Class II TCP TFs interact with FD and act directly at the AP1 promoter to control flowering.
Abstract
The appropriate timing of flowering is critical for plant reproductive success. Although the FLOWERING LOCUS T (FT)–FD module plays crucial roles in the photoperiodic flowering pathway, the underlying mechanisms and signaling pathways involved still remain elusive. Here, we demonstrate that class II TCP transcription factors (TFs) integrate into the FT–FD complex to control floral initiation in Arabidopsis (Arabidopsis thaliana). Class II CINCINNATA (CIN) TCP TFs function as transcriptional activators by directly binding to the promoters of downstream floral meristem identity genes, such as APETALA1 (AP1). In addition, these TCPs directly interact with FD, a basic Leu zipper TF that plays a critical role in photoperiodic flowering, which further activates AP1 expression. Genetic analyses indicated that class II CIN TCP TFs function synergistically with FT and FD, to positively regulate flowering in an AP1-dependent manner. Thus, our results provide compelling evidence that class II CIN TCP TFs act directly at the AP1 promoter to enhance its transcription, thus further elucidating the molecular mechanisms underlying the regulation of photoperiodic flowering in Arabidopsis.
The correct phase transition from vegetative to reproductive growth determines the reproductive success of flowering plants because the timing of this must correlate with suitable conditions for fertilization and seed dispersal (Huijser and Schmid, 2011; Yamaguchi and Abe, 2012). Intricate gene regulatory networks consisting of multiple overlapping, cross-regulating pathways have evolved to coordinate this developmental switch. The environmental conditions and endogenous developmental cues are integrated in these networks, which then converge to modulate the expression of a set of key floral integrators, including FLOWERING LOCUS T (FT), LEAFY (LFY), and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1; Han et al., 2008; Kumar et al., 2012; Riboni et al., 2013; Wang, 2014). This subsequently activates several floral meristem identity genes, including LFY, APETALA1 (AP1), CAULIFLOWER, and FRUITFULL (FUL), leading to the switch from vegetative to reproductive growth (Han et al., 2008; Davis, 2009; Wang, 2014). As the timing of flowering has a considerable effect on both plant fitness and crop yield, a comprehensive understanding of the regulatory mechanisms controlling flowering time is crucial for continued improvements in agricultural practices (Huijser and Schmid, 2011; Srikanth and Schmid, 2011).
Arabidopsis (Arabidopsis thaliana) is a model long-day (LD) plant, and thus its flowering is promoted by LD conditions and repressed by short-day (SD) conditions via their effect on the photoperiod pathway. The FT–FD module functions as a key component of the photoperiodic flowering pathway. This model depicts the FT protein moving through the vasculature to the plant apex (Turck et al., 2008), where it interacts with the basic Leu zipper transcription factor (TF) FD to activate floral meristem identity genes, such as AP1 and FUL (Abe et al., 2005; Wigge et al., 2005; Jung et al., 2016). Mutations in the residues of FT or FD that mediate their interactions disrupt the transcriptional activation activity of FD (Abe et al., 2005; Wigge et al., 2005). The formation of the FT–FD complex appears to be bridged by 14-3-3 proteins and to depend on the phosphorylation of FD (Abe et al., 2005; Taoka et al., 2011; Kawamoto et al., 2015). Recently, SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE3 (SPL3), SPL4, and SPL5 was demonstrated to act synergistically with the FT–FD module to induce flowering under LD conditions through interactions with FD (Jung et al., 2016). As a key factor that links FT and initiation of floral development, the FD protein alone does not directly bind to the promoter of AP1 (Benlloch et al., 2011), raising the question of how the FT–FD complex regulates downstream floral meristem identity genes. Thus, to understand how the FT–FD module participates in the photoperiod pathway, it is still necessary to identify their interacting proteins that may contribute to the reconstruction of signaling pathways that involve the FT–FD module.
By positively or negatively regulating the floral transcriptome, TFs control a significant proportion of the floral transition. The roles of TFs include the involvement of MADS-box TFs in floral transition (Becker and Theissen, 2003). The TCP protein family was named based on the first characterized members, namely TEOSINTE BRANCHED1 in maize (Zea mays), CYCLOIDEA in snapdragon (Antirrhinum majus), and PROLIFERATING CELL FACTORS1 (PCF1) and PCF2 in rice (Oryza sativa), which each contain a noncanonical basic helix–loop–helix motif referred to as the “TCP” domain (Cubas et al., 1999; Kosugi and Ohashi, 2002; Navaud et al., 2007). The TCP domain is responsible for the mediation of DNA binding or interactions with other proteins (Cubas et al., 1999). In Arabidopsis, the TCP transcription family comprises 24 members that can be classified into two subfamilies, class I (PCF or TCP-P) and class II (TCP-C), based on the structure of the TCP domain (Cubas et al., 1999; Navaud et al., 2007). The class II TCPs can be further divided into two clades, namely the CINCINNATA (CIN)-like TCP (CIN-TCP) clade and the CYCLOIDEA/TEOSINTE BRANCHED1 clade (Navaud et al., 2007). The CIN-TCP clade consists of eight members (TCP2/3/4/5/10/13/17/24), five of which are regulated by microRNA319 (miRNA319; Palatnik et al., 2003). The TCP members play essential roles in plant growth and development by influencing cell proliferation and cell differentiation, with class I TCP members promoting, and class II TCP members inhibiting, cell proliferation and growth (Cubas et al., 1999; Nath et al., 2003; Palatnik et al., 2003; Hervé et al., 2009). In addition to their involvement in cell proliferation and expansion, the TCP members contribute to leaf-shape control, axillary meristem development, plant height determination, floral organ asymmetry, hormone synthesis and signaling, and both biotic and abiotic stress responses (Martín-Trillo and Cubas, 2010; Davière et al., 2014; Kim et al., 2014; Nicolas and Cubas, 2016; Viola et al., 2016).
Recent functional analyses demonstrated that TCP proteins also participate in flowering time regulation. In Arabidopsis, the jaw-d mutants, which overexpress miRNA319 and downregulate TCP2/3/4/10/24, were first found to show a modest delayed-flowering phenotype (Palatnik et al., 2003; Schommer et al., 2008; Sarvepalli and Nath, 2011). These miRNA319-regulated TCPs also physically interact with both FLOWERING BHLH and PHYTOCHROME AND FLOWERING TIME1 to facilitate CONSTANS (CO) transcription and function as positive regulators of Arabidopsis photoperiodic flowering (Liu et al., 2017). TCP4 can also physically interact with GIGANTEA and promoter CO expression in a GIGANTEA-dependent manner (Kubota et al., 2017). The class I TCP protein TCP15 promotes flowering by acting upstream of the flowering integrator SOC1 (Lucero et al., 2017). On the contrary, class I TCP proteins TCP20 and TCP22 interact with clock proteins LWD1 and LWD2 and delay flowering by regulating the expression of the clock gene CIRCADIAN CLOCK ASSOCIATED1 (Wu et al., 2016).
In this work, we used molecular and genetic approaches to investigate the roles of class II TCP TFs in flowering time regulation. Our results demonstrate that altered expression levels of the class II CIN TCP TF genes affect floral initiation. The class II CIN TCP TFs act as transcriptional activators to control flowering by directly binding the promoter of AP1. Moreover, these TCPs directly interact with FD causing additive activation of AP1 expression, and these TCPs function redundantly among themselves or synergistically with FT and FD to positively regulate flowering in an AP1-dependent manner. Our results thus provide compelling evidence that class II TCP TFs integrate with the FT–FD complex to control flowering in Arabidopsis.
RESULTS
TCP5/13/17 Expression Knockdown or Ectopic Expression Affect Floral Initiation
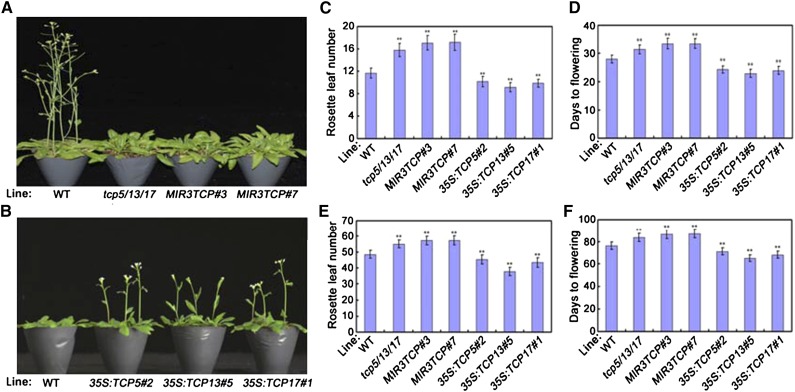
Previous studies have demonstrated that TCP5/13/17 function as important regulators in leaf development (Efroni et al., 2008). To investigate the possible regulatory roles of these three TCP TFs in flowering, we first obtained one tcp5/13/17 triple T-DNA insertion mutant and two lines using the Cauliflower mosaic virus (CaMV) 35S promoter, 35S:miR-TCP5/13/17 (MIR3TCP#3 and MIR3TCP#7), to determine their possible involvement in flowering time regulation. As shown in Supplemental Figure S1, reverse transcription quantitative PCR (RT-qPCR) analysis demonstrated that the expression levels of TCP5/13/17 were effectively knocked down by the T-DNA insertion and the artificial miRNA. Then, to determine the possible functions of the three TCPs in flowering time regulation, we analyzed the flowering phenotypes of tcp5/13/17, MIR3TCP#3, and MIR3TCP#7. Wild-type, tcp5/13/17, MIR3TCP#3, and MIR3TCP#7 seeds were germinated simultaneously and grown in soil under the same growth conditions. As shown in Figure 1, A–D, under LD (16-h light/8-h dark) conditions, the tcp5/13/17 mutant plants exhibited a delayed flowering phenotype, as measured by the total rosette leaf number (RLN) and days from germination to flowering (DTF). Under SD conditions, the tcp5/13/17 mutant plants showed a similar delayed flowering phenotype compared with their wild-type counterparts (Fig. 1, E and F). However, their single or double T-DNA insertion mutant plants exhibited similar flowering phenotypes as the wild type (Supplemental Fig. S2). Thus, the knockdown of tcp5/13/17 caused a delay in flowering time in Arabidopsis and their function in flowering time control may be redundant.
Figure 1.
