Deep learning combined with multiple object tracking detects ear tips in images and videos of field-grown wheat and can recover simple movement patterns caused by wind.
Abstract
Understanding the relationships between local environmental conditions and plant structure and function is critical for both fundamental science and for improving the performance of crops in field settings. Wind-induced plant motion is important in most agricultural systems, yet the complexity of the field environment means that it remained understudied. Despite the ready availability of image sequences showing plant motion, the cultivation of crop plants in dense field stands makes it difficult to detect features and characterize their general movement traits. Here, we present a robust method for characterizing motion in field-grown wheat plants (Triticum aestivum) from time-ordered sequences of red, green, and blue images. A series of crops and augmentations was applied to a dataset of 290 collected and annotated images of ear tips to increase variation and resolution when training a convolutional neural network. This approach enables wheat ears to be detected in the field without the need for camera calibration or a fixed imaging position. Videos of wheat plants moving in the wind were also collected and split into their component frames. Ear tips were detected using the trained network, then tracked between frames using a probabilistic tracking algorithm to approximate movement. These data can be used to characterize key movement traits, such as periodicity, and obtain more detailed static plant properties to assess plant structure and function in the field. Automated data extraction may be possible for informing lodging models, breeding programs, and linking movement properties to canopy light distributions and dynamic light fluctuation.
Understanding the relationships between local environmental conditions and plant structure and function is critical for both fundamental science and for improving the performance of crops in field settings. Wind is a given aspect of most agricultural settings; yet its effect on crop productivity has often been overlooked. In particular, we have a poor understanding of how movement influences yield or determines traits such as photosynthesis (Caldwell, 1970; Burgess et al., 2016, 2019; Kaiser et al., 2018). The effect of wind on plant performance depends upon its speed, duration, and structural features of the crop canopy. Wind can affect crop development, disease, and insect incidence; cause structural or mechanical damage; and alter the light environment inside the canopy, thus affecting photosynthesis (Caldwell, 1970; Roden and Pearcy, 1993a, 1993b; Roden, 2003; Smith and Ennos, 2003; de Langre, 2008; Moser et al., 2009; Onoda and Anten, 2011; Shaw, 2012; Burgess et al., 2016, 2019; Kaiser et al., 2018). Specifically, movement in low wind speeds should have an impact on canopy carbon gain by relieving photosynthetic limitations through increased light penetration to lower canopy layers (Burgess et al., 2016, 2019). However, as a stochastic process, wind-induced canopy movement is difficult to measure, particularly in dense crop stands, and there is currently no method that permits quantitative assessment.
Characterizing simple movement patterns could be used as a means to link local conditions to the altered light environment and photosynthetic performance. Furthermore, the characteristics of movement can be linked to the susceptibility of plants to withstand adverse environmental conditions, or to lodge, resulting in yield loss (Berry et al., 2003, 2007). The effect of movement on light dynamics and lodging susceptibility has been shown to be dependent on the frequency of motion, which can be determined from videos (Doaré et al., 2004; de Langre, 2008; Der Loughian et al., 2014; Burgess et al., 2016). Image sequences showing plant motion are readily available, and the ability to isolate and detect plant features of interest, such as the leaves and reproductive parts in cereal species, and to characterize simple movement patterns via tracking will be an essential step toward this goal. This linkage will support assessment of crop performance and the identification of traits for crop improvement, which are linked to mechanical canopy excitation. There are further implications for field phenotyping and precision agriculture, which must cope with moving plants, when quantifying plant features such as leaves and wheat ears (Cai et al., 2018).
Plant canopies are complex three-dimensional (3D) ‘scenes’ with movement involving many structures and organs possessing different physical and dynamic properties. Visual tracking, and specifically multiple object tracking (MOT), can be applied to time-ordered image sequences to count features and characterize movement within field-grown crop plants. Given an input video, objects, in this instance the plant features, must be successfully detected, identities maintained across multiple frames, and trajectories determined. Previous applications of MOT include the movement of people (i.e. pedestrians or sports players), vehicles, animals, or cells (Betke et al., 2000; Spampinato et al., 2008, 2012; Meijering et al., 2009; Pellegrini et al., 2009; Yang et al., 2011; Lu et al., 2013). The success of MOT is confounded by several key issues including (1) high levels of occlusion, (2) the movement of objects in and out of view leading to initialization and termination of trajectories, (3) similarity in appearance of features, and (4) interactions between objects. Image and video capture within the field setting can further complicate tracking through the production of highly challenging datasets. This is because of the inability to control illumination, the dynamic and unpredictable nature of wind, and the complexity of field-grown plants leading to dense scenes. Moreover, plant features may differ in appearance according to their developmental stage or the camera position (i.e. depth or orientation). Alternatively, distinct organs may look highly similar, making matching across multiple frames particularly challenging.
In MOT, objects must first be detected in each frame and then linked between frames to attain a trajectory. The first step, detection, may be achieved through deep learning, a broad category of machine learning. Machine learning techniques are expected to take a dominant role in the future of high throughput plant phenotyping and plant feature detection, with the ability to yield automated, unbiased, and consistent measurements over rapid timescales (Mohanty et al., 2016; Pound et al., 2017; Ubbens and Stavness, 2017; Kamilaris and Prenafeta-Boldú, 2018). Furthermore, the application of deep learning for detection in MOT can help improve tracking performance (Wang et al., 2015; Lee et al., 2016; Yu et al., 2016). In particular, deep convolutional neural networks (CNNs) offer the ability to learn to classify images or perform object recognition within scenes if provided with a carefully annotated dataset (Krizhevsky et al., 2012). A CNN is a powerful discriminative method that can take a series of annotated images (a training dataset) and train a network to identify features in images it has not previously seen (test images). A well-trained CNN can obtain accuracies of 97% and above (Pound et al., 2017)—a rate comparable with humans if not higher, and are considerably faster, particularly for complicated images, or so-called ‘dense scenes.’ Although many CNNs exist for plant phenotyping, they often work on simple scenes, for example, counting leaves in Arabidopsis (Arabidopsis thaliana) rosettes (Aich and Stavness, 2018; Dobrescu et al., 2018) or identifying root or shoot features from isolated wheat plants (Pound et al., 2017). A pipeline that aims to identify features from more complex images, such as the detection of wheat ear tips from images taken in the field, is much more difficult; manually annotating these features is time consuming, and the individual annotating the images requires training or expert knowledge of the plants to annotate the features consistently to avoid human error. More powerful network architectures may also be needed.
The ability to detect, and so count, harvestable organs from images could be used as a means to predict yields from a given stand of crops. Methods for the counting of ears within field-grown wheat (Triticum aestivum) already exist such as segmentation from mobile laser scanner data, or via image processing techniques (Cointault and Gouton, 2007; Velumani et al., 2017; Fernandez-Gallego et al., 2018). However, natural conditions such as wind produce significant challenges, for example, the blurring of images or appearance that ears merge because of image distortion. Moreover, errors in counting from static images are common such as the duplicate counting of wheat ears if image sets are overlapping, or the inability to count ears when they are occluded or distorted. The use of deep learning to detect distorted ears, and multiple object tracking to maintain ear identity across frames, is able to address these difficulties. Ear counting also provides a real and well-defined task against which to assess motion analysis tools.
Here we propose the use of detection-based tracking to characterize movement patterns of wheat ears in field-grown plants. Detection will be performed using a bounding box regression network applied to images taken in the field. Bounding box regressors are an end-to-end deep learning model that predict both the identity of objects in a scene, as well as the rectangular bounds of these objects. Several notable works exist in this area, for example, Faster-RCNN (Regional Convolutional Neural Network; Girshick, 2015); however, we utilized the YOLO (You Only Look Once) v3 network (Redmon and Farhadi, 2018) because of its reported accuracy and applicability to the problem domain. Here, we use high resolution images to train the network as they are easier to manually annotate. Finally, an MOT algorithm will be applied using characteristics of the detected bounding boxes to reconstruct the trajectories of individual ear tips in videos obtained in the field.
We use wheat ears as an easily identifiable complete target plant feature that is highly responsive to wind in terms of movement and provides a means to help develop more complex tracking procedures for leaves and other canopy features. Furthermore, the natural frequency and periodicity of ear tips are commonly used to infer movement characteristics of crops (i.e. for the assessment of lodging; Berry et al., 2003, 2007). The aim is to create a broad feature detection method (i.e. without the need for camera calibration or a fixed imaging position or strategy) and combined tracking algorithm to improve feature detection and enable the determination of movement traits.
