment, mice were weighted and assessed for disease signs every 24 h. The mice without therapy showed significantly less body weight gain whereas the rAgNAs-treated group effectively recovered from the loss of weight after infection (Figure S14). Clinical scores were assessed according to the following scale: 0, no disease signs; 1, ruffled fur; 2, very mild hind limb weakness; 3, mild hind limb weakness; 4, moderate hind limb weakness; 5, severe hind limb weakness/dragging; 6, complete loss of hind limb function; 7, moribund; and 8, death.88 After infection, mice of every group exhibited very mild or mild hind limb weakness. Without treatment (blank group), mice suffered from deteriorated symptoms rapidly, and severe hind limb weakness was presented, with the hind limb function being completely lost in several mice on day 1. The treatment with PEG-PSB-PALA, vancomycin, or uAgNAs was not very effective at suppressing the progression, whereas rAgNAs were able to suppress the symptoms of deterioration (Figure 5d), which was further verified by the real-time ultrasound images (Figure 5e). Notably, the biofilm was disrupted by rAgNAs as confirmed by the pathological examination (Figure 5f), revealing less accumulated bacteria in the rAgNAs-treated group. Interestingly, the rAgNAs-treated group preserved the tight junctions of muscle cells and maintained the complete muscle structure. On day 10, after treatment, the mice were sacrificed, and the infected thighs were harvested and the muscle was homogenized in phosphate-buffered saline (PBS). rAgNAs could significantly reduce the amount of bacteria in the infectious site according to Figure 5g. The superiority of rAgNAs was also confirmed by the assessment of the serum TNF-α levels via the enzyme-linked immunosorbent assay (ELISA). All of these therapeutic agents themselves would not induce systemic inflammation (Figure S15), whereas the mice with severe pyomyositis in the rAgNAs-treated group had significantly lower systemic inflammation levels (Figure 5h), indicating the inhibition of the further progression of infection. In addition, the serum biochemical analysis (Figure S16) and the histopathological examination of major organs (Figure S17) demonstrated that rAgNAs highly biocompatible.
Conclusions
Biofilm-microenvironment-responsive nanoantibiotics (rAgNAs) were successfully fabricated by the self-assembly of ultrasmall AgNCs and biofilm-responsive polymeric ligands. The acidic microenvironment of the biofilm triggered ligand protonation and the disassembly of rAgNAs, allowing for enhanced retention and penetration in the biofilm. Furthermore, the accelerated silver ions leaching from isolated AgNCs efficiently killed the bacteria inside the biofilm by damaging the cell membrane. In contrast to conventional antimicrobial agents with limited penetration and inhibited activity, the therapeutic activity of rAgNAs is boosted by the acidic biofilm microenvironment. The outstanding therapeutic effect of rAgNAs was verified on a methicillin-resistant Staphylococcus aureus infection model both in vitro and in vivo. Importantly, the mortality rate of mice with biofilm-induced severe pyomyositis can be significantly reduced by rAgNAs in vivo. The rAgNAs-treated group preserved the tight junctions and structure of the muscle, revealing no toxicity to healthy tissues. Overall, we have developed a biofilm-responsive nanoantibiotic with simultaneous biofilm homing and deep penetration as well as locally amplified bactericidal activity in the biofilm microenvironment, showing immense therapeutic potential in the treatment of drug-resistant bacterial biofilm-associated infectious diseases.
Methods
Materials
All reagents and solvents obtained commercially were used without further purification. Triphosgene, ι-aspartate acid β-benzyl ester (Bla), tetrahydrofuran (THF), alanine, silver nitrate, oleylamine (C18, 80%–90%), oleic acid (technical grade, 90%), crystal violet, propidium iodide (PI), ethyl acetate (EA), dichloromethane (CH2Cl2), N,N-dimethylformamide (DMF), and dimethyl sulfoxide (DMSO) were purchased from Aladdin Inc. (Shanghai, China). Sodium hydroxide, sodium chloride, diethyl ether, hydrochloric acid (HCl), toluene, ethanol, chloroform, and acetic acid were purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). Rhodamine B isothiocyanate (RITC) and 1-(3-aminopropyl) imidazole (API) were obtained from Sigma-Aldrich Co. (St. Louis, MO). Methoxypolyethylene glycol amine (mPEG-NH2, MW = 5000 Da) was produced by Ziqi Biotechnology Co., Ltd. (Shanghai, China). Tryptone soy broth (TSB) was obtained from Hangzhou Microbial Reagent Co., Ltd. (Hangzhou, China). Syto green and Hoechst 33258 were purchased from KeyGEN BioTECH Co., Ltd. (Jiangsu, China).
Preparation of pH-Responsive Silver Nanoassemblies and Unresponsive Silver Nanoassemblies
A film dispersion method was adopted to prepare silver nanoassemblies. Briefly, polymer (PEG-PSB-PALA for rAgNAs and PEG-PIB-PALA for uAgNAs, 3.3 mg) was dissolved in the mixture solvents, which were composed of ethanol (1 mL) and PBS (0.01 M, pH 7.4, 0.1 mL). The solution of polymer (1 mL) was added to chloroform solution (1 mL) containing silver nanoclusters (1.5 mg) and stirred for 1 h at room temperature. Then the mixture was evaporated under vacuum to form a film. Finally, PBS (2 mL) was added and water-bath sonication was adopted to obtained the products.
