miR166 and its target PHB monitor SPL/NZZ gene dynamically in construction of internal boundary and formation of dehiscence zones in anthers.
Abstract
The internal boundary between inner and outer microsporangia within anthers is essential for male fertility of vascular plants. Dehiscence zones embedded in the boundary release pollen for fertilization. However, the molecular mechanism underlying boundary formation in anthers remains poorly understood. Here, we report that microRNA166 (miR166) and its target PHABULOSA (PHB) regulate SPOROCYTELESS/NOZZLE (SPL/NZZ), which controls microsporogenesis. In developing anthers of Arabidopsis (Arabidopsis thaliana), the expression domains of miR165/6 and SPL/NZZ are overlapped and rearranged synchronously. Dominant mutation of PHB suppresses SPL/NZZ expression on the adaxial sides of stamens, resulting in a thickened boundary, whereas activation of MIR166g up-regulates SPL/NZZ expression, leading to ectopic microsporogenesis in the boundary. PHB limits the expression domains of SPL/NZZ to facilitate construction of the boundary, while miR166 preserves the expression domains of SPL/NZZ by inhibiting PHB to allow the inner microsporangia to take shape. Subsequently, PHB activates the key stem cell maintainer WUSCHEL in anthers to restrict the stomium cells to the boundary so that dehiscence zones develop and release pollen properly. These findings link adaxial/abaxial polarity to microsporogenesis in building of the internal boundary of anthers and thus advance the concepts underlying the establishment of the internal structure of male organs.
The stamen is an important reproductive lateral organ consisting of a four-lobed anther that is anchored to the third whorl of a flower by a filament in many flowering plants, including Arabidopsis (Arabidopsis thaliana; Smyth et al., 1990; Goldberg et al., 1993; Sanders et al., 1999; Scott et al., 2004). The four-lobed structure is mainly composed of two inner microsporangia and two outer ones, also known as pollen sacs, which are separated by a boundary where stomium regions and dehiscence zones differentiate successively. Male fertility in many flowering plants relies on pollen release from dehiscence zones in the boundary between inner and outer microsporangia.
The formation of haploid cells (spores) in microsporangia by meiosis is called sporogenesis (Yuan and Sundaresan, 2015). Pollen grains are produced in the locules of microsporangia (Ma, 2005; Feng and Dickinson, 2007, 2010). Anther development in Arabidopsis is divided into 14 stages (Sanders et al., 1999). Stamen/anther primordia initiate from the floral apex at stage 1. Archesporial cells then emerge in the four corners of the L2 layer at stage 2 and differentiate into parietal and sporogenous cells in the four corners at stage 3. Concentrically organized microsporangia are then generated in each corner of the anther at stage 5. At the same time, the stomium region (also called the dehiscence zone) emerges between two microsporangia in each half of the anther (theca) starting at stage 4. At stage 12, the anther dehisces along the stomium regions, and pollen grains are released for pollination at anthesis.
The specification of the sporocytes represents a critical step during sporogenesis. The SPOROCYTELESS/NOZZLE (SPL/NZZ) gene, which encodes a nuclear protein, was identified as a key regulator of this sporogenesis stage in Arabidopsis (Schiefthaler et al., 1999; Yang et al., 1999; Ito et al., 2004; Hord et al., 2006). The loss-of-function mutant spl produces anthers lacking microsporangia. Two Leu-rich repeat receptor-like protein kinases encoded by BARELY ANY MERISTEM1 (BAM1) and BAM2 function in a regulatory loop with SPL/NZZ to restrict SPL expression to the inner region of the locule (Zhao, 2009). Additionally, the JAGGED and NUBBIN transcription factors function redundantly in adaxial cell proliferation and differentiation in anthers (Dinneny et al., 2006).
Along with germline determination, organ polarity establishment is a critical issue that has recently been thoroughly investigated (Schiefthaler et al., 1999; Yang et al., 1999). In some lateral organs, adaxial-abaxial polarity identity is precisely regulated by two classes of antagonistic genes. The adaxial identity genes include the HD-ZIP III family genes REVOLUTA (REV), PHABULOSA (PHB), PHAVOLUTA (PHV), CORONA (CNA), and ATHB8, which are repressed by microRNA165/6 (miR165/6; McConnell et al., 2001; Emery et al., 2003; Williams et al., 2005). In leaves and some other lateral organs, HD-ZIP III genes are essential for the adaxial identity and function antagonistically toward the abaxial identity genes, including the KANADI family members, which positively regulate the expression of the YABBY group of abaxial identity genes (Zhong and Ye, 1999; Eshed et al., 2001; Bowman, 2004; Wu et al., 2007; Liu et al., 2011; Yang et al., 2014). The HD-ZIP III genes are also responsible for apical identity during embryogenesis and are antagonistic to PLETHORA1/2 (Smith and Long, 2010).
Although the effect of PHB and the other HD-ZIP III genes on leaf development has been extensively studied, it remains poorly understood in anthers. In the stamen of rice (Oryza sativa), the expression of PHB3 (ortholog of Arabidopsis PHB) is rearranged during anther development (Zhong and Ye, 2004; Toriba et al., 2010). HYPONASTIC LEAF1 is an important player in miRNA processing (Wu et al., 2007) and is required for the establishment of stamen architecture with four microsporangia in Arabidopsis (Lian et al., 2013).
Between inner and outer microsporangia is the boundary where stomium cells are maintained to form dehiscence zones. WUSCHEL (WUS), originally identified as a central regulator of stem cell maintenance, plays an important role in the specification of stomium cells (Deyhle et al., 2007). However, the molecular mechanism underlying the formation of the boundary and dehiscence zone in anthers remains poorly understood. Here, we report the dynamic expression programs of miR165/6 and miR165/6-tageted PHB genes and their regulation to the SPL/NZZ gene during the formation of the boundary and determination of stomium regions in anthers.
RESULTS
MIR166g Activation Causes Ectopic Microsporogenesis in the Boundary between Inner and Outer Microsporangia
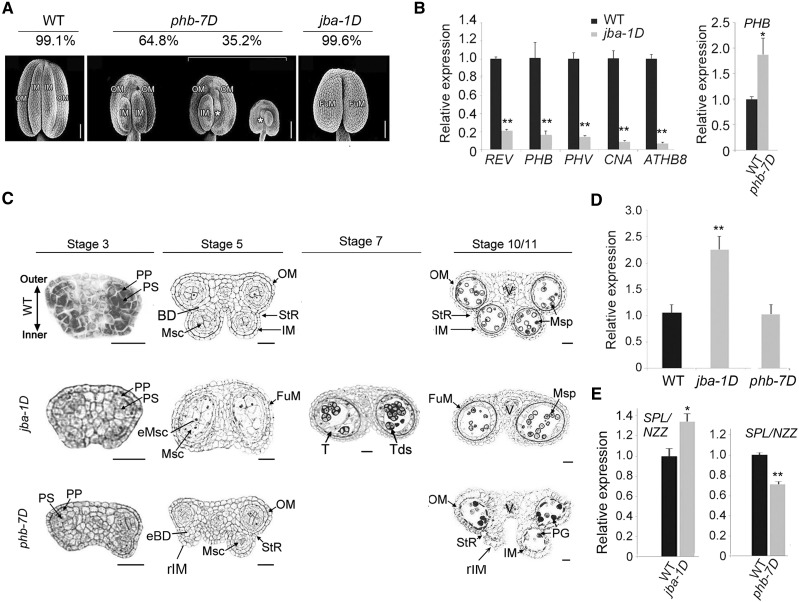
To investigate whether and how miR165/6 regulate the formation of the boundary between microsporangia, we chose the jba-1D mutant, in which the MIR166g gene was activated (Williams et al., 2005). As expected, miR166 was up-regulated and all miR166-targeted HD-ZIP III genes were down-regulated in the young inflorescences (Supplemental Fig. S1, A and B) and anthers (Fig. 1B) of the jba-1D mutant. In the wild-type anthers, the four microsporangia were separated into two pairs (theca), each of which contained an inner microsporangium and an outer microsporangium with a boundary in between (Fig. 1A). Surprisingly, jba-1D anthers appeared to have only two microsporangia instead of four. The boundary between inner and outer microsporangia and dehiscence zones were not seen. However, the epidermal cells of their connective tissues were not distinct from the wild type (Supplemental Fig. S1C). Homozygous jba-1D plants were male sterile.
