Wheat TaSPL8 regulates lamina joint development and leaf angle through auxin signaling and the brassinosteroid biosynthesis pathway.
Abstract
In grass crops, leaf angle is determined by development of the lamina joint, the tissue connecting the leaf blade and sheath, and is closely related to crop architecture and yield. In this study, we identified a mutant generated by fast neutron radiation that exhibited an erect leaf phenotype caused by defects in lamina joint development. Map-based cloning revealed that the gene TaSPL8, encoding a SQUAMOSA PROMOTER BINDING-LIKE (SPL) protein, is deleted in this mutant. TaSPL8 knock-out mutants exhibit erect leaves due to loss of the lamina joint, compact architecture, and increased spike number especially in high planting density, suggesting similarity with its LIGULESS1 homologs in maize (Zea mays) and rice (Oryza sativa). Hence, LG1 could be a robust target for plant architecture improvement in grass species. Common wheat (Triticum aestivum, 2n = 6× = 42; BBAADD) is an allohexaploid containing A, B, and D subgenomes and the homeologous gene of TaSPL8 from the D subgenome contributes to the length of the lamina joint to a greater extent than that from the A and B subgenomes. Comparison of the transcriptome between the Taspl8 mutant and the wild type revealed that TaSPL8 is involved in the activation of genes related to auxin and brassinosteroid pathways and cell elongation. TaSPL8 binds to the promoters of the AUXIN RESPONSE FACTOR gene and of the brassinosteroid biogenesis gene CYP90D2 and activates their expression. These results indicate that TaSPL8 might regulate lamina joint development through auxin signaling and the brassinosteroid biosynthesis pathway.
Leaf angle, defined as the inclination between the leaf blade midrib and the stem, directly influences canopy structure and consequentially affects yield (Mantilla-Perez and Salas Fernandez, 2017). Plants with erect leaves have an increased capacity to intercept light and higher photosynthetic efficiency, which results in improved grain filling (Sinclair and Sheehy, 1999). More efficient grain filling enables planting of larger populations with a greater leaf area index. Hence, using plants with more erect leaves generally improves the yield per unit of cultivated area (Pendleton et al., 1968; Duvick, 2005; Lee and Tollenaar, 2007; Lauer et al., 2012). For example, the maize (Zea mays) liguleless2 mutant has erect leaves and produces 40% more grain than its counterpart with horizontal-type leaves due to the relative efficiency of CO2 fixation per unit of incoming sunlight, which increases as the leaf angle decreases (Pendleton et al., 1968). In rice (Oryza sativa), the Osdwarf4-1 mutant exhibits an erect leaf phenotype that is associated with brassinosteroid (BR) deficiency and has enhanced grain yields under dense planting populations (Sakamoto et al., 2006). In wheat (Triticum sp), the important characteristics of the ideal plant architecture (ideotype) include short and strong stems with few, small, and erect leaves (Donald, 1968). Wheat genotypes with erect leaves also have a superior net carbon fixation capacity during grain filling (Austin et al., 1976). Therefore, breeding grass crops for more erect leaves is a reasonable strategy for improving crop productivity.
Leaf angle depends on cell division, expansion, and cell wall composition in the lamina joint (including the auricle and ligule), which connects the leaf blade and sheath (Kong et al., 2017; Zhou et al., 2017). Genetic and functional studies indicated that various factors are involved in regulating lamina joint development, and consequentially affect leaf angle. Phytohormones, such as BR and auxin, play crucial roles in regulating the lamina inclination (Luo et al., 2016). In rice, loss of function of BR biosynthetic genes, such as dwarf4-1 (Sakamoto et al., 2006), ebisu dwarf (Hong et al., 2003), brassinosteroid-deficient dwarf1 (brd1; Hong et al., 2002), and chromosome segment deleted dwarf1 (Li et al., 2013) results in reduced leaf inclination. Lamina joint development is also associated with BR signaling, as reported by studies of mutants less sensitive to BR, such as the BR-defective mutant d61 (Yamamuro et al., 2000) and transgenic rice plants with suppressed expression of BRASSINOSTEROID INSENSITIVE1-Associated receptor Kinase1 (Li et al., 2009) and BRASSINAZOLE-RESISTANT1 (Bai et al., 2007). Similarly, auxin biosynthesis and signaling pathways influence lamina joint development. Both the gain-of-function rice LEAF INCLINATION1 (Zhao et al., 2013) mutant and plants overexpressing Gretchen Hagen3 acyl acid amido synthetases (GH3) family members, including OsGH3-1, -2, -5, and -13 (Du et al., 2012; Zhao et al., 2013; Zhang et al., 2015), have reduced auxin levels and show an enlarged leaf angle due to stimulated cell elongation at the lamina joint region. Rice transgenic lines overexpressing the auxin-responsive factor OsARF19 exhibit an enlarged lamina inclination related to the increase of adaxial cell division (Zhang et al., 2015). Furthermore, high concentrations of the auxin indole-3-acetic acid (IAA) influence leaf inclination by interacting with BR (Wada et al., 1981; Cao and Chen, 1995) and ethylene, which also participates in BR-induced leaf inclination (Jang et al., 2017). Therefore, crosstalk occurs between different phytohormones in regulating lamina joint development and leaf inclination.
Genetic approaches have identified several genes that affect lamina joint development in rice and maize (Mantilla-Perez and Salas Fernandez, 2017). For instance, rice BRASSINOSTEROID UPREGULATED1-LIKE1, encoding a typical basic helix-loop-helix (bHLH) transcription factor, is preferentially expressed in the lamina joint and affects leaf angle by controlling cell elongation (Jang et al., 2017). Similarly, overexpression or ectopic expression of other bHLH transcription factor genes, such as LAX PANICLE, OsbHLH153, OsbHLH173, OsbHLH174, BRASSINOSTEROID UPREGULATED1, INCREASED LAMINA INCLINATION1, and POSITIVE REGULATOR OF GRAIN LENGTH1, results in a larger lamina inclination (Komatsu et al., 2003; Tanaka et al., 2009; Zhang et al., 2009; Dong et al., 2018). LC2, a VIN3-like protein, regulates lamina inclination by repressing adaxial cell division in the collar region (Zhao et al., 2010). An increased leaf angle1 mutant shows increased leaf angle due to abnormal vascular bundle formation and cell wall composition in the lamina joint (Ning et al., 2011). It was suggested that rice SPX1, named after the suppressor of yeast gpa1, yeast cyclin-dependent kinase inhibitor, and human xenotropic and polytropic retrovirus receptor 1 (SYG1/Pho81/XPR1), could interact with Regulator of Leaf Inclination1 and negatively regulate leaf inclination by affecting lamina joint cell elongation (Ruan et al., 2018). Mutations in LIGULELESS1 (LG1), which encodes a conserved SQUAMOSA PROMOTER BINDING-LIKE (SPL) transcription factor in maize and rice, result in an erect leaf phenotype with a complete loss of the auricle and ligule (Moreno et al., 1997; Lee et al., 2007). A single-nucleotide polymorphism in the upstream region of OsLG1 may be responsible for its expression and affects panicle closure or spreading by controlling cell proliferation at the panicle pulvinus (Ishii et al., 2013; Zhu et al., 2013). However, the contributions of LG1 to potential yield and its mechanism in affecting lamina joint development are still unknown.
In wheat, genetic approaches have identified several quantitative trait loci (QTL) that control leaf inclination. For example, QTL analysis of the markers for flag leaf angle from durum wheat (Triticum durum) populations indicated the presence of large-effect QTL on chromosomes 2A, 2B, 3A, 3B, 4B, 5B, and 7A (Isidro et al., 2012). Several environmentally stable QTL for flag leaf angle were mapped on chromosomes 1B, 3D, 5A, 6B, 7B, and 7D using two recombinant inbred line populations (Wu et al., 2015; Liu et al., 2018b). However, to date, no genes have been shown to be directly involved in the control of leaf angle in wheat. In this study, we characterized a common wheat (Triticum aestivum) mutant with a multigene deleted region, which exhibits erect leaves and compact plant architecture. We demonstrate that the TaSPL8 gene, which encodes a SQUAMOSA PROMOTER BINDING-LIKE (SPL) protein and is homologous to LG1 in maize and rice, is responsible for this mutant phenotype. Our results indicate that TaSPL8 regulates lamina joint development through affecting auxin response and BR biogenesis pathways, thus providing information about the mechanisms linking the activity of SPL family members and hormone-mediated regulation of leaf angle.
RESULTS
The cpa Mutant Exhibits Defects in Lamina Joint Development
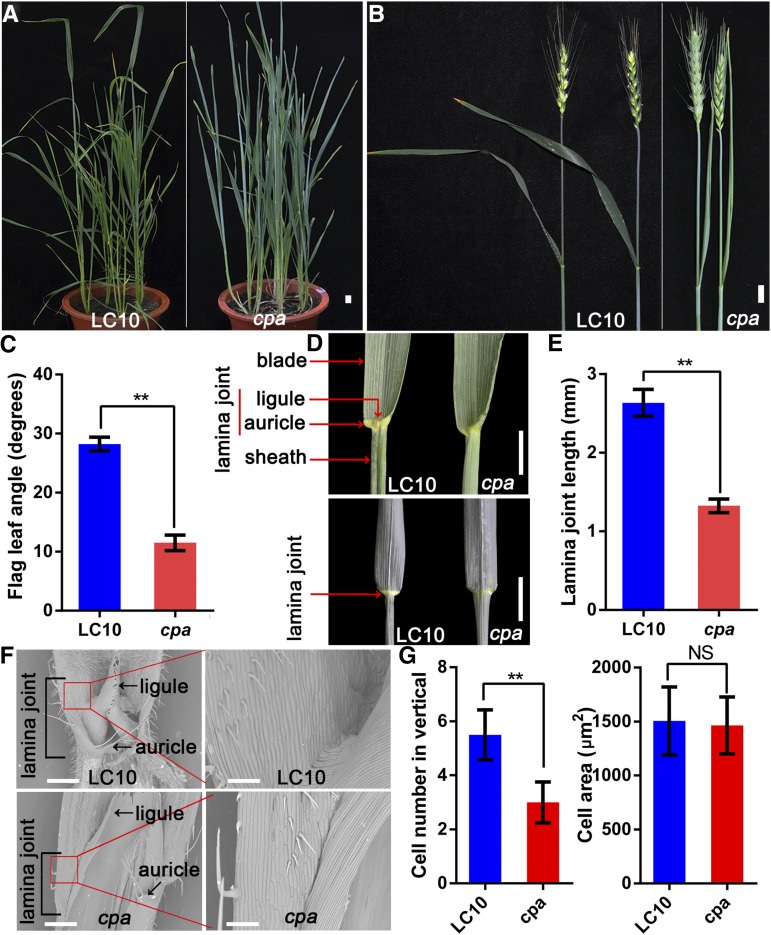
To study the molecular mechanism that controls leaf angle in wheat, we screened for mutants with erect leaves from a wheat population mutagenized by fast neutron radiation. We identified a mutant, named compact plant architecture (cpa), with a smaller leaf angle and a more-compact plant architecture compared to wheat cultivar Liaochun10 (LC10; Fig. 1A). We measured the flag leaf angle between the midrib of the flag leaf and the internode below the spike in the cpa mutant and LC10 and found that the flag leaf angle was 11.50° and 28.23° in the cpa mutant and LC10, respectively (Fig. 1, B and C). The smaller flag leaf angle of the cpa mutant is mainly caused by abnormal development of the lamina joint (Fig. 1D). In particular, the length of the lamina joint of the flag leaf in the cpa mutant is 1.33 mm, whereas in LC10 it is 2.64 mm (Fig. 1E). Further morphological observation through scanning electron microscopy showed that the lamina joint is much smaller in the cpa mutant than in LC10 due to a decrease in cell number, although no changes in cell size were detected (Fig. 1, F and G). These results indicate that the cpa mutant exhibits defects in lamina joint development.
