Insertion of a nonfunctional DcMYB7 duplication or transposons into the promoter region of DcMYB7 in nonpurple carrots blocks anthocyanin biosynthesis and modification.
Abstract
The original domesticated carrots (Daucus carota) are thought to have been purple, accumulating large quantities of anthocyanins in their roots. A quantitative trait locus associated with anthocyanin pigmentation in purple carrot roots has been identified on chromosome 3 and includes two candidate genes, DcMYB6 and DcMYB7. Here, we characterized the functions of DcMYB6 and DcMYB7 in carrots. Overexpression of DcMYB7, but not DcMYB6, in the orange carrot ‘Kurodagosun’ led to anthocyanin accumulation in roots. Knockout of DcMYB7 in the solid purple (purple periderm, phloem, and xylem) carrot ‘Deep Purple’ using the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 system resulted in carrots with yellow roots. DcMYB7 could activate the expression of its DcbHLH3 partner, a homolog of the anthocyanin-related apple (Malus × domestica) bHLH3, and structural genes in the anthocyanin biosynthetic pathway. We determined that the promoter sequence of DcMYB7 in nonpurple carrots was interrupted either by DcMYB8, a nonfunctional tandem duplication of DcMYB7, or by two transposons, leading to the transcriptional inactivation of DcMYB7 in nonpurple carrot roots. As a result, nonpurple carrots fail to accumulate anthocyanins in their roots. Our study supports the hypothesis that another genetic factor suppresses DcMYB7 expression in the phloem and xylem of purple peridermal carrot root tissues. DcMYB7 also regulated the glycosylation and acylation of anthocyanins by directly activating DcUCGXT1 and DcSAT1. We reveal the genetic factors conditioning anthocyanin pigmentation in purple versus nonpurple carrot roots. Our results also provide insights into the mechanisms underlying anthocyanin glycosylation and acylation.
Carrot (Daucus carota ssp. sativus; 2n = 2x = 18) provides rich health-promoting nutrients to humans. Carrots are classified into two groups according to exact botanical determination: the carotene group (variety sativus) and the anthocyanin group (variety atrorubens; Kammerer et al., 2004). Carotene group members, also known as nonpurple carrots, accumulate massive amounts of carotenoids in their roots (Clotault et al., 2008; Arscott and Tanumihardjo, 2010); anthocyanin group members, also known as purple carrots, accumulate high levels of anthocyanins in their roots (Kammerer et al., 2004; Montilla et al., 2011). Anthocyanins are water-soluble flavonoid compounds and confer red, blue, and purple pigmentation to plants. In addition to their important roles in plants, they are also beneficial human nutrients (He and Giusti, 2010).
Nonpurple carrots are considered to have arisen from purple carrots that acquired mutations (Banga, 1963; Arscott and Tanumihardjo, 2010; Iorizzo et al., 2013, 2016; Leja et al., 2013). In addition, purple carrot root pigmentation extensively varies across different carrot genotypes, ranging from the purple peridermal carrot type (purple periderm but nonpurple phloem and xylem) to the solid purple carrot type (purple periderm, phloem, and xylem). The genetic control of anthocyanin pigmentation in purple carrots has been investigated (Simon, 1996; Yildiz et al., 2013; Cavagnaro et al., 2014; Xu et al., 2014, 2016, 2017; Kodama et al., 2018; Iorizzo et al., 2019). Two genes that condition the anthocyanin pigmentation of carrot roots from different genetic backgrounds, P1 and P3, have been identified and genetically mapped within 28.2- and 12-centimorgan regions, respectively, on chromosome 3, supporting the theory of two independent mutation and human selection events during the domestication of purple carrots (Cavagnaro et al., 2014). P1 controls anthocyanin pigmentation in carrots with purple roots but nonpurple petioles, whereas P3 is an inherited dominant gene conditioning anthocyanin biosynthesis in carrots with purple roots (both solid purple and purple peridermal carrot types) and petioles (Cavagnaro et al., 2014).
Anthocyanin biosynthesis involves a number of structural and regulatory genes in many plant species. The structural genes, encoding enzymes that directly catalyze the production of anthocyanin, are regulated by transcription factors. The expression levels of the previously tested structural genes correlate with anthocyanin biosynthesis in carrots and none of them mapped to the same location as P1 or P3, indicating that these structural genes are not the key genetic factors controlling anthocyanin pigmentation in purple carrots (Yildiz et al., 2013; Cavagnaro et al., 2014; Xu et al., 2014). Thus, genetic mutations that cause pigmentation changes in carrot root may occur in regulatory genes. In all the plant species studied to date, the anthocyanin biosynthetic structural genes are directly regulated by R2R3-MYB, basic helix-loop-helix (bHLH), and WD-repeat proteins, in the form of the MBW complex (Espley et al., 2007; Chagné et al., 2013; Jin et al., 2016). Mutations in R2R3-MYB often lead to aberrant anthocyanin biosynthesis and result in color changes. The mutational types include nucleotide sequence changes, transposon or microsatellite insertion, and methylation (Kobayashi et al., 2004; Morita et al., 2006; Espley et al., 2009; Wang et al., 2013b).
DcMYB6, corresponding to DCAR_000385 in the carrot genome, encodes an R2R3-MYB that can induce anthocyanin biosynthesis in Arabidopsis (Arabidopsis thaliana; Xu et al., 2017). Very recently, DcMYB6 was anchored to the P3 region (Iorizzo et al., 2019). Using phylogenetic analyses, these authors also identified five additional MYB transcription factors of the anthocyanin biosynthesis-related subgroup of MYBs, DcMYB7 (DCAR_010745), which was designated DcMYB113-like in the carrot genome (Iorizzo et al., 2016), DcMYB8 (DCAR_010746), DcMYB9 (DCAR_010747), DcMYB10 (DCAR_010749), and DcMYB11 (DCAR_010751), within the P3 genomic region (Iorizzo et al., 2019). Among these R2R3-MYBs, the expression of DcMYB7 was consistently associated with purple root pigmentation in all the purple carrots tested, while the expression of DcMYB6 was only associated with anthocyanin pigmentation in the solid purple carrots. The authors speculated that DcMYB6 controls anthocyanin pigmentation in the inner root tissues while DcMYB7 determines anthocyanin pigmentation in the root outer phloem. The expression levels of the other four R2R3-MYBs were not associated with anthocyanin pigmentation in the roots of purple carrots according to the transcriptome analysis. Previously, we also obtained DcMYB7-knockout (originally DcMYB113-like-knockout) ‘Deep Purple’ (DPP) carrot plantlets (Xu et al., 2019). To date, however, the functions of DcMYB6 and DcMYB7 have still not been systematically studied in carrots, and the genetic mechanism behind pigmentation-related mutations in purple carrot versus nonpurple carrot roots is still unclear.
In purple carrots, cyanidin-based anthocyanins are almost exclusively responsible for the purple pigment, while trace amounts of derivatives of peonidin- and pelargonidin-based anthocyanins are also present in some purple carrot cultivars (Kammerer et al., 2004; Montilla et al., 2011). In carrot, UCGalT1 catalyzes the first glycosylation step of anthocyanidins, generating stable anthocyanins (Xu et al., 2016). Anthocyanins further undergo several glycosylation and acylation steps (Glässgen et al., 1998; Cavagnaro et al., 2014). These changes increase their stability and water solubility. Owing to their stable and water-soluble characteristics, as well as their health-promoting properties, anthocyanins from purple carrots serve as excellent natural colors in beverages, candies, and ice cream (Netzel et al., 2007). Progress in understanding the genetic control of anthocyanin glycosylation and acylation in purple carrot has been reported (Cavagnaro et al., 2014). Several quantitative trait loci (QTLs), distributed across six chromosomes, were proposed by Cavagnaro et al. (2014) as being associated with anthocyanin glycosylation and acylation. However, the genes conditioning anthocyanin glycosylation and acylation in purple carrots have still not been identified and characterized.
In this study, we characterized the functions of DcMYB6 and DcMYB7 using stable plant transformations and clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9)-based genome editing. The mechanism of purple pigment loss in nonpurple carrot roots and the roles of DcMYB7 in anthocyanin glycosylation and acylation were also analyzed. Our results not only improve our understanding of the molecular mechanism behind the origin of the nonpurple root phenotype during carrot domestication but are also valuable for breeding programs aimed at manipulating and modifying anthocyanin biosynthesis in carrot and other plant species.
