WRINKLED1 regulates genes for fatty acid synthesis as well as inhibitors of fatty acid synthesis (BADCs) in a homeostatic mechanism.
Abstract
WRINKLED1 (WRI1) is a transcriptional activator that binds to a conserved sequence (designated as AW box) boxes in the promoters of many genes from central metabolism and fatty acid (FA) synthesis, resulting in their transcription. BIOTIN ATTACHMENT DOMAIN-CONTAINING (BADC) proteins lack a biotin-attachment domain and are therefore inactive, but in the presence of excess FA, BADC1 and BADC3 are primarily responsible for the observed long-term irreversible inhibition of ACETYL-COA CARBOXYLASE, and consequently FA synthesis. Here, we tested the interaction of WRI1 with BADC genes in Arabidopsis (Arabidopsis thaliana) and found purified WRI1 bound with high affinity to canonical AW boxes from the promoters of all three BADC genes. Consistent with this observation, both expression of BADC1, BADC2, and BADC3 genes and BADC1 protein levels were reduced in wri1-1 relative to the wild type, and elevated upon WRI1 overexpression. The double mutant badc1 badc2 phenocopied wri1-1 with respect to both reduction in root length and elevation of indole-3-acetic acid-Asp levels relative to the wild type. Overexpression of BADC1 in wri1-1 decreased indole-3-acetic acid-Asp content and partially rescued its short-root phenotype, demonstrating a role for BADCs in seedling establishment. That WRI1 positively regulates genes encoding both FA synthesis and BADC proteins (i.e. conditional inhibitors of FA synthesis), represents a coordinated mechanism to achieve lipid homeostasis in which plants couple the transcription of their FA synthetic capacity with their capacity to biochemically downregulate it.
Lipids are primary metabolites in cells, acting as structural components of cell membranes, energy-dense storage compounds, and cell signaling molecules. Fatty acids (FAs) are major components of lipids and triacylglycerols (TAGs), which are storage lipids that accumulate mostly in oil bodies in plant seeds (Li-Beisson et al., 2013). De novo synthesis of FAs occurs in the plastid via a well-established pathway (Ohlrogge and Browse, 1995; Rawsthorne, 2002), In Arabidopsis (Arabidopsis thaliana), transcription factors (TFs) LEAFY COTYLEDON1 (LEC1) and LEC2, FUSCA3, and ABSCISIC ACID INSENSITIVE3 serve as the master regulators of embryogenesis and seed maturation (Parcy et al., 1997; Lotan et al., 1998; To et al., 2006) and also regulate accumulation of TAG (Kagaya et al., 2005; Santos Mendoza et al., 2005; Braybrook et al., 2006). WRINKLED1 (WRI1), an APETALA2-type TF, functions downstream of LEC1 and LEC2 (Baud et al., 2007; Mu et al., 2008) and is a master regulator of FA synthesis (FAS) because seeds of the WRI1 mutant show an 80% reduction in TAGs compared to the wild type (Focks and Benning, 1998). More than 20 WRI1 target genes have been identified by comparing gene expression in the wild type with that of wri1 and WRI1 overexpression-lines, and electrophoretic mobility shift assays confirmed WRI1-gene promoter binding. Based on promoter sequence comparisons, [CnTnG](n)7[CG] was identified as the consensus WRI1 binding site (designated as AW box). WRI1 target sequences are found upstream of genes coding for enzymes involved in glycolysis (Suc synthase, pyruvate kinase, and pyruvate dehydrogenase), Glu-6P and phosphoenolpyruvate plastidial transporters (Glu-6P/phosphate translocator and phosphoenolpyruvate/phosphate translocator), subunits of acetyl-CoA carboxylase (ACCase; biotin carboxyl carrier protein 2 [BCCP2], biotin carboxylase, and carboxyltransferase), FAS (malonyl-CoA:acyl carrier protein [ACP] malonytransferase, ketoacyl-ACP synthase, hydroxyacyl-ACP dehydrase, enoyl-ACP reductase, ACPs, and oleoyl-ACP thioesterase), and genes involved in synthesis of lipoic acid, a cofactor of pyruvate dehydrogenase (Ruuska et al., 2002; Baud et al., 2007; Maeo et al., 2009; Fukuda et al., 2013; Li et al., 2015). Interestingly, genes involved in TAG synthesis and oil body assembly, such as diacylglycerol acyltransferase 1, which catalyzes the final step of TAG synthesis, and oleosins and caleosins appear not to be regulated by WRI1, though some species-specific variation has been reported (Maeo et al., 2009; Grimberg et al., 2015). Characterizing gene expression of subunits of ACCase revealed that the distance between the AW site and the translational initiation site (TIS) strongly influences the function of the AW box (Fukuda et al., 2013) in that the majority of AW sites fall within 200 bp of the TIS in genuine WRI1 target genes (Maeo et al., 2009; Fukuda et al., 2013).
A class of proteins annotated as biotin attachment domain containing (BADC; Olinares et al., 2010) have been recently identified as inhibitors of FAS (Salie et al., 2016; Keereetaweep et al., 2018). BADC proteins are homologs of BCCP, sharing 25% to 30% sequence similarity, but they lack a biotin attachment site, rendering them inactive. All three Arabidopsis BADC isoforms interact with BCCP. Displacement of functional BCCP subunits in ACCase by inactive BADC subunits is believed to reduce the ACCase catalytic activity (Salie et al., 2016). ACCase is subject to feedback regulation upon exposure to exogenously supplied FAs in the form of Tween esters (Tween-80 containing predominantly oleic acid [18:1]). Short-term exposure results in reversible inhibition of ACCase, whereas longer-term exposure results in irreversible inhibition (Andre et al., 2012). Oleoyl-ACP mediates the reversible phase of inhibition (Andre et al., 2012), whereas BADC1 and BADC3 are primarily responsible for the phase of irreversible inhibition (Keereetaweep et al., 2018).