Characterization
The hydrodynamic size, zeta potential, transmittance, and UV absorption of rAgNAs were characterized in PBS at different pH values. The stability of rAgNAs in serum-containing media was evaluated in both 10% fetal bovine serum (FBS) containing PBS (FBS/PBS) and 10% FBS containing Dulbecco’s Modified Eagle’s Medium (FBS/DMEM) via a dynamic light scattering (DLS) study. Moreover, silver ion release profiles were obtained using a dialysis method. Assemblies (rAgNAs or uAgNAs, 1 mL) were put inside a dialysis bag (MW = 8000–14 000 Da), and PBS (0.01 M, 9 mL) at pH 7.4 or 5.5 was applied as the releasing medium. At set time points, the releasing medium was replaced and the released silver ions were evaluated by inductively coupled plasma–mass spectrometry (ICP–MS, PerkinElmer NexION 300X).
In Vitro Antibacterial Tests on Planktonic Bacteria
No unexpected or unusually high safety hazards were encountered during the antibacterial and antibiofilm tests’ in vitro and in vivo evaluations. The bacterial strain tested in this study was a multi-drug-resistant Staphylococcus aureus (MRSA) isolated from the clinical samples. In brief, 100 μL of bacterial suspensions with OD = 0.04 and 100 μL of the rAgNAs with different concentrations was added to each well of a 96-well plate. During coincubation for a period of up to 18 h, turbidity measurements were conducted every hour to monitor the growth of bacteria in real time. After 18 h, bacterial cultures were plated on TSB agar and incubated for another 24 h to observe the number of colonies. To further check the bacterial morphology, bacteria were fixed with 2.5% glutaraldehyde in phosphate buffer overnight after incubation with the assemblies for 6 h, and then the sample was postfixed with 1% OsO4 for 1.5 h. Following fixation, the samples were dehydrated with a graded series of ethanol (from 30 to 100%). The morphology of the bacteria was observed with Hitachi H7650 TEM and Hitachi SU8010 SEM.
In Vitro Antibiofilm Tests
The bacterial strain being tested was grown from frozen stock in TSB for 24 h at 37 °C in 5% CO2. A bacterial suspension (200 μL) diluted 1:100 (OD600 = 0.01) in TSB + 1% glucose was added to each well and incubated for 48 h to form a mature biofilm. rAgNAs in TSB (100 μL) + 1% glucose were prepared in a flat-bottomed 96-well microplate (Costar, Corning). Control wells without any therapeutic agent were also prepared. In addition to bare pH-responsive polymer, uAgNAs and vancomycin were applied as controls. After incubation for 24 h at 37 °C in 5% CO2 under aerobic conditions, spent media and free-floating bacteria were removed by turning over the plates. The wells were vigorously rinsed a minimum of three times with doubly distilled water (DDW). Next, 0.05% crystal violet (50 μL) was added to each well after fixation with methanol for 15 min. After 5 min, wells were vigorously rinsed three times with DDW to remove the unbound dye. After 33% acetic acid (200 μL) was added to each well, the plate was shaken for 15 min to release the dye. Biofilm formation was quantified by measuring the difference between the absorbance of untreated and treated bacterial samples for each tested concentration of the compounds and the absorbance of appropriate blank wells at 570 nm (A570) using a plate reader. For bacterial activity in the biofilm after treatment, spent media and free-floating bacteria were removed by turning the plates over. The wells were vigorously rinsed a minimum of three times with DDW. Then the bacteria were collected and bacterial cultures were plated on TSB agar and incubated for another 24 h to observe the number of colonies. To visualize the bacterial killing effect, live/dead staining was carried out. Propidium iodide (PI) was used to stain the dead bacteria, and Syto green was applied to stain the live bacteria with red and green fluorescence, respectively. Briefly, after incubation with therapeutic agents for 24 h, the biofilm was thoroughly washed and the medium was replaced with fresh PBS, which contained PI (0.05 mg mL–1) and Syto green (4.5 μM). After incubation for 20 min at 37 °C, the fluorescent dye was washed away and the fluorescence of bacteria was observed by confocal laser scanning microscopy (CLSM). The ratio of green/red fluorescence was calculated with ImageJ. The penetration ability of the assemblies was explored with RITC-labeled assemblies. Similarly, the mature biofilm was incubated with RITC-labeled assemblies (200 μg mL–1) for 2 h, followed by fixation with 4% paraformaldehyde for 10 min. After fixation, the biofilm was washed twice and then stained with Hoechst 33258 for 10 min. Next, the dye was washed away and the biofilm was observed by CLSM to explore the penetration of assemblies into the biofilm.
In Vivo Antibiofilm Tests
Animals were maintained under the guidelines of the National Institute Guide for the Care and Use of Laboratory Animals, and all of the animal studies were approved by the Institutional Animal Care and Use Committee of Zhejiang University. In vivo antibiofilm tests were performed on 6-week-old female balb/c mice (purchased from Shanghai SLAC Laboratory Animal Co., Ltd.) that were first rendered neutropenic by cyclophosphamide pretreatment (150 mg kg-1 and 100 mg kg-1 96 and 24 h before infection, respectively). Infection was introduced by injecting bacteria (1 × 1012 cells in 100 μL of saline solution) intramuscularly on the right thigh of a mouse and was allowed to develop for 24 h before therapy. Then these model mice were treated with rAgNAs, uAgNAs, vancomycin, PEG-PSB-PALA, or PBS. During the 10 days of treatment, mice were weighed and scored every 24 h. Moreover, ultrasonic imaging was performed to analyze the development of the infected thighs. Mice were sacrificed after treatment, and thighs were removed and homogenized in PBS. Homogenates were serially diluted, and samples were plated on TSB agar and incubated for another 24 h. The number of colony-forming units on the agar was counted. The serum of mice was collected to evaluate the serum biochemicals including alanine aminotransferase (ALT), aspartate aminotransferase (AST), and blood urine nitrogen (BUN) along with the serum TNF-α level. Thighs and major organs were collected after treatment and fixed with 10% formalin solution. The fixed tissues were embedded into paraffin and cut into slices. The slices of thighs were stained with Gram stain to check the morphology of bacteria. Hematoxylin and eosin (H&E) staining was performed for the histopathology evaluation.