RESULTS
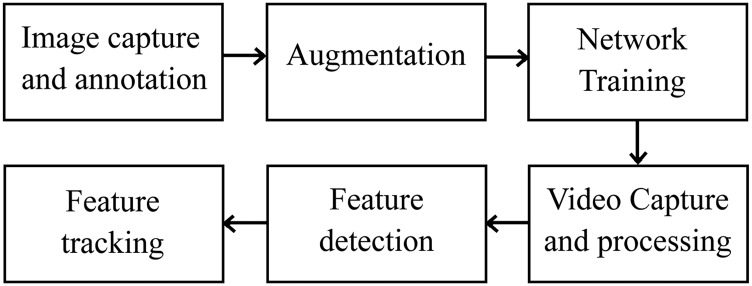
Analyzing the movement of crops within the field setting requires accurate detection of key features and the tracking of these features between frames in a video. An overview of the suggested pipeline for obtaining movement patterns is given in Figure 1.
Figure 1.
Overview of the pipeline from feature detection to multiple object tracking.
Accuracy of Feature Detection
To quantify the accuracy of feature detection, a convolutional neural network was trained using test images of native resolution, and of cropped resolution, with and without augmentations applied. Each network was trained for 100 epochs. The results of each trained network are given in Table 1, where mean Average Precision (mAP) is the measure of the accuracy of an object detector, where the higher the value (< 1), the more likely the network will correctly perform detection. Loss is calculated as the sum-squared error between the predictions and the ground truth and is composed of classification loss, the localization loss (errors between the predicted boundary box and the ground truth), and the confidence loss (the objectness of the box; Redmon et al., 2015; Redmon and Farhadi, 2016, 2018), where the lower the value, the more accurate the detector is considered to be. Finally, detection, is calculated as the percentage of detected plant features in comparison with those which were annotated across all test images. The cropped images achieved a higher accuracy both in terms of mAP and loss. This is because of the size of features when passed through the network and production of white space in training sets of uneven resolution.
Table 1. Results of the YOLO v3 network for ear tip detection depending on whether images were cropped and presence of augmentations.
Full resolution images are 3456 × 2304 pixels, whereas cropped images were either 576 × 576 or 684 × 684 pixels. A maximum of three augmentations were applied to the image. mAP and loss are measures of the accuracy of feature detection where the higher and the lower the values, respectively, indicate a more accurately trained network. Detection represents the amount of ear tips detected by the network compared the amount that were manually annotated.
| Cropping | Augmentations | mAP | Loss | Detection (%) |
|---|---|---|---|---|
| No | No | 0.0159 | 15.075 | 29.03 |
| No | Yes | 0.0553 | 7.614 | 45.82 |
| Yes | No | 0.57 | 3.735 | 84.16 |
| Yes | Yes | 0.58 | 3.127 | 89.26 |
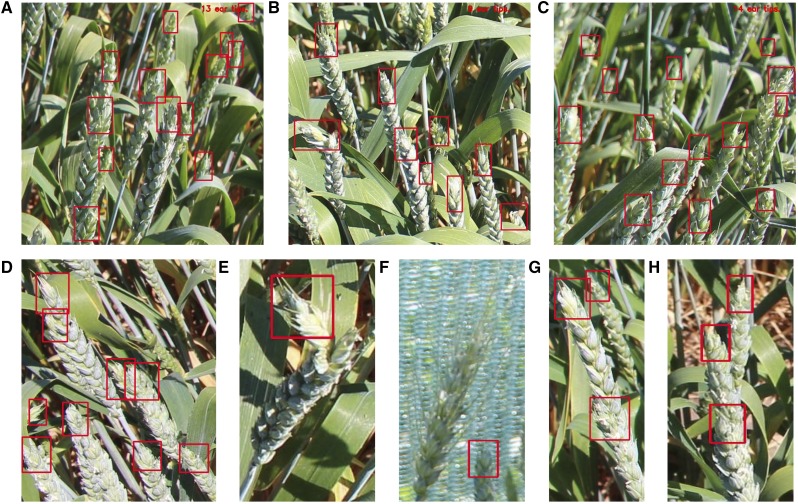
The efficacy of detection can be visualized in Figure 2 where ear tips have been detected in relatively difficult scenes. This includes images in which the ears are bunched together (Fig. 2, A–D); semi-occluded ear tips (Fig. 2A); the correct annotation of bent ear tips (Fig. 2E); the omission of a long-awned phenotype (Fig. 2F); and the correct detection of ear tips that are lined up in a row and thus overlapping (Fig. 2, G and H). Furthermore, the bounding boxes are accurately sized and orientated for each of the features present.
Figure 2.
Example detection (red boxes) of ear tips from field-grown wheat plants using the YOLO v3 network. A–C, Detection on image segment of 576 × 576 pixels. The bounding box size and orientation is adjusted according to the size of the feature, and total number of ear tips is given in the top right corner. Interesting examples include images in which the ears are bunched together (A–D); semi-occluded ear tips (A); the correct annotation of bent ear tips (E); the omission of a long-awned phenotype (F); and the correct detection of ear tips that are lined up in a row and thus overlapping (G) and (H).
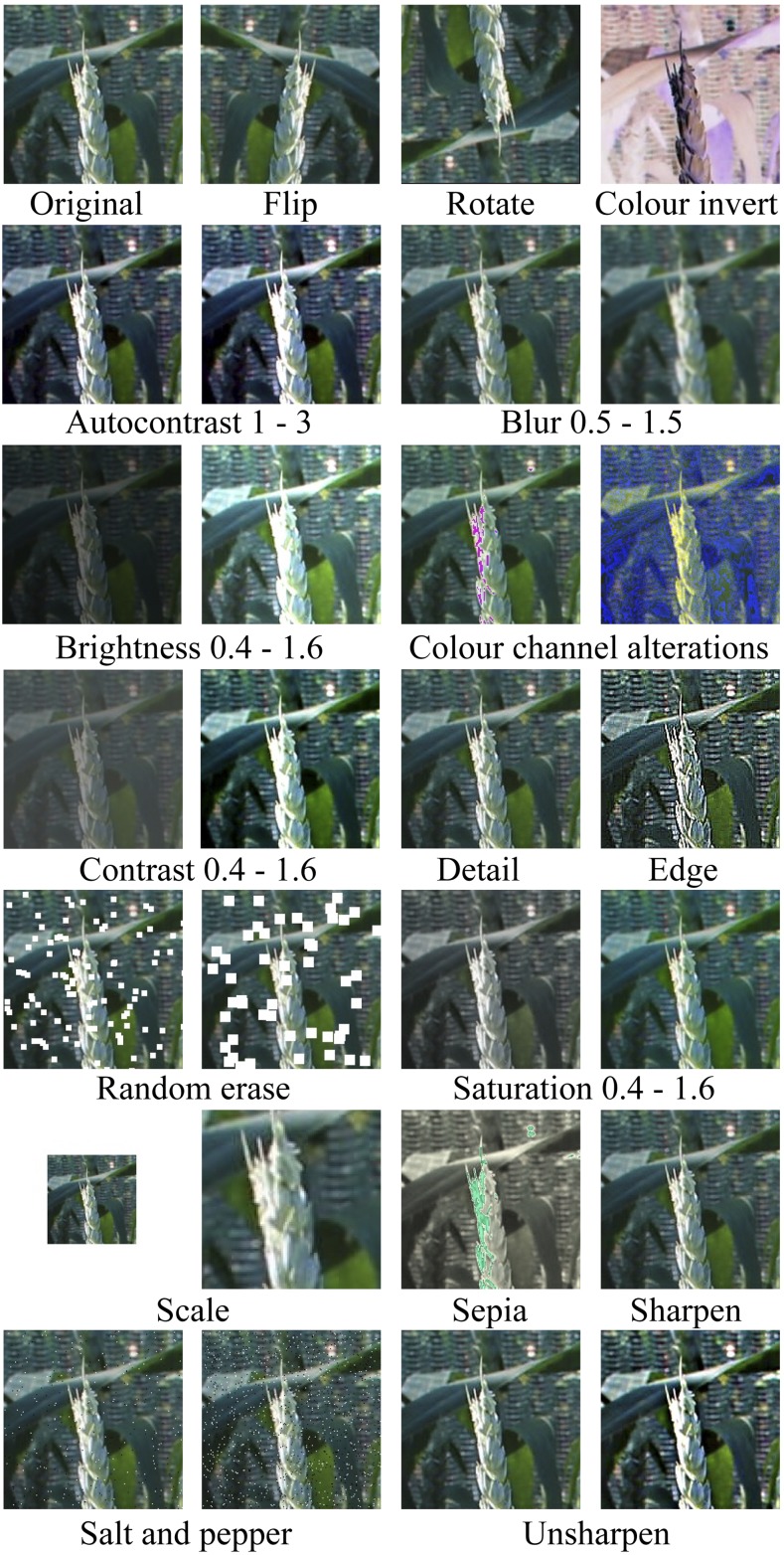
Low efficacy of deep learning is often related to the size or quality of the available training data. Within this work, the 290 native images (∼30,000 cropped images) represents a relatively small dataset. To overcome this, on the fly augmentations were applied to the images as they pass through the network to increase the range and variation of training data (Fig. 3). Presence of augmentations also improved the accuracy of feature detection because of the increase in the variability of the dataset. This is because alterations to the images allow them to mimic moving plants (i.e. video frames), through the reduction in resolution, quality, and the representation of motion via blur.
