Abstract
Background:
The fecal microbiome is associated with prostate cancer risk factors (obesity, inflammation) and can metabolize and produce various products that may influence cancer but have yet to be defined in prostate cancer.
Objective:
To investigate gut bacterial diversity, identify specific metabolic pathways associated with disease, and develop a microbiome risk profile for prostate cancer.
Design, setting, and participants:
After prospective collection of 133 rectal swab samples 2 wk before the transrectal prostate biopsy, we perform 16S rRNA amplicon sequencing on 105 samples (64 with cancer, 41 without cancer). Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) was applied to infer functional categories associated with taxonomic composition. The p values were adjusted using the false discovery rate. The α- and β-diversity analyses were performed using QIIME. The Mann-Whitney U test was employed to evaluate the statistical significance of β-diversity distances within and between groups of interest, and least absolute shrinkage and selection operator (LASSO) regression analysis was used to determine pathway significance.
Outcome measurements and statistical analysis:
The detection of prostate cancer on transrectal prostate needle biopsy and 16s microbiome profile.
Results and limitations:
We identified significant associations between total community composition and cancer/non-cancer status (Bray-Curtis distance metric, p < 0.01). We identified significant differences in enrichments of Bacteroides and Streptococcus species in cancer (all p < 0.04). Folate (LDA 3.8) and arginine (LDA 4.1) were the most significantly altered pathways. We formed a novel microbiome-derived risk factor for prostate cancer based on 10 aberrant metabolic pathways (area under curve = 0.64, p = 0.02).
Conclusions:
Microbiome analyses on men undergoing prostate biopsy noted mostly similar bacterial species diversity among men diagnosed with and without prostate cancer. The microbiome may have subtle influences on prostate cancer but are likely patient-specific and would require paired analysis and precise manipulation, such as improvement of natural bacterial folate production.
Patient summary:
Microbiome evaluation may provide patients with personalized data regarding the presence or absence of particular bacteria that have metabolic functions and implications regarding prostate cancer risk. The study provides a basis to investigate the manipulation of aberrant microbiomes to reduce prostate cancer risk.
Keywords: Microbiome, Biomarker, Prostate cancer, Folate, Biotin, B vitamins
1. Introduction
Each year, there are approximately 1 million prostate cancer diagnoses worldwide and attributed to genetics, race, age, and lifestyle factors (diet/obesity) [1]. The intestinal microbiome has been implicated in lifestyle factors and may impact the availability of various micronutrients on cancer risk [2]. Although some publications report on the urinary microbiome, we focus on the systemic effects from the fecal microbiome to provide insight regarding metabolic pathways to manipulate in the diet for prostate cancer [3-5].
Intestinal bacteria contain a wealth of information due to their diverse properties, including the ability to produce influential products on cancer development and progression. A reduced diversity profile in the intestinal microbiome can lead to overgrowth of bacteria causing mild systemic inflammation called endotoxemia, contributing to an overall inflammatory state promoting neoplasia in many organ sites [6,7]. A recent case-control study (n = 20) did suggest differences in fecal microbiome in those men with cancer compared with benign prostate hypertrophy. They found a higher abundance of Bacteroides massiliensis in prostate cancer and higher Faecalibacterium prausnitzii and Eubacterium rectalie in controls and suggested an influence on micronutrients [8]. Groups of other bacteria could also play a role in prostate cancer development by producing and manipulating various proteins, vitamins, minerals, and other substances.
We anticipate differences in those men diagnosed with cancer compared with those without cancer and hypothesize that there could be a particular set of bacteria to target in probiotics for future studies regarding prostate cancer prevention. Herein, we investigate bacterial diversity in patients with or without prostate cancer. Additionally, we examine particular bacterial species differences in the fecal microbiome and their related metabolic pathways associated with prostate cancer.
2. Patients and methods
2.1. Patients
We prospectively collected rectal swab samples at least 2 wk prior to a transrectal prostate biopsy. We excluded patients who had any antibiotic therapy in the last 6 mo. All samples were analyzed to identify cancer versus no cancer; however, for the association of future risk of prostate cancer, we excluded men with prostate-specific antigen (PSA) higher than 20.