Flowering phenotype of TCP5/13/17 expression knockdown or ectopic expression plants. A, Representative images of TCP5/13/17 knockdown plants showing their flowering phenotype under LD conditions. Three independent experiments were performed with each replica containing >30 plants for each line. Representative plants were photographed. B, Representative images of 35S:TCP5/13/17 transgenic plants showing their flowering phenotype under LD conditions. Three independent experiments were performed with each replica containing >30 plants for each line. Representative plants were photographed. C and D, Flowering phenotype of TCP5/13/17 knockdown or ectopic expression plants assessed by RLN (C) and DTF (D) under LD conditions. E and F, Flowering phenotype of TCP5/13/17 knockdown or ectopic expression plants assessed by RLN (E) and DTF (F) under SD conditions. C–F, Values are means ± sd of ∼30 plants. Asterisks indicate significant difference compared to that of wild type (WT); Student’s t test, **P < 0.01. The experiments were repeated at least three times with similar results.
To further investigate the roles of TCP5/13/17 in flowering time regulation, we generated transgenic Arabidopsis plants constitutively expressing TCP5/13/17 under the control of the CaMV 35S promoter. Three representative transgenic lines, one from each construct type, were selected from 21 primary T1 35S:TCP5, 15 primary T1 35S:TCP13, and 19 primary T1 35S:TCP17 lines (Supplemental Fig. S3, A–C). In contrast to tcp5/13/17 mutant plants, flowering was clearly accelerated in the 35S:TCP5, 35S:TCP13, and 35S:TCP17 plants compared with wild-type plants, as measured by total RLN and DTF under both LD and SD conditions (Fig. 1, B–F; Supplemental Fig. S3, D and E). Thus, the constitutive overexpression of TCP5/13/17 accelerated flowering time in Arabidopsis. These results confirmed that TCP5/13/17 function as positive regulators of flowering initiation.
In Vivo Interactions of TCP5/13/17 with the AP1 Promoter
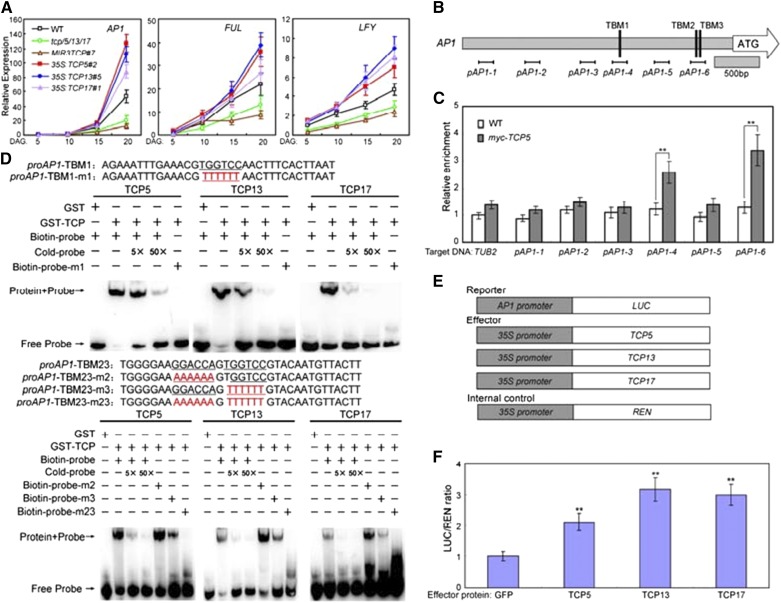
To determine the mechanisms underlying TCP5/13/17 modulation of flowering time regulation, we analyzed the expression patterns of both flowering time-related genes and floral meristem identity genes. As shown in Supplemental Figure S4, the expression levels of both CO and FT were slightly affected by altered expression of TCP5/13/17. Furthermore, consistent with the flowering phenotypes, the expression levels of several downstream floral meristem identity genes, including AP1, FUL, and LFY, were all lower in tcp5/13/17 mutant plants compared with those in wild type (Fig. 2A). By contrast, the expression levels of these genes were all greater in 35S:TCP5, 35S:TCP13, and 35S:TCP17 plants (Fig. 2A). Thus, the three TCPs may regulate flowering through both flowering time-related genes and floral meristem identity genes.
Figure 2.
TCP5/13/17 promotes flowering by activating AP1 transcription. A, Expression of AP1, FUL, and LFY in the indicated genotypes and time points. Whole plants were harvested for total RNA extraction at the indicated time points. Transcript levels of floral meristem identity genes in Col-0 plants were arbitrarily set to 1. ACTIN2 expression levels were used as an internal control. Values are means ± sd of three independent biological replicates. DAG, days after germination. B, The promoter structure of the AP1 gene and fragment used in the chromatin immunoprecipitation (ChIP) assay. (Upper) Schematic representation of the AP1 promoter regions containing TCP-binding motif (TBM) elements. The diagram indicates the number and relative position of the TBMs in the respective promoters relative to the ATG start codon. In the promoter fragment names, the prefix “p” indicates promoter. Black lines indicate the position of sequences detected by ChIP assays. C, ChIP-qPCR analysis of the relative binding of TCP5 to the promoter of AP1.ChIP assays were performed with chromatin prepared from Myc-TCP5 plants using an anti-Myc antibody. Gray bars indicate the enrichment fold changes normalized to TUB2. Values are means ± sd of three independent biological replicates. Asterisks indicate significant differences; Student’s t test, **P < 0.01. D, The electrophoretic mobility shift assays (EMSA) analysis of the binding of recombinant TCP5/13/17 proteins to the promoter of AP1. The oligonucleotides (proAP1-TBM1/2/3 and proAP1-TBM1/2/3-m) were used as the probes. TBMs sequences and their mutated forms (depicted in red) are underlined. glutathione S-transferase (GST), GST-TCP5/13/17, biotin-probe, labeled mutated probe, and unlabeled probe at a 5× and 50× molar excess were present (+) or absent (−) in each reaction. E, Schematic of the reporter and effectors used in the transient transactivation assays. F, Transient dual-luciferase (LUC) reporter assays show that TCP5/13/17 activates the expression of AP1. Values are means ± sd of three independent biological replicates. Asterisks indicate significant difference compared to that of GFP; Student’s t test, **P < 0.01. WT, wild type.
TCP5/13/17 belong to class II TCP TFs and may bind to the common TBM (GGACCA) to regulate the expression levels of target genes (Schommer et al., 2008). Interestingly, a search of the Arabidopsis genome sequence uncovered several putative TBM elements in the promoter of AP1 (Fig. 2B). The presence of these elements indicated that the modulations we observed may have been caused by the direct interaction of TCP5/13/17 with the AP1 promoter. To examine whether AP1 is a direct target of TCP5/13/17, in vivo ChIP assays were performed using a transgenic line expressing a TCP5 cDNA construct with an N-terminal Myc tag under the control of its native promoter in the tcp5/13/17 triple mutant (designated “Myc-TCP5”). The ChIP-qPCR results showed that TCP5 could bind to the promoter of AP1 through the TBM sequence (pAP1-4 and pAP1-6; Fig. 2C). Additionally, we conducted EMSAs with the GST-TCP5/13/17 recombinant proteins to determine the in vitro binding of these three TCPs to these regions. As shown in Figure 2D, TCP5/13/17 were all capable of binding to the probes containing TBM1, TBM2 and TBM3. The binding signals decreased after the addition of unlabeled wild-type competitors. In contrast, the TCP5/13/17 proteins did not bind to the probes containing the mutated TBMs (Fig. 2D). The GST protein alone also did not bind to the TBMs (Fig. 2D). These data suggest that TCP5/13/17 directly binds to the promoter of AP1.
To confirm the positive regulatory functions of TCP5/13/17, we performed transient expression assays in Columbia-0 wild-type Arabidopsis mesophyll protoplasts (Yoo et al., 2007). Because AP1 is a direct target of TCP5/13/17, the AP1 promoter was fused to the LUC gene as a reporter (AP1:LUC; Fig. 2E). The effector constructs contained TCP5/13/17 genes driven by the CaMV35S promoter (35S:TCP5/13/17; Fig. 2E). The coexpression of TCP5/13/17 with the reporter plasmid resulted in the activated expression of LUC compared with the control (Fig. 2F). This result further supports the hypothesis that TCP5/13/17 act as positive regulators of flowering time control through the direct activation of AP1.
TCP5/13/17 Promote Flowering in an AP1-Dependent Manner
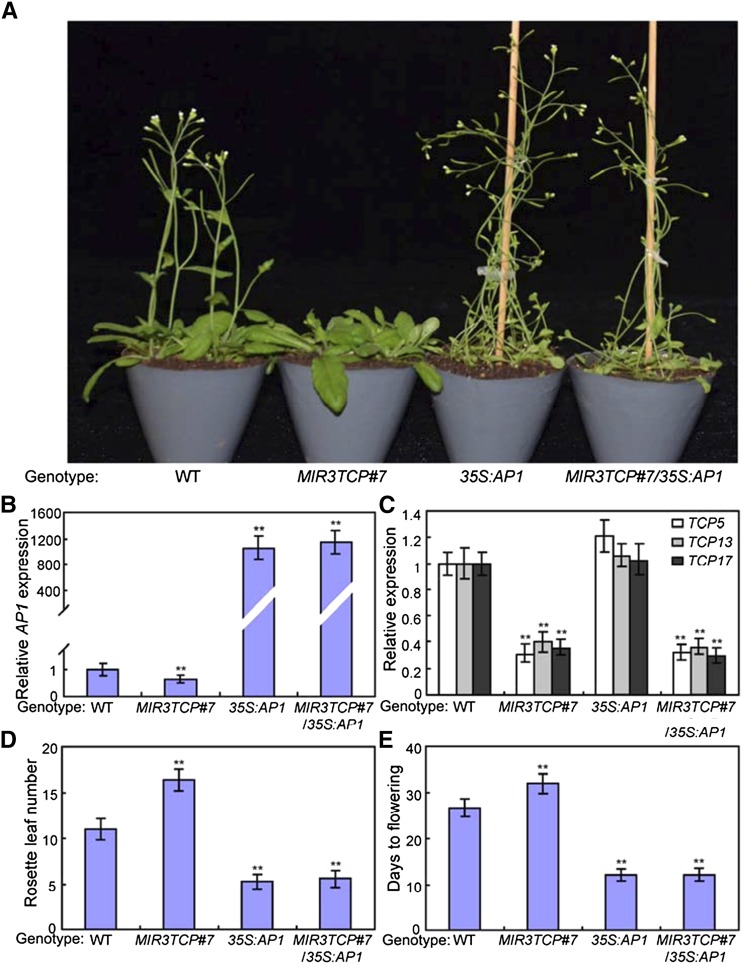
The phenotypic analysis, and biochemical and molecular evidence demonstrated that the TFs TCP5/13/17 positively regulate flowering time through the direct activation of AP1 expression. To further confirm this conclusion, the genetic relationships between TCP5/13/17 and AP1 were explored. MIR3TCP#7 was crossed with 35S:AP1 transgenic plants to produce MIR3TCP#7/35S:AP1, and their flowering phenotypes were examined. Under our experimental conditions, we detected an early flowering phenotype in 35S:AP1 plants, and a knockdown of TCP5/13/17 did not change the RLN and DTF values, although MIR3TCP#7 plants showed a delay in the floral transition (Fig. 3). Thus, TCP5/13/17 act upstream of AP1 and function as positive regulators of flowering time in an AP1-dependent manner.