Figure 3.
Example augmentations applied to training images. As the training images pass through the YOLO v3 network, augmentations are randomly applied to increase the size and variation of the available dataset. A random value is selected between a set range for each filter.
Another measure of the accuracy of the trained network is a comparison of the number of features counted via the deep learning approach with those manually counted by the annotators. This was performed on a subset of images that were not used to train the network (Detection; Table 1). Similarly to the mAP and loss values discussed above, the use of cropping and application of augmentations to the training data leads to the highest detection relative to the manual approach. In most cases the accuracy of deep learning can overcome that of human, manually detecting, and counting from an image. The training of CNNs requires high-quality annotations from which to train the network. Furthermore, the complex images, combined with the relatively small dataset size, will have contributed to the reduced detection reported here compared with other plant feature detection networks (Pound et al., 2017; Dobrescu et al., 2018).
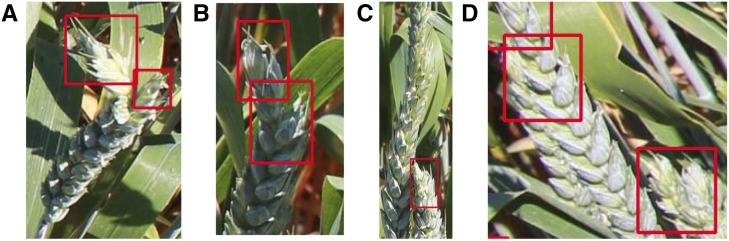
Although the mAP and loss values for the network trained on cropped images without augmentations are very similar to those trained with augmentations, the accuracy of detection in test images was lower (Fig. 4). For example, the network incorrectly detected bent ear tips as two separate detections (Fig. 4, A and B); failed to detect ear tips that were overlapping (Fig. 4C); and grouped closely located ear tips within a single bounding box (Fig. 4D). Low mAP and loss values can occur in instances where the network is overfitted, that is, it fits the training data too well and is unable to generalize the model to fit unknown, or unseen, data; where the network is overtrained, that is, it has been run for too long; or a combination of both. This issue can be overcome by increasing the amount of variation within the dataset, for example, by the addition of further augmentations.
Figure 4.
Examples of incorrect detections (red boxes) of ear tips from field-grown wheat plants using the YOLO v3 network on cropped images with no augmentations applied. For example, two separate detections made for a bent ear tip (A) and (B); failure to detect overlapping ear tips (C); and the grouping of closely located ear tips within a single bounding box (D).
We further validated the training of the network by detecting ear tips from images of wheat obtained from publicly available web-based sources (Fig. 5; Table 2). Images were selected that had a resolution between 1280 × 720 and 1920 × 1080 as these are common image resolutions obtainable through most commercial cameras. The images were selected based on the criterion that they must be in a natural environment, the features of the plant must be clearly visible, and the number of features must be countable so that we are able to evaluate the performance. Ear tips were accurately detected for short-awned wheat lines (Fig. 5, top) including images of plants at a later stage of development and thus of different color. The network was unable to detect long-awned phenotypes or ear tips of different species (Fig. 5, bottom). The same images were also passed through the network trained on cropped images without augmentations applied. Although the network was able to detect most of the ear tips from these images, it was not able to detect as many as the network trained with augmentations applied, particularly for the ears at a later stage of development. This is attributed to the potential overfitting of the network, with detection being restricted to images that are very similar to the training set. Whereas the presence of augmentations enables these different ear tips to be correctly detected, even though they do not exist in the original, training, dataset. This indicates the applicability of deep learning techniques for application in phenotyping platforms given sufficient training data.
Figure 5.
Example detection (red boxes) of ear tips from from publicly available web-based sources of cereals using the YOLO v3 network. Top: detection of ear tips in short-awned wheat varieties. Bottom: ear tips are correctly not detected in images of different cereal species (left) or long-awned wheat varieties (right). Images are open source (creative commons license) from www.pexels.com.
Table 2. Results of the YOLO v3 network for ear tip detection on images of wheat plants sourced from internet databases (Fig. 9).
Left, left middle, right middle, and right correspond to the position of the image on the top row of Figure 9. The images from the bottom row of Figure 9 are not included because no ears were found, which is the expected result of the network as ears with awns are classified as incorrect. The All images row includes all images that were used. “Detected” refers to the number of found ears, whereas “undetected” are the total number of ears missed. “Accuracy” is the percent of detected ears.
| Image | Detected | Undetected | Actual Ears | Accuracy (%) |
|---|---|---|---|---|
| Left | 4 | 3 | 7 | 57 |
| Left-middle | 11 | 2 | 13 | 85 |
| Right-middle | 16 | 4 | 20 | 80 |
| Right | 8 | 2 | 10 | 80 |
| All images | 229 | 47 | 276 | 83 |
Because of the higher detection accuracy, the remainder of the article will focus on results obtained using the network trained on cropped images with augmentations applied.
Evaluation of Tracking
The output of the multiple object tracking algorithm is a series of trajectories (given as sequences of coordinates) for each of the identified features and a video combining the original frames with mapped trajectories to visualize movement paths of the ears (Fig. 6; Supplemental Movie S1). To determine the accuracy of the tracking, a subset (∼100) of randomly selected frames were visually inspected to determine whether the identification number of each feature was correctly maintained. Videos were manually evaluated for accuracy comparing the number of correctly tracked features to those incorrectly tracked or unidentified. The tracking algorithm correctly identified and maintained the identification of 88.61% of features; 0.83% were incorrectly identified, 4.44% were occluded, and 6.11% were not detected by the CNN.
Figure 6.
Feature tracking of wheat ear tips in video frames (full video available in Supplemental Movie S1). A, Detection of ear tips using the YOLO v3 network in the first video frame (t = 0). B, Trajectory of ear tips across all frames (t = n) where darker shades of the same color indicate the most recent position of an identified ear tip. C, Reduced section of image with detected ear tips in the first video frame. D, Position of ear tips and trajectory after 8 frames (t = 8). E, Trajectory of ear tips across all frames.
Here it is important to note that the accuracy of the tracker is dependent upon the accuracy of detection by the CNN. Therefore, increasing detection accuracy, for example, by increasing the size and variation of the dataset, would also increase the accuracy of the tracker. Equally, a poor training dataset would reduce the quality of tracking. To further test the accuracy of the tracker, single ear tips that move in and out of view can be assessed to see if identity is maintained. An example can be seen in Figure 7, where a single ear tip (ID:1) maintains identification over time, despite being occluded by an overlapping leaf. This is important as it provides a means to more accurately count features from videos by preventing duplicate counts of features that move in and out of view.
Figure 7.
Frames of a video indicating the occlusion of an ear tip behind a leaf and its reappearance, where the identification number of each feature is given above. The tracking algorithm enables the identity of the ear tip to be maintained, despite its disappearance and reappearance, based on properties of the bounding box. This can be used to improve counting accuracy by preventing duplicate counts of features.