Figure 1.
Formation of microsporangia and gene expression of HD-ZIP III genes and SPL/NZZ in the anthers of phb-7D and jba-1D mutants. A, Scanning electron microscopy images showing the numbers and positions of microsporangia in anthers. Percentages of the anthers with the mutant phenotypes are shown. White asterisks indicate the remnants of shriveled microsporangia. Scanning electron micrographs of phb-7D were digitally abstracted and made into a composite for comparison. Bars = 100 μm. B, Reverse transcription quantitative PCR (RT-qPCR) showing relative expression of HD-ZIP III genes in the jba-1D and phb-7D anthers (mild category) at stage 3. C, Cross sections of anthers of the wild type (WT) and jba-1D and phb-7D mutants. BD, boundary tissue; eBD, ectopic boundary tissue; eMsc, ectopic microsporocyte; FuM, fused microsporangium; IM, inner microsporangium; Msc, microsporocytes; Msp, microspores; OM, outer microsporangium; PG, pollen grains; PP, primary parietal cells; PS, primary sporogenous cells; rIM, remnants of inner microsporangia; StR, stomium region; T, tapetum; Tds, tetrads. Bars = 20 μm. D, The ratio of the number of microsporocytes per locule in the mutants to the wild type. More than 30 locules for the phb-7D mutant with mild phenotype or the jba-1D mutant were observed. E, RT-qPCR showing relative expression of the SPL/NZZ gene in jba-1D and phb-7D anthers at stage 3. Three biological replicates were analyzed in B and E. Error bars indicate the sd, while black asterisks show significant differences at the 0.05 (*) and 0.01 (**) levels in Student’s t test.
We observed the internal structures of anthers and microsporangia using transverse sections of early stamens (Fig. 1C). In the wild-type anther, the parietal and sporogenous cells at stage 3 appeared in the two lateral regions, which were bilaterally symmetrical, while each was composed of one inner locule close to the carpel and one outer locule close to the petal. The internal boundary between the inner and outer locules was not observed at stage 2 but was seen at stage 3. The stomium regions were located in the boundary furrow. The pollen grains were observed in the four locules at stage 10/11. In jba-1D anthers, the two locules were much bigger than those of the wild type (Fig. 1, A and C; Table 1), and each of them contained more than twice the number of microsporocytes than were in the wild-type locules (Fig. 1D), indicating ectopic microsporogenesis in the enlarged microsporangia.
Table 1. Microsporangium size in phb-7D and jba-1D mutants.
Microsporangia of stage 12 anthers were measured; n ≥ 66 for each wild type and mutant. Each value is the mean ± sd. Dashes indicate data is not available. Asterisks indicate statistical significance (Student’s t test, *, P < 0.05 and **, P < 0.01). FuM, Fused microsporangium; IM, inner microsporangium; OM, outer microsporangium.
| Name | Parameter | IM | OM | FuM |
|---|---|---|---|---|
| C24 | Length | 329.3 ± 25.9 | 372.8 ± 30.3 | – |
| Width | 87.3 ± 6.4 | 103.5 ± 10.2 | – | |
| phb-7D | Length | 236.47 ± 43.9** | 338.0 ± 43.4* | – |
| Width | 88.4 ± 11.1 | 100.7 ± 12.0 | – | |
| Col-7 | Length | 391.1 ± 51.0 | 410.0 ± 54.1 | – |
| Width | 94.9 ± 11.9 | 105.0 ± 15.2 | – | |
| jba-1D | Length | – | – | 373.4 ± 47.5* |
| Width | – | – | 161.5 ± 22.7** |
Close observation of the anthers at stage 7 showed that narrow creases still developed down the center of some enlarged locules, which is where the two locules would normally be separated and where anther dehiscence would occur. The presence of the narrow creases marks the remnants of the boundary between inner and outer microsporangia and indicates that the place of the boundary is occupied by microsporocytes. These results suggest that silencing of HD-ZIP III genes by MIR166g activation causes ectopic microsporocytes in the boundary.
We noticed that the boundary between inner and outer locules in the wild-type anthers formed after initiation of the parietal and sporogenous cells in the two lateral regions at stage 3 (Fig. 1C), meaning that inner and outer microsporangia are separated by the boundary after formation of the two lateral regions. The defects of the boundary in jba-1D anthers appeared at stage 3 or earlier.
PHB Up-Regulation Leads to the Ectopic Boundary Tissues in Inner Microsporangia
To test the effects of miR166-targeted genes on the boundary and microsporogenesis, we selected phb-7D, which is a miR166-resistant gain-of-function allele of PHB (Carlsbecker et al., 2010), as a representative mutant of the HD-ZIP III genes targeted by miR166. Compared with the wild-type microsporangia, the outer microsporangia in all phb-7D anthers were normal while the inner microsporangia were varied between the anthers. Among the phb-7D anthers, 64.8% of anthers were categorized as having a mild mutant phenotype because they had two small inner microsporangia (Table 1; Fig. 1A), 30.2% of anthers were designated as having a medium phenotype, since they lost one inner microsporangium, and about 5% of anthers were regarded as having a severe phenotype, as they lost two inner microsporangia.
To examine the internal structure of inner microsporangia, we observed the cross sections of the phb-7D anthers with the medium phenotype. In these anthers, one inner locule was aberrant (Fig. 1C) and the boundary between this inner locule and its outer locule was thickened, thus showing small and/or empty inner locules. Numbers of microsporocytes in these inner locules were much fewer than those of the wild type (Fig. 1D). The anthers with mild and medium phenotypes were able to open and released fewer pollen grains than the wild type; however, the anthers without inner microsporangia failed to release pollen. These results suggest that up-regulation of PHB induces ectopic boundary tissues inside the inner microsporangia, causing a thickened boundary and an aberrance or absence of inner microsporangia.
Expression Domains of miR165/6 and SPL/NZZ Are Overlapped in Anthers
SPL/NZZ is a key regulator of sporogenesis, especially regarding the specification of microsporocytes derived from archesporial cells at stage 3 (Sanders et al., 1999). We hypothesized that the expression of this gene was regulated by miR165/6. To test this hypothesis, we compared expression levels of SPL/NZZ in wild-type and jba-1D anthers. The expression levels of SPL/NZZ in jba-1D anthers increased significantly (Fig. 1E), coinciding with the down-regulation of HD-ZIP III genes. These data suggest that miR165/6 positively regulate the expression of SPL/NZZ in anthers.
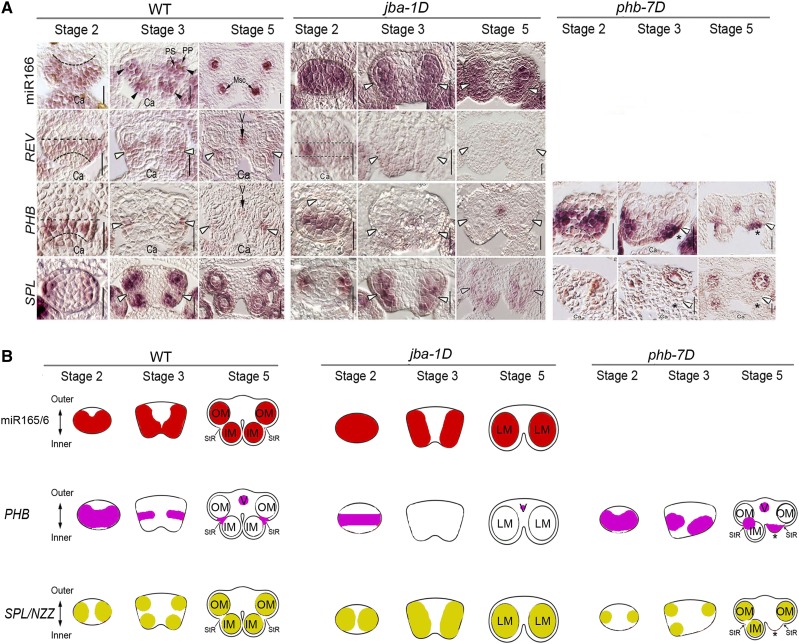
To test whether miR166 regulates the expression domains of SPL/NZZ in anthers, we conducted an RNA in situ hybridization experiment involving miR165/6 and SPL/NZZ. In the wild-type anthers, miR165/6 accumulated in the lateral-adaxial regions of stamens at stage 2 (Fig. 2), forming two expression domains; then their expression domains appeared in the four corners of stamens at stage 3 and were localized in the four microsporangia at stage 5. It thus appeared that the miR165/6 domains were rearranged during anther development.