Figure 3.
The genetic analysis of TCP5/13/17 and AP1. MiR3TCP#7/35S:AP1 was generated by genetic crossing, and then the flowering time phenotype of these genotypes was examined. A, Representative images of the indicated genotypes showing their flowering phenotype under LD conditions. Three independent experiments were performed with each replica containing >30 plants for each line. Representative plants were photographed. B and C, Expression levels of AP1 and TCP5/13/17 in the indicated genotypes. RNA was isolated from 2-week–old plants of the indicated genotype. ACTIN2 was used as an internal control. Values are means ± sd of three independent biological replicates. Asterisks indicate significant differences compared to that in the wild type (WT); Student’s t test, **P < 0.01. D and E, Flowering phenotype of the indicated genotypes assessed by RLN (D) and DTF (E) under LD conditions. Values are means ± sd of ∼30 plants. Asterisks indicate significant differences compared to that in wild type; Student’s t test, **P < 0.01.
Physical Interactions of TCP5/13/17 with FD
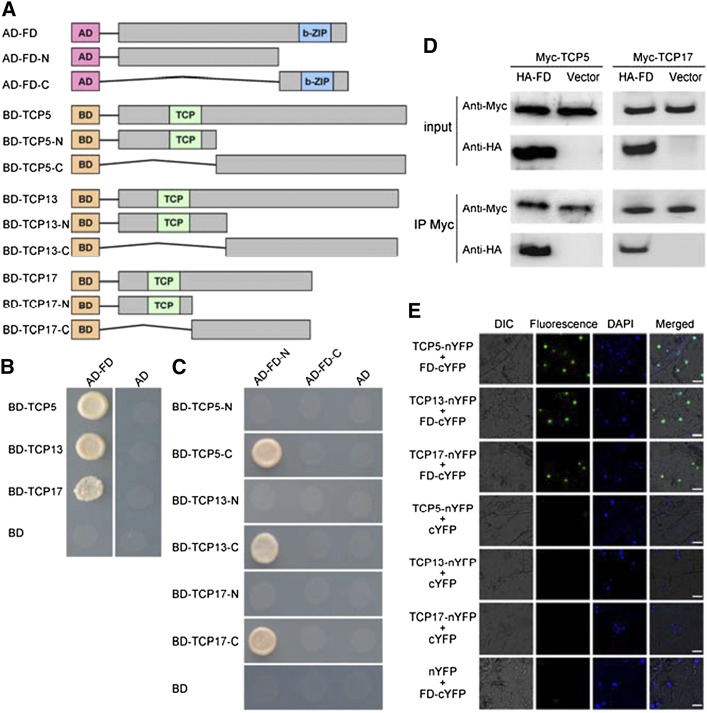
To understand how TCP5/13/17 participate in flowering time regulation, we used the yeast two-hybrid system to identify their potential interaction partners. The full-length TCP5 coding sequence (CDS) was fused to the Gal4 DNA-binding domain of the bait vector pGBKT7 (BD-TCP5). Yeast cells harboring the bait were transformed with a library of cDNAs containing inserts of prey proteins fused to GAL4-AD. After screening, three independent clones encoding FD were identified by prototrophy for His and Ade. To confirm the interaction, the open reading frame sequence of FD was fused with the AD domain of the pGADT7 vector and used in further interaction experiments with TCP5 (Fig. 4A). The bait and prey vectors were cotransformed into yeast, and the protein–protein interactions were reconstructed. We further investigated the interactions of TCP13 and TCP17 with FD in the yeast two-hybrid system. In addition to TCP5, TCP13, and TCP17 also interacted with FD (Fig. 4, A and B).
Figure 4.
Interaction between TCP5/13/17 and FD. A, Schematic representation of full-length and truncated FD and TCP5/13/17 constructs with specific deletions. B, Yeast two-hybrid assay analysis. Interaction was indicated by the ability of cells to grow on synthetic dropout medium lacking Leu, Trp, His, and Ade. The GAL4 activation domain expressed by pGADT7 (shown as “AD”) was used as negative controls. The experiment was repeated three times with similar results and representative photos are displayed. C, C-termini of TCP5/13/17 and the N terminus of FD are required for their interactions. Interactions were indicated by the ability of yeast cells to grow on synthetic dropout medium lacking Leu, Trp, His, and Ade. The GAL4 activation domain expressed by pGADT7 (shown as “AD”) was used as negative controls. The experiment was repeated three times with similar results and representative photos are displayed. D, Coimmunoprecipitation (CoIP) analysis. Myc-fused TCP5 and TCP17 were immunoprecipitated using anti-Myc antibody, and coimmunoprecipitated HA-FD was then detected using anti-HA antibody. Protein input for Myc-TCP5/17 and HA-FD in immunoprecipitated complexes were also detected and are shown. The experiment was repeated three times with similar results and representative photos are displayed. E, bimolecular fluorescence complementation (BiFC) analysis. Fluorescence was observed in nuclear compartments of N. benthamiana leaf epidermal cells; the fluorescence resulted from complementation of the C-terminal portion of yellow fluorescent protein (YFP) fused to FD (FD-cYFP) with the N-terminal portion of YFP fused to TCP5/13/17 (TCP5/13/17-nYFP). No signal was observed from negative controls. DAPI, 49,6-diamidino-2-phenylindole; DIC, Differential interference contrast. The experiment was repeated three times with similar results and representative photos are displayed. Scale bars = 25 μm.
To investigate which regions of TCP5/13/17 and FD were required for their interactions, we independently fused the N- and C-termini of FD to the AD domain of the pGADT7 vector and the N- and C-termini of TCP5/13/17 to the BD domain of the pGBKT7 vector (Fig. 4A). The interactions between these derivatives were then assayed using the yeast two-hybrid system. As shown in Figure 4C, the C-termini of TCP5/13/17 and the N terminus of FD were specifically responsible for the interactions. Thus, these results show that the C-termini of TCP5/13/17 and the N terminus of FD are important for the interactions between TCP5/13/17 and FD.
The interactions of TCP5/13/17 with FD were further corroborated by CoIP and BiFC assays. TCP5 and TCP17 were used as representative in the CoIP assay. For the CoIP analysis, Myc-TCP5 and Myc-TCP17 were coexpressed with HA-FD in Nicotiana benthamiana leaves. The protein complexes were incubated with anti-Myc and A/G-agarose beads, and then separated by SDS-PAGE for immunoblotting with an anti-HA antibody. Both TCP5 and TCP17 pulled down FD (Fig. 4D). To determine whether these interactions also occurred in plant cells, we then used BiFC. Full-length TCP5/13/17 and FD proteins were fused to the N-terminal and C-terminal fragments of YFP, yielding TCP5/13/17-nYFP and FD-cYFP, respectively. Agrobacterium cells harboring each interaction pair were infiltrated into N. benthamiana leaves. In parallel, empty vectors in combination with each fusion construct were coinfiltrated into N. benthamiana leaves as controls. After an 48-h incubation, the resultant YFP signals were observed by fluorescence microscopy. The samples coinfiltrated with an interaction pair showed YFP fluorescence in the cell nuclei, whereas no YFP fluorescence was seen in the control samples (Fig. 4E). Thus, TCP5/13/17 and its partners colocalize and interact in plant cell nuclei. TCP5/13/17 appear to physically interact with FD, and may form complexes with FD to positively regulate flowering in Arabidopsis.
TCP5/13/17 Facilitate the DNA Binding of FD
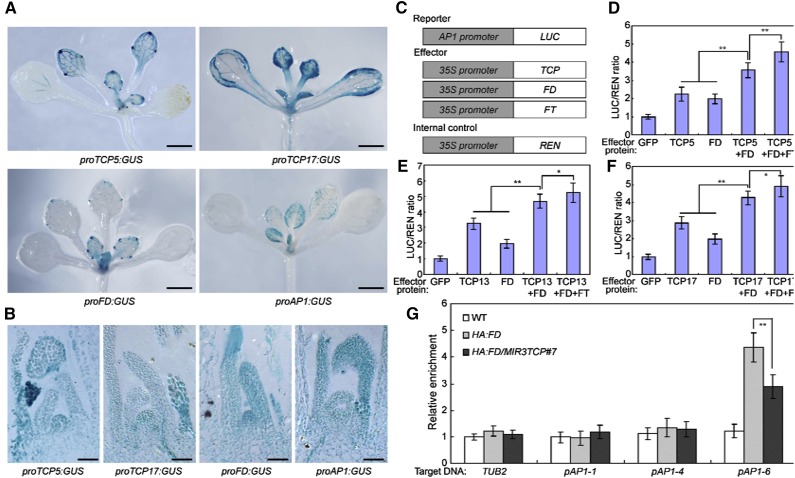
FD can form a complex with FT to function as a critical component of the photoperiodic flowering pathway, and FT binding enables FD to act as a transcriptional activator of floral meristem identity genes, such as AP1 (Abe et al., 2005; Wigge et al., 2005). Based on the repressed and induced expression of AP1 in TCP5/13/17 knockdown and overexpression plants, respectively, the direct regulation of AP1 by TCP5/13/17, and the physical interactions between TCP5/13/17 and FD, we hypothesized that TCP5/13/17 may be incorporated into the FT–FD module where they positively regulate flowering time control. Because both FD and AP1 show strong expression levels in the shoot apex (Abe et al., 2005), which was confirmed in this study (Fig. 5, A and B), we then tested whether TCP5/13/17 are also expressed in the shoot apex. An expression analysis of theGUS reporter in transgenic plants expressing GUS under the control of the TCP5/17 promoters revealed that these two promoters were active in the shoot apex (Fig. 5, A and B). Thus, these TCPs may function synergistically with the FT–FD module to promote flowering through the activation of AP1.
Figure 5.