Two-Dimensional Motion Determination: Calculation of Periodicity
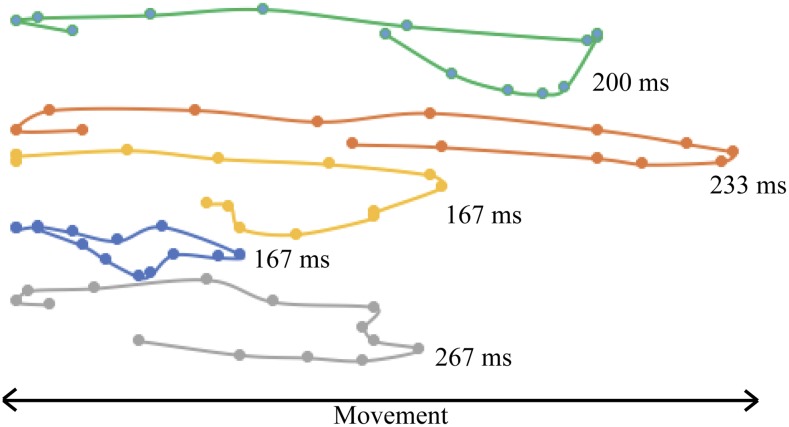
Characterization of movement can be achieved through the analysis of periodicity of features. Key biomechanical characteristics of plants include stem strength, stiffness, and weight and together these determine periodicity of movement. Hence tracking methods can in principle be used to screen plants and their component organs for new properties that relate to movement. Here we demonstrate proof of concept by tracking the motion of wheat ears. Within wheat, different ears will have different periodicity depending on growth stage and structural characteristics of the plant, which in turn can be related to functional properties of the canopy structure such as the ability to intercept radiation, or the susceptibility to lodge. The movement path of each individual feature can be used as a means to infer periodicity and can be presented as the distance traveled in a given amount of time or, alternatively, as the time taken for a feature to travel between two extreme values (i.e. in this instance, from a leftmost position to a rightmost position). An example of the movement paths of five different ears over the same time period (433 ms) is given in Figure 8. This indicates wide variation in movement characteristics, despite being obtained from the same wheat variety and same growth stage.
Figure 8.
Example movement paths of five randomly selected wheat ears during the same period of time, where each point on the path indicates the position of the ear tip in a single frame, located by deep learning. The amount of time taken for each feature to move from its leftmost position to the rightmost position is given.
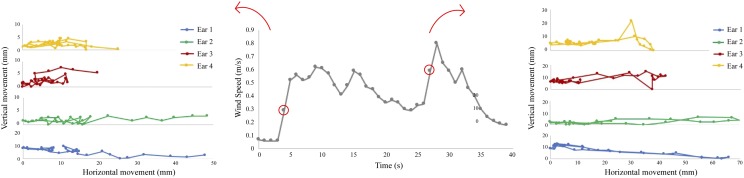
Moreover, locally recorded wind speed values can be directly linked to ear movement distances as shown in Figure 9. This was performed in a controlled environment using a hot bulb anenometer (Testo) to record wind speed and a domestic fan as wind speed, and direction was not available during the field trial. The controlled environment consisted of multiple cameras such that real world measurements can be obtained, for example, millimeters, which is often difficult using a single camera as depth is unknown. The same method described here for detecting and tracking was used, using a dataset obtained within the glasshouse. Because controlled environments contain less variation, the network was able to obtain 100% of the ear tips in the video despite the video being out of sample data. This allows the correlation between wind speed and actual distance traveled (Fig. 9), which would not be possible without camera calibration.
Figure 9.
Movement paths of four ear tips with associated wind speed. The movement paths of four ear tips during one second of wind-induced displacement were captured using a calibrated camera set up and deep learning approaches. Simultaneous wind speed measurements were calculated (middle) and linked to movement at two selected wind speeds: 0.3 m/s (left) and 0.6 m/s (right).
DISCUSSION
Movement of plants has been well studied with respect to traits such as lodging, which are associated with high wind speeds. In these scenarios biomechanical models exist to predict failure events. However, there is little knowledge concerning the types of movement that occur at lower wind speeds where in fact movement may provide beneficial canopy properties including better gas and light distribution (Burgess et al., 2016, 2019). Furthering this understanding is essential for improvement of fundamental knowledge but also application to crop improvement. However, the methodology barely exists to (1) quantify movement, (2) screen plants for contrasting movement types, and (3) simulate accurate movement in mechanical models from plant 3D reconstructions. Here we make a substantial step to address this and show a method for the application of deep learning to identify and track plant organs (wheat ears). Moreover, we show the potential for quantification of movement types.
Deep learning offers unparalleled discriminative performance in terms of locating features of interest from images. Here, deep learning was applied to images of field-grown wheat plants, where the characteristics of each of the images (i.e. the camera angle, position, lighting, number, and location of features of interest in each scene) were highly inconsistent. The ability to locate and then track these features between frames provides the first step in assessing real life patterns of movement in the field environment. The success demonstrated here reflects the success seen when applying deep learning to related image analysis tasks such as the detection and counting of other plant features from controlled growth facilities or leaf segmentation (Romera-Paredes and Torr, 2016; Pound et al., 2017)
The work presented here is not constrained by illumination affects, provided a sufficient training dataset is provided; this is further improved by the implementation of augmentations. The layout of the crop field or the camera setup has little impact on the ability to accurately detect features in scenes because of the variation in data. Moreover, the images do not need to be taken at specific angles or distances and can vary in terms of complexity, that is, the density of the scene. Finally, the number of tips in an image can be accurately counted at a much higher speed in comparison with counting by humans.
The application of a bounding box regressor and multiple object tracking has several advantages over previous methods. First, using characteristics of the bounding box can offer more information than tracking based on a single point. The detection of features via a CNN means that feature size (i.e. bounding box area) is likely to be highly similar in corresponding frames, or at least within a given range, even if changes in feature orientation and distance from the camera (i.e. movement forward or backward) occurs. Furthermore, the bounding box offers the ability to use further descriptions of the feature such as histogram correlation, which would not be possible if the feature boundary was not known. Feature tracking can also be applied as a means to improve the counting of features within complex scenes. The ability to maintain identity of a given feature across multiple frames, even if it moves in and out of view, prevents the duplicate counting of the same feature. This is particularly relevant to counting wheat ear tips within the field where wind-induced movement is a regular occurrence.
Biological Context and Future Applications
The ability to link crop structure and function to the local environmental conditions is the first step in optimizing cropping systems and improving crop performance. The presence of wind has numerous effects on the biotic and abiotic environment (see Introduction); thus analyzing simple movement patterns from videos taken in the field will enable the assessment of crop performance across a range of different environments. This could be used as a means to match crop ideotypes to local environmental conditions or study the effect of altered or extreme weather events (in this case changes in wind speed and duration) on crop performance.
An obvious and immediate example for the application of these tracking-based methods for assessing crop movement is the analysis of lodging susceptibility in cereal crops. Lodging is dependent upon morphological traits as well as local environmental conditions. One key determinant of lodging susceptibility is the height at the center of gravity, and corresponding natural frequency of movement. This can be inferred from trajectories of individual plant ears within videos, and thus provide a screening method to inform lodging models (Berry et al., 2003, 2007). The ability to track movement automatically and quantify frequency would enable the screening of plants in the field with no need to resort to manual time-consuming methodologies such as the manual measurements obtained for lodging analysis (Berry et al., 2003, 2007). It is possible to envisage the installation of field cameras that are able to track and record movement of key plant features. We have used ears, but leaves and stems may be possible using the same approach.
Linking movement trajectories with local wind conditions will enable key plant properties to be inferred. Previous studies indicate that local movement depends upon the combined influence of several structural traits including plant height, stem and stem wall thickness, material strength, and organ weight. The stem can be viewed as a rod with the center of gravity part way up (Doaré et al., 2004). The center of gravity will be dependent upon the weight of the ear (including individual kernels) and any attached leaves, and the distribution of weight about the stem. When the stem is displaced, it will be subject to both gravity and stem elasticity, which, in combination with wind speed and direction, will determine the overall trajectory of the stem and thus the ear tip. In general, the center of gravity is positively correlated with plant height, which in turn is negatively correlated with natural frequency (Piñera-Chavez et al., 2016). Improved lodging resistance has been achieved through the use of dwarfing genes to reduce height, thus increasing the natural frequency of movement. The characteristics required to increase lodging resistance are a short stature (0.7 m), large root spread, and a specific diameter/material strength combination to provide sufficient strength with the minimal investment of biomass so as not to limit yield potential (Berry et al., 2007).