Figure 2.
Expression domains of miR165/6, miR165/6-targeted genes, and SPL/NZZ in anthers. A, RNA in situ hybridization showing the expression domains of miR166, REV, PHB, and SPL/NZZ. Ca, Carpel; Msc, microsporocytes; PP, primary parietal cells; PS, primary sporogenous cells; V, vasculature; WT, wild type. White arrowheads show the stomium regions. Asterisks indicate the remnants of shriveled microsporangia. Bars = 20 μm. B, Schematic diagrams indicating the rearranged expression domains of PHB and SPL/NZZ during anther development. IM, Inner microsporangium; LM, Large microsprorangium; OM, outer microsporangium; StR, stomium region.
The expression domains of SPL/NZZ were largely overlapped with the miR165/6 domains in the wild type. They were rearranged from the two lateral regions of anthers at stage 2, similar to miR165/6 domains, to the four corners at stage 3 and the microsporangia at stage 5. The difference is that SPL/NZZ expression domains in the two thecae appeared clearly in the regions surrounding microsporangia, in contrast with the miR165/6 domains that are strictly localized inside locules. This result suggests that SPL/NZZ is involved in the development of septum while miR165/6 is not.
Expression Domains of PHB and SPL/NZZ Are Opposite in Anthers
miR165/6 silence PHB in shoot and floral meristems. To investigate the regulation of the HD-ZIP III genes by SPL/NZZ in anthers, we compared expression levels of SPL/NZZ in wild-type and phb-7D anthers. The expression levels of SPL/NZZ in phb-7D anthers declined significantly (Fig. 1E), coinciding with the up-regulation of HD-ZIP III genes. These data suggest that PHB and/or the other HD-ZIP III genes negatively regulate the expression of SPL/NZZ in anthers.
In situ hybridization was adopted to show the expression patterns of REV and PHB in developing anthers. In the wild-type anthers, REV and PHB were preferentially expressed in the middle regions of the thecae as well as vascular tissue in anthers at stage 2 (Fig. 2); then their expression domains appeared in the boundary between inner and outer locules at stage 3 and were subsequently localized in the stomium regions at stage 5. Therefore, the REV and PHB domains appear to have rearranged during anther development.
The Mutation of MIR166g and PHB Affects SPL/NZZ Domains
We wondered whether miR165/6 and the HD-ZIP III genes regulate the rearrangement of SPL/NZZ domains. In jba-1D anthers, the miR165/6 domains were expanded and occupied the whole locules at stage 2 (Fig. 2). Then they formed the two domains in the two thecae at stage 3 and subsequently became enlarged in the four locules at stage 5 compared with the wild type. By contrast, the expression domains of PHB and REV were smaller in the lateral regions at stage 2 compared with those of the wild type and disappeared in anthers at stages 3 and 5. The miR165/6 domains of jba-1D anthers at stage 3 are consistent with their enlarged microsporangia. Interestingly, SPL/NZZ domains at stage 3 were seen in two lateral regions of early stamens and dispersed in the two locules at stage 5. SPL/NZZ domains were matched with the enlarged microsporangia. This observation suggests that miR166 maintains SPL/NZZ domains and facilitates microsporogenesis in inner microsporangia.
In phb-7D anthers, PHB domains were expanded and occupied most of the thecae in the adaxial regions of anthers at stage 2 (Fig. 2). Then they formed the two clear domains in the boundary at stage 3 and subsequently became enlarged in the stomium regions at stage 5 compared with the wild type. Interestingly, the enlarged PHB domains of phb-7D anthers at stage 3 are matched with their thickened boundary. Correspondingly, SPL/NZZ domains were smaller in stamen primordia at stages 2, 3, and 5 compared with the wild type. Together, these results suggest that PHB limits SPL/NZZ domains and promotes the formation of stomium in the boundary.
SPL/NZZ Acts Downstream of miR166 and the HD-ZIP III Genes
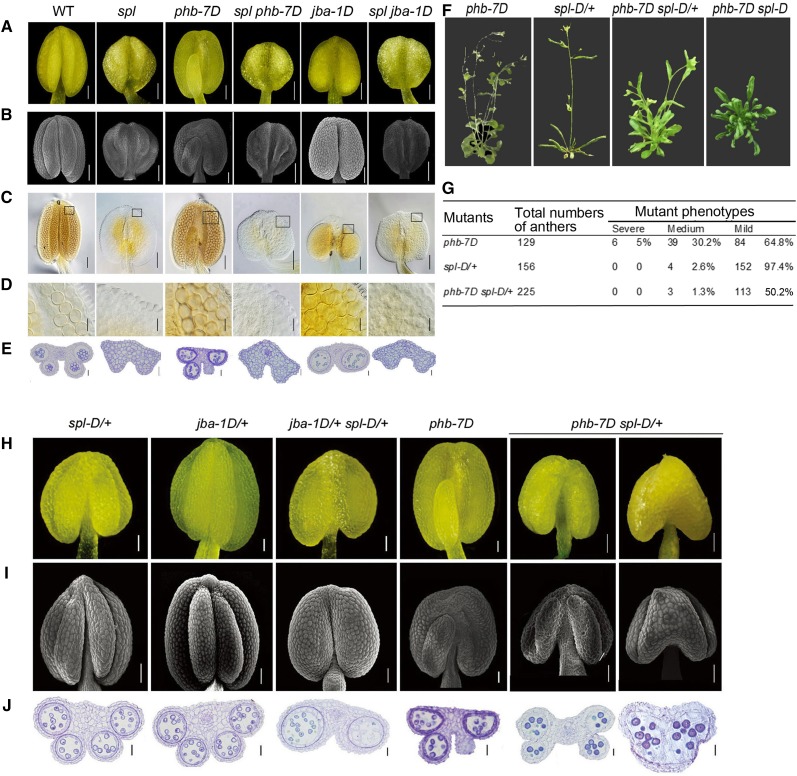
To verify the genetic interaction between HD-ZIP III genes and SPL/NZZ, we crossed the spl mutant with the phb-7D and jba-1D mutants. The spl seedlings and flowers looked similar to the wild type (Supplemental Fig. S2, A and B), but their anthers did not form microsporocytes as previously described (Yang et al., 1999) with the similar cells on the adaxial and abaxial surfaces (Supplemental Fig. S2, C and E). At anthesis, the spl anthers consisted of highly vacuolated parenchyma cells, without any microsporangia or pollen (Fig. 3, A–E). The anthers of the spl phb-7D double mutants resembled those of the spl mutant (i.e. microsporangia and locules were absent). Similar to the spl anthers, the spl jba-1D anthers completely lacked microsporangia. These results suggest that SPL/NZZ acts downstream of the HD-ZIP III genes.
Figure 3.
Anther morphology and anatomy in the single and double mutants of SPL/NZZ, MIR166g, and PHB. A, Anthers of spl, jba-1D, and phb-7D mutants at stage 12. B, Scanning electron microscopy images showing the appearance of spl, jba-1D, and phb-7D anthers at stage 12. C, Whole-mount clearing images showing the internal structures of spl, jba-1D, and phb-7D locules at stage 12. D, Magnified images of the boxes in C showing the pollen grains in spl, jba-1D, and phb-7D locules at stage 12. E, Cross sections of spl, jba-1D, and phb-7D anthers. F, Plant phenotypes of phb-7D, spl-D/+, and jba-1D single and double mutants at flowering stage. G, The distribution of phb-7D spl-D/+ anthers with the mutant phenotypes of different severity. H, Anthers of phb-7D, spl-D/+, and jba-1D single and double mutants at stage 11 imaged with an anatomical microscope. I, Scanning electron microscopy images showing the appearance of anthers of the phb-7D, spl-D/+, and jba-1D single and double mutants at stage 11. J, Cross sections of the anthers of phb-7D, spl-D/+, and jba-1D single and double mutants at stage 11. Bars = 50 μm (A–C, H, and I) and 20 μm (D, E, and J).