Statistical Analysis
Data are presented as the mean ± SD unless otherwise specified. One-way ANOVA analysis is used for multiple-group comparison by Prism 6. P < 0.05 is considered to be a statistically significant difference.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (2016YFA0203600), the National Natural Science Foundation of China (31822019, 51703195, 91859116, 81571743, and 51502251), the One Belt and One Road International Cooperation Project from the Key Research and Development Program of Zhejiang Province (2019C04024), the National Science & Technology Major Project “Key New Drug Creation and Manufacturing Program”, China (2018ZX09711002), the Ten-thousand Talents Program of Zhejiang Province (2018R52049), the Zhejiang Provincial Natural Science Foundation of China (LGF19C100002) and the Fundamental Research Funds for the Central Universities (2018QNA7020).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acscentsci.9b00359.
Additional results as described in the main text including the characterization of PEG-PSB-PALA, the extra characterization of rAgNAs, the characterization of uAgNAs, and in vivo exploration and biocompatibility studies (PDF)
Author Contributions
D.L. and J.G. supervised the project and commented on the project. J.W. constructed and characterized the nanoassemblies, performed in vitro experiments, and collected and analyzed the data. J.W. and X.H. performed in vivo experiments. F.L., J.L., and X.S. offered valuable suggestions for the presented work. J.W., F.L., and D.L. wrote the manuscript. All authors contributed to the discussion during the whole project.
Author Contributions
⊥ These authors contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- Fauci A. S.; Morens D. M. The Perpetual Challenge of Infectious Diseases. N. Engl. J. Med. 2012, 366, 454–461. 10.1056/NEJMra1108296. [DOI] [PubMed] [Google Scholar]
- GBD 2015 Mortality and Causes of Death Collaborators . Global, Regional, and National Life Expectancy, All-Cause Mortality, and Cause-Specific Mortality for 249 Causes of Death, 1980–2015: A Systematic Analysis for the Global Burden of Disease Study 2015. Lancet 2016, 388, 1459–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom D. E.; Black S.; Rappuoli R. Emerging Infectious Diseases: A Proactive Approach. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, 4055–4059. 10.1073/pnas.1701410114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubes G. The Bacteria Fight Back. Science 2008, 321, 356–361. 10.1126/science.321.5887.356. [DOI] [PubMed] [Google Scholar]
- Bruniera F. R.; Ferreira F. M.; Saviolli L. R. M.; Bacci M. R.; Feder D.; da Luz Goncalves Pedreira M.; Sorgini Peterlini M. A.; Azzalis L. A.; Campos Junqueira V. B.; Fonseca F. L. A. The Use of Vancomycin with Its Therapeutic and Adverse Effects: A Review. Eur. Rev. Med. Pharmacol. Sci. 2015, 19, 694–700. [PubMed] [Google Scholar]
- Brooks B. D.; Brooks A. E. Therapeutic Strategies to Combat Antibiotic Resistance. Adv. Drug Delivery Rev. 2014, 78, 14–27. 10.1016/j.addr.2014.10.027. [DOI] [PubMed] [Google Scholar]
- Ren C.; Wang H.; Zhang X.; Ding D.; Wang L.; Yang Z. Interfacial Self-Assembly Leads to Formation of Fluorescent Nanoparticles for Simultaneous Bacterial Detection and Inhibition. Chem. Commun. 2014, 50, 3473–3475. 10.1039/C3CC48807A. [DOI] [PubMed] [Google Scholar]
- Cowger T. A.; Yang Y.; Rink D. E.; Todd T.; Chen H.; Shen Y.; Yan Y.; Xie J. Protein-Adsorbed Magnetic-Nanoparticle-Mediated Assay for Rapid Detection of Bacterial Antibiotic Resistance. Bioconjugate Chem. 2017, 28, 890–896. 10.1021/acs.bioconjchem.7b00016. [DOI] [PubMed] [Google Scholar]
- Joseph R.; Naugolny A.; Feldman M.; Herzog I. M.; Fridman M.; Cohen Y. Cationic Pillararenes Potently Inhibit Biofilm Formation without Affecting Bacterial Growth and Viability. J. Am. Chem. Soc. 2016, 138, 754–757. 10.1021/jacs.5b11834. [DOI] [PubMed] [Google Scholar]