RESULTS
Differential Expression Profiles of DcMYB6 and DcMYB7 in Purple and Nonpurple Carrot Roots
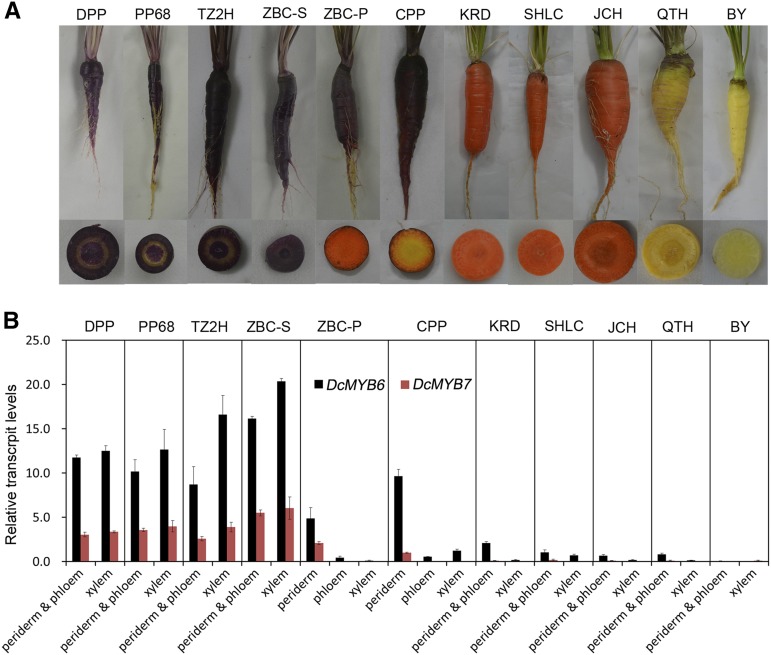
The transcript levels of DcMYB6 and DcMYB7 were detected in different root tissues of six purple carrot and five nonpurple carrot cultivars (Fig. 1A). DPP, ‘Purple 68’ (PP68), and ‘Tianzi2hao’ (TZ2H) are solid purple carrot cultivars, accumulating anthocyanins in root periderm, phloem, and xylem tissues. ‘Zibacun’ (ZBC) is a carrot cultivar with two types of roots: ZBC-S is a solid purple carrot, accumulating anthocyanins across the entire root section, while ZBC-P is a purple peridermal carrot, accumulating anthocyanins in root peridermal tissue. ‘Cosmic Purple’ (CPP) is also a carrot cultivar with purple root peridermal tissue. ‘Kurodagosun’ (KRD), ‘Sanhongliucun’ (SHLC), and ‘Junchuanhong’ (JCH) are orange carrot cultivars, while ‘Qitouhuang’ (QTH) and ‘Baiyu’ (BY) are yellow carrot cultivars.
Figure 1.
Expression patterns of DcMYB6 and DcMYB7 in carrot roots. A, Four-month-old purple and nonpurple carrots of the 11 cultivars used in this study. Cultivar abbreviations are defined in “Results.” Separate images are shown as a composite for comparison. B, Relative transcript levels of DcMYB6 and DcMYB7 in different root tissues of 11 purple and nonpurple carrot cultivars. Data are means of biological triplicate RT-qPCRs ± sd.
Reverse transcription quantitative PCR (RT-qPCR) analyses indicated that DcMYB6 and DcMYB7 were coexpressed in purple carrot roots (Fig. 1B). DcMYB6 showed high transcript levels in the anthocyanin-pigmented tissues of purple carrot roots. The transcript levels of DcMYB6 were relatively lower but still detectable in the nonpurple root tissues of purple and nonpurple carrot cultivars. DcMYB7 was positively correlated with anthocyanin pigmentation in carrot roots, showing high transcript levels in the anthocyanin-pigmented root tissues of the purple carrot cultivars but almost undetectable transcript levels in the nonpurple root tissues of purple and nonpurple carrot cultivars. Compared with DcMYB7, DcMYB6 displayed higher transcript levels in all the root tissues of purple and nonpurple carrot cultivars.
DcMYB6 and DcMYB7 Functional Assays
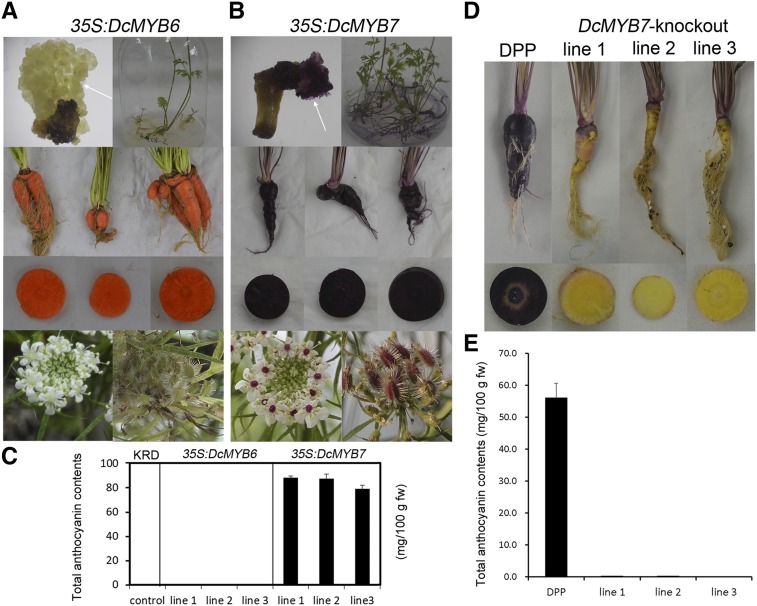
In our previous study, the heterologous overexpression of DcMYB6 in Arabidopsis induced anthocyanin accumulation (Xu et al., 2017). Here, DcMYB6 was introduced into the orange carrot KRD under the control of the cauliflower mosaic virus (CaMV) 35S promoter to investigate its function in the homologous system. No purple pigmentation was observed at either the callus or plantlet stage after transformation (Fig. 2A). The 4-month-old 35S:DcMYB6 transgenic carrot lines had no purple pigmentation in their roots or petioles. After bolting, 35S:DcMYB6 transgenic carrots generated no purple-pigmented inflorescences or seeds (Fig. 2A).
Figure 2.
Functional detection of DcMYB6 and DcMYB7 in carrots. A and B, Overexpression of DcMYB6 (A) and DcMYB7 (B) in orange KRD carrots. Separate images are shown as composites for comparison. C, Total anthocyanin contents in roots of untransformed, 35S:DcMYB6, and 35S:DcMYB7 transgenic KRD carrots. Values are means of three biological replicates with error bars representing sd. D, Untransformed and DcMYB7-knockout DPP carrots. Separate images are shown as a composite for comparison. E, Total anthocyanin contents in roots of untransformed and DcMYB7-knockout DPP carrots. Data are means of three technical replicates ± sd. fw, Fresh weight.
DcMYB7 from the solid purple carrot DPP was also introduced into Arabidopsis plants and the orange carrot KRD under the control of the CaMV 35S promoter to investigate its functions. The heterologous overexpression of DcMYB7 in Arabidopsis induced anthocyanin accumulation in both vegetative and reproductive organs (Supplemental Fig. S1). When the orange carrot KRD explants were transformed with 35S:DcMYB7, deep purple calli were produced and regenerated to produce purple plantlets (Fig. 2B). The 35S:DcMYB7 transgenic KRD carrots accumulated high levels of anthocyanins across the entire root section (Fig. 2, B and C). After bolting, 35S:DcMYB7 transgenic KRD carrots generated purple-pigmented ovaries and seeds but no purple-pigmented petals or stamens.
DcMYB7 was knocked out in the solid purple carrot DPP using the CRISPR/Cas9 system (Xu et al., 2019). Four DcMYB7-knockout plantlet lines with two or three edited target sites were previously generated. After growing for 4 months, three DcMYB7-knockout DPP carrot lines (line 1, line 2, and line 3) were chosen for study (Fig. 2D). The roots of these DcMYB7-knockout DPP carrot lines were yellow across the entire section. In contrast to normal DPP carrots, which accumulate high levels of anthocyanins in their roots, the three DcMYB7-knockout DPP carrot lines accumulated undetectable or trace amounts of anthocyanins in their roots (Fig. 2E). These results together suggest that DcMYB7 is the P3 gene that conditions anthocyanin biosynthesis in purple carrot roots. However, petioles of the three DcMYB7-knockout DPP carrot lines remained purple pigmented, supporting the hypothesis that, in addition to DcMYB7, another genetic factor also controls anthocyanin production in purple carrot petioles.