2.2. Rectal swab collection
The rectal swab technique involved the urology provider by using sterile Medline E-Z lubricating jelly and placing the swab approximately 5.08 cm into the rectum and turning the swab 360 degrees. Once the collection was complete, the rectal swab was removed and examined for stool. The physician re-inserted the swab if there was no material collected. Next, the physician placed the swab in a 15-ml sterile centrifuge tube containing 1 ml of phosphate-buffered saline (PBS). We stored the swab specimen at 4 °C during transport to the laboratory within 4 h of collection. We saved the swab and PBS material at −20 °C and then performed DNA isolation.
2.3. Microbiome analysis
Raw paired-end 16S rRNA reads (V1V2 region) were merged into consensus fragments by FLASH [9] and subsequently filtered for quality (target error rate <0.5%) and length (minimum 200 bp) using Trimmomatic [10] and QIIME [11,12]. Spurious hits to the PhiX control genome were identified using BLASTN and removed. Passing sequences were trimmed of primers, evaluated for chimeras with UCLUST (de novo mode) [13], and screened for human-associated contaminant using Bowtie2 [14], followed by a more sensitive BLASTN search against the GreenGenes 16S rRNA database [15]. Chloroplast and mitochondrial contaminants were detected and filtered using the Ribosomal Database Project classifier [16] with a confidence threshold of 80%. High-quality 16S rRNA sequences were assigned to a high-resolution taxonomic lineage using Resphera Insight [17-21]. Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) analysis was performed to identify Kyoto Encyclopedia of Genes and Genomes (KEGG) metabolic pathways potentially affected by groups of bacteria. PICRUSt is a bioinformatics software package designed to predict metagenome functional content from marker gene (eg, 16S rRNA) surveys and full genomes. PICRUSt was applied to infer functional categories associated with taxonomic composition [22]. Differential abundance analysis utilized the nonparametric permutation difference test for α diversity and PICRUSt values or the negative binomial test (DESeq [23]) for taxonomic count data. Then, p values were adjusted using the false discovery rate [24]. The α- and β-diversity analyses were performed using QIIME. The Mann-Whitney U test was employed to evaluate the statistical significance of β-diversity distances within and between groups of interest.
2.4. Statistical analysis
We utilized t tests for continuous variables and chi-square tests for categorical variables. Using the PICRUSt data, we compared continuous variables for microbial metabolic function sets in cancer versus non-cancer groups. Univariate and binary logistic regression analyses were performed on standard predictor variables of cancer to include in the microbiome metabolic profile. An area under the receiver operator curve (AUC) was used to compare the predictive ability of the microbiome compared with other risk factors. An AUC of 1.0 is a perfect prediction, and an AUC of 0.5 is equivalent to a random, uncorrelated predictor.
2.5. Development of the microbiome profile
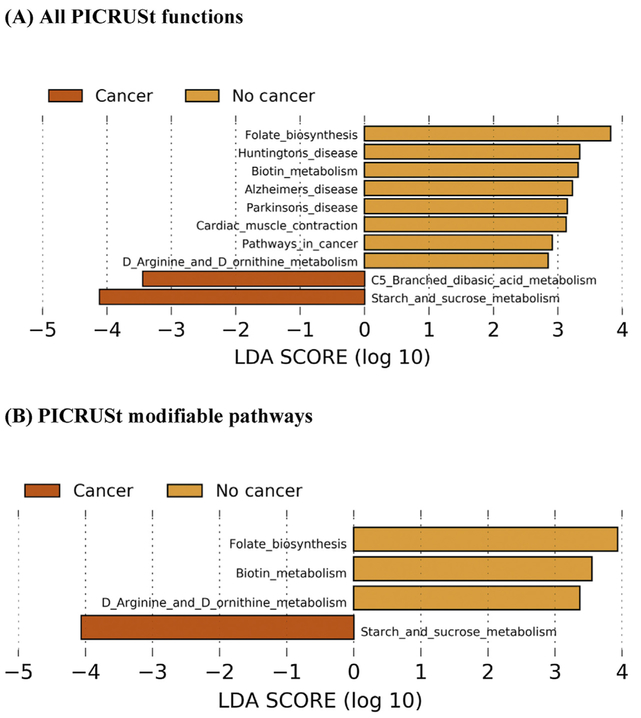
Due to a large number of possible individual patterns from this analysis, we identified a list of KEGG biologic pathways to use in our overall microbiome profile targeting specific biological pathways. We utilized least absolute shrinkage and selection operator (LASSO) analysis as a form of linear regression modeling to limit (shrink) the absolute value toward the mean and to perform variable selection when there is a high likelihood of multicollinearity. We used a LASSO regression analysis to identify the pathways most likely to differentiate between prostate cancer and no cancer disease using a cross-validated model-fitting procedure. Once we identified the microbiome metabolic pathways, we calculated quintiles for each metabolic process derived from PICRUSt values. If the value was positively (or negatively) correlated to cancer, then values in the top (or lowest) quintile (20th percentile) were coded as 1.0, indicating positive, and all other values were coded as 0, indicating null. We compared utilizing only the LASSO regression-derived variables (“LASSO microbiome profile”) to the secondary profile by including the B vitamins and carbohydrate profiles. Additional profiles were identified by LDA effect size (LefSe) analysis (Fig. 3A and 3B) in a biased approach to give preference to those pathways that are modifiable and not human-associated functional categories (such as “cardiac muscle contraction” or “pathways in cancer” identified in univariate analysis). After the selection of functional pathways, we also coded them by quintile to provide the 10 pathway-based overall “microbiome score.”