- Czuban M.; Srinivasan S.; Yee N. A.; Agustin E.; Koliszak A.; Miller E.; Khan I.; Quinones I.; Noory H.; Motola C.; Volkmer R.; Di Luca M.; Trampuz A.; Royzen M.; Oneto J. M. M. Bio-Orthogonal Chemistry and Reloadable Biomaterial Enable Local Activation of Antibiotic Prodrugs and Enhance Treatments against Staphylococcus aureus Infections. ACS Cent. Sci. 2018, 4, 1624–1632. 10.1021/acscentsci.8b00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebeaux D.; Chauhan A.; Rendueles O.; Beloin C. From in vitro to in vivo Models of Bacterial Biofilm-Related Infections. Pathogens 2013, 2, 288–356. 10.3390/pathogens2020288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlmayer F.; Jaeger T.; Jenal U.; Fussenegger M. Quorum-Quenching Human Designer Cells for Closed-Loop Control of Pseudomonas aeruginosa Biofilms. Nano Lett. 2017, 17, 5043–5050. 10.1021/acs.nanolett.7b02270. [DOI] [PubMed] [Google Scholar]
- Davies D. Understanding Biofilm Resistance to Antibacterial Agents. Nat. Rev. Drug Discovery 2003, 2, 114–122. 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- McDougald D.; Rice S. A.; Barraud N.; Steinberg P. D.; Kjelleberg S. Should We Stay or should We Go: Mechanisms and Ecological Consequences for Biofilm Dispersal. Nat. Rev. Microbiol. 2012, 10, 39–50. 10.1038/nrmicro2695. [DOI] [PubMed] [Google Scholar]
- Sahoo P. K.; Janissen R.; Monteiro M. P.; Cavalli A.; Murillo D. M.; Merfa M. V.; Cesar C. L.; Carvalho H. F.; de Souza A. A.; Bakkers E. P. A. M.; Cotta M. A. Nanowire Arrays as Cell Force Sensors to Investigate Adhesin-Enhanced Holdfast of Single Cell Bacteria and Biofilm Stability. Nano Lett. 2016, 16, 4656–4664. 10.1021/acs.nanolett.6b01998. [DOI] [PubMed] [Google Scholar]
- Flemming H. C.; Wingender J.; Szewzyk U.; Steinberg P.; Rice S. A.; Kjelleberg S. Biofilms: An Emergent Form of Bacterial Life. Nat. Rev. Microbiol. 2016, 14, 563–575. 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- Lee H. S.; Dastgheyb S. S.; Hickok N. J.; Eckmann D. M.; Composto R. J. Targeted Release of Tobramycin from a pH-Responsive Grafted Bilayer Challenged with S. aureus. Biomacromolecules 2015, 16, 650–659. 10.1021/bm501751v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilton M.; Charron-Mazenod L.; Moore R.; Lewenza S. Extracellular DNA Acidifies Biofilms and Induces Aminoglycoside Resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2016, 60, 544–553. 10.1128/AAC.01650-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siala W.; Mingeot-Leclercq M. P.; Tulkens P. M.; Hallin M.; Denis O.; Van Bambeke F. Comparison of the Antibiotic Activities of Daptomycin, Vancomycin, and the Investigational Fluoroquinolone Delafloxacin against Biofilms from Staphylococcus aureus Clinical Isolates. Antimicrob. Agents Chemother. 2014, 58, 6385–6639. 10.1128/AAC.03482-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings L. K.; Storek K. M.; Ledvina H. E.; Coulon C.; Marmont L. S.; Sadovskaya I.; Secor P. R.; Tseng B. S.; Scian M.; Filloux A.; Wozniak D. J.; Howell P. L.; Parsek M. R. Pel is a Cationic Exopolysaccharide that Cross-Links Extracellular DNA in the Pseudomonas aeruginosa Biofilm Matrix. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 11353–11358. 10.1073/pnas.1503058112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A.; Das R.; Tonga G. Y.; Mizuhara T.; Rotello V. M. Charge-Switchable Nanozymes for Bioorthogonal Imaging of Biofilm-Associated Infections. ACS Nano 2018, 12, 89–94. 10.1021/acsnano.7b07496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radovic-Moreno A. F.; Lu T. K.; Puscasu V. A.; Yoon C. J.; Langer R.; Farokhzad O. C. Surface Charge-Switching Polymeric Nanoparticles for Bacterial Cell Wall-Targeted Delivery of Antibiotics. ACS Nano 2012, 6, 4279–4287. 10.1021/nn3008383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H.; Allan R. N.; Howlin R. P.; Stoodley P.; Hall-Stoodley L. Targeting Microbial Biofilms: Current and Prospective Therapeutic Strategies. Nat. Rev. Microbiol. 2017, 15, 740–755. 10.1038/nrmicro.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z. W.; Yan R.; Yi X.; Li J. L.; Rao J. M.; Guo Z. Q.; Yang Y. M.; Li W. F.; Li Y. Q.; Chen C. Y. Bacteria-Activated Theranostic Nanoprobes against Methicillin-Resistant Staphylococcus aureus Infection. ACS Nano 2017, 11, 4428–4438. 10.1021/acsnano.7b00041. [DOI] [PubMed] [Google Scholar]
- Pelgrift R. Y.; Friedman A. J. Nanotechnology as A Therapeutic Tool to Combat Microbial Resistance. Adv. Drug Delivery Rev. 2013, 65, 1803–1815. 10.1016/j.addr.2013.07.011. [DOI] [PubMed] [Google Scholar]