Figure 1.
Morphological comparison of the wheat cultivar LC10 and the cpa mutant. A, Morphologies of LC10 and cpa plants at the heading stage. Bar = 2 cm. B, Morphologies of the flag leaf in LC10 and cpa plants at the flowering stage. Bar = 2 cm. C, Flag leaf angle of LC10 and cpa plants at the flowering stage. The double asterisk represents significant differences determined by the two-tailed Student’s t test at P < 0.01. Error bars = sd (n = 10). D, The lamina joint phenotype of LC10 and cpa plants. Bars = 2 cm. E, Lamina joint length of LC10 and cpa plants. The double asterisk represents significant differences determined by two-tailed Student’s t test at P < 0.01. Error bars = sd (n = 10). F, Morphology of the adaxial surface by a scanning electron microscope of the lamina joint of the LC10 and cpa flag leaf. Image magnification of the red box in the left (Bars = 200 μm) is shown on the right (Bars = 60 μm). G, Quantitative cell number and cell size of lamina joint from LC10 and cpa flag leaf. The cell number from the bottom to the top of the lamina joint was counted manually (n = 10) and cell size measured by the software ImageJ (n = 15). The double asterisk represents significant differences determined by two-tailed Student’s t test at P < 0.01. Error bars = sd. NS, not significant.
Map-Based Cloning of Mutated Gene
A genetic linkage analysis of 450 F2 individuals derived from a cross between LC10 and the cpa mutant was performed. A total of 335 individual plants showed a large lamina joint like LC10, while 115 plants showed a small lamina joint like the cpa mutant. This result indicated that the lamina joint defect of the cpa mutant is caused by a single recessive gene with a segregation ratio of approximately 3:1 (χ2 = 0.074 < χ2[0.05,1] = 3.84) of the normal phenotype versus the mutant phenotype. We designated the dominant wild-type gene as TaCPA.
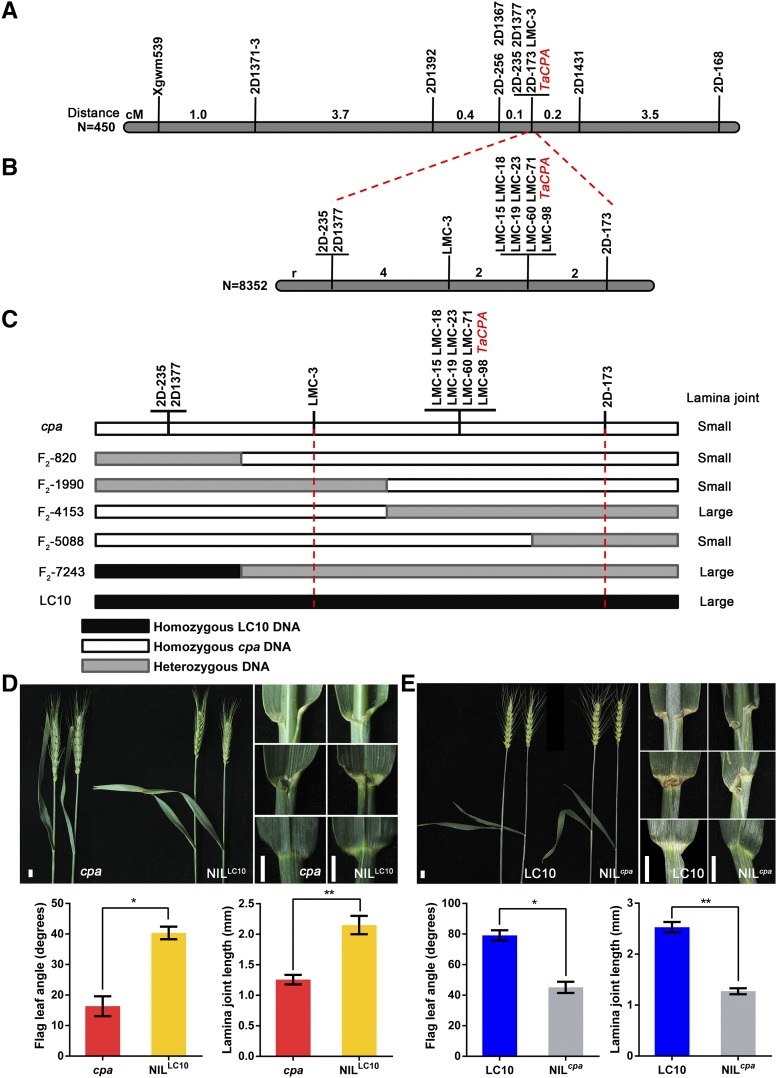
Following a map-based cloning approach using 450 F2 individuals, TaCPA was found to cosegregate with markers 2D-235, 2D1377, LMC-3, and 2D-173 on chromosome 2D (Fig. 2A). By a larger segregation population consisting of 8,352 individuals we narrowed down the candidate region of TaCPA to ∼19 Mb between LMC-3 and 2D-173 (Fig. 2, B and C). Generation of additional markers in this region allowed the mapping of TaCPA as being completely linked to LMC-15, -18, -19, -23, -60, -71, and -98 (Fig. 2, B and C). Near-isogenic lines (NILs) in the cpa genetic background containing the LC10 allele (NILLC10) and NILs in the LC10 genetic background containing the cpa allele (NILcpa) were generated through backcrossing of heterozygous F1 plants to LC10 and the cpa mutant for three generations (BC3), respectively (Supplemental Fig. S1). The backcrossed line containing a heterozygous fragment from markers LMC-3 and 2D-173 was selected. We found that in NILLC10 plants, the flag leaf angle and lamina joint length is significantly increased compared to the cpa mutant (Fig. 2D), while these parameters are decreased in the NILcpa plants compared to LC10 (Fig. 2E). This confirmed that TaCPA is located in the interval we have identified.
Figure 2.
Fine mapping of the TaCPA locus and complementation test using NILs. A, Coarse linkage map of TaCPA on chromosome 2D. Numbers indicate the genetic distance between the adjacent markers. Thick black line indicates partial region of 2D chromosome. B, High-resolution linkage map of TaCPA. The number of recombinants (labeled as “r”) between the molecular markers and TaCPA is indicated. Thick black line indicates partial region of 2D chromosome. C, Fine mapping of TaCPA. Genotypes and phenotypes of recombinants of five F2 plants including (F2-820, F2-1990, F2-4153, F2-5088, and F2-7243) are reported. The name and phenotype of F2 individual plants were labeled in the left and right parts, respectively. Black and white blocks indicate genomic region from LC10 and cpa, respectively. Gray blocks indicate the heterozygous region. D and E, Phenotype of the flag leaf angle and lamina joint in cpa and NILLC10 (D) and NILcpa (E) plants. Bars = 1 cm. Single and double asterisks represent statistically significant differences determined by the two-tailed Student’s t test at P < 0.05 and P < 0.01, respectively. Error bars = sd (n = 10).
The region between markers LMC-3 and 2D-173 contains a total of 257 predicted genes in the wheat Chinese Spring inbred line genome IWGSCv1(Appels et al., 2018). To map TaCPA at a higher resolution, we performed bulked segregant RNA sequencing (RNA-Seq), which makes use of the quantitative reads of RNA-Seq to efficiently map genes (Liu et al., 2012). RNA samples were extracted from 30 homozygous F2-derived F3 lines with normal or mutant phenotypes and from cpa and LC10 plants and were combined into four separate pools used for RNA-Seq. Reads located in the region between markers LMC-3 and 2D-173 were analyzed and the results indicated that 83 genes, distributed within this region, were expressed in both LC10 and homozygous normal phenotype pools, but were absent in the cpa mutant and mutant phenotype pools. These 83 genes could have been deleted in the cpa mutant by fast neutron radiation. To test this hypothesis, 10 genes located within and two genes located outside of the region (LMC-3 to 2D-173) were randomly selected and subjected to PCR amplification using genomic DNA of cpa and LC10 plants (Supplemental Fig. S2A). We found that all 10 genes within the LMC-3-2D-173 region are absent in the cpa genome, while the two external genes are present (Supplemental Fig. S2B). The specificity of the primers used for the experiments was further assessed using Chinese Spring null-tetrasomic lines. PCR products were not obtained in the lines with a chromosome 2D deletion, but were present in the other null-tetrasomic lines (Supplemental Fig. S2B). These results indicate that the chromosome fragment containing TaCPA is deleted in the cpa mutant.
TaSPL8, the Homolog of LG1 in Maize and Rice, Affects Lamina Joint Development and Plant Architecture in Wheat
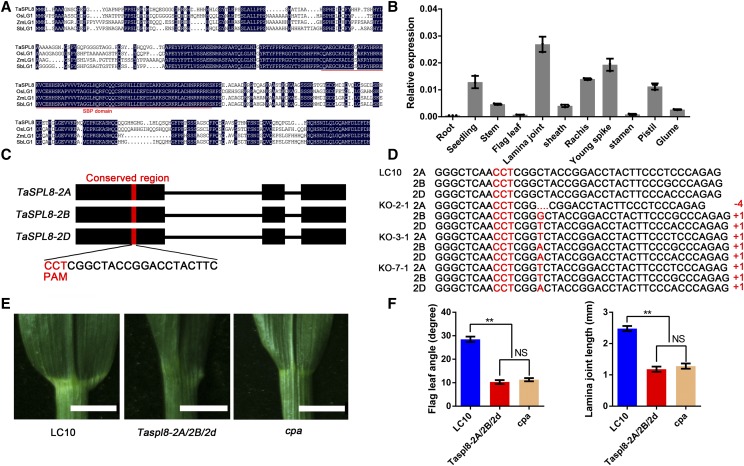
Among the 83 genes deleted in the cpa mutant, one gene (TraesCS2D01G502900), containing three exons and two introns, encodes a plant-specific SPL domain-containing transcription factor, SPL, with a total protein length of 407 amino acids. TraesCS2D01G502900 shows high amino acid similarity to OsLG1(SPL8) in rice, ZmLG1 in maize, and SbLG1 in sorghum (Sorghum bicolor; Fig. 3A), which are involved in controlling auricle, ligule, and lamina joint development (Moreno et al., 1997; Lee et al., 2007). Therefore, we renamed TaCPA as TaSPL8, a candidate gene for lamina joint development in wheat. The expression of TaSPL8 in various wheat tissues were detected by reverse transcription quantitative PCR (RT-qPCR), and we found that the TaSPL8 transcript is highly expressed in the lamina joint and young spikes (Fig. 3B).
Figure 3.