DcMYB7 Increased the Expression Levels of DcbHLH3 and Anthocyanin Biosynthesis-Related Structural Genes in Carrots
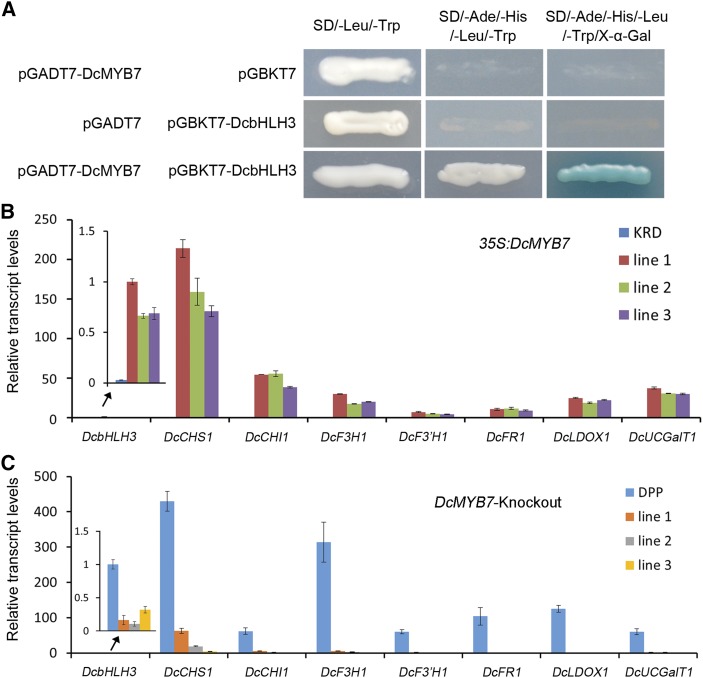
MYBs often interact with bHLHs to coregulate anthocyanin biosynthesis in many plant species. MYBs control corresponding bHLH interactor expression in Arabidopsis (Tohge et al., 2005). Iorizzo et al. (2019) identified several differentially expressed genes within the P3 genomic regions of chromosome 3, but none grouped with anthocyanin-related bHLHs from other species. Two carrot bHLHs, both located on chromosome 1, corresponding to the predicted carrot genes DCAR_002739 and DCAR_004632, were identified by referring to orthologous apple (Malus × domestica) bHLH3 and through phylogenetic analyses. MdbHLH3 is a bHLH that can enhance anthocyanin accumulation (Espley et al., 2007). DCAR_002739 and DCAR_004632 clustered together with anthocyanin-related bHLHs from other plant species in phylogenetic analyses (Supplemental Fig. S2). However, RT-qPCR analyses indicated that DCAR_004632 was not coexpressed with anthocyanin pigmentation in carrots and showed much lower transcript levels than DCAR_002739 (hereafter DcbHLH3) in the purple-pigmented tissues of all the tested purple carrot roots. DcbHLH3, which shared higher amino acid sequence identity with MdbHLH3 than DCAR_004632, was positively correlated with anthocyanin biosynthesis in carrot roots, displaying high transcript levels in the purple pigmented tissues of all the tested purple carrot roots but very low or undetectable transcript levels in the nonpurple root tissues of purple and nonpurple carrot cultivars (Supplemental Fig. S3). Yeast (Saccharomyces cerevisiae) two-hybrid assays indicated that DcMYB7 could interact with DcbHLH3 (Fig. 3A).
Figure 3.
Role of DcMYB7 in regulating DcbHLH3 and structural genes in the anthocyanin pathway. A, Yeast two-hybrid assays validating the interaction of DcMYB7 with DcbHLH3. SD, Synthetic defined; X-α-Gal, 5-bromo-4-chloro-3-indolyl-α-D-galactopyranoside. B, Relative transcript levels of DcbHLH3 and structural genes in roots of untransformed and 35S:DcMYB7 transgenic KRD carrots. Data represent means of biological triplicate RT-qPCRs ± sd. C, Relative transcript levels of DcbHLH3 and structural genes in roots of untransformed and DcMYB7-knockout DPP carrot lines. Data are means of three technical replicates with error bars representing sd.
To determine whether DcMYB7 regulates DcbHLH3, RT-qPCR analyses were performed in roots of untransformed KRD carrots and three 35S:DcMYB7 transgenic KRD carrot lines at 4 months of age. Compared with the untransformed KRD carrots, DcbHLH3 transcript levels were greatly increased in the three 35S:DcMYB7 KRD carrot lines (Fig. 3B). Transcriptional analysis of anthocyanin biosynthetic structural genes (DcCHS1, DcCHI1, DcF3H1, DcF3′H1, DcDFR1, DcLDOX1, and DcUCGalT1) indicated that these genes were all up-regulated in the roots of the three 35S:DcMYB7 transgenic KRD carrot lines compared with untransformed KRD carrots (Fig. 3B). In addition, we analyzed the transcript levels of DcbHLH3 and the structural genes in the untransformed DPP carrot and the three DcMYB7-knockout DPP carrot lines. Compared with the untransformed DPP carrot, DcbHLH3 and all the tested structural genes were down-regulated in the roots of the three DcMYB7-knockout DPP carrots (Fig. 3C). These results together suggest that DcMYB7 controls the expression of its partner DcbHLH3 and the anthocyanin biosynthetic structural genes.
Functional Tests of DcMYB7 from Different Purple and Nonpurple Carrot Cultivars
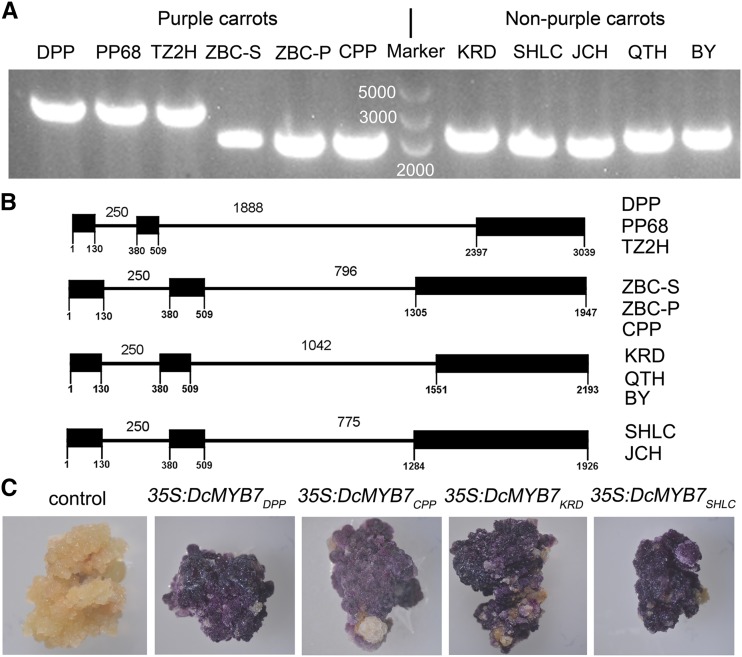
DcMYB7 was cloned from genomic DNA (gDNA) and cDNA libraries constructed using 11 different purple and nonpurple carrot cultivars. DcMYB7 gDNA sequences were different lengths among the carrot cultivars (Fig. 4A). On the basis of the sequencing results, four variant gDNA sequences were identified among the 11 different purple and nonpurple carrot cultivars (Fig. 4B). The DcMYB7 gDNA sequences from three solid purple carrot cultivars (DPP, PP68, and TZ2H) are identical and 3,039 bp long, which is much longer than the lengths of the gene in the other eight carrot cultivars. The DcMYB7 gDNA sequences from solid purple carrot ZBC-S and purple peridermal carrots ZBC-P and CPP are identical and 1,947 bp long. The DcMYB7 gDNA sequences from the orange carrot KRD and yellow carrots QTH and BY are 2,193 bp long and share 100% identity with DcMYB7 sequence from the DH1 (orange) carrot genome (Iorizzo et al., 2016). The DcMYB7 gDNA sequences from the orange carrots SHLC and JCH are identical and 1,926 bp long.
Figure 4.
Functional investigation of DcMYB7 gDNA sequences from various carrot cultivars. A, PCR amplification of DcMYB7 gDNA from 11 different carrot cultivars. B, Schematic representation of full-length DcMYB7 from 11 different carrot cultivars having three exons (black boxes) and two introns (lines between exons). C, Overexpression of DcMYB7 gDNA from different carrot cultivars in the orange KRD carrot under the control of the CaMV 35S promoter. KRD explants transformed with the pCAMBIA 1301 vector were used as the control.
The cDNA sequence of DcMYB7 was also cloned from the 11 different purple or nonpurple carrot cultivars. Four variant cDNA sequences were identified among the 11 different purple and nonpurple carrot cultivars. Based on the sequencing and alignment analyses, DcMYB7 cDNA from the 11 tested carrot cultivars were 903 bp long. The alignment analysis of DcMYB7 cDNA and gDNA sequences indicated that the genomic structure consisted of three exons and two introns. The different lengths of DcMYB7 gDNA products from various carrot cultivars were caused by variation of intron II (Fig. 4B).
The gDNA sequences of DcMYB7 from the 11 different purple and nonpurple carrot cultivars differed. To determine whether the variation among DcMYB7 gDNA sequences affected their function in inducing anthocyanin biosynthesis, the four variant gDNAs were amplified from the solid purple carrot DPP, purple peridermal carrot CPP, and orange carrots KRD and SHLC. They were then independently introduced into the orange carrot KRD for expression under the control of the CaMV 35S promoter. Deep purple calli were induced from explants transformed with DcMYB7 gDNAs from the solid purple carrot DPP (35S:DcMYB7DPP), purple peridermal carrot CPP (35S:DcMYB7CPP), and orange carrots KRD (35S:DcMYB7KRD) and SHLC (35S:DcMYB7SHLC), whereas nonpurple-pigmented calli were produced from orange carrot KRD explants transformed with the pCAMBIA 1301 vector (control; Fig. 4C). Thus, gDNA sequences of DcMYB7 from different nonpurple carrot cultivars appear to have retained their anthocyanin-induction function.