Fig. 3 -.
LefSe analyses including the top differentially abundant PICRUSt categories. Predicted KEGG functional pathway differences between cancer (dark orange) and no cancer (orange) participants. PICRUSt was used to predict functional potential of microbiomes using 16s rRNA gene sequence data. (A) Indicates all PICRUSt variables and (B) indicates those specifically modifiable factors after removal of all human-associated functional categories. Differentially enriched bacterial functions among groups were identified using LefSe. Pathway differences plot as LDA score (log 10). Bars to the right of zero represent bacterial functions enriched in the microbiome of patients without prostate cancer, and bars to the left of zero represent bacterial functions in those diagnosed with prostate cancer with prostate needle biopsy.
KEGG = Kyoto Encyclopedia of Genes and Genomes; LDA = linear discriminant analysis; LefSe = LDA effect size; PICRUSt = Phylogenetic Investigation of Communities by Reconstruction of Unobserved States.
3. Results
3.1. Demographic data
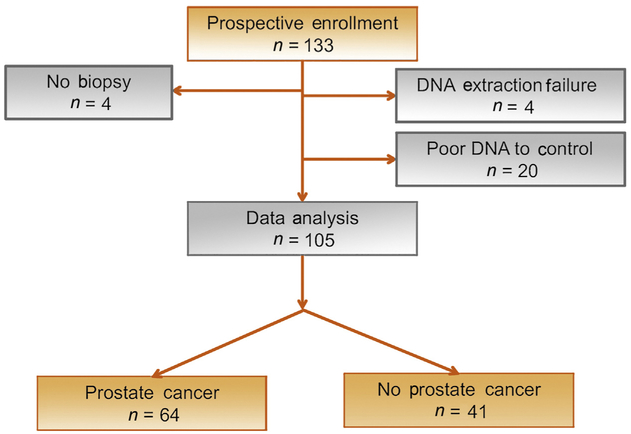
We prospectively collected 133 rectal swab samples at least 2 wk prior to a transrectal prostate biopsy. After excluding patients with any antibiotic therapy in the last 6 mo and low-quality samples, we analyzed 105 samples. To identify the specific target population that could be utilized to alter the microbiome, we were left with 64 men with prostate cancer and 41 without cancer (Fig. 1). Demographics are displayed in Table 1.
Fig. 1 -.
Consort diagram.
Table 1 -.
Univariate and multivariate analyses based on demographics obtained prior to transrectal prostate needle biopsy
| Univariate analysis | Prostate cancer n = 64 n (%) or median (IQR) |
No prostate cancer n = 41 n (%) or median (IQR) |
p value |
|---|---|---|---|
| Age | 66.5 (62–70) | 65 (60–68.5) | 0.1 |
| Body mass index | 29.7 (27.5–34.9) | 28.7 (25.9–31.8) | 0.7 |
| Diabetes mellitus | 19 (18.1) | 14 (13.3) | 0.6 |
| Race | 0.4 | ||
| White | 35 (35) | 28 (28) | |
| African-American | 18 (18.2) | 8 (8.1) | |
| Latino/other | 5 (5.1) | 5 (5.1) | |
| PSA | 6.1 (4.3–8.3) | 6.1 (4.6–8.2) | 0.7 |
| Abnormal prostate examination | 18 (18.4) | 8 (8.4) | 0.2 |
| Family history | 13 (13.4) | 12 (12.4) | 0.4 |
| Lower urinary tract symptoms a | 26 (24.8) | 15 (14.3) | 0.7 |
| Number of negative prostate biopsies | |||
| Active surveillance | 29 (29) | 0 | |
| Gleason score | NA | ||
| Group 1 (3 + 3) | 28 (48.3) | ||
| Group 2 (3 + 4) | 20 (34.5) | ||
| Group 3 (4 + 3) | 6 (10.3) | ||
| Group 4 (4 + 4) | 2 (3.4) | ||
| Group 5 > (4 + 5) | 2 (3.4) | ||
| Number of aberrant microbiome profiles | 2 (0–4) | 1 (0–2) | 0.03 |
| Multivariate analysis b | OR (95% CI) | ||
| PSA | 0.99 (0.89–1.11) | 0.9 | |
| Age | 1.04 (0.98–1.11) | 0.2 | |
| Number of aberrant microbiome profiles | 1.25 (1.01–1.55) | 0.04 | |
CI = confidence interval; IQR = interquartile range; OR = odds ratio; PSA = prostate-specific antigen.
