Villins are required for the action of an auxin transport inhibitor on actin cytoskeleton and polar auxin transport in Arabidopsis.
Abstract
Auxin transport inhibitors are essential tools for understanding auxin-dependent plant development. One mode of inhibition affects actin dynamics; however, the underlying mechanisms remain unclear. In this study, we characterized the action of 2,3,5-triiodobenzoic acid (TIBA) on actin dynamics in greater mechanistic detail. By surveying mutants for candidate actin-binding proteins with reduced TIBA sensitivity, we determined that Arabidopsis (Arabidopsis thaliana) villins contribute to TIBA action. By directly interacting with the C-terminal headpiece domain of villins, TIBA causes villin to oligomerize, driving excessive bundling of actin filaments. The resulting changes in actin dynamics impair auxin transport by disrupting the trafficking of PIN-FORMED auxin efflux carriers and reducing their levels at the plasma membrane. Collectively, our study provides mechanistic insight into the link between the actin cytoskeleton, vesicle trafficking, and auxin transport.
The phytohormone auxin plays critical roles in various plant developmental programs by controlling cell expansion and polarity, as well as organ patterning. Auxin action relies on polar transport through different plant tissues (Benjamins and Scheres, 2008; Bennett and Leyser, 2014). Our knowledge of the mechanisms of polar auxin transport (PAT) has been extended by the use of synthetic auxin transport inhibitors (ATIs). ATIs inhibit auxin efflux and block polar auxin flow between cells (Snyder, 1949; Niedergang-Kamien and Leopold, 1957). However, the mechanism of ATI action remains largely elusive. It has long been recognized that one mode of ATI action is its impact on actin cytoskeleton (Rahman et al., 2007; Dhonukshe et al., 2008; Higaki et al., 2010; Zhu et al., 2016). Treatments with ATIs such as 1-naphthylphthalamic acid (NPA), 2,3,5-triiodobenzoic acid (TIBA), and 2-(1-pyrenoyl) benzoic acid lead to actin rearrangements in Arabidopsis (Arabidopsis thaliana). However, different ATIs have distinct impacts on actin organization. Long-term treatment with NPA reduces the abundance of F-actin and generates punctate structures in root epidermal cells, whereas TIBA tends to increase the abundance of actin filaments and enhances filament bundling (Rahman et al., 2007). Following short-term treatment with TIBA or 2-(1-pyrenoyl) benzoic acid, Dhonukshe et al. (2008) observed a more bundled and less dynamic actin array. Using live-cell imaging techniques, NPA-induced actin rearrangement has recently been revisited by Zhu et al. (2016), who found that actin arrays display significantly decreased filament density and increased bundling after long-term treatment with NPA. Quantitative analyses of actin filament dynamics suggest that NPA lowered actin filament dynamics by increasing actin filament lengths and lifetimes, as well as decreasing filament severing; it also reduced the frequency of actin filament bundling and debundling (Zhu et al., 2016). The different impacts on actin cytoskeleton by various ATIs suggest distinct underlying mechanisms. However, except for NPA, the precise mechanism of actin remodeling in response to other ATIs remains unclear.
One major role of actin cytoskeleton in auxin transport is to regulate vesicular trafficking of auxin efflux carriers, PIN-FORMED (PIN) family (Dhonukshe et al., 2008; Blancaflor, 2013; Zhu and Geisler, 2015). PIN proteins show a polar localization that determines the direction of auxin flow, and PIN targeting is a highly dynamic process with constitutive cycling between the plasma membrane (PM) and endosomal compartments (Friml, 2010). Actin cytoskeleton is required for both endocytosis and recycling of PINs (Zhu and Geisler, 2015). Disrupting actin arrays with latrunculin B or cytochalasin D inhibited brefeldin A (BFA)–induced internalization of PIN1 as well as its relocalization to the PM when BFA was washed out. Similar effects were observed when cells were treated with ATIs (Geldner et al., 2001). Additionally, genetic disruption of actin cytoskeleton also impairs PIN trafficking (Wu et al., 2015; Zhu and Geisler, 2015; Zhu et al., 2016). Mutants of ACTIN genes in Arabidopsis show abnormal accumulation of PIN2 in the intracellular compartments (Lanza et al., 2012; Zhu et al., 2016). Rice morphology determinant, a type II formin, was shown to regulate PM localization and internalization of PIN2 (Li et al., 2014a). In the rice (Oryza sativa) villin2 (vln2) mutant, increased internalization and inhibited recycling of PIN2 led to reduced abundance of PIN2 on the PM (Wu et al., 2015). These pharmacological and genetic perturbations of actin dynamics often cause defects in PAT (Dhonukshe et al., 2008; Li et al., 2014a; Wu et al., 2015; Zhu et al., 2016), further demonstrating the importance of actin cytoskeleton and actin-mediated PIN trafficking for auxin transport.