PHB Binds the Promoter of SPL/NZZ
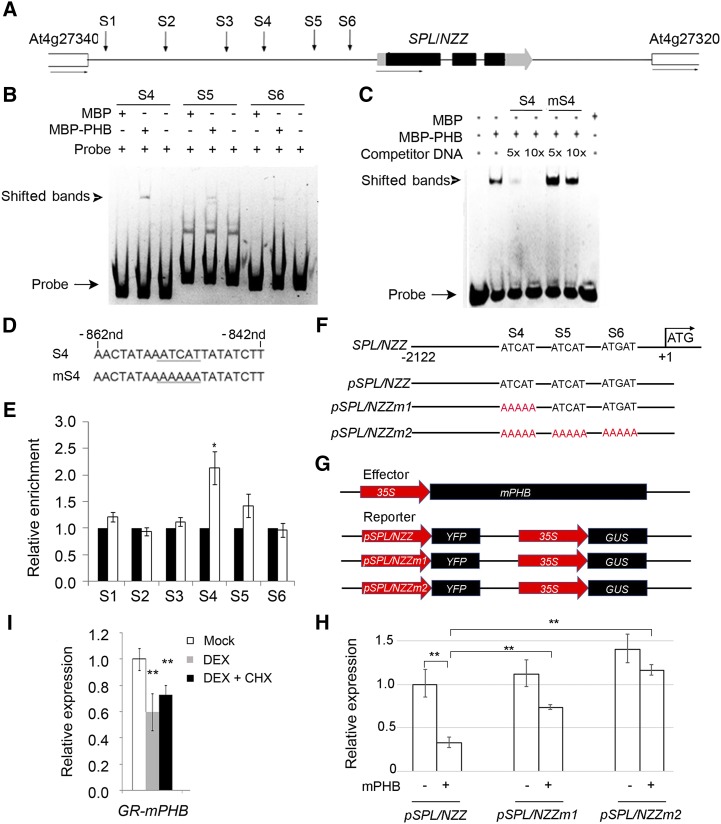
The HD-ZIP III genes regulate the downstream genes by directly binding to the conserved motifs in the promoters of auxin biosynthesis genes (Brandt et al., 2012). Within the upstream and 5′ untranslated regions of SPL/NZZ, there are six putative binding sites (Fig. 4A). To test the binding affinity between PHB and these motifs, we purified MBP-PHB fusion proteins (Supplemental Fig. S3A). Electrophoretic mobility shift assay (EMSA) data show that PHB was bound to the motif (Supplemental Fig. S3, B and C). Moreover, the binding signal between PHB and the motif is enhanced with increased concentrations of probe (Supplemental Fig. S3D). Then we used this reaction system to verify the binding possibility between PHB and the SPL/NZZ promoter. S4, S5, and S6 sites showed binding affinity (Fig. 4B). Among them, site S4 exhibited specific binding affinity with PHB under the competitive condition with nonlabeled probes and mutated probes (Fig. 4, C and D).
Figure 4.
Binding of PHB in the promoter of SPL/NZZ. A, Diagram of SPL/NZZ genomic regions showing the positions of putative binding sites (S1–S6, indicated by arrows). The gray and black boxes indicate the untranslated region and exon regions, respectively, while the lines indicate intergenic regions and introns. B, EMSA showing the binding affinity of the putative sites of SPL/NZZ promoter regions by PHB. The positions of PHB-DNA complexes are marked by shifted bands. C, Competitor DNA-binding assay showing the binding affinity of the S4 site of the SPL/NZZ promoter regions by PHB. D, Sequences of the S4 site and its competitor DNA. E, ChIP assays showing PHB binding to the SPL/NZZ promoter. F, DNA sequences of the wild-type SPL/NZZ promoter and mutant SPL/NZZ promoters. G, Structure of the SPL/NZZ promoter-driven YFP reporter gene. The SPL/NZZ promoter, 35S promoter, YFP, GUS, and PHB genes are indicated. H, Relative reporter (YFP) expression in plants with different SPL/NZZ promoters. N. benthamiana leaves were transfected with the reporters (pSPL/NZZ, pSPL/NZZm1, and pSPL/NZZm2) and the effector (mPHB). I, RT-qPCR showing relative expression of SPL/NZZ in p35S::GR-mPHB lines treated with DEX and DEX + CHX solutions. Three biological replicates were analyzed. Error bars indicate the sd, while asterisks show significant differences at the 0.05 (*) and 0.01 (**) levels in Student’s t test.
To confirm the direct interaction between PHB and SPL/NZZ loci in vivo, we conducted a chromatin immunoprecipitation (ChIP) assay using a transgenic line expressing the 3×Flag-mPHB fusion construct under the control of the 35S promoter. The chromatin extracted from the inflorescences was immunoprecipitated with anti-Flag antibodies, and the abundance of approximately 150-bp regions containing PHB-binding sequences was measured by quantitative PCR (qPCR). The site S4 in the SPL/NZZ promoter was enriched nearly 2-fold in the plants with 3×Flag-mPHB after being immunoprecipitated by the corresponding antibody (Fig. 4E). However, the other sites were not enriched significantly. These data suggest that PHB directly binds the SPL/NZZ promoter in vivo.
SPL/NZZ Expression Is Suppressed by PHB Up-Regulation
To verify the function of PHB in its regulation to SPL/NZZ, a reporter transaction assay in Nicotiana benthamiana leaves was performed. The vectors that harbor pSPL/NZZ (the promoter of SPL/NZZ), pSPL/NZZm1 (pSPL/NZZ with mutated binding site S4), and pSPL/NZZm2 (pSPL/NZZ with mutated binding sites S4, S5, and S6) were designed to drive YFP reporters (Fig. 4, F and G). In the presence or absence of PHB, YFP was transiently expressed in N. benthamiana plants with pSPL/NZZ, pSPL/NZZm1, and pSPL/NZZm2. The YFP expression levels declined in pSPL/NZZ plants when coexpressed with PHB, and YFP expression levels were much higher in pSPL/NZZm1 and pSPL/NZZm2 plants compared with pSPL/NZZ plants (Fig. 4H). This result revealed that PHB negatively regulated SPL/NZZ by directly binding to the SPL/NZZ promoter.
Next, we explored the possibility that PHB represses SPL/NZZ expression in Arabidopsis using an inducible expression system based on the posttranscriptional activation of the rat Glucocorticoid Receptor (GR). A GR-mPHB construct was transformed into Arabidopsis plants. In the inflorescences of the transgenic plants expressing GR-mPHB, SPL/NZZ expression levels declined significantly following a 6-h treatment with dexamethasone (DEX) and a 6-h treatment with DEX and cycloheximide (CHX; Fig. 4I). We conclude that SPL/NZZ transcription is repressed following PHB induction.
PHB Activates WUS to Maintain Stomium Cells
The key regulator of stem cell homeostasis, WUS, is the only known regulator that engages in the initiation and development of stomium cells in Arabidopsis (Bäurle and Laux, 2005). To examine how MIR166g activation affects the formation of the stomium region in the boundary, we performed in situ hybridization of WUS using the anthers at different stages. In the wild-type anthers, the WUS signal was faint in the boundary furrow between inner and outer locules at stage 3 (Fig. 5A) and became stronger and expanded at stage 5. This experiment reveals that the stomium is in the boundary furrow between inner and outer microsporangia. In jba-1D anthers, WUS signal was not detected at stages 2, 3, and 5. In phb-7D anthers, by contrast, WUS domains were expanded beyond the internal boundary at stage 2 but became weaker and smaller at stages 3 and 5. This result implies that PHB restricts stomium to the boundary between inner and outer microsporangia.
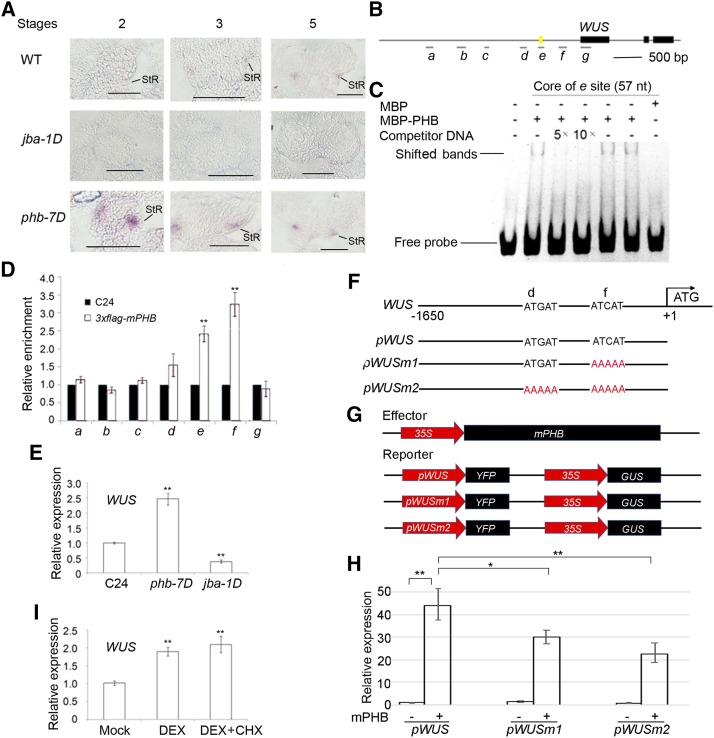
Figure 5.