Lower urinary tract symptoms are indicated by the medication report stating five a reductase inhibitors or a blocker use.
We chose age and PSA as the strongest predictors of prostate cancer and forced into the model. This model excludes PSA >20 (n = 99).
3.2. Microbiome results
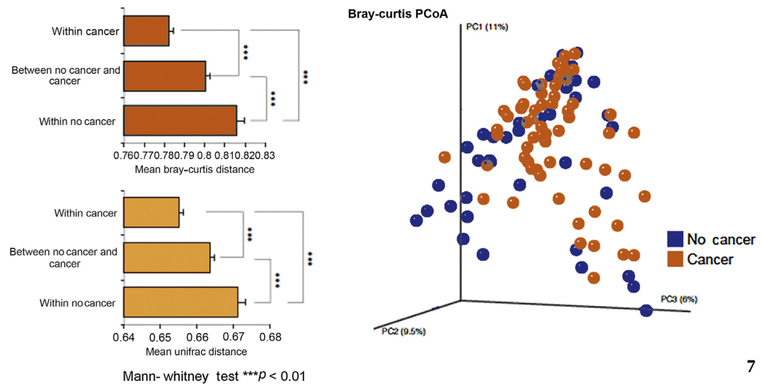
We computed a variety of α-diversity estimators to measure within-sample diversity and compared our two groups using the nonparametric difference test. We did not identify any significant associations between cancer status and α diversity. We computed Bray-Curtis and unweighted Uni-Frac distances to assess β diversity (shared/unique diversity) across samples and by group membership. We also identified significant associations between total community composition and cancer/non-cancer status, which led to associations between total community composition and cancer/non-cancer status (Bray-Curtis distance metric, p < 0.01, Fig. 2). We identified a significant overlap between cancer and non-cancer microbiome profiles using principal component analysis. Additionally, the overall differential abundance analysis indicated that the vast majority of taxa were statistically similar between cancer and non-cancer groups (Supplementary Fig. 1). Bifidobacterium, usually associated with a healthy flora, was infrequent and was not differentially abundant between cancer and non-cancer groups (adj p = 0.68). At the species level, we found significant differences between cancer and non-cancer groups for some well-represented members including enrichments of Bacteroides and Streptococcus species in cancer compared with non-cancer (all p < 0.04). When we examined differentially abundant microbial gene content using PICRUSt analysis, we identified significant differences between cancer group and non-cancer group microbiome profiles (Table 2) and associated LefSe analysis (Fig. 3A and 3B). We performed a correlation plot of the functional PICRUSt variables to include age and PSA and noted that the B vitamins (folate, biotin, and riboflavin) were correlated (Supplementary Fig. 2).
Fig. 2 -.
Microbial diversity. Beta-diversity analysis reveals an altered intestinal microbiome composition associated with prostate cancer. Left: Mean beta-diversity distances (Bray-Curtis/Unweighted UniFrac) within and between groups by cancer status shows significant associations. Patients with prostate cancer are significantly more similar in taxonomic composition compared with the non-cancer group. Right: Principal coordinates analysis plot showing patients with and without cancer (based on Bray-Curtis distances).
PCoA = principal coordinate analysis.
Table 2 -.