- Lemire J. A.; Harrison J. J.; Turner R. J. Antimicrobial Activity of Metals: Mechanisms, Molecular Targets and Applications. Nat. Rev. Microbiol. 2013, 11, 371–384. 10.1038/nrmicro3028. [DOI] [PubMed] [Google Scholar]
- Xin X. X.; Li P. F.; Zhu Y. Y.; Shi L. G.; Yuan J.; Shen J. Mussel-Inspired Surface Functionalization of PET with Zwitterions and Silver Nanoparticles for the Dual-Enhanced Antifouling and Antibacterial Properties. Langmuir 2019, 35, 1788–1797. 10.1021/acs.langmuir.8b01603. [DOI] [PubMed] [Google Scholar]
- Eshed M.; Lellouche J.; Matalon S.; Gedanken A.; Banin E. Sonochemical Coatings of ZnO and CuO Nanoparticles Inhibit Streptococcus mutans Biofilm Formation on Teeth Model. Langmuir 2012, 28, 12288–12295. 10.1021/la301432a. [DOI] [PubMed] [Google Scholar]
- Naha P. C.; Liu Y.; Hwang G.; Huang Y.; Gubara S.; Jonnakuti V.; Simon-Soro A.; Kim D.; Gao L.; Koo H.; Cormode D. P. Dextran-Coated Iron Oxide Nanoparticles as Biomimetic Catalysts for Localized and pH-Activated Biofilm Disruption. ACS Nano 2019, 13, 4960. 10.1021/acsnano.8b08702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X.; Radovic-Moreno A. F.; Wu J.; Langer R.; Shi J. J. Nanomedicine in the Management of Microbial Infection - Overview and Perspectives. Nano Today 2014, 9, 478–498. 10.1016/j.nantod.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.; Xie X.; Zhao W. T.; Liu N.; Maraccini P. A.; Sassoubre L. M.; Boehm A. B.; Cui Y. Conducting Nanosponge Electroporation for Affordable and High-Efficiency Disinfection of Bacteria and Viruses in Water. Nano Lett. 2013, 13, 4288–4293. 10.1021/nl402053z. [DOI] [PubMed] [Google Scholar]
- Wijnhoven S. W. P.; Peijnenburg W. J. G. M.; Herberts C. A.; Hagens W. I.; Oomen A. G.; Heugens E. H. W.; Roszek B.; Bisschops J.; Gosens I.; Van de Meent D.; Dekkers S.; De Jong W. H.; Van Zijverden M.; Sips A. J. A. M.; Geertsma R. E. Nano-Silver - A Review of Available Data and Knowledge Gaps in Human and Environmental Risk Assessment. Nanotoxicology 2009, 3, 109–138. 10.1080/17435390902725914. [DOI] [Google Scholar]
- Chernousova S.; Epple M. Silver as Antibacterial Agent: Ion, Nanoparticle, and Metal. Angew. Chem. Int. Ed. 2013, 52, 1636–1653. 10.1002/anie.201205923. [DOI] [PubMed] [Google Scholar]
- Pal S.; Tak Y. K.; Song J. M. Does the Antibacterial Activity of Silver Nanoparticles Depend on the Shape of the Nanoparticle? A Study of the Gram-Negative Bacterium Escherichia coli. Appl. Environ. Microb. 2007, 73, 1712–1720. 10.1128/AEM.02218-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q. L.; Wu J.; Chen G. Q.; Cui F. Z.; Kim T. N.; Kim J. O. A Mechanistic Study of the Antibacterial Effect of Silver Ions on Escherichia coli and Staphylococcus aureus. J. Biomed. Mater. Res. 2000, 52, 662–668. . [DOI] [PubMed] [Google Scholar]
- Liu J. Y.; Hurt R. H. Ion Release Kinetics and Particle Persistence in Aqueous Nano-Silver Colloids. Environ. Sci. Technol. 2010, 44, 2169–2175. 10.1021/es9035557. [DOI] [PubMed] [Google Scholar]
- Peretyazhko T. S.; Zhang Q. B.; Colvin V. L. Size-Controlled Dissolution of Silver Nanoparticles at Neutral and Acidic pH Conditions: Kinetics and Size Changes. Environ. Sci. Technol. 2014, 48, 11954–11961. 10.1021/es5023202. [DOI] [PubMed] [Google Scholar]
- Kim B. H.; Hackett M. J.; Park J.; Hyeon T. Synthesis, Characterization, and Application of Ultrasmall Nanoparticles. Chem. Mater. 2014, 26, 59–71. 10.1021/cm402225z. [DOI] [Google Scholar]
- Zhang W.; Yao Y.; Sullivan N.; Chen Y. S. Modeling the Primary Size Effects of Citrate-Coated Silver Nanoparticles on Their Ion Release Kinetics. Environ. Sci. Technol. 2011, 45, 4422–4428. 10.1021/es104205a. [DOI] [PubMed] [Google Scholar]
- Lok C. N.; Zou T. T.; Zhang J. J.; Lin I. W. S.; Che C. M. Controlled-Release Systems for Metal-Based Nanomedicine: Encapsulated/Self-Assembled Nanoparticles of Anticancer Gold(III)/Platinum(II) Complexes and Antimicrobial Silver Nanoparticles. Adv. Mater. 2014, 26, 5550–5557. 10.1002/adma.201305617. [DOI] [PubMed] [Google Scholar]