Binding of PHB in the promoter of WUS. A, In situ hybridization showing the expression patterns of WUS in developing anthers of jba-1D and phb-7D mutants. StR, Stomium regions; WT, wild type. Bars = 50 μm. B, Putative binding sites of PHB in WUS. Letters a, b, c, d, e, f, and g represent seven putative binding sites in WUS. The yellow mark indicates a 57-bp regulatory region (−655 to −712 bp) conferring WUS transcription in the shoot apical meristem stem cell niche. C, EMSA showing the binding affinity of the putative sites of WUS promoter regions by PHB. The positions of PHB-DNA complexes are marked by shifted bands. D, ChIP assays showing PHB binding to the WUS promoter. E, RT-qPCR showing relative expression of WUS in phb-7D and jba-1D. F, DNA sequences of the wild-type WUS promoter and mutant WUS promoters with mutations in different binding sites. G, Schematic diagram of the reporter (pWUS, pWUSm1, and pWUSm2) and effector (mPHB) constructs. H, RT-qPCR showing relative expression of YFP measured after transient transformation of the reporter (pWUS, pWUSm1, and pWUSm2) and effector (mPHB) constructs in N. benthamiana leaves. I, RT-qPCR showing relative expression of WUS in p35S::GR-mPHB lines treated with DEX and DEX + CHX solutions. Three biological replicates were analyzed. Error bars indicate the sd, while asterisks show significant differences at the 0.05 (*) and 0.01 (**) levels in Student’s t test.
To determine whether PHB directly binds to the WUS promoter, we performed EMSA experiments. There are seven putative binding sites for PHB within the promoter region of WUS (Fig. 5B). A 57-bp regulatory region (−655 to −712 bp) confers WUS transcription in the shoot apical meristem stem cell niche (Bäurle and Laux, 2005). As expected, PHB shows binding affinity with this regulatory region (Fig. 5C). Then, a ChIP experiment showed that two DNA fragments are enriched two to four times in the transgenic lines with 3×Flag-mPHB (Fig. 5D). These results confirm that PHB binds the promoter of WUS.
RT-qPCR shows that WUS was up-regulated in phb-7D anthers but down-regulated in jba-1D anthers (Fig. 5E). We wondered whether the induction of PHB expression affects WUS expression level.
The transient transcription assay was applied to verify the regulation of PHB to WUS. The YFP gene was driven by pWUS (the wild-type WUS promoter), pWUSm1 (mutant WUS promoter with mutant binding site f), and pWUSm2 (mutant binding sites d and f; Fig. 5, F and G). YFP expression was up-regulated in pWUS plants upon cotransformation with p35S::mPHB (Fig. 5H) and was reduced in pWUSm1 and pWUSm2 plants when PHB was coexpressed with WUS. This result indicated that PHB positively regulated WUS (Fig. 5H).
We next used an inducible expression system based on posttranscriptional activation of the rat GR. A GR-mPHB construct was transformed into Arabidopsis plants. In the inflorescences of the transgenic plants expressing GR-mPHB, the expression levels of WUS were significantly increased following a 6-h treatment with DEX and a 6-h treatment with DEX and CHX (Fig. 5I). We conclude that PHB acts upstream of WUS to maintain stomium stem cells.
spl-D Aggravates the Phenotype of jba-1D/+ Plants
spl-D is a dominant mutant of the SPL/NZZ gene. Homozygous spl-D plants produced upwardly curved leaves (Supplemental Fig. S4A) and never bolted, as noted in earlier studies (Li et al., 2008). Therefore, we used the anthers of heterozygous spl-D plants (spl-D/+; Supplemental Fig. S4B) for the following study. The spl-D/+ plants flowered (Fig. 3F) and set seeds because they had normal anthers with four microsporangia (Fig. 3H). To further define the relationship between the HD-ZIP III genes and SPL/NZZ, we crossed jba-1D/+ with spl-D/+ plants. Homozygous jba-1D anthers were male sterile and could not be used for genetic analysis, while the heterozygotic jba-1D/+ anthers resembled the wild-type anthers with four microsporangia. The heterozygotic jba-1D/+ spl-D/+ plants were individually identified by PCR and self-pollinated to generate F2 segregating populations, some of which were selected for genetic analysis. The jba-1D/+ spl-D/+ plants formed anthers with only two enlarged microsporangia (Fig. 3, H and I). These anthers had ectopic microsporangia in the place of an internal boundary (Fig. 3J), just like the homozygous jba-1D anthers. Meanwhile, the expression levels of SPL/NZZ in jba-1D/+ spl-D/+ anthers increased substantially compared with those of jba-1D/+ anthers (Supplemental Fig. S5). Our observations provide molecular support that spl-D/+ aggravates the mutant phenotype of jba-1D/+ in terms of boundary defects.
spl-D Partially Rescues the Mutant Phenotype of phb-7D
To verify the epistatic effect of SPL/NZZ on PHB, we used the phb-7D mutant as the female parent to cross with the spl-D/+ mutant. The phb-7D/+ spl-D/+ plants in the F1 population were selected using phb-7D derived cleaved-amplified polymorphic sequence primers and spl-D genotyping primers. Similarly, the F1 plants were self-fertilized to obtain F2 segregating populations, which included phb-7D, spl-D/+, phb-7D spl-D/+, and phb-7D spl-D plants. Like the spl-D mutant, phb-7D spl-D plants did not flower. Hence, we were unable to analyze the phb-7D spl-D anthers. The phb-7D spl-D/+ plants flowered (Fig. 3F) like the phb-7D mutant, but their flowers lacked petals. These plants produced upwardly curved leaves like the spl-D mutant plants. Surprisingly, the anthers without inner microsporangia in phb-7D spl-D/+ plants disappeared, while almost all inner microsporangia contained microsporocytes (Fig. 3G). Fifty percent of the anthers of the phb-7D spl-D/+ plants showed the thinner boundary than phb-7D anthers (Fig. 3, H–J), with inner microsporangia smaller than those of the wild-type anthers but larger than those of the phb-7D anthers. Meanwhile, the expression levels of SPL/NZZ in phb-7D spl-D/+ anthers increased substantially compared with those of phb-7D anthers (Supplemental Fig. S5). The changes in phb-7D spl-D/+ plants suggest that the mutant phenotypes of the phb-7D allele were partially recovered in the thickened boundary and aberrant inner microsporangia. The remaining anthers contained two enlarged inner microsporangia that were partially joined. A subsequent analysis of a semithin section revealed that the joined inner microsporangia were separated by a septum. Interestingly, the anthers with joined inner microsporangia opened and released pollen grains even though their seed-set rate was much lower than that of the wild-type anthers. These results suggest that the mutant phenotypes of phb-7D plants are partially rescued by spl-D/+.
DISCUSSION
miR165/6 Monitors SPL/NZZ for Boundary Building
HD-ZIP III genes such as PHB and REV are essential for the adaxial identity of lateral organs. In Arabidopsis, expression domains of PHB and REV are found to be rearranged during stamen development. The inference is that the rearrangement of the expression domain of HD-ZIP III genes during anther development is a general event in vascular plants. Importantly, this type of rearrangement of expression domains is controlled by miR165/6 through posttranscriptional gene silencing. Originally, miR165/6 accumulates in the adaxial regions of stamen primordia (stage 2), is redistributed on the lateral sides of early stamen (stage 3), and then is rearranged to the microsporangia in the late stamen (stage 5). Meanwhile, PHB and REV expression domains were moved to the boundary regions between inner and outer microsporangia in the early stamen and then located in stomium regions within the late stamen. Therefore, temporal and spatial expression patterns of PHB and REV expression domains show an opposite trend to that of miR165/6 accumulation.