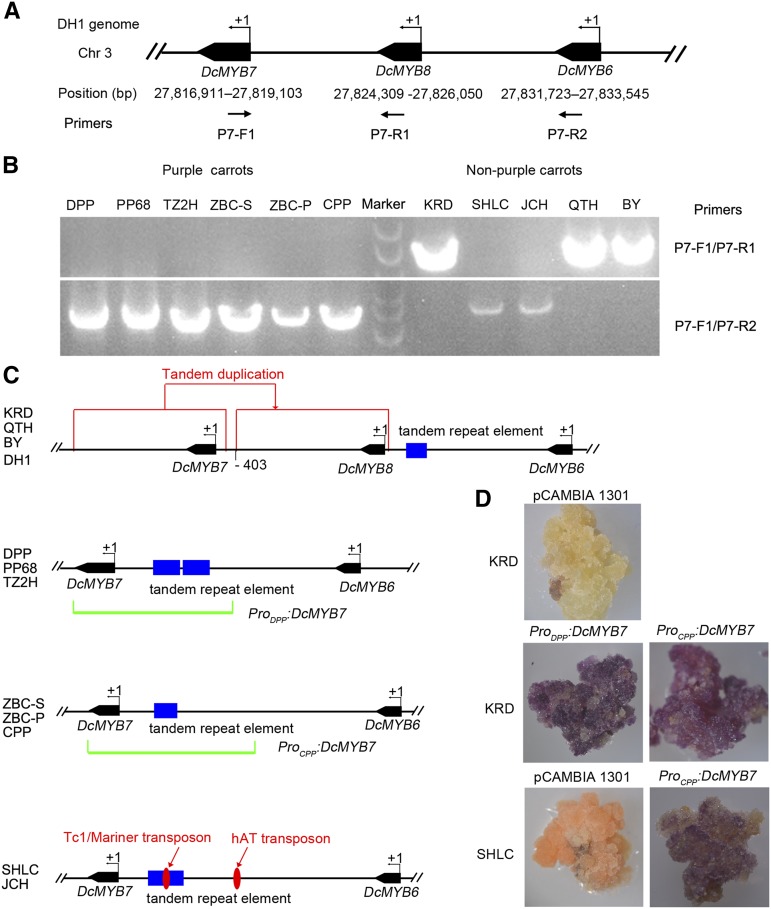
Insertion of a Nonfunctional DcMYB7 Duplication or Transposons into the Promoter Region of DcMYB7 in Nonpurple Carrots
To identify the molecular mechanism underlying the differential expression patterns of DcMYB7 in 11 different purple and nonpurple carrots, we cloned the upstream sequence of DcMYB7 from the 11 carrot cultivars. According to the carrot genome sequence (Iorizzo et al., 2016) and a recent report (Iorizzo et al., 2019), DcMYB7, DcMYB8, and DcMYB6 are organized in tandem within the 27,816,911- to 27,833,545-bp region of chromosome 3 (Fig. 5A). The P7-F1 and P7-R1 primers were designed based on the DNA sequences of DcMYB7 and DcMYB8 to amplify the sequence upstream of DcMYB7 (Fig. 5A). The upstream sequence of DcMYB7 was successfully amplified from the orange carrot KRD and yellow carrots QTH and BY but not from the other eight carrot cultivars (Fig. 5B). Sequencing results showed that the sequences upstream of DcMYB7 from KRD, QTH, and BY are identical to that from the DH1 orange carrot genome (Supplemental Fig. S4). We further analyzed the region containing DcMYB7, DcMYB8, and DcMYB6 on chromosome 3 of the DH1 orange carrot genome. The full-length predicted protein sequences of DcMYB7 (300 amino acids) and DcMYB8 (292 amino acids) were highly similar (over 90%; Supplemental Fig. S5). However, DcMYB8 expression was undetectable in all the purple carrots tested in a previous report (Iorizzo et al., 2019). Thus, DcMYB8 was regarded as a nonfunctional duplication of DcMYB7 and inserted between DcMYB7 and DcMYB6 at 403 bp upstream of the putative DcMYB7 start codon (Fig. 5C; Supplemental Fig. S4).
Figure 5.
Functional investigation of the DcMYB7 promoter from various carrot cultivars. A, Physical locations of DcMYB6, DcMYB7, and DcMYB8 on chromosome 3 of the DH1 orange carrot genome. B, PCR amplification of the sequence upstream of DcMYB7 from 11 different carrot cultivars. C, Schematic representation of DcMYB6, DcMYB7, and DcMYB8 from 11 different carrot cultivars. D, Functional investigation of the DcMYB7 promoter from different carrots in KRD and SHLC carrots.
The sequence upstream of DcMYB7 was successfully amplified from the other eight carrot cultivars using the P7-F1 and P7-R2 primers designed based on the DNA sequences of DcMYB7 and DcMYB6 (Fig. 5B). Sequencing results indicated that DcMYB8 was absent in the regions between DcMYB7 and DcMYB6 in these eight carrot cultivars. Sequencing results also showed that the sequences upstream of DcMYB7 from DPP, PP68, and TZ2H carrots are identical (MK637849), those from ZBC-S, ZBC-P, and CPP carrots are identical (MK637850), and those from SHLC and JCH are identical (MK637851; Fig. 5C).
To investigate why the promoters of DcMYB7 were not functional in nonpurple carrots, an alignment of the DcMYB7 promoter regions from DPP, CPP, SHLC, and KRD was conducted. The promoter region of DcMYB7 from KRD (Pro-DcMYB7-KRD) shared a high sequence identity with those from the other carrot cultivars within the 1- to 281-bp upstream region of the putative start codon, but there were no common sequences in the region 403 bp upstream of the putative start codon owing to the insertion of DcMYB8 (Supplemental Fig. S6). The DcMYB7 promoter sequence from DPP (Pro-DcMYB7-DPP) shared a high identity with those from CPP (Pro-DcMYB7-CPP) and SHLC (Pro-DcMYB7-SHLC) within the 1- to 1,392-bp upstream region of the putative start codon, designated as the consensus sequences region. A poly(dA-dT) element and a tandem repeat element were found in the consensus sequences region of DPP. The latter element was not repeated in the DcMYB7 promoters from CPP and SHLC, and it was interrupted by another unknown element in SHLC (Fig. 5C; Supplemental Fig. S6). This unknown element was widely present in the carrot genome on the basis of BLAST search analysis (E ≤ 10−120) against the carrot genome sequence. On the basis of BLAST search analysis against the GIRI repeat database (score = 236 [http://www.girinst.org/]; Kohany et al., 2006), it belongs to the Tc1/Mariner transposon family. Another transposon belonging to the hAT family was also identified in DcMYB7 promoter sequences of SHLC on the basis of BLAST search analysis against the GIRI repeat database (score = 480). This transposon was also widely present in the carrot genome on the basis of BLAST search analysis (E ≤ 10−150) against the carrot genome sequence. In addition, the DcMYB7 promoter sequences from SHLC and CPP were nearly identical, except that the former was interrupted by Tc1/Mariner and hAT transposons.
The entire coding DNA sequence (CDS) and promoter region of DcMYB7 were cloned from DPP and CPP to construct ProDPP:DcMYB7 and ProCPP:DcMYB7, respectively. These constructs and the pCAMBIA 1301 vector were then independently genetically transformed into the orange carrot KRD. After transformation, purple-pigmented calli were produced from explants transformed with ProDPP:DcMYB7 and ProCPP:DcMYB7 (Fig. 5D). However, nonpurple-pigmented calli were produced from explants transformed with pCAMBIA 1301 vector (Fig. 5D). These results suggested that inactivation of DcMYB7 from KRD, QTH, and BY resulted from the insertion of DcMYB8, a nonfunctional duplication of DcMYB7, in its promoter region. ProCPP:DcMYB7 and the pCAMBIA 1301 vector were also independently genetically transformed into orange carrot SHLC. After transformation, purple-pigmented calli were produced from explants transformed with ProCPP:DcMYB7, but nonpurple-pigmented calli were produced from explants transformed with the pCAMBIA 1301 vector (Fig. 5D). The promoter sequences of SHLC and CPP were nearly identical except for the region interrupted by transposons, supporting the idea that the transposon insertions in the DcMYB7 promoter of the former have resulted in the loss of function.
DcMYB7 Regulates Further Anthocyanin Modifications by Activating DcUCGXT1 and DcSAT1 in Carrot
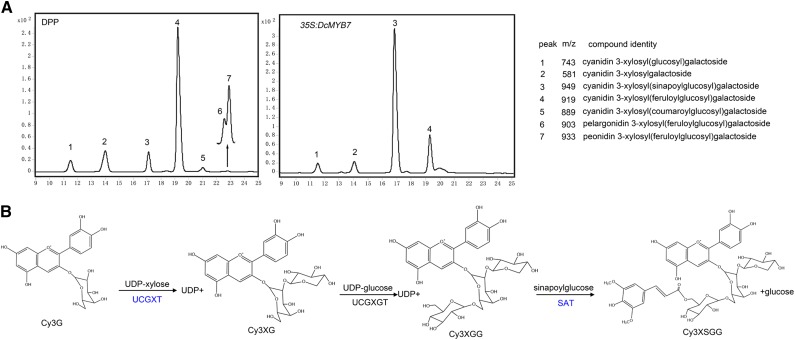
In purple carrots, anthocyanins are usually stored in glycosylated and acylated forms (Kammerer et al., 2004; Montilla et al., 2011; Cavagnaro et al., 2014). In this study, the anthocyanin compositions of DPP and 35S:DcMYB7 transgenic KRD carrot (line 1) roots were measured using HPLC-mass spectrometry. DPP carrot produced seven peaks that were confirmed to be those of anthocyanins (Fig. 6A). The major peak, 4, was identified as cyanidin 3-xylosyl (feruloylglucosyl) galactoside (Cy3XFGG). The other six peaks, 1, 2, 3, 5, 6, and 7, were identified as cyanidin 3-xylosyl (glucosyl) galactoside (Cy3XGG), cyanidin 3-xylosylgalactoside (Cy3XG), cyanidin 3-xylosyl (sinapoylglucosyl) galactoside (Cy3XSGG), cyanidin 3-xylosyl (coumaroylglucosyl) galactoside, pelargonidin 3-xylosyl (feruloylglucosyl) galactoside, and peonidin 3-xylosyl (feruloylglucosyl) galactoside. The 35S:DcMYB7 transgenic KRD carrot only produced four peaks, 1 to 4, corresponding to Cy3XGG, Cy3XG, Cy3XSGG, and Cy3XFGG, respectively (Fig. 6A). Through observation of the peak areas from the HPLC measurements, Cy3XSGG was determined to be the predominant anthocyanin in 35S:DcMYB7 transgenic KRD carrot roots. A reaction scheme for the biosynthesis of Cy3XSGG from cyanidin-3-galactoside (Cy3G) in carrot has been proposed (Glässgen et al., 1998). Cy3G undergoes two glycosylation steps and one acylation step in the formation of Cy3XSGG (Fig. 6B). UDP-xylose:cyanidin 3-galactoside xylosyltransferase (UCGXT), UDP-glucose:cyanidin 3-xylosylgalactoside glucosyltransferase (UCGXGT), and sinapoyl-glucose:anthocyanin acyltransferase (SAT) participate in Cy3XSGG biosynthesis.