The addition of 3D movement capture combined with camera calibration enables quantitative plant traits to be obtained including organ dimensions, angles, and physical displacement of visible plant material. This will enable rapid screening assessment of biomechanical traits within the field setting and could be used to inform prebreeding trials to select the optimal combination of biomechanical traits across a range of varieties without the need for cumbersome manual measurements.
A second important example concerns the improvement of canopy microclimate, which has been discussed at length in recent reviews, for example, Burgess et al. (2019). Wind-induced movement is critical in determining the dynamics of light reaching photosynthetic organs (Roden and Pearcy, 1993a, 1993b; Roden, 2003; Burgess et al., 2016). The spatial arrangement of plant material within the canopy leads to a complex pattern of light components (direct, diffused, and scattered light of different wavelengths). Typically, light levels become progressively lower further down in the canopy; however, these periods of low light are interspersed with periods of high light, called sun flecks, the duration of which will depend upon both solar angle and the displacement of the canopy brought about by wind. Recently models have shown that plant movement probably enhances canopy photosynthesis by increasing the probability of photon penetration into the canopy (Burgess et al., 2016). Moreover, the spatio-temporal variation in light in a moving canopy, which is likely to provide rapid switching between low and high light, may be suited to maintaining high photosynthetic induction states, typically limited by factors such as Rubisco activation state, stomatal conductance, and photoprotection. It was pointed out that biomechanical properties of crop plants might be used to generate an idealized phenotype that distributed light more effectively, relieving photosynthetic limitations (Burgess et al., 2019). However, any theorized optimal property would need to be tested, and there would be a strong need to develop models and screen germplasm for the right properties.
Another application arises from organ identification in a complex environment. The ability to count harvestable organs from images of crowded (and moving) scenes, in this case the number of ears of wheat, could be used as a means to predict yields from a given stand of crops. Although this is easily feasible with use of the method demonstrated here, there are several considerations that may limit the accuracy of predictions. First, to allow predictions of yield based on area, care must be taken to image a select and consistent area of land every time. However, the use of a fixed camera setup, for example, a fixed camera stand that enables the same area to be captured in every image, would get around this issue. Second, the structure of the crop plants of interest may preclude accurate predictions of yield. Occlusion caused by overlapping leaves, or the placement of harvestable organs lower down on the plant, and thus not visible to the camera view, could limit accuracy. However, given sufficiently large image datasets are available, this error is likely to be consistent across a wide range of different crop plants. Future work will involve the estimation of crop yield using a combination of deep learning and feature tracking and the improvement of network accuracy by increases in the size of the training dataset.
The substantial increase in throughput offered by deep learning approaches will be critical in underpinning and revolutionizing plant phenotyping methods. This approach can be applied not only to feature location and counting tasks but also to the characterization of further traits. It enables links to be made between growth, development, and the local environmental conditions, as demonstrated here with the analysis of movement patterns. Deep learning can also be applied to other data tasks, for example, identifying quantitative trait loci for linked traits (Pound et al., 2017). However, the benefits and high accuracy of deep learning approaches can be achieved only given a sufficiently large training dataset. The quality of the training data (e.g. the range available data) and the process of training the network (including the accuracy of their annotation) are also likely to alter the final results.
In the case of feature detecting in the field setting, the future predictive ability and success of deep learning will revolve around increasing the range of available images and training datasets, including the crop plants of interest, their developmental stage, and the location/conditions of the field setting (e.g. to achieve a range of images with different lighting conditions). Here, ∼ 300 images constitute a relatively small dataset; yet detection is still possible on internet obtained images of plants with a similar phenotype (Fig. 9). For more specific phenotyping tasks, datasets will be required that are tailored toward a specific feature detection, for example, the length of awns or the curvature of leaves. This will be aided by the creation and use of databases for annotated images such as the Annotated Crop Image Database (ACID, 2018). Increasing interest in deep learning approaches to image analysis or other data processing tasks is likely to lead to the creation of online systems for processing data. This will negate the need for a high-powered computer, which is a current requirement for deep learning tasks, and thus will make deep learning approaches more accessible, both in terms of cost and expertise, for a wider range of users.
CONCLUSION
Here, a method for detection-based tracking was presented to characterize movement patterns of wheat ears via their accurate detection within movie frames. This provides the first stage toward linking plant growth and function to the local environmental conditions will be critical in determining performance of crop plants. Although training image sets taken at different growth stages and under different environmental conditions may be time-consuming, there are clear data gains to be made by using deep learning approaches to plant phenotyping in the field setting. Any future increases in computing power and databases will increase the predictive ability of this approach further.
MATERIALS AND METHODS
Imaging and Annotation of the Training Dataset
The accuracy of deep learning is highly dependent on the accuracy of annotation of the training set, along with the quality and variation of images included. For a network to be efficiently and effectively trained, the data must be wholly and accurately annotated. Moreover, images captured from different angles, during different weather conditions, or at different developmental stages (i.e. capturing color change during senescence) can provide a more robust network capable of detecting features under varying conditions. If insufficient images are provided, the network will fail to learn an accurate model of the task at hand, resulting in poor performance on new images.
The acquisition of large annotated datasets may be facilitated by obtaining freely available datasets released by other researchers and projects. However, this has been uncommon in practice on plant phenotyping tasks because of their specificity, and the lack of suitable datasets for field-based phenotyping. Although tracking requires the plant features to be detected from video frames, high resolution photographs constitute the training image set within this work. This is because the individual frames of a standard video recording often suffer significantly from motion blur, whereas higher resolution of photographs are easier to manually annotate. Augmentations can then be applied to photographs to decrease their resolution and make them applicable to different detection scenarios. An undistorted image can be distorted, although in most instances it is not possible to reverse this process.
A key practical contribution of this work is a new dataset containing 290 images of wheat (Triticum aestivum) plants, variety Willow, with accurately labeled ear tips. Images were taken from an ongoing field trial at the University of Nottingham farm (Sutton Bonington Campus; (52.834 N, 1.243 W) on a sandy loam soil type (Dunnington Heath Series) in July 2018. The wheat was grown in plots of 6.00 × 1.65 m, with 0.13 m between rows and at a seed rate of 300 seeds m−2. The training images were taken postanthesis, equivalent to GS70 (Zadoks et al., 1974), and were captured using a 650D consumer grade 12MP camera, with a pixel resolution of 3456 × 2304 (Canon). The camera was held by hand, and care was taken to capture images from varying positions and under different lighting conditions to increase variability. These included top-down and side views of varying distances. Example images are shown in Figure 10. The variation in light, distance, and angle all add variety to the dataset.
Figure 10.
Example images used to train YOLO v3 to detect ear tips from field-grown wheat.
Each of the images contains 10–100 ear tips that have been manually and accurately annotated using bounding boxes by two experts at their native resolution using the annotation software LabelImg (LabelImg, 2018). Care was taken to consistently place the bounding box so that it covered the awns and top one or two spikelets, depending on visibility, to ensure consistency across images and allow the center of the bounding box to represent the center part of the ear tip. Figure 11 shows some examples of the annotated ear tips. Semi-occluded tips were included given sufficient features for recognition (i.e. the awns and part of a spikelet) were considered to be present.
Figure 11.
Example annotation (red boxes) of wheat ears used for network training. Ears were manually annotated using bounding boxes from full resolution images (Fig. 1).
Network Architecture and Training
A CNN framework typically consists of an input and an output, as well as multiple hidden layers, comprising convolutional layers, pooling layers, and fully connected layers. Convolution layers are used to extract useful features such as edges, corners, and boundaries, which are output as several feature maps. The number of feature maps increases deeper in the network to improve the accuracy of classification. Pooling layers, or convolution layers with a stride greater than 1, function in between the convolution layers to downsample the size of feature maps and thus improve overall efficiency of the network, whereas the fully connected (neural network) layers perform classification of the images. CNNs have consistently been shown to outperform traditional machine learning methods, for example, in image classification (He et al., 2016) and instance detection and segmentation (Girshick, 2015), and work directly on raw red, green, and blue images without the need for any complex preprocessing.