Significant mean PICRUSt values comparing prostate cancer to no prostate cancer
| Prostate cancerNo prostate cancer | |||
|---|---|---|---|
| Mean (SD) PICRUSt value |
Mean (SD) PICRUSt value |
p value | |
| Negative association | |||
| Biotin | 1.595 (0.149) | 1.689 (0.209) | 0.01 |
| Folate | 4.138 (0.393) | 4.455 (0.643) | 0.007 |
| Riboflavin | 2.660 (0.252) | 2.827 (0.402) | 0.02 |
| Arginine | 0.021 (0.014) | 0.040 (0.040) | 0.005 |
| Glycan | 0.006 (0.010) | 0.021 (0.041) | 0.03 |
| Alzheimer's disease | 0.491 (0.081) | 0.571 (0.156) | 0.004 |
| Huntington's disease | 0.364 (0.128) | 0.460 (0.209) | 0.01 |
| Positive association | |||
| Carbohydrate (unclassified) | 1.958 (0.322) | 1.756 (0.373) | 0.005 |
| C5 Branched dibasic acid | 3.004 (0.212) | 2.856 (0.364) | 0.02 |
| Starch/sucrose | 12.054 (1.386) | 9.470 (1.765) | 0.03 |
PICRUSt = Phylogenetic Investigation of Communities by Reconstruction of Unobserved States; SD = standard deviation.
3.3. Microbiome score
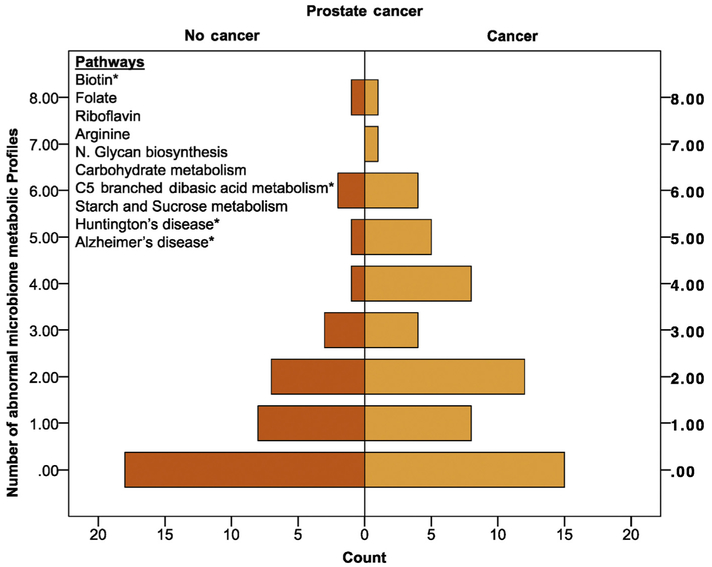
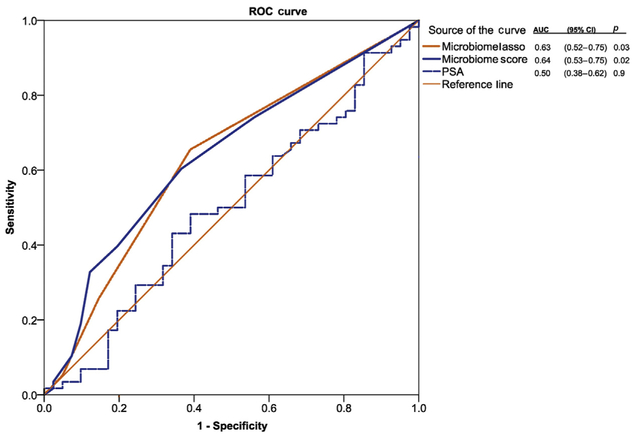
We generated the associated KEGG metabolic pathways as part of the PICRUSt bioinformatics platform. We then performed an unbiased selection using LASSO linear regression analysis and subsequent biased selection using LefSe analysis based on the ability to target the pathway. In LASSO regression analysis, the four pathways identified were not individually significant in logistic regression, but they were predictive in AUC curve analysis (AUC = 0.63; 95% confidence interval [CI] = 0.52–0.75), p = 0.03]. While disease-based pathways such as Alzheimer's and Huntington's disease may perform well for predictive markers, they do not provide the ability for modification of the microbiome. Therefore, using LefSe, we identified six additional metabolic profiles that could be added to the already defined LASSO regression analysis to develop a microbiome score. After identifying 10 microbiome metabolic pathways of interest, we computed the microbiome score (0–10; Fig. 4). A number of aberrant microbiome-associated gut metabolic pathways are associated with cancer compared with no cancer on biopsy (Fig. 4; Kendell’s Tau for ordinal assessment, p = 0.01). We test the microbiome score to predict future risk of prostate cancer; therefore, we excluded six men with a high likelihood of prostate cancer (eg, PSA >20) for the final multivariate statistical analysis because their risk would outweigh most other factors. Additionally, the microbiome score was the only significant factor in a univariate analysis; therefore, we selected PSA and age in the logistic regression model as the most historically influential factors for prostate cancer. Controlling for age and PSA, the microbiome score remained significant (odds ratio [OR] = 1.25 [1.01–1.55]; p = 0.04; Table 1]. The microbiome score also had a higher AUC over the LASSO approach (AUC = 0.64; 95% CI = 0.53–0.75); p = 0.02; Fig. 5).