In this work we present evidence that the expression of all three BADC genes is under the control of the WRI1 TF. The short-root phenotype and elevated conjugated indole-3-acetic acid (IAA) levels common to the wri1-1 and the badc1 badc2 double mutant led us to hypothesize that BADC deficiency is responsible for the observed short-root phenotype. Restoration of near-wild type root length upon overexpression of BADC1 in both wri1 and badc1 badc2 is consistent with a yet-to-be identified role for BADCs in seedling development.
RESULTS
The badc1 badc2 Double Mutant Shows Reduced Primary Root Growth and Fewer Lateral Roots Compared to the Wild Type
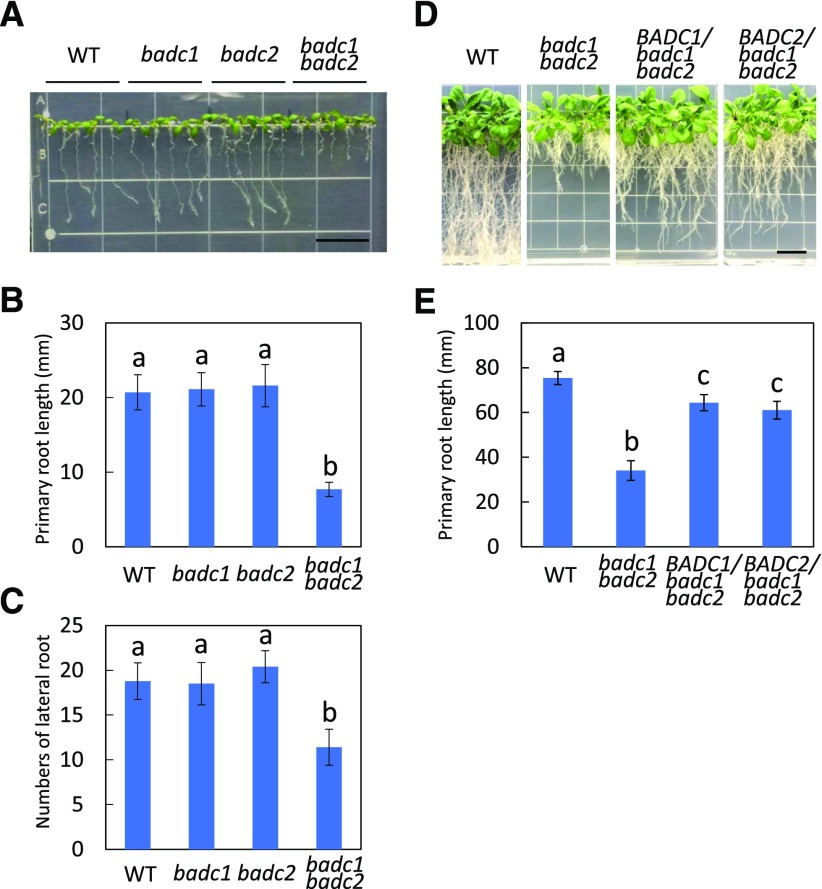
Seeds of the badc1 badc2 double mutant generated in our previous study contained 17% and 23% increases in total FA and TAG accumulation, respectively, compared to the wild type (Keereetaweep et al., 2018). When grown on vertical Murashige and Skoog (MS) plates, badc1 badc2 exhibited a 62% reduction in primary root length and a 39% decrease in lateral root number compared with the wild type (Fig. 1, A–C). In contrast, root growth of badc1 or badc2 single mutants showed no differences from that of the wild type. A complementation experiment was performed in which either BADC1 or BADC2 was expressed under the control of the constitutive 35S promoter in the badc1 badc2 mutant. More than 10 independent lines were generated for each of BADC1 and BADC2 constructs into badc1 badc2 (Supplemental Fig. S1). All transgenic lines demonstrated restored root growth to varying degrees, indicating the involvement of BADC1 and BADC2 in seedling development (Fig. 1, D and E). Because the wri1 mutant (wri1-1) was recently shown to have a short-root phenotype during seedling establishment (Kong et al., 2017), we compared the growth of badc1 badc2 and wri1-1 under the same conditions and show that their phenotypes are visually indistinguishable (Supplemental Fig. S2).
Figure 1.
The badc1 badc2 double mutant shows reduced primary root growth and fewer lateral roots than the wild type (WT). A, Nine-day-old wild-type, badc1, badc2, and badc1 badc2 seedlings grown vertically on one-half strength MS media supplemented with 1% Suc. Bar = 13 mm. B, Primary root length measurement of each genotype in A. C, Lateral root numbers for each genotype in A. Values in B and C represent means ± sd from 10 individual plants for each genotype. D and E, Root growth (D) and primary root length (E) of 20-d-old wild-type, badc1 badc2, and representative transgenic badc1 badc2 plants overexpressing BADC1 or BADC2 lines. Bar = 13 mm. Values in E are means ± sd from 10 individual plants for each indicated genotype. Lowercase letters above histogram bars indicate significant difference (Student's t test for all pairs of genotypes, P < 0.01). Data are representative of three independent repetitions.
Levels of Conjugated Auxin Were Elevated in badc1 badc2 Plants Compared to Wild-Type Plants
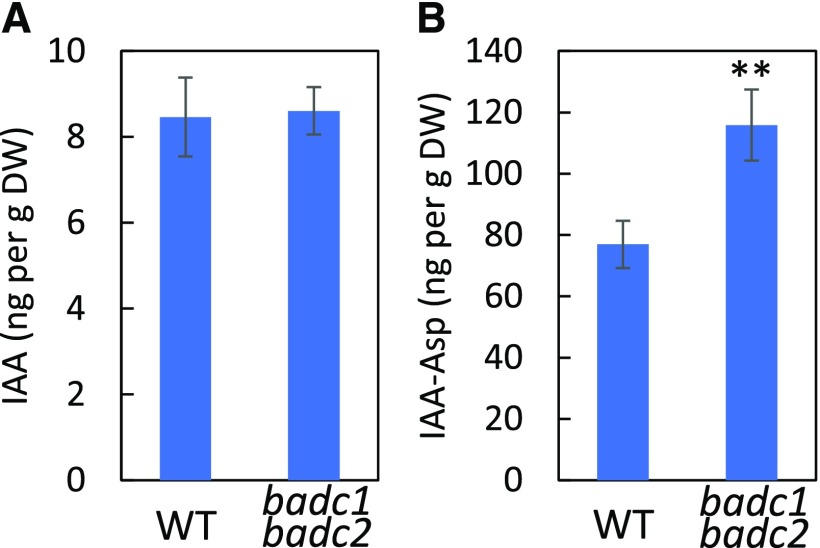
In wri1-1 the levels of IAA-Asp (a conjugated form of auxin thought to be destined for degradation [Ludwig-Müller, 2011]) were reported to be significantly elevated compared to the wild type (Kong et al., 2017). The similarity between the root phenotypes of badc1 badc2 and wri1-1 during seedling establishment prompted us to measure the levels of several growth hormones. As shown in Figure 2, IAA-Asp levels in 7-d-old seedlings showed a highly significant ∼50% increase relative to levels found in the wild type. In contrast, the levels of free IAA and other plant hormones (i.e. abscisic acid, jasmonic acid, and salicylic acid) were not significantly different between badc1 badc2 and the wild type (Fig. 2A; Supplemental Table S1).