Cloning and characterization of TaSPL8 and the phenotype of its knockout (KO) lines by the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein9 (Cas9) system. A, Amino acid sequence alignment of TaSPL8 and its orthologs in rice (Os04g0656500), maize (Zm00001d002005), and sorghum (SORBI_3006G247700). The conserved SQUAMOSA-PROMOTER BINDING PROTEIN domain is indicated by a red line. B, Expression pattern of TaSPL8 in various tissues. The expression levels were normalized to wheat TaACTIN. Each bar in the graph corresponds to the mean value of three data points, shown by dots. C, Sequence of a single-guide RNA (sgRNA) designed to target a conserved region of exon 1 among the three TaSPL8 homeologs. The protospacer-adjacent motif (PAM) sequence is highlighted in red. D, The genotypes of three mutant lines were identified by sequencing. The symbols “+” and “−” indicate the insertion or deletion caused by CRISPR/Cas9-induced mutations, respectively. Numbers indicate the length of the insertion or deletion. E, Flag leaf phenotypes of mutant Taspl8-2A/2B/2d, cpa and LC10. Bars = 1 cm. F, Flag leaf angle and lamina joint length of Taspl8-2A/2B/2d, cpa and LC10 plants. The double asterisk represents significant differences determined by two-tailed Student’s t test at P < 0.01. Error bars = sd (n = 10).
Wheat is a hexaploid species that possesses three subgenomes originating from the fusion of genomes from three diploid ancestral species (Triticum urartu, Aegilops speltoides, and Aegilops tauschii; El Baidouri et al., 2017). Accordingly, three TaSPL8 homeologs were identified from the International Wheat Genome Sequencing Consortium database (IWGSCv1; Appels et al., 2018) and were designated TaSPL8-2A, TaSPL8-2B, and TaSPL8-2D, respectively. To determine whether the loss of TaSPL8-2D is responsible for the cpa mutant phenotype, we knocked out this gene using the CRISPR/Cas9 system. Due to the nucleotide similarity among TaSPL8-2A, TaSPL8-2B, and TaSPL8-2D, sgRNA specifically targeting to TaSPL8-2D cannot be designed. Thus, a sgRNA targeting a conserved region within the first exon of the TaSPL8-2A, TaSPL8-2B, and TaSPL8-2D was designed and the CRISPR/Cas9 vector pBUE411::sgRNA was introduced into LC10 (Fig. 3C). A total of 10 independent transgenic T0 plants (KO lines) were obtained and three lines with all three homeologous genes mutated simultaneously were selected for further analysis. Frameshift mutations in all three subgenomes were also detected in the T1 offspring (Fig. 3D). The KO-2-1 line was backcrossed to LC10 to generate the TaSPL8-2A/2B/2d single mutant, which showed a smaller lamina joint and flag leaf angle similar to the cpa mutant, compared with LC10 (Fig. 3, E and F). These results indicated that the loss of TaSPL8-2D is responsible for the cpa mutant phenotype.
Three Homeologous Genes of TaSPL8 Differentially Contribute to Lamina Joint Growth
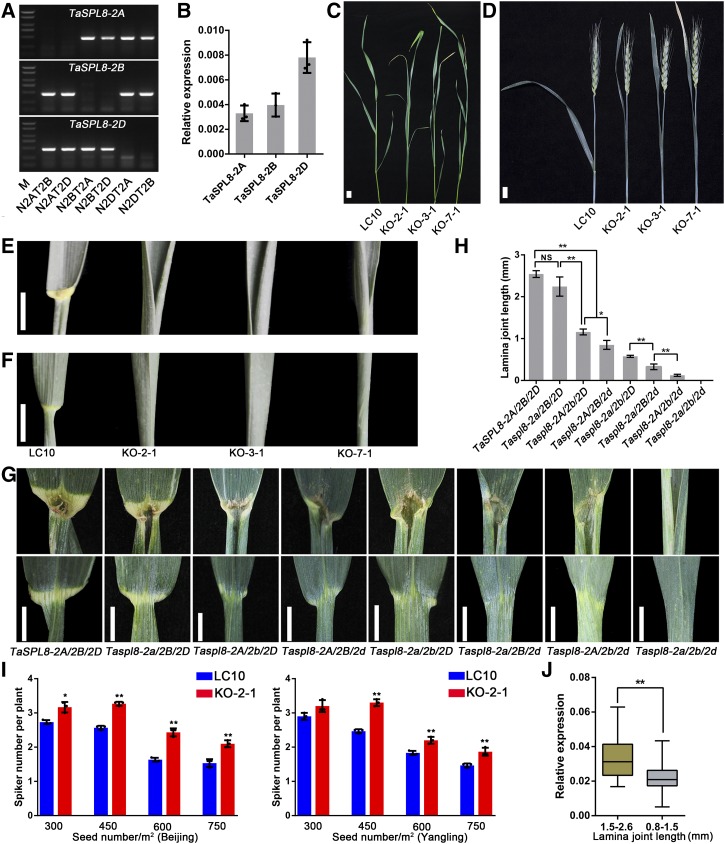
One extraordinary feature of polyploid evolution is genomic asymmetry, which is the predominant control of a variety of morphological, physiological, and molecular traits by one subgenome over the others, which prompted us to determine the functional divergence of TaSPL8 from 2A, 2B, and 2D subgenomes (Wang et al., 2016). We firstly compared the sequence similarity of open reading frames (ORFs) and protein sequences of the three TaSPL8 homeologous genes, and they showed 97.2% and 97.5% similarities for nucleotide and amino acid sequences, respectively. Primer combinations located in the coding sequence region specific for each homeolog were designed and verified using the Chinese Spring null-tetrasomic lines of chromosome group 2. We found that the PCR fragments could not be amplified when the corresponding chromosome was deleted (Fig. 4A). Using these primers, the transcript level of TaSPL8-2D was observed to be higher than that of TaSPL8-2A and TaSPL8-2B in the lamina joint (Fig. 4B).
Figure 4.
Three homeologous genes of TaSPL8 differentially contribute to lamina joint growth. A, The specificity of primers for three TaSPL8 homeologous genes were verified by amplifying the genomic DNA of the wheat cultivar Chinese Spring null-tetrasomic lines of chromosome group 2, where N2DT2B represents nullisomic2D-tetrasomic2B, for example. B, Relative abundance of three homeologous genes, TaSPL8-2A, TaSPL8-2B, and TaSPL8-2D, in the lamina joint. DNA of the Chinese Spring cultivar was used to analyze the amplification efficiency of the three pairs of primers. The RT-qPCR data were normalized to wheat TaACTIN and each bar in the graph corresponds to the mean value of three data points, shown by dots. C and D, Leaf phenotype of LC10 and mutant lines at heading (C) and flowering stages (D). Bars = 2 cm. E and F, Adaxial (E) and abaxial side (F) phenotypes of the lamina joint region of LC10 and mutant lines. Bars = 2 cm. G, Lamina joint phenotype of TaSPL8 single, double, and triple homeologous mutant plants. Bars = 1 cm. H, The lamina joint length of TaSPL8 single, double, and triple homeologous mutants. The values shown are averages from three plants and the sd is indicated. NS, nonsignificant. Single and double asterisks represent statistically significant differences determined by the two-tailed Student’s t test at *P < 0.05 and **P < 0.01, respectively. I, Spike number per plant in LC10 and mutant lines in different planting densities (300, 450, 600, and 750 seeds/m2) in Beijing and Yangling. The values shown are averages from three replicates and the sd is reported. Single and double asterisks represent statistically significant differences determined by the two-tailed Student’s t test at *P < 0.05 and **P < 0.01, respectively. J, Comparison of TaSPL8 expression levels between the wheat cultivars with larger (1.5–2.6 mm; n = 29) and smaller (0.8–1.5 mm; n = 49) lamina joints. The level of TaSPL8 mRNA was normalized to wheat ACTIN. Lines in the box plots indicate the median. The 10th/90th percentiles of outliers are shown. The double asterisk represents statistically significant differences as determined by Student’s t test at **P < 0.05.
Possible divergence among these homeologs in their contribution to the observed phenotype was investigated by generating single, double, and triple mutants for each homeolog. As mentioned above (Fig. 3D), we firstly obtained the triple mutant TaSPL8-2a/2b/2d by gene editing. The KO-2-1, KO-3-1, and KO-7-1 mutants with three TaSPL8 homeologs knocked out simultaneously showed erect leaves at both vegetative and reproductive stages (Fig. 4, C and D), with a notable decrease in flag leaf angle compared to the wild-type LC10 (Fig. 4D). Additionally, the lamina joint was severely affected and the boundary between the blade and sheath was absent in these KO lines (Fig. 4, E and F). Similar phenotypes were also observed for the KO lines in CB037 and Bobwhite recipients (Supplemental Fig. S3). The triple mutant KO-2-1 was crossed to wild-type LC10 to generate single and double mutants. Compared to LC10, the TaSPL8-2a/2B/2D single mutant did not show differences in lamina joint length, while the TaSPL8-2A/2b/2D and TaSPL8-2A/2B/2d single mutants had a shorter lamina joint, with the major effect detected in TaSPL8-2A/2B/2d (Fig. 4, G and H). Accordingly, the double mutant TaSPL8-2a/2B/2d had a smaller lamina joint length compared with the TaSPL8-2a/2b/2D double mutant, indicating that the TaSPL8-2D homeolog provides the major contribution for the lamina joint length phenotype (Fig. 4, G and H). Observations from double mutants also corroborated the information from single mutants that TaSPL8-2B affects lamina joint length to a greater extent than TaSPL8-2A (Fig. 4, G and H). These results indicated that the homeologous gene from the D subgenome contributed to lamina joint length to a greater extent than those from the A and B subgenomes.
To evaluate the potential of manipulating TaSPL8 for optimizing wheat leaf architecture and improving grain yield, we compared the yield-related traits of the TaSPL8 mutant KO-2-1 and the wild-type LC10 in different planting populations of 300, 450, 600, and 750 seeds/m2 in two different locations (Beijing and Yangling). Spike numbers per plant for the KO-2-1 line were higher compared to LC10, especially at dense planting populations (450, 600, and 750 seeds/m2; Fig. 4I), which could contribute to higher grain yield. Other traits, including plant height, spike length, grain number per spike, grain weight, heading date (flowering time), and the grain-filling period (time to maturity) did not differ between the KO-2-1 line and LC10 (Supplemental Table S1).
Natural Variation of TaSPL8-2D
To determine the variation of TaSPL8-2D in a natural wheat population, 192 wheat cultivars were screened for nucleotide polymorphisms. No single nucleotide polymorphism was identified in the coding and promoter region, suggesting that the TaSPL8-2D sequence is conserved because of its pivotal role in controlling lamina joint development. The TaSPL8 expression level and lamina joint length was also assessed in 78 wheat cultivars, including 29 cultivars with larger lamina joints (1.5–2.6 cm) and 49 cultivars with smaller lamina joints (0.8–1.5 cm; Supplemental Table S2). The results indicated that the TaSPL8 mRNA level in the lamina joint is higher in cultivars with larger lamina joints compared to those with smaller lamina joints (Fig. 4J). These results further corroborated the role of TaSPL8 in lamina joint development.