Figure 6.
Anthocyanin modifications in carrots. A, Anthocyanin composition profiles from roots of DPP carrots and 35S:DcMYB7 transgenic KRD carrots (line 1). B, Schematic of the proposed biosynthetic pathway of Cy3XSGG. UCGXT and SAT were identified in this study.
In a previous report, Cavagnaro et al. (2014) identified three QTLs (Q1, Q2, and Q3) associated with Cy3XSGG accumulation in carrots. Q1 was genetically mapped within the same locus on chromosome 3 as P3 (Cavagnaro et al., 2014), a gene confirmed to be DcMYB7 in this study. Q2 and Q3 were genetically mapped to chromosomes 6 and 3, respectively (Cavagnaro et al., 2014). Within the Q3 locus associated with Cy3XSGG, a gene encoding UCGXT, DcUCGXT1 (DCAR_021269), was identified by referring to orthologous F3GGT1 (FG404013) from kiwifruit (Actinidia chinensis) and UGT79B1 (At5g54060) from Arabidopsis (Montefiori et al., 2011; Yonekura-Sakakibara et al., 2012). The F3GGT1 and UGT7291 proteins catalyze the glycosylation of Cy3G to produce Cy3XG. Within the Q2 locus, a gene encoding SAT, DcSAT1 (no corresponding predicted gene in the carrot genome), was identified by referring to the orthologous SAT (At2g23000) from Arabidopsis. At2g23000 encodes a protein capable of adding sinapoylglucose to anthocyanins to synthesize sinapoylated anthocyanins (Fraser et al., 2007).
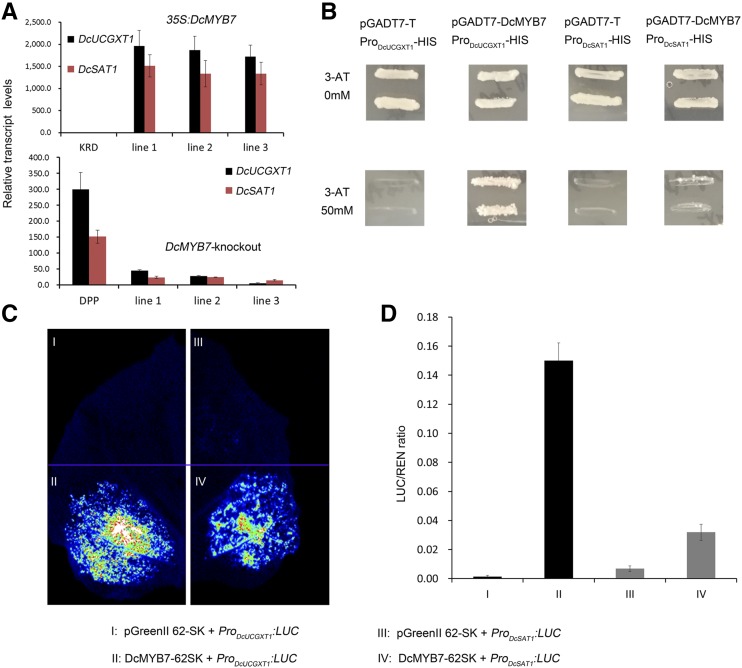
An RT-qPCR analysis indicated that DcUCGXT1 and DcSAT1 showed high transcript levels in roots of DPP but visually undetectable transcript levels in roots of KRD (Fig. 7A). Compared with untransformed KRD carrot, DcUCGXT1 and DcSAT1 showed greatly increased transcript levels in the three 35S:DcMYB7 KRD carrot lines. In addition, transcript levels of DcUCGXT1 and DcSAT1 in the roots of the three DcMYB7-knockout DPP carrot lines were obviously decreased compared with untransformed DPP carrots. Thus, DcMYB7 probably conditions anthocyanin glycosylation and acylation by regulating DcUCGXT1 and DcSAT1 in carrots.
Figure 7.
Role of DcMYB7 in regulating DcUCGXT1 and DcSAT1. A, Relative transcript levels of DcUCGXT1 and DcSAT1 in roots of untransformed and 35S:DcMYB7 transgenic KRD (top) as well as untransformed and DcMYB7-knockout DPP (bottom) carrot lines. Data are means of three biological (top) or technical (bottom) replicates with error bars representing sd. B, Yeast one-hybrid assays showing that DcMYB7 binds to the promoter fragments of DcUCGXT1 and DcSAT1. 3-AT, 3-amino-1,2,4-triazole. C, Transient expression assays showing that DcMYB7 promotes the expression of DcUCGXT1 and DcSAT1. D, Promoter activities of DcUCGXT1 and DcSAT1 expressed as a ratio of firefly LUC to Renilla luciferase (REN) activity. Data are means of six replicate reactions with error bars representing sd.
DcMYB7 Directly Regulates the Expression Levels of DcUCGXT1 and DcSAT1
Yeast one-hybrid assays were performed to detect the binding of DcMYB7 to the promoters of DcUCGXT1 and DcSAT1. The yeast cells cotransformed with pGADT7-DcMYB7 and ProDcUCGXT1-HIS or ProDcSAT1-HIS showed greater resistance to 50 mm 3-amino-1,2,4-triazole than the negative control (Fig. 7B), indicating that DcMYB7 could bind the promoters of DcUCGXT1 and DcSAT1.
A dual luciferase (LUC) reporter system was used to further confirm the interaction of DcMYB7 with the DcUCGXT1 and DcSAT1 promoters. DcMYB7 was cloned into the pGreenII 62-SK vector to generate DcMYB7-62SK, and DcUCGXT1 and DcSAT1 promoter sequences were independently fused to LUC of the pGreenII 0800 vector to generate ProDcUCGXT1:LUC and ProDcSAT1:LUC, respectively. DcMYB7-62SK was coinfiltrated with ProDcUCGXT1:LUC or ProDcSAT1:LUC into Nicotiana benthamiana leaves for expression. As a result, the coexpression of DcMYB7-62SK with either ProDcUCGXT1:LUC or ProDcSAT1:LUC exhibited strong luminescence intensity, whereas the coexpression of pGreenII 62-SK with ProDcUCGXT1:LUC or ProDcSAT1:LUC exhibited no visible or weak luminescence signals (Fig. 7C).
The ratio of luminescence produced by ProDcUCGXT1:LUC or ProDcSAT1:LUC to that produced by 35S:Renilla was calculated to determine the transactivation activity (Fig. 7D). There was a greater than 100-fold increase in the transactivation activity of DcMYB7-62SK on ProDcUCGXT1:LUC compared with pGreenII 62-SK on ProDcUCGXT1:LUC. Additionally, the transactivation activity of DcMYB7-62SK on ProDcSAT1:LUC increased more than 4-fold compared with that of pGreenII 62-SK on ProDcSAT1:LUC. Thus, DcMYB7 appears to directly activate the promoters of DcUCGXT1 and DcSAT1.
DISCUSSION
Carrots with purple or yellow root were first cultivated as a root crop in Central Asia ∼1,100 years ago and spread to the East and West (Iorizzo et al., 2013). The yellow carrot was concluded to be a color mutant of the purple carrot (Banga, 1963). The color of carrot roots has significantly changed during the domestication process. White and red carrots originated in Europe and China, respectively, between the 11th and 15th centuries (Arscott and Tanumihardjo, 2010). The Europeans preferred yellow carrot over purple and white carrot until orange carrot arrived in the 16th century (Simon, 2000). Then, the latter gradually spread throughout Europe and other continents worldwide. Nowadays, cultivated carrot roots are purple, orange, yellow, red, and white, resulting from anthocyanin, carotene, lutein, and lycopene accumulations and the lack of pigments, respectively (Surles et al., 2004; Clotault et al., 2008; Montilla et al., 2011).