- Xie X. Z.; Mao C. Y.; Liu X. M.; Tan L.; Cui Z. D.; Yang X. J.; Zhu S. L.; Li Z. Y.; Yuan X. B.; Zheng Y. F.; Yeung K. W. K.; Chu P. K.; Wu S. L. Tuning the Bandgap of Photo-Sensitive Polydopamine/Ag3PO4/Graphene Oxide Coating for Rapid, Noninvasive Disinfection of Implants. ACS Cent. Sci. 2018, 4, 724–738. 10.1021/acscentsci.8b00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durmus N. G.; Webster T. J. Eradicating Antibiotic-Resistant Biofilms with Silver-Conjugated Superparamagnetic Iron Oxide Nanoparticles. Adv. Healthcare Mater. 2013, 2, 165–171. 10.1002/adhm.201200215. [DOI] [PubMed] [Google Scholar]
- Chaw K. C.; Manimaran M.; Tay F. E. Role of Silver Ions in Destabilization of Intermolecular Adhesion Forces Measured by Atomic Force Microscopy in Staphylococcus epidermidis Biofilms. Antimicrob. Agents Chemother. 2005, 49, 4853–4859. 10.1128/AAC.49.12.4853-4859.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudi M.; Serpooshan V. Silver-Coated Engineered Magnetic Nanoparticles are Promising for the Success in the Fight Against Antibacterial Resistance Threat. ACS Nano 2012, 6, 2656–2664. 10.1021/nn300042m. [DOI] [PubMed] [Google Scholar]
- Pauksch L.; Hartmann S.; Rohnke M.; Szalay G.; Alt V.; Schnettler R.; Lips K. S. Biocompatibility of Silver Nanoparticles and Silver Ions in Primary Human Mesenchymal Stem Cells and Osteoblasts. Acta Biomater. 2014, 10, 439–449. 10.1016/j.actbio.2013.09.037. [DOI] [PubMed] [Google Scholar]
- Nor Y. A.; Niu Y. T.; Karmakar S.; Zhou L.; Xu C.; Zhang J.; Zhang H. W.; Yu M. H.; Mahony D.; Mitter N.; Cooper M. A.; Yu C. Z. Shaping Nanoparticles with Hydrophilic Compositions and Hydrophobic Properties as Nanocarriers for Antibiotic Delivery. ACS Cent. Sci. 2015, 1, 328–334. 10.1021/acscentsci.5b00199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.; Huang P.; Chen X. Y. Hierarchical Targeting Strategy for Enhanced Tumor Tissue Accumulation/Retention and Cellular Internalization. Adv. Mater. 2016, 28, 7340–7364. 10.1002/adma.201601498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R.; Wei T.; Goldberg H.; Wang W. P.; Cullion K.; Kohane D. S. Getting Drugs across Biological Barriers. Adv. Mater. 2017, 29, 1606596. 10.1002/adma.201606596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q.; Zhou Z.; Qiu N.; Shen Y. Rational Design of Cancer Nanomedicine: Nanoproperty Integration and Synchronization. Adv. Mater. 2017, 29, 1606628. 10.1002/adma.201606628. [DOI] [PubMed] [Google Scholar]
- Chen J. J.; Ding J. X.; Wang Y. C.; Cheng J. J.; Ji S. X.; Zhuang X. L.; Chen X. S. Sequentially Responsive Shell-Stacked Nanoparticles for Deep Penetration into Solid Tumors. Adv. Mater. 2017, 29, 1701170. 10.1002/adma.201701170. [DOI] [PubMed] [Google Scholar]
- Qi G. B.; Zhang D.; Liu F. H.; Qiao Z. Y.; Wang H. An “On-Site Transformation” Strategy for Treatment of Bacterial Infection. Adv. Mater. 2017, 29, 1703461. 10.1002/adma.201703461. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Busscher H. J.; Zhao B.; Li Y.; Zhang Z.; van der Mei H. C.; Ren Y.; Shi L. Surface-Adaptive, Antimicrobially Loaded, Micellar Nanocarriers with Enhanced Penetration and Killing Efficiency in Staphylococcal Biofilms. ACS Nano 2016, 10, 4779–4789. 10.1021/acsnano.6b01370. [DOI] [PubMed] [Google Scholar]
- Hu D.; Li H.; Wang B.; Ye Z.; Lei W.; Jia F.; Jin Q.; Ren K. F.; Ji J. Surface-Adaptive Gold Nanoparticles with Effective Adherence and Enhanced Photothermal Ablation of Methicillin-Resistant Staphylococcus aureus Biofilm. ACS Nano 2017, 11, 9330–9339. 10.1021/acsnano.7b04731. [DOI] [PubMed] [Google Scholar]
- Horev B.; Klein M. I.; Hwang G.; Li Y.; Kim D.; Koo H.; Benoit D. S. W. pH-Activated Nanoparticles for Controlled Topical Delivery of Farnesol To Disrupt Oral Biofilm Virulence. ACS Nano 2015, 9, 2390–2404. 10.1021/nn507170s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri K.; Yepes L. R.; Duncan B.; Parameswaran P. K.; Yan B.; Jiang Y.; Bilska M.; Moyano D. F.; Thompson M.; Rotello V. M.; Prakash Y. S. Targeting Bacterial Biofilms via Surface Engineering of Gold Nanoparticles. RSC Adv. 2015, 5, 105551–105559. 10.1039/C5RA16305F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.; Wang S.; Hu X.; Li F.; Ling D. Controlled Synthesis and Assembly of Ultra-Small Nanoclusters for Biomedical Applications. Biomater. Sci. 2019, 7, 480–489. 10.1039/C8BM01200H. [DOI] [PubMed] [Google Scholar]