TaSPL8 Regulates Lamina Joint Development Through the Auxin and BR Pathways
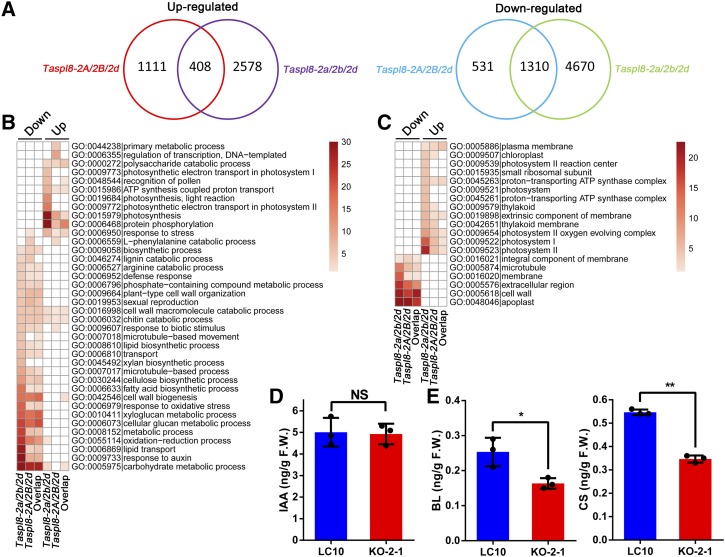
To understand the mechanisms behind TaSPL8-mediated lamina joint regulation, we performed a transcriptome analysis using RNA-seq in the lamina joint of main tiller from TaSPL8-2a/2b/2d (two independent KO lines: KO-2-1 and KO-7-1) and TaSPL8-2A/2B/2d (single KO line) mutants, as well as of LC10 wild-type plants at the heading stage. For each sample, three biological replicates were employed. The TaSPL8-2A/2B/2d mutant was included in the experiments because the triple TaSPL8-2a/2b/2d mutant lacks the lamina joint and the cell type between the sheath and blade are completely different with respect to LC10, while the single mutant exhibits a phenotype more similar to the wild type, but with a noticeable decrease in lamina joint length. The results indicated that 5,980 genes are downregulated and 2,986 genes are upregulated in the TaSPL8-2a/2b/2d mutants compared to LC10 (Fig. 5A; Supplemental Tables S3 and S4). Furthermore, a comparison between the TaSPL8-2A/2B/2d single mutant and the wild type indicated that 1,841 genes are downregulated and 1,509 are upregulated (P values < 0.05, |Fold Change| > 2). Among the differentially expressed genes (DEGs), 408 and 1,310 genes are upregulated and downregulated, respectively, in both the triple and single TaSPL8-2D mutant (Fig. 5A).
Figure 5.
Gene Ontology (GO) term enrichment analysis and IAA/BR level measurement in TaSPL8 mutants and wild-type plants. A, Venn diagram analysis of DEGs in TaSPL8-2A/2B/2d and TaSPL8-2a/2b/2d mutants compared to the wild type (LC10). B, GO terms for biological processes exhibiting statistically significant enrichment in DEGs (only DEGs in common for both single and triple TaSPL8 mutants were considered). C, GO terms for cell components exhibiting statistically significant enrichment in DEGs. D and E, IAA, Brassinolide (BL), and Castasterone (CS) levels were measured in the lamina joint tissue of LC10 and KO-2-1 (TaSPL8-2a/2b/2d) plants. Single and double asterisks represent statistically significant differences determined by the two-tailed Student’s t test at *P < 0.05 and **P < 0.01, respectively. Error bars = sd (n = 3). NS, not significant.
GO functional annotation of the DEGs shared between the triple and single TaSPL8 mutants revealed statistically significant enrichment in downregulated genes for pathways related to response to auxin, metabolic process for carbohydrate, cellular glucan, xyloglucan, biosynthetic process for fatty acid, cellulose, and cell wall (Fig. 5B). Among the upregulated genes, we found an enrichment in pathways associated with protein phosphorylation and photosynthesis (Fig. 5B). For cell component ontology, downregulated genes were enriched in the extracellular region and apoplast and cell wall, whereas upregulated genes were enriched in PSI and PSII (Fig. 5C). These observations suggest TaSPL8 involvement in cell wall development through the auxin pathway.
Among the DEGs in the single and triple TaSPL8 mutants, we found 188 downregulated genes related to auxin signaling, including the auxin-responsive Aux/IAA gene family (IAAs), ARFs, auxin efflux carrier components PIN-FORMED (PIN), small auxin-upregulated RNA, and GH3 family (Supplemental Fig. S4; Supplemental Tables S3 and S4). Measurement of the IAA content in the lamina joint region (tissue between the blade and sheath) of LC10 and the TaSPL8-2a/2b/2d mutant indicated that the IAA content was similar between the mutant and the wild type (Fig. 5D), indicating that TaSPL8 does not affect IAA biogenesis but may act through auxin response signaling.
Another interesting category of DEGs that were downregulated in the single and triple TaSPL8 mutants include genes involved in cell division, cell elongation, and the cytoskeleton (Supplemental Fig. S4; Supplemental Tables S3 and S4). Downregulation of these genes may lead to failure of cell elongation and result in the lamina joint defects observed in the mutants. These effect can be envisaged especially for the 128 genes encoding glycosyl hydrolase (which function in primary cell wall structure; López-Bucio et al., 2002), the 48 genes for expansins (which regulate cell wall extension; Cosgrove, 2015), and seven genes for pectinacetylesterase (which function in cell wall extensibility by enhancing aggregation processes between galacturonan chains inside the apoplasm; Sénéchal et al., 2014).
Finally, among the downregulated DEGs, we found three genes with high similarity to rice BRD1/OsDWARF/OsBR6ox, BRD2, and Cytochrome P450 90D2 (D2/CYP90D2), which are implicated in BR biosynthesis processes (Supplemental Fig. S4; Supplemental Tables S3 and S4). Moreover, 14 genes encoding BRASSINOSTEROID INSENSITIVE1-associated proteins and BRASSINAZOLE-RESISTANT1 homolog proteins, which are responsive to the BR signaling pathway, were also among the downregulated DEGs (Supplemental Fig. S4; Supplemental Tables S3 and S4). In agreement with these observations, we found that the brassinolide and castasterone content in the TaSPL8-2a/2b/2d mutant is reduced in comparison with LC10 (Fig. 5E), indicating that TaSPL8 affects BR biosynthesis and signaling pathways. Altogether, our findings indicate that TaSPL8 might regulate lamina joint development through BR and auxin pathways, which are essential for both cell elongation and cell proliferation.
TaSPL8 Acts as a Transcriptional Activator of AUXIN RESPONSE FACTOR and TaD2
Because maize LG1 is expressed in ligule and activates PIN1a expression as a transcription activator (Johnston et al., 2014), we screened the downregulated DEGs identified in our transcriptome analysis for the presence of the 16-nt sequence containing a GTAC core motif (Motif ID: MA0578.1) in their putative promoters, which was previously identified as the cis-element of AtSPL8 targets in Arabidopsis (Arabidopsis thaliana; Kropat et al., 2005). Among the 5,980 downregulated genes in the TaSPL8-2a/2b/2d mutant, 1,687 have this motif in their putative promoter sequence, as tested using the software FIMO (http://meme-suite.org/tools/fimo). The ability of TaSPL8 to bind to the sequence containing the GTAC core motif was assessed using electrophoretic mobility shift assays (EMSAs). To this end, nine probes were synthesized from the promoters of three auxin-responsive genes and four cell expansin genes. The results indicated that TaSPL8 can bind to these probes (Supplemental Fig. S5).
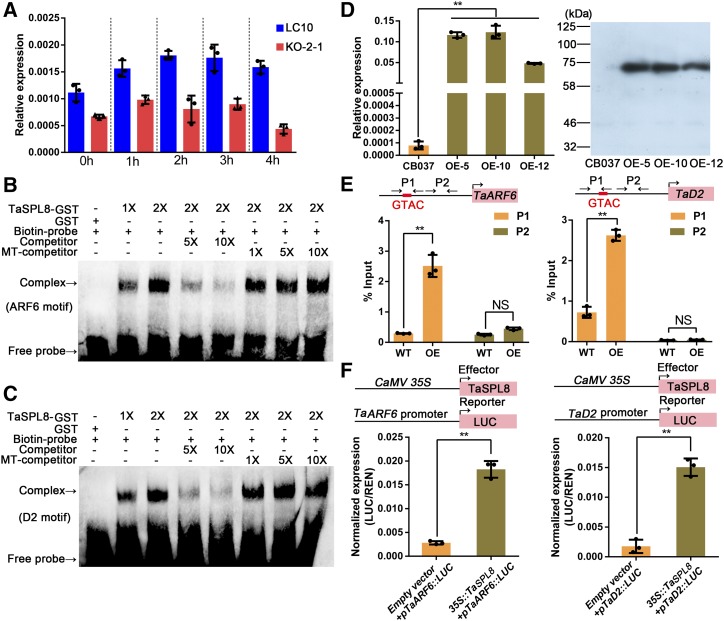
Among the DEGs downregulated in the TaSPL8 mutant, one gene TraesCS5B01G039800 (TaARF6) is the putative ortholog of Arabidopsis AtARF6 (Supplemental Fig. S6). ARFs are key players in mediating auxin-induced gene activation (Lau et al., 2008). In addition, we found that TraesCS5B01G039800 is upregulated in wild-type LC10 plants after exogenous auxin treatment, although it does not respond to auxin in the KO-2-1 (TaSPL8-2a/2b/2d) mutant (Fig. 6A). Therefore, we investigated whether TaSPL8 acts in auxin signaling by directly activating the expression of ARF genes. Similarly, we also tested whether TaSPL8 activates the expression of genes involved in BR biosynthesis because TraesCS3A01G103800 (TaD2), the putative ortholog of rice D2, is strongly downregulated in TaSPL8 mutants. Rice D2 encodes a cytochrome P450, which catalyzes the steps from 6-deoxoteasterone to 3-dehydro-6-deoxoteasterone and from teasterone to 3-dehydroteasterone in the late BR biosynthesis pathway. The results of our EMSAs revealed that binding of a TaSPL8 recombinant protein to the putative promoters of TaARF6 and TaD2 occurs in vitro (Fig. 6, B and C).
Figure 6.
TaSPL8 binds and activates TaARF6 and TaD2. A, TaARF expression analysis after auxin treatment (10−4 mol/L) in the wild-type LC10 plant and a TaSPL8 mutant (KO-2-1). Each bar in the graph corresponds to the mean value of three data points, shown by dots. B and C, EMSAs to assess the TaSPL8 binding to the promoter region of TaARF6 (B) and TaD2 (C) containing the GTAC motif. The symbols “+” and “−” indicate the presence and absence, respectively, of corresponding probes, proteins, or competitors. The terms “1X” and “2X” indicate amounts of protein in the reaction. The terms “5X” and “10X” indicate 5- and 10-fold molar excess, respectively, of competitor or mutated competitor (MT) probes relative to biotin-labeled probes. D, Generation of wheat transgenic lines overexpressing TaSPL8-Myc chimeric protein (overexpressing [OE] lines) in the CB037 genetic background. The level of TaSPL8 transcripts (left) and TaSPL8-Myc protein (right) were measured. RT-qPCR data were normalized to wheat TaACTIN and each bar in the graph corresponds to the mean value of three data points, shown by dots. Double asterisks represent statistically significant differences determined by the two-tailed Student’s t test at **P < 0.01. E, ChIP assays to detect TaSPL8 binding of TaARF6 and TaD2 in vivo. Arrows indicate the position (P1 and P2) of primer combinations used in qPCR after ChIP assays. “P1” indicates the region where GTAC is located and “P2” indicates the region without GTAC with more than 400-bp distance from P1. Each bar in the graph corresponds to the mean value of three data points, shown by dots. Double asterisks represent statistically significant differences determined by the two-tailed Student’s t test at **P < 0.01. WT, wild type. F, Transactivation assays with tobacco leaves infiltrated with different combinations of effector and reporter constructs. Individual values (black dots) and means (bars) of three independent biological replicates are shown. The double asterisks represent significant differences determined by the two-tailed Student’s t test at **P < 0.01. LUC/REN (Renilla luciferase) indicates the ratio of the firefly luciferase activity and the renilla reniformis activity.
