Growth defects arising from lignin modification are associated with the extent, but not the nature, of ectopic expression of defense pathways.
Abstract
Lignin provides essential mechanical support for plant cell walls but decreases the digestibility of forage crops and increases the recalcitrance of biofuel crops. Attempts to modify lignin content and/or composition by genetic modification often result in negative growth effects. Although several studies have attempted to address the basis for such effects in individual transgenic lines, no common mechanism linking lignin modification with perturbations in plant growth and development has yet been identified. To address whether a common mechanism exists, we have analyzed transposon insertion mutants resulting in independent loss of function of five enzymes of the monolignol pathway, as well as one double mutant, in the model legume Medicago truncatula. These plants exhibit growth phenotypes from essentially wild type to severely retarded. Extensive phenotypic, transcriptomic, and metabolomics analyses, including structural characterization of differentially expressed compounds, revealed diverse phenotypic consequences of lignin pathway perturbation that were perceived early in plant development but were not predicted by lignin content or composition alone. Notable phenotypes among the mutants with severe growth impairment were increased trichome numbers, accumulation of a variety of triterpene saponins, and extensive but differential ectopic expression of defense response genes. No currently proposed model explains the observed phenotypes across all lines. We propose that reallocation of resources into defense pathways is linked to the severity of the final growth phenotype in monolignol pathway mutants of M. truncatula, although it remains unclear whether this is a cause or an effect of the growth impairment.
Lignin is a complex aromatic polymer deposited in plant cell walls to provide mechanical support and facilitate the transport of water and solutes through the vascular system (Boerjan et al., 2003). Lignin waterproofs plant cells by providing a hydrophobic environment by chemical bonding with cellulose and hemicellulose (Reina et al., 2001; Boerjan et al., 2003). This lignin-polysaccharide matrix underlies the recalcitrance that limits chemical, enzymatic, and microbial digestion of plant cell walls, thereby hindering the conversion of lignocellulosic biomass to liquid biofuels (Chen and Dixon, 2007; Fu et al., 2011; Li et al., 2016).
In addition to providing a mechanical function, lignification plays an important role in passive and active defense in plants (Bhuiyan et al., 2009; Moura et al., 2010; Xu et al., 2011; Zhao and Dixon, 2014). Lignin functions as a preexisting physical barrier against pathogen invasion and decreases accessibility of cellulose to herbivores. Furthermore, lignification following pathogen challenge and wounding restricts growth of pathogens and decreases herbivore feeding and sometimes herbivore fertility (Kawasaki et al., 2006; Rinaldi et al., 2007; Moura et al., 2010; Bennett et al., 2015).
Lignin polymers generally consist of p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) units, derived from the three primary monolignols, p-coumaryl, coniferyl, and sinapyl alcohols, respectively (Boerjan et al., 2003). Currently, 11 enzymes are thought to be involved in monolignol biosynthesis from the aromatic amino acid l-Phe in dicot species. These enzymes are l-Phe ammonia-lyase, trans-cinnamate 4-hydroxylase, 4-coumarate:coenzyme A ligase (4CL), hydroxycinnamoyl-coenzyme A shikimate/quinate hydroxycinnamoyl transferase (HCT), p-coumaroyl-shikimate-3′-hydroxylase (C3′H), caffeoyl shikimate esterase (CSE), caffeoyl-coenzyme A 3-O-methyltransferase (CCoAOMT), cinnamoyl-coenzyme A reductase (CCR), ferulate 5-hydroxylase (F5H), caffeic acid/5-hydroxyconiferaldehyde-3-O-methyltransferase (COMT), and cinnamyl alcohol dehydrogenase (CAD; Boerjan et al., 2003; Bonawitz and Chapple, 2010; Vanholme et al., 2013; Supplemental Fig. S1). After their formation in the cytosol with species, tissue, and developmental specificity, the monolignols are transported to the cell wall, are there transformed into radicals by laccases and peroxidases, and incorporated into the lignin polymer via oxidative coupling (Boerjan et al., 2003).
Many studies have reported genetic modification to reduce lignin amount or alter its structure to facilitate paper pulping, improve forage digestibility, or overcome recalcitrance for processing of bioenergy feedstocks (Guo et al., 2001b; Chen and Dixon, 2007; Vanholme et al., 2008; Barros et al., 2019). However, modification of the lignin pathway can result in diverse and unpredictable impacts on plant physiology, especially the development of dwarf plants (Franke et al., 2002; Reddy et al., 2005; Besseau et al., 2007; Do et al., 2007; Zhou et al., 2010; Vanholme et al., 2013; Bonawitz et al., 2014; Xue et al., 2015; Ha et al., 2016). In spite of extensive studies on individual dwarf lignin mutants in several different species, there is no clear agreement as to whether a common mechanism is responsible for these dwarf phenotypes.
One commonly held view is that reduced lignin levels impact growth by affecting solute transport through the formation of abnormal vessel walls (Anterola and Lewis, 2002; Bonawitz and Chapple, 2010), and this is supported by vessel-specific genetic complementation studies in the Arabidopsis (Arabidopsis thaliana) ccr1 mutant (De Meester et al., 2018). A second general hypothesis is that lignin modification reduces growth through the accumulation of one or more toxic or bioactive pathway intermediates. For example, flavonoid accumulation leading to inhibition of auxin transport was proposed as the main cause for impaired development in dwarf HCT-RNA interference (RNAi) Arabidopsis lines (Besseau et al., 2007). However, it was subsequently shown that the growth inhibition observed in both HCT-RNAi and ref8 (c3′h) mutant Arabidopsis plants is independent of flavonoids (Li et al., 2010a). Hyperaccumulation of the defense signal salicylic acid (SA) was proposed to be the reason for dwarfing in the Arabidopsis hct mutant, as blocking SA biosynthesis in this mutant substantially overcame the dwarf phenotype (Gallego-Giraldo et al., 2011a). However, when the ref8 mutant was crossed with the mediator 5a/5b double mutant in Arabidopsis, the homozygous triple mutant showed phenotypic rescue even though the SA level was still very high (Bonawitz et al., 2014). Depletion of SA did not rescue the dwarf phenotype in the ref8 mutant (Kim et al., 2014), and further genetic studies led to the hypothesis that transcriptional regulation by the Mediator complex, rather than accumulation of defense signaling molecules per se, may underlie the dwarf phenotypes in at least some lignin pathway mutants (Dolan et al., 2017).
Previous studies addressing growth phenotypes in plants with modified lignin content and composition have mainly involved single gene targets, and few if any studies present either pathway-wide modifications in a single species or controlled cross-species comparisons. A comparison of multiple monolignol pathway mutants in Arabidopsis at the transcriptome and metabolome levels avoided mutants with severely dwarf phenotypes (Vanholme et al., 2012). We have now carried out a combined transcriptome and metabolome analysis in a range of lignin pathway mutants in the model legume Medicago truncatula, targeting lines with growth phenotypes from normal to highly dwarfed, to address whether there could be a common mechanism underlying the growth defects resulting from lignin modification. Analyses were performed on lignifying stems and on young seedlings prior to the onset of the major lignification program. Our data fail to support models that ascribe altered solute transport or accumulation of specific toxic compounds as general reasons for impaired growth in this species. Rather, based on the observation that the various dwarf mutants exhibit large but different alterations in defense-related responses at both the transcriptome and metabolome levels, we suggest that impaired growth in the M. truncatula lignin mutants results from alterations in resource allocation in these plants.
RESULTS
Phenotypic Characterization and Sampling of Monolignol Pathway Mutants
To study the effects of perturbing lignin biosynthesis at multiple independent steps in the monolignol pathway, we performed reverse genetic screening of Tnt1 retrotransposon insertional mutant seed pools of M. truncatula in the ecotype R108 (wild-type) background (Supplemental Table S1), followed by self-pollination and genotypic selection to obtain homozygous hct, ccr1, cse, comt, and ccoaomt mutants. Two independent alleles were obtained for each target gene (Supplemental Table S1), and all Tnt1 insertions were found in exon regions in every mutant studied, resulting in RNA null mutations (Ha et al., 2016; Supplemental Fig. S2). A homozygous comt ccoaomt double mutant was generated by crossing comt and ccoaomt single mutants. The overall appearance of the mutants is shown in Figure 1, and their detailed growth phenotypes are summarized in Table 1. The cse and ccr1 mutants were dwarf plants (Fig. 1, A and B). In addition, cse mutants produced few seeds, all of reduced size (Ha et al., 2016), whereas ccr1 mutants did not produce any seeds (Table 1). The hct mutant plants exhibited an early seedling-lethal phenotype, with growth cessation at the two- to three-leaf stage after approximately 3 weeks of growth (Fig. 1, C and D). Single mutation in either the COMT or CCOAOMT gene resulted in no phenotypic change and a semidwarf phenotype, respectively (Fig. 1, E and F; Table 1). In contrast, comt ccoaomt double mutant plants showed growth arrest at 3 weeks, similar to the hct mutants (Fig. 1, G and H).
Figure 1.
Phenotypes of M. truncatula monolignol biosynthesis pathway mutants. A, cse mutant plants. B, ccr1 mutant plants. C and D, hct mutant plants. E, comt mutant plants. F, ccoaomt mutant plants. G and H, comt ccoaomt mutant plants. Photographs were taken from 72-d plants in A, B, E, and F and from 24-d plants in C, D, G, and H. D and H are magnified views of the red rectangles in C and G, respectively. cse and ccr1 mutant plants show dwarf phenotypes (A and B), and hct and comt ccoaomt mutants show growth arrest (C, D, G, and H). WT, Wild type. Bars = 10 cm (A, B, E, and F) and 10 mm (D and H).
Table 1. Phenotypes of monolignol biosynthesis pathway mutants in M. truncatula.
Data are averages ± sd for 16 biological replicates of each of two alleles (growth and reproduction) or three biological replicates of each of two alleles (lignin measurements). N/A, Not available due to developmental arrest. Stem tissue was harvested from plants that were beginning to produce their second flower. *, 0.001 < P < 0.01; **, 0.001 > P, Student’s t test.
| Parameter | Subparameter | Wild Type | cse | ccr1 | comt | ccoaomt | comt ccoaomt | hct |
|---|---|---|---|---|---|---|---|---|
| Growth | Plant height (cm) | 112.7 ± 11.2 | 12.2 ± 1.4** | 14.4 ± 1.3** | 109.6 ± 10.7 | 89.3 ± 8.5* | N/A | N/A |
| Leaf size (first leaf; mm) | 6.1 ± 0.4 | 4.2 ± 0.3** | 3.0 ± 0.3** | 6.1 ± 0.2 | 4.8 ± 0.8** | 3.0 ± 0.2** | 2.5 ± 0.2** | |
| Leaf size (cauline leaf; mm) | 17.5 ± 1.4 | 7.9 ± 0.4** | 5.5 ± 0.7** | 17.5 ± 1.3 | 10.9 ± 0.7** | N/A | N/A | |
| Reproduction | Flowering time (d after sowing) | 58.4 ± 1.9 | 75.9 ± 1.9** | 71.0 ± 2.9** | 63.6 ± 2.1** | 66.5 ± 2.5** | No flowering | No flowering |
| Seeds per pod | 7.1 ± 0.5 | 0.8 ± 0.6** | 0 | 7.0 ± 0.4 | 4.6 ± 1.2** | N/A | N/A | |
| Lignin amount | 10d plant (μmol g−1 dry wt) | 7.8 ± 0.1 | 3.9 ± 0.7** | 2.9 ± 0.7** | 3.4 ± 1.3** | 2.9 ± 0.6** | 1.9 ± 0.8** | 2.1 ± 0.0** |
| Stem tissue (μmol g−1 dry wt) | 161.9 ± 2.5 | 11.7 ± 1.0** | 6.6 ± 0.9** | 84.0 ± 7.3 | 73.3 ± 14.1** | N/A | N/A | |
| Lignin composition | 10d plant (H:G:S) | 6:80:14 | 39:44:17 | 9:78:13 | 8:89:3 | 13:70:16 | 20:76:4 | 65:30:4 |
| Stem tissue (H:G:S) | 0:87:13 | 55:33:11 | 4:87:8 | 1:99:0 | 2:81:17 | N/A | N/A | |
| Overall phenotype | Normal | Dwarf (strong) | Dwarf (strong) | Normal | Semidwarf | Dwarf (very strong) | Dwarf (very strong) | |
| Growth arrest | No | No | No | No | No | Yes | Yes | |
Scanning electron microscopy (SEM) revealed that the number of trichomes developing on the leaf surface of the dwarf mutants (hct, ccr1, and cse) was significantly greater per unit of leaf surface area than the number on the wild-type control (Fig. 2, A, B, I, and J; Supplemental Table S2). Whereas the comt and ccoaomt single mutants exhibited no change in trichome development, comt ccoaomt double mutant plants showed increased numbers of trichomes (Fig. 2, K–M). We also observed a large reduction in the size of epidermal cells in the cse, ccr1, hct, and comt ccoaomt mutants (Fig. 2, E, F, N, O, and R). The ccoaomt mutant showed a smaller reduction in epidermal cell size, whereas the comt mutant exhibited wild-type epidermal cell morphology (Fig. 2, P and Q).
Figure 2.
SEM of leaf surfaces of M. truncatula mutants. SEM images of adaxial leaf blades are from control (Ctrl; A and E), cse mutant (B and F), β-as mutant (C and G), cse β-as double mutant (D and H), ccr1 mutant (I and N), hct mutant (J and O), comt mutant (K and P), ccoaomt mutant (L and Q), and comt ccoaomt double mutant (M and R) plants. Photographs were taken from the second developing leaves of 15-d plants. Analyses were repeated on leaves from 10 individual plants of each genotype (five from each allele), and representative results are shown. Bars = 200 μm (A–D and I–M) and 50 μm (E–H and N–R).
To compare system-wide molecular changes in these mutants, we collected samples at two different developmental stages, 10-d-old (10d) seedling plants, which had not yet developed stem tissue and showed very little lignification, and mature stem tissue, which was collected from plants harvested at the stage between development of the first and second flowers, to compensate for developmental differences in the various lines. The overall appearance of the plants at 10 d is shown in Supplemental Figure S3, and the reduced growth phenotypes were most clear in the ccr1, hct, and comt ccoaomt mutants. To determine whether monolignol biosynthesis was active at or before 10 d postgermination, we measured the levels of lignin pathway gene transcripts (CCR1 and CCR2, CSE, COMT, CCoAOMT, CAD, and F5H) and lignin content and composition in wild-type seedlings at 1, 2, and 3 d postgermination and compared them with the values in mature stem tissue. CCR2 was highly expressed in very young seedlings, and the other transcript levels were between 10% and 45% of those recorded in stem tissue (Supplemental Fig. S4). Therefore, monolignol biosynthesis starts at early developmental stages in M. truncatula.