Anthocyanins have protective functions against biotic and abiotic stresses in plants, such as insect attack in Arabidopsis (Johnson and Dowd, 2004), blast fungus infection in rice (Oryza sativa; Gandikota et al., 2001), and cold stress in sweet potato (Ipomoea batatas) storage roots (Wang et al., 2013a). We speculate that anthocyanins in purple carrot roots may also have protective functions. In addition, anthocyanin can protect humans against diseases (Netzel et al., 2007; He and Giusti, 2010; Tsutsumi et al., 2019). The intake of anthocyanin-rich food decreases the risks of several kinds of diseases. Purple carrots are still popular and cultivated in China, Japan, India, and other Asiatic countries. With the human health benefits of anthocyanins, purple carrots have been rediscovered by breeders that are aiming to improve the nutrient content.
However, to date, the molecular basis of the mutation leading to anthocyanin pigment loss in carrot has still not been revealed. Two inherited genes, P1 and P3, controlling anthocyanin pigmentation in purple carrot roots were genetically mapped within 28.2- and 12-centimorgan regions, respectively, on chromosome 3 in carrots from different genetic backgrounds (Simon, 1996; Cavagnaro et al., 2014). P1 is responsible for anthocyanin pigmentation in carrots with purple roots but green petioles, whereas P3 controls anthocyanin pigmentation in carrots with purple roots and petioles (Cavagnaro et al., 2014). Previously, we found that the expression level of DcMYB6, a gene anchored in the P3 region (Iorizzo et al., 2019), was associated with anthocyanin pigmentation in purple carrot roots (Xu et al., 2017). Iorizzo et al. (2019) also identified another MYB gene, DcMYB7, in the P3 region. Because the expression of DcMYB7 was correlated with anthocyanin pigmentation in all the tested purple carrot roots and DcMYB6 was only expressed in solid purple carrots, they speculated that DcMYB6 and DcMYB7 control anthocyanin biosynthesis in the inner root tissues and the root outer phloem, respectively.
Here, the expression levels of DcMYB6 and DcMYB7 were both consistent with anthocyanin pigmentation in different purple carrot roots. However, the expression of DcMYB6, which could lead to anthocyanin accumulation in Arabidopsis plants (Xu et al., 2017), was unable to induce anthocyanin biosynthesis in the orange carrot KRD. The overexpression of DcMYB7 in the orange carrot KRD resulted in the reconstitution of anthocyanin accumulation in vegetative and reproductive tissues, including roots, by activating its partner DcbHLH3 and all the tested anthocyanin biosynthetic structural genes. The DcMYB7-knockout DPP carrots produced yellow roots, which suggests that DcMYB7 is the P3 gene that controls anthocyanin biosynthesis both in inner and outer root tissues of purple carrots. The petioles of DcMYB7-knockout DPP carrots retain their purple pigmentation, indicating that another dominant gene, probably DcMYB11 identified in a previous study (Iorizzo et al., 2019), conditions anthocyanin pigmentation in carrot petioles.
R2R3-MYB mutations can lead to anthocyanin pigment losses in some plant species (Kobayashi et al., 2004; Ban et al., 2007; Espley et al., 2009; Wang et al., 2013b). Although the gDNA sequence of DcMYB7 varies among different purple and nonpurple carrots, the CDS regions of DcMYB7 gDNAs from orange and yellow carrots retained their abilities to induce anthocyanin biosynthesis. Our data indicated that the promoter sequence of DcMYB7 was interrupted either by DcMYB8, a nonfunctional tandem duplication of DcMYB7, or by transposons, leading to the transcriptional inactivation of DcMYB7 in nonpurple carrot roots. As a result, nonpurple carrots were unable to accumulate anthocyanins in their roots.
In this study, we unraveled the genetic factors conditioning anthocyanin pigmentation in purple versus nonpurple carrot roots, providing new insight into carrot domestication. However, the CDS and promoter region of DcMYB7 from the solid purple carrot ZBC-S and the purple peridermal carrots ZBC-P and CPP are identical. DcMYB7 showed high expression levels across the entire root section in ZBC-S but was only specifically expressed in the root peridermal tissue of ZBC-P and CPP carrots, suggesting that there is another genetic factor that suppresses the expression of DcMYB7 in phloem and xylem tissues of ZBC-P and CPP carrot roots. In future work, we will focus on determining the genetic factors contributing to the differential expression pattern of DcMYB7 in solid purple and purple peridermal carrots.
In plants, anthocyanin usually undergoes modifications that increase its stability and water solubility (Cheng et al., 2014). Anthocyanins from purple carrots undergo a series of glycosylation and acylation events, resulting in relatively higher temperature and pH stability levels than other plant species (Kirca et al., 2007). In purple carrots, anthocyanins are dominated by glycosylated and acylated cyanidin, with Cy3XSGG and Cy3XFGG being the predominant anthocyanin compositions (Kammerer et al., 2004; Montilla et al., 2011). In this study, the overexpression of DcMYB7 in the orange carrot KRD induced anthocyanin accumulation in their roots, with Cy3XSGG being detected as the predominant anthocyanin.
Three QTLs (Q1, Q2, and Q3) were detected as associated with Cy3XSGG on chromosomes 3 and 6 by Cavagnaro et al. (2014). Q1 was anchored to the same region on chromosome 3 as P3, which was confirmed to be DcMYB7 in this study. Within the other two QTLs (Q2 and Q3), DcUCGXT1 and DcSAT1 involved in anthocyanin glycosylation and acylation were identified. UCGXT and SAT had been identified previously in Arabidopsis plants (Fraser et al., 2007; Yonekura-Sakakibara et al., 2012). In carrot, DcUCGXT1 and DcSAT1 are coexpressed with DcMYB7 in anthocyanin pigmented roots, supporting the hypothesis that DcMYB7 could trigger the expression of both DcUCGXT1 and DcSAT1, which was shown in this study. These data suggest that DcMYB7 conditions the route from Cy3G to acylated Cy3XSGG in the anthocyanin modification pathways of carrot.
Acylation increases anthocyanin stability but reduces it bioavailability (Charron et al., 2009). Carrot breeders can determine breeding objectives for anthocyanin composition based on market demand, with preferences for acylated anthocyanins as stable food colorants and nonacylated anthocyanins as bioavailable nutraceuticals. Our study will aid attempts to manipulate anthocyanin composition in carrots and other root crops.
CONCLUSION
Functional analysis confirmed that DcMYB7 is the P3 gene that controls purple pigmentation in carrot roots by regulating its DcbHLH3 partner and the tested anthocyanin biosynthetic structural genes. The promoter sequence of DcMYB7 in nonpurple carrots has been interrupted by a tandem duplication event or transposon insertion, leading to undetectable levels of anthocyanins in their roots. Our study suggests that there is another genetic factor that suppresses DcMYB7 expression in phloem and xylem of purple peridermal carrot root tissues. DcMYB7 also conditions anthocyanin modifications by directly activating DcUCGXT1 and DcSAT1. Our determination of these genetic factors involved in anthocyanin biosynthesis and modification in carrots will aid in the breeding of carrot as well as that of other root crops.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
All the carrots (Daucus carota) used in this study were grown in artificial climatic chambers as previously described by Xu et al. (2014). DPP, PP68, and TZ2H are carrot cultivars with solid purple roots (purple periderm, phloem, and xylem). ZBC is a carrot cultivar with two types of roots: solid purple (ZBC-S) and purple periderm but nonpurple phloem and xylem (ZBC-P). CPP is a carrot cultivar with purple peridermal root tissue. KRD, SHLC, and JCH are orange carrot cultivars. QTH and BY are yellow cultivars. Young leaves of carrots were used for gDNA extraction. Different tissues of 4-month-old carrot roots were used for anthocyanin and total RNA extraction. Periderm, phloem, and xylem tissues of roots from CPP and ZBC-P were collected separately as samples. The ability to distinguish and excise the periderm from the phloem was impossible for roots of the other carrot cultivars. For these carrot cultivars, root periderm and phloem tissues were collected together as periderm and phloem samples, and xylem tissue was individually collected. All the samples were immediately frozen in liquid nitrogen after harvest and then stored at −80°C before further analyses.
Anthocyanin Measurement
Total anthocyanin was extracted from carrot roots using the methanol-HCl method (Li et al., 2012), with a modified extraction buffer (50% [v/v] methanol, 49.9% [v/v] distilled, deionized water, and 0.1% [v/v] HCl), and was then quantitatively analyzed as described in Li et al. (2012). The anthocyanin composition was analyzed using HPLC-mass spectrometry with the method described by Feng et al. (2018).
gDNA and Total RNA Extraction, Gene Cloning, and RT-qPCR Assays
The gDNA and total RNA were isolated using a DNAsecure Plant Kit or RNAsimple Total RNA Kit (Tiangen) according to the manufacturer’s instructions. First-strand cDNA was synthesized using the HiScript II Q RT SuperMix for qPCR kit (Vazyme Biotech) following the manufacturer’s protocol. Gene cloning was performed using PrimeSTAR HS DNA polymerase (Takara). RT-qPCR assays were conducted as described previously by Xu et al. (2017). The relative gene transcript levels were normalized to DcActin1 (Wang et al., 2016) and calculated using the 2−ΔΔCT method (Schmittgen and Livak, 2008). The primers used for RT-qPCR assays of DcCHS1, DcCHI1, DcF3H1, DcF3′H1, DcDFR1, DcLDOX1, DcUCGalT1, and DcActin1 are the same as described in our previous report (Xu et al., 2014). All the other primers used are listed in Supplemental Table S1.