To determine if the direct binding also takes place in vivo, we first generated a overexpression transgenic wheat line (OE lines) in which TaSPL8 was fused to Myc tag and driven by promoter of maize UBIQUTIN1 (ProUbi::TaSPL8-9MYC) and then performed chromatin immunoprecipitation (ChIP) assays with a Myc-specific antibody. We achieved three independent transgenic lines (OE-5, OE-10, and OE-12) with high levels of TaSPL8 mRNA and TaSPL8-9Myc protein (Fig. 6D). Compared to the wild type of reference (CB037), the OE lines were smaller and displayed fewer tiller numbers at the seedling stage, and decreased spike length and grain size (Supplemental Fig. S7). ChIP followed by qPCR was performed from the lamina joint tissue of the OE lines and wild-type CB037. The results indicated that TaSPL8-MYC enrichment of the GTAC core motif of TaARF6 and TaD2 in OE lines is significantly higher than that in wild-type CB037 and its enrichment in another region (outside GTAC core motif) is very low, suggesting that TaSPL8 specifically binds to the promoter region of TaARF6 and TaD2 with the GTAC core motif (Fig. 6E). The effect of TaSPL8 binding to the TaARF6 and TaD2 promoter was investigated using a transient expression assay. Specifically, a plasmid containing the promoter of TaARF6 (pTaARF6::LUC) or the TaD2 promoter (pTaD2::LUC) driving the expression of the luciferase (LUC) reporter gene was cotransformed in tobacco (Nicotiana tabacum) leaves with the effector plasmid (35S::TaSPL8) expressing TaSPL8. The results indicated that TaSPL8 can activate the expression of the reporter for both promoters (Fig. 6F). Overall, these findings indicate that TaSPL8 directly binds to the promoter of TaARF6 and TaD2 in the region containing the GTAC motif, which is also associated with their transcriptional activation. Therefore, we can envisage that one of the mechanisms underlying TaSPL8-mediated regulation of lamina joint development involves the direct activation of key genes of auxin signaling and BR biogenesis.
DISCUSSION
TaSPL8 and Its Homolog LG1 in Maize and Rice Could Be a Robust Target for Plant Architecture Improvement in Grass
Erect leaves improve light interception and biomass production under field conditions, resulting in increased grain yield in grass crops (Sakamoto et al., 2006). The erect-leaf phenotype of a rice BR-deficient mutant, osdwarf4-1, is associated with enhanced grain yields under dense planting populations, even without extra fertilizer (Sakamoto et al., 2006). Therefore, it can be assumed that a compact plant architecture mediated by erect leaves is also an important goal for wheat breeding, but no potential target genes for breeding programs have been identified to date in wheat. This study provides information about the role and mechanisms of TaSPL8, which encodes an SPL-like transcription factor, in controlling leaf angle by regulating lamina joint development. Lamina joint formation is impaired in TaSPL8 KO mutants, resulting in erect leaves and a more compact architecture. Our findings suggest that TaSPL8 is an important regulator of compact architecture in wheat, and therefore, could be used as a target to achieve elite wheat cultivars with erect leaves through genetic engineering and molecular breeding.
TaSPL8 plays a similar function in lamina joint development as LG1 in maize and rice. The mutation of LG1 in maize and rice also resulted in an erect leaf phenotype with a complete loss of the auricle and ligule (Moreno et al., 1997; Lee et al., 2007). Although the contribution of maize and rice LG1 to potential yield in high density planting is still unknown, we speculated that this gene might be a robust target for compact plant architecture in grass, considering the highly conserved function in lamina joint development. However, we have to consider the functional variation among LG1 genes of maize, rice, and wheat. Beside affecting ligule development, OsLG1 regulates panicle closure or spreading by controlling cell proliferation at the panicle pulvinus (Ishii et al., 2013; Zhu et al., 2013). ZmLG1 also controls the branch angle of the tassel and its KO mutants result in a more compact tassel than their wild-type counterparts (Bai et al., 2012). Different from OsLG1 and ZmLG1, TaSPL8 does not affect the spike structure, while directly or indirectly leading to the alteration of spike numbers. TaSPL8 mutants show increased spike numbers especially under dense planting condition. This phenotype may be due to erect leaves that allow greater penetration of light to the lower leaves, thereby improving canopy photosynthesis, which, in turn, improves the formation of tillers or the ability to generate a spike. A similar phenotype was observed in the osdwarf4-1 rice mutant with erect leaves due to BR deficiency (Sakamoto et al., 2006). Alternatively, TaSPL8 may directly regulate tiller and spike formation. TaSPL8 is highly expressed both in the lamina joint and young spikes and overexpressing plants show a clear reduction in tiller number and spike size. However, an overexpression transgenic line driven by the Ubi promoter actually shows an ectopic effect from the early growth stage. Ubiquitous overexpression may affect the natural function of a gene/protein, for example by altering transcription factor binding specificity when binding is sensitive to doses, or by altering the stoichiometric composition of a multiprotein complex, thus affecting its possible activity and specificity. Therefore, ubiquitous overexpression mutants are not the best for judging a gene’s function, because pleiotropic and “artificial” effects are more likely to be observed. The specific upregulation of TaSPL8 in spikes or tillers will help us to understand its biological function.
A wild species can be changed to a new form to meet human needs through crop domestication. OsLG1 has experienced substantial artificial selection during rice domestication (Ishii et al., 2013; Zhu et al., 2013). The selection of a single-nucleotide polymorphism residing in the cis-regulatory region of the OsLG1 gene was responsible for the transition from a spread panicle typical of ancestral wild rice to the compact panicle of present cultivars during rice domestication (Zhu et al., 2013). This selection led to the reduced expression of OsLG1 specifically at the panicle pulvinus while maintaining normal levels during ligule formation (Zhu et al., 2013). In wheat, we found that the level of TaSPL8 mRNA in the lamina joint is higher in modern cultivars with larger lamina joints (n = 49) compared to those with smaller lamina joints (n = 29; Fig. 4J), indicating that TaSPL8 is also selected for leaf angle during wheat breeding. However, we did not find sequence variation in the cis-regulatory region of TaSPL8 among these cultivars. Hence, genetic variation in possible distal regulatory elements or epigenetic variation in the TaSPL8 promoter may account for the observed difference in the transcript level.
TaSPL8-2D Is a Major Contributor to Erect Leaf in Wheat
Common wheat is an allohexaploid (BBAADD) derived from a hybridization between a cultivated form of allotetraploid wheat Triticum turgidum (BBAA) and diploid goat grass A. tauschii (DD). T. turgidum originates from the hybridization and polyploidization of the diploid species T. urartu (AA) and A. speltoides (SS genomes, most probably the donor of the B genome). The combination and interaction of subgenomes in the allohexaploid give rise to phenotypic variation during wheat evolutionary history. The evolution of duplicated gene loci in allopolyploid plants contribute either to a favorable effect of extra gene dosage or to built-up asymmetric expression divergence: only one genome is active, whereas the homoeoalleles on the other genome(s) are either eliminated or partially or completely suppressed by genetic or epigenetic means (Feldman et al., 2012). However, there is no evidence showing how asymmetric expression divergence affects the trait. We found that the expression level of homoeoallele TaSPL8-2D was higher in the lamina joint compared to the expression level of TaSPL8-2A and TaSPL8-2B. Observations from single, double, and triple mutants also corroborated the information that the TaSPL8-2D homeolog provides the major contribution for the lamina joint length phenotype and TaSPL8-2B affects lamina joint length to a greater extent than TaSPL8-2A. These data expand our knowledge about genome asymmetry during allopolyploid wheat evolution and suggest a possible mechanism and evolutionary advantage of this divergence, but addressing this requires further work.
The Utilization of TaSPL8 for Compact Plant Architecture in Wheat Breeding
Simultaneous mutations of all three TaSPL8 homeologs lead to the absence of the lamina joint, sometimes resulting in flag leaves wrapping around the spikes, which blocks spike extension. Thus, potential routes for TaSPL8 utilization in wheat breeding include: (1) using a single mutation of TaCAP-2D or TaCAP-2B to obtain a shorter lamina joint without affecting spike extension; and (2) using alleles associated with lower expression of TaSPL8 and shorter lamina joints, which could be used in marker-assisted selection to fine-tune lamina joint length and leaf angle.
TaSPL8 Modulates Leaf Angle through Auxin and BR Signaling
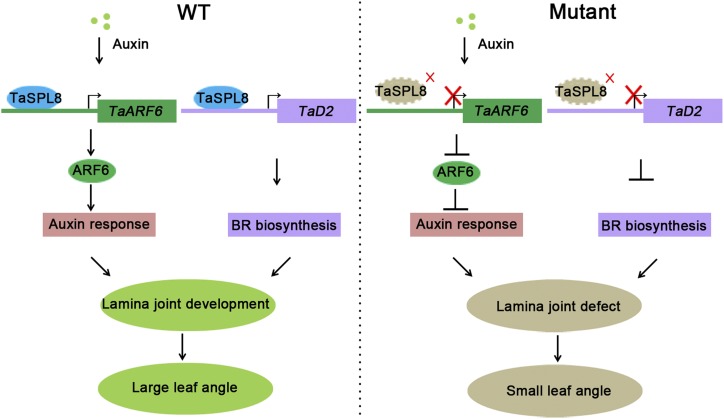
LG1 orthologs in maize, rice, and wheat function as key regulators of lamina joint development. ZmLG1 was found to function downstream of maize TEOSINTE BRANCHED 1/CYCLOIDEA/PCF transcription factor (Lewis et al., 2014) and to regulate a series of downstream genes such as PIN1 and gene encoding for a homeobox-transcription factor 48, which were identified by comparing transcript accumulation between an lg1 mutant and wild type in the preligule region (Johnston et al., 2014). Our study provides evidence that TaSPL8 affects lamina joint development by directly activating the expression of genes involved in auxin and BR pathways. Previous findings indicated that these hormones and various transcription factors contribute to cell division and elongation at the lamina joint region (Sakamoto et al., 2006; Bian et al., 2012; Zhao et al., 2013). Here, we have identified the mechanistic link between one transcription factor and the auxin and BR pathways, which are involved in lamina joint development and leaf architecture. Specifically, we found that TaSPL8 directly binds and activates TaARF6, which is involved in the auxin signaling pathway, and TaD2, which regulates BR biosynthesis (Fig. 7). Auxin is essential for cell elongation in nearly all developmental processes. The two Arabidopsis TaARF6 orthologs, ARF6 and ARF8, are required for cell elongation in the inflorescence stem, stamen filament, stigmatic papillae, and hypocotyls, and in anther dehiscence (Nagpal et al., 2005; Oh et al., 2014; Liu et al., 2018a). TaARF6 may similarly act as a regulator of cell elongation in the lamina joint. This scenario is supported by findings that other members of the ARF gene family regulate leaf angle. OsARF19-overexpressing rice lines show multiple phenotypes, including increased leaf angles (Zhang et al., 2015), and an OsARF11 loss-of-function line exhibits a reduced angle of the flag leaves (Sakamoto et al., 2013). Similarly, rice mutants with impaired expression of ebisu dwarf, the ortholog of wheat TaD2, are deficient in BR biogenesis and have erect leaves (Hong et al., 2003). This strongly supports our findings that another form of TaSPL8-mediated regulation of lamina joint development occurs through transcriptional activation of TaD2 and the subsequent increase in BR biosynthesis. Although rice BR-deficient mutants are usually dwarfed with short internodes, TaSPL8 mutants do not exhibit other phenotypic alterations except for lamina joint development. We speculate that high expression of TaSPL8 in lamina joint tissue specifically regulates BR biogenesis in this region. Our results indicate that TaSPL8 also affects the expression of genes involved in cell elongation and expansion. For example, we found that 48 genes for expansins are downregulated in the TaSPL8-2a/2b/2d mutant, and TaSPL8 can bind to the GTAC motif of the promoters of four cell expansin genes in vitro (Supplemental Fig. S5). This suggests that TaSPL8 regulates lamina joint development by directly targeting expansin genes. An alternative and not mutually exclusive possibility is that TaSPL8 regulates expansin genes through auxin signaling, because cell elongation genes are also responsive to auxin (Pacheco-Villalobos et al., 2016).