Figure 2.
IAA-Asp content is significantly higher in the badc1 badc2 double mutant than in the wild type (WT). A, Quantification of IAA in 7-d-old seedlings of wild type and badc1 badc2. B, Quantification of IAA-Asp in wild type and badc1 badc2. Values in this figure are means ± sd (n = 4) for each sample of 30 seedlings. Asterisks denote a statistically significant difference from the wild type (**P < 0.01; Student's t test). Data in this figure are representative of two independent repetitions.
WRI1 Interacts with AW Boxes from the Promoters of BADCs
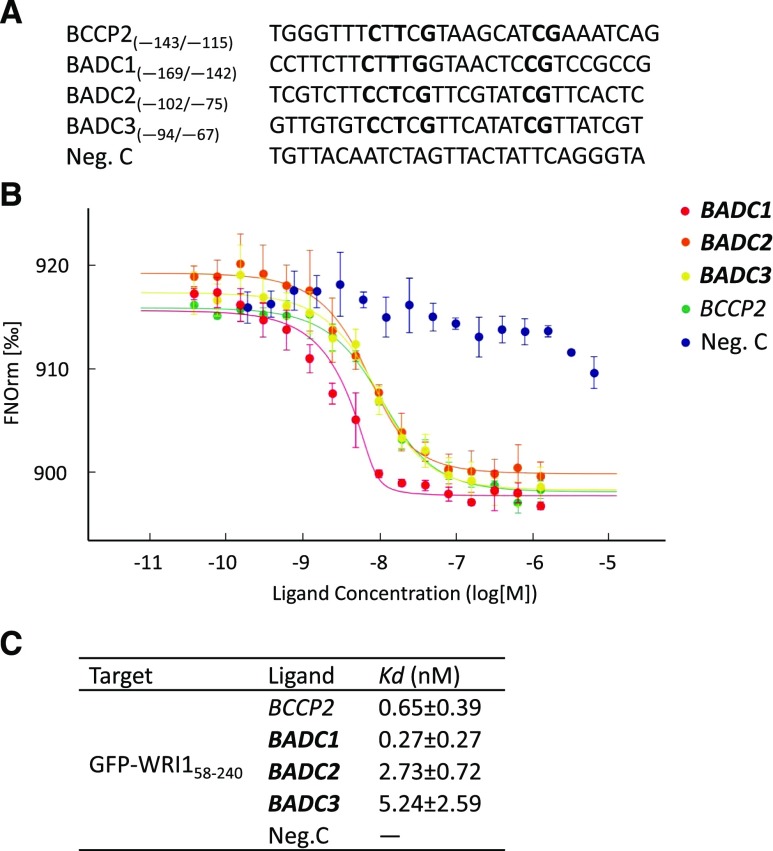
That badc1 badc2 mimics wri1-1 in both its short-root phenotype and IAA-Asp content led us to hypothesize that BADCs are under transcriptional control of WRI1. Sequence analysis identified a canonical AW box consensus sequence within 200 bp upstream of the TIS in the promoters of all three BADC isoforms (Fig. 3A; Supplemental Table S2). To test whether WRI1 binds directly to these AW boxes, microscale thermophoresis (MST) assays were performed, in which a 28-bp double-stranded DNA fragment (Supplemental Table S2) containing each of the AW boxes from BADCs was titrated against purified WRI1 DNA binding domain (amino acids 58 to 240) fused with GFP (Supplemental Fig. S3). As shown in Figure 3, B and C, WRI1 exhibited low equilibrium dissociation constants (K), i.e. tight binding affinities for all three putative AW boxes, with K values between 0.27 and 5.24 nm, comparable to the affinity of WRI1 for the genuine AW box1 from the promoter of the WRI1 target gene BCCP2 (0.65 nm; Maeo et al., 2009). Among the three BADC genes, WRI1 showed the highest affinity for the BADC1 AW box at 0.27 ± 0.27 nm. The AW box from the BCCP1 promoter was also tested, because although BCCP1 contains an AW box (−60/−47 relative to the TIS), the gene was shown not to be regulated by WRI1 (Fukuda et al., 2013). Consistent with this, the K value for double-stranded DNA that includes the BCCP1 AW box and WRI1 showed a K value ∼37-fold higher than those for BCCP2 and WRI1 (Supplemental Fig. S4). To test whether the consensus sequence [CnTnG](n)7[CG] in BADC AW boxes is critical for WRI1 binding, three different mutant sequences were generated for each BADC AW box (Supplemental Table S2) and tested in MST. As shown in Supplemental Table S3, mutations in conserved nucleotides (m1 and m3) dramatically decreased their affinities to WRI1. Mutations in the seven nucleotides (m3) between also decreased their affinity, but to a lesser degree because they are in a less conserved region (Supplemental Table S3).
Figure 3.