TCP5/13/17 facilitate the DNA binding of FD. A, TCP5/17:GUS, FD:GUS, and AP1:GUS expression in 12-day–old seedlings. The expression of TCP5/17, FD, and AP1 was detected by GUS staining. Representative seedlings were photographed. Scale bars = 500 μm. B, Sections of shoot apexes of 12-d–old TCP5/17:GUS, FD:GUS, and AP1:GUS transgenic seedlings. Scale bars = 50 μm. C, Schematic of the reporter and effectors used in the transient transactivation assays. D to F, Transient dual-LUC reporter assays show that TCP5/13/17 and FT-FD module synergistically activate the expression of AP1. Values are means ± sd of three independent biological replicates. Asterisks indicate significant differences; Student’s t test, *P < 0.05, **P < 0.01. G, TCP5/13/17 facilitate the DNA binding of FD to its target gene AP1 (showed in Fig. 2B). MIR3TCP#7 was crossed with 35S:HA-FD to obtain HA-FD/MIR3TCP#7 plants. ChIP assays were performed with chromatin prepared from HA-FD/MIR3TCP#7 plants, using an anti-HA antibody. Gray bars indicate the enrichment fold changes normalized to TUB2. Values are means ± sd of three independent biological replicates. Asterisks indicate significant differences; Student’s t test, **P < 0.01. WT, wild type.
We next examined the functional relationships between TCP5/13/17 and the FT–FD module by transient LUC expression in Arabidopsis mesophyll protoplasts. In addition to the previously used reporter plasmid (AP1:LUC) and effector constructs (35S:TCP5/13/17), we designed two additional effector constructs, namely 35S:FD and 35S:FT (Fig. 5C). As shown in Figure 5, D–F, the coexpression of TCP5/13/17 or FD alone with the reporter plasmid elevated LUC expression by ∼2- to 3-fold relative to the effector control. The LUC expression was further elevated when TCP5/13/17 were coexpressed with FD or with both FD and FT, indicating that TCP5/13/17 may act synergistically with the TF–FD module to activate AP1 expression (Fig. 5, D–F). These results support the hypothesis that TCP5/13/17 and the FT–FD module act additively to activate AP1 expression.
Although the ChIP assays demonstrated that FD can strongly bind to the sequence region containing the C-box in the AP1 promoter (Supplemental Fig. S5; Wigge et al., 2005; Jung et al., 2016), the EMSAs revealed that FD alone does not bind to this conserved sequence motif in the AP1 promoter (Supplemental Fig. S6; Benlloch et al., 2011; Jung et al., 2016). The sequence analysis uncovered two putative TBMs (2/3) near the C-box in the AP1 promoter (Fig. 2B; Supplemental Fig. S5), raising the possibility that TCP5/13/17 bind to the TBM motifs and facilitate the recruitment of FD to the AP1 promoter. To examine whether TCP5/13/17 mediate the DNA binding of FD, we investigated whether knockdowns of TCP5/13/17 could repress the binding of FD to the floral meristem identity genes. We then crossed MIR3TCP#7 with 35S:HA-FD to produce HA-FD/MIR3TCP. ChIP assays using two-week–old HA-FD/MIR3TCP plants revealed that knockdown of TCP5/13/17 can reduce the binding of FD to floral meristem identity genes (Fig. 5G), indicating that TCP5/13/17 can potentiate the FT–FD module in photoperiodic flowering by facilitating the binding of FD to its target loci.
TCP5/13/17 and the FT–FD Module Synergistically Control Flowering Initiation
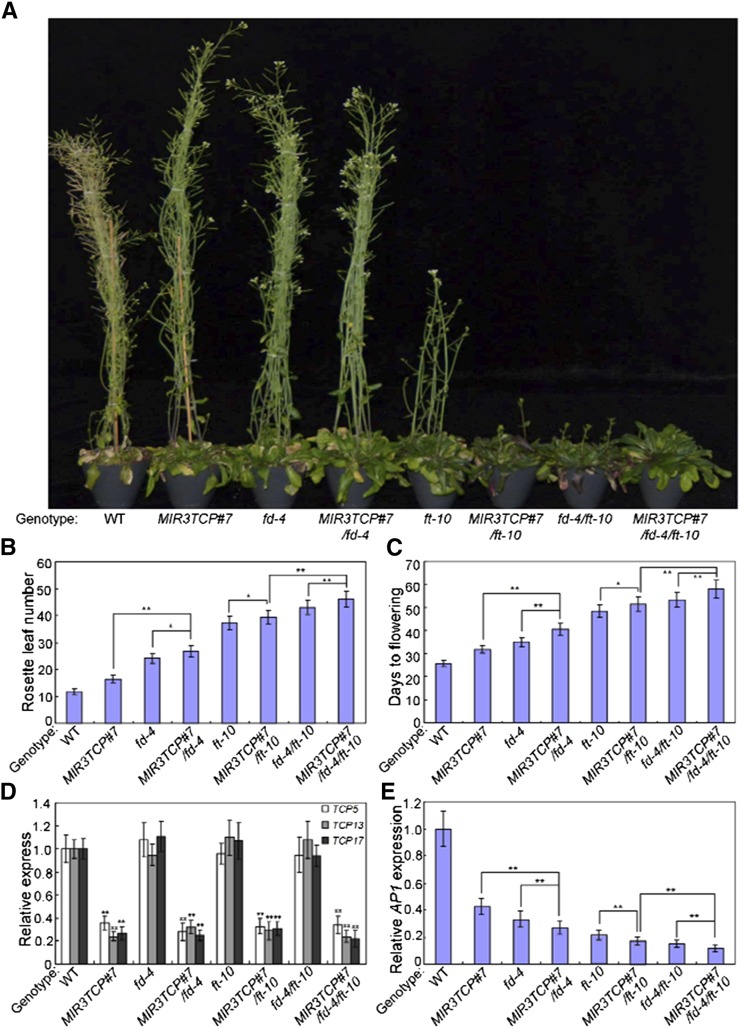
To investigate the possible signaling connections between TCP5/13/17 and the FT–FD module in photoperiodic flowering, we crossed 35S:TCP5/13/17 with the FT-defective ft-10 mutant. The early flowering phenotype of 35S:TCP5/13/17 plants was largely compromised in the 35S:TCP5/13/17/ft-10 plants (Supplemental Fig. S7, A and B). Accordingly, the high-level expression of AP1 in 35S:TCP5/13/17 plants was also suppressed by the ft-10 mutation (Supplemental Fig. S7C). The early flowering time phenotype of 35S:TCP5/13/17 plants was also delayed by the fd-4 mutation, and AP1 expression was suppressed in the 35S:TCP5/13/17/fd-4 plants (Supplemental Fig. S7, D–F). Thus, the promotion of flowering by TCP5/13/17 is closely associated with FT and FD.
TCP5/13/17 may be transcriptional activators of AP1. To further analyze the functional relationships between TCP5/13/17 and the FT–FD module, we conducted genetic crossings among MIR3TCP#7, fd-4 and ft-10 to produce MIR3TCP#7/fd-4, MIR3TCP#7/ft-10, fd-4/ft-10 and MIR3TCP#7/fd-4/ft-10. As shown in Figure 6, A–D, knockdowns of TCP5/13/17 can delay the flowering times of both fd-4 and ft-10, and can further delay the flowering time of fd-4/fd-10. Consistent with the flowering phenotypes, the expression of AP1 was also synergistically suppressed by the knockdown of both TCP5/13/17 and FT–FD module (Fig. 6E). Thus, TCP5/13/17 and the FT–FD module act synergistically to control flowering in Arabidopsis.
Figure 6.
TCP5/13/17 and FT–FD module synergistically control flowering initiation. A, Representative images of the indicated genotypes showing their flowering phenotype under LD conditions. Three independent experiments were performed with each replica containing more than 30 plants for each line. Representative plants were photographed. B and C, Flowering phenotype of the indicated genotypes assessed by RLN (B) and DTF (C) under LD conditions. Values are means ± sd of ∼30 plants. Asterisks indicate significant differences; Student’s t test, *P < 0.05, **P < 0.01. The experiments were repeated at least three times with similar results. D, Relative expression of TCP5/13/17 in the indicated genotypes. Transcript levels of TCP5/13/17 in Col-0 leaves were arbitrarily set to “1.” ACTIN2 was used as an internal control. Values are means ± sd of three independent biological replicates. Asterisks indicate significant differences compared with that in wild type (WT); Student’s t test, **P < 0.01. E, Expression of AP1 in the indicated genotypes. RNA was isolated from 2-week–old plants of the indicated genotype. ACTIN2 gene was used as an internal control. Values are means ± sd of three independent biological replicates. Asterisks indicate significant differences; Student’s t test, *P < 0.05, **P < 0.01.