Figure 7.
Molecular model for the regulation of wheat lamina joint development by TaSPL8. TaSPL8 directly binds to the TaARF6 and TaD2 promoters to activate their expression. TaARF6 mediates the auxin signaling pathway, whereas TaD2 is involved in BR biogenesis. Both hormones are well known for regulating cell elongation, lamina joint development, and leaf angle. WT, wild type.
In conclusion, this study reveals the mechanisms that link one member of the SPL transcription factor family with hormones in regulating lamina joint development, in that TaSPL8 acts upstream of genes involved in the regulation of lamina joint cell elongation, such as TaARF6 and TaD2, which mediate the auxin signaling pathway and BR biosynthesis, respectively. In addition, our results identify additional genes that may function in lamina joint development (e.g. expansin). These findings will be useful for programs aimed at manipulating wheat architecture for improved grain yield.
MATERIALS AND METHODS
Plant Materials
Names and geographical origins of all wheat (Triticum aestivum) cultivars used in this study are listed in Supplemental Table S2. Plant materials used for visible phenotypic, microscopic, and molecular analyses were grown in a greenhouse at a relative humidity of 75% and 26°C/20°C day/night temperatures, with a light intensity of 3,000 lx (Master GreenPower CG T 400W E40; Philips). Wheat cultivars used for seed reproduction and phenotypic analysis were grown in the experimental field of China Agricultural University in Beijing (39°57ʹN, 116°17ʹE), Yangling in the Shaanxi province (34°16'N, 108°04'E). Lamina joints for gene expression analysis were harvested at the heading stage. For each sample, the lamina joint from three different planting pots was harvested, representing three biological replicates. All samples were immediately frozen in liquid nitrogen and stored at −80°C. The natural wheat population consisted of 192 wheat cultivars including Chinese landraces (111), Chinese modern cultivars (42), and foreign cultivars from other countries in Europe, Asia, America, and Australia (39).
Phenotypic Analysis
The length of the lamina joint and auricle were measured as the widest section between the leaf blade and the sheath with a Vernier caliper. Flag leaf angle was measured using a protractor between the stem under the spike and the flag leaf midrib. Plant height and the length of the main spike were measured before harvesting, and 10 main spikes were bagged at the same time to measure fertile grain number per spike and grain weight per 1,000 grains. Spike number per plant was calculated as the average value of 10 plants. A randomized complete block design with three replicates was employed for phenotyping in open field experiments.
Mapping of TaSPL8
Fine mapping was performed with an F2 and F2-derived F3 population from a cross between cpa and LC10. Primers used for fine mapping were designed based on the sequence of Chinese Spring (IWGSCv1; http://www.wheatgenome.org/) and Aegilops tauschii (Luo et al., 2017) and listed in Supplemental Table S5. NILs in the cpa genetic background containing the LC10 allele (NILLC10) and NILs in the LC10 genetic background containing the cpa allele (NILcpa) were generated through backcrossing LC10 × cpa F1 plants to LC10 and the cpa mutant for BC3, respectively. The introgression fragment between markers 2D-173 and LMC-3 was screened at each generation. The BC3F1 plant was self-crossed to generate NILLC10 and NILcpa, respectively.
Plasmid Construction and Transformation
For genome editing via CRISPR/Cas9, a sgRNA was designed in the first exon sequence of TaSPL8 using the E-CRISP Design Web site (http://www.e-crisp.org/E-CRISP/designcrispr.html). Two reverse complementary sgRNA sequences with the BsaI cohesive ends were synthesized. Double-stranded oligonucleotides were annealed by cooling from 100°C to room temperature, followed by insertion into the expression cassette of the pBUE411 vector (Xing et al., 2014). Plasmids were transformed into three wheat cultivars (CB037, LC10, and Bobwhite) using Agrobacterium-mediated (strain EHA105) transformation (Ishida et al., 2015). For TaSPL8 overexpression, the ORF of TaSPL8-2D from CB037 and the nine repeated Myc domain sequences were amplified and linked by PCR, then the TaSPL8-9Myc fragment was inserted into the pMWB122 vector (Wang et al., 2017) using SmaI and SpeI cloning sites to achieve the ProUbi::TaSPL8-9Myc construct. Plasmids were transformed into the wheat cultivar CB037 using Agrobacterium-mediated (strain EHA105) transformation (Ishida et al., 2015).
RT-qPCR Analysis
Total RNA was extracted from different tissues using TRIzol reagent (Takara) according to the manufacturer’s instructions and cDNA was synthesized by a reverse transcription kit (Vazyme Biotech). Wheat TaACTIN (TraesCS5B01G124100) was used as an internal control. For RT-qPCR, the reaction mixture was composed of the cDNA first-strand template, primers, and SYBR Green Mix (Takara) to a final volume of 10 μL. Reactions were performed using the CFX96 real-time system (Bio-Rad). For technical replicates, the RT-qPCR for each sample was replicated three times. Three biological replicates were performed (one replication is shown). The average values of 2−ΔCT were used to determine the differences in gene expression (https://assets.thermofisher.com/TFS-Assets/LSG/manuals/cms_040980.pdf).
ChIP-qPCR Assay
Wheat leaves were cross-linked in fixative buffer (1% [v/v] volume/volume formaldehyde) by vacuum infiltration. Cross-linking reactions were stopped by adding Gly at a final concentration of 0.17 M. Chromatin extracts were sonicated and precleared with Dynabeads protein A agarose (Merck Millipore) for 1 h. For immunoprecipitation, anti-Myc antibody (cat. no. ab9132; Abcam) was added and samples were incubated at 4°C overnight. The chromatin-antibody complex was recovered with Dynabeads protein A+G and beads were washed sequentially with low salt wash buffer, high salt wash buffer, LiCl wash buffer, and Tris-EDTA buffer for 5 min each (Yang et al., 2016). The protein/DNA complex was eluted with elution buffer (1% [w/v] weight/volume SDS and 0.1 m of NaHCO3). Cross-linking was reversed by adding NaCl at a final concentration of 200 mM, followed by incubation for 5 h at 65°C. Proteins in the resulting complex were removed by proteinase K at 45°C for 1 h. DNA was precipitated in the presence of three volumes of ethanol and one-tenth volume of 3-m sodium acetate at pH 5.2. Precipitated DNA was dissolved in 30 μL of 10-mm Tris-HCl at pH 7.5, and treated with RNase (DNase-free). Two independent ChIP assays were performed for each sample. qPCR was carried out using the CFX96 real-time system (Bio-Rad). ChIP values were normalized to their respective DNA input values, and the fold changes in concentration were calculated based on the relative enrichment in the overexpression lines compared with wild-type immunoprecipitates. Primer sequences used in the qPCR analysis are listed in Supplemental Table S5.
Transient Expression in Tobacco Leaves
Dual luciferase reporter assays were performed as described in Guan et al. (2014). A fragment corresponding to 1,800 bp upstream of the transcription start site of TaARF6 was PCR-amplified from LC10 genomic DNA and cloned into the pGreenII 0800-LUC vector (Hellens et al., 2005), generating the reporter pGreenII pTaARF::LUC and pTaD2::LUC plasmids. The ORF of TaSPL8 was cloned into the pUC18-35S vector generating the effector 35S::TaSPL8 plasmid. Young leaves of 4-week–old tobacco (Nicotiana tabacum) plants were coinfiltrated with Agrobacterium (strain GV3101) harboring the desired plasmids in the following combination: pTaARF6::LUC + pUC18-35S (empty vector), pTaARF6::LUC + 35S::TaSPL8, pTaD2::LUC + pUC18-35S (empty vector), and pTaD2::LUC + 35S::TaSPL8. The activities of firefly luciferase under the control of the TaARF6 (pTaARF6::LUC) or TaD2 (pTaD2::LUC) promoter and Renilla reniformis under the control of the 35S promoter (35S::REN) were quantified using the Dual-Luciferase Reporter Assay system (Promega) with the Synergy 2 Multi-Detection Microplate Reader (BioTek Instruments). Normalized data are presented as the ratio of luminescent signal intensity for the pTaARF6::LUC or pTaD2::LUC reporter and luminescent signal intensity for the internal control reporter 35S::REN from three independent biological samples.
Western Blot
Harvested leaves were ground in liquid nitrogen and proteins were extracted with 4× loading buffer (200 mm of Tris-HCl at pH 6.8, 40% [v/v] glycerol, 8% [w/v] SDS, and 20% [v/v] β-mercaptoethanol) at 100°C for 5 min. The solution was placed on ice for 5 min and centrifuged for 10 min at 12,000g at room temperature. Protein extracts were separated on 10% (v/v) SDS-PAGE and transferred to a polyvinylidene fluoride membrane (cat. no. 162-0177; Bio-Rad) for 60 min at 300 mA in temperature-controlled conditions. The membrane was blocked using 5% (w/v) defatted milk for 1 h. The membrane and primary antibody (1:1,000), dissolved in 5% (w/v) defatted milk and 1×Tris-buffered saline plus TWEEN 20 (TBST; 1×TBS with 0.1% [v/v] TWEEN 20), were incubated at 4°C overnight. After washing three times for 10 min with 1×TBST, the secondary antibody (1:3,000) was added in 5% (w/v) defatted milk-1×TBST solution and incubated for 1 h at room temperature, followed by three washing steps of 10 min with 1×TBST. Two-component reagent-clarity western ECL Substrate (cat. no. 170-5060; Bio-Rad) was used for detection. The signal was detected with x-ray film.
RNA-Seq
Total RNA was isolated as reported above for RT-qPCR analysis. The lamina joint from TaSPL8-2A/2B/2d mutant and LC10, and the corresponding part between the leaf blade and sheath from the TaSPL8-2a/2b/2d mutant at the heading stage, were collected. Three replicates of 10 plants each were used for each tissue type. Barcoded cDNA libraries were constructed using Poly-A Purification TruSeq library reagents (Illumina) and sequenced on HiSeq 2500 or NovaSeq platforms (Illumina). Approximately 10 Gb of high-quality 125-bp or 150-bp paired-end reads were generated from each library. The overall sequencing quality of the reads in each sample was evaluated with the software FastQC (v0.11.5; http://www.bioinformatics.babraham.ac.uk). Poor-quality bases were removed using Trimmomatic (v0.36; Bolger et al., 2014) in its paired-end model with the parameters “LEADING:3 TRAILING:3 HEADCROP:6 SLIDINGWINDOW:4:15 MINLEN:40.” The remaining reads were aligned to the wheat reference genome sequence IWGSCv1 (http://www.wheatgenome.org/) using the software TopHat2 v2.11 (https://ccb.jhu.edu/software/tophat/index.shtml) with the parameters “–b2-D 20–b2-R 3 -N 3–read-edit-dist 3” (Kim et al., 2013), and only the uniquely mapped reads were retained for the following analysis. The tool FeatureCounts (v1.5.2; http://subread.sourceforge.net/) with the parameters “–readExtension5 70–readExtension3 70 –p –C –s 0” was used to estimate the number of reads mapped to respective high-confidence genes annotated from IWGSCv1 in each sample. The Bioconductor package “DESeq2” (Love et al., 2014) was used to perform differential expression analysis between TaSPL8 mutant lines and the wild type, and only the genes with an absolute value of log2 (fold change) ≥ 1 and P value < 0.05 were considered as DEGs.