Purified WRI1 shows specific binding with AW boxes in the promoters of BADCs. In the thermophoresis experiment, a 28-bp oligonucleotide containing a putative AW box for each gene (A) was titrated against the purified WRI1 DNA binding domain (amino acids 58–240). Dissociation curves for each pair are shown (B) and fitted to the data for calculation of the K values (C). An AW box in the promoter of BCCP2 was used as a positive control. A random sequence is used as a negative control (Neg. C). AW box consensus sequences are marked in bold. Data shown are the means ± sd, n = 3 independent repetitions.
BADC Expression and BADC1 Protein Levels Are Lower in Developing Siliques of wri1-1 than in Those of the Wild Type
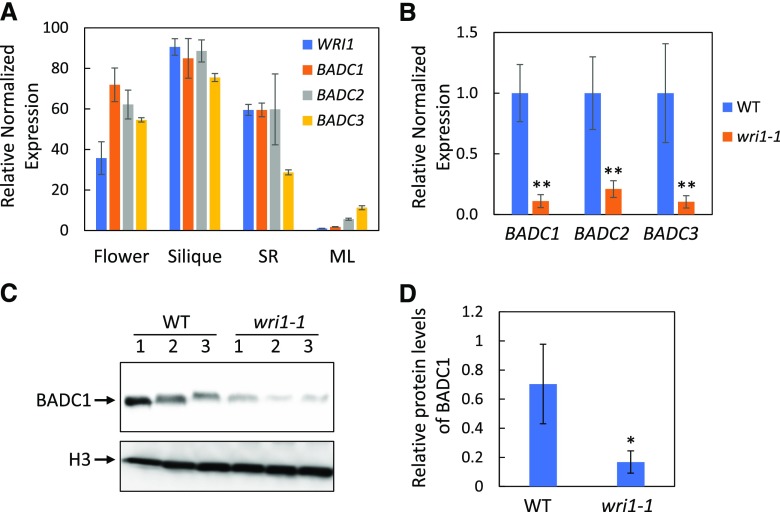
Gene expression quantification of WRI1 and BADCs in different tissues (flower, silique, root, and mature leaf) showed tissue-specific expression patterns qualitatively similar to that of WRI1 (Fig. 4A), i.e. high expression in siliques (developing seeds) and low expression in mature leaves. Because of the highest expression of WRI1 and BADCs in developing seeds among analyzed tissues (Fig. 4A), siliques were selected for investigating the relationship between WRI1 and BADCs. Gene expression of BADCs in developing siliques (7 d after flowering) of wri1-1 were only 10% to 20% of that in the wild type (Fig. 4B). We next used a BADC1 antibody that showed high specificity toward BADC1 protein (Supplemental Fig. S5) to probe BADC1 levels in wri1-1. Consistent with the observed decreased expression of BADC1 in wri1-1 relative to the wild type, the level of BADC1 protein in wri1-1 observed by immunoblotting was only 23% of that in the wild type (Fig. 4, C and D). Since relatively high gene expression of WRI1 and BADCs was also observed in seedling roots (Fig. 4A), BADC1 protein level was quantified in seedling roots and was significantly lower in wri1-1 seedling roots (12 d old) than in those of the wild type (Supplemental Fig. S6).
Figure 4.
BADC expression and BADC1 protein level are significantly lower in wri1-1 developing siliques than in the wild type (WT). A, Tissue expression pattern of WRI1 and BADCs in flowers, siliques (7 d after flowering), seedling roots (SR; 10 d old) and mature leaves (ML). B, Reverse transcription quantitative PCR (RT-qPCR) results of BADC1, BADC2, and BADC3 expression in siliques (7 d after flowering) of wild-type and wri1-1 plants. For A and B, values are means ± sd from three independent experiments. Expression of each gene in the wild type was set to 1. For each experiment, total RNA was isolated from pooled siliques from wild-type and wri1-1 plants. Fbox (At5g15710) and UBQ10 (At4g05320) were used as reference genes. Asterisks denote a statistically significant difference from the wild type (using mean crossing point deviation analysis computed by the relative expression [REST] software algorithm; **P < 0.01). C, BADC1 protein levels in siliques (7 d after flowering) of wild type or wri1-1 are shown by immunoblot with BADC1-specific antibody. Siliques were collected from three independent wild-type or wri1-1 mutant plants. Protein loading is shown by histone H3 in the same protein samples. D, Relative BADC1 protein levels in C quantified with GelAnalyzer2010 and normalized against corresponding protein levels of histone H3. The asterisk denotes a statistically significant difference from the wild type (*P < 0.05; Student's t test). Data in this figure are representative of three independent repetitions.
BADC Gene Expression and BADC1 Protein Levels Are Significantly Higher in Developing Siliques of WRI1 Overexpressing Transgenic Plants than in Those of the Wild Type
To test whether overexpression of WRI1 would increase gene expression of BADCs and the abundance of BADC1 polypeptide, we used two ethanol-inducible WRI1 transgenic lines that were generated in our previous study (Zhai et al., 2017). Both lines demonstrated elevated WRI1 accumulation after induction with 2% (v/v) ethanol treatment (Zhai et al., 2017). As shown in Figure 5A, BADC, WRI1, and BCCP2 expression in developing siliques of WRI1 transgenic plants that were induced by irrigation with 2% (v/v) ethanol for 4 d was significantly higher than either the wild type or the corresponding WRI1 transgenic plant lines that were not exposed to ethanol treatment (Fig. 5A). Consistent with the elevated expression of BADC1, BADC1 protein levels were also significantly higher upon WRI1 overexpression (Fig. 5, B and C). To test whether global regulation of WRI1 on BADCs, gene expression, and protein levels were also quantified in the seedling roots of WRI1 transgenics, as shown in Supplemental Figure S7, both gene expression and protein levels were higher in WRI1 transgenics induced by ethanol treatment than either nonethanol treatment control or wild type (Supplemental Fig. S7).
Figure 5.
BADC expression and BADC1 protein levels are elevated in inducible WRI1 transgenic plants. A, Relative gene expression of BADCs, BCCP2, and WRI1 in siliques (7 d after flowering) of wild-type (WT) or two independent ethanol inducible WRI1 transgenic lines (AlcA:WRI1 line 1 and line 4) treated (+) or not treated (−) with 2% ethanol for 4 d. B, BADC1 protein levels. C, Relative BADC1 protein levels in B, quantified with GelAnalyzer2010 and normalized against corresponding protein levels of histone H3. Asterisks denote statistically significant difference from the wild type (*P < 0.05; Student's t test). Data in this figure are representative of three independent repetitions.