Class II TCP TFs Interact with FD to Control Flowering
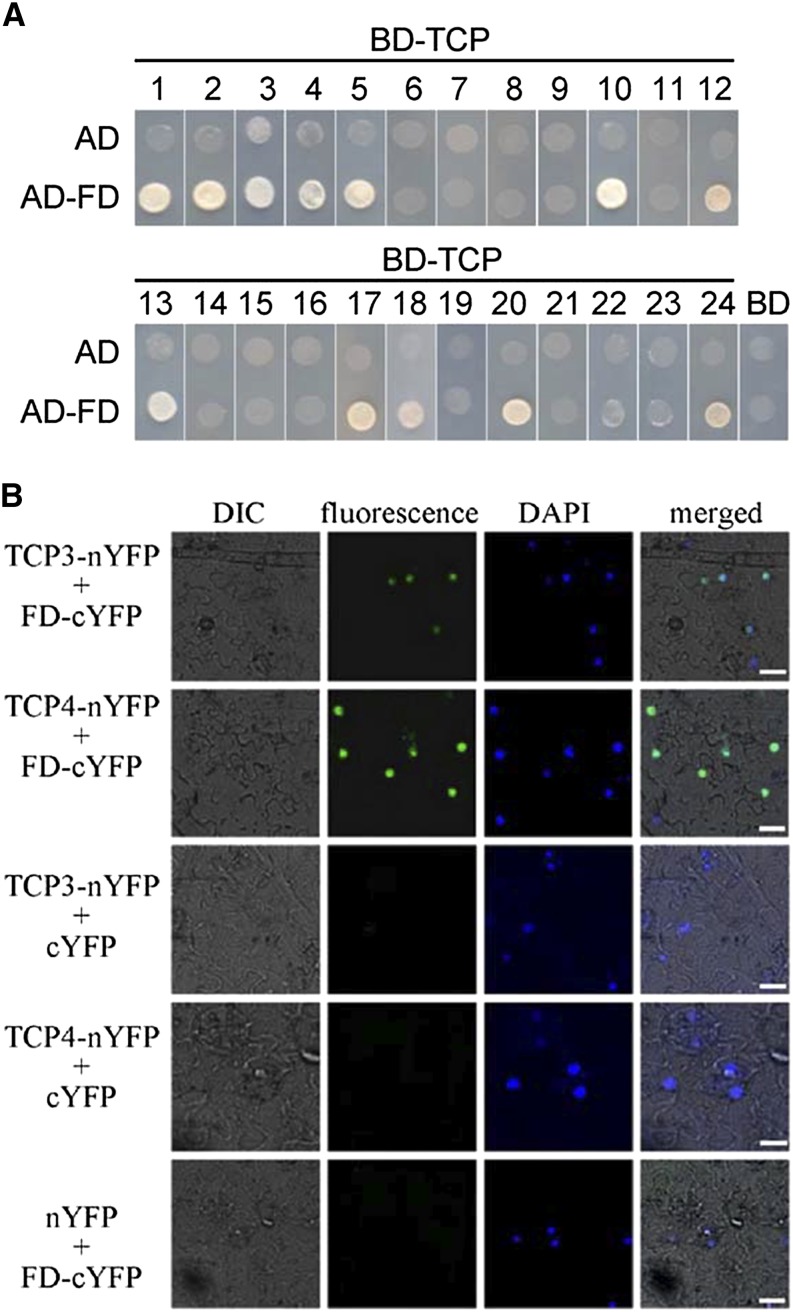
Because TCP5/13/17 formed complexes with FD, we investigated whether all members of the TCP family can interact with FD in flowering time regulation. We further examined the interactions of the TCP members with FD using the yeast two-hybrid system. To easily distinguish the classification of each TCP member, we constructed a phylogenetic tree (Supplemental Fig. S8). In addition to TCP5/13/17, TCP1/2/3/4/10/12/18/20/24 can also interact with FD (Fig. 7A). The interactions of TCP3/4 with FD were further corroborated by BiFC assays (Fig. 7B). The TCP members that interact with FD are all class II TCP TFs, except TCP20 (Supplemental Fig. S8), raising the possibility that class II TCP TFs may incorporate into the FT–FD module to control photoperiodic flowering.
Figure 7.
Interaction between Class II TCP TFs and FD. A, Yeast two-hybrid assay analysis. Interaction was indicated by the ability of cells to grow on synthetic dropout medium lacking Leu, Trp, His, and Ade. For TCP1/2/4/6/10/12/18/20/24, autoactivation was observed and thus 10 mm of 3-aminotriazole was added to the synthetic dropout medium to suppress false positives. The GAL4 activation domain expressed by pGADT7 (shown as “AD”) was used as a negative control. The experiment was repeated three times with similar results and representative photos are displayed. B, BiFC analysis. Fluorescence was observed in nuclear compartments of N. benthamiana leaf epidermal cells; the fluorescence resulted from complementation of the C-terminal portion of YFP fused to FD (FD-cYFP) with the N-terminal portion of YFP fused to TCP3/4 (TCP3/4-nYFP). No signal was observed from negative controls. DAPI, 49,6-diamidino-2-phenylindole; DIC, Differential interference contrast. The experiment was repeated three times with similar results and representative photos are displayed. Scale bars = 25 μm.
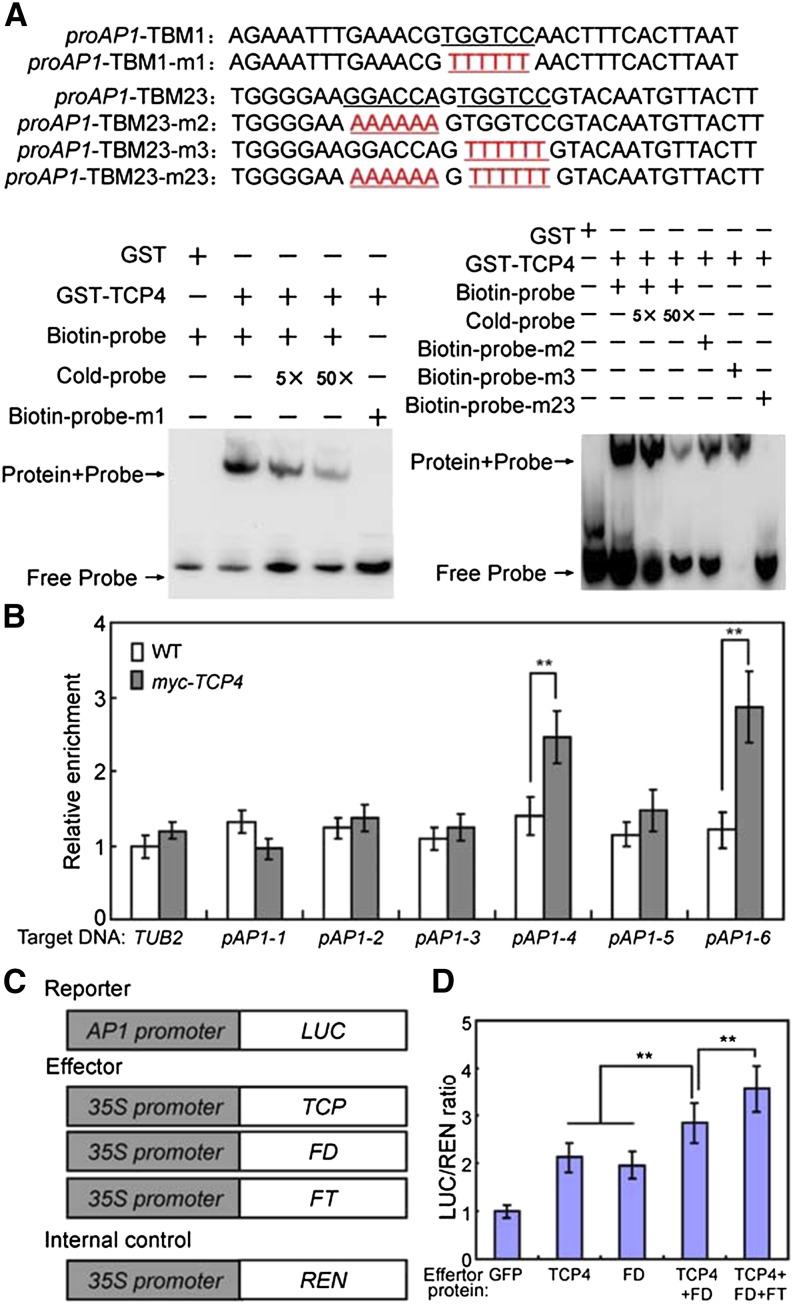
The miRNA319-regulated TCPs, including TCP2/3/4/10/24, play roles in photoperiodic flowering (Kubota et al., 2017; Liu et al., 2017). In our study, we demonstrated that these TCPs also interact with FD. Thus, we determined whether they also participate in photoperiodic flowering regulation through the direct regulation of downstream floral meristem identity genes. To test this possibility, we conducted a serious of experiments using 35S:miR319 and 35S:TCP4. Consistent with previous studies (Kubota et al., 2017; Liu et al., 2017), expression knockdown of miRNA319-regulated TCPs delayed flowering, whereas the overexpression of TCP4 accelerated it (Supplemental Fig. S9, A and B). Furthermore, the expression levels of AP1, FUL, and LFY were all lower in 35S:miR319 but higher in 35S:TCP4 compared with that in wild type (Supplemental Fig. S9C). Both ChIP-qPCR and EMSA results showed that TCP4 could also bind to the promoter of AP1 through the TBM elements (pAP1-4 and pAP1-6; Figure 8, A and B). Furthermore, TCP4 could function synergistically with FD and FT to activate the expression of AP1 (Fig. 8, C and D). Thus, class II TCP TFs may function together with the FT–FD module to control photoperiodic flowering by directly targeting downstream floral meristem identity genes.
Figure 8.
TCP4 directly regulates AP1 expression. A, The EMSA analysis of the binding of recombinant TCP4 protein to the promoter of AP1. The oligonucleotides (proAP1-TBM1/2/3 and proAP1-TBM1/2/3-m) were used as the probes. TBMs sequences and their mutated forms (red letters) are underlined. GST, GST-TCP4, biotin-probe, labeled mutated probe, and unlabeled probe at a 5× and 50× molar excess were present (+) or absent (−) in each reaction. B, ChIP-qPCR analysis of the relative binding of TCP4 to the promoter of AP1 (as shown in Fig 2B). ChIP assays were performed with chromatin prepared from Myc-TCP4 plants, using an anti-Myc antibody. Gray bars indicate the enrichment fold changes normalized to TUB2. Values are means ± sd of three independent biological replicates. Asterisks indicate significant differences; Student’s t test, **P < 0.01. WT, wild type. C, Schematic of the reporter and effectors used in the transient transactivation assays. D, Transient dual-LUC reporter assays show that TCP4 and FT-FD module synergistically activate the expression of AP1. Values are means ± sd of three independent biological replicates. Asterisks indicate significant differences; Student’s t test, **P < 0.01.
