Abstract
Murine typhus is a flea-borne typhus group rickettsiosis caused by Rickettsia typhi. Once a prevalent disease in the United States, the use of dichlorodiphenyltrichloroethane in the 1940s broke the classic rat-rat flea cycle of transmission, and the remaining endemic foci are now believed to be associated with opossums and the cat flea (Ctenocephalides felis). In Galveston, Texas murine typhus has re-emerged as a cause of febrile illness, and 7% of fleas collected from opossums are infected with R. typhi. In this study, we sought to explore the prevalence of rickettsiae associated with fleas on cats, as these animals have been speculated to play a role in the epidemiology of murine typhus. Fleas were collected from feral cats entering a local veterinary clinic as part of a trap, spay, neuter, and release program. Fleas were identified and subjected to analysis by PCR and sequencing. An estimation of the minimum infection rate (MIR) of pooled samples was performed. Three hundred fourteen fleas (all C. felis) were collected from 24 cats. Sequences for the outer membrane protein B gene revealed R. typhi in one pool (MIR 0.3%), Rickettsia felis in four pools (MIR 1.3%), Rickettsia asembonensis in one pool (MIR 0.3%), and “Candidatus R. senegalensis” in six pools (MIR 2.0%). Results were confirmed by sequencing portions of the rickettsial citrate synthase and 17-kD protein gene. In this study, the presence of R. typhi in fleas from cats suggests that in Galveston, there exists a small but measurable risk to humans who come into contact with flea-infested cats. Despite this, we believe that the low prevalence from cat-collected fleas, compared with that previously detected from opossums, makes cats less likely to play a role in the maintenance of R. typhi in this region. The significance of other identified flea-borne rickettsiae is yet to be elucidated.
Keywords: murine typhus, endemic typhus, opossums, fleas, Ctenocephalides felis, Rickettsia typhi
Introduction
Rickettsia typhi is an obligately intracellular Gram-negative flea-borne organism that causes murine typhus in humans. The classic reservoir and vector cycle involves Rattus species and their fleas, Xenopsylla cheopis. An alternate cycle of disease transmission is presumed to be related to opossums and Ctenocephalides felis (the cat flea) (Azad et al. 1997). In Galveston, Texas, murine typhus is again recognized as a cause of febrile illness. Decades after its apparent disappearance—a phenomenon attributed to measures to control populations of rats and their fleas—it has re-emerged (Blanton et al. 2015). As in southern California and more southern parts of Texas, recent cases of murine typhus in Galveston have been attributed to R. typhi-infected C. felis collected from opossums (Blanton et al. 2016). In the past, several reports epidemiologically linked cats to cases of murine typhus (Lepine and Lorando 1935, Irons et al. 1944, Older 1970), and C. felis, collected from kittens, had been found to be infected with R. typhi (Irons et al. 1944). As in many municipalities, Galveston has an abundant population of wild, nonsocialized cats that are often considered a nuisance to citizens. Since C. felis are ubiquitous among cats (Rust 2017), in addition to opossums, we sought to explore the role of cats as a vehicle for R. typhi-infected fleas. We hypothesized that in Galveston, cats play a role in the transmission of R. typhi by harboring infected fleas. To explore this hypothesis, we sought to determine the prevalence of rickettsiae within fleas collected from cats.
Materials and Methods
In Galveston, feral cats are trapped, spayed, or neutered, and returned to their habitat as part of a program to control their population. Fleas were collected from these cats entering a local veterinary clinic before surgical sterilization. The study was approved by the University of Texas Medical Branch Institutional Animal Care and Use Committee. The sex and location of collection of each cat were recorded. After the cats were anesthetized for surgery, fleas were collected with a flea comb; they remained alive and were placed in plastic sealable bags for transport. Fleas were then transferred to the laboratory, examined through a dissecting microscope, and identified using a taxonomic key (Centers for Disease Control and Prevention 2006). The fleas underwent surface decontamination by washing them in 5% bleach solution for 5 min (to eliminate exogenous DNA template), then in 70% ethanol for 5 min, and then in three subsequent 1-min rinses with phosphate-buffered saline (PBS). The washed fleas were pooled in groups of 2–5 fleas. These pool sizes were chosen based on experience from a previous study on fleas collected from opossums in Galveston (Blanton et al. 2016). Flea pools were placed in microcentrifuge tubes with 100 μL of PBS and homogenized using a pipet tip. DNA was extracted from these homogenates using the DNeasy Blood and Tissue Kit (QIAGEN, Valencia, CA) per the manufacturer's instructions.