SPL/NZZ expression domains overlapped with the domain of miR165/6 accumulation. Both domains are rearranged in different directions from the PHB domains. miR165/6 and SPL/NZZ domains are moved from the lateral regions of stamen primordia to stamen corners and then to microsporangia, while the PHB domain migrated from the middle regions of stamen primordia to the boundary between inner and outer microsporangia and then to stomium regions. MIR166g activation causes the misplacement of SPL/NZZ, leading to the ectopic formation of microsporocytes in the place of boundary. Importantly, the absence of the boundary undoes the formation of stomium and dehiscence zones, resulting in male sterility. The formation of the boundary is therefore a prerequisite for the differentiation of the stomium region.
The regulation of miR65/6 to SPL/NZZ is accomplished by HD-ZIP III genes. PHB inhibits SPL/NZZ in inner microsporangia by binding to the promoter to build the boundary. The dominant mutation of PHB causes shrunken domains of SPL/NZZ in the whole or part of the inner microsporangia and disrupts two important developmental events: thickening of the boundary between inner and outer microsporangia and microsporogenesis in inner microsporangia. By inhibiting SPL activity, PHB blocks cell formation of microsporocytes in the boundary and facilitates its formation. The spl-D allele aggravates the jba-1D mutant phenotype in terms of boundary defects but partially rescues the phb-7D phenotype in terms of boundary thickening. Considering that miR165/6 and PHB act upstream of SPL/NZZ, we conclude that miR165/6 monitors SPL/NZZ domains for building of the boundary.
Archesporial cells emerge in the four corners of early stamen (Feng and Dickinson, 2007, 2010). We observed the parietal and sporogenous cells in the two lateral regions of early stamens at stage 2 before the stamen primordia are separated into the four corners. The occupation of miR165/6 and SPL/NZZ domains in the lateral regions of early stamens implies that there is meristem-like tissue in the boundary from where an archesporial cell is initiated and divides, timely forming two lateral regions of early stamens. This meristem-like tissue could be the stomium region. The stomium would thus assume some aspect of apical meristem function (Deyhle et al., 2007). It is interesting to uncover a molecular connection between embryonic meristem and the stomium region.
PHB Determines the Formation of Stomium in Anthers
In rice, the expression of PHB3 (rice homolog of PHB) marks the stomium region as an adaxial one (Toriba et al., 2010). We asked whether PHB of Arabidopsis determines the formation of stomium. Notably, PHB domains in stomium regions of Arabidopsis anthers at stages 3 and 5 are the same as PHB3 domains. The inference is that monocots and dicots share similar adaxial regions in the boundary. WUS expression indicated that stomium cells are localized in the boundary furrow between inner and outer microsporangia. WUS expression is not expressed in anther primordium a stage 1 but appears at stage 2 and continues until stage 11, when the stomium cells start to display specific differentiation (Deyhle et al., 2007). We detected WUS expression in boundary furrows at stage 2. This suggests that stomium regions are initiated at stage 3 or before.
In the jba-1D mutant, the SPL/NZZ domain is expanded to the place of the boundary containing the stomium regions, concurrent with the appearance of ectopic microsporocytes and the absence of the dehiscence zones. In the phb-7D mutant, SPL/NZZ expression domains were shrunk, concurrent with the thickening of the boundary and the presence of dehiscence zones. Given that PHB acts upstream of WUS, we propose that PHB determines the formation of stomium through WUS.
Our genetic evidence indicates that PHB is required for boundary building. In Arabidopsis, adaxial thecae and abaxial connectives differentiate from the stamen primordia. During theca development, the boundary between inner and outer microsporangia is the adaxial region, as marked by PHB domains; stomium embedded in the boundary is the adaxial region, as indicated by PHB. PHB domains in anthers become centralized progressively from stages 2 to 5. We suggest that PHB restricts the stomium to the boundary, so that the inner microsporangia take shape for the differentiation of microsporocyte cells.
Cross Talk between the miR165/6 Pathway and SPL/NZZ Links Polarity to Microsporogenesis
The internal structure of an anther with four microsporangia largely relies on the balance between the HD-ZIP III genes and SPL/NZZ. As such, there is considerable cross talk between genes that control adaxial identity and microsporogenesis. In the stamen primordia (stage 2), miR165/6 accumulates in the lateral regions while SPL/NZZ is preferentially expressed in the lateral regions. Then, miR165/6 accumulation and SPL/NZZ domains are rearranged simultaneously to the corners of stamen primordia at stage 3 and to the microsporangia at stage 5. Meanwhile, the expression domains of miR165/6-targeted PHB are redistributed to the boundary between inner and outer microsporangia at stage 3 and to stomium at stage 5. During the translocation of the adaxial regions in anthers, miR165/6 acts to control the expression domains and activity of SPL/NZZ by silencing HD-ZIP III genes and maintaining the balance between PHB and SPL/NZZ for programming of the two developmental events: microsporogenesis in microsporangia and formation of the boundary between inner and outer microsporangia. During this process, PHB determines boundary formation by inhibiting the SPL/NZZ gene. Thus, the cross talk between the genes that control adaxial polarity and microsporogenesis constitutes an important link between adaxial polarity in the internal boundary of anthers and microsporogenesis in microsporangia.
Arabidopsis and rice are different in anther shape and size, but they program similar developmental events. To illustrate the regulatory roles of miR165/6 and SPL/NZZ in the establishment of internal structure in anthers, we refine the early stages of anther development as follows. Initially, two adaxial regions are established in the place adjacent to the floral meristem (at stage 1 and earlier) and develop into the two thecae, while abaxial regions form in the place opposite to it and develop into the connectives (Feng and Dickinson, 2007, 2010; Toriba et al., 2010) Then, the newly formed adaxial regions are formed in the middle regions of two thecae and subsequently turn into the boundary between inner and outer microsporangia. In the boundary furrows, stomium regions are maintained and become another newly formed adaxial region. According to these developmental events, we propose a model for the regulation of miR165/6 to SPL/NZZ in the formation of the boundary between microsporangia in anthers (Fig. 6). In general, there are three dimensions of anther formation: miR165/6 modulates SPL/NZZ by PHB; miR165/6, PHB, and SPL/NZZ domains are rearranged in the developing anthers; and PHB controls the building of the internal boundary by inhibiting SPL/NZZ and determines the formation of stomium by activating WUS. Therefore, miR165/6 balances the expression of PHB and SPL/NZZ to link adaxial polarity to microsporogenesis in anthers
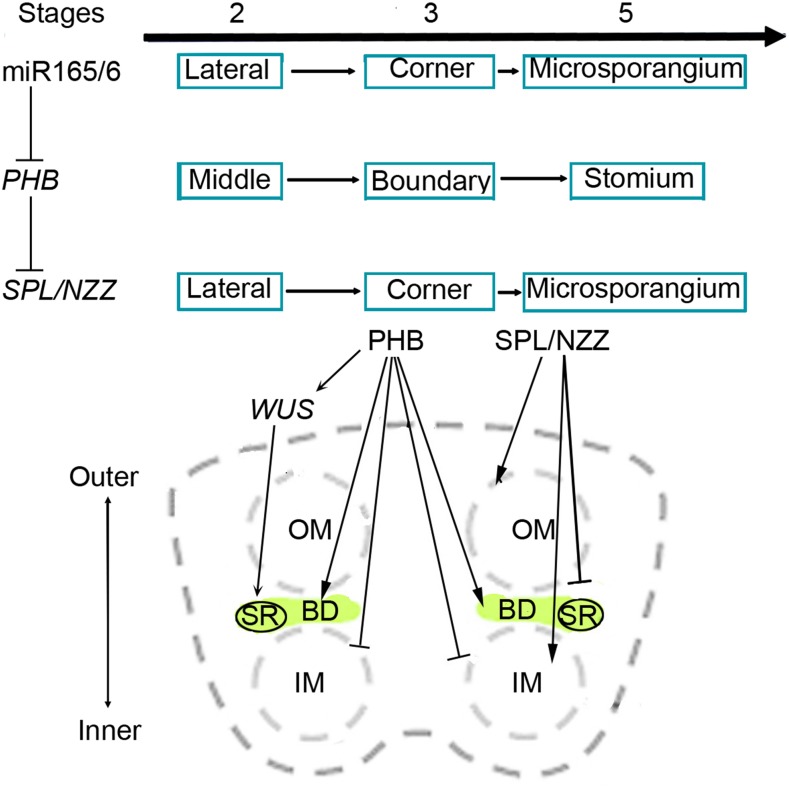
Figure 6.