- Hu X.; Li F.; Wang S.; Xia F.; Ling D. Biological Stimulus-Driven Assembly/Disassembly of Functional Nanoparticles for Targeted Delivery, Controlled Activation, and Bioelimination. Adv. Healthcare Mater. 2018, 7, 1800359. 10.1002/adhm.201800359. [DOI] [PubMed] [Google Scholar]
- Li F.; Lu J.; Kong X.; Hyeon T.; Ling D. Dynamic Nanoparticle Assemblies for Biomedical Applications. Adv. Mater. 2017, 29, 1605897. 10.1002/adma.201605897. [DOI] [PubMed] [Google Scholar]
- Wang T.; LaMontagne D.; Lynch J.; Zhuang J. Q.; Cao Y. C. Colloidal Superparticles from Nanoparticle Assembly. Chem. Soc. Rev. 2013, 42, 2804–2823. 10.1039/C2CS35318K. [DOI] [PubMed] [Google Scholar]
- Stolarczyk J. K.; Deak A.; Brougham D. F. Nanoparticle Clusters: Assembly and Control Over Internal Order, Current Capabilities, and Future Potential. Adv. Mater. 2016, 28, 5400–5424. 10.1002/adma.201505350. [DOI] [PubMed] [Google Scholar]
- Yi C. L.; Zhang S. Y.; Webb K. T.; Nie Z. H. Anisotropic Self-Assembly of Hairy Inorganic Nanoparticles. Acc. Chem. Res. 2017, 50, 12–21. 10.1021/acs.accounts.6b00343. [DOI] [PubMed] [Google Scholar]
- Cai Q.; Fei Y.; An H. W.; Zhao X. X.; Ma Y.; Cong Y.; Hu L. M.; Li L. L.; Wang H. Macrophage-Instructed Intracellular Staphylococcus aureus Killing by Targeting Photodynamic Dimers. ACS Appl. Mater. Interfaces 2018, 10, 9197–9202. 10.1021/acsami.7b19056. [DOI] [PubMed] [Google Scholar]
- Li L. L.; Ma H. L.; Qi G. B.; Zhang D.; Yu F. Q.; Hu Z. Y.; Wang H. Pathological-Condition-Driven Construction of Supramolecular Nanoassemblies for Bacterial Infection Detection. Adv. Mater. 2016, 28, 254–262. 10.1002/adma.201503437. [DOI] [PubMed] [Google Scholar]
- Cai Q.; Fei Y.; Hu L. M.; Huang Z. J.; Li L. L.; Wang H. Chemotaxis-Instructed Intracellular Staphylococcus aureus Infection Detection by a Targeting and Self-Assembly Signal-Enhanced Photoacoustic Probe. Nano Lett. 2018, 18, 6229–6236. 10.1021/acs.nanolett.8b02286. [DOI] [PubMed] [Google Scholar]
- Li F.; Liang Z.; Liu J.; Sun J.; Hu X.; Zhao M.; Liu J.; Bai R.; Kim D.; Sun X.; Hyeon T.; Ling D.. Dynamically Reversible Iron Oxide Nanoparticle Assemblies for Targeted Amplification of T1-Weighted Magnetic Resonance Imaging of Tumors. Nano Lett. 2019, 10.1021/acs.nanolett.8b04411. [DOI] [PubMed] [Google Scholar]
- Li F.; Du Y.; Liu J.; Sun H.; Wang J.; Li R.; Kim D.; Hyeon T.; Ling D. Responsive Assembly of Upconversion Nanoparticles for pH-Activated and Near-Infrared-Triggered Photodynamic Therapy of Deep Tumors. Adv. Mater. 2018, 30, 1802808. 10.1002/adma.201802808. [DOI] [PubMed] [Google Scholar]
- Lu J. X.; Sun J. H.; Li F. Y.; Wang J.; Liu J. N.; Kim D.; Fan C. H.; Hyeon T.; Ling D. Highly Sensitive Diagnosis of Small Hepatocellular Carcinoma Using pH-Responsive Iron Oxide Nanocluster Assemblies. J. Am. Chem. Soc. 2018, 140, 10071–10074. 10.1021/jacs.8b04169. [DOI] [PubMed] [Google Scholar]
- Hu X.; Sun J. H.; Li F. Y.; Li R. Q.; Wu J. H.; He J.; Wang N.; Liu J. A.; Wang S. F.; Zhou F.; Sun X. L.; Kim D.; Hyeon T.; Ling D. Renal-Clearable Hollow Bismuth Subcarbonate Nanotubes for Tumor Targeted Computed Tomography Imaging and Chemoradiotherapy. Nano Lett. 2018, 18, 1196–1204. 10.1021/acs.nanolett.7b04741. [DOI] [PubMed] [Google Scholar]
- Ling D.; Park W.; Park S. J.; Lu Y.; Kim K. S.; Hackett M. J.; Kim B. H.; Yim H.; Jeon Y. S.; Na K.; Hyeon T. Multifunctional Tumor pH-Sensitive Self-Assembled Nanoparticles for Bimodal Imaging and Treatment of Resistant Heterogeneous Tumors. J. Am. Chem. Soc. 2014, 136, 5647–5655. 10.1021/ja4108287. [DOI] [PubMed] [Google Scholar]
- Ling D.; Xia H.; Park W.; Hackett M. J.; Song C.; Na K.; Hui K. M.; Hyeon T. pH-Sensitive Nanoformulated Triptolide as a Targeted Therapeutic Strategy for Hepatocellular Carcinoma. ACS Nano 2014, 8, 8027–8039. 10.1021/nn502074x. [DOI] [PubMed] [Google Scholar]
- Li X. N.; Yeh Y. C.; Giri K.; Mout R.; Landis R. F.; Prakash Y. S.; Rotello V. M. Control of Nanoparticle Penetration into Biofilms through Surface Design. Chem. Commun. 2015, 51, 282–285. 10.1039/C4CC07737G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.; Kwon S. G.; Jun S. W.; Kim B. H.; Hyeon T. Large-Scale Synthesis of Ultra-Small-Sized Silver Nanoparticles. ChemPhysChem 2012, 13, 2540–2543. 10.1002/cphc.201101035. [DOI] [PubMed] [Google Scholar]