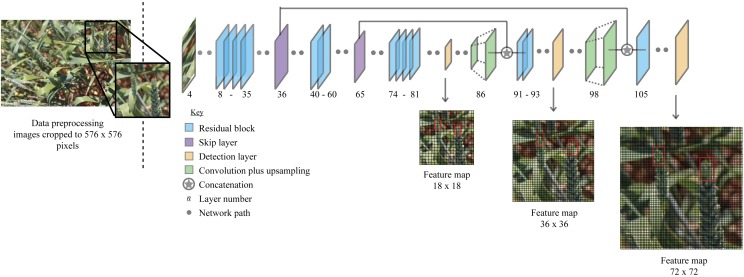
The YOLO v3 network (Redmon and Farhadi, 2018) was trained using the annotated dataset discussed in the previous section. An overview of the network is given in Figure 12. Tiling, the cropping of images, was used to prevent the downsampling of whole images, ensuring that small objects such as wheat ears are sufficiently resolved. Each image was cropped into ∼ 80 images of 576 × 576 and 684 × 684 pixels (hereby known as training set), before training (Fig. 13; crop 1) using a sliding window approach. Images are fed into the network at 576 × 576 pixels (the network resolution), because of memory limitations and as a compromise between computational efficiency and an adequate receptive field (field of view). Typically, in object detection, the cropping of images is not performed because of object size resulting in larger bounding boxes, and instead full resolution images are resized to the network resolution. However, within this case at full image resolution (3456 × 2304), the ear tips and associated annotations are too small for accurate detection and become pixelated when resized to the network resolution. For example, at native resolution, some annotated ear tips comprise a 30 × 30 pixel bounding box, which when condensed into the network will become 5 × 5 pixels. Moreover, the height and width of the image crops were kept identical to ensure that the aspect ratio matched that expected by the network architecture (Fig. 13; crop 2).
Figure 12.
A simplified overview of the YOLO v3 network architecture used to detect ear tips from images of field-grown wheat plants. The network is split into several layers. The size of the feature maps increases deeper in the network, and detection is performed at three different points to improve classification accuracy.
Figure 13.
Example image cropping from native resolution (3456 × 2304 pixels) to network input (576 × 576 pixels). Test images were made by cropping full images to either 576 × 576 or 684 × 684 pixels (Crop 1). If images are randomly cropped to be uneven width:height (Crop 2; i.e. 1029 × 769), white space is inserted into the test image input (bottom right). For this reason, uneven width:height crops are not used in this work.
The original dataset consists of 290 full resolution images and annotations, indicating the bounding box of the ear tips, presented as XML files. The image set used for training consists of 29,993 cropped images generated from the full resolution images, resulting in ∼ 211,000 annotated ear tips containing duplicates (caused by tiling) in which the original number of ears was ∼ 40,000. Within this network, duplicates do not pose issues because of the application of augmentations (see below), which modifies each image so that the training set no longer contains duplicates.
The YOLO v3 network is composed of 16 residual blocks, each of which contain convolution layers, batch normalization, an activation function—a leaky rectified linear unit (ReLU)—plus optional skip layers (Fig. 12; Table 3). Convolutions apply a convolution, an image processing operation, to the input to extract a specific type of image feature such as edges or corners. YOLO uses a convolution of stride 1 or of 2 to downsample, as opposed to a pooling layer, thus reducing dimensionality. Skip layers (skip connections) are used to pass feature information to deeper layers by skipping layers in between, preventing learning issues associated with vanishing gradients. Skip layers have been shown to reduce training time and improve performance, by ensuring that earlier layers of the network train more quickly. Batch normalization normalizes the input, and an activation function defines the output given some input, mapping the results of the input to a value between 0 and 1. Three YOLO layers (fully connected detection layers) are present and two upsampling layers; this enables detection of features of multiple sizes (Fig. 12). The YOLO v3 framework has a total of 106 layers [see (Redmon and Farhadi, 2018) for more details]. In total, the network contains 61,576,342 parameters constituting 61,523,734 trainable parameters and 52,608 nontrainable parameters.
Table 3. Structure of the YOLO v3 Network.
Blank cells indicate no data.
| Layer(s) | Filter | Kernel | Stride |
|---|---|---|---|
| 0–4 | [32, 64, 32, 64, Skip] | [3, 3, 1, 3] | [1, 2, 1, 1] |
| 5–8 | [128, 64, 128, Skip] | [3, 1, 3] | [2, 1, 1] |
| 9–11 | [64, 128, Skip] | [1, 3] | [1, 1] |
| 12–15 | [256, 128, 256, Skip] | [3, 1, 3] | [2, 1, 1] |
| 16–35 | [128, 256, Skip] | [1, 3] | [1, 1] |
| 36 | Skip Layer | ||
| 37–40 | [512, 256, 512, Skip] | [3, 1, 3] | [2, 1, 1] |
| 41–60 | [256, 512, Skip, ..] | [1, 3, ..] | [1, 1, ..] |
| 61 | Skip Layer | ||
| 62–65 | [1024, 512, 1024, Skip] | [3, 1, 3] | [2, 1, 1] |
| 66–74 | [512, 1024, Skip, ..] | [1, 3, ..] | [1, 1, ..] |
| 75–79 | [512, 1024, 512, 1024, 512] | [1, 3, 1, 3, 1] | [1, 1, 1, 1, 1] |
| 80–81 | [1024, (3*(5+nb_class))] | [3, 1] | [1, 1] |
| 82–83 | Detection Layer - 1 | ||
| 84 | [256] | [1] | [1] |
| 85 | Upsampling Layer - 1 | ||
| 86 | Concatenation | ||
| 87–91 | [256, 512, 256, 512, 256] | [1, 3, 1, 3, 1] | [1, 1, 1, 1, 1] |
| 92–93 | [512, (3*(5+nb_class))] | [3, 1] | [1, 1] |
| 94–95 | Detection Layer - 2 | ||
| 96 | [128] | [1] | [1] |
| 97 | Upsampling Layer - 2 | ||
| 98 | Concatenation | ||
| 99–105 | [128, 256, 128, 256, 128, 256, 3*(5+nb_class)] | [1, 3, 1, 3, 1, 3, 1] | [1, 1, 1, 1, 1, 1, 1] |
| 106 | Detection Layer - 3 |
At each epoch, the training set was passed through the network twice. Each time, on the fly augmentations were applied randomly, whereby each image in the training set had a 95% chance of being selected for augmentations. Here, several common data augmentations were applied such as rotating and flipping images. However, to further maximize the variability when training the network further augmentations were included in the form of several image processing techniques, a series of filters (i.e. gaussian blur, unsharpening, and auto-contrast), color alterations (i.e. red, green, and blue channel reduction, swap channels, sepia and sliding window convolutions), distortions (i.e. random erase, pixelate, and salt and pepper), and alterations to lighting (i.e. saturation, brightness, contrast, and sharpness). Examples of these augmentations are shown in Figure 3. For each distortion a random value is selected between a set minimum and maximum, for example, a random size and frequency for random erase, thus further increasing variability within the dataset. A maximum of 3 augmentations can be applied to a single image. The YOLO v3 network and on the fly augmentation is available at Github (Gibbs, 2018).
The image dataset was split, where 80% of the images constitute the training data, used to train the network, and the remaining 20% constitute the validation and test images (50:50), which are used to test the accuracy of the network. For training, a total of 100 epochs, with small batches of 4, because of memory limitations, were performed. The network was trained for ∼3 d on a personal computer with the following specification: Intel Core i7 6820HK, 32GB DDR4, 8GB GTX 1070. The resolution of the network was set to 576 × 576 pixels, unlike traditional object detection that may shrink images after x amount of epochs. We maintain the size because of the size of the bounding boxes and the fact that ears can significantly differ in appearance; thus a dataset of varying sizes is already in use. A learning rate of 1e−4 is used with three warmup epochs, which allow the network time to get used to the data, and the Adam optimizer (Kingma and Ba, 2015) ) is applied, which performs gradient descent. The output and accuracy of the network is discussed further in the next section.
Two-Dimensional Motion Determination: Tracking
An algorithm for detecting the motion of ear tips in a field environment is proposed. Videos of wheat crops were obtained during the field imaging stage and recorded using the Canon 650D at a pixel resolution of 1920 × 1088 and with a frame rate of 30 frames per second (FPS). Each video is split into its constituent frames. As for the YOLO training dataset, frames were cropped to maintain appropriate feature size. It is important to note that these frames act as test images and have not been annotated for ear tips; thus they have not been used to train the network. Individual frames from videos often consist of blurred plant features and other distortions, making annotation unreliable.