Real-time PCR (qPCR) using primers to amplify a conserved region of the rickettsial citrate synthase gene was used to detect rickettsial DNA from pools of fleas (Labruna et al. 2004). Samples detected positive by qPCR were subjected to conventional PCR to amplify a portion of the outer membrane protein B gene (sca5) using the forward and reverse primers designated 120.2788 and 120.3599, respectively (Roux and Raoult 2000). Genomic DNA of R. sibirica was used as a positive control, and molecular grade water was used as a negative control in all PCRs. These amplicons were then cloned in chemically competent Escherichia coli cells using the TOPO TA Cloning Kit (Life Technologies). Plasmid DNA was purified with the PureLink Plasmid Miniprep Kit (Life Technologies) and sequenced in triplicate using a 3130xl Genetic Analyzer (Life Technologies). The sca5 sequences were compared with others in the GenBank database. Further characterization of flea pools (representatives were selected to correspond to each of the various rickettsial species noted on sequencing of sca5) was carried out through conventional PCR of the 17-kDa antigen gene (htrA) and citrate synthase gene (gltA) (Webb et al. 1990, Regnery et al. 1991). Amplicons were cloned and sequenced in triplicate as described above. The primer sequences were trimmed using EditSeq of the Lasergene Core Suite software version 12.0 (DNASTAR, Madison, WI) and submitted to the GenBank database.
An estimation of the minimum infection rate (MIR) of pooled flea samples was performed using computational methods as previously described (Biggerstaff 2008) with downloadable software for use in Microsoft Excel 2010 (Redmond, WA) (Centers for Disease Control and Prevention 2015).
Results
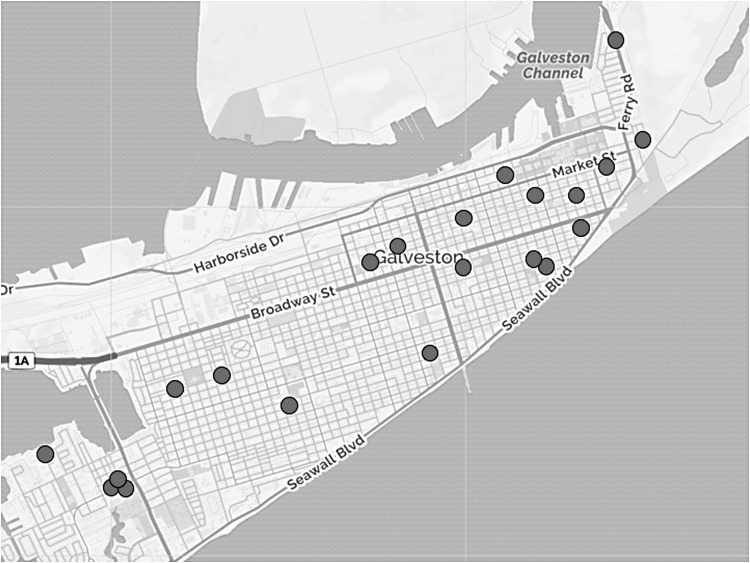
Fleas were collected from 24 cats. Fifteen (62.5%) of the cats were female. The cats were trapped from neighborhoods across Galveston Island (Fig. 1). The exact location of three cats trapped within the city of Galveston was not recorded. All cats were infested with fleas. A total of 314 fleas were collected from these animals and grouped into 87 pools (Table 1). The median number of fleas collected per animal was 11.5 (min–max: 2–28). The fleas were all identified as C. felis. Seventeen pools (19.5%) had evidence of rickettsial DNA as amplified by qPCR. Conventional PCR for sca5 successfully amplified 12 of these pools. Sequences for the sca5 products (see Table 2 for accession numbers) demonstrated R. typhi in one pool (MIR 0.3%), Rickettsia felis in four pools (MIR 1.3%), Rickettsia asembonensis in one pool (MIR 0.3%), and “Candidatus R. senegalensis” in six pools (MIR 2.0%). All of these positive pools demonstrated the presence of only one rickettsial agent. Further sequencing of gltA and htrA performed on a representative pool containing each identified flea-borne species confirmed the results of sca5 sequencing (Table 2).