Sequence Alignment and Phylogenetic Analyses
Sequence alignments were performed using ClustalW (Chenna et al., 2003). Phylogenetic analysis was conducted with the deduced amino acid sequences of bHLHs from carrot and other plant species. The phylogenetic tree was constructed using MEGA version 5.0 software with the neighbor-joining method (bootstrap value = 1,000; Tamura et al., 2011).
Generation of Transgenic Carrot and Arabidopsis
The CDS regions of DcMYB6 and DcMYB7 were amplified from a cDNA library of DPP and inserted independently into the pCAMBIA 1301 vector between the CaMV 35S promoter and the pea (Pisum sativum) rbcSE9 terminator to create 35S:DcMYB6 and 35S:DcMYB7 constructs, respectively. DcMYB7 was also cloned from gDNA libraries of DPP, CPP, KRD, and SHLC to create 35S:DcMYB7DPP, 35S:DcMYB7CPP, 35S:DcMYB7KRD, and 35S:DcMYB7SHLC constructs, respectively. The gDNA fragments of DcMYB7, including the promoter region and entire CDS region, were amplified from DNA libraries of DPP and CPP to prepare ProDPP:DcMYB7 and ProCPP:DcMYB7 constructs, respectively. The recombined vectors were independently transformed into Agrobacterium tumefaciens (GV3101). The transformation of carrot was performed using a previously described method (Xu et al., 2019). The orange carrot KRD and SHLC were chosen for transformation. The 35S:DcMYB7 construct was also transformed into Arabidopsis (Arabidopsis thaliana) using the A. tumefaciens-mediated floral dip method (Clough and Bent, 1998). All of the primers used in this procedure are listed in Supplemental Table S1.
Generation of DcMYB7-Knockout Mutant Plants
DcMYB7, which was designated DcMYB113-like previously (Xu et al., 2019), was successfully knocked out in DPP carrot using the CRISPR/Cas9 system. After growing in an artificial climatic chamber for 4 months, three DcMYB7-knockout plant lines (line 1, line 2, and line 3) were chosen for study.
Yeast Two-Hybrid Assay
The CDS regions of DcMYB7 and DcbHLH3 from DPP and 35S:DcMYB7 transgenic KRD carrots, respectively, were separately cloned into the pGADT7 and pGBKT7 vectors (Clontech) to generate pGADT7-DcMYB7 and pGBKT7-DcbHLH3, respectively. These two recombined constructs were cotransformed into yeast (Saccharomyces cerevisiae) strain Y2HGold cells following the manufacturer’s manual (Clontech). The pGADT7 and pGBKT7 vectors were used as negative controls. The transformants were selected on synthetically defined/−Leu/−Trp medium at 30°C for 3 to 4 d. The interactions were tested on synthetically defined/−Ade/−His/−Leu/−Trp medium with or without 5-bromo-4-chloro-3-indolyl-α-D-galactopyranoside.
Yeast One-Hybrid Assay
The yeast one-hybrid assay was performed using the method described previously by Feng et al. (2018). The promoter fragments of DcUCGXT1 and DcSAT1 corresponding to the regions −1,349 to −1 and −1,662 to −1, respectively, relative to the translation initiation site were cloned from gDNA of KRD and were then fused to HIS to generate ProDcUCGXT1-HIS and ProDcSAT1-HIS, respectively. These two constructs were separately cotransformed with pGADT7-DcMYB7 into Y1H Gold yeast strain. The pGADT7-T vector was used as a negative control. The interactions were detected on medium supplemented with 50 mm 3-amino-1,2,4-triazole.
Dual LUC Reporter Assay of Transient Expression
The CDS of DcMYB7 was cloned into the pGreenII 62-SK vector to generate the DcMYB7-62SK effector. The promoters of DcUCGXT1 (2,431 bp upstream of the putative start codon) and DcSAT1 (2,568 bp upstream of the putative start codon) were introduced into the pGreenII 0800-LUC vector to generate the ProDcUCGXT1:LUC and ProDcSAT1:LUC reporter constructs, respectively. The constructs were transformed into A. tumefaciens strain GV3101 (pMP90). A. tumefaciens was mixed and coinfiltrated into Nicotiana benthamiana leaves for transient expression. The luminescence of firefly LUC was detected using a live imaging system (Tanon-5500Multi) according to the method described by Li et al. (2017). A Dual-Luciferase Reporter Assay System (Promega; catalog no. E1910) was used to measure the ratio of luminescence of firefly LUC to Renilla LUC according to the manufacturer’s instructions.
Accession Numbers
Sequence data from this article can be found in GenBank under the following accession numbers: DcbHLH3 (MK572822), DcMYB7 gDNA and cDNA sequences from DPP, CPP, KRD, and SHLC (MK572814–MK572817 and MK572818–MK572821, respectively), DcUCGXT1 (MK572822), DcSAT1 (MK572823), promoter sequences of DcUCGXT1 and DcSAT1 (MK572825 and MK572826, respectively), and sequences upstream of DcMYB7 from DPP, CPP, and SHLC (MK637849–MK637851, respectively).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Overexpression of DcMYB7 in Arabidopsis plants.
Supplemental Figure S2. Phylogenetic relationships among DCAR_002739 (DcbHLH3), DCAR_004632, and bHLHs involved in anthocyanin biosynthesis in other plant species.
Supplemental Figure S3. Expression of DCAR_002739 (DcbHLH3) and DCAR_004632 in different root tissues of 11 purple and nonpurple carrot cultivars.
Supplemental Figure S4. Region containing DcMYB6 (green highlight), DcMYB8 (red highlight), and DcMYB7 (blue highlight) on chromosome 3 of the DH1 orange carrot genome.
Supplemental Figure S5. Alignment analysis of the predicted protein sequences of DcMYB7 and DcMYB8 from the carrot genome.
Supplemental Figure S6. Alignment analysis of the promoter sequences of DcMYB7 from DPP (3,000 bp), CPP (2,775 bp), SHLC (3,500 bp), and KRD (3,000 bp).
Supplemental Table S1. List of primers used in this study.
Footnotes
This work was supported by the National Natural Science Foundation of China (31501775 and 31872098), the Open Project of State Key Laboratory of Crop Genetics and Germplasm Enhancement (ZW201710), the China Postdoctoral Science Foundation (2016M590467), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
References
- Arscott SA, Tanumihardjo SA (2010) Carrots of many colors provide basic nutrition and bioavailable phytochemicals acting as a functional food. Compr Rev Food Sci 9: 223–239 [Google Scholar]
- Ban Y, Honda C, Hatsuyama Y, Igarashi M, Bessho H, Moriguchi T (2007) Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol 48: 958–970 [DOI] [PubMed] [Google Scholar]
- Banga O. (1963) Origin and distribution of the western cultivated carrot. Genet Agrar 17: 357–370 [Google Scholar]
- Cavagnaro PF, Iorizzo M, Yildiz M, Senalik D, Parsons J, Ellison S, Simon PW (2014) A gene-derived SNP-based high resolution linkage map of carrot including the location of QTL conditioning root and leaf anthocyanin pigmentation. BMC Genomics 15: 1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagné D, Lin-Wang K, Espley RV, Volz RK, How NM, Rouse S, Brendolise C, Carlisle CM, Kumar S, De Silva N, et al. (2013) An ancient duplication of apple MYB transcription factors is responsible for novel red fruit-flesh phenotypes. Plant Physiol 161: 225–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron CS, Kurilich AC, Clevidence BA, Simon PW, Harrison DJ, Britz SJ, Baer DJ, Novotny JA (2009) Bioavailability of anthocyanins from purple carrot juice: Effects of acylation and plant matrix. J Agric Food Chem 57: 1226–1230 [DOI] [PubMed] [Google Scholar]
- Cheng J, Wei G, Zhou H, Gu C, Vimolmangkang S, Liao L, Han Y (2014) Unraveling the mechanism underlying the glycosylation and methylation of anthocyanins in peach. Plant Physiol 166: 1044–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31: 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clotault J, Peltier D, Berruyer R, Thomas M, Briard M, Geoffriau E (2008) Expression of carotenoid biosynthesis genes during carrot root development. J Exp Bot 59: 3563–3573 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC (2007) Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J 49: 414–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espley RV, Brendolise C, Chagné D, Kutty-Amma S, Green S, Volz R, Putterill J, Schouten HJ, Gardiner SE, Hellens RP, et al. (2009) Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell 21: 168–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng K, Xu ZS, Liu JX, Li JW, Wang F, Xiong AS (2018) Isolation, purification, and characterization of AgUCGalT1, a galactosyltransferase involved in anthocyanin galactosylation in purple celery (Apium graveolens L.). Planta 247: 1363–1375 [DOI] [PubMed] [Google Scholar]