In addition to cycling between PM and endosomes, PINs also enter endocytic sorting to the vacuole for protein degradation (Petrásek et al., 2006; Kleine-Vehn et al., 2008; Laxmi et al., 2008). These two trafficking pathways have been proposed to enable changes in PIN polarity or to control the abundance of PINs at the PM, and thus determine auxin transport in response to environmental stimuli (Friml, 2010). Treatment with latrunculin B inhibits vacuole degradation of PIN2, suggesting the requirement of actin cytoskeleton in this process (Kleine-Vehn et al., 2008).
The use of ATIs provides insights into the mechanisms of actin-dependent PIN trafficking for auxin transport and is useful to identify responsible players. TWISTED DWARF1 (TWD1) has been characterized as a target of NPA and mediates NPA action on actin remodeling (Zhu et al., 2016). However, the mechanism that controls actin remodeling by TWD1 is rather indirect, because it does not bind to actin and directly regulate actin dynamics (Zhu et al., 2016). Therefore, the direct connection between ATIs and actin cytoskeleton remains obscure. Here, using live-cell imaging and quantitative analyses of actin dynamics, the effect of TIBA on actin organization and dynamics was studied in detail. Following treatment with TIBA, increases in actin filament abundance and bundling were detected in root epidermal cells. Changes in actin organization resulted from reduced actin turnover and enhanced actin bundle formation. We further found that Arabidopsis villin isovariants are cytoskeletal targets of TIBA. VLN4 has the highest affinity to this inhibitor. It has been reported that recombinant VLN4 exhibits actin filament bundling, Ca2+-dependent severing, and barbed-end-capping activities (Zhang et al., 2011). Loss of VLN4 in Arabidopsis leads to impaired actin array in root hairs and reduces root hair growth (Zhang et al., 2011). Here, we show that mutants of VLN4 are less sensitive to TIBA. TIBA-induced actin dynamics were altered in vln4 knockout mutants. VLN4-dependent actin remodeling is involved in the action of TIBA on trafficking of auxin exporters and PAT. TIBA directly binds to the C-terminal headpiece domain of VLN4 (VHP) and promotes oligomerization of VLN4, leading to enhanced formation of actin bundles. TIBA is less effective in rearranging actin filaments and inhibiting PAT in plants with VLN4 headpiece domain deletion. Our data uncover the molecular mechanism by which villins contribute to the action of TIBA on actin cytoskeleton, supporting the important role of actin dynamics in the mechanism of auxin transport.