FIG. 1.
Feral cats were trapped from various parts of Galveston. The exact trapping location of three cats was not recorded and therefore not represented in this figure.
Table 1.
Molecular Evidence of Rickettsiae Infecting Fleas Collected from Feral Cats
| No. positive pools/MIR (95% CI) | |||||
|---|---|---|---|---|---|
| Cat (n = 24) | No. pools (total fleas) | Rickettsia typhi | Rickettsia felis | Rickettsia asembonensis | “R. sengalensis” |
| 4 | 2 (4) | 0 | 1/24.9% (1.6–82.0) | 0 | 0 |
| 7 | 2 (6) | 0 | 0 | 1/16.2% (1.1–68.1) | 0 |
| 8 | 5 (23) | 0 | 0 | 0 | 2/9.7% (1.8–31.0) |
| 9 | 5 (10) | 0 | 1/10.0% (0.6–40.6) | 0 | 1/10% (0.6–40.6) |
| 11 | 5 (20) | 0 | 1/5.0% (0.3–22.9) | 0 | 0 |
| 13 | 4 (17) | 0 | 0 | 0 | 1/6.0% (0.4–28.2) |
| 16 | 3 (11) | 0 | 1/9.1% (0.6–41.2) | 0 | 1/9.1% (0.6–41.2) |
| 17 | 4 (12) | 1/8.3% (0.5–36.0) | 0 | 0 | 0 |
| 21 | 3 (11) | 0 | 0 | 0 | 1/9.1% (0.6–41.2) |
| All others | 54 (200) | 0 | 0 | 0 | 0 |
| Total | 87 (314) | 1/0.3% (0.1–1.5) | 4/1.3% (0.4–3.1) | 1/0.3% (0.1–1.5) | 6/2.0% (0.8–4.0) |
MIR, minimum infection rate.
Table 2.
GenBank Accession Numbers for Sequenced Amplicons
| Accession no. for rickettsial gene products | ||||
|---|---|---|---|---|
| Flea pool | sca5 | gltAa | htrAa | Species identification |
| 4.2 | MH325367 | R. felis | ||
| 7.2 | MH325368 | MH325379 | MH325383 | R. asembonensis |
| 8.3 | MH325369 | Candidatus R. senegalensis | ||
| 8.5 | MH325370 | Candidatus R. senegalensis | ||
| 9.1 | MH325371 | R. felis | ||
| 9.2 | MH325372 | MH325380 | MH325384 | Candidatus R. senegalensis |
| 11.5 | MH325373 | MH325381 | MH325385 | R. felis |
| 13.1 | MH325374 | Candidatus R. senegalensis | ||
| 16.1 | MH325375 | Candidatus R. senegalensis | ||
| 16.2 | MH325376 | R. felis | ||
| 17.4 | MH325377 | MH325382 | MH325386 | R. typhi |
| 21.1 | MH325378 | Candidatus R. senegalensis | ||
Amplification and sequencing of gltA and htrA were performed to confirm sca5 results on a representative specimen corresponding to each of the four flea-borne rickettsial species detected.
The R. typhi sequences were 768/770 (99.7%), 341/342 (99.7%), and 394/394 (100%) base pairs similar to R. typhi Wilmington (type strain) [GenBank accession AE017187.1] for portions of the corresponding sca5, gltA, and htrA genes, respectively. The characterized flea pool containing R. felis was 770/770 (100%), 341/342 (99.7%), and 391/393 (99.5%) base pairs similar to sca5, gltA, and htrA genes, respectively, for the R. felis URRWXcal2 strain [accession CP000053.1]. The R. asembonensis sequences were 769/770 (99.9%), 342/342 (100%), and 394/394 (100%) base pairs similar to sca5 [accession KY650699.1], gltA [accession KY650697.1], and htrA [accession KY650696.1] gene submissions from a cultivated isolate of R. asembonensis. Respectively, the “Candidatus R. senegalensis” sequences were 770/770 (100%) and 342/342 (100%) similar to sca5 [accession KF666470.1] and gltA [accession KF666478.1] sequences from an isolate of the organism from Senegal and 391/394 (99.2%) base pairs similar to an htrA [accession AY953285.1] sequence derived from the DNA extracted from a flea in South Carolina.