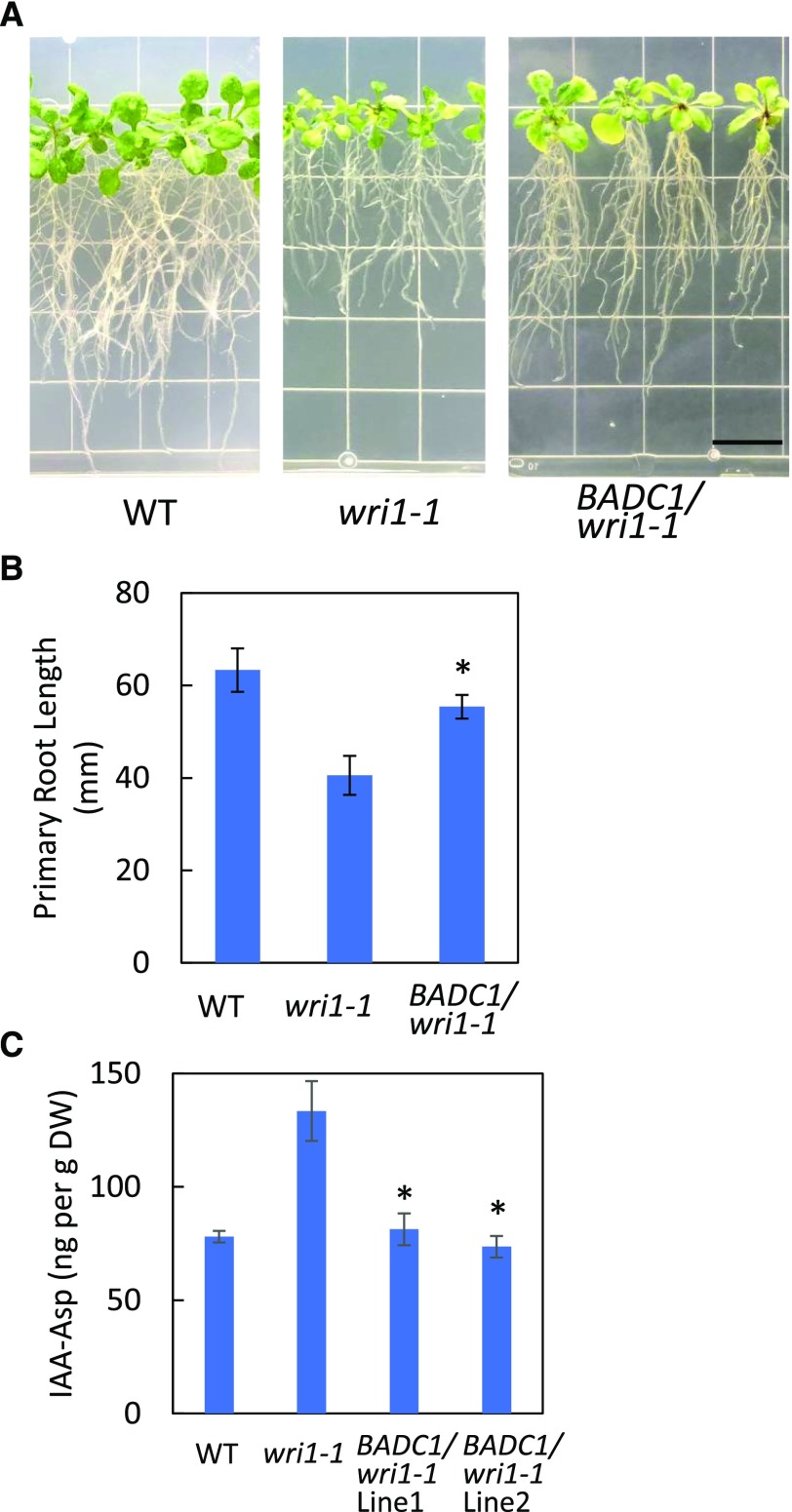
Overexpression of BADC1 in wri1-1 Partially Rescues the wri1-1 Short-Root Phenotype
The data presented above support the hypothesis that the expression of BADC genes is under the control of WRI1 and that the short-root phenotype of wri1-1 primarily results from a deficiency of BADC1 and BADC2. To test this hypothesis, the coding sequence of BADC1 was placed under the control of the 35S promoter and transformed into wri1-1. Fifteen of 17 independent transgenics demonstrated significantly longer primary root length than that of wri1-1 when germinated and grown vertically on one-half strength MS medium plates for 16 d (Fig. 6, A and B). Immunoblotting showed BADC1 protein levels were significantly increased in the BADC1/wri1-1 transgenics (Supplemental Fig. S8). To test whether the levels of IAA-Asp relate with the short-root phenotype of wri1-1, IAA contents were measured in 7-d-old seedlings of BADC1/wri1-1, wri1-1, and the wild type. The levels of IAA-Asp showed a significant decrease in BADC1/wri1-1 relative to wri1-1 (Fig. 6C), consistent with BADC1 playing a downstream role in WRI1-dependent root development.
Figure 6.
Overexpression of BADC1 in wri1-1 partially rescues wri1-1 short-root phenotype. A and B, Root growth (A) and primary root length (B) measurement of 16-d-old wild-type (WT), wri1-1, and representative transgenic wri1-1 overexpressing BADC1 lines. Bar = 13 mm. Values in B are means ± sd from 10 individual plants for each indicated genotype. The asterisk denotes a statistically significant difference from the wri1-1 mutant (*P < 0.05; Student's t- test). C, IAA-Asp was quantified in 7-d-old seedlings of wild-type, wri1-1, and BADC1/wri1-1 transgenic lines 1 and 2. Asterisks denote statistically a significant difference from wri1-1 (*P < 0.05; Student's t test). Data in this figure are representative of three independent repetitions.