For each frame 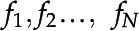 , where
, where 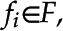 , and F is a full set of frames from some video, each set of features
, and F is a full set of frames from some video, each set of features 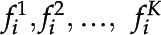 is detected. For any given frame, K is the total number of features detected by the CNN. Each feature
is detected. For any given frame, K is the total number of features detected by the CNN. Each feature 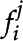 , corresponding to the
, corresponding to the 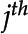 feature in the
feature in the 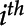 frame, is referred to as a detected feature, and is assigned an identification number, a bounding box,
frame, is referred to as a detected feature, and is assigned an identification number, a bounding box, 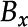 , a position (the centroid of the bounding box),
, a position (the centroid of the bounding box), 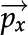 , a histogram representation,
, a histogram representation, 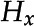 , a label,
, a label, 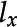 , and a size.
, and a size.
For each feature in the first frame where i = 0, a series of tracked features, 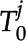 , is initialized. For the remaining frames,
, is initialized. For the remaining frames, 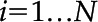 , the probability that some new feature,
, the probability that some new feature, 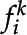 , is the same feature as one that is already being tracked is calculated, and the feature is assigned to the trajectory if the probability is greater than some defined threshold, here set to 0.8. Across each frame feature identification is maintained and tracked and is mapped into a series of movements or trajectories as online tracking, where only information from past frames up to the current frame is used (Luo et al., 2014). The identification of each tracked feature is maintained and kept in memory for all frames, N, such that if the feature moves out of view- (i.e. it becomes occluded or it leaves the frame), the trajectory can be terminated and restarted once it is back in view. The probability of
, is the same feature as one that is already being tracked is calculated, and the feature is assigned to the trajectory if the probability is greater than some defined threshold, here set to 0.8. Across each frame feature identification is maintained and tracked and is mapped into a series of movements or trajectories as online tracking, where only information from past frames up to the current frame is used (Luo et al., 2014). The identification of each tracked feature is maintained and kept in memory for all frames, N, such that if the feature moves out of view- (i.e. it becomes occluded or it leaves the frame), the trajectory can be terminated and restarted once it is back in view. The probability of 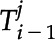 being the same feature as
being the same feature as 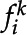 is calculated based on the following information:
is calculated based on the following information:
Euclidean distance
Euclidean distance measures distance between centroids in corresponding frames and is expected to be smaller for the same feature. Distances greater than 100 pixels apart in corresponding frames are automatically omitted from analysis to reduce computing time. Distance has a weight of 0.3 and is calculated according to Equation 1:
where px is a two-dimensional position vector.
Histogram correlation
Histogram correlation is used to compare two histograms. Histograms are commonly used to represent color appearance. A feature is more likely to have the same histogram in corresponding frames than any other feature. In this instance the histogram has 32 bins, which was found to produce more accurate probabilities than higher or lower values. Histogram correlation has a weight of 0.2 and is calculated according to Equation 2:
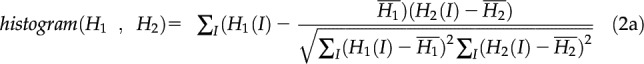 |
where Hx is the histogram of x and N is the number of bins and where
 |
Bounding box area
Because the trained network is used for feature detection, each feature is expected to be consistently labeled with a high degree of accuracy, that is, the bounding box of some feature is more likely to be of similar size in adjacent frames. However, this may not hold for instances where a feature becomes occluded. Consequently, A has a weight of 0.1 and is calculated according to Equation 3:
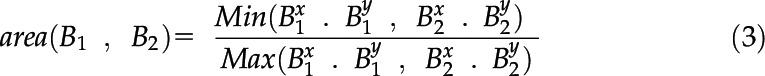 |
where Bi is the bounding box of x.
Movement direction
The direction of movement of a feature, k, is likely to be consistent across frames; however, stochastic changes in wind speed and direction, combined with the anchoring of a plant structure to the ground, mean that the direction of movement of the ear tip can change. The direction of movement for the previous frames is stored and used as a basis. Direction has a weight of 0.1 and can be calculated according to Equation 4:
where
 |
Label
An object will maintain the same identification across frames, that is, an ear tip cannot become a leaf tip. If only one feature is present (i.e. ear tips), probability = 1; if more than one feature is present the probability is either 0.5 or 1 depending on whether the labels are equal. Label has a weighting of 0.3 and can be calculated by Equation 5:
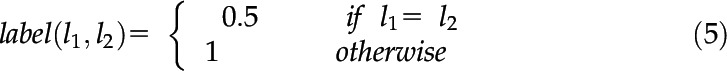 |
where lx is the feature label.
The probability, Pi, that fi + 1 is the same feature as  is then calculated according to Equation 6:
is then calculated according to Equation 6:
If the probability is lower than the threshold of 0.8, it is considered that the detected feature is not assigned to an existing trajectory; a new identification is assigned to the feature and tracked from this point onward. Once all frames have been processed, the mean number of frames per tracked feature is determined. If the tracked feature comprises fewer frames than the mean, the tracked feature is re-evaluated starting at t = N to merge trajectories. This is computed by comparing all identified features that are less than the mean probability to higher probability features to determine whether they can be merged, that is, to see if they are the same feature.
The output is a series of trajectories (given as sequences of coordinates) for each of the identified features and a video combining the original frames with mapped trajectories to visualize movement paths of the ears (Fig. 6; Supplemental Movie S1).
SUPPLEMENTAL DATA
The following supplemental material is available:
Supplemental Movie S1. Feature tracking of wheat ear tips in a video.
Footnotes
This work was supported by the Biotechnology and Biological Sciences Research Council (BBSRC) (grant no. BB/R004633/1).
[CC-BY]: Article free via Creative Commons CC-BY 4.0 license.