Cross-sectioned petiole tissue from 10d plants was observed with UV light, under which lignin and wall-bound phenolic compounds exhibit blue autofluorescence. Young petioles from cse, ccr1, comt, and ccoaomt mutant plants did not exhibit noticeable differences from the wild-type control, except for a small decrease in petiole size (Supplemental Fig. S5, A–C, E, and F). However, petiole tissue from hct single and comt ccoaomt double mutant plants possessed somewhat reduced fluorescence and lack of organization in vascular tissues (Supplemental Fig. S5, D and G).
For subsequent analysis of 10d seedlings, all aboveground tissue was collected, whereas the bottom first through sixth internodes were selected for analysis of mature stem tissue. Because hct single mutant and comt ccoaomt double mutant plants do not develop stems, only 10d seedlings were analyzed for these mutants. The growth of stems in cse and ccr1 mutants was delayed compared with that of wild-type plants (Table 1). To compensate for this developmental shift, we sampled stems between development of the first and second flowers in each line.
Lignin Content and Composition in Monolignol Pathway Mutants
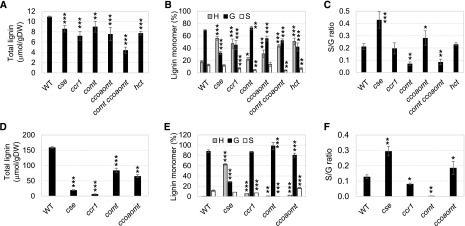
Lignin content and composition in 10d seedlings and stem tissue was determined by thioacidolysis (Lapierre et al., 1985). Analysis of monomeric thioacidolysis products by gas chromatography-mass spectrometry (GC-MS) showed that 10d wild-type plants had very low amounts of total lignin (estimated as H + G + S, approximately 15-fold less than that in mature stem tissue) and that levels were reduced compared with that in the wild-type control in all the mutants analyzed, with particularly low levels in the comt ccoaomt double mutant (Fig. 3A). On the basis of UV autofluorescence and staining with phloroglucinol, the latter of which is considered to reveal aldehydes in the polymer (Pomar et al., 2002), lignin in the leaves of 10d plants was mainly found in xylem tissue (Supplemental Fig. S6). The percentage of H-derived monomers was increased in all mutants at 10 d postgermination, most highly in the cse, ccr1, comt ccoaomt, and hct mutants (Fig. 3B), and S/G ratio was elevated in the cse mutant and strongly reduced in mutants with COMT loss of function as the result of a large reduction in S units.
Figure 3.
Lignin content and composition of M. truncatula monolignol pathway mutants. Cell wall material was prepared from aboveground whole tissue from 10d seedlings (A–C) and mature stem tissue (D–F) and analyzed by thioacidolysis. A and D, Total lignin thioacidolysis yields (H + G + S units). B and E, Lignin monomer compositions. C and F, Lignin S/G ratios. Data are means ± sd (n = 6) of three replicates of each of the two independent alleles. WT, Wild type. *, 0.01 < P < 0.05; **, 0.001 < P < 0.01; and ***, 0.001 > P, Student’s t test.
In stems, H monomers were increased in all mutants and appeared at highest percentage levels in the cse mutant (Fig. 3E). In contrast, the percentage of G monomers was greatly decreased both in 10d and mature stems of the cse mutant, whereas the percentage of S monomers was only modestly changed (Fig. 3, B and E), resulting in a significantly increased S/G ratio in the cse mutant at both early seedling and mature stages (Fig. 3, C and F). The comt single mutant exhibited a large decrease in the percentage of S monomers, both in mature stem tissue, as previously described (Zhou et al., 2010), and also in 10d seedlings. In contrast, the ccoaomt mutant exhibited increased S/G ratio in both 10d and mature stem tissue (Fig. 3, C and F). In summary, the results with mature stems reflect previous observations of lignin composition in monolignol pathway mutants. In 10d seedlings, all mutants showed larger changes in lignin composition than in lignin content.
Hormone Levels in Monolignol Pathway Mutants
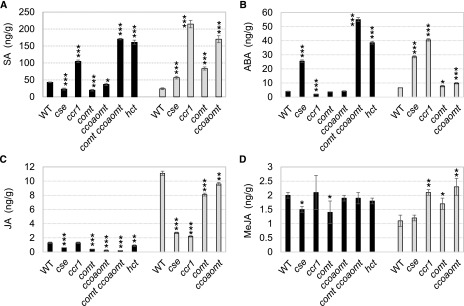
The more severe phenotypes among the monolignol pathway mutants were reminiscent of plant hormone signaling mutants (Heinrich et al., 2013; Westfall et al., 2013; Ahmad et al., 2016; Wang et al., 2018). We therefore determined the levels of SA, abscisic acid (ABA), jasmonic acid (JA) and methyl jasmonate (MeJA), the major hormones associated with plant stress responses (Verma et al., 2016), by a targeted ultra-high-pressure liquid chromatography-tandem mass spectrometry (UHPLC-MS/MS) method (Watson et al., 2015; Fig. 4).
Figure 4.
Phytohormone levels in monolignol pathway mutants. Phytohormone levels are shown in lignin biosynthesis pathway mutants as determined by UHPLC. A, SA. B, ABA. C, JA. D, MeJA. Fresh samples were prepared from aboveground whole tissue from 10d seedlings (black bars) and mature stem tissue (gray bars). Data are means ± sd (n = 3 biological replicates) of results from one allele for each target gene. WT, Wild type. *, 0.01 < P < 0.05; **, 0.001 < P < 0.01; and ***, 0.001 > P, Student’s t test.
In 10d seedlings, SA levels in the ccr1, hct, and comt ccoaomt mutants were twofold to fourfold higher than the wild-type level but were lower in the cse and comt mutants than in the wild type (Fig. 4A). In stem tissue, SA levels were higher in all mutants examined than in the wild type, particularly in ccr1 and ccoaomt (Fig. 4A). ABA levels in 10d seedlings of the cse, hct, and comt ccoaomt mutants were sixfold to 15-fold higher than in the wild type, whereas there was a small decrease in the ccr1 mutant compared with that in the wild type (Fig. 4B). In stem tissue, ABA levels were significantly higher than in the wild type in every mutant examined, especially in cse and ccr1 (Fig. 4B). JA and MeJA were present at low abundance in both 10d plants and stem tissue. JA accumulation was significantly decreased in both 10d seedlings and stem tissue of all mutants except ccr1 (Fig. 4C). MeJA levels were also slightly decreased in 10d cse and comt plants, with no change observed in the other mutants (Fig. 4D). MeJA levels slightly increased in stem tissue of the ccr1, comt, and ccoaomt, but not cse, mutants (Fig. 4D).
Nontargeted Metabolomics Analyses of Monolignol Pathway Mutants
Metabolomics analyses of the monolignol pathway mutants using GC-MS revealed a total of 333 compounds from 10d seedlings and 235 compounds from stem tissue, which showed significant changes in mutant compared with wild-type plant tissues (Table 2; Supplemental Data Set S1). Among the hydroxycinnamic acids in, or derived from, the monolignol pathway, levels of p-coumaric acid were significantly elevated above wild-type levels at 10 d in the comt ccoaomt double mutant. Both coumaric acid and its hexose conjugate were elevated in the hct mutant and in stems of the cse and ccoaomt mutants (Table 3; Supplemental Data Set S1). In stems, the level of ferulic acid was significantly elevated in cse, ccr1, and ccoaomt mutants. Levels of caffeic acid and its glucose conjugate were significantly elevated in every mutant examined at 10 d, whereas, in stems, they were elevated in the cse, ccr1, and ccoaomt mutants, and the glucose conjugate was strongly reduced in the comt mutant. Overall, the mutant plants with the most impaired growth phenotypes accumulated the highest hydroxycinnamic acid levels (sum of cinnamic, coumaric, caffeic, and ferulic acids) at both 10 d and in stems.
Table 2. Numbers of transcripts and metabolites that showed significant changes in M. truncatula monolignol pathway mutants compared with the wild type.
Microarray analysis was performed on 10d cse, comt, ccoaomt, and comt ccoaomt mutant plants, whereas RNA-seq analysis was performed on 10d hct and ccr1 mutant plants and on all whole stem samples. n = 6 (three replicates of each allele). ND, Not determined, as hct and comt ccoaomt mutant plants do not develop stem tissues; –, GC-MS analysis was not carried out on 10d ccr1 mutant plants.
| Genotype | 10d Seedlings | Mature Stem Tissue | ||||
|---|---|---|---|---|---|---|
| No. of Transcripts | No. of Compounds by GC-MS | No. of Compounds by UPLC-MS | No. of Transcripts | No. of Compounds by GC-MS | No. of Compounds by UPLC-MS | |
| hct | 6,750 | 89 | 33 | ND | ND | ND |
| cse | 2,837 | 52 | 20 | 9,659 | 83 | 119 |
| ccr1 | 1,272 | – | 14 | 14,286 | 70 | 111 |
| comt ccoaomt | 3,057 | 97 | 15 | ND | ND | ND |
| comt | 1,377 | 46 | 21 | 12,820 | 35 | 10 |
| ccoaomt | 777 | 49 | 22 | 10,753 | 47 | 1 |
Table 3. Comparison of metabolite concentrations in 10d seedlings and stems of M. truncatula determined by GC-MS analysis.
The fold change (log2 scale) of mutant/wild type (each with four biological replicates per plant genotype) is shown. The full list of compounds determined by GC-MS is given in Supplemental Data Set S1. Asterisks designate that the fold change is statistically significant (P < 0.05) based on Student’s t test. RT, retention time; m/z, mass to charge ratio of major ion.
| 10d | Stem | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| cse | hct | comt ccoaomt | comt | ccoaomt | Metabolite (RT: Key m/z) | cse | ccr1 | comt | ccoaomt |
| Lignin pathway | |||||||||
| 5.3* | −9.0* | 1.0* | −0.8 | 0.2 | 3-O-Caffeoylshikimate | 9.3* | 4.0* | 0.0 | 2.8* |
| 2.1* | 1.8* | 3.5* | 1.4* | 2.5* | Caffeic acid | 1.1* | 0.8 | −0.5 | 2.8* |
| 2.1* | −6.6 | 6.9* | −6.6 | 5.2* | Caffeic acid 3-O-β-d-glucoside | 2.4* | 2.7* | −6.3* | 6.8* |
| 1.1* | −0.5 | 0.0 | 0.0 | 0.1 | Cis-ferulic acid 4-O-glucoside | 6.3* | 8.9* | 0.0 | 4.7* |
| −0.1 | 0.2 | 0.7 | 0.1 | 0.3 | Coniferyl alcohol | −1.7* | −1.4* | 0.0 | −0.4 |
| 1.2* | 0.5 | 1.2 | 0.5 | 0.7 | Ferulic acid | 2.0* | 4.1* | −0.6 | 1.4 |
| 0.5 | 2.4* | 1.4* | 0.2 | 0.5 | p-Coumaric acid | 1.5* | 0.7 | −0.1 | 1.9* |
| 0.5 | 2.6* | 0.5 | 0.0 | 0.3 | p-Coumaric acid 4-O-glucoside | 4.4* | 4.9* | −5.0* | 3.0* |
| 0.6 | 0.8 | 0.7 | 0.1 | 0.6 | t-Cinnamic acid | 0.7 | −0.1 | 0.8 | −0.6 |
| Lignan | |||||||||
| 3.4* | 3.1* | 0.9* | 0.1 | 0.2 | 15.94: 311 Hydroxyphenyl lignan | 11.2* | 6.9* | 0.0 | 3.4 |
| 4.3* | 5.7* | 2.8* | −6.6 | −6.6 | 16.11: 411 Hydroxyphenyl lignan | 8.1* | 0.0 | 0.0 | 0.0 |
| 2.5* | 3.7* | 1.5* | −6.6 | −6.6 | p-Coumarylresinol | 10.8* | 5.7* | 0.0 | 4.2 |
| SA and derivative | |||||||||
| −0.4 | 1.0* | 1.9* | −0.9* | 1.6* | SA | 3.5* | 5.8* | 3.5* | 2.2 |
| 0.1 | 0.7 | 1.5* | −0.5 | 1.6* | SA 2-O-glucoside | 1.9* | 4.7* | 1.7* | 0.9 |
| 0.1 | 1.4* | 1.1* | −0.5 | 0.2 | 4-Hydroxybenzoic acid | 3.6* | 2.3* | 0.3* | 1.2 |
| Amino acid | |||||||||
| 0.3 | 1.3* | 1.4* | 0.0 | −0.2 | Ala | 1.1* | 1.5* | 1.2* | 1.3 |
| 0.3 | 1.1* | 1.2* | 0.2 | 0.5 | Asn | 1.2* | 1.0* | 1.3* | 1.3 |
| 0.6 | 3.3* | 2.5* | 0.2 | 0.4 | Phe | −1.1* | −0.3 | 0.6 | −1.3 |
| −0.6 | 6.9* | 4.8* | −0.4 | 0.0 | Pro | 4.7* | 3.5* | 0.2 | −0.3 |
| 1.1* | 3.3* | 2.8* | 0.4 | 0.8 | Tyr | 1.0* | 1.2* | 0.1 | 0.3 |
| Sugar | |||||||||
| 0.4 | 0.0 | 0.5 | 0.0 | 0.2 | Fru | 1.1* | −0.7 | −1.4* | −0.1 |
| 0.3 | 0.2 | 0.3 | 0.0 | 0.3 | Glc | 1.1* | 0.8 | 0.0 | 0.4 |
| 0.4 | 0.3 | 0.3 | 0.0 | 0.3 | Suc | 0.3 | 0.0 | 0.3 | 0.2 |
| Fatty acid | |||||||||
| 0.2 | 1.1* | 0.4 | 0.0 | 0.1 | Hexacosanoic acid | 1.3* | 2.5* | 0.4 | 0.8 |
| −0.3 | −0.1 | −0.2 | −0.4 | −0.4 | Linoleic acid | 0.6 | −0.1 | 0.3 | 0.3 |
| −0.1 | 0.1 | −0.2 | −0.3 | −0.2 | Palmitic acid | 0.5 | 0.1 | 0.3 | 0.1 |
Levels of dimeric H lignans, derived from the oxidative coupling of H monolignol units, were highly elevated at 10 d in the cse and hct mutants and less highly in the comt ccoaomt double mutant (Table 3; Supplemental Data Set S1). The changes in lignans are consistent with the parallel accumulation of H lignin in these mutants (Fig. 3). No change in H lignan levels was observed at 10 d in the comt and ccoaomt single mutants. In stems, the cse mutant exhibited strongly elevated levels of H lignans, with smaller increases in the ccr1 and ccoaomt mutants and again no change in the comt mutant. Levels of two dimeric G lignans decreased significantly in every monolignol pathway mutant examined at 10 d, and all four G lignans identified were also decreased in stems of the cse, ccr1, and ccoaomt mutants but exhibited no significant change in the comt mutant.