Discussion
Once a common occurrence in the United States, the incidence of murine typhus greatly decreased as a result of efforts to control rats and their fleas. The decrease in reported cases of murine typhus (5401 in 1944 to <100 by 1956) coincided with the use of dichlorodiphenyltrichloroethane on rat runs and harborages during the mid-1940s (Hill and Morlan 1948, Pratt 1958). In the continental United States, opossums and their fleas (C. felis) have been linked to the remaining endemic foci of murine typhus, and have also been linked to outbreaks occurring in areas where the disease had long been unrecognized (Adams et al. 1970, Adjemian et al. 2010, Blanton et al. 2016, Maina et al. 2016). Galveston is located on a barrier island off the upper Texas coast where the re-emergence of murine typhus has prompted investigation of opossums and their fleas—two-thirds of opossums demonstrated antibodies reactive against typhus group rickettsia, and 7.0% of C. felis collected from these animals were infected with R. typhi (Blanton et al. 2016).
C. felis, also known as the cat flea, is ubiquitous around the world in association of its feline host, the domestic cat (Rust 2017). Cats have been epidemiologically linked to murine typhus for some time (Lepine and Lorando 1935, Irons et al. 1944, Raynal 1947, Older 1970), and some of these historical reports have demonstrated R. typhi reactive sera and R. typhi-infected fleas collected from cats (Irons et al. 1944). The seroprevalence of typhus group antibodies within cats studied in areas endemic for murine typhus has varied widely based on the study (0–46%) (Sorvillo et al. 1993, Eremeeva et al. 2012, Gracia et al. 2015, Mullins et al. 2018, Nelson et al. 2018). In a study that specifically reported serology from cats located in a suburban area of Los Angeles where murine typhus cases were reported, the seroprevalence of typhus group antibodies in cats reached 90% (Sorvillo et al. 1993). Experimentally, cats inoculated intraperitoneally with an agent thought to cause murine typhus became infected (demonstrated by guinea pig passage of the triturated organs), showed little clinical effect, and maintained infectious organisms for <30 days (Lepine and Lorando 1935). The presence of R. typhi DNA amplified from the blood of five cats has been reported in Spain (Nogueras et al. 2013) but not in cats from southern California (Eremeeva et al. 2012, Mullins et al. 2018). To our knowledge, as has been demonstrated with Rattus spp. and Xenopsylla cheopis, the ability of domestic cats to act as a reservoir or amplifying host for R. typhi, through the transmission to feeding uninfected fleas, has not been experimentally demonstrated. Thus, the true role that cats play in the spread of murine typhus is only speculated.
In Galveston, feral cats are considered a nuisance to many and have been blamed for preying on migratory birds that are often reveled by tourists and residents. To help control their population, trapped feral cats are spayed or neutered and returned to the wild. We chose to study fleas from this population as we assumed these cats would be flea infested, rather than their more socialized counterparts, which are more likely to be given antiflea treatments. We found the prevalence of R. typhi-infected fleas to be low compared with the 7% found in fleas collected from area opossums (Blanton et al. 2016). In other areas where murine typhus is endemic, the presence of R. typhi-infected fleas from cats is similarly low or absent. Studies in southern California and Colombia have detected other flea-borne rickettsiae (R. felis and R. felis-like organisms) but failed to detect R. typhi (Hidalgo et al. 2013, Maina et al. 2016, Mullins et al. 2018, Nelson et al. 2018). A notable exception is the 55% of R. typhi-infected C. felis reported in Spain (it is unclear if these fleas were collected from the aforementioned cats with R. typhi DNA-containing blood) (Nogueras et al. 2013). It should be noted that the low prevalence of infected fleas from cats does not preclude the possibility that cats act as a vehicle to bring infected fleas into contact with humans—especially in regard to flea-infested companion cats that are likely to come into close contact with their owners.