Fig. 4 -.
Pyramid plot of microbiome-associated gut metabolic pathways (KEGG). Pyramid plot displaying the number of microbiome-associated gut metabolic pathways (KEGG) associated with cancer compared with no cancer on biopsy (Kendell’s Tau for ordinal assessment, p = 0.01). Pyramid plot in which the X axis is the number of cases (individual patients) and the Y axis is the number of aberrant microbiome metabolic profiles. The 10 important microbiome PICRUSt pathways are represented, and the asterisk (*) indicates those important on the LASSO regression analysis.
KEGG = Kyoto Encyclopedia of Genes and Genomes; LASSO = least absolute shrinkage and selection operator; PICRUSt = Phylogenetic Investigation of Communities by Reconstruction of Unobserved States.
Fig. 5 -.
Microbiome score. Receiver operating curve for the presence of prostate cancer on prostate biopsy. We utilized PSA as there were no other known markers that are predictive in this cohort for prostate cancer. We excluded men with a PSA >20 for prediction purposes in this particular analysis. The two microbiome-associated lines are comparing two techniques to identify the most important microbiome KEGG metabolic pathways. The LASSO regression analysis identified four metabolic pathways with a point given for each association (score 0–4). The microbiome profile is a larger number of KEGG pathways associated with prostate cancer (n = 10) including vitamin B pathway and other pathways that could be targeted with probiotics or dietary interventions.
AUC = area under the curve; CI = confidence interval; KEGG = Kyoto Encyclopedia of Genes and Genomes; LASSO = least absolute shrinkage and selection operator; PSA = prostate-specific antigen; ROC = receiver operating curve.
Discussion
We have identified several factors in which the intestinal microbiome could be involved in men diagnosed with prostate cancer:
Microbiome analysis on men undergoing prostate biopsy noted mostly overlapping bacterial communities between those with and without prostate cancer.
Bacteria associated with carbohydrate metabolism pathways were in abundance, and natural B-vitamin production was lacking in patients with prostate cancer.
The identification of a new 10-microbiome metabolic pathway score may provide an additional risk factor for prostate cancer, which is potentially modifiable. We noted overlap in bacterial community analysis in principal component analysis between cancer and non-cancer patients. We found subtle differences within groups of bacteria regarding specific metabolism pathways, which may be informative to individual patients at high risk. Unfortunately, as a microbiome score, the AUC for prostate cancer prediction is suboptimal (AUC = 0.64) despite being statistically significant; therefore, while the findings are of biological interest, the microbiome score or individual bacterial analysis is unlikely to lead to biomarkers of risk.
The intestinal microbiome assists in digestion and provides a source of various bioactive substances such as folate, riboflavin, biotin, arginine, and butyrate [25]. Folate is the most intriguing metabolic pathway according to the research regarding folate and the risk of prostate cancer [26,27]. In the aspirin and folate polyp prevention trial, a secondary analysis revealed that moderate folic acid supplementation resulted in an age-adjusted hazard ratio = 2.63 (95% CI = 1.23–5.65, Wald test p = 0.01) for developing prostate cancer [28]. Interestingly, baseline dietary folate intake and plasma folate in non-supplement users are protective of prostate cancer, which may implicate that supplements may equate to an increased risk of cancer and natural sources of folate may be preventative. Our study noted a higher prevalence of microbiota involved in natural folate production in men without prostate cancer. Therefore, one could potentially encourage increased natural folate production in high-risk men by using probiotics and reducing supplementation [29]. An interaction exists between folate and riboflavin (vitamin B2) because riboflavin is a co-factor of methylenetetrahydrofolate reductase [30]. In the Melbourne Collaborative Cohort Study with more than 40 000 participants, dietary intake of riboflavin and folate seemed to have a U-shaped curve concerning the incidence of prostate cancer [31].