GC-MS analyses revealed differences in levels of proteinogenic and nonproteinogenic amino acids in the various mutants (Table 3; Supplemental Data Set S1). The number of amino acids with significantly changed levels was greater in 10d seedlings than in stems (Supplemental Data Set S1). Levels of all five amino acids that showed significant changes in at least one line were higher in hct and comt ccoaomt mutant seedlings at 10 d than in the wild type, but they were not elevated in the cse (except for Tyr), comt, or ccoaomt mutants. Ala and Asn were the only amino acids significantly higher in all mutants in stem tissues. Levels of Pro were highly elevated at 10 d in the hct and comt ccoaomt mutants but not in the cse mutant, whereas mature stems of the cse mutant showed highly elevated Pro levels (Table 3; Supplemental Data Set S1). There were nonsignificant decreases in Pro levels in the comt and ccoaomt mutants in both 10d seedlings and stems.
Monolignol pathway mutants exhibited little or no changes in levels of other primary metabolites, including hexose sugars, fatty acids, cyclitols, organic acids, nucleotide precursors, vitamins, glycerol and its derivatives, and amines (Supplemental Data Set S1).
Metabolite Profiling by Ultra-High-Performance Liquid Chromatography-Mass Spectrometry
To address potential changes in more complex polar metabolites in the mutants, we performed nontargeted ultra-high-performance liquid chromatography-mass spectrometry (UHPLC-MS) analyses. Based on these analyses, levels of 125 compounds from 10d seedlings and 241 compounds from stems changed significantly in mutants compared with those in wild-type plants (Table 2). In stems, most of the identified compounds were triterpene saponins, especially oleanane-type saponins derived from β-amyrin (i.e. soyasapogenol, medicagenic acid, and zhanic acid; Table 4; Supplemental Fig. S7, A–C; Supplemental Data Set S2). Levels of saponins were highly elevated in the stem tissue of cse and ccr1 mutants but not in the stems of comt or ccoaomt mutants (Table 4; Supplemental Data Set S2). Zhanic acid-derived saponins exhibited the highest fold change (∼7-fold) in stems of the ccr1 mutant compared with that in the wild type, whereas soyasapogenol-derived saponins exhibited the highest fold change (∼6-fold) in stems of the cse mutant. In contrast, only a few saponins accumulated differentially in 10d seedlings, most with decreased levels in the mutants (Supplemental Data Set S3).
Table 4. Summary of nontargeted UHPLC-MS/MS metabolite profiling of M. truncatula monolignol pathway mutants.
| Mutant | Total Compoundsa | No. of Compounds | No. of Saponins | ||
|---|---|---|---|---|---|
| Increaseda | Decreaseda | Increasedb | Decreasedb | ||
| 10d seedlings | |||||
| hct | 33 | 18 (54.5%) | 15 (45.5%) | 0/13 (0%) | 2/10 (20%) |
| cse | 20 | 9 (45%) | 11 (55%) | 0/4 (0%) | 0/2 (0%) |
| ccr1 | 14 | 9 (64.3%) | 5 (35.7%) | 1/4 (25%) | 0/2 (0%) |
| ccoaomt | 22 | 8 (36.4%) | 14 (63.6%) | 0/2 (0%) | 1/7 (14.3%) |
| comt | 21 | 2 (9.5%) | 19 (90.5%) | 0/0 (0%) | 2/19 (10.5%) |
| comt ccoaomt | 15 | 6 (40%) | 9 (60%) | 0/4 (0%) | 4/7 (57.1%) |
| Stems | |||||
| cse | 119 | 89 (74.8%) | 30 (25.2%) | 18/29 (62.0%) | 0/6 (0%) |
| ccr1 | 111 | 97 (87.4%) | 14 (12.6%) | 17/18 (94.4%) | 1/3 (33.3%) |
| comt | 10 | 8 (80%) | 2 (20%) | 0/0 (0%) | 0/0 (0%) |
| ccoaomt | 1 | 1 (100%) | 0 | 0/0 (0%) | 0/0 (0%) |
Number of compounds that have significantly altered level (±2-fold change and P < 0.05, Student’s t test) in mutant plants compared with the wild type. Four independent plant tissue samples of each of the two independent alleles were used for UHPLC-MS/MS analysis. Numbers include saponins.
Number of saponins/total number of identified compounds.
The major saponins identified were glycosides of medicagenic acid, soyasapogenol B, and zhanic acid, with side chains of up to five different sugars (Supplemental Fig. S7, A–C; Supplemental Data Sets S2 and S3; Supplemental Table S3). Many of these identifications were based upon previous analyses of standards and incorporation of standard spectra within custom libraries (Huhman and Sumner, 2002; Huhman et al., 2005; Farag et al., 2007; Lei et al., 2015). Many of the identifications have also been verified by other groups (Pollier et al., 2011). We also used a custom computational tool to predict the identity of other saponins and validated that our prediction tool had a high degree of accuracy based upon empirical structural determinations obtained by coupled liquid chromatography-mass spectrometry and automated solid-phase extraction and concentration followed by NMR spectroscopy (i.e. UHPLC-MS-SPE-NMR; see “Materials and Methods”; Supplemental Fig. S7, A–C; Qiu et al., 2016). The differences between the saponin compositions of the different mutants resided primarily in the glycosylation pattern, although a soyaspogenol E-derived saponin was found only in the cse mutant.
Two glycosylated isoflavonoid derivatives were identified by UHPLC-MS-SPE-NMR as differentially accumulated only in the cse mutant; these were the related compounds 6′-malonyl-3-Glu-medicarpin and 6′-malonyl-ononin, and levels of both were decreased (Supplemental Data Set S2). In contrast, 7-O-(2′-feruloylglucuronyl)-(1-2)-glucuronyl apigenin, an acylated flavone glycoside, accumulated in stems of both cse and ccr1 mutants (Supplemental Fig. S7D; Supplemental Data Set S2). Finally, levels of an unusual fatty acyl substituted diglycoside, oct-1-en-3-yl arabinose-(1,6)-Glc, decreased in the cse mutant (Supplemental Fig. S7E; Supplemental Data Set S2). We believe this to be the first report of this compound; full details of the structural elucidation of this, and the characterized saponins referenced above, will be provided elsewhere.
Overall Transcriptomic Changes in M. truncatula Monolignol Pathway Mutants
To compare the transcriptomic profiles of the different monolignol pathway mutants, we performed microarray analyses on 10d cse, comt, and ccoaomt single mutants and the comt ccoaomt double mutant, and RNA sequencing (RNA-seq) analyses on 10d ccr1 and hct mutants and stems from all available lines, alongside the corresponding wild-type controls. The Affymetrix Medicago GeneChip genome array was used for microarray analyses. It contains 61,103 probe sets, most of which (50,902) are from M. truncatula gene sequences. For RNA-seq analyses, we generated approximately 1.23 billion high-quality 75-bp reads, which were aligned with the M. truncatula genome sequence (Mt4.0 version). Because copies of the tobacco (Nicotiana tabacum) Tnt1 retrotransposon are inserted into more than one site in the M. truncatula genome (Tadege et al., 2008), we analyzed the transcriptomes of two independent alleles for each monolignol pathway gene.
Transcriptomic analyses of 10d plants showed that 777 to 6,737 genes had significantly different expression patterns (P < 0.05 and fold change of ±1.5) in monolignol pathway mutants compared with wild-type plants (Table 2). In stems, 9,659 to 14,286 genes were differentially expressed in the mutants compared with the wild type (Table 2). Overall, there was a positive correlation between the strength of the mutant phenotype (dwarfing) and the total number of transcripts with significant expression changes in mutants compared with the wild type. However, Venn diagram analyses (Supplemental Fig. S8) revealed that there were, for some mutants, more differentially expressed genes (DEGs) common to a particular mutant than shared among mutants in 10d seedlings (Supplemental Fig. S8, A–D). Thus, in qualitative terms, the transcriptional reprogramming was more characteristic of genotype than phenotype at the early growth stage. In mature stems, the proportion of shared DEGs between mutants was greater (Supplemental Fig. S8, E and F).
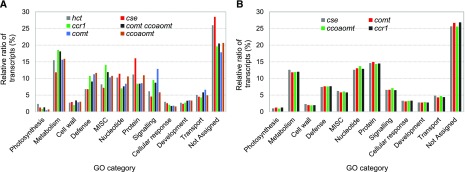
Gene Ontology (GO) enrichment analysis of DEGs and comparison of the number of genes assigned into each category were used to evaluate convergence in the distribution of gene categories in the mutants (Fig. 5; Supplemental Tables S4 and S5; Supplemental Data Set S4). The distribution pattern of each functional category was generally uniform across all mutants studied in both 10d seedlings and stems. There were, however, large differences in the transcriptomes of individual mutants at the two developmental stages. For example, in the cse mutant, approximately 58% of DEGs had different expression patterns (up versus down) between stems and 10d seedlings (Supplemental Fig. S9A). These values were 56.3% and 48.1%, respectively, for the comt and ccoaomt mutants (Supplemental Fig. S9, B and C). Among the expressed defense response-related transcripts, more than 40% had differential expression patterns in the cse, comt, and ccoaomt mutants at the two developmental stages (Supplemental Fig. S9, D–F). Furthermore, the absolute expression level of DEGs was overall lower in 10d seedlings compared with that in stems (Supplemental Fig. S9). The extent of differential expression of transcripts between 10d plants and stem tissue was also visualized by MapMan analysis (Thimm et al., 2004). The magnitude of reprogramming of defense response-related genes was much greater in stem tissue than in 10d plants (Supplemental Figs. S10 and S11). Furthermore, mature stem tissue showed more transcriptional changes associated with cell wall metabolism, secondary metabolism, and hormone metabolism than did 10d seedlings (Supplemental Figs. S10 and S11).
Figure 5.
GO enrichment analysis of DEGs in M. truncatula monolignol pathway mutants. The DEGs were identified with the criteria ±1.5-fold change and P < 0.05. A, GO enrichment analysis in 10d seedlings. Transcriptomes of cse, comt, ccoaomt, and comt ccoaomt mutants were determined by DNA microarray analysis, whereas RNA-seq analysis was performed on the hct and ccr1 mutants. B, GO enrichment analysis in stems. Transcriptomes were determined by RNA-seq analysis.
Next, we compared the transcriptomes of the comt and ccoaomt single mutants with that of the comt ccoaomt double mutant at 10 d. The difference between the double and single mutants was most noticeable in terms of the large number of genes that were down-regulated in the double mutant (Supplemental Fig. S12). In the comt mutant, approximately 78% and 85% of all DEGs and defense response-related genes showed a different expression pattern (up versus down) compared with that in the comt ccoaomt double mutant, respectively (Supplemental Fig. S12, A and C). This is consistent with the strong differences in visible, molecular, and biochemical phenotypes of the single and double mutants. In contrast, the ccoaomt single and comt ccoaomt double mutants displayed closer correlations in their expression tendency of DEGs. Approximately 67% of all DEGs between the different genotypes, and 82% of defense response-related genes, had the same expression tendency in the ccoaomt mutant compared with the comt ccoaomt double mutant (Supplemental Fig. S12, B and D). This was unexpected because of the large difference in the growth phenotype between ccoaomt single and comt ccoaomt double mutant plants. However, the actual transcript levels of commonly expressed genes were much more strongly altered in the comt ccoaomt double mutant compared with either of the single mutant plants (Supplemental Fig. S12).
Specific Transcriptomic Changes in M. truncatula Monolignol Pathway Mutants
In considering transcriptomic changes associated with the monolignol pathway itself, all mutant lines analyzed were homozygous nulls for the one particular lignin biosynthetic gene targeted. Interestingly, in 10d seedlings, levels of transcripts encoding several of the 17 COMT-LIKE and four CCOAOMT-LIKE genes in the M. truncatula genome were highly increased in the hct, ccr1, and comt ccoaomt mutants and less strongly increased in the cse, comt, and ccoaomt mutants (Supplemental Data Set S5). In stem tissue, fewer COMT-LIKE and CCOAOMT-LIKE genes were highly expressed in the mutants (Supplemental Data Set S6), although one COMT-LIKE gene (Medtr4g038440) was highly expressed in the stem, but not in 10d seedlings, of the comt mutant (Supplemental Data Sets S5 and S6). Because loss of function of the single Medtr1g036490 COMT gene results in a nearly complete loss of S lignin, Medtr4g038440 is unlikely to be a true COMT and encode a protein with 5-hydroxyconiferaldehyde O-methyltransferase activity.
In addition to altered expression of COMT and CCOAOMT homologs, there was extensive reprogramming of gene expression associated with monolignol oxidation (laccase and peroxidase genes), other cell wall-related pathways, transport processes, transcriptional control, and cytochrome P450s (Supplemental Data Sets S5 and S6).
Transcriptome analyses revealed striking changes in the expression of genes related to SA, JA, ethylene, and ABA signaling, all known to function in general responses against various stresses (Verma et al., 2016). In 10d seedlings, the strongest changes in expression were seen in hct, ccr1, and comt ccoaomt mutants (all dwarf), with much smaller changes observed in cse, comt, and ccoaomt lines (Supplemental Data Set S5). In contrast, the expression of these hormone signaling genes in stems was, with a few exceptions (e.g. higher expression of jasmonate zim-domain proteins in the comt and ccoaomt mutants), similar in the cse, ccr1, comt, and ccoaomt mutants (Supplemental Data Set S6).
Genes encoding a number of proteins associated with biotic and abiotic stress responses were induced strongly in some of the mutant lines. These included PATHOGENESIS-RELATED (PR) proteins, defensins, and ABA-inducible LATE EMBRYOGENESIS ABUNDANT (LEA) proteins including dehydrins. At 10 d, the largest increases in PR gene transcripts were observed in the hct, ccr1, and comt ccoaomt mutants (Supplemental Data Set S5). PR genes were more highly induced in stems than in 10d seedlings of the cse, ccr1, comt, and ccoaomt mutants (Supplemental Data Set S6). In 10d seedlings, the ccr1, comt ccoaomt, and especially hct mutants, with the highest levels of SA (Fig. 4), exhibited the highest levels of defensin gene transcripts, whereas no induction of defensin genes was observed in the comt, ccoaomt, or cse mutants with decreased accumulation of SA (Fig. 4; Supplemental Data Set S5). In stems, defensin genes were induced in the cse, ccr1, and ccoaomt mutants but not in the comt mutant (Supplemental Data Set S5). In this case, the correlation with SA levels (Fig. 4) was less clear.
In 10d seedlings, the transcript levels of many LEA genes, including those encoding the group 2 LEA protein DEHYDRIN, were most highly elevated in the hct mutant, moderately elevated in the ccr1 and comt ccoaomt mutants, but not altered in the cse, comt, and ccoaomt mutants (Supplemental Data Set S5). In stems, enhanced expression of several LEA genes was observed in all monolignol pathway mutants examined, but only one (Medtr4g011270) was strongly induced across all four mutant lines (Supplemental Data Set S6). At 10 d, accumulation of transcripts encoding known ABA response-related genes was highest in the dwarf mutants (Supplemental Data Sets S5 and S6). However, in stems, the expression of abiotic stress response-related genes was similar in the cse, ccr1, comt, and ccoaomt mutants (Supplemental Data Set S6).