For GO analysis, cDNAs of high-confidence genes were aligned to the protein sequences of Oryza sativa and Arabidopsis (Arabidopsis thaliana) using the program BLASTX (https://blast.ncbi.nlm.nih.gov/Blast.cgi?LINK_LOC=blasthome&PAGE_TYPE=BlastSearch&PROGRAM=blastx) with an e-value cutoff of 1e-05. Only the best alignment with identity ≥ 50% and aligned length ≥ 30 amino acids were considered. GO annotations were obtained based on the orthologs with the priority: O. sativa > Arabidopsis. GO enrichment analysis of DEGs was implemented using the GOseq “R” package (https://www.rdocumentation.org/packages/goseq/versions/1.24.0/topics/goseq), which is based on the Wallenius noncentral hypergeometric distribution and can adjust for gene length bias (Young et al., 2010). GO terms with false discovery rate < 0.01 were considered as statistically significantly enriched.
EMSA
The full-length ORF sequence of TaSPL8 was cloned into the BamHI site of the pGEX6P-1 vector to produce the chimeric GST-TaSPL8 protein (primer sequences are listed in Supplemental Table S5). Expression of recombinant protein in Transetta (DE3) Escherichia coli (TransGen, Beijing) was induced with 0.1 mm of isopropyl-b-d-thiogalactopyranoside in Luria Bertani buffer overnight at 16°C. Cells were subsequently harvested, washed, and suspended in 30 mL of phosphate-buffered saline (137 mm of NaCl, 2.7 mm of KCl, 10 mm of Na2HPO4, and 2 mm of KH2PO4) containing 1 mm of PMSF and 1/2 a tablet of protease inhibitor cocktail (Roche). Then, cells were sonicated for 30 min and centrifuged at 13,000g for 45 min at 4°C. The supernatant was filtered through a 0.22-μm membrane into a 50-mL tube. The supernatant was mixed with 1 mL of GST MAG Agarose Beads (Novagen) and shaken overnight at 4°C. GST beads were washed four times with 5 mL of phosphate-buffered saline and the recombinant proteins were eluted by incubation at 4°C for more than 4 h with 50 mm of Tris-HCl at pH 8.0 supplemented with 10 mm of reduced glutathione. Protein concentration was determined using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific).
The biotin-probe was 5′ end-labeled with biotin. Double-stranded oligonucleotides used in the assays were annealed by cooling from 100°C to room temperature in annealing buffer (Guo et al., 2018). DNA-binding reactions were performed in 20 μL of 1×binding buffer (100 mm of Tris, 500 mm of KCl, and 10 mm of dithiothreitol at pH 7.5), 10% (v/v) glycerol, 0.5 mm of EDTA, 10 mm of ZnCl2, and 50-ng μL−1 poly dI-dC (Thermo Scientific). Competition analysis was done by adding 5- and 10- fold molar excess of the unlabeled DNA fragment (same sequence used for the labeled probe) to the binding reaction, 5 min before the labeled probe. After incubation at room temperature for 30 min, samples were loaded onto a 6% native polyacrylamide gel. Electrophoretic transfer to a nylon membrane and detection of the biotin-labeled DNA was performed with the Light Shift Chemiluminescent EMSA Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions.
Quantification of Endogenous IAA and BR
The lamina joint of the wild type and the TaSPL8-2a/2b/2d line were harvested at the heading stage. Quantification of endogenous IAA and BR were performed as described in Chen et al. (2012) and Ding et al. (2013). Each experiment was performed using three replicates.
Statistical Analysis
The statistical analyses, including Student’s t test and correlation, were performed with the software SPSS v21 (SPSS/IBM). All barcharts and boxplot results were graphically presented using the software Prism v6.0 (GraphPad).
Phylogenetic Analysis
For phylogenetic analysis, related protein sequences to TaARF6 and TaD2 in Arabidopsis and rice were selected by employing a National Center for Biotechnology Information (NCBI) BLASTp search (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins) using the nonredundant protein sequences database. Their corresponding accession numbers were shown in Supplemental Figure S6. The phylogenetic tree was generated by the “neighbor joining tree” method in MEGA5 software.
Primers for RT-qPCR and Vector Construction
All primers used in this research are listed in Supplemental Table S5.
Accession Numbers
Accession code: the genomic DNA sequence of TaSPL8 in LC10 has been deposited in GenBank with the accession no. MH765574. AUXIN RESPONSE FACTOR TaARF6 gene: TraesCS5B01G039800.1; CYP90D2 TaD2 gene: TraesCS3A01G103800.
RNA sequencing data are available at the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under accession no. SRP157960.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Schematic of genotype of LC10, cpa, NILLC10, and NILcpa.
Supplemental Figure S2. A fragment on chromosome 2D was deleted in the cpa mutant.
Supplemental Figure S3. CRISPR/Cas9 knock-out TaSPL8 mutants in CB037 and Bobwhite cultivars.
Supplemental Figure S4. Heat map for the DEGs involved in auxin and BR signaling and cell elongation in the wild type and TaSPL8 mutants.
Supplemental Figure S5. TaSPL8 in vitro binding of sequences containing the GTAC motif.
Supplemental Figure S6. Phylogenetic tree of TraesCS5B01G039800.1 (TaARF6) and TraesCS3A01G103800.1 (TaD2) with Arabidopsis or rice proteins.
Supplemental Figure S7. The phenotypes of TaSPL8-Myc overexpression (OE) and wild-type CB037 lines.
Supplemental Table S1. Morphological characterization of wild type and KO-2-1 (TaSPL8-2a/2b/2d) mutant plants.
Supplemental Table S2. The names and geographical origins of all wheat cultivars used in this research.
Supplemental Table S3. Differentially expressed transcripts in the Taspl8-2a/2b/2d mutant and the wild type in lamina joint tissue.
Supplemental Table S4. Differentially expressed transcripts in the Taspl8-2A/2B/2d mutant and the wild type in lamina joint tissue.
Supplemental Table S5. Primers and Gene IDs for sequences used in this study.
Acknowledgments
We thank Dr. Qijun Chen (China Agricultural University), Dr. Rentao Song (China Agricultural University), and Dr. Xingguo Ye (Chinese Academy of Agricultural Sciences) for providing the CRISPR-Cas9 vector (pBUE11), the transient expression vectors (pUC18-35S and pGreenII 0800-LUC), and the overexpression vector (pMWB122), respectively. We thank Prof. Mingcheng Luo (University of California, Davis) for providing the A. tauschii genome sequence. We thank Prof. Guihua Bai (Kansas State University) for critical reading of the article. We thank Dr. Yuqi Feng (Wuhan University) for assistance in IAA and BR quantification and Dr. Hongxia Li and Dr. Yu Wang (Northwest A&F University) for help in phenotypic measurements of yield-related traits. We also thank the International Wheat Genome Sequencing Consortium for providing the wheat reference sequence.
Footnotes
This work was supported by the National Key Research and Development Program of China (grant nos. 2016YFD0100801 and 2016YFD0101004).
Articles can be viewed without a subscription.
References
- Appels R, Eversole K, Feuillet C, Keller B, Rogers J, Stein N, Pozniak CJ, Stein N, Choulet F, Distelfeld A, et al. (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361: eaar7191. [DOI] [PubMed] [Google Scholar]
- Austin R, Ford M, Edrich J, Hooper B (1976) Some effects of leaf posture on photosynthesis and yield in wheat. Ann Appl Biol 83: 425–446 [Google Scholar]
- Bai F, Reinheimer R, Durantini D, Kellogg EA, Schmidt RJ (2012) TCP transcription factor, BRANCH ANGLE DEFECTIVE 1 (BAD1), is required for normal tassel branch angle formation in maize. Proc Natl Acad Sci USA 109: 12225–12230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai MY, Zhang LY, Gampala SS, Zhu SW, Song WY, Chong K, Wang ZY (2007) Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc Natl Acad Sci USA 104: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian H, Xie Y, Guo F, Han N, Ma S, Zeng Z, Wang J, Yang Y, Zhu M (2012) Distinctive expression patterns and roles of the miRNA393/TIR1 homolog module in regulating flag leaf inclination and primary and crown root growth in rice (Oryza sativa). New Phytol 196: 149–161 [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Chen S (1995) Brassinosteroid-induced rice lamina joint inclination and its relation to indole-3-acetic acid and ethylene. Plant Growth Regul 16: 189–196 [Google Scholar]
- Chen ML, Fu XM, Liu JQ, Ye TT, Hou SY, Huang YQ, Yuan BF, Wu Y, Feng YQ (2012) Highly sensitive and quantitative profiling of acidic phytohormones using derivatization approach coupled with nano-LC-ESI-Q-TOF-MS analysis. J Chromatogr B Analyt Technol Biomed Life Sci 905: 67–74 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. (2015) Plant expansins: Diversity and interactions with plant cell walls. Curr Opin Plant Biol 25: 162–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Mao LJ, Wang ST, Yuan BF, Feng YQ (2013) Determination of endogenous brassinosteroids in plant tissues using solid-phase extraction with double layered cartridge followed by high-performance liquid chromatography-tandem mass spectrometry. Phytochem Anal 24: 386–394 [DOI] [PubMed] [Google Scholar]
- Donald CM. (1968) The breeding of crop ideotypes. Euphytica 17: 385–403 [Google Scholar]
- Dong H, Zhao H, Li S, Han Z, Hu G, Liu C, Yang G, Wang G, Xie W, Xing Y (2018) Genome-wide association studies reveal that members of bHLH subfamily 16 share a conserved function in regulating flag leaf angle in rice (Oryza sativa). PLoS Genet 14: e1007323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Wu N, Fu J, Wang S, Li X, Xiao J, Xiong L (2012) A GH3 family member, OsGH3-2, modulates auxin and abscisic acid levels and differentially affects drought and cold tolerance in rice. J Exp Bot 63: 6467–6480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvick D. (2005) Genetic progress in yield of United States maize (Zea mays L.). Maydica 50: 193–202 [Google Scholar]
- El Baidouri M, Murat F, Veyssiere M, Molinier M, Flores R, Burlot L, Alaux M, Quesneville H, Pont C, Salse J (2017) Reconciling the evolutionary origin of bread wheat (Triticum aestivum). New Phytol 213: 1477–1486 [DOI] [PubMed] [Google Scholar]
- Feldman M, Levy AA, Fahima T, Korol A (2012) Genomic asymmetry in allopolyploid plants: Wheat as a model. J Exp Bot 63: 5045–5059 [DOI] [PubMed] [Google Scholar]
- Guan Q, Yue X, Zeng H, Zhu J (2014) The protein phosphatase RCF2 and its interacting partner NAC019 are critical for heat stress-responsive gene regulation and thermotolerance in Arabidopsis. Plant Cell 26: 438–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, Liu X, Sun F, Cao J, Huo N, Wuda B, Xin M, Hu Z, Du J, Xia R, et al. (2018) Wheat miR9678 affects seed germination by generating phased siRNAs and modulating abscisic acid/gibberellin signaling. Plant Cell 30: 796–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Ueguchi-Tanaka M, Shimizu-Sato S, Inukai Y, Fujioka S, Shimada Y, Takatsuto S, Agetsuma M, Yoshida S, Watanabe Y, et al. (2002) Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J 32: 495–508 [DOI] [PubMed] [Google Scholar]