Other exciting pathways include biotin and arginine. Mammalian cells cannot produce the vitamin biotin (vitamin B7) and, therefore, depend on the intestinal microbiota to maintain healthy levels. Again, this protein is not enriched (is missing) in men diagnosed with prostate cancer. Prostate cancer cells deficient in argininosuccinate synthetase are sensitive to arginine deprivation by arginine deiminase in pre-clinical models [32].
The microbiome score is an addition of aberrant microbiome functional profiles associated with prostate cancer. In the LASSO regression analysis, two neurodegenerative pathways (Alzheimer's and Huntington's) were noted to be abnormal in cancer patients. Usually PICRUSt disease-based pathways are not reported; however, in this case, a risk of neurodegenerative diseases has been shown to be inversely related to all cancers including prostate [33]. The pathways are used for their predictive ability as a prostate cancer risk factor rather than their ability to be modified. Therefore, we added the B vitamins that were significant in our analysis that also have a modifiable potential to slightly improve the AUC. We specifically note that folate and riboflavin may have implications for modifiable risk factors. The lack of a biotin–associated microbiome was observed to be one of the most robust indicators associated with prostate cancer in LASSO analysis. As with high levels of other B vitamins, biotin could be harmful if taken in highly concentrated supplements [34].
Our study has limitations. We collected specimens by rectal swab rather than stool collection techniques. Stool provides superior DNA yield at the expense of collection compliance. The rectal swabs were very easy to obtain in our clinic because all men undergo a rectal culture screening protocol for the presence of fluoroquinolone-resistant bacteria. We then use the extra swab for microbiome research. This methodology has a practical advantage of increasing compliance and, thus, can become part of a routine clinic visit at the time of biopsy. We did not use magnetic resonance imaging-targeted biopsy, which may have helped with the location of tumors in patients where we discovered “no cancer.” The no-cancer group did have larger prostates and could lead to sampling error. We could not evaluate a causal inference between folate and prostate cancer as we did not collect plasma B vitamin levels or food frequency questionnaires. Several publications associated inflammation with a benign or malignant disease. Based on a retrospective review of the pathology reports, we documented that prostate inflammation was more common in non-cancer cases (10/41, 24%) compared with prostate cancer cases (5/64, 7.8%; p = 0.018). To provide an appropriate inflammatory assessment, pathologists need to examine each slide for inflammation irrespective of the diagnosis, which was beyond the scope of this project.
5. Conclusions
The fecal microbiome of men undergoing prostate biopsy is similar between cancer and non-cancer groups. The microbiome metabolic pathways provide interesting biological insights; however, it is unlikely to produce a “cancer” microbiome predictive risk profile. The most interesting bacterial metabolic pathways are ones that create natural folate, and other B-vitamins that are more common among those without prostate cancer warrant further investigation.
Supplementary Material
Acknowledgments:
We thank all the patients, physicians, residents, and research coordinators who worked together to accomplish this project.
Funding/Support and role of the sponsor: Roger L. and Laura D. Zeller Charitable Foundation and Los Padres, a San Antonio-based prostate cancer foundation.
Footnotes
Financial disclosures: Michael A. Liss certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: James Robert White is the founder of Resphera Biosciences that performed the microbiome analysis but did not inform the statistical analysis of the biomarker component of the manuscript. Dr. White was paid as a consultant on the microbiome analysis.
Appendix A. Supplementary data
Supplementary material related to this article can be found, in the online version, at https://doi.org/10.1016/j.eururo.2018.06.033.