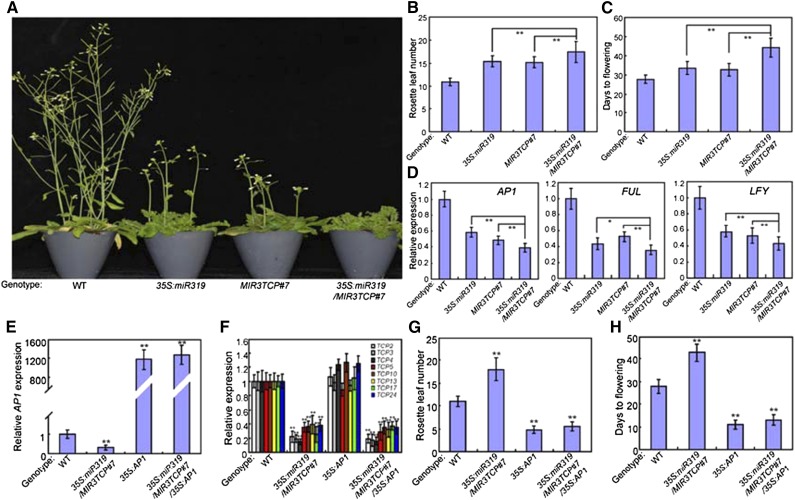
To further determine the roles of class II TCP TFs in photoperiodic flowering time control, we then crossed MIR3TCP#7 with 35S:miR319 to produce 35S:miR319/MIR3TCP#7. This was a knockdown of eight class II TCP members, TCP2/3/4/10/24/5/13/17 (Supplemental Fig. S10), which also belonged to the class II CIN-TCP clade, and we then conducted a flowering phenotype analysis. As shown in Figure 9, A–C, both 35S:miR319 and MIR3TCP#7 displayed delayed flowering phenotypes, and the delayed flowering phenotype of the 35S:miR319/MIR3TCP#7 construct was much greater than that of wild type. Consistently, the expression levels of AP1, FUL, and LFY in 35S:miR319/MIR3TCP#7 were all remarkably lower than that in wild type (Fig. 9D). To determine whether class II CIN-TCP TFs positively regulate flowering time through the direct activation of AP1 expression, the genetic relationships between class II CIN-TCP TFs and AP1 were explored. 35S:miR319/MIR3TCP#7/ was crossed with 35S:AP1 transgenic plants to produce 35S:miR319/MIR3TCP#7/35S:AP1, and their flowering phenotypes were examined. Under our experimental conditions, we detected an early flowering phenotype in 35S:AP1 plants, and a knockdown of class II CIN-TCP TFs did not change the RLN and DTF values, although 35S:miR319/MIR3TCP#7 plants showed a great delay in the floral transition (Fig. 9, E–H). Thus, class II CIN TCP TFs may have overlapping functions in the regulation of photoperiodic flowering and participate in flowering time control by modulating the transcriptional expression of floral meristem identity genes.
Figure 9.
Class II CIN TCP TFs control photoperiodic flowering. A, Representative images of the indicated genotypes showing their flowering phenotype under LD conditions. Three independent experiments were performed with each replica containing more than 30 plants for each line. Representative plants were photographed. B and C, Flowering phenotype of the indicated genotypes assessed by RLN (B) and DTF (C) under LD conditions. Values are means ± sd of ∼30 plants. Asterisks indicate significant differences; Student’s t test, **P < 0.01. The experiments were repeated at least three times with similar results. D, Expression of AP1, FUL, and LFY in the indicated genotypes. Two-week–old plants were harvested for total RNA extraction. Transcript levels of floral meristem identity genes in Col-0 plants were arbitrarily set to 1. ACTIN2 was used as an internal control. Values are means ± sd of three independent biological replicates. E and F, Expression of AP1 and TCP2/3/4/5/10/13/17/24 in the indicated genotypes. RNA was isolated from 2-week–old plants. ACTIN2 was used as an internal control. Values are means ± sd of three independent biological replicates. Asterisks indicate significant differences compared with that in wild type (WT); Student’s t test, **P < 0.01. G and H, Flowering phenotype of the indicated genotypes assessed by RLN (G) and DTF (H) under LD conditions. Values are means ± sd of ∼30 plants. Asterisks indicate significant differences compared with that in wild type; Student’s t test, **P < 0.01.
DISCUSSION
Class II TCPs Mediate Photoperiodic Flowering Time Control
The TCP family is a group of phylogenetically related, plant-specific TFs. Whereas functional research on TCP TFs has progressed over the past 20 years (Martín-Trillo and Cubas, 2010; Nicolas and Cubas, 2016), little is known about their possible involvement in flowering time regulation. In Arabidopsis, several TCP TFs, such as TCP4 (Kubota et al., 2017; Liu et al., 2017), TCP15 (Lucero et al., 2017), TCP20, and TCP22 (Wu et al., 2016) participate in the regulation of flowering through different pathways.
As a key floral meristem identity gene, the transcription of AP1 is regulated by only few TFs (Abe et al., 2005; Wigge et al., 2005; Jung et al., 2016). In our study, AP1 expression decreased in class II CIN TCP knockdown plants and increased in 35S:TCP4/5/13/17 transgenic plants (Figs. 2A and 9D; Supplemental Fig. S9C). This expression model is perfectly consistent with the flowering time behaviors of these lines. This observation suggests that class II CIN TCP TFs are able to positively regulate the expression of AP1. Furthermore, genetic analyses demonstrated that 35S:miR319/MIR3TCP#7/35S:AP1 had a markedly early flowering phenotype (Fig. 9, G and H), which mimicked that of 35S:AP1 plants, implying that class II CIN TCP TFs may act to accelerate flowering in an AP1-dependent manner. Analyses of LFY and FUL expression levels indicated that both genes were also positively regulated by class II CIN TCP TFs (Figs. 2A and 9D). Furthermore, the expression levels of CO and FT were lower in both MIR3TCP#7 and 35S:miR319, and were further reduced in 35S:miR319/MIR3TCP#7 compared with that in wild type (Supplemental Fig. S4). Thus, class II CIN TCP TFs may participate in flowering time control through the regulation of both flowering time-related genes and floral meristem identity genes.
The class II TCP proteins appear to perform their biological functions by directly binding to the TBM (GGACCA) present in their target promoters (Schommer et al., 2008). AP1 contains several TBM cis-elements in its promoter. Our ChIP experiments revealed that TCP4/5/13/17 can directly bind to the AP1 promoter (Figs. 2, B–D, and 8, A and B), suggesting that AP1 is a direct target of class II CIN TCPs. The opposite expression patterns of class II CIN TCPs and AP1 in class II CIN TCPs knockdown plants and overexpression lines (Figs. 2A and 9D), and the upregulation of LUC expression in transient expression assays (Figs. 2F and 8D), further suggest that class II CIN TCPs are positive regulators of AP1. Our results also demonstrate that LFY is not a direct target of TCP5 in flowering initiation (Supplemental Fig. S11). Thus, our results provide evidence that class II CIN TCPs function as promoters of flowering and trigger flowering through the direct activation of AP1. Because TCP4 promotes flowering under LD conditions by the direct regulation of CO (Kubota et al., 2017; Liu et al., 2017) and TCP15 accelerates flowering through the direct activation of SOC1 (Lucero et al., 2017), TCP proteins may participate in their distinct flowering pathways by modulating the expression levels of different floral integrators and/or floral meristem identity genes. TCP proteins can act as both positive and negative regulators. For example, the miR319-targetted TCP TFs play negative roles in leaf growth, but have positive roles during leaf senescence (Schommer et al., 2008). Furthermore, TCP8, TCP14, and TCP15 act redundantly to positively regulate effector-triggered immunity (Kim et al., 2014), and TCP15 also functions as a negative regulator of anthocyanin accumulation under high-light conditions (Viola et al., 2016). Thus, TCP TFs can act as both positive and negative regulators, and they participate in the fine-tuning and control of the complex signaling and transcriptional networks that mediate plant growth and stress responses.
Class II TCP TFs Facilitate the Binding of the FD–FT Complex To Target Gene Loci
The proper timing of the floral transition is controlled by sophisticated and elaborate regulatory mechanisms that monitor various environmental conditions and endogenous developmental cues, ensuring that flowering is initiated under the conditions most likely to maximize reproductive success and seed production. In nature, photoperiod, as a critical environmental factor, is used by many plants to guide flowering initiation. The FT–FD module integrates photoperiodic signals into the flowering networks by activating floral meristem identity genes, such as AP1, LFY, and FUL. However, the exact molecular mechanisms underlying how the FT–FD module controls the photoperiod pathway remain largely unknown. Identifying new key components that incorporate into the FT–FD module will provide new insights into our understanding of the floral transition.
Neither the transcriptional regulator FD nor its interacting partner FT directly bind to the regulatory regions of the AP1 gene (Benlloch et al., 2011), which raises the question of how the FT–FD module functions in inducing photoperiodic flowering. Certain TFs may recruit the FT–FD complex to the AP1, LFY, or FUL locus, allowing FD or both FD and its interaction partner to synergistically activate gene expression under favorable conditions. Indeed, miR156-targeted SPL3/4/5 may form a transcriptional coactivator complex with FD to regulate flowering (Jung et al., 2016). In our study, class II TCP TFs could directly bind to the conserved sequence element (GGACCA) in the promoter of the floral meristem identity gene AP1 (Figs. 2, B–D and 8, A and B). Interestingly, these TCPs also interacted with FD (Fig. 4), and the activation of the target genes by the FT–FD module required TCP5/13/17 (Fig. 5G). Genetic analyses indicated that TCP5/13/17-mediated floral inductions were dramatically compromised by ft and fd mutations (Supplemental Fig. S7) and knockdowns of TCP5/13/17 further delayed the flowering time of the fd/ft double mutant (Fig. 6). Thus, class II CIN TCP TFs may act synergistically and additively with the FT–FD module to positively regulate flowering initiation in Arabidopsis. Class II CIN TCP TFs and FD may form transcriptional complexes for the photoperiodic activation of floral meristem identity genes. Interestingly, TCP18/BRANCHED1 can interact with FT and modulates its activity in the axillary buds to prevent premature floral transition of the axillary meristem (Niwa et al., 2013). Meanwhile, several other TCP TFs, especially the class I TCPs, also interact with FT to form potential candidate complexes to modulate its activity (Ho and Weigel, 2014). These results indicate that TCP proteins can form complexes with both FT and FD to control flowering or floral meristem identity.
Functional Relationships between Class II TCPs and the FT–FD Module
Class II TCPs appear to incorporate into the FT–FD module by facilitating the accessibility of FD to its target loci, which controls flowering initiation. AP1 was used as an indicator of FT–FD activity, because it is a direct target of the FT–FD module (Abe et al., 2005; Wigge et al., 2005). However, it also plays an important role in the regulation of floral meristem identity (Weigel et al., 1992; Ferrándiz et al., 2000). Therefore, it is possible that class II TCPs, like FD, participate in modulating both flowering initiation and floral meristem identity (Abe et al., 2005). LFY plays an important role in the integration of flowering signals, in parallel with FD, to activate floral meristem identity genes (Abe et al., 2005). Mutations of FD alone only cause very weak defects in flower development, whereas mutations of both FD and LFY cause a strong inflorescence phenotype and severely reduce AP1 expression (Abe et al., 2005; Wang et al., 2009). Thus, knockdowns or knockouts of both the class II TCPs and LFY may help to verify the possible roles of class II TCPs in the floral meristem identity transition.