- Hong Z, Ueguchi-Tanaka M, Umemura K, Uozu S, Fujioka S, Takatsuto S, Yoshida S, Ashikari M, Kitano H, Matsuoka M (2003) A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 15: 2900–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida Y, Tsunashima M, Hiei Y, Komari T (2015) Wheat (Triticum aestivum L.) transformation using immature embryos. Methods Mol Biol 1223: 189–198 [DOI] [PubMed] [Google Scholar]
- Ishii T, Numaguchi K, Miura K, Yoshida K, Thanh PT, Htun TM, Yamasaki M, Komeda N, Matsumoto T, Terauchi R, et al. (2013) OsLG1 regulates a closed panicle trait in domesticated rice. Nat Genet 45: 462–465, e1–e2 [DOI] [PubMed] [Google Scholar]
- Isidro J, Knox R, Clarke F, Singh A, DePauw R, Clarke J, Somers D (2012) Quantitative genetic analysis and mapping of leaf angle in durum wheat. Planta 236: 1713–1723 [DOI] [PubMed] [Google Scholar]
- Jang S, An G, Li HY (2017) Rice leaf angle and grain size are affected by the OsBUL1 transcriptional activator complex. Plant Physiol 173: 688–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston R, Wang M, Sun Q, Sylvester AW, Hake S, Scanlon MJ (2014) Transcriptomic analyses indicate that maize ligule development recapitulates gene expression patterns that occur during lateral organ initiation. Plant Cell 26: 4718–4732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu K, Maekawa M, Ujiie S, Satake Y, Furutani I, Okamoto H, Shimamoto K, Kyozuka J (2003) LAX and SPA: Major regulators of shoot branching in rice. Proc Natl Acad Sci USA 100: 11765–11770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong F, Zhang T, Liu J, Heng S, Shi Q, Zhang H, Wang Z, Ge L, Li P, Lu X, et al. (2017) Regulation of leaf angle by auricle development in maize. Mol Plant 10: 516–519 [DOI] [PubMed] [Google Scholar]
- Kropat J, Tottey S, Birkenbihl RP, Depège N, Huijser P, Merchant S (2005) A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc Natl Acad Sci USA 102: 18730–18735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S, Jürgens G, De Smet I (2008) The evolving complexity of the auxin pathway. Plant Cell 20: 1738–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer S, Hall BD, Mulaosmanovic E, Anderson SR, Nelson B, Smith S (2012) Morphological changes in parental lines of pioneer brand maize hybrids in the U.S. central corn belt. Crop Sci 52: 1033–1043 [Google Scholar]
- Lee EA, Tollenaar M (2007) Physiological basis of successful breeding strategies for maize grain yield. Crop Sci 47: S202–S215 [Google Scholar]
- Lee J, Park J-J, Kim SL, Yim J, An G (2007) Mutations in the rice liguleless gene result in a complete loss of the auricle, ligule, and laminar joint. Plant Mol Biol 65: 487–499 [DOI] [PubMed] [Google Scholar]
- Lewis MW, Bolduc N, Hake K, Htike Y, Hay A, Candela H, Hake S (2014) Gene regulatory interactions at lateral organ boundaries in maize. Development 141: 4590–4597 [DOI] [PubMed] [Google Scholar]
- Li D, Wang L, Wang M, Xu YY, Luo W, Liu YJ, Xu ZH, Li J, Chong K (2009) Engineering OsBAK1 gene as a molecular tool to improve rice architecture for high yield. Plant Biotechnol J 7: 791–806 [DOI] [PubMed] [Google Scholar]
- Li H, Jiang L, Youn JH, Sun W, Cheng Z, Jin T, Ma X, Guo X, Wang J, Zhang X, et al. (2013) A comprehensive genetic study reveals a crucial role of CYP90D2/D2 in regulating plant architecture in rice (Oryza sativa). New Phytol 200: 1076–1088 [DOI] [PubMed] [Google Scholar]
- Liu K, Li Y, Chen X, Li L, Liu K, Zhao H, Wang Y, Han S (2018a) ERF72 interacts with ARF6 and BZR1 to regulate hypocotyl elongation in Arabidopsis. J Exp Bot 69: 3933–3947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Xu H, Liu G, Guan P, Zhou X, Peng H, Yao Y, Ni Z, Sun Q, Du J (2018b) QTL mapping of flag leaf-related traits in wheat (Triticum aestivum L.). Theor Appl Genet 131: 839–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Yeh CT, Tang HM, Nettleton D, Schnable PS (2012) Gene mapping via bulked segregant RNA-Seq (BSR-Seq). PLoS One 7: e36406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bucio J, Hernández-Abreu E, Sánchez-Calderón L, Nieto-Jacobo MF, Simpson J, Herrera-Estrella L (2002) Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol 129: 244–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo MC, Gu YQ, Puiu D, Wang H, Twardziok SO, Deal KR, Huo N, Zhu T, Wang L, Wang Y, et al. (2017) Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 551: 498–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Zheng J, Huang R, Huang Y, Wang H, Jiang L, Fang X (2016) Phytohormones signaling and crosstalk regulating leaf angle in rice. Plant Cell Rep 35: 2423–2433 [DOI] [PubMed] [Google Scholar]
- Mantilla-Perez MB, Salas Fernandez MG (2017) Differential manipulation of leaf angle throughout the canopy: Current status and prospects. J Exp Bot 68: 5699–5717 [DOI] [PubMed] [Google Scholar]
- Moreno MA, Harper LC, Krueger RW, Dellaporta SL, Freeling M (1997) liguleless1 encodes a nuclear-localized protein required for induction of ligules and auricles during maize leaf organogenesis. Genes Dev 11: 616–628 [DOI] [PubMed] [Google Scholar]
- Nagpal P, Ellis CM, Weber H, Ploense SE, Barkawi LS, Guilfoyle TJ, Hagen G, Alonso JM, Cohen JD, Farmer EE, et al. (2005) Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132: 4107–4118 [DOI] [PubMed] [Google Scholar]
- Ning J, Zhang B, Wang N, Zhou Y, Xiong L (2011) Increased leaf angle1, a Raf-like MAPKKK that interacts with a nuclear protein family, regulates mechanical tissue formation in the Lamina joint of rice. Plant Cell 23: 4334–4347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu J-Y, Bai M-Y, Arenhart RA, Sun Y, Wang Z-Y (2014) Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife 3: e03031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco-Villalobos D, Díaz-Moreno SM, van der Schuren A, Tamaki T, Kang YH, Gujas B, Novak O, Jaspert N, Li Z, Wolf S, et al. (2016) The effects of high steady state auxin levels on root cell elongation in brachypodium. Plant Cell 28: 1009–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendleton JW, Smith GE, Winter SR, Johnston TJ (1968) Field investigations of the relationships of leaf angle in corn (Zea mays L.) to grain yield and apparent photosynthesis1. Agron J 60: 422–424 [Google Scholar]
- Ruan W, Guo M, Xu L, Wang X, Zhao H, Wang J, Yi K (2018) An SPX-RLI1 module regulates leaf inclination in response to phosphate availability in rice. Plant Cell 30: 853–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Morinaka Y, Ohnishi T, Sunohara H, Fujioka S, Ueguchi-Tanaka M, Mizutani M, Sakata K, Takatsuto S, Yoshida S, et al. (2006) Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat Biotechnol 24: 105–109 [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Morinaka Y, Inukai Y, Kitano H, Fujioka S (2013) Auxin signal transcription factor regulates expression of the brassinosteroid receptor gene in rice. Plant J 73: 676–688 [DOI] [PubMed] [Google Scholar]
- Sénéchal F, Wattier C, Rustérucci C, Pelloux J (2014) Homogalacturonan-modifying enzymes: Structure, expression, and roles in plants. J Exp Bot 65: 5125–5160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair TR, Sheehy JE (1999) Erect leaves and photosynthesis in rice. Science 283: 1455 [Google Scholar]
- Tanaka A, Nakagawa H, Tomita C, Shimatani Z, Ohtake M, Nomura T, Jiang C-J, Dubouzet JG, Kikuchi S, Sekimoto H, et al. (2009) BRASSINOSTEROID UPREGULATED1, encoding a helix-loop-helix protein, is a novel gene involved in brassinosteroid signaling and controls bending of the lamina joint in rice. Plant Physiol 151: 669–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada K, Marumo S, Ikekawa N, Morisaki M, Mori K (1981) Brassinolide and homobrassinolide promotion of lamina inclination of rice seedlings. Plant Cell Physiol 22: 323–325 [Google Scholar]
- Wang K, Liu H, Du L, Ye X (2017) Generation of marker-free transgenic hexaploid wheat via an Agrobacterium-mediated co-transformation strategy in commercial Chinese wheat varieties. Plant Biotechnol J 15: 614–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhang H, Li Y, Zhang Z, Li L, Liu B (2016) Transcriptome asymmetry in synthetic and natural allotetraploid wheats, revealed by RNA-sequencing. New Phytol 209: 1264–1277 [DOI] [PubMed] [Google Scholar]
- Wu Q, Chen Y, Fu L, Zhou S, Chen J, Zhao X, Zhang D, Ouyang S, Wang Z, Li D, et al. (2015) QTL mapping of flag leaf traits in common using an integrated high-density SSR and SNP genetic linkage map. Euphytica 208: 337–351 [Google Scholar]
- Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14: 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamuro C, Ihara Y, Wu X, Noguchi T, Fujioka S, Takatsuto S, Ashikari M, Kitano H, Matsuoka M (2000) Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12: 1591–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Liu X, Xin M, Du J, Hu Z, Peng H, Rossi V, Sun Q, Ni Z, Yao Y (2016) Genome-wide mapping of targets of maize histone deacetylase HDA101 reveals its function and regulatory mechanism during seed development. Plant Cell 28: 629–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol 11: R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L-Y, Bai M-Y, Wu J, Zhu J-Y, Wang H, Zhang Z, Wang W, Sun Y, Zhao J, Sun X, et al. (2009) Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 21: 3767–3780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Wang S, Xu Y, Yu C, Shen C, Qian Q, Geisler M, Jiang A, Qi Y (2015) The auxin response factor, OsARF19, controls rice leaf angles through positively regulating OsGH3-5 and OsBRI1. Plant Cell Environ 38: 638–654 [DOI] [PubMed] [Google Scholar]
- Zhao S-Q, Hu J, Guo L-B, Qian Q, Xue H-W (2010) Rice leaf inclination2, a VIN3-like protein, regulates leaf angle through modulating cell division of the collar. Cell Res 20: 935–947 [DOI] [PubMed] [Google Scholar]
- Zhao S-Q, Xiang J-J, Xue H-W (2013) Studies on the rice LEAF INCLINATION1 (LC1), an IAA-amido synthetase, reveal the effects of auxin in leaf inclination control. Mol Plant 6: 174–187 [DOI] [PubMed] [Google Scholar]
- Zhou LJ, Xiao LT, Xue HW (2017) Dynamic cytology and transcriptional regulation of rice lamina joint development. Plant Physiol 174: 1728–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Tan L, Fu Y, Liu F, Cai H, Xie D, Wu F, Wu J, Matsumoto T, Sun C (2013) Genetic control of inflorescence architecture during rice domestication. Nat Commun 4: 2200. [DOI] [PMC free article] [PubMed] [Google Scholar]