Auxin, cytokinin (CK), and GA also function in coordinating responses to biotic and abiotic stress (Robert-Seilaniantz et al., 2011), and the expression of genes involved in auxin, CK, and GA signaling and/or biosynthesis was massively changed in the lignin pathway mutants. As with the other hormone pathways, many of these genes showed highest expression at 10 d in hct, ccr1, and comt ccoaomt mutants, with smaller changes at 10 d in cse, comt, and ccoaomt mutants (Supplemental Data Set S5). In stems, the expression of genes associated with auxin, CK, and GA response/biosynthesis was generally similar in the cse, ccr1, comt, and ccoaomt mutants, although the SAUR-like auxin-responsive family proteins were notably not induced in the comt mutant (Supplemental Data Set S6).
Comparisons of Transcriptome and Metabolome across Monolignol Pathway Mutants
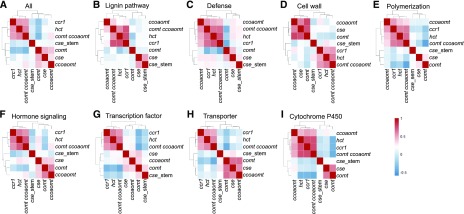
Pearson correlation coefficient (PCC) analysis of the above omics data between the different mutants was performed to obtain an integrated picture of the plants' responses to altered monolignol biosynthesis. When all DEGs from 10d plants were compared, transcripts from the dwarf ccr1, hct, and comt ccoaomt mutants constituted one group, whereas the comt and ccoaomt mutants with an unaltered or weakly impaired growth phenotype constituted another group (Fig. 6A). Surprisingly, transcripts from 10d cse mutant seedlings that also have a dwarf phenotype were organized with those in the comt and especially the ccoaomt mutant subgroups. To check whether this was the case for each functional category of transcripts, we performed PCC analysis in each GO class. The transcripts involved in lignin biosynthesis and polymerization, cytochrome P450s, and response to biotic and abiotic stress were organized into one group in 10d seedlings of ccoaomt, hct, ccr1, and comt ccoaomt mutants. Transcripts from 10d seedlings of the comt and cse mutants, and also from stems of the cse mutant, formed a second group (Fig. 6, B, C, E, and I). Transcripts associated with hormone signaling pathways, transcription factors, and transport processes were clustered together for 10d seedlings of the hct, ccr1, and comt ccoaomt mutants, with 10d seedlings of cse, comt, and ccoaomt and cse stem constituting a second cluster for this ontology group (Fig. 6, F–H). The same relationships were seen in the cell wall-related functional ontology category for transcripts expressed in 10d seedlings (Fig. 6D).
Figure 6.
PCC analysis based on the abundance of DEGs at 10 d postgermination in M. truncatula monolignol pathway mutants. A, Profiles of all DEGs. B to I, Profiles of DEGs in each GO class as indicated. All gene lists involved in this analysis are available in Supplemental Data Set S5. Red color indicates that a set of genes is expressed in the same direction (up-up, down-down), and blue color indicates that a set of genes is expressed in the opposite direction (up-down, down-up).
When all DEGs in stems were compared (Fig. 7), transcripts from the cse and ccoaomt mutants constituted one group, whereas, surprisingly, the ccr1 and comt mutants, which have very different phenotypes, constituted another group (Fig. 7A). PCC analysis showed that transcripts involved in plant defense pathways and transport mechanisms grouped together in the ccr1 and comt mutants (Fig. 7, C and H), whereas transcripts associated with the lignin pathway, cell wall-related responses, and hormone signaling grouped together in the cse, ccr1, and ccoaomt mutants (Fig. 7, B, D, and F). In the transcription factor and P450-related transcript categories, comt and ccoaomt constituted one group, whereas cse and ccr1 constituted another group (Fig. 7, G and I). Lignin biosynthesis and polymerization genes exhibited close grouping in the phenotypically different ccr1 and ccoaomt mutants (Fig. 7, B and E).
Figure 7.
PCC analysis based on the abundance of DEGs in mature stem tissue of M. truncatula monolignol pathway mutants. A, Profiles of all DEGs. B to I, Profiles of DEGs in each GO class as indicated. All gene lists involved in this analysis are available in Supplemental Data Set S6. Red color indicates that a set of genes is expressed in the same direction (up-up, down-down), and blue color indicates that a set of genes is expressed in the opposite direction (up-down, down-up).
To determine whether similar correlations exist between the mutants at the metabolite level, we compared GC-MS profiles for the compounds that were common in every lignin pathway mutant examined. PCC analysis was performed with (1) all compounds including those related to the lignin pathway and (2) compounds excluding those related to the lignin pathway (Fig. 8). In 10d seedlings, comt, ccoaomt, and comt ccoaomt mutants constituted one group if the monolignol pathway-related compounds were included, and cse and hct mutants constituted another group (Fig. 8A). This grouping reflects the points in the monolignol pathway at which the pathway is perturbed. However, when compounds excluding those related to the monolignol pathway were compared, the comt ccoaomt double mutant grouped with the hct and cse mutants, with the second group comprising the comt and ccoaomt mutants (Fig. 8B). cse, ccr1, and ccoaomt mutants constituted one group in the stem samples, and comt alone constituted another group for both 10d seedlings and stems (Fig. 8, C and D).
Figure 8.
PCC analysis based on the levels of accumulation of metabolites determined by GC-MS analysis of monolignol pathway mutants of M. truncatula. A and B, Accumulation profiles of compounds from 10d seedlings. C and D, Accumulation profiles of compounds from mature stem tissue. A and C represent accumulation profiles of compounds including lignin pathway compounds, whereas B and D show accumulation profiles of compounds excluding lignin pathway compounds. All compound lists involved in this analysis are given in Supplemental Data Set S1. Red color means that accumulation or decrease of compounds in monolignol pathway mutants occurs in the same direction (up-up, down-down), and blue color indicates the opposite direction (up-down, down-up).
Taken together, the above analyses indicate that, qualitatively, the transcriptional reprogramming occurring in 10d seedlings does not necessarily reflect the pattern seen in mature stems and that metabolic and transcriptional reprogramming can be qualitatively different among mutants with similar growth phenotypes.
Relationship between Transcripts and Metabolites in Specific Pathways
SA is synthesized by two pathways in plants: one from Phe, the other from isochorismate (Chen et al., 2009). In 10d seedlings, levels of transcripts encoding isochorismate synthase, the key enzyme of the plastidial isochorismate pathway for SA biosynthesis, were decreased in hct and comt ccoaomt mutants that exhibited the highest SA levels (Fig. 4; Supplemental Data Set S5), suggesting that SA biosynthesis in these mutants occurs via the alternative phenylpropanoid pathway.
Levels of transcripts encoding DELTA-1-PYRROLINE-5-CARBOXYLATE SYNTHETASE (P5CS), which plays a key role in Pro biosynthesis, were high in 10d seedlings of every monolignol pathway mutant examined except comt, with a massive increase in the hct mutant (Supplemental Data Set S5), which accumulated the highest levels of Pro (Table 3; Supplemental Data Set S1). Conversely, levels of transcripts encoding the PROLINE DEHYDROGENASE (PRODH) that functions in Pro catabolism were decreased in 10d seedlings of hct and cse mutants, with little or no change in the other mutants (Supplemental Data Set S5). In stems of the cse and ccr1 mutants, expression of P5CS was strongly increased, whereas that of PRODH was strongly decreased (Supplemental Data Set S6), again correlating with elevated Pro levels in these plants (Table 3; Supplemental Data Set S1). Finally, the transcript level of P5CS was slightly decreased and the transcript level of PRODH was highly increased in stem tissue of the comt mutant (Supplemental Data Set S6), consistent with the low levels of free Pro (Table 3; Supplemental Data Set S1).
To examine the basis for the unexpectedly high levels of saponins recorded in stem tissue of several of the mutants, we determined the transcript levels of BETA-AMYRIN SYNTHASE (β-AS), the product of which serves as the first intermediate in triterpene saponin biosynthesis in plants (Zhao et al., 2010). Although saponin levels were increased in stems of cse and ccr1 mutants, expression of β-AS was not increased in the same tissues (Supplemental Data Sets S2 and S6). However, β-AS transcript levels were decreased in 10d seedlings of hct single mutant and comt ccoaomt double mutant plants (Supplemental Data Set S5), and levels of saponins were also decreased in these plants (Supplemental Data Set S3).
Supplemental Figure S13 summarizes the specific metabolic and transcriptomic changes discussed mapped onto the diagrams of phenylpropanoid metabolism, amino acid metabolism, and SA, JA, and ABA signaling (defense gene changes are collected in Supplemental Data Set S7). Clearly, increased SA and ABA biosynthesis and signaling are common responses among the dwarf mutants.
Genetic Analysis of Saponin Accumulation
In the M. truncatula monolignol pathway mutants, saponin levels were strongly increased in the stems of the most severely dwarfed lines. Genetic modification of the saponin pathway, through either up-regulation or alteration of the state of sapogenin glycosylation, can cause severe growth defects in plants (Naoumkina et al., 2010; Moses et al., 2014). To determine whether the accumulation of saponins could be responsible for the dwarf phenotypes, we carried out genetic analysis to determine whether β-AS loss of function could alleviate such growth defects. Tnt1 retrotransposon insertions in the β-AS gene of M. truncatula resulted in a semidwarf growth phenotype (Supplemental Fig. S14), and targeted UHPLC-MS analyses showed that levels of various saponins were highly decreased in stems of β-as mutant plants (Supplemental Data Set S2). When the cse mutant was crossed with the β-as mutant, homozygous cse β-as double mutant plants showed the same growth phenotype as the cse single mutant, indicating an epistatic relationship. Assuming that CSE loss of function cannot restore saponin levels in the absence of the major entry point enzyme into the triterpene pathway, the genetic data therefore suggest that elevated saponin levels do not contribute to the dwarf phenotype of the cse mutant. Interestingly, like the monolignol pathway mutant plants, β-as single and cse β-as double mutants also showed an increased number of trichomes and decreased leaf epidermal cell size (Fig. 2, C, D, G, and H).
DISCUSSION
A number of different models have been proposed to explain the negative growth phenotypes encountered following loss-of-function or knockdown expression of certain monolignol pathway genes. To focus discussion of the large amount of data we have now collected across multiple M. truncatula mutants, we here consider the multiple phenotypic changes observed in the plants for consistency with these different models.
Leaf Epidermal Cell Phenotypes in Monolignol Pathway Mutants
A clear correlation was observed between leaf cell size and overall growth phenotype in 10d mutant plants, with smaller epidermal cells in all the dwarf mutants. This phenotype was also associated with increased density and number of trichomes. Besides functioning as a physical barrier to prevent pathogen attack, trichomes secrete defensive compounds including organic acids, acyl sugars, polysaccharides, terpenes, nectar, or salt (Calo et al., 2006; Schilmiller et al., 2010; Wang, 2014; Huchelmann et al., 2017). It is possible that defects in monolignol biosynthesis result in alteration of trichome development as a compensatory mechanism to enhance plant defense (see below).
There is increasing evidence that changes in the cell wall are perceived by plasma membrane receptors. For example, some wall-associated kinases recognize oligogalacturonides released from pectin that induce defense responses such as the synthesis of PR proteins (Kohorn, 2015), whereas the FERONIA receptor kinase senses cell wall polysaccharides in the control of cell size and shape (Lin et al., 2018). It is possible that reduced leaf cell size favors redirection of resources toward defense responses (see "Discussion"). However, the epidermal cell wall is essentially nonlignified, so the initial signal for cell wall remodeling, if originating from a cell wall polymer, would likely have to be generated elsewhere, presumably in a tissue type with lignified secondary cell walls.
Loss of Lignin Function as a Determinant of Plant Growth Phenotype
Overall, our data show that the effects on lignin composition of the loss of function of specific enzymes in the monolignol pathway are generally similar in M. truncatula to those results reported in Arabidopsis (Besseau et al., 2007; Gallego-Giraldo et al., 2011a; Vanholme et al., 2012, 2013; Van Acker et al., 2013). Lignin composition itself is not a predictor of growth phenotype (Gallego-Giraldo et al., 2018). In this study, the lignin composition of the dwarf ccr1 mutant is close to that of the wild type, the nearly total loss of S units in the comt mutant is not associated with growth reduction, and the stems of the dwarf cse mutant have very high levels of H units, which are not observed in the equally dwarf ccr1 mutant.
In general, the level of lignin in the 10d plants correlated with the growth phenotype (the less lignin, the more severe the phenotype), with the exception of the dwarf cse mutant, which had no less lignin at this stage than the comt mutant with no growth defect. This suggests that lignin level at 10 d postgermination is not per se a determinant of subsequent growth phenotype. However, the cse mutant, along with the ccr1 mutant, exhibited strongly reduced lignin levels in mature stems, consistent with their dwarf phenotypes.
Impacts of severe reductions in lignin levels include lodging (particularly from loss of lignin in fibers) and collapse of xylem vessels with corresponding impairment of water and solute conductance. It has recently been suggested, on the basis of experiments in which lignin expression was restored in the xylem of a ccr mutant of Arabidopsis, that impairment of xylem function is likely to be the cause of the dwarf phenotype in this instance (De Meester et al., 2018). As previously shown by RNAi in alfalfa (Medicago sativa; Nakashima et al., 2008) and supported by our data here, strong reduction of HCT expression causes severe loss of integrity of vascular elements and other lignified structures in M. truncatula, with similar effects in the comt ccoaomt double mutant seen in this work.
Impaired water transport, as may arise from reduced lignin levels in xylem, could lead to drought responses via ABA signaling. Increased ABA levels were observed in 10d seedlings of the cse, hct, and comt ccoaomt mutants but not in the ccr1 mutant. We found this latter observation surprising, since the Arabidopsis ccr1 mutant has defective xylem (Jones et al., 2001), and the negative impact on transpiration could perhaps lead to water deficit resulting in elevated ABA levels in the corresponding mutant. Typical drought stress responses also include the production of osmocompatible solutes such as Pro (Ashraf and Foolad, 2007) and the synthesis of dehydrins and other LEA proteins (Verslues et al., 2006; Verbruggen and Hermans, 2008; Cutler et al., 2010; Battaglia and Covarrubias, 2013; Bhaskara et al., 2015). Pro levels were high in 10d seedlings of the hct and comt ccoaomt mutants but not in the cse mutant, which, along with the ccr1 mutant, did exhibit high Pro levels in stems. Parallel with Pro levels, dehydrin transcripts were high in 10d seedlings of the hct and comt ccoaomt mutants but not in the cse mutant. Various LEA protein transcripts were highly expressed in stems of all the mutants, irrespective of growth phenotype.
Overall, these data fail to support a model in which altered water conductance leading to osmotic imbalance, with sensing through ABA and resultant expression of drought response genes/metabolites, is a common, direct cause of the negative growth phenotypes across all lines.
Changes in Specialized Metabolism in Monolignol Pathway Mutants
Perturbation of the monolignol pathway may cause metabolic imbalances that result in loss of a molecule or molecules that are required for normal growth, or accumulation of a molecule or molecules that are inhibitory to growth. The phenylpropanoid/lignin biosynthesis pathway plays a critical role in the production of many specialized metabolites as well as providing the monomeric units for incorporation into the lignin polymer (Vogt, 2010). Interestingly, most of the accumulating specialized metabolites detected by GC-MS in our set of monolignol pathway mutants have been previously reported to have defense functions.