DISCUSSION
In this work we establish that expression of BADC1, BADC2, and BADC3 is under the transcriptional control of WRI1. Evidence to support this comes from: 1) identification of AW boxes within 200 bp upstream of the TIS; 2) MST measurement demonstrating tight binding, i.e. K values in the low nanomolar range between WRI1 and the AW boxes from all three BADC genes; 3) expression of each of the three BADC transcripts is reduced (by ∼85%) in the wri1-1 mutant; 4) protein levels of BADC1 are reduced similarly to reductions in the level of its transcript in the wri1-1 mutant; and 5) both gene expression of BADCs and protein levels of BADC1 are increased upon overexpression of WRI1.
That WRI1, the master transcriptional activator of genes involved in FAS, simultaneously activates the transcription of BADCs, which have been shown to act as negative regulators of ACCase, might at first appear paradoxical. However, FAS is metabolically expensive in terms of both energy and reducing power, creating an imperative for tight metabolic control. For example, ACCase is generally acknowledged to be rate limiting with respect to FAS under tight genetic and biochemical control (Salie et al., 2016). When nonesterified FA levels in the cell were increased beyond demand by supplementation with Tween-esters, two phases of feedback inhibition were reported, one short term, the second longer term (Andre et al., 2012). Short-term inhibition occurs when the levels of oleoyl-ACP, the terminal product of the ACP track of FAS in the plastid, increase. Oleoyl-ACP inhibits ACCase by binding to it in a reversible manner (Andre et al., 2012). If, however, excess FA levels persist for 2 d or more, an irreversible phase of inhibition of ACCase is initiated that is primarily mediated by BADC1 and BADC3 (Keereetaweep et al., 2018). In this context, BADC subunits should not be regarded as a simple inhibitor, but rather as a conditional inhibitor of ACCase. Thus, coexpression of the ∼20 genes that promote FAS, along with the three BADC subunits, makes biological sense. As WRI1 increases cellular metabolic demand by activating FAS, it concomitantly increases the capacity to downregulate ACCase, and thereby FAS, by increasing abundance of the conditionally inhibitory BADC subunits. Coregulation of FAS and the BADCs thus allows the cell to match synthetic capacity with its regulatory capacity. The biochemical regulation of ACCase described herein is complementary to the transcriptional regulation of FAS we recently reported, in which levels of the WRI1 polypeptide are tightly coupled to the availability of cellular carbon in the form of sugars. When the carbon/energy status of the cell decreases, the SnRK1 carbon-sensing kinase becomes activated and phosphorylates WRI1, leading to its selective proteasomal degradation, thereby reducing activation of FAS genes (Zhai et al., 2017). Conversely, when carbon/energy abundance increases, levels of the SnRK1 kinase inhibitor trehalose 6-phosphatase increase, reducing WRI1 phosphorylation/degradation, thereby increasing the amount of WRI1 available to activate transcription of FAS genes (Zhai et al., 2018).
In this study we also observed that the badc1 badc2 double mutant exhibits reduced primary root growth, a phenotype that resembles that of the wri1-1 mutant (Kong et al., 2017), which in combination with our finding that WRI1 controls BADC transcription led us to ask whether the short-root phenotype characteristic of both wri1-1 and badc1 badc2 mutants results from a deficit of BADC. If correct, overexpression of BADC in wri1-1 would mitigate the wri1-1 short-root phenotype. This view is supported by the observation that overexpression of BADC1 in wri1-1 partially rescued the short-root phenotype in a manner similar to its overexpression in the badc1 badc2 mutant background (and as overexpression of BADC2 had in the badc1 badc2 mutant background). Consistent with this, the elevated levels of IAA-Asp observed in wri1-1 were restored to wild-type levels in BADC1/wri1-1. Taken together, these observations suggest that deficiency of BADC levels, resulting either directly via mutation of badc genes, or indirectly through the mutation of their upstream transcriptional activator, wri1, result in a short-root phenotype via a mechanism yet to be determined. Further work will be required to identify details of the mechanism, which is likely related to increased levels of conjugated IAA that were observed for both wri1-1 and badc1 badc2 mutants.
Taken together, we show that all three BADC genes are direct target genes of WRI1 and can play a role in root development. The observation that WRI1 upregulates not only genes contributing to FAS, but also BADCs, which are conditional regulators of FAS, identifies an additional layer of regulation with respect to FAS.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Wild-type Arabidopsis (Arabidopsis thaliana ecotype Columbia-0) and wri1-1 (CS69538), badc1 (Salk 000817C), badc2 (Salk 021108C), and badc3 (CS2103834) were used in this study. The double mutant badc1 badc2 was generated as described previously by Keereetaweep et al. (2018). For growth on plates, seeds were surface sterilized with 70% (v/v) ethanol, followed by 30% (v/v) bleach containing 0.01% Tween20, and rinsed three times with sterile water. Seeds were stratified for 3 d at 4°C in the dark and germinated on one-half strength MS medium supplemented with 1% (w/v) Suc plates in an incubator with a light/dark cycle of 18h/6h at 23°C, photosynthetic photon flux density of 250 μmol m−2 s−1, and 75% relative humidity.
Genetic Constructs
Coding sequences of BADC1 and BADC2 were amplified by PCR from cDNAs using the primers listed in Supplemental Table S4. The PCR products were then cloned into the Invitrogen GATEWAYM pDONR/Zeo vector (Thermo Fisher Scientific) using the BP reaction and subcloned (LR reaction) into the plant GATEWAY binary vector pGWB414 (Nakagawa et al., 2007). BADC1/pGWB414 and BADC2/pGWB414 were introduced into badc1 badc2 double mutants by Agrobacterium tumefaciens (AgL0)-mediated transformation. For rescuing the short-root phenotype of wri1-1, BADC1/pGWB414 was transformed into wri1-1.