References
- [1].Wong MC, Goggins WB, Wang HH, et al. Global incidence and mortality for prostate cancer: analysis of temporal patterns and trends in 36 countries. Eur Urol 2016;70:862–74. [DOI] [PubMed] [Google Scholar]
- [2].Amirian ES, Petrosino JF, Ajami NJ, Liu Y, Mims MP, Scheurer ME. Potential role of gastrointestinal microbiota composition in prostate cancer risk. Infect Agent Cancer 2013;8:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Cavarretta I, Ferrarese R, Cazzaniga W, et al. The microbiome of the prostate tumor microenvironment. Eur Urol 2017;72:625–31. [DOI] [PubMed] [Google Scholar]
- [4].Sfanos KS, Yegnasubramanian S, Nelson WG, De Marzo AM. The inflammatory microenvironment and microbiome in prostate cancer development. Nat Rev Urol 2018;15:11–24. [DOI] [PubMed] [Google Scholar]
- [5].Shrestha E, White JR, Yu SH, et al. Profiling the urinary microbiome in men with positive versus negative biopsies for prostate cancer. J Urol 2018;199:161–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hullar MA, Burnett-Hartman AN, Lampe JW. Gut microbes, diet, and cancer. Cancer Treat Res 2014;159:377–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer 2013;13:800–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Golombos DM, Ayangbesan A, O’Malley P, et al. The role of gut microbiome in the pathogenesis of prostate cancer: a prospective, pilot study. Urology 2018;111:122–8. [DOI] [PubMed] [Google Scholar]
- [9].Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011;27:2957–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kuczynski J, Stombaugh J, Walters WA, Gonzalez A, Caporaso JG, Knight R. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr Protoc Bioinformatics 2012,1E–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 2010;26:266–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011;27:2194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].McDonald D, Price MN, Goodrich J, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 2012;6:610–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wang Q Garrity GM, Tiedje JM Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 2007;73:5261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Grim CJ, Daquigan N, Lusk Pfefer TS, Ottesen AR, White JR, Jarvis KG. High-resolution microbiome profiling for detection and tracking of Salmonella enterica. Front Microbiol 2017;8:1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ottesen A, Ramachandran P, Reed E, et al. Enrichment dynamics of Listeria monocytogenes and the associated microbiome from naturally contaminated ice cream linked to a listeriosis outbreak. BMC Microbiol 2016;16:275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Abernethy MG, Rosenfeld A, White JR, Mueller MG, Lewicky-Gaupp C, Kenton K. Urinary microbiome and cytokine levels in women with interstitial cystitis. Obstet Gynecol 2017;129:500–6. [DOI] [PubMed] [Google Scholar]
- [20].Drewes JL, White JR, Dejea CM, et al. High-resolution bacterial 16S rRNA gene profile meta-analysis and biofilm status reveal common colorectal cancer consortia. NPJ Biofilms Microbiomes 2017;3:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Daquigan N, Seekatz AM, Greathouse KL, Young VB, White JR. High-resolution profiling of the gut microbiome reveals the extent of Clostridium difficile burden. NPJ Biofilms Microbiomes 2017;3:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Langille MG, Zaneveld J, Caporaso JG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 2013;31:814–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 2010;11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res 2001;125:279–84. [DOI] [PubMed] [Google Scholar]
- [25].Paul B, Barnes S, Demark-Wahnefried W, et al. Influences of diet and the gut microbiome on epigenetic modulation in cancer and other diseases. Clin Epigenetics 2015;7:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Price AJ, Travis RC, Appleby PN, et al. Circulating folate and vitamin B12 and risk of prostate cancer: a collaborative analysis of individual participant data from six cohorts including 6875 cases and 8104 controls. Eur Urol 2016;70:941–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wang R, Zheng Y, Huang JY, Zhang AQ, Zhou YH, Wang JN. Folate intake, serum folate levels, and prostate cancer risk: a meta-analysis of prospective studies. BMC Public Health 2014;14:1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Figueiredo JC, Grau MV, Haile RW, et al. Folic acid and risk of prostate cancer: results from a randomized clinical trial. J Natl Cancer Inst 2009;101:432–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Pompei A, Cordisco L, Amaretti A, Zanoni S, Matteuzzi D, Rossi M. Folate production by bifidobacteria as a potential probiotic property. Appl Environ Microbiol 2007;73:179–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Powers HJ. Interaction among folate, riboflavin, genotype, and cancer, with reference to colorectal and cervical cancer. J Nutr 2005;135:2960S–6S. [DOI] [PubMed] [Google Scholar]
- [31].Bassett JK, Severi G, Hodge AM, et al. Dietary intake of B vitamins and methionine and prostate cancer incidence and mortality. Cancer Causes Control 2012;23:855–63. [DOI] [PubMed] [Google Scholar]
- [32].Hsueh EC, Knebel SM, Lo WH, Leung YC, Cheng PN, Hsueh CT. Deprivation of arginine by recombinant human arginase in prostate cancer cells. J Hematol Oncol 2012;5:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Plun-Favreau H, Lewis PA, Hardy J, Martins LM, Wood NW. Cancer and neurodegeneration: between the devil and the deep blue sea. PLoS Genet 2010;6:e1001257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Rodriguez-Melendez R, Griffin JB, Zempleni J. Biotin supplementation increases expression of the cytochrome P450 1B1 gene in Jurkat cells, increasing the occurrence of single-stranded DNA breaks. J Nutr 2004;134:2222–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.