Among the compounds that showed decreased levels in monolignol pathway mutants, G lignans are of interest because at least one member of this class of compounds, dehydrodiconiferyl alcohol, has been ascribed a role in the control of cell division (Teutonico et al., 1991), and changes in lignan levels could theoretically underlie the alterations in leaf cell size. However, dehydrodiconiferyl alcohol was not detected in this study. The changes in lignan levels essentially reflected the changes in lignin composition across the mutant lines, with elevated levels of H lignans in the cse and hct mutants in which the monolignol pathway is disrupted prior to G monolignol biosynthesis and reductions in G lignans in all lines except comt, where the ablated gene functions primarily in S lignin biosynthesis.
The level of hydroxycinnamic acids was elevated in both young seedlings and mature stems of the M. truncatula monolignol pathway mutants that exhibited growth defects. 4-Hydroxybenzoic acid accumulated only in the hct and comt ccoaomt mutants at the 10d stage, in parallel with the strong growth defects in these mutants. Hydroxybenzoic acid and its derivatives have been shown to confer tolerance against both biotic and abiotic stresses (Horvath et al., 2007; Dempsey and Klessig, 2017).
Saponins are a group of natural products that consist of an isoprenoidal (triterpene)-derived aglycone covalently linked to one or more sugar moieties (Augustin et al., 2011; Moses et al., 2014). Plant-derived saponins are toxic to pathogens and herbivores (Faizal and Geelen, 2013; Moses et al., 2014). Furthermore, plant growth and development are impacted strongly by both overaccumulation and decreased accumulation of triterpene metabolites (Naoumkina et al., 2010; Moses et al., 2014). In mature stems, saponins accumulated only in the cse and ccr1 mutants where they were qualitatively similar, comprising a range of complex glycosides of zhanic acid, medicagenic acid, and soyaspogenol B. However, apart from one zhanic acid derivative that exhibited slightly elevated levels in the ccr1 mutant, levels of saponins were generally decreased in all the other mutants in 10d plants, irrespective of growth phenotype. This finding, along with the inability to restore growth to the cse mutant through loss of function of β-AS, suggests that alteration of saponins is not in itself a determinant of reduced growth in monolignol pathway mutants.
Isoflavonoids are often associated with defense in legumes (Dixon and Steele, 1999). However, few were differentially expressed in the M. truncatula monolignol pathway mutants. Levels of two isoflavonoid glycosides, 6′-malonyl-3-Glu-medicarpin and 6′-malonyl-ononin, decreased below wild-type levels in the cse mutant but were not differentially expressed in the other lines. The increase in the acylated flavone glycoside 7-O-(2′-feruloylglucuronyl)-(1-2)-glucuronyl apigenin in stems of both cse and ccr1 mutants did correlate with the growth phenotypes of these mutants; it is unfortunate that the other two highly dwarf mutants could not develop stems for further analysis of flavonoid levels.
The monolignol pathway mutants also exhibited small but significant changes in primary metabolism. Once plants are exposed to stress, they experience more requirements for energy, reducing equivalents, and carbon skeletons than nonstressed plants, which can lead to massive reprogramming of plant primary metabolism toward supplying such requirements to make secondary metabolites (Bolton, 2009; Rojas et al., 2014).
Overall, the changes in both primary and specialized metabolites in the various mutants point toward the sum of the changes in defense metabolites, rather than any one particular metabolite, as being associated with the observed growth phenotypes.
Defense Signaling in the Phenotypic Response of M. truncatula to Monolignol Pathway Perturbations
The JA family of signaling molecule plays a central role in the coordination of a remarkably diverse set of defensive traits in different plant species. These include interference with cell division, cell expansion, and seed development, induction of trichome development, and induction of a range of defensive phytochemicals, particularly terpenoids (Wasternack and Hause, 2013; Havko et al., 2016; Züst and Agrawal, 2017; Li et al., 2018). In view of the striking changes in development, seed number, trichome number, and terpene saponin production, as well as changes in some JA signaling gene transcripts in our monolignol pathway mutants, it is perhaps surprising that JA levels were relatively unaffected, at least at the growth stages measured.
In contrast to the lack of a clear JA response, perturbation of the monolignol pathway led to large increases in levels of SA, a phenolic defense hormone required for both local defense and systemic acquired resistance (Vlot et al., 2009; Dempsey et al., 2011). In addition, and in coordination with other plant hormones, SA contributes to the regulation of plant growth and development, including seed germination, seedling establishment, cell growth, photosynthesis, respiration, flowering, stomatal closure, nodulation in legumes, leaf senescence, and fruit yield (Vlot et al., 2009; Rivas-San Vicente and Plasencia, 2011; Liu et al., 2015). Overaccumulation of SA leads to dwarfing and altered cell division and expansion (Bowling et al., 1997; Vanacker et al., 2001; Li et al., 2010b; Gallego-Giraldo et al., 2011a, 2011b; Sašek et al., 2014). The effects of SA on cell division and cell expansion can be positive or negative depending on the cellular context and the level of signal transduction (Vanacker et al., 2001). Treatment with SA resulted in enhanced production of trichomes in Arachis hypogaea (War et al., 2013), an increase in saponin production in multiple plant species (Shabani et al., 2009; Lambert et al., 2011), and reduced seed yield and fitness (Heidel et al., 2004).
All of the above responses were seen in the dwarf monolignol pathway mutants and correlated with the elevated SA levels in both early seedling development and mature stems. Furthermore, levels of SA and transcript levels of genes involved in SA signaling and response positively correlated with phenotypic severity across the group of monolignol pathway mutants studied at 10 d postgermination, from high levels in hct and comt ccoaomt to reduced levels in the comt mutant with no negative growth phenotype. SA induces systemic expression of PR genes to protect uninfected tissues, a process called systemic acquired resistance (Dempsey and Klessig, 2017), and, in 10d plants, the largest increases in PR gene transcripts were observed in the hct, ccr1, and comt ccoaomt mutants. Plant defensins are small, highly stable, Cys-rich peptides with antifungal, antibacterial, proteinase inhibitory, and insect amylase inhibitory activities (Stotz et al., 2009). Defensin genes can be induced by MeJA or high levels of SA in M. truncatula (Hanks et al., 2005). In 10d seedlings, the mutants with the highest levels of SA exhibited the highest levels of defensin gene transcripts, whereas no induction of defensin genes was observed in the mutants with decreased accumulation of SA. Interestingly, although the comt mutant does exhibit elevated SA levels in mature stems, the corresponding defensin and some other PR genes are not induced, although others are. Perhaps the SA level (Fig. 4A) falls below a threshold value for the induction of all PR genes.
Depleting accumulated SA rescues the dwarf phenotype of the Arabidopsis HCT-RNAi line (Gallego-Giraldo et al., 2011a), suggesting that SA-mediated defense pathways limit growth rather than being a consequence of growth inhibition. However, removal of SA does not rescue the dwarf phenotype of the Atref8 (c3′h) mutant (Kim et al., 2014). It remains to be determined whether depletion of accumulated SA can also rescue the dwarf phenotypes of the M. truncatula hct, comt ccoaomt, ccr1, and cse mutants.
Growth Retardation in the cse Mutant
GO enrichment analysis of DEGs indicated that, irrespective of growth phenotype, each mutant showed a similar qualitative distribution pattern for the relative ratios of transcripts in different GO classes. However, the individual mutants differed in the level of gene expression, especially in the expression of defense response-related genes (i.e. the lignin pathway mutants with the stronger growth phenotypes generally showed stronger expression of defense response genes). This was particularly apparent when comparing the responses of the comt, ccoaomt, and comt ccoaomt mutants. The pattern of transcriptional changes in ccoaomt and the double mutant were quite similar, but quantitatively much greater in the double mutant. This suggests that loss of function of COMT somehow amplifies the response arising from the loss of function of CCoAOMT. However, growth retardation in the cse mutant is associated with less reprograming of metabolome and transcriptome than observed in other dwarf mutants.
In PCC analyses of total transcripts in 10d seedlings, the cse mutant grouped with the comt and ccoaomt mutants rather than with the hct, ccr1, and comt ccoaomt mutants that have similar growth phenotypes. Transcripts from stem tissue of the cse mutant also grouped with the ccoaomt mutant rather than with the ccr1 mutant, which is the only other mutant with a severe growth phenotype that survived to maturity. These results could be explained by functional equivalency of CSE and COMTs/CCoAOMTs. In spite of the suggested preference of COMT for 5-hydroxyferulic acid toward S lignin biosynthesis in semidwarf alfalfa (Guo et al., 2001a), COMT may also function as a 3-O-methyltransferase at the level of caffeic acid in M. truncatula (Inoue et al., 1998). Caffeoyl shikimate is converted to caffeic acid by CSE and subsequently converted to caffeoyl CoA by 4CL (Vanholme et al., 2013; Ha et al., 2016). As COMT, CCoAOMT, and CSE function in consecutive biosynthetic steps, related changes in the transcriptomes of their loss-of-function mutants might reflect similar metabolic perturbations rather than downstream effects on lignin content and composition, particularly at early developmental stages.
The overall expression level of defense-related genes was very low in the cse mutant at 10 d postgermination, with many expressed below wild-type levels. In contrast, defense-related genes were highly expressed in 10d hct, ccr1, and comt ccoaomt mutant seedlings. It should be noted that CSE is different from most other lignin pathway enzymes in two important respects: it is not essential for lignification, as its gene is absent in some species (Ha et al., 2016), and it possesses an alternative biochemical function, as a lysophospholipase involved in phospholipid repair under oxidative stress (Gao et al., 2010). The latter feature might explain why its loss of function leads to different phenotypes than that predicted from its positioning in the monolignol pathway.
Tradeoffs between Growth and Defense in Monolignol Pathway Mutants
Plants are sessile organisms and, therefore, are permanently exposed to a wide range of both biotic and abiotic stresses in their natural environments (Suzuki et al., 2014). Waxy cuticles, rigid cell walls, thorns, needles, and trichomes, and various phytoanticipins contribute a first, constitutive barrier that pathogens need to overcome to successfully colonize plant tissues (Hückelhoven, 2007; Malinovsky et al., 2014; Zhao and Dixon, 2014). Among these first constitutive barriers, the cell wall has traditionally been considered a passive defense barrier. However, an increasingly large body of evidence suggests that the cell wall, through its associated receptor kinases, provides active signaling toward mounting defense responses against a variety of stresses and controlling plant development (Gallego-Giraldo et al., 2011b, 2018; Wolf et al., 2012, 2014; Miedes et al., 2014; Kohorn, 2015; Tenhaken, 2015; Bethke et al., 2016; Voxeur and Höfte, 2016; Feng et al., 2018; Zhao et al., 2018). Modification of lignin quantity and quality can lead to cell wall remodeling and ectopic release of signal molecules that are recognized by cell wall receptors and turn on a variety of defense responses (Gallego-Giraldo et al., 2011b, 2018). Redirection of limited resources toward defense responses can make plants autotoxic (Baldwin and Callahan, 1993), and many reports indicate that plants with constitutively induced resistance lose fitness and become dwarfed (Bowling et al., 1997; Clarke et al., 1998; Clough et al., 2000; Baldwin et al., 2002; Genger et al., 2008). Tradeoffs between plant growth and plant defense appear to result from plant allocation decisions that are intended to maintain optimal plant fitness while responding to variable environmental stresses (Havko et al., 2016; Takatsuji, 2017; Züst and Agrawal, 2017).
The hct single and comt ccoaomt double mutant plants that have the most severe phenotypes and cannot complete the reproductive stage showed much stronger alteration in expression level of defense-related genes and accumulation of defense-related metabolites than the cse and ccr1 mutant plants that had somewhat milder dwarf mutant phenotypes, although defective in seed production (Supplemental Fig. S12). This is consistent with autotoxic effects of constitutively expressed defense mechanisms. Plants with induced resistance often exhibit enhanced trichome development (a defense mechanism) associated with reduction in cell division and expansion (Havko et al., 2016; Züst and Agrawal, 2017). Terpenoids, which accumulate in stem tissues of cse and ccr1 mutants, have a higher metabolic cost than carbohydrates and phenolic metabolites, even though they consist largely of carbon, which is not typically a limiting resource in plants (Havko et al., 2016; Züst and Agrawal, 2017).
We propose that the overall extent of the redirection of metabolism toward defense, rather than any one specific response, is an underlying cause of the growth phenotypes in the mutants studied here. This concept was recently proposed based on a review of previous findings across plant species (Xie et al., 2018) and here gains direct experimental support from the analyses of multiple mutants in the same species. Although we have not performed flux analysis to examine directly the flow of carbon into defense-related metabolites (a difficult task in our case because of the different qualitative responses in the different mutants), we believe that the size and direction of the transcriptomic changes alone support this conclusion. The model in Figure 9 describes how allocation of limited resource between growth, reproduction, and defense affects overall plant phenotypes. Very similar hormonal and transcriptomic changes to those observed in the M. truncatula hct mutant are also seen in semidwarf alfalfa lines in which HCT is targeted by RNAi (Gallego-Giraldo et al., 2011b), and HCT-RNAi Arabidopsis lines also highly express PR protein genes (Gallego-Giraldo et al., 2011a). It will be interesting to perform comparative transcriptomics across species for other monolignol pathway targets to determine whether ectopic defense gene expression is a conserved response to lignin modification
Figure 9.
Diagram showing the relationship between resource allocation and phenotype in monolignol pathway mutants of M. truncatula. In the phenotypic depictions, black circles represent reproductive development and gray circles represent defective reproductive development. hct and comt ccoaomt mutant plants have developmental arrest without bolting. The number of arrows in the resource allocation categories indicates the strength of the allotted resource.
The qualitative differences in defense responses in the different lines may result from differences in cell wall signaling as the changes in lignin are perceived and cell walls remodeled (Gallego-Giraldo et al., 2018), perhaps alone or in concert with alterations in signaling molecules derived from the monolignol pathway itself. Further differences may result from the loss of potential additional functions for the targeted monolignol pathway enzymes (e.g. the proposed roles of CCR in defense signaling and development [Kawasaki et al., 2006; Xue et al., 2015] and CSE in lipid repair [Gao et al., 2010]). It is also important to state that, although we could not find support for any other proposed model to explain the results across genotypes, the pleiotropic nature of the responses to lignin modification in all the mutants examined here makes it likely that additional factors may contribute to the ultimate growth phenotypes.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Tnt1 retrotransposon insertion mutants of Medicago truncatula (Tadege et al., 2008) were reverse screened to find Mtccr1, Mthct, Mtcomt, and Mtccoaomt mutant plants. To confirm insertion lines, genotyping was conducted using Tnt1-specific primers and gene-specific primers (Supplemental Table S6). The double mutants comt ccoaomt and cse β-as were generated by crossing of the individual single mutants (both of which were true nulls with no transcripts), and homozygosity of both loci in the double mutants was confirmed by DNA genotyping using primers listed in Supplemental Table S6. Seed scarification, planting, and plant growth conditions (in a growth chamber set at a 16-h/8-h day/night cycle at a 22°C [day]/20°C [night] photoperiod [120 μmol m−2 s−1] and 70% to 80% relative humidity) were performed as described in Ha et al. (2016). Isolation of the Mtcse mutant was described in Ha et al. (2016). Plants were harvested at 10 d after germination and (for mature stems) at the time between development of the first and second flowers. This equated to 45 to 55 d after germination for most plants except for the cse and ccr1 mutants (75–85 d after germination). All analyses (except for hormone analysis) included multiple replicates of each of two independent alleles per gene targeted.