The sequence analysis revealed that both the TBMs and a putative SPL-binding motif (GTAC) are present near the C-box in the AP1 promoter (Supplemental Fig. S5; Cardon et al., 1997; Wigge et al., 2005; Yamaguchi et al., 2009; Jung et al., 2016), raising the possibility that both TCPs and SPL3/4/5 can bind to their recognized motifs and may synergistically recruit FD to the AP1 promoter. MiR319-targeted TCP4 and miR156-regulated SPL9 can form a complex to regulate age-dependent increases in leaf complexity (Rubio-Somoza et al., 2014). Thus, TCPs may also form complexes with certain SPLs and together recruit FD to control flowering or even inflorescence phenotype by targeting the floral meristem identity gene AP1. Simultaneous knockdowns or knockouts of the TCPs, SPLs, and FD may help verify their roles in both floral initiation and the floral meristem identity transition.
FD forms a complex with FT and a 14-3-3 protein, triggering floral initiation through the activation of key floral meristem identity genes, such as AP1, at the shoot apex (Abe et al., 2005; Wigge et al., 2005; Taoka et al., 2011). Phosphorylation of Thr-282 in FD is required for its interactions with FT and 14-3-3 (Kawamoto et al., 2015). Here, class II TCPs were shown to physically interact with FD and facilitate its binding to target gene loci. It will be interesting to determine whether FD phosphorylation affects its interactions with class II TCPs. Meanwhile, FD also participates in the repression of the expression of both AP1 and LFY through the FT-related floral repressor TERMINAL FLOWER1 (Jaeger et al., 2013), indicating that FD mediates both repressive and stimulative flowering signals. The incorporation of class II TCPs into the complicated FD-mediated regulatory network increases our understanding of the dual roles of FD during flowering initiation. Overall, we have provided evidence that class II TCPs function as key components of the regulatory network that modulates the onset and progression of floral initiation via their role in the FT–FD-controlled flowering pathway.
MATERIALS AND METHODS
Materials and Arabidopsis thaliana Growth Conditions
Taq DNA polymers was purchased from Takara Biotechnology. Other chemicals were obtained from Sangon Biotechnology. Arabidopsis (Arabidopsis thaliana) wild-type ecotype Columbia (Col-0) was used for all experiments. The tcp5/13/17, 35S:miR-3TCP (MIR3TCP), and ft-10 lines were kindly provided by Yuval Eshed (Weizmann Institute of Science) and Liangyu Liu (Capital Normal University), respectively. The tcp5 (CS116350), tcp13 (CS110881), tcp17 (Salk_147288), and fd-4 (SALK_118487) lines were obtained from the Arabidopsis Biological Resource Center. Arabidopsis plants were grown in an artificial growth chamber at 22°C under LD (16-h light/8-h dark cycle) or SD (8-h light/16-h dark cycle) conditions. Primers used for the identification of mutants or clones are listed in Supplemental Table S1.
Construction of Transgenic Overexpression Lines
To generate TCP4/5/13/17, FD, and AP1 overexpression transgenic plants, full-length cDNAs of these genes were cloned into a pOCA30, pOCA30-HA, or pOCA30-Myc vector in the sense orientation behind a CaMV 35S promoter (Chen and Chen, 2002; Jiang et al., 2014). Arabidopsis transformation was performed by the floral dip procedure. The seeds were collected from the infiltrated plants and selected on half-strength Murashige and Skoog medium containing 50 μg/mL of kanamycin. Kanamycin-resistant plants were transferred to soil 8 d after germination and were grown in a growth chamber. Primers used for identification of clones are listed in Supplemental Table S1.
RT-qPCR Analysis
For RT-qPCR analysis, total RNA was extracted using TRIzol reagent (Invitrogen) and was treated with RNase-free DNase (Fermentas), according to the manufacturer’s instructions. Total RNA (1 μg) was reverse-transcribed in a 20-μL reaction mixture using the Superscript II (Invitrogen). After the reaction, 1-μL aliquots were used as templates for RT-qPCR. Half reactions (10 μL each) were performed with the Lightcycler FastStart DNA Master SYBR Green I Kit (Roche) on a Roche LightCycler 480 real-time PCR machine, according to the manufacturer’s instructions. ACT2 (AT3G18780) was used as a control. The gene-specific primers are listed in Supplemental Table S1.
GUS Staining
To generate GUS reporter transgenic plants, the promoter sequences of TCP5/17, FD, and AP1 were separately amplified from genomic DNA and cloned into vector pOCA28-GUS (Honma et al., 1993). Transgenic plants were subjected to GUS staining as described in Chen et al. (2013). Chlorophyll was removed using several changes of 70% (v/v) ethanol, and the tissues were photographed. Primers used for generating various clones for GUS staining are listed in Supplemental Table S1.
Yeast Two-Hybrid Screening and Confirmation
The full-length or truncated CDS of TCPs were cloned into the bait vector pGBKT7 and the full-length FD CDS were cloned into the prey vector pGADT7. Two-hybrid screening was performed via the mating protocol described in the Clontech Matchmaker Gold Yeast Two-Hybrid user manual. Primers used for generating various clones for yeast two-hybrid assays are listed in Supplemental Table S1.
BiFC Assays
The cDNA sequences of enhanced YFP fragments, 173 amino acids located in the nYFP, and 64 amino acids located in the cYFP, were PCR-amplified and cloned into the XbaI-XhoI and BamHI-XhoI sites of pFGC5941 to generate pFGC-nYFP and pFGC-cYFP, respectively (Kim et al., 2008). The full-length CDS of TCP3/4/5/13/17 were inserted into pFGC-nYFP vector to generate N-terminal in-frame fusions with N-YFP, whereas FD CDS were cloned into pFGC-cYFP vector to form C-terminal in-frame fusions with C-YFP. All plasmids were introduced into Agrobacterium tumefaciens (strain EHA105), and then infiltration of Nicotiana benthamiana leaves was performed as described in Chen et al. (2017). Infected tissues were analyzed 48 h after infiltration under a confocal laser-scanning microscope (Olympus). The primers used for BiFC are listed in Supplemental Table S1.
CoIP Assays
For CoIP assays, the full-length CDS of TCP5/17 or FD were individually cloned into tagging plasmids behind the Myc- or HA-tag sequence. Myc-fused TCP5/17 and HA-fused FD were then transiently coexpressed in N. benthamiana leaves. After 48 h, the infected leaves were homogenized in an extraction buffer containing 500 mm of 400 Tris-HCl at pH 7.5, 150 mm of NaCl, 1 mm of EDTA, 0.1% (w/v) Trition-X-100, and 1× complete protease inhibitor cocktail (Roche). Then Myc-fused TCP5/17 were immunoprecipitated using an anti-Myc Affinity Gel (Sigma-Aldrich), and coimmunoprecipitated proteins were detected using an anti-HA antibody (Santa Cruz).
ChIP Assays
For the ChIP assay, 15-day–old seedlings of Myc-TCP5, HA-FD, or Col-0 seedlings were used as materials. The ChIP experiment was performed as described in Saleh et al. (2008). The Myc and HA antibody was used to immunoprecipitate the protein–DNA complex, and the precipitated DNA was purified using a PCR purification kit for RT-qPCR analysis. The ChIP experiments were performed three times. The primers used for RT-qPCR amplification of different promoters are listed in Supplemental Table S1.
EMSAs
The EMSA was conducted using a Chemiluminescent EMSA Kit (Beyotime) following the manufacturer’s protocol. The recombinant GST-TCP4/5/13/17 or FD protein and GST protein were purified from Escherichia coli. The DNA fragments of the AP1 promoter were synthesized and the 5′ termini were labeled with biotin. Biotin-unlabeled fragments of the same sequences or mutated sequences were used as competitors, and the GST protein alone was used as the negative control.
Transient Expression Assays
To generate reporter constructs, a 2,331-bp region upstream of the start codon of AP1 was amplified and cloned into a pGreenII 0800-LUC vector (Hellens et al., 2005). To create the effector constructs, the corresponding cDNAs of TCP4/5/13/17, FD, and FT were amplified and cloned into pGreenII 62-SK vectors (Hellens et al., 2005). All primers used for generating these constructs are listed in Supplemental Table S1. Preparation of Arabidopsis mesophyll protoplasts from wild-type (Col-0) leaves and subsequent transfections were performed as described by Yoo et al. (2007). A dual-LUC reporter assay system (Promega) was used to measure firefly LUC and renilla luciferase (REN) activities. The REN gene under the control of the CaMV 35S promoter and the LUC gene were in the pGreenII 0800-LUC vector (Hellens et al., 2005). Relative REN activity was used as an internal control, and LUC/REN ratios calculated.
Accession Numbers
The Arabidopsis Genome Initiative identifiers for the genes described in this article are as follows: CO (At5g15840), TCP1 (At1g67260), TCP2 (At4g18390), TCP3 (At1g53230), TCP4 (At3g15030), TCP5 (At5g60970), TCP6 (At5g41030), TCP7 (At5g23280), TCP8 (At1g58100), TCP9 (At2g45680), TCP10 (At2g31070), TCP11 (At2g37000), TCP12 (At1g68800), TCP13 (At3g02150), TCP14 (At3g47620), TCP15 (At1g69690), TCP16 (At3g45150), TCP17 (At5g08070), TCP18 (At3g18550), TCP19 (At5g51910), TCP20 (At3g27010), TCP21 (At5g08330), TCP22 (At1g72010), TCP23 (At1g35560), TCP24 (At1g30210), FT (AT1G65480), FD (AT4G35900), AP1 (AT1G69120), FUL (AT5G60910), LFY (AT5G61850), ACTIN2 (AT3G18780), and IPP2 (AT3G02780).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Relative expression of TCP5/13/17 in tcp5/13/17 triple T-DNA insertion mutant and two miRNA overexpression lines.
Supplemental Figure S2. The flowering phenotype of single, double, and triple mutant of/among TCP5/13/17.
Supplemental Figure S3. RT-qPCR analysis of 35S:TCP5/13/17 transgenic plants and their flowering phenotype.
Supplemental Figure S4. The expression of CO and FT in 35S:miR319, MIR3TCP#7, and 35S:miR319/MIR3TCP#7.
Supplemental Figure S5. The promoter structure of the AP1 gene.
Supplemental Figure S6. EMSA on FD binding to AP1 promoter.
Supplemental Figure S7. Flowering times of ft-10 and fd-4 mutants overexpressing TCP5/13/17.
Supplemental Figure S8. Phylogeny of Arabidopsis TCP genes.
Supplemental Figure S9. Flowering times of 35S:miR319 and 35S:TCP4 transgenic plants.
Supplemental Figure S10. Expression of class II CIN-TCP TFs in 35S:miR319/MIR3TCP#7.
Supplemental Figure S11. ChIP-qPCR analysis of the relative binding of TCP5 to the genomic sequence of LFY.
Supplemental Table S1. Primers used in this article.