Quantification of IAA, IAA-Asp in badc1 badc2 Double Mutant Seedlings
Seven-day-old Arabidopsis wild-type and badc1 badc2 seedlings grown vertically on one-half strength MS medium supplemented with 1% (w/v) Suc were harvested (20–30 seedlings per replicate, ∼100 mg fresh weight) and frozen in liquid nitrogen. Frozen samples were sent to the Proteomics & Mass Spectrometry Facility at the Donald Danforth Plant Science Center for acidic hormone analysis, where frozen plant material was extracted twice with ice-cold acetonitrile/methanol (1:1 v/v) using a bead beater and resulting samples were dried with the use of a speedvac. Samples were reconstituted in 30% MeOH and separated via HPLC using a Waters Acquity UPLC BEH C18 1.0 × 100 mm, 1.7 µm column kept at 50°C with a flow rate of 15 µL/min coupled to an AB Sciex 6500 QTrap mass spectrometer operated in multiple reaction monitoring mode employing polarity switching for simultaneous detection of positive and negative ions. Data analysis was completed using MultiQuant 3.0.2 (AB Sciex) with unlabeled compound peak areas normalized to their respective labeled internal standard peak areas. Labeled standards for IAA and IAA-Asp were obtained from Sigma-Aldrich. Calibration curves were linear (r > 0.99) within the ranges provided above and applying 1/x weighting.
RNA Isolation and RT-qPCR
To quantify gene expression in mutants and transgenic plants, total RNA was isolated using the RNeasy Plant Mini Kit (Qiaqen). cDNA was prepared using SuperScript IV VILO Master Mix with ezDNase Enzyme (Invitrogen). RT-qPCR reactions were set up with SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) and gene-specific primers (primer sequences are listed in Supplemental Table S4) and performed with the CFX96 qPCR Detection System (Bio-Rad). Statistical analysis of RT-qPCR data were carried out with REST2009.
Expression and Purification of Recombinant GFP-WRI158–240
The DNA sequence of full-length WRI1 fused with GFP on its N terminus (GFP-WRI1) was amplified from an OWD5 (Oleosin+WRI1+DAGT1) vector previously described in Zhai et al. (2017). GFP-WRI1 then was inserted into pet28b between XhoI and NcoI by in-fusion cloning. Full-length WRI1 in the resulting GFP-WRI1/pet28b was replaced with the DNA sequence corresponding to the WRI1 DNA binding domain (WRI158–240) between XhoI and BsrGI (Kong et al., 2017). The primer pairs used in building this construct are listed in Supplemental Table S4. Recombinant GFP-WRI158–240 was expressed in Escherichia coli BL21(DE3). Protein purification was performed as reported by Zhai et al. (2017).
MST
Thermophoretic assays were conducted using a Monolith NT.115 apparatus (NanoTemperTechnologies). For determining K values, 8 nm of GFP-WRI158–240 was incubated with a serial dilution of the ligand (double-stranded DNA) from 1.25 µm to 38.81 pm. Samples of ∼10 μL were loaded into capillaries and inserted into the MST instrument loading tray (Monolith NT.115). The thermophoresis experiments were carried out using 40% MST power and 80% LED power at 25°C.
Antibody and Immunoblotting
Anti-BADC1 and antihistone H3 antibodies were used in this study. Anti-BADC1 antibodies were requested from Dr. Jay Thelen (Salie et al., 2016). Histone H3 polyclonal antibodies were purchased from Agrisera (catalog no. AS10710). A total of 50 mg of siliques (7 d after flowering) of wild-type and wri1-1 mutant plants were ground in liquid nitrogen and then mixed with 200 mL of preheated protein extraction buffer (8 m urea, 2% SDS, 0.1 m dithiothreitol, 20% glycerol, 0.1 m Tris-HCl, pH 6.8, and 0.004% Bromophenol Blue). Samples were centrifuged at 17,000 × g before transferring the supernatant to a new microcentrifuge tube and then loading into SDS-PAGE. Twenty μL of supernatant was loaded and resolved in SDS-PAGE and transferred onto polyvinylidene difluoride membrane for immunoblot analysis. Primary antibodies of anti-BADC1 were used at a 1:3,000 dilution, and the anti-H3 antibodies were used at a 1:5,000 dilution. Immunoblots of targeted proteins were visualized using horseradish peroxidase-conjugated secondary antibodies (catalog no. AP187P, Millipore) with SuperSignal West Femto Maximum Sensitivity Substrate (Catalog No. 34095, ThermoFisher). Immunoblot signals were detected and digitalized with Image Quant LAS4000 and quantified with GelAnalyzer2010a.
Accession Numbers
Sequence data from this article can be found in The Arabidopsis Information Resource under the following accession numbers: AT3G54320 (WRI1), AT3G56130 (BADC1), AT1G52670 (BADC2), AT3G15690 (BADC3), AT5G16390 (BCCP1), AT5G15530 (BCCP2), AT5G15710 (Fbox), and AT4G05320 (UBQ10).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. BADC1 expression in transgenic badc1 badc2 double mutants overexpressing BADC1 lines.
Supplemental Figure S2. badc1 badc2 shows a short-root phenotype similar to that of wri1-1.
Supplemental Figure S3. Purified recombinant GFP-WRI158-240.
Supplemental Figure S4. Putative AW boxes show varied binding affinity with WRI1.
Supplemental Figure S5. The BADC1 antibody used in this study specifically recognizes BADC1.
Supplemental Figure S6. BADC1 protein levels are lower in the roots of seedlings of wri1-1 than in the wild type.
Supplemental Figure S7. Both BADC1 expression and protein levels are elevated in the roots of WRI1-inducible expression transgenic plants.
Supplemental Figure S8. BADC1 protein levels in the BADC1/wri1-1 transgenic lines.
Supplemental Table S1. Quantification of plant hormones in badc1 badc2 double mutant seedlings.
Supplemental Table S2. Nucleotide sequences of AW boxes in the thermophoretic experiment.
Supplemental Table S3. Binding affinity of BADC AW boxes with and without mutations to WRI1 were quantified as equilibrium dissociation constants (K) using MST.
Supplemental Table S4. Oligonucleotide sequences of primers used in this study.
Acknowledgments
We thank Dr. Jay Thelen (University of Missouri) for the gift of the BADC1 antibody.