- Tong S.; Hou S. J.; Ren B. B.; Zheng Z. L.; Bao G. Self-Assembly of Phospholipid-PEG Coating on Nanoparticles through Dual Solvent Exchange. Nano Lett. 2011, 11, 3720–3726. 10.1021/nl201978c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.; Feng Y. G.; Wang X.; Li T. C.; Zhang J. Y.; Qian D. J. Silver Nanoparticles Capped by Oleylamine: Formation, Growth, and Self-Organization. Langmuir 2007, 23, 5296–5304. 10.1021/la700553d. [DOI] [PubMed] [Google Scholar]
- Kim D. H.; Larson A. C. Deoxycholate Bile Acid Directed Synthesis of Branched Au Nanostructures for Near Infrared Photothermal Ablation. Biomaterials 2015, 56, 154–164. 10.1016/j.biomaterials.2015.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam J.; Won N.; Jin H.; Chung H.; Kim S. pH-Induced Aggregation of Gold Nanoparticles for Photothermal Cancer Therapy. J. Am. Chem. Soc. 2009, 131, 13639–13645. 10.1021/ja902062j. [DOI] [PubMed] [Google Scholar]
- Mahmoud M. A.; Tabor C. E.; El-Sayed M. A. Surface-Enhanced Raman Scattering Enhancement by Aggregated Silver Nanocube Monolayers Assembled by the Langmuir-Blodgett Technique at Different Surface Pressures. J. Phys. Chem. C 2009, 113, 5493–5501. 10.1021/jp900648r. [DOI] [Google Scholar]
- Jung S.; Nam J.; Hwang S.; Park J.; Hur J.; Im K.; Park N.; Kim S. Theragnostic pH-Sensitive Gold Nanoparticles for the Selective Surface Enhanced Raman Scattering and Photothermal Cancer Therapy. Anal. Chem. 2013, 85, 7674–7681. 10.1021/ac401390m. [DOI] [PubMed] [Google Scholar]
- Hussain S.; Joo J.; Kang J.; Kim B.; Braun G. B.; She Z. G.; Kim D.; Mann A. P.; Molder T.; Teesalu T.; Carnazza S.; Guglielmino S.; Sailor M. J.; Ruoslahti E. Antibiotic-Loaded Nanoparticles Targeted to the Site of Infection Enhance Antibacterial Efficacy. Nat. Biomed. Eng. 2018, 2, 95–103. 10.1038/s41551-017-0187-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boda S. K.; Broda J.; Schiefer F.; Weber-Heynemann J.; Hoss M.; Simon U.; Basu B.; Jahnen-Dechent W. Cytotoxicity of Ultrasmall Gold Nanoparticles on Planktonic and Biofilm Encapsulated Gram-Positive Staphylococci. Small 2015, 11, 3183–3193. 10.1002/smll.201403014. [DOI] [PubMed] [Google Scholar]
- Liebeke M.; Dorries K.; Zuhlke D.; Bernhardt J.; Fuchs S.; Pane-Farre J.; Engelmann S.; Volker U.; Bode R.; Dandekar T.; Lindequist U.; Hecker M.; Lalk M. A Metabolomics and Proteomics Study of the Adaptation of Staphylococcus aureus to Glucose Starvation. Mol. BioSyst. 2011, 7, 1241–1253. 10.1039/c0mb00315h. [DOI] [PubMed] [Google Scholar]
- Morones-Ramirez J. R.; Winkler J. A.; Spina C. S.; Collins J. J. Silver Enhances Antibiotic Activity against Gram-Negative Bacteria. Sci. Transl. Med. 2013, 5, 190ra81. 10.1126/scitranslmed.3006276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming H. C.; Wingender J. The Biofilm Matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- Chen Z. W.; Wang Z. Z.; Ren J. S.; Qu X. G. Enzyme Mimicry for Combating Bacteria and Biofilms. Acc. Chem. Res. 2018, 51, 789–799. 10.1021/acs.accounts.8b00011. [DOI] [PubMed] [Google Scholar]
- Gupta A.; Mumtaz S.; Li C. H.; Hussain I.; Rotello V. M. Combatting Antibiotic-Resistant Bacteria using Nanomaterials. Chem. Soc. Rev. 2019, 48, 415–427. 10.1039/C7CS00748E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon B. P.; Nakayasu E. S.; Fleck L. E.; LaFleur M. D.; Isabella V. M.; Coleman K.; Leonard S. N.; Smith R. D.; Adkins J. N.; Lewis K. Activated ClpP Kills Persisters and Eradicates a Chronic Biofilm Infection. Nature 2013, 503, 365–370. 10.1038/nature12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang X.; Xiao Q.; Chen Y.; Ren E.; Lian L.; Zhang Y.; Gao H.; Wang X.; Leung W.; Chen X.; Liu G.; Xu C. Bacteria-Responsive Nanoliposomes as Smart Sonotheranostics for Multidrug Resistant Bacterial Infections. ACS Nano 2019, 13, 2427–2438. 10.1021/acsnano.8b09336. [DOI] [PubMed] [Google Scholar]
- Herrero L. J.; Nelson M.; Srikiatkhachorn A.; Gu R.; Anantapreecha S.; Fingerle-Rowson G.; Bucala R.; Morand E.; Santos L. L.; Mahalingam S. Critical Role for Macrophage Migration Inhibitory Factor (MIF) in Ross River Virus-Induced Arthritis and Myositis. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 12048–12053. 10.1073/pnas.1101089108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.