Model for miR165/6 regulation of SPL/NZZ in the formation of the boundary between inner and outer microsporangia in anthers. During anther development, miR165/6 silence PHB by complementary sequences. PHB binds to SPL/NZZ to inhibit the activity of SPL/NZZ. miR165/6 and SPL/NZZ domains move from the two lateral regions of stamens to the four corners of anthers and then to microsporangia, while PHB domains migrate from the middle regions of thecae to the boundary between inner and outer microsporangia and then to the stomium regions. PHB facilitates the formation of the boundary, where it determines the formation of stomium through WUS but inhibits the differentiation of microsporocytes. SPL/NZZ promotes the initiation of microsporocytes but obstructs the formation of the boundary. BD, Boundary between inner and outer microsporangia; IM, inner microsporangia; OM, outer microsporangia; SR, stomium region.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) mutants used in this study included jba-1D, phb-7D, spl, and spl-D (Supplemental Table S1). The seeds of wild-type and mutant plants were surface sterilized in 70% (v/v) ethanol for 1 min and then in 1% (v/v) NaOCl for 10 min, after which they were washed four times in sterile distilled water. Seeds were then placed on the surface of 1% (w/v) agar-solidified Murashige and Skoog medium. Plates were sealed with Parafilm, incubated at 4°C in darkness for 3 to 4 d, and then moved to a growth chamber at 22°C with a 16-h photoperiod. The spl jba-1D, spl phb-7D, jba-1D/+ spl-D/+, phb-7D spl-D/+, and phb-7D jba-1D double mutants were generated by crossing the corresponding mutants and then characterized by culturing in a medium supplemented with the appropriate antibiotic as well as by PCR and phenotypic analyses.
Scanning Electron Microscopy
Flowers and inflorescences were fixed in FAA (50% [v/v] ethanol, 5% [v/v] acetic acid, and 3.7% [v/v] formaldehyde) and dried. They were then dissected using a stereomicroscope and mounted on sample stubs for scanning electron microscopy. Mounted anthers were coated with palladium-gold and then examined using a JSM-6360LV scanning electron microscope (JEOL) with an acceleration voltage of 7 to 15 kV.
Histology and Light Microscopy
Flowers (inflorescences) of 5- to 6-week-old wild-type and mutant plants were fixed in FAA and embedded in paraffin (Sigma-Aldrich), after which 7-µm sections were stained with 0.05% (w/v) Toluidine Blue (Sigma-Aldrich) at 37°C for 15 min. The stained sections were washed in water and treated with Histo-Clear (National Diagnostics), which is a nontoxic histological clearing agent, to remove paraffin. To analyze semithin sections, samples fixed in FAA were embedded in epoxy resin. Glass knives were used to prepare 2-µm sections, which were then affixed to glass slides and stained with 0.05% (w/v) Toluidine Blue.
Samples and sections were observed using a BX 51 wide-field microscope equipped with UPlanSAPO series objectives and a cooled DP71 camera (Olympus) and with a Stemi 2000 stereomicroscope (Zeiss). To observe the hybridization signal after in situ hybridization, slides were mounted in water and analyzed by differential interference contrast microscopy. For anther imaging, Image-Pro Express version 5.1 software (Media Cybernetics) was used to extend the depth of field.
For whole-mount clearing analysis, anthers were collected using a stereomicroscope and immersed in clearing solution (2 g of phenol, 2 g of chloral hydrate, 2 mL of 75% [v/v] lactic acid, 2 g of oil of clove, and 1 mL of xylene) for several minutes. The specimens were analyzed by differential interference contrast microscopy.
In Situ Hybridization
Flower sections (7 μm thick) from wild-type and mutant plants were prepared using previously described pretreatment and hybridization methods (Lian et al., 2013). The primers used to generate hybridization probes specific for REV, PHB, and SPL/NZZ are listed in Supplemental Table S2. Digoxigenin-labeled probes were prepared by in vitro transcription (Roche) according to the manufacturer’s recommended protocol. Additionally, LNA (Locked Nucleic Acid)-modified probes specific for miR166 were synthesized and labeled with digoxigenin at the 3′ end by TaKaRa.
RT-qPCR
Stage 3 anthers were carefully harvested from 6-week-old wild-type and mutant plants using a stereomicroscope and stored immediately in liquid nitrogen. Total RNA was extracted from anthers with the RNeasy Plant Mini Kit (Qiagen) and then reverse transcribed using oligo(dT) primers. RT-qPCR analysis was completed with the Rotor-Gene 3000 system (Corbett Research) using SYBR Premix Ex Taq (TaKaRa). ACTIN mRNA was used as an internal control, and relative mRNA abundance was calculated as previously described by Liu et al. (2011). Primers specific for ACTIN, SPL/NZZ, REV, PHB, PHV, CNA, and ATHB8 (Supplemental Table S2) were used to detect gene expression in the mutants.
ChIP Assay
The 35S:3×Flag-mPHB transgenic plants were analyzed using a ChIP assay. Inflorescence was cross-linked with 1% (v/v) formaldehyde, and chromatin was isolated and precipitated as described in Zhang et al. (2017). The crude chromatin extract was divided into three parts. One part was used as the input control. The other two parts were used for immunoprecipitations with 5 µL of Flag antibodies (Sigma-Aldrich). After several washes, chromatin cross-linking was reversed and DNA was purified. qPCR analysis was completed using a MyiQ2 two-color real-time PCR detection system, with immunoprecipitated DNA as the template. Values for the ChIP samples were first normalized against that of the input and then divided by the normalized Flag signal values to calculate the fold enrichment. The sequences of the primers used to amplify different promoter regions are listed in Supplemental Table S2.
GR Induction and RNA Quantification
The expression of the gene encoding the GR was induced as previously described by Zhang et al. (2017). Seeds carrying the 35S:GR-mREV and 35S:GR-mPHB constructs were plated on agar-solidified Murashige and Skoog medium, and the resulting seedlings were transplanted in soil. When the plants started to bolt, the inflorescences were immersed in 0.1% (v/v) ethanol (mock), 10 µm DEX in 0.1% (v/v) ethanol, 10 µm CHX in 0.1% (v/v) ethanol, or 10 µm DEX and 10 µm CHX in 0.1% (v/v) ethanol for 1 min. Posttreatment inflorescences were harvested after 6 h of treatment. Total RNA was extracted using Trizol reagent (Invitrogen) and then treated with RNase-free DNase (TaKaRa). RT-qPCR was performed as described above using primers listed in Supplemental Table S2. The experiment was repeated three times.
YFP Expression Assay for Protein-DNA Interactions in Nicotiana benthamiana Leaves
To measure the effects of PHB on the transcriptional regulation of SPL/NZZ and WUS, the coding sequence of PHB was cloned into pCAMBIA1300 as an effector. The 2,122-bp sequences upstream of the SPL/NZZ coding regions and the 1,650-bp sequence upstream of the WUS coding regions were cloned into pCAMBIA3301 as reporters. Type IIS restriction enzyme BsmBI was used for the construction of SPL/NZZ and WUS promoters with binding site mutation. Agrobacterium tumefaciens GV3101 strains carrying the above-verified constructs were introduced into N. benthamiana leaves. As negative controls, leaves were infiltrated with strains carrying reporter with the different SPL/NZZ and WUS promoters and empty effector. RNA was extracted from the leaves 3 d after infiltration, and YFP expression was measured by RT-qPCR.
EMSA
The PHB and REV genes were cloned into the pMAL-c2x vector (New England Biolabs) for the expression of a fusion protein with an N-terminal MBP peptide. The prepared expression vectors as well as empty pMAL-c2x vector (control) were inserted into competent Escherichia coli cells. The fusion proteins were purified according to the manufacturer’s recommended procedure (E8021S). Additionally, DNA fragments from the SPL/NZZ promoter were end labeled with Cy5. The recombinant fusion protein (500 ng) in binding buffer (10 mm Tris-HCl, pH 7.6, 25 mm KCl, 2.5 mm MgCl2, 1 mm EDTA, 1 mm DTT, and 10% [v/v] glycerol) was supplemented with 10 ng of Cy5-labeled DNA and incubated for 30 min at 25°C. The reaction mixtures were electrophoresed on 6% native polyacrylamide gels in 0.5× TBE buffer (45 mm Tris, 45 mm boric acid, and 1 mm EDTA, pH 8) at 200 V for 1.5 h. Gels were analyzed using a Tanon 4200SF chemiluminescent imaging system.