Acquisition of Gene Sequences
Gene sequences for MtCCR1, MtCSE, MtCOMT, and MtCCoAOMT were obtained from previous reports (Zhou et al., 2010; Ha et al., 2016). To obtain the single MtHCT sequence, the protein sequence of AtHCT (At5g48930) was used as a query for BLAST (BLASTP) analysis in the M. truncatula genome database at the J. Craig Venter Institute Web site (http://blast.jcvi.org/er-blast/index.cgi?project=mtbe).
Gene Expression Analysis
RNA isolation and reverse transcription quantitative PCR were performed as described previously by Ha et al. (2016). In brief, quantitative PCR was performed in a QuantStudio6 Flex machine (Applied Biosystems). Cycles were as follows: for hold stage, 50°C for 2 min, 95°C for 10 min; for PCR stage, 95°C for 15 s, 60°C for 1 min, repeat 40 cycles; for melt-curve stage, 95°C for 15 s, 60°C for 1 min, 95°C for 15 s. Three independent biological replicates were analyzed in each experiment. For microarray analyses, samples were evaluated for purity with a Bioanalyzer 2100 (Agilent Technologies). The Affymetrix GeneChip-Medicago Genome Array (Affymetrix) was used for expression analyses. Probe labeling, array hybridization, and scanning were performed according to the manufacturer’s instructions (Affymetrix) for eukaryotic RNA, using a one-cycle protocol for cDNA synthesis. Functional analysis of DEGs from microarray data was performed using the GOEAST program, which by default adjusts the raw P values into a false discovery rate using the Benjamini-Yekutieli method (Zheng and Wang, 2008). The enriched GO annotation results were then classified using WEGO (Ye et al., 2006).
For RNA-seq analyses, sequencing libraries were generated using an Illumina TruSeq Total RNA sample preparation kit (Illumina) following the manufacturer’s recommendations. The library preparations were sequenced on an Illumina NextSeq 500 platform (Illumina), and 75-bp paired-end reads were generated. The run produced over 43.39 GB of data with 1,298.8 million paired-end reads with 92.89% ≥ Q30. Sequencing was performed at the University of North Texas BioDiscovery Institute Genomics Center in Denton, Texas. For gene annotation from RNA-seq, low-quality reads were discarded to ensure that the retained reads were longer than 40 bp with quality scores above 20 from the end, as previously described by Rao et al. (2014). Clean reads were mapped to M. truncatula version 4.0 (Tang et al., 2014) obtained from the Phytozome Web site (http://phytozome.jgi.doe.gov/) using Bowtie v2.3.2.0 (Langmead and Salzberg, 2012) and TopHat v2.1.1 (Kim et al., 2013) with default parameters. The expression levels of each gene were normalized and estimated as fragments per kilobase of exon model per million mapped fragments using Cufflinks v2.2.1 (Trapnell et al., 2013). Differential expression analysis between two samples was performed using Cuffdiff in the cufflinks package (Trapnell et al., 2013). The significantly DEGs were identified using adjusted P ≤ 0.05. Gene annotation and biological pathways were obtained, respectively, from the Phytozome Web site and from MapMan (Thimm et al., 2004) by mapping to the Arabidopsis (Arabidopsis thaliana) homologs as described previously by Rao et al. (2016).
PCC Analysis
Pairwise PCCs were calculated and clustered with the Euclidean distance method using R software.
Histochemical Analysis
The first leaf from 10d M. truncatula seedlings was cut and embedded as reported previously in Ha et al. (2003). Embedded plant samples were cut to 10-µm-thick sections using a rotary microtome (MICROM International). Images showing lignin UV autofluorescence were taken with an AMG-EVOS microscope (AMEX-4304; Thermo Scientific).
SEM Analysis
SEM analysis was performed immediately after sampling at an accelerating voltage of 15 kV using a scanning electron microscope (model TM-1000; Hitachi).
Determination of Lignin Content and Composition
Determination of lignin content and composition was performed as described previously by Ha et al. (2016).
Phytohormone Analyses
Tissues were harvested from NF13103 (cse), NF5145 (ccr1), NF17882 (comt), NF14405 (ccoaomt), NF17882 NF 14405 (comt ccoaomt double), and NF9055 (hct) mutants, frozen in liquid nitrogen, and stored at −80°C. The frozen tissues were ground with a mortar and pestle in liquid nitrogen to a fine powder and quickly weighed into an Eppendorf tube. The extraction method was based on Almeida Trapp et al. (2014) with modifications. In brief, 1 mL of cold methanol:water (70:30, v/v) plus 50 pmol each of 2H-labeled ABA, JA, MeJA, and SA were immediately added to the samples. The samples were vortexed and sonicated, then extracted for 1 h in the cold and centrifuged at 16,000g at 0°C for 5 min. The supernatant was removed and dried with nitrogen. Each sample was redissolved in 100% methanol, and the supernatant was injected into an Agilent 1290 UHPLC device connected to an Agilent 6430 Triple Quad mass spectrometer (Agilent Technologies). Separations were carried out using a Waters BEH C18 column (1.7 µm, 2.1 × 150 mm; Waters) and a gradient separation using 0.05% (v/v) formic acid/water (solvent A) and acetonitrile with 0.05% (v/v) formic acid (solvent B). Gradient separations were performed by starting with 5% solvent B, linear ramping from 5% to 46% solvent B over 19 min, followed by a steep increase to 90% B in 0.1 min, then a hold at 90% B for 2 min, and a steep decrease to 5% solvent B in 0.1 min at a flow rate of 0.4 mL min–1. The temperature of the UPLC column was set to 40°C. The gas temperature was 300°C, gas flow was 9 mL min–1, nebulizer was 25 pounds per square inch. Fragmentor and collision energies were optimized for each compound individually. The single reaction monitoring (SRM) analysis conditions were as follows: ABA and stable isotope-labeled [2H6]ABA (negative ion mode), capillary = 4,000 V, fragmentor voltage = 100 V, collision energy = 4 V, dwell time = 200 ms, and SRM transition (m/z) = 263/153 for unlabeled ABA and 269/159 for [2H6]ABA; JA and [2H]JA (negative ion mode), capillary = 4,000 V, fragmentor voltage = 100 V, collision energy = 6 V, dwell time = 200 ms, and SRM transition (m/z) = 209/59 for unlabeled JA and 211/59 for [2H]JA; MeJA and Me[2H]JA (positive ion mode), capillary = 4,000 V, fragmentor voltage = 70 V, collision energy = 8 V, dwell time = 200 ms, and SRM transition (m/z) = 225/151 for unlabeled MeJA and 227/153 for Me[2H]JA; SA and [2H6]SA (negative ion mode), capillary = 4,000 V, fragmentor voltage = 80 V, collision energy = 14 V, dwell time = 200 ms, and SRM transition (m/z) = 137/93 for unlabeled SA and 141/97 for [2H6]SA. Relative amounts of ABA, JA, MeJA, and SA were based on comparisons with the labeled hormones.
Metabolite Extraction and GC-MS Analysis
Metabolite profiling of methanol extracts was performed as reported in Tschaplinski et al. (2012), with 10 mL of the extracts being dried under nitrogen. Sorbitol (15 μg) was added as an internal standard, the extracts were silylated for 2 d as described previously by Tschaplinski et al. (2012), and 0.5 μL of the 1-mL reaction volume was analyzed by GC-MS.
Metabolite Extraction and UHPLC-MS Profiling
Metabolite extraction and profiling were performed according to protocols described elsewhere (Khan et al., 2016). Briefly, lyophilized samples were ground and extracted for 2 h in 80% (v/v) methanol containing 18 µg mL–1 umbelliferone as an internal standard. Samples were sonicated for 10 s and agitated for 2 h at 150 rpm. Then, samples were centrifuged, and the supernatant was analyzed using a Waters Aquity UPLC I-class device coupled to a Bruker Impact QTof-MS/MS apparatus operating at a mass resolution of approximately 40,000. At least three replicates were analyzed for each sample. Separations were achieved using a Waters 2.1 150-mm BEH C18 column, mobile phases of 0.1% (v/v) aqueous formic acid (A) and acetonitrile (B), and a linear gradient of 95%:5% to 30%:70% eluents A/B in 30 min, 30% to 5% A over 3 min, and 95% to 95% A over 3 min with a flow rate of 0.56 mL min–1 and column temperature of 60°C. The mass spectrometer was operated in negative electrospray ionization mode. Tandem mass data were acquired for targeted masses using a stepped collision energy that was dependent upon precursor mass. The MS system was calibrated using sodium formate clusters, and hexakis-(2,2-difluoroethoxy)-phosphazene was used as the lock-mass compound.
Initial metabolite identifications were based upon spectral and chromatographic matching to authentic compounds using orthogonal data; however, commercial standards and database information are not available for many metabolites, especially triterpene saponins, and a large number of the detected peaks could not be identified using spectral matching. Thus, analytes targeted for identification were concentrated and purified using UHPLC-MS coupled to a Bruker/Spark Holland Prospekt II solid-phase extraction device (UHPLC-MS-SPE). The column eluent from the UHPLC was split between the SPE and Bruker maXis Impact QTofMS with a split ratio of approximately 20:1, with the majority of eluent directed toward the SPE. A supplemental makeup flow of 1.5 mL min–1 water containing 0.05% (v/v) formic acid from a Knauer K120 pump was added to the larger split prior to trapping. SPE trapping was performed using 1- × 10-mm Waters Oasis HLB cartridges that were conditioned with 1 mL of acetonitrile and then equilibrated with 1 mL of water containing 0.05% (v/v) formic acid at 1 mL min–1. Typically, 20 to 40 injections of extract were performed, and the target analytes were repetitively collected from each of the injections onto individual SPE cartridges in an automated manner. After trapping, the cartridges were dried with nitrogen for 5 min. The extracted compounds were eluted from the cartridges using 250 µL of methanol-d4 into vials with a Gilson 215 liquid handler. The solutions were concentrated to 30 µL before being transferred into a 1.7-mm NMR tube.
Compound Identification by NMR
Compound identification by NMR was performed as described elsewhere (Khan et al., 2016). Briefly, all 1D and 2D NMR spectra were recorded on a Bruker Avance III HD 600 MHz spectrometer equipped with a SAMPLEJET autosampler and 1.7-mm TCI Micro-CryoProbe at 298 K. All spectra were recorded in methanol-d4 and referenced to residual solvent (1H, 3.31 ppm; and 13C, 49.15 ppm). 1D 1H spectra were recorded in Bruker’s defined WETDC pulse program with 3-s relaxation delays, 16,384 data points, and multiplied with an exponential function for a line broadening of 0.3 Hz before Fourier transformation. Multiplicity-edited Heteronuclear Single Quantum Coherence (pulse sequence, hsqcedetgpsp.3) spectra were acquired with spectral widths of 9 ppm for 1H and 200 ppm for 13C, 2,048 × 128 data points, and a relaxation delay of 1 s. 1D gradient-selected Total Correlation Spectroscopy (pulse sequence, selmlgp) spectra were recorded as a series of four spectra with mixing times of 0.02, 0.04, 0.08, and 0.16 s each, with a relaxation delay of 2 s, and multiplied with an exponential function for a line broadening of 1 Hz before Fourier transformation. All NMR data were processed using Bruker’s TopSpin 3.2 software.
Statistical Analysis
Two-tailed Student’s t test was used to compare pairs of data values (mutant versus wild-type control).
Data Deposition
All data needed to evaluate the conclusions in this article are present in the article and/or the supplementary materials. The data sets supporting the transcriptomic analyses in this article are publicly available in the National Center for Biotechnology Information Sequence Read Archive repository, accession numbers SRR9306568 to SRR9306614, and in Supplemental Data Sets S4 to S7.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the accession numbers listed in Supplemental Data Set S4 (for both M. truncatula genes and their best-hit Arabidopsis genes).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Diagrammatic representation of the currently accepted monolignol pathway.
Supplemental Figure S2. Identification of monolignol pathway mutants in M. truncatula.
Supplemental Figure S3. Photographs of monolignol pathway mutants in M. truncatula.
Supplemental Figure S4. Relative expression of monolignol pathway genes in 1- to 3-d seedlings of wild-type M. truncatula.
Supplemental Figure S5. Histological analysis of monolignol pathway mutants in M. truncatula.
Supplemental Figure S6. Histological analysis of early leaf development in wild-type M. truncatula.
Supplemental Figure S7. Structures of compounds from stems of M. truncatula cse and ccr1 mutants identified by LC-MS-NMR analysis.
Supplemental Figure S8. Venn diagram analysis of DEGs in M. truncatula monolignol pathway mutants.
Supplemental Figure S9. Comparison of the expression of DEGs between stems and 10d seedlings for monolignol pathway mutants of M. truncatula.
Supplemental Figure S10. MapMan representations of metabolic and associated transcriptomic changes in 10d monolignol pathway mutants of M. truncatula.
Supplemental Figure S11. MapMan representations of metabolic and associated transcriptomic changes in mature stem tissue from monolignol pathway mutants of M. truncatula.
Supplemental Figure S12. Comparison of the expression of DEGs in 10d seedlings of comt, ccoaomt, and comt ccoaomt mutants of M. truncatula.
Supplemental Figure S13. Representations of metabolic and transcriptomic changes related to the phenylpropanoid pathway and defense signal transduction in monolignol pathway mutants of M. truncatula.
Supplemental Figure S14. Genetic analysis of the involvement of saponins in the growth phenotype of the M. truncatula cse mutant.
Supplemental Table S1. Mutants analyzed in this study.
Supplemental Table S2. Numbers of leaf epidermal cells per trichome in SEM scans of lignin biosynthesis pathway mutants.
Supplemental Table S3. Structures of the side groups of saponins listed in Supplemental Figure S7.
Supplemental Table S4. GO enrichment analysis of DEGs in 10d plants.
Supplemental Table S5. GO enrichment analysis of DEGs in stem tissue.
Supplemental Table S6. Primers used in this study.
Supplemental Data Set S1. List of metabolites determined by GC-MS analysis in 10d plants, stems, and common between 10d plants and stems of M. truncatula monolignol pathway mutants.
Supplemental Data Set S2. List of metabolites determined by UHPLC-MS/MS analysis in stem tissue of M. truncatula monolignol pathway mutants.
Supplemental Data Set S3. List of metabolites determined by UHPLC-MS/MS analysis in 10-d seedlings of M. truncatula monolignol pathway mutants.