R. felis, demonstrated within fleas collected in this study, has a worldwide distribution within the ubiquitous C. felis. Its prevalence within fleas of various geographic locations ranges greatly though. In studies in which at least 100 C. felis specimens were studied (data as summarized in a systematic review by Brown), the prevalence of R. felis ranges from 0.3 to 75% (Brown and Macaluso 2016). In our study, the 1.3% MIR of R. felis is relatively low, although within the bounds of what has been described elsewhere. Rickettsia felis is considered a human pathogen, but numerous reports challenging the known paradigms of rickettsial infection raise skepticism to its true pathogenic nature and role in human disease (Billeter and Metzger 2017, Blanton and Walker 2017). Currently, the pathogenic nature of R. asembonensis and Candidatus R. senegalensis to humans is unknown. The ability of animals to act as a reservoir or amplifying host for these organisms is also unknown. These recently discovered rickettsiae have been termed R. felis-like organisms because of their close phylogenic relationship to R. felis. Their prevalence among fleas is increasingly described and indicates, as with R. felis, an extensive geographic distribution (Blanton and Walker 2017). Their role in modulating the vector acquisition and maintenance of known pathogenic flea-borne rickettsiae—R. typhi—is unknown.
This study is limited as it only shows the prevalence of flea-borne rickettsiae as a snapshot in time rather than a dynamic process. Although the real-time PCR assay amplified rickettsial DNA from 17 flea pools, only 12 amplified by conventional PCR (a possible consequence of inhibitors of PCR or the more limited analytic sensitivity of the latter technique). Thus, our data may underestimate the prevalence of flea-borne rickettsiae in this study. Furthermore, typhus group seroprevalence, which was not able to be performed in this study, would give more clues to the burden of exposure that these cats exhibit to R. typhi and their potential to act as an amplifying host.
Conclusions
In Galveston, C. felis collected from cats contain R. typhi, although at a much lower prevalence than that found in C. felis collected from opossums. Although we believe this indicates that cats are not as important in the maintenance of R. typhi in this region, it is possible that exposure to these animals, hence their fleas, puts people at risk of murine typhus. More studies are needed to delineate the role of domestic animals and other flea-borne rickettsiae in the epidemiology and ecology of R. typhi infection in humans.
Acknowledgments
This study was conducted with the support of the Carmage and Martha Walls Distinguished University Chair in Tropical Diseases Endowment. Dr. L.S.B. is supported by the Institute for Translational Sciences at the University of Texas Medical Branch, supported in part by a CTSA Mentored Career Development (KL2) Award (KL2TR001441) from the National Center for Advancing Translational Sciences, National Institutes of Health. The authors acknowledge and thank the staff of The Animal Clinic for their efforts in collecting fleas.
Author Disclosure Statement
No conflicting financial interests exist.
References
- Adams WH, Emmons RW, Brooks JE. The changing ecology of murine (endemic) typhus in Southern California. Am J Trop Med Hyg 1970; 19:311–318 [DOI] [PubMed] [Google Scholar]
- Adjemian J, Parks S, McElroy K, Campbell J, et al. Murine typhus in Austin, Texas, USA, 2008. Emerg Infect Dis 2010; 16:412–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad AF, Radulovic S, Higgins JA, Noden BH, et al. Flea-borne rickettsioses: Ecologic considerations. Emerg Infect Dis 1997; 3:319–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggerstaff BJ. Confidence intervals for the difference of two proportions estimated from pooled samples. J Agric Biol Environ Stat. 2008; 13:478–496 [Google Scholar]
- Billeter SA, Metzger ME. Limited Evidence for Rickettsia felis as a cause of zoonotic flea-Borne rickettsiosis in southern California. J Med Entomol 2017; 54:4–7 [DOI] [PubMed] [Google Scholar]