References
- ACID (2018) University of Nottingham; Nottingham, United Kingdom. https://plantimages.nottingham.ac.uk (March 3, 2019)
- Aich S, Stavness I (2018) Leaf counting with deep convolutional and deconvolutional networks. In: 2017 IEEE International Conference on Computer Vision Workshops (ICCVW). IEEE, pp 2080–2089 [Google Scholar]
- Berry P, Spink J, Foulkes M, Wade A (2003) Quantifying the contributions and losses of dry matter from non-surviving shoots in four cultivars of winter wheat. F Crop Res 80: 111–121 [Google Scholar]
- Berry P, Sylvester-Bradley R, Berry S (2007) Ideotype design for lodging-resistant wheat. Euphytica 154: 165–179 [Google Scholar]
- Betke M, Haritaoglu E, Davis L (2000) Real-time multiple vehicle detection and tracking from a moving vehicle. Mach Vis Appl 12: 69–83 [Google Scholar]
- Burgess AJ, Retkute R, Preston SP, Jensen OE, Pound MP, Pridmore TP, Murchie EH (2016) The 4-dimensional plant: Effects of wind-induced canopy movement on light fluctuations and photosynthesis. Front Plant Sci 7: 1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess A, Gibbs J, Murchie E (2019) A canopy conundrum: Can wind-induced movement help to increase crop productivity by relieving photosynthetic limitations? J Exp Bot 70: 2371–2380. [DOI] [PubMed] [Google Scholar]
- Cai J, Kumar P, Chopin J, Miklavcic SJ (2018) Land-based crop phenotyping by image analysis: Accurate estimation of canopy height distributions using stereo images. PLoS One 13: e0196671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell MM. (1970) Plant gas exchange at high wind speeds. Plant Physiol 46: 535–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cointault F, Gouton P (2007) Texture or color analysis in agronomic images for wheat ear counting. In: International IEEE Conference on Signal-Image Technologies and Internet-Based System, 2007. SITIS 2007. IEEE, pp 696–701 [Google Scholar]
- de Langre E. (2008) Effects of wind on plants. Annu Rev Fluid Mech 40: 141–168 [Google Scholar]
- Der Loughian C, Tadrist L, Allain J, Diener J, Moulia B, De Langre E (2014) Measuring local and global vibration modes in model plants. C R Mec 342: 1–7 [Google Scholar]
- Doaré O, Moulia B, de Langre E (2004) Effect of plant interaction on wind-induced crop motion. J Biomech Eng 126: 146–151 [DOI] [PubMed] [Google Scholar]
- Dobrescu A, Giuffrida M, Tsaftaris S (2018) Leveraging multiple datasets for deep leaf counting. In: IEEE International Conference on Computer Vision Workshops (ICCVW). IEEE, pp 2072–2079 [Google Scholar]
- Fernandez-Gallego JA, Kefauver SC, Gutiérrez NA, Nieto-Taladriz MT, Araus JL (2018) Wheat ear counting in-field conditions: High throughput and low-cost approach using RGB images. Plant Methods 14: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs J. (2018) An implementation of the YOLO framework for a custom dataset (Wheat). https://github.com/pszjg/YoloV3_Wheat (November 19, 2018)
- Girshick R. (2015) Fast R-CNN. In: Proceedings of the IEEE International Conference on Computer Vision. IEEE, pp 1440–1448 [Google Scholar]
- He K, Zhang X, Ren S, Sun J (2016) Deep Residual Learning for Image Recognition. In: IEEE Conference on Computer Vision and Pattern Recognition (CVPR). IEEE, pp 770–778 [Google Scholar]
- Kaiser E, Morales A, Harbinson J (2018) Fluctuating light takes crop photosynthesis coaster ride. Plant Physiol 176: 977–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamilaris A, Prenafeta-Boldú F (2018) Deep learning in agriculture: A survey. Comput Electron Agric 147: 70–90 [Google Scholar]
- Kingma D, Ba J (2015) Adam: Method for Stochastic Optimization . International Conference for Learning Representations, San Diego, CA. https://arxiv.org/abs/1412.6980 [Google Scholar]
- Krizhevsky A, Sutskever I, Hinton G (2012) ImageNet Classification with Deep Convolutional Neural Networks. In Pereira F, Burges CJC, Bottou L, Weinberger KQ (eds) Adv. Neural Inf. Process. Syst. Curran Associates, Inc, vol 25 pp 1097–1105 [Google Scholar]
- LabelImg (2018) LabelImg is a graphical image annotation tool and label object bounding boxes in images. https://github.com/tzutalin/labelImg (July 15, 2018)
- Lee B, Erdenee E, Jin S, Nam M, Jung Y, Rhee P (2016) Multi-class multi-object tracking using changing point detection.In: Hua G., Jégou H (eds) Computer Vision – ECCV 2016 Workshops. ECCV 2016. Lecture Notes in Computer Science, vol 9914 Springer, Cham, pp 68–83 [Google Scholar]
- Lu WL, Ting JA, Little JJ, Murphy KP (2013) Learning to track and identify players from broadcast sports videos. IEEE Trans Pattern Anal Mach Intell 35: 1704–1716 [DOI] [PubMed] [Google Scholar]
- Luo W, Xing J, Milan A, Zhang X, Liu W, Zhao X, Kim T-K (2014) Multiple Object Tracking: A Review. https://arxiv.org/abs/1409.7618 (December 01, 2018)
- Meijering E, Dzyubachyk O, Smal I, van Cappellen WA (2009) Tracking in cell and developmental biology. Semin Cell Dev Biol 20: 894–902 [DOI] [PubMed] [Google Scholar]
- Mohanty SP, Hughes DP, Salathé M (2016) Using deep learning for image-based plant disease detection. Front Plant Sci 7: 1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser D, Drapela T, Zaller J, Frank T (2009) Interacting effects of wind direction and resource distribution on insect pest densities. Basic Appl Ecol 10: 208–215 [Google Scholar]
- Onoda Y, Anten NP (2011) Challenges to understand plant responses to wind. Plant Signal Behav 6: 1057–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini S, Ess A, Schindler K, Van Gool L (2009) You’ll never walk alone: Modeling social behavior for multi-target tracking. In: 2009 IEEE 12th International Conference on Computer Vision. IEEE, pp 261–268 [Google Scholar]
- Piñera-Chavez FJ, Berry PM, Foulkes MJ, Jesson MA, Reynolds MP (2016) Avoiding lodging in irrigated spring wheat. I. Stem and root structural requirements. Field Crops Res 196: 325–336 [Google Scholar]
- Pound M, Atkinson J, Townsend A, Wilson M, Griffiths M, Jackson A, Bulat A, Tzimiropoulos G, Wells D, Murchie E, et al. (2017) Deep machine learning provides state-of-the-art performance in image-based plant phenotyping. Gigascience 6: 1–10; erratum Pound M, Atkinson J, Townsend A, Wilson M, Griffiths M, Jackson A, Bulat A, Tzimiropoulos G, Wells D, Murchie E, et al. (2018) Gigascience 7:giy042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmon J, Farhadi A (2016) YOLO9000: Better, faster, stronger. https://arxiv.org/abs/1612.08242 (September 01, 2018)
- Redmon J, Farhadi A (2018) YOLOv3: An incremental improvement. https://arxiv.org/abs/1804.02767 (September 01, 2018)
- Redmon J, Divvala S, Girshick R, Farhadi A (2015) You Only Look Once: Unified, Real-Time Object Detection. https://arxiv.org/abs/1506.02640 (September 01, 2018)
- Roden J. (2003) Modeling the light interception and carbon gain of individual fluttering aspen (Populus tremuloides Michx) leaves. Trees (Berl) 17: 117–126 [Google Scholar]
- Roden JS, Pearcy RW (1993a) Effect of leaf flutter on the light environment of poplars. Oecologia 93: 201–207 [DOI] [PubMed] [Google Scholar]
- Roden JS, Pearcy RW (1993b) Photosynthetic gas exchange response of poplars to steady-state and dynamic light environments. Oecologia 93: 208–214 [DOI] [PubMed] [Google Scholar]
- Romera-Paredes B, Torr P (2016) Recurrent instance segmentation. In: Computer Vision – ECCV Workshops. ECCV 2016. Lecture Notes in Computer Science. Leibe B., Matas J., Sebe N., Welling M. (eds) 9910. Springer, Cham: pp9910: 312–329 [Google Scholar]
- Shaw R. (2012) Wind movement within canopies. In Biometeorology in Integrated Pest Management. Hatfield J. (ed). Elsevier, London, New York, pp 17–41 [Google Scholar]
- Smith VC, Ennos AR (2003) The effects of air flow and stem flexure on the mechanical and hydraulic properties of the stems of sunflowers Helianthus annuus L. J Exp Bot 54: 845–849 [DOI] [PubMed] [Google Scholar]
- Spampinato C, Chen-Burger Y-H, Nadarajan G, Fisher R (2008) Detecting, Tracking and Counting Fish in Low Quality Unconstrained Underwater Videos. Proc. 3rd Int. Conf. Comput. Vis. Theory Appl. (VISAPP), vol 2 pp 514–519 [Google Scholar]
- Spampinato C, Palazzo S, Giordano D, Kavasidis I, Lin F-P, Lin Y (2012) Covariance based fish tracking in real-life underwater environment. Proc. Int. Conf. Comput. Vis. Theory Appl. pp 409–414 [Google Scholar]
- Ubbens JR, Stavness I (2017) Deep plant phenomics: A deep learning platform for complex plant phenotyping tasks. Front Plant Sci 8: 1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velumani K, Oude Elberink S, Yang M, Baret F (2017) Wheat ear detection in plots by segmenting mobile laser scanner data. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci, pp 149–156 [Google Scholar]
- Wang L, Ouyang W, Wang X, Lu H (2015) Visual tracking with fully convolutional networks. In: ICCV '15 Proceedings of the 2015 IEEE International Conference on Computer Vision (ICCV). IEEE Computer Society, Washington, DC, pp 3119–3127 [Google Scholar]
- Yang H, Shao L, Zheng F, Wang L, Song Z (2011) Recent advances and trends in visual tracking: A review. Neurocomputing 74: 3823–3831 [Google Scholar]
- Yu F, Li W, Li Q, Liu Y, Shi X, Yan J (2016) POI: Multiple object tracking with high performance detection and appearance feature. In: Computer Vision – ECCV 2016 Workshops. ECCV 2016. Lecture Notes in Computer Science, Hua G., Jégou H (eds). Springer, Cham, 9914: 36–42 [Google Scholar]
- Zadoks C, Chang TT, Konzak CF (1974) A decimal code for the growth stages of cereals. Weed Res 14: 415–421 [Google Scholar]