Footnotes
This work was supported by grants from the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences, U.S. Department of Energy (DOE KC0304000 to J.Sh. and DE-SC0012704 to J.Sc.) and by a National Science Foundation grant under which initial discussions took place and several reagents were made (DBI 1117680 to J.Sh. and J.Sc.).
Articles can be viewed without a subscription.
References
- Andre C, Haslam RP, Shanklin J (2012) Feedback regulation of plastidic acetyl-CoA carboxylase by 18:1-acyl carrier protein in Brassica napus. Proc Natl Acad Sci USA 109: 10107–10112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Mendoza MS, To A, Harscoët E, Lepiniec L, Dubreucq B (2007) WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J 50: 825–838 [DOI] [PubMed] [Google Scholar]
- Braybrook SA, Stone SL, Park S, Bui AQ, Le BH, Fischer RL, Goldberg RB, Harada JJ (2006) Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc Natl Acad Sci USA 103: 3468–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focks N, Benning C (1998) wrinkled1: A novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol 118: 91–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda N, Ikawa Y, Aoyagi T, Kozaki A (2013) Expression of the genes coding for plastidic acetyl-CoA carboxylase subunits is regulated by a location-sensitive transcription factor binding site. Plant Mol Biol 82: 473–483 [DOI] [PubMed] [Google Scholar]
- Grimberg Å, Carlsson AS, Marttila S, Bhalerao R, Hofvander P (2015) Transcriptional transitions in Nicotiana benthamiana leaves upon induction of oil synthesis by WRINKLED1 homologs from diverse species and tissues. BMC Plant Biol 15: 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagaya Y, Okuda R, Ban A, Toyoshima R, Tsutsumida K, Usui H, Yamamoto A, Hattori T (2005) Indirect ABA-dependent regulation of seed storage protein genes by FUSCA3 transcription factor in Arabidopsis. Plant Cell Physiol 46: 300–311 [DOI] [PubMed] [Google Scholar]
- Keereetaweep J, Liu H, Zhai Z, Shanklin J (2018) Biotin attachment domain-containing proteins irreversibly inhibit acetyl CoA carboxylase. Plant Physiol 177: 208–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Q, Ma W, Yang H, Ma G, Mantyla JJ, Benning C (2017) The Arabidopsis WRINKLED1 transcription factor affects auxin homeostasis in roots. J Exp Bot 68: 4627–4634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Shao J, Tang S, Shen Q, Wang T, Chen W, Hong Y (2015) Wrinkled1 accelerates flowering and regulates lipid homeostasis between oil accumulation and membrane lipid anabolism in Brassica napus. Front Plant Sci 6: 1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y, Shorrosh B, Beisson F, Andersson MX, Arondel V, Bates PD, Baud S, Bird D, DeBono A, Durrett TP (2013) Acyl-lipid metabolism. The Arabidopsis Book 8: e0161, doi:10.1199/tab.0133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan T, Ohto M, Yee KM, West MA, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ (1998) Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93: 1195–1205 [DOI] [PubMed] [Google Scholar]
- Ludwig-Müller J. (2011) Auxin conjugates: Their role for plant development and in the evolution of land plants. J Exp Bot 62: 1757–1773 [DOI] [PubMed] [Google Scholar]
- Maeo K, Tokuda T, Ayame A, Mitsui N, Kawai T, Tsukagoshi H, Ishiguro S, Nakamura K (2009) An AP2-type transcription factor, WRINKLED1, of Arabidopsis thaliana binds to the AW-box sequence conserved among proximal upstream regions of genes involved in fatty acid synthesis. Plant J 60: 476–487 [DOI] [PubMed] [Google Scholar]
- Mu J, Tan H, Zheng Q, Fu F, Liang Y, Zhang J, Yang X, Wang T, Chong K, Wang X-J, et al. (2008) LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol 148: 1042–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, Niwa Y, et al. (2007) Improved gateway binary vectors: High-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem 71: 2095–2100 [DOI] [PubMed] [Google Scholar]
- Ohlrogge J, Browse J (1995) Lipid biosynthesis. Plant Cell 7: 957–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olinares PDB, Ponnala L, van Wijk KJ (2010) Megadalton complexes in the chloroplast stroma of Arabidopsis thaliana characterized by size exclusion chromatography, mass spectrometry, and hierarchical clustering. Mol Cell Proteomics 9: 1594–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy F, Valon C, Kohara A, Miséra S, Giraudat J (1997) The ABSCISIC ACID-INSENSITIVE3, FUSCA3, and LEAFY COTYLEDON1 loci act in concert to control multiple aspects of Arabidopsis seed development. Plant Cell 9: 1265–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawsthorne S. (2002) Carbon flux and fatty acid synthesis in plants. Prog Lipid Res 41: 182–196 [DOI] [PubMed] [Google Scholar]
- Ruuska SA, Girke T, Benning C, Ohlrogge JB (2002) Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 14: 1191–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salie MJ, Zhang N, Lancikova V, Xu D, Thelen JJ (2016) A family of negative regulators targets the committed step of de novo fatty acid biosynthesis. Plant Cell 28: 2312-2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos Mendoza M, Dubreucq B, Miquel M, Caboche M, Lepiniec L (2005) LEAFY COTYLEDON 2 activation is sufficient to trigger the accumulation of oil and seed specific mRNAs in Arabidopsis leaves. FEBS Lett 579: 4666–4670 [DOI] [PubMed] [Google Scholar]
- To A, Valon C, Savino G, Guilleminot J, Devic M, Giraudat J, Parcy F (2006) A network of local and redundant gene regulation governs Arabidopsis seed maturation. Plant Cell 18: 1642–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Z, Liu H, Shanklin J (2017) Phosphorylation of WRINKLED1 by KIN10 results in its proteasomal degradation, providing a link between energy homeostasis and lipid biosynthesis. Plant Cell 29: 871–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Z, Keereetaweep J, Liu H, Feil R, Lunn JE, Shanklin J (2018) Trehalose 6-phosphate positively regulates fatty acid biosynthesis by stabilizing WRINKLED1. Plant Cell 30: 2616–2627 [DOI] [PMC free article] [PubMed] [Google Scholar]