- Blanton LS, Idowu BM, Tatsch TN, Henderson JM, et al. Opossums and cat fleas: New insights in the ecology of murine typhus in Galveston, Texas. Am J Trop Med Hyg 2016; 95:457–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanton LS, Vohra RF, Bouyer DH, Walker DH. Reemergence of murine typhus in Galveston, Texas, USA, 2013. Emerg Infect Dis 2015; 21:484–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanton LS, Walker DH. Flea-borne rickettsioses and rickettsiae. Am J Trop Med Hyg 2017; 96:53–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown LD, Macaluso KR. Rickettsia felis, an emerging flea-borne rickettsiosis. Curr Trop Med Rep 2016; 3:27–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Mosquito Surveillance Software. 2015. Available at https://www.cdc.gov/westnile/resourcepages/mosqSurvSoft.html
- Centers for Disease Control and Prevention. Pictorial keys to arthropods, reptiles, birds, and mammals of public health significance. Atlanta, GA: Department of Health and Human Services; 2006. Available at www.cdc.gov/ehs/docs/pictoral_keys/fleas.pdf [Google Scholar]
- Eremeeva ME, Karpathy SE, Krueger L, Hayes EK, et al. Two pathogens and one disease: Detection and identification of flea-borne rickettsiae in areas endemic for murine typhus in California. J Med Entomol 2012; 49:1485–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracia MJ, Marcen JM, Pinal R, Calvete C, et al. Prevalence of Rickettsia and Bartonella species in Spanish cats and their fleas. J Vector Ecol 2015; 40:233–239 [DOI] [PubMed] [Google Scholar]
- Hidalgo M, Montoya V, Martinez A, Mercado M, et al. Flea-borne rickettsioses in the north of Caldas province, Colombia. Vector Borne Zoonotic Dis 2013; 13:289–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill EL, Morlan HB. Evaluation of county-wide DDT dusting operations in murine typhus control. Public Health Rep 1948; 63:1635–1653 [PMC free article] [PubMed] [Google Scholar]
- Irons JV, Bohls SW, Thurman DC, McGregor T. Probable role of the cat flea, Ctenocephalides felis, in transmission of murine typhus. Am J Trop Med 1944; 24:359–362 [Google Scholar]
- Labruna MB, Whitworth T, Horta MC, Bouyer DH, et al. Rickettsia species infecting Amblyomma cooperi ticks from an area in the state of Sao Paulo, Brazil, where Brazilian spotted fever is endemic. J Clin Microbiol 2004; 42:90–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepine PP, Lorando N. Le typhus exanthematique du chat. Bulletin de la Societe de la Pathologie Exotique et de ses Filiales 1935; 28:356–360 [Google Scholar]
- Maina AN, Fogarty C, Krueger L, Macaluso KR, et al. Rickettsial infections among Ctenocephalides felis and host animals during a flea-borne rickettsioses outbreak in Orange County, California. PLoS One 2016; 11:e0160604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins KE, Maina AN, Krueger L, Jiang J, et al. Rickettsial infections among cats and cat fleas in Riverside County, California. Am J Trop Med Hyg 2018; 99:291–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson K, Maina AN, Brisco A, Foo C, et al. A 2015 outbreak of flea-borne rickettsiosis in San Gabriel Valley, Los Angeles County, California. PLoS Negl Trop Dis 2018; 12:e0006385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueras MM, Pons I, Ortuno A, Miret J, et al. Molecular detection of Rickettsia typhi in cats and fleas. PLoS One 2013; 8:e71386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Older JJ. The epidemiology of murine typhus in Texas, 1969. JAMA 1970; 214:2011–2017 [PubMed] [Google Scholar]
- Pratt HD. The changing picture of murine typhus in the United States. Ann N Y Acad Sci 1958; 70:516–527 [DOI] [PubMed] [Google Scholar]
- Raynal JH. Le chat dans l'epidemiologie du typhus exanthematique murin. Bull Soc Pathol Exot Filiales 1947; 40:367–375 [PubMed] [Google Scholar]
- Regnery RL, Spruill CL, Plikaytis BD. Genotypic identification of rickettsiae and estimation of intraspecies sequence divergence for portions of two rickettsial genes. J Bacteriol 1991; 173:1576–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux V, Raoult D. Phylogenetic analysis of members of the genus Rickettsia using the gene encoding the outer-membrane protein rOmpB (ompB). Int J Syst Evol Microbiol 2000; 50 Pt 4:1449–1455 [DOI] [PubMed] [Google Scholar]
- Rust MK. The biology and ecology of cat fleas and advancements in their pest management: A review. Insects 2017; 8:pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorvillo FJ, Gondo B, Emmons R, Ryan P, et al. A suburban focus of endemic typhus in Los Angeles County: Association with seropositive domestic cats and opossums. Am J Trop Med Hyg 1993; 48:269–273 [DOI] [PubMed] [Google Scholar]
- Webb L, Carl M, Malloy DC, Dasch GA, et al. Detection of murine typhus infection in fleas by using the polymerase chain reaction. J Clin Microbiol 1990; 28:530–534 [DOI] [PMC free article] [PubMed] [Google Scholar]