Statistical Analysis
The statistical analyses including Student’s t test were performed by Excel 2010 software. Both RT-qPCR and qPCR for each sample were replicated three times, the average values of 2−ΔCT were used to determine the differences, and the data were expressed as means ± sd. A significant difference was considered at P < 0.05 and extremely significant at P < 0.01.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: PHB (AT2G34710), SPL/NZZ (AT4G27330), WUS (AT2G17950), and MIR166g (AT5G63715).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Expression of miR166 and HD-ZIP III in the jba-1D mutant with the abaxial side of the anther.
Supplemental Figure S2. The plant and flower of the spl mutant with the abaxial side of the anther.
Supplemental Figure S3. Interaction between PHB and SPL/NZZ.
Supplemental Figure S4. The plant and flower of the spl-D mutant.
Supplemental Figure S5. Expression levels of SPL/NZZ in the anthers of jba-1D, phb-7D, and spl-D single and double mutants.
Supplemental Table S1. Mutants included in this study.
Supplemental Table S2. Probes and primers used in this study.
Acknowledgments
We thank Dr. Nina V. Fedoroff (Pennsylvania State University), Dr. Jennifer C. Fletcher (Plant Gene Expression Center), Dr. Ykä. Helariutta (Department of Biosciences, University of Helsinki), Dr. Li Jia Qu (College of Life Sciences, Peking University), and Dr. Wei Cai Yang (Institute of Genetics and Development, Chinese Academy of Sciences) for providing mutant seeds. We thank Ji Qin Li (Shanghai Institute of Plant Physiology and Ecology) for helping us develop the analyzed semi-sections.
Footnotes
This work was supported by grants from the National Key Research and Development Program of China (2016YFD0101900) and the National Natural Science Foundation of China (31771442 and 31571261).
References
- Bäurle I, Laux T (2005) Regulation of WUSCHEL transcription in the stem cell niche of the Arabidopsis shoot meristem. Plant Cell 17: 2271–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL. (2004) Class III HD-Zip gene regulation, the golden fleece of ARGONAUTE activity? BioEssays 26: 938–942 [DOI] [PubMed] [Google Scholar]
- Brandt R, Salla-Martret M, Bou-Torrent J, Musielak T, Stahl M, Lanz C, Ott F, Schmid M, Greb T, Schwarz M, et al. (2012) Genome-wide binding-site analysis of REVOLUTA reveals a link between leaf patterning and light-mediated growth responses. Plant J 72: 31–42 [DOI] [PubMed] [Google Scholar]
- Carlsbecker A, Lee JY, Roberts CJ, Dettmer J, Lehesranta S, Zhou J, Lindgren O, Moreno-Risueno MA, Vatén A, Thitamadee S, et al. (2010) Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465: 316–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyhle F, Sarkar AK, Tucker EJ, Laux T (2007) WUSCHEL regulates cell differentiation during anther development. Dev Biol 302: 154–159 [DOI] [PubMed] [Google Scholar]
- Dinneny JR, Weigel D, Yanofsky MF (2006) NUBBIN and JAGGED define stamen and carpel shape in Arabidopsis. Development 133: 1645–1655 [DOI] [PubMed] [Google Scholar]
- Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL (2003) Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol 13: 1768–1774 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Perea JV, Bowman JL (2001) Establishment of polarity in lateral organs of plants. Curr Biol 11: 1251–1260 [DOI] [PubMed] [Google Scholar]
- Feng X, Dickinson HG (2007) Packaging the male germline in plants. Trends Genet 23: 503–510 [DOI] [PubMed] [Google Scholar]
- Feng X, Dickinson HG (2010) Cell-cell interactions during patterning of the Arabidopsis anther. Biochem Soc Trans 38: 571–576 [DOI] [PubMed] [Google Scholar]
- Goldberg RB, Beals TP, Sanders PM (1993) Anther development: Basic principles and practical applications. Plant Cell 5: 1217–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hord CL, Chen C, Deyoung BJ, Clark SE, Ma H (2006) The BAM1/BAM2 receptor-like kinases are important regulators of Arabidopsis early anther development. Plant Cell 18: 1667–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Wellmer F, Yu H, Das P, Ito N, Alves-Ferreira M, Riechmann JL, Meyerowitz EM (2004) The homeotic protein AGAMOUS controls microsporogenesis by regulation of SPOROCYTELESS. Nature 430: 356–360 [DOI] [PubMed] [Google Scholar]
- Li LC, Qin GJ, Tsuge T, Hou XH, Ding MY, Aoyama T, Oka A, Chen Z, Gu H, Zhao Y, et al. (2008) SPOROCYTELESS modulates YUCCA expression to regulate the development of lateral organs in Arabidopsis. New Phytol 179: 751–764 [DOI] [PubMed] [Google Scholar]
- Lian H, Li X, Liu Z, He Y (2013) HYL1 is required for establishment of stamen architecture with four microsporangia in Arabidopsis. J Exp Bot 64: 3397–3410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Jia L, Wang H, He Y (2011) HYL1 regulates the balance between adaxial and abaxial identity for leaf flattening via miRNA-mediated pathways. J Exp Bot 62: 4367–4381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H. (2005) Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu Rev Plant Biol 56: 393–434 [DOI] [PubMed] [Google Scholar]
- McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK (2001) Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411: 709–713 [DOI] [PubMed] [Google Scholar]
- Sanders PM, Bui AQ, Weterings K, McIntire KN, Hsu YC, Lee PY, Truong MT, Beals TP, Robert B, Goldberg RB (1999) Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex Plant Reprod 11: 297–322 [Google Scholar]
- Schiefthaler U, Balasubramanian S, Sieber P, Chevalier D, Wisman E, Schneitz K (1999) Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proc Natl Acad Sci USA 96: 11664–11669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RJ, Spielman M, Dickinson HG (2004) Stamen structure and function. Plant Cell (Suppl) 16: S46–S60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZR, Long JA (2010) Control of Arabidopsis apical-basal embryo polarity by antagonistic transcription factors. Nature 464: 423–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toriba T, Suzaki T, Yamaguchi T, Ohmori Y, Tsukaya H, Hirano HY (2010) Distinct regulation of adaxial-abaxial polarity in anther patterning in rice. Plant Cell 22: 1452–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams L, Grigg SP, Xie M, Christensen S, Fletcher JC (2005) Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD-ZIP target genes. Development 132: 3657–3668 [DOI] [PubMed] [Google Scholar]
- Wu F, Yu L, Cao W, Mao Y, Liu Z, He Y (2007) The N-terminal double-stranded RNA binding domains of Arabidopsis HYPONASTIC LEAVES1 are sufficient for pre-microRNA processing. Plant Cell 19: 914–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WC, Ye D, Xu J, Sundaresan V (1999) The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev 13: 2108–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Ren W, Zhao Q, Zhang P, Wu F, He Y (2014) Homodimerization of HYL1 ensures the correct selection of cleavage sites in primary miRNA. Nucleic Acids Res 42: 12224–12236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Sundaresan V (2015) Spore formation in plants: Sporocyteless and more. Cell Res 25: 7–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang TQ, Lian H, Zhou CM, Xu L, Jiao Y, Wang JW (2017) A two-step model for de novo activation of WUSCHEL during plant shoot regeneration. Plant Cell 29: 1073–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D. (2009) Control of anther cell differentiation: A teamwork of receptor-like kinases. Sex Plant Reprod 22: 221–228 [DOI] [PubMed] [Google Scholar]
- Zhong R, Ye ZH (1999) IFL1, a gene regulating interfascicular fiber differentiation in Arabidopsis, encodes a homeodomain-leucine zipper protein. Plant Cell 11: 2139–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye ZH (2004) Amphivasal vascular bundle 1, a gain-of-function mutation of the IFL1/REV gene, is associated with alterations in the polarity of leaves, stems and carpels. Plant Cell Physiol 45: 369–385 [DOI] [PubMed] [Google Scholar]