Acknowledgments
We thank the Arabidopsis Resource Center at Ohio State University for providing mutants. We also thank Yuval Eshed (Weizmann Institute of Science) and Liangyu Liu (Capital Normal University) for sharing research materials and Jianghua Chen (Xishuangbanna Tropical Botanical Garden, CAS) for technical support.
Footnotes
This work was supported by the Natural Science Foundation of China (31671275 to L.C.), the National Key R & D Plan (2016YFD0101006 to L.C.), the Candidates of the Young and Middle Aged Academic Leaders of Yunnan Province (2015HB094 to L.C.), the Natural Science Foundation of Yunnan Province of China (2017FB047 to L.C.), and Core Botanical Gardens, Chinese Academy of Sciences.
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056 [DOI] [PubMed] [Google Scholar]
- Becker A, Theissen G (2003) The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol Phylogenet Evol 29: 464–489 [DOI] [PubMed] [Google Scholar]
- Benlloch R, Kim MC, Sayou C, Thévenon E, Parcy F, Nilsson O (2011) Integrating long-day flowering signals: A LEAFY binding site is essential for proper photoperiodic activation of APETALA1. Plant J 67: 1094–1102 [DOI] [PubMed] [Google Scholar]
- Cardon GH, Höhmann S, Nettesheim K, Saedler H, Huijser P (1997) Functional analysis of the Arabidopsis thaliana SBP-box gene SPL3: A novel gene involved in the floral transition. Plant J 12: 367–377 [DOI] [PubMed] [Google Scholar]
- Chen C, Chen Z (2002) Potentiation of developmentally regulated plant defense response by AtWRKY18, a pathogen-induced Arabidopsis transcription factor. Plant Physiol 129: 706–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Zhang L, Li D, Wang F, Yu D (2013) WRKY8 transcription factor functions in the TMV-CG defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proc Natl Acad Sci USA 110: E1963–E1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Xiang S, Chen Y, Li D, Yu D (2017) Arabidopsis WRKY45 interacts with the DELLA protein RGL1 to positively regulate age-triggered leaf senescence. Mol Plant 10: 1174–1189 [DOI] [PubMed] [Google Scholar]
- Cubas P, Lauter N, Doebley J, Coen E (1999) The TCP domain: A motif found in proteins regulating plant growth and development. Plant J 18: 215–222 [DOI] [PubMed] [Google Scholar]
- Davière JM, Wild M, Regnault T, Baumberger N, Eisler H, Genschik P, Achard P (2014) Class I TCP-DELLA interactions in inflorescence shoot apex determine plant height. Curr Biol 24: 1923–1928 [DOI] [PubMed] [Google Scholar]
- Davis SJ. (2009) Integrating hormones into the floral-transition pathway of Arabidopsis thaliana. Plant Cell Environ 32: 1201–1210 [DOI] [PubMed] [Google Scholar]
- Efroni I, Blum E, Goldshmidt A, Eshed Y (2008) A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 20: 2293–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrándiz C, Gu Q, Martienssen R, Yanofsky MF (2000) Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 127: 725–734 [DOI] [PubMed] [Google Scholar]
- Han P, García-Ponce B, Fonseca-Salazar G, Alvarez-Buylla ER, Yu H (2008) AGAMOUS-LIKE 17, a novel flowering promoter, acts in a FT-independent photoperiod pathway. Plant J 55: 253–265 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervé C, Dabos P, Bardet C, Jauneau A, Auriac MC, Ramboer A, Lacout F, Tremousaygue D (2009) In vivo interference with AtTCP20 function induces severe plant growth alterations and deregulates the expression of many genes important for development. Plant Physiol 149: 1462–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho WW, Weigel D (2014) Structural features determining flower-promoting activity of Arabidopsis FLOWERING LOCUS T. Plant Cell 26: 552–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma MA, Baker BJ, Waddell CS (1993) High-frequency germinal transposition of DsALS in Arabidopsis. Proc Natl Acad Sci USA 90: 6242–6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijser P, Schmid M (2011) The control of developmental phase transitions in plants. Development 138: 4117–4129 [DOI] [PubMed] [Google Scholar]
- Jaeger KE, Pullen N, Lamzin S, Morris RJ, Wigge PA (2013) Interlocking feedback loops govern the dynamic behavior of the floral transition in Arabidopsis. Plant Cell 25: 820–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Liang G, Yang S, Yu D (2014) Arabidopsis WRKY57 functions as a node of convergence for jasmonic acid- and auxin-mediated signaling in jasmonic acid-induced leaf senescence. Plant Cell 26: 230–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JH, Lee HJ, Ryu JY, Park CM (2016) SPL3/4/5 integrate developmental aging and photoperiodic signals into the FT–FD module in Arabidopsis flowering. Mol Plant 9: 1647–1659 [DOI] [PubMed] [Google Scholar]
- Kawamoto N, Sasabe M, Endo M, Machida Y, Araki T (2015) Calcium-dependent protein kinases responsible for the phosphorylation of a bZIP transcription factor FD crucial for the florigen complex formation. Sci Rep 5: 8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KC, Lai Z, Fan B, Chen Z (2008) Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell 20: 2357–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Son GH, Bhattacharjee S, Kim HJ, Nam JC, Nguyen PDT, Hong JC, Gassmann W (2014) The Arabidopsis immune adaptor SRFR1 interacts with TCP transcription factors that redundantly contribute to effector-triggered immunity. Plant J 78: 978–989 [DOI] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y (2002) DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J 30: 337–348 [DOI] [PubMed] [Google Scholar]
- Kubota A, Ito S, Shim JS, Johnson RS, Song YH, Breton G, Goralogia GS, Kwon MS, Laboy Cintrón D, Koyama T, et al. (2017) TCP4-dependent induction of CONSTANS transcription requires GIGANTEA in photoperiodic flowering in Arabidopsis. PLoS Genet 13: e1006856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar SV, Lucyshyn D, Jaeger KE, Alós E, Alvey E, Harberd NP, Wigge PA (2012) Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484: 242–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Cheng X, Liu P, Li D, Chen T, Gu X, Sun J (2017) MicroRNA319-regulated TCPs interact with FBHs and PFT1 to activate CO transcription and control flowering time in Arabidopsis. PLoS Genet 13: e1006833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucero LE, Manavella PA, Gras DE, Ariel FD, Gonzalez DH (2017) Class I and Class II TCP transcription factors modulate SOC1-dependent flowering at multiple levels. Mol Plant 10: 1571–1574 [DOI] [PubMed] [Google Scholar]
- Martín-Trillo M, Cubas P (2010) TCP genes: A family snapshot ten years later. Trends Plant Sci 15: 31–39 [DOI] [PubMed] [Google Scholar]
- Nath U, Crawford BC, Carpenter R, Coen E (2003) Genetic control of surface curvature. Science 299: 1404–1407 [DOI] [PubMed] [Google Scholar]
- Navaud O, Dabos P, Carnus E, Tremousaygue D, Hervé C (2007) TCP transcription factors predate the emergence of land plants. J Mol Evol 65: 23–33 [DOI] [PubMed] [Google Scholar]
- Nicolas M, Cubas P (2016) TCP factors: New kids on the signaling block. Curr Opin Plant Biol 33: 33–41 [DOI] [PubMed] [Google Scholar]
- Niwa M, Daimon Y, Kurotani K, Higo A, Pruneda-Paz JL, Breton G, Mitsuda N, Kay SA, Ohme-Takagi M, Endo M, et al. (2013) BRANCHED1 interacts with FLOWERING LOCUS T to repress the floral transition of the axillary meristems in Arabidopsis. Plant Cell 25: 1228–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D (2003) Control of leaf morphogenesis by microRNAs. Nature 425: 257–263 [DOI] [PubMed] [Google Scholar]
- Riboni M, Galbiati M, Tonelli C, Conti L (2013) GIGANTEA enables drought escape response via abscisic acid-dependent activation of the florigens and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS. Plant Physiol 162: 1706–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Somoza I, Zhou CM, Confraria A, Martinho C, von Born P, Baena-Gonzalez E, Wang JW, Weigel D (2014) Temporal control of leaf complexity by miRNA-regulated licensing of protein complexes. Curr Biol 24: 2714–2719 [DOI] [PubMed] [Google Scholar]
- Saleh A, Alvarez-Venegas R, Avramova Z (2008) An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat Protoc 3: 1018–1025 [DOI] [PubMed] [Google Scholar]
- Sarvepalli K, Nath U (2011) Hyper-activation of the TCP4 transcription factor in Arabidopsis thaliana accelerates multiple aspects of plant maturation. Plant J 67: 595–607 [DOI] [PubMed] [Google Scholar]
- Schommer C, Palatnik JF, Aggarwal P, Chételat A, Cubas P, Farmer EE, Nath U, Weigel D (2008) Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol 6: e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth A, Schmid M (2011) Regulation of flowering time: All roads lead to Rome. Cell Mol Life Sci 68: 2013–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoka K, Ohki I, Tsuji H, Furuita K, Hayashi K, Yanase T, Yamaguchi M, Nakashima C, Purwestri YA, Tamaki S, et al. (2011) 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476: 332–335 [DOI] [PubMed] [Google Scholar]
- Turck F, Fornara F, Coupland G (2008) Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol 59: 573–594 [DOI] [PubMed] [Google Scholar]
- Viola IL, Camoirano A, Gonzalez DH (2016) Redox-dependent modulation of anthocyanin biosynthesis by the TCP transcription factor TCP15 during exposure to high light intensity conditions in Arabidopsis. Plant Physiol 170: 74–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JW. (2014) Regulation of flowering time by the miR156-mediated age pathway. J Exp Bot 65: 4723–4730 [DOI] [PubMed] [Google Scholar]
- Wang JW, Czech B, Weigel D (2009) miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138: 738–749 [DOI] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM (1992) LEAFY controls floral meristem identity in Arabidopsis. Cell 69: 843–859 [DOI] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059 [DOI] [PubMed] [Google Scholar]
- Wu JF, Tsai HL, Joanito I, Wu YC, Chang CW, Li YH, Wang Y, Hong JC, Chu JW, Hsu CP, et al. (2016) LWD-TCP complex activates the morning gene CCA1 in Arabidopsis. Nat Commun 7: 13181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A, Abe M (2012) Regulation of reproductive development by non-coding RNA in Arabidopsis: To flower or not to flower. J Plant Res 125: 693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A, Wu MF, Yang L, Wu G, Poethig RS, Wagner D (2009) The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev Cell 17: 268–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]