Supplemental Data Set S4. List of differentially expressed transcripts in 10d seedlings and stem tissues of M. truncatula monolignol pathway mutants.
Supplemental Data Set S5. List of DEGs in 10d seedlings of M. truncatula monolignol pathway mutants arranged according to gene class.
Supplemental Data Set S6. List of differentially expressed transcripts from stem tissue of M. truncatula monolignol pathway mutants arranged according to gene class.
Supplemental Data Set S7. List of differentially expressed transcripts from 10d seedlings and stem tissue of M. truncatula monolignol pathway mutants associated with defense-related pathways.
Acknowledgments
We thank Bonnie Watson, Noble Research Institute, for targeted hormone analysis.
Footnotes
This work was supported by the U.S. National Science Foundation (1639618) and the U.S. Department of Energy (Center for Bioenergy Innovation).
C.M.H. generated all mutants, performed phenotypic analyses, determined lignin content and composition, and analyzed transcriptomic and metabolomics data; D.F., A.B., and D.J.W. performed UHPLC-MS/MS and UHPLC-MS-SPE-NMR analyses; X.R. analyzed RNA-seq data; N.L.E., M.Z.M., and T.J.T. conducted the GC-MSbased metabolomics; R.A.D. and L.W.S. conceived the study; R.A.D. and C.M.H. wrote the article with editing assistance from L.W.S. and T.J.T.
Articles can be viewed without a subscription.
References
- Ahmad P, Rasool S, Gul A, Sheikh SA, Akram NA, Ashraf M, Kazi AM, Gucel S (2016) Jasmonates: Multifunctional roles in stress tolerance. Front Plant Sci 7: 813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida Trapp M, De Souza GD, Rodrigues-Filho E, Boland W, Mithöfer A (2014) Validated method for phytohormone quantification in plants. Front Plant Sci 5: 417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anterola AM, Lewis NG (2002) Trends in lignin modification: A comprehensive analysis of the effects of genetic manipulations/mutations on lignification and vascular integrity. Phytochemistry 61: 221–294 [DOI] [PubMed] [Google Scholar]
- Ashraf M, Foolad MR (2007) Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environ Exp Bot 59: 206–216 [Google Scholar]
- Augustin JM, Kuzina V, Andersen SB, Bak S (2011) Molecular activities, biosynthesis and evolution of triterpenoid saponins. Phytochemistry 72: 435–457 [DOI] [PubMed] [Google Scholar]
- Baldwin IT, Callahan P (1993) Autotoxicity and chemical defense: Nicotine accumulation and carbon gain in solanaceous plants. Oecologia 94: 534–541 [DOI] [PubMed] [Google Scholar]
- Baldwin IT, Kessler A, Halitschke R (2002) Volatile signaling in plant-plant-herbivore interactions: What is real? Curr Opin Plant Biol 5: 351–354 [DOI] [PubMed] [Google Scholar]
- Barros J, Temple S, Dixon RA (2019) Development and commercialization of reduced lignin alfalfa. Curr Opin Biotechnol 56: 48–54 [DOI] [PubMed] [Google Scholar]
- Battaglia M, Covarrubias AA (2013) Late Embryogenesis Abundant (LEA) proteins in legumes. Front Plant Sci 4: 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett AE, Grussu D, Kam J, Caul S, Halpin C (2015) Plant lignin content altered by soil microbial community. New Phytol 206: 166–174 [DOI] [PubMed] [Google Scholar]
- Besseau S, Hoffmann L, Geoffroy P, Lapierre C, Pollet B, Legrand M (2007) Flavonoid accumulation in Arabidopsis repressed in lignin synthesis affects auxin transport and plant growth. Plant Cell 19: 148–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke G, Thao A, Xiong G, Li B, Soltis NE, Hatsugai N, Hillmer RA, Katagiri F, Kliebenstein DJ, Pauly M, et al. (2016) Pectin biosynthesis is critical for cell wall integrity and immunity in Arabidopsis thaliana. Plant Cell 28: 537–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskara GB, Yang TH, Verslues PE (2015) Dynamic proline metabolism: Importance and regulation in water limited environments. Front Plant Sci 6: 484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhuiyan NH, Selvaraj G, Wei Y, King J (2009) Role of lignification in plant defense. Plant Signal Behav 4: 158–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Bolton MD. (2009) Primary metabolism and plant defense: Fuel for the fire. Mol Plant Microbe Interact 22: 487–497 [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Chapple C (2010) The genetics of lignin biosynthesis: Connecting genotype to phenotype. Annu Rev Genet 44: 337–363 [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Kim JI, Tobimatsu Y, Ciesielski PN, Anderson NA, Ximenes E, Maeda J, Ralph J, Donohoe BS, Ladisch M, et al. (2014) Disruption of Mediator rescues the stunted growth of a lignin-deficient Arabidopsis mutant. Nature 509: 376–380 [DOI] [PubMed] [Google Scholar]
- Bowling SA, Clarke JD, Liu Y, Klessig DF, Dong X (1997) The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9: 1573–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo L, García I, Gotor C, Romero LC (2006) Leaf hairs influence phytopathogenic fungus infection and confer an increased resistance when expressing a Trichoderma alpha-1,3-glucanase. J Exp Bot 57: 3911–3920 [DOI] [PubMed] [Google Scholar]
- Chen F, Dixon RA (2007) Lignin modification improves fermentable sugar yields for biofuel production. Nat Biotechnol 25: 759–761 [DOI] [PubMed] [Google Scholar]
- Chen Z, Zheng Z, Huang J, Lai Z, Fan B (2009) Biosynthesis of salicylic acid in plants. Plant Signal Behav 4: 493–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke JD, Liu Y, Klessig DF, Dong X (1998) Uncoupling PR gene expression from NPR1 and bacterial resistance: Characterization of the dominant Arabidopsis cpr6-1 mutant. Plant Cell 10: 557–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Fengler KA, Yu IC, Lippok B, Smith RK Jr, Bent AF (2000) The Arabidopsis dnd1 “defense, no death” gene encodes a mutated cyclic nucleotide-gated ion channel. Proc Natl Acad Sci USA 97: 9323–9328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR (2010) Abscisic acid: Emergence of a core signaling network. Annu Rev Plant Biol 61: 651–679 [DOI] [PubMed] [Google Scholar]
- De Meester B, de Vries L, Özparpucu M, Gierlinger N, Corneillie S, Pallidis A, Goeminne G, Morreel K, De Bruyne M, De Rycke R, et al. (2018) Vessel-specific reintroduction of CINNAMOYL-COA REDUCTASE1 (CCR1) in dwarfed ccr1 mutants restores vessel and xylary fiber integrity and increases biomass. Plant Physiol 176: 611–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey DA, Klessig DF (2017) How does the multifaceted plant hormone salicylic acid combat disease in plants and are similar mechanisms utilized in humans? BMC Biol 15: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey DA, Vlot AC, Wildermuth MC, Klessig DF (2011) Salicylic acid biosynthesis and metabolism. The Arabidopsis Book 9: e0156, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RA, Steele CL (1999) Flavonoids and isoflavonoids: A gold mine for metabolic engineering. Trends Plant Sci 4: 394–400 [DOI] [PubMed] [Google Scholar]
- Do CT, Pollet B, Thévenin J, Sibout R, Denoue D, Barrière Y, Lapierre C, Jouanin L (2007) Both caffeoyl coenzyme A 3-O-methyltransferase 1 and caffeic acid O-methyltransferase 1 are involved in redundant functions for lignin, flavonoids and sinapoyl malate biosynthesis in Arabidopsis. Planta 226: 1117–1129 [DOI] [PubMed] [Google Scholar]
- Dolan WL, Dilkes BP, Stout JM, Bonawitz ND, Chapple C (2017) Mediator complex subunits MED2, MED5, MED16, and MED23 genetically interact in the regulation of phenylpropanoid biosynthesis. Plant Cell 29: 3269–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faizal A, Geelen D (2013) Saponins and their role in biological processes in plants. Phytochem Rev 12: 877–893 [Google Scholar]
- Farag MA, Huhman DV, Lei Z, Sumner LW (2007) Metabolic profiling and systematic identification of flavonoids and isoflavonoids in roots and cell suspension cultures of Medicago truncatula using HPLC-UV-ESI-MS and GC-MS. Phytochemistry 68: 342–354 [DOI] [PubMed] [Google Scholar]
- Feng W, Kita D, Peaucelle A, Cartwright HN, Doan V, Duan Q, Liu MC, Maman J, Steinhorst L, Schmitz-Thom I, et al. (2018) The FERONIA receptor kinase maintains cell-wall integrity during salt stress through Ca2+ signaling. Curr Biol 28: 666–675.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke R, Hemm MR, Denault JW, Ruegger MO, Humphreys JM, Chapple C (2002) Changes in secondary metabolism and deposition of an unusual lignin in the ref8 mutant of Arabidopsis. Plant J 30: 47–59 [DOI] [PubMed] [Google Scholar]
- Fu C, Mielenz JR, Xiao X, Ge Y, Hamilton CY, Rodriguez M Jr, Chen F, Foston M, Ragauskas A, Bouton J, et al. (2011) Genetic manipulation of lignin reduces recalcitrance and improves ethanol production from switchgrass. Proc Natl Acad Sci USA 108: 3803–3808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Giraldo L, Escamilla-Trevino L, Jackson LA, Dixon RA (2011a) Salicylic acid mediates the reduced growth of lignin down-regulated plants. Proc Natl Acad Sci USA 108: 20814–20819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Giraldo L, Jikumaru Y, Kamiya Y, Tang Y, Dixon RA (2011b) Selective lignin downregulation leads to constitutive defense response expression in alfalfa (Medicago sativa L.). New Phytol 190: 627–639 [DOI] [PubMed] [Google Scholar]
- Gallego-Giraldo L, Posé S, Pattathil S, Peralta AG, Hahn MG, Ayre BG, Sunuwar J, Hernandez J, Patel M, Shah J, et al. (2018) Elicitors and defense gene induction in plants with altered lignin compositions. New Phytol 219: 1235–1251 [DOI] [PubMed] [Google Scholar]
- Gao W, Li HY, Xiao S, Chye ML (2010) Acyl-CoA-binding protein 2 binds lysophospholipase 2 and lysoPC to promote tolerance to cadmium-induced oxidative stress in transgenic Arabidopsis. Plant J 62: 989–1003 [DOI] [PubMed] [Google Scholar]
- Genger RK, Jurkowski GI, McDowell JM, Lu H, Jung HW, Greenberg JT, Bent AF (2008) Signaling pathways that regulate the enhanced disease resistance of Arabidopsis “defense, no death” mutants. Mol Plant Microbe Interact 21: 1285–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D, Chen F, Inoue K, Blount JW, Dixon RA (2001a) Downregulation of caffeic acid 3-O-methyltransferase and caffeoyl CoA 3-O-methyltransferase in transgenic alfalfa: Impacts on lignin structure and implications for the biosynthesis of G and S lignin. Plant Cell 13: 73–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D, Chen F, Wheeler J, Winder J, Selman S, Peterson M, Dixon RA (2001b) Improvement of in-rumen digestibility of alfalfa forage by genetic manipulation of lignin O-methyltransferases. Transgenic Res 10: 457–464 [DOI] [PubMed] [Google Scholar]
- Ha CM, Kim GT, Kim BC, Jun JH, Soh MS, Ueno Y, Machida Y, Tsukaya H, Nam HG (2003) The BLADE-ON-PETIOLE 1 gene controls leaf pattern formation through the modulation of meristematic activity in Arabidopsis. Development 130: 161–172 [DOI] [PubMed] [Google Scholar]
- Ha CM, Escamilla-Trevino L, Yarce JC, Kim H, Ralph J, Chen F, Dixon RA (2016) An essential role of caffeoyl shikimate esterase in monolignol biosynthesis in Medicago truncatula. Plant J 86: 363–375 [DOI] [PubMed] [Google Scholar]
- Hanks JN, Snyder AK, Graham MA, Shah RK, Blaylock LA, Harrison MJ, Shah DM (2005) Defensin gene family in Medicago truncatula: Structure, expression and induction by signal molecules. Plant Mol Biol 58: 385–399 [DOI] [PubMed] [Google Scholar]
- Havko NE, Major IT, Jewell JB, Attaran E, Browse J, Howe GA (2016) Control of carbon assimilation and partitioning by jasmonate: An accounting of growth-defense tradeoffs. Plants (Basel) 5: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidel AJ, Clarke JD, Antonovics J, Dong X (2004) Fitness costs of mutations affecting the systemic acquired resistance pathway in Arabidopsis thaliana. Genetics 168: 2197–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich M, Hettenhausen C, Lange T, Wünsche H, Fang J, Baldwin IT, Wu J (2013) High levels of jasmonic acid antagonize the biosynthesis of gibberellins and inhibit the growth of Nicotiana attenuata stems. Plant J 73: 591–606 [DOI] [PubMed] [Google Scholar]
- Horvath E, Pal M, Szalai G, Paldi E, Janda T (2007) Exogenous 4-hydroxybenzoic acid and salicylic acid modulate the effect of short-term drought and freezing stress on wheat plants. Biol Plant 51: 480–487 [Google Scholar]
- Huchelmann A, Boutry M, Hachez C (2017) Plant glandular trichomes: Natural cell factories of high biotechnological interest. Plant Physiol 175: 6–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hückelhoven R. (2007) Cell wall-associated mechanisms of disease resistance and susceptibility. Annu Rev Phytopathol 45: 101–127 [DOI] [PubMed] [Google Scholar]
- Huhman DV, Sumner LW (2002) Metabolic profiling of saponins in Medicago sativa and Medicago truncatula using HPLC coupled to an electrospray ion-trap mass spectrometer. Phytochemistry 59: 347–360 [DOI] [PubMed] [Google Scholar]
- Huhman DV, Berhow MA, Sumner LW (2005) Quantification of saponins in aerial and subterranean tissues of Medicago truncatula. J Agric Food Chem 53: 1914–1920 [DOI] [PubMed] [Google Scholar]
- Inoue K, Sewalt VJH, Murray GB, Ni W, Stürzer C, Dixon RA (1998) Developmental expression and substrate specificities of alfalfa caffeic acid 3-O-methyltransferase and caffeoyl coenzyme A 3-O-methyltransferase in relation to lignification. Plant Physiol 117: 761–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L, Ennos AR, Turner SR (2001) Cloning and characterization of irregular xylem4 (irx4): A severely lignin-deficient mutant of Arabidopsis. Plant J 26: 205–216 [DOI] [PubMed] [Google Scholar]
- Kawasaki T, Koita H, Nakatsubo T, Hasegawa K, Wakabayashi K, Takahashi H, Umemura K, Umezawa T, Shimamoto K (2006) Cinnamoyl-CoA reductase, a key enzyme in lignin biosynthesis, is an effector of small GTPase Rac in defense signaling in rice. Proc Natl Acad Sci USA 103: 230–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan BR, Wherritt DJ, Huhman D, Sumner LW, Chapman KD, Blancaflor EB (2016) Malonylation of glucosylated N-lauroylethanolamine. A new pathway that determines N-acylethanolamine metabolic fate in plants. J Biol Chem 291: 27112–27121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JI, Ciesielski PN, Donohoe BS, Chapple C, Li X (2014) Chemically induced conditional rescue of the reduced epidermal fluorescence8 mutant of Arabidopsis reveals rapid restoration of growth and selective turnover of secondary metabolite pools. Plant Physiol 164: 584–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohorn BD. (2015) The state of cell wall pectin monitored by wall associated kinases: A model. Plant Signal Behav 10: e1035854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert E, Faizal A, Geelen D (2011) Modulation of triterpene saponin production: In vitro cultures, elicitation, and metabolic engineering. Appl Biochem Biotechnol 164: 220–237 [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre C, Monties B, Rolando C (1985) Thioacidolysis of lignin: Comparison with acidolysis. J Wood Chem Technol 5: 277–292 [Google Scholar]