- Fraser CM, Thompson MG, Shirley AM, Ralph J, Schoenherr JA, Sinlapadech T, Hall MC, Chapple C (2007) Related Arabidopsis serine carboxypeptidase-like sinapoylglucose acyltransferases display distinct but overlapping substrate specificities. Plant Physiol 144: 1986–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandikota M, de Kochko A, Chen L, Ithal N, Fauquet C, Reddy AR (2001) Development of transgenic rice plants expressing maize anthocyanin genes and increased blast resistance. Mol Breed 7: 73–83 [Google Scholar]
- Glässgen WE, Rose A, Madlung J, Koch W, Gleitz J, Seitz HU (1998) Regulation of enzymes involved in anthocyanin biosynthesis in carrot cell cultures in response to treatment with ultraviolet light and fungal elicitors. Planta 204: 490–498 [DOI] [PubMed] [Google Scholar]
- He J, Giusti MM (2010) Anthocyanins: Natural colorants with health-promoting properties. Annu Rev Food Sci Technol 1: 163–187 [DOI] [PubMed] [Google Scholar]
- Iorizzo M, Senalik DA, Ellison SL, Grzebelus D, Cavagnaro PF, Allender C, Brunet J, Spooner DM, Van Deynze A, Simon PW (2013) Genetic structure and domestication of carrot (Daucus carota subsp. sativus) (Apiaceae). Am J Bot 100: 930–938 [DOI] [PubMed] [Google Scholar]
- Iorizzo M, Ellison S, Senalik D, Zeng P, Satapoomin P, Huang J, Bowman M, Iovene M, Sanseverino W, Cavagnaro P, et al. (2016) A high-quality carrot genome assembly provides new insights into carotenoid accumulation and asterid genome evolution. Nat Genet 48: 657–666 [DOI] [PubMed] [Google Scholar]
- Iorizzo M, Cavagnaro PF, Bostan H, Zhao Y, Zhang J, Simon PW (2019) A cluster of MYB transcription factors regulates anthocyanin biosynthesis in carrot (Daucus carota L.) root and petiole. Front Plant Sci 9: 1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Wang H, Li M, Wang J, Yang Y, Zhang X, Yan G, Zhang H, Liu J, Zhang K (2016) The R2R3 MYB transcription factor PavMYB10.1 involves in anthocyanin biosynthesis and determines fruit skin colour in sweet cherry (Prunus avium L.). Plant Biotechnol J 14: 2120–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ET, Dowd PF (2004) Differentially enhanced insect resistance, at a cost, in Arabidopsis thaliana constitutively expressing a transcription factor of defensive metabolites. J Agric Food Chem 52: 5135–5138 [DOI] [PubMed] [Google Scholar]
- Kammerer D, Carle R, Schieber A (2004) Quantification of anthocyanins in black carrot extracts (Daucus carota ssp sativus var. atrorubens Alef.) and evaluation of their color properties. Eur Food Res Technol 219: 479–486 [Google Scholar]
- Kirca A, Ozkan M, Cemeroglu B (2007) Effects of temperature, solid content and pH on the stability of black carrot anthocyanins. Food Chem 101: 212–218 [Google Scholar]
- Kobayashi S, Goto-Yamamoto N, Hirochika H (2004) Retrotransposon-induced mutations in grape skin color. Science 304: 982. [DOI] [PubMed] [Google Scholar]
- Kodama M, Brinch-Pedersen H, Sharma S, Holme IB, Joernsgaard B, Dzhanfezova T, Amby DB, Vieira FG, Liu S, Gilbert MTP (2018) Identification of transcription factor genes involved in anthocyanin biosynthesis in carrot (Daucus carota L.) using RNA-Seq. BMC Genomics 19: 811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohany O, Gentles AJ, Hankus L, Jurka J (2006) Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics 7: 474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leja M, Kamińska I, Kramer M, Maksylewicz-Kaul A, Kammerer D, Carle R, Baranski R (2013) The content of phenolic compounds and radical scavenging activity varies with carrot origin and root color. Plant Foods Hum Nutr 68: 163–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Xu Y, Zhang L, Ji Y, Tan D, Yuan H, Wang A (2017) The jasmonate-activated transcription factor MdMYC2 regulates ETHYLENE RESPONSE FACTOR and ethylene biosynthetic genes to promote ethylene biosynthesis during apple fruit ripening. Plant Cell 29: 1316–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YY, Mao K, Zhao C, Zhao XY, Zhang HL, Shu HR, Hao YJ (2012) MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light-induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiol 160: 1011–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montefiori M, Espley RV, Stevenson D, Cooney J, Datson PM, Saiz A, Atkinson RG, Hellens RP, Allan AC (2011) Identification and characterisation of F3GT1 and F3GGT1, two glycosyltransferases responsible for anthocyanin biosynthesis in red-fleshed kiwifruit (Actinidia chinensis). Plant J 65: 106–118 [DOI] [PubMed] [Google Scholar]
- Montilla EC, Arzaba MR, Hillebrand S, Winterhalter P (2011) Anthocyanin composition of black carrot (Daucus carota ssp. sativus var. atrorubens Alef.) cultivars Antonina, Beta Sweet, Deep Purple, and Purple Haze. J Agric Food Chem 59: 3385–3390 [DOI] [PubMed] [Google Scholar]
- Morita Y, Saitoh M, Hoshino A, Nitasaka E, Iida S (2006) Isolation of cDNAs for R2R3-MYB, bHLH and WDR transcriptional regulators and identification of c and ca mutations conferring white flowers in the Japanese morning glory. Plant Cell Physiol 47: 457–470 [DOI] [PubMed] [Google Scholar]
- Netzel M, Netzel G, Kammerer DR, Schieber A, Carle R, Simons L, Bitsch I, Bitsch R, Konczak L (2007) Cancer cell antiproliferation activity and metabolism of black carrot anthocyanins. Innov Food Sci Emerg 8: 365–372 [Google Scholar]
- Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Simon PW. (1996) Inheritance and expression of purple and yellow storage root color in carrot. J Hered 1: 63–66 [Google Scholar]
- Simon PW. (2000) Domestication, historical development, and modern breeding of carrot. Plant Breed Rev 19: 157–190 [Google Scholar]
- Surles RL, Weng N, Simon PW, Tanumihardjo SA (2004) Carotenoid profiles and consumer sensory evaluation of specialty carrots (Daucus carota, L.) of various colors. J Agric Food Chem 52: 3417–3421 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T, Nishiyama Y, Hirai MY, Yano M, Nakajima J, Awazuhara M, Inoue E, Takahashi H, Goodenowe DB, Kitayama M, et al. (2005) Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J 42: 218–235 [DOI] [PubMed] [Google Scholar]
- Tsutsumi A, Horikoshi Y, Fushimi T, Saito A, Koizumi R, Fujii Y, Hu QQ, Hirota Y, Aizawa K, Osakabe N (2019) Acylated anthocyanins derived from purple carrot (Daucus carota L.) induce elevation of blood flow in rat cremaster arteriole. Food Funct 10: 1726–1735 [DOI] [PubMed] [Google Scholar]
- Wang GL, Tian C, Jiang Q, Xu ZS, Wang F, Xiong AS (2016) Comparison of nine reference genes for real-time quantitative PCR in roots and leaves during five developmental stages in carrot (Daucus carota L.). J Hortic Sci Biotechnol 91: 264–270 [Google Scholar]
- Wang H, Fan W, Li H, Yang J, Huang J, Zhang P (2013a) Functional characterization of dihydroflavonol-4-reductase in anthocyanin biosynthesis of purple sweet potato underlies the direct evidence of anthocyanins function against abiotic stresses. PLoS ONE 8: e78484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Meng D, Wang A, Li T, Jiang S, Cong P, Li T (2013b) The methylation of the PcMYB10 promoter is associated with green-skinned sport in Max Red Bartlett pear. Plant Physiol 162: 885–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu ZS, Huang Y, Wang F, Song X, Wang GL, Xiong AS (2014) Transcript profiling of structural genes involved in cyanidin-based anthocyanin biosynthesis between purple and non-purple carrot (Daucus carota L.) cultivars reveals distinct patterns. BMC Plant Biol 14: 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu ZS, Ma J, Wang F, Ma HY, Wang QX, Xiong AS (2016) Identification and characterization of DcUCGalT1, a galactosyltransferase responsible for anthocyanin galactosylation in purple carrot (Daucus carota L.) taproots. Sci Rep 6: 27356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu ZS, Feng K, Que F, Wang F, Xiong AS (2017) A MYB transcription factor, DcMYB6, is involved in regulating anthocyanin biosynthesis in purple carrot taproots. Sci Rep 7: 45324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu ZS, Feng K, Xiong AS (2019) CRISPR/Cas9-mediated multiply targeted mutagenesis in orange and purple carrot plants. Mol Biotechnol 61: 191–199 [DOI] [PubMed] [Google Scholar]
- Yildiz M, Willis DK, Cavagnaro PF, Iorizzo M, Abak K, Simon PW (2013) Expression and mapping of anthocyanin biosynthesis genes in carrot. Theor Appl Genet 126: 1689–1702 [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Fukushima A, Nakabayashi R, Hanada K, Matsuda F, Sugawara S, Inoue E, Kuromori T, Ito T, Shinozaki K, et al. (2012) Two glycosyltransferases involved in anthocyanin modification delineated by transcriptome independent component analysis in Arabidopsis thaliana. Plant J 69: 154–167 [DOI] [PMC free article] [PubMed] [Google Scholar]