- Lei Z, Jing L, Qiu F, Zhang H, Huhman D, Zhou Z, Sumner LW (2015) Construction of an ultrahigh pressure liquid chromatography-tandem mass spectral library of plant natural products and comparative spectral analyses. Anal Chem 87: 7373–7381 [DOI] [PubMed] [Google Scholar]
- Li C, Wang P, Menzies NW, Lombi E, Kopittke PM (2018) Effects of methyl jasmonate on plant growth and leaf properties. J Plant Nutr Soil Sci 181: 409–418 [Google Scholar]
- Li M, Pu Y, Ragauskas AJ (2016) Current understanding of the correlation of lignin structure with biomass recalcitrance. Front Chem 4: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Bonawitz ND, Weng JK, Chapple C (2010a) The growth reduction associated with repressed lignin biosynthesis in Arabidopsis thaliana is independent of flavonoids. Plant Cell 22: 1620–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li S, Bi D, Cheng YT, Li X, Zhang Y (2010b) SRFR1 negatively regulates plant NB-LRR resistance protein accumulation to prevent autoimmunity. PLoS Pathog 6: e1001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Tang W, Anderson C, Yang Z (2018) FERONIA’s sensing of cell wall pectin activates ROP GTPase signaling in Arabidopsis. bioRxiv [Google Scholar]
- Liu X, Rockett KS, Kørner CJ, Pajerowska-Mukhtar KM (2015) Salicylic acid signalling: New insights and prospects at a quarter-century milestone. Essays Biochem 58: 101–113 [DOI] [PubMed] [Google Scholar]
- Malinovsky FG, Fangel JU, Willats WG (2014) The role of the cell wall in plant immunity. Front Plant Sci 5: 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miedes E, Vanholme R, Boerjan W, Molina A (2014) The role of the secondary cell wall in plant resistance to pathogens. Front Plant Sci 5: 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses T, Papadopoulou KK, Osbourn A (2014) Metabolic and functional diversity of saponins, biosynthetic intermediates and semi-synthetic derivatives. Crit Rev Biochem Mol Biol 49: 439–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura JCMS, Bonine CAV, de Oliveira Fernandes Viana J, Dornelas MC, Mazzafera P (2010) Abiotic and biotic stresses and changes in the lignin content and composition in plants. J Integr Plant Biol 52: 360–376 [DOI] [PubMed] [Google Scholar]
- Nakashima J, Chen F, Jackson L, Shadle G, Dixon RA (2008) Multi-site genetic modification of monolignol biosynthesis in alfalfa (Medicago sativa): Effects on lignin composition in specific cell types. New Phytol 179: 738–750 [DOI] [PubMed] [Google Scholar]
- Naoumkina MA, Modolo LV, Huhman DV, Urbanczyk-Wochniak E, Tang Y, Sumner LW, Dixon RA (2010) Genomic and coexpression analyses predict multiple genes involved in triterpene saponin biosynthesis in Medicago truncatula. Plant Cell 22: 850–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollier J, Morreel K, Geelen D, Goossens A (2011) Metabolite profiling of triterpene saponins in Medicago truncatula hairy roots by liquid chromatography Fourier transform ion cyclotron resonance mass spectrometry. J Nat Prod 74: 1462–1476 [DOI] [PubMed] [Google Scholar]
- Pomar F, Merino F, Barceló AR (2002) O-4-Linked coniferyl and sinapyl aldehydes in lignifying cell walls are the main targets of the Wiesner (phloroglucinol-HCl) reaction. Protoplasma 220: 17–28 [DOI] [PubMed] [Google Scholar]
- Qiu F, Fine DD, Wherritt DJ, Lei Z, Sumner LW (2016) PlantMAT: A metabolomics tool for predicting the specialized metabolic potential of a system and for large-scale metabolite identifications. Anal Chem 88: 11373–11383 [DOI] [PubMed] [Google Scholar]
- Rao X, Krom N, Tang Y, Widiez T, Havkin-Frenkel D, Belanger FC, Dixon RA, Chen F (2014) A deep transcriptomic analysis of pod development in the vanilla orchid (Vanilla planifolia). BMC Genomics 15: 964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao X, Lu N, Li G, Nakashima J, Tang Y, Dixon RA (2016) Comparative cell-specific transcriptomics reveals differentiation of C4 photosynthesis pathways in switchgrass and other C4 lineages. J Exp Bot 67: 1649–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy MS, Chen F, Shadle G, Jackson L, Aljoe H, Dixon RA (2005) Targeted down-regulation of cytochrome P450 enzymes for forage quality improvement in alfalfa (Medicago sativa L.). Proc Natl Acad Sci USA 102: 16573–16578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina JJ, Domínguez E, Heredia A (2001) Water sorption-desorption in conifer cuticles: The role of lignin. Physiol Plant 112: 372–378 [DOI] [PubMed] [Google Scholar]
- Rinaldi C, Kohler A, Frey P, Duchaussoy F, Ningre N, Couloux A, Wincker P, Le Thiec D, Fluch S, Martin F, et al. (2007) Transcript profiling of poplar leaves upon infection with compatible and incompatible strains of the foliar rust Melampsora larici-populina. Plant Physiol 144: 347–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas-San Vicente M, Plasencia J (2011) Salicylic acid beyond defence: Its role in plant growth and development. J Exp Bot 62: 3321–3338 [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Grant M, Jones JD (2011) Hormone crosstalk in plant disease and defense: More than just jasmonate-salicylate antagonism. Annu Rev Phytopathol 49: 317–343 [DOI] [PubMed] [Google Scholar]
- Rojas CM, Senthil-Kumar M, Tzin V, Mysore KS (2014) Regulation of primary plant metabolism during plant-pathogen interactions and its contribution to plant defense. Front Plant Sci 5: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sašek V, Janda M, Delage E, Puyaubert J, Guivarc’h A, López Maseda E, Dobrev PI, Caius J, Bóka K, Valentová O, et al. (2014) Constitutive salicylic acid accumulation in pi4kIIIβ1β2 Arabidopsis plants stunts rosette but not root growth. New Phytol 203: 805–816 [DOI] [PubMed] [Google Scholar]
- Schilmiller A, Shi F, Kim J, Charbonneau AL, Holmes D, Daniel Jones A, Last RL (2010) Mass spectrometry screening reveals widespread diversity in trichome specialized metabolites of tomato chromosomal substitution lines. Plant J 62: 391–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabani L, Ehsanpour AA, Asghari G, Emami J (2009) Glycyrrhizin production by in vitro cultured Glycyrrhiza glabra elicited by methyl jasmonate and salicylic acid. Russ J Plant Physiol 56: 621–626 [Google Scholar]
- Stotz HU, Spence B, Wang Y (2009) A defensin from tomato with dual function in defense and development. Plant Mol Biol 71: 131–143 [DOI] [PubMed] [Google Scholar]
- Suzuki N, Rivero RM, Shulaev V, Blumwald E, Mittler R (2014) Abiotic and biotic stress combinations. New Phytol 203: 32–43 [DOI] [PubMed] [Google Scholar]
- Tadege M, Wen J, He J, Tu H, Kwak Y, Eschstruth A, Cayrel A, Endre G, Zhao PX, Chabaud M, et al. (2008) Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J 54: 335–347 [DOI] [PubMed] [Google Scholar]
- Takatsuji H. (2017) Regulating tradeoffs to improve rice production. Front Plant Sci 8: 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Krishnakumar V, Bidwell S, Rosen B, Chan A, Zhou S, Gentzbittel L, Childs KL, Yandell M, Gundlach H, et al. (2014) An improved genome release (version Mt4.0) for the model legume Medicago truncatula. BMC Genomics 15: 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenhaken R. (2015) Cell wall remodeling under abiotic stress. Front Plant Sci 5: 771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teutonico RA, Dudley MW, Orr JD, Lynn DG, Binns AN (1991) Activity and accumulation of cell division-promoting phenolics in tobacco tissue cultures. Plant Physiol 97: 288–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L (2013) Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31: 46–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschaplinski TJ, Standaert RF, Engle NL, Martin MZ, Sangha AK, Parks JM, Smith JC, Samuel R, Jiang N, Pu Y, et al. (2012) Down-regulation of the caffeic acid O-methyltransferase gene in switchgrass reveals a novel monolignol analog. Biotechnol Biofuels 5: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacker H, Lu H, Rate DN, Greenberg JT (2001) A role for salicylic acid and NPR1 in regulating cell growth in Arabidopsis. Plant J 28: 209–216 [DOI] [PubMed] [Google Scholar]
- Van Acker R, Vanholme R, Storme V, Mortimer JC, Dupree P, Boerjan W (2013) Lignin biosynthesis perturbations affect secondary cell wall composition and saccharification yield in Arabidopsis thaliana. Biotechnol Biofuels 6: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R, Morreel K, Ralph J, Boerjan W (2008) Lignin engineering. Curr Opin Plant Biol 11: 278–285 [DOI] [PubMed] [Google Scholar]
- Vanholme R, Storme V, Vanholme B, Sundin L, Christensen JH, Goeminne G, Halpin C, Rohde A, Morreel K, Boerjan W (2012) A systems biology view of responses to lignin biosynthesis perturbations in Arabidopsis. Plant Cell 24: 3506–3529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R, Cesarino I, Rataj K, Xiao Y, Sundin L, Goeminne G, Kim H, Cross J, Morreel K, Araujo P, et al. (2013) Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 341: 1103–1106 [DOI] [PubMed] [Google Scholar]
- Verbruggen N, Hermans C (2008) Proline accumulation in plants: A review. Amino Acids 35: 753–759 [DOI] [PubMed] [Google Scholar]
- Verma V, Ravindran P, Kumar PP (2016) Plant hormone-mediated regulation of stress responses. BMC Plant Biol 16: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues PE, Agarwal M, Katiyar-Agarwal S, Zhu J, Zhu JK (2006) Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J 45: 523–539 [DOI] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DA, Klessig DF (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol 47: 177–206 [DOI] [PubMed] [Google Scholar]
- Vogt T. (2010) Phenylpropanoid biosynthesis. Mol Plant 3: 2–20 [DOI] [PubMed] [Google Scholar]
- Voxeur A, Höfte H (2016) Cell wall integrity signaling in plants: “To grow or not to grow that’s the question.” Glycobiology 26: 950–960 [DOI] [PubMed] [Google Scholar]
- Wang GD. (2014) Recent progress in secondary metabolism of plant glandular trichomes. Plant Biotechnol 31: 353–361 [Google Scholar]
- Wang B, Smith SM, Li J (2018) Genetic regulation of shoot architecture. Annu Rev Plant Biol 69: 437–468 [DOI] [PubMed] [Google Scholar]
- War AR, Hussain B, Sharma HC (2013) Induced resistance in groundnut by jasmonic acid and salicylic acid through alteration of trichome density and oviposition by Helicoverpa armigera (Lepidoptera: Noctuidae). AoB Plants 5: plt053 [Google Scholar]
- Wasternack C, Hause B (2013) Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot 111: 1021–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson BS, Bedair MF, Urbanczyk-Wochniak E, Huhman DV, Yang DS, Allen SN, Li W, Tang Y, Sumner LW (2015) Integrated metabolomics and transcriptomics reveal enhanced specialized metabolism in Medicago truncatula root border cells. Plant Physiol 167: 1699–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall CS, Muehler AM, Jez JM (2013) Enzyme action in the regulation of plant hormone responses. J Biol Chem 288: 19304–19311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S, Hématy K, Höfte H (2012) Growth control and cell wall signaling in plants. Annu Rev Plant Biol 63: 381–407 [DOI] [PubMed] [Google Scholar]
- Wolf S, van der Does D, Ladwig F, Sticht C, Kolbeck A, Schürholz AK, Augustin S, Keinath N, Rausch T, Greiner S, et al. (2014) A receptor-like protein mediates the response to pectin modification by activating brassinosteroid signaling. Proc Natl Acad Sci USA 111: 15261–15266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Zhang J, Tschaplinski TJ, Tuskan GA, Chen JG, Muchero W (2018) Regulation of lignin biosynthesis and its role in growth-defense tradeoffs. Front Plant Sci 9: 1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Zhu L, Tu L, Liu L, Yuan D, Jin L, Long L, Zhang X (2011) Lignin metabolism has a central role in the resistance of cotton to the wilt fungus Verticillium dahliae as revealed by RNA-Seq-dependent transcriptional analysis and histochemistry. J Exp Bot 62: 5607–5621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J, Luo D, Xu D, Zeng M, Cui X, Li L, Huang H (2015) CCR1, an enzyme required for lignin biosynthesis in Arabidopsis, mediates cell proliferation exit for leaf development. Plant J 83: 375–387 [DOI] [PubMed] [Google Scholar]
- Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, Wang J, Li S, Li R, Bolund L, et al. (2006) WEGO: A web tool for plotting GO annotations. Nucleic Acids Res 34: W293–W297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Dixon RA (2014) Altering the cell wall and its impact on plant disease: From forage to bioenergy. Annu Rev Phytopathol 52: 69–91 [DOI] [PubMed] [Google Scholar]
- Zhao CL, Cui XM, Chen YP, Liang Q (2010) Key enzymes of triterpenoid saponin biosynthesis and the induction of their activities and gene expressions in plants. Nat Prod Commun 5: 1147–1158 [PubMed] [Google Scholar]
- Zhao F, Chen W, Traas J (2018) Mechanical signaling in plant morphogenesis. Curr Opin Genet Dev 51: 26–30 [DOI] [PubMed] [Google Scholar]
- Zheng Q, Wang XJ (2008) GOEAST: A web-based software toolkit for Gene Ontology enrichment analysis. Nucleic Acids Res 36: W358–W363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Jackson L, Shadle G, Nakashima J, Temple S, Chen F, Dixon RA (2010) Distinct cinnamoyl CoA reductases involved in parallel routes to lignin in Medicago truncatula. Proc Natl Acad Sci USA 107: 17803–17808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Züst T, Agrawal AA (2017) Trade-offs between plant growth and defense against insect herbivory: An emerging mechanistic synthesis. Annu Rev Plant Biol 68: 513–534 [DOI] [PubMed] [Google Scholar]