RESULTS
Cortical Actin Arrays in Root Epidermal Cells Remodel Rapidly following TIBA Treatment
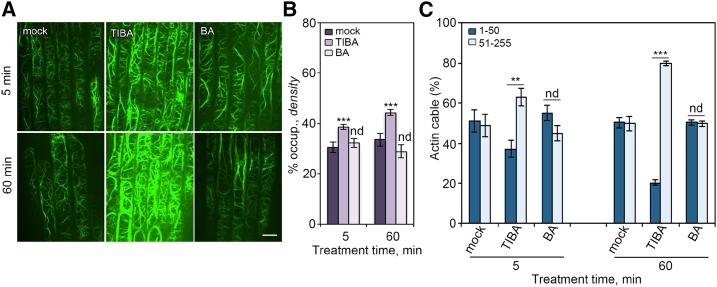
TIBA was previously shown to affect actin organization in plant cells (Rahman et al., 2007; Dhonukshe et al., 2008; Higaki et al., 2010); however, the molecular mechanism of TIBA-induced actin rearrangement remains unclear. Here, we revisited TIBA and studied its effect on actin organization and dynamics in detail. Arabidopsis seedlings expressing the actin reporter (GFP)-tagged Fimbrin Actin Binding Domain 2 (fABD2; Sheahan et al., 2004) were treated with TIBA at various concentrations and time points. Cortical actin arrays in epidermal cells from root transition and elongation zone were imaged with spinning disk confocal microscopy. A more dense and robustly bundled actin array was observed following treatment with TIBA (Fig. 1A), which is similar to previous data shown by Dhonukshe et al. (2008). To verify the changes to the actin network, the optical densities of actin filament structures were analyzed by measuring the percentage of occupancy of actin filaments (Fig. 1B; Higaki et al., 2010; Henty et al., 2011). Additionally, intensity profiles of GFP fluorescence were created (Fig. 1C; Martin et al., 2007; van der Honing et al., 2012). In these intensity profiles, high peaks represent brightly labeled actin bundles, while low peaks represent weakly labeled actin filament bundles (or perhaps single actin filaments). We distributed these peaks in two classes: high (51∼255) and low (1∼50) gray levels (Fig. 1C).
Figure 1.
Cortical actin array rearranges in response to TIBA treatment. A, Representative images of the cortical actin array in epidermal cell from root tips. Five-day-old Arabidopsis seedlings were treated with 10 µm TIBA or 50 µm benzoic acid (BA) for indicated times. Bar = 10 µm. B and C, Actin architecture was measured on images shown in A. B, Percentage of occupancy, or density, is a measure of the abundance of actin filaments in the cortical array. C, Quantification of the fluorescence intensity of actin cables. We measured the peaks of the fluorescence profile along a line drawn across actin cables and subtracted the background value. For ease of comparison, populations of fluorescence intensities that were lower than 50 and higher than 50 were binned. Values given are means ± se (n > 300 images from 25 seedlings for each treatment; **P < 0.01; ***P < 0.001; nd, no significant difference between mock and treatment; t test; Pearson’s χ2 test was applied to evaluate significant differences in the frequency distribution across intensity classes between mock and treatment).
The optical density value was significantly higher in TIBA-treated cells, as shown in Figure 1B, confirming the observation that TIBA treatment results in a more crowded actin array. The optical density measures the occupancy of GFP signal, not the actual actin filament density. We further estimated the relative level of actin filaments by analyzing the total intensity in filamentous structures. The total intensities were then normalized to the intensity of single actin filaments to account for variance in actin reporter expression or optical efficiency during imaging. As shown in Supplemental Figure S1, the mean fluorescence intensity values for more than 300 single filaments from each treatment showed no significant differences (Supplemental Fig. S1A). Additionally, the relative amount of actin filaments was significantly increased after TIBA treatment (Supplemental Fig. S1B), which is consistent with the results from the optical density analysis. The frequency distribution of the number of peaks across the two classes was clearly different between mock- and TIBA-treated cells (Fig. 1C). In mock-treated cells, the peaks belonging to each class were equally distributed, whereas the peaks with high fluorescence intensity were more abundant in TIBA-treated cells (63%, 79% after 5- and 60-min treatment, respectively), representing thicker actin bundles. Pearson’s χ2 test showed that the frequency distribution across the two classes was significantly different between mock- and TIBA-treated cells (Fig. 1C). TIBA-induced actin responses were both dose- and time-dependent (Fig. 1, B and C; Supplemental Fig. S2). The actin rearrangements were specific because treatment with an inactive analog, BA, had no noticeable effect on actin organization (Fig. 1). These data demonstrate that TIBA treatment results in a high abundance of thick actin filament bundles in root epidermal cells.
To investigate the underlying mechanisms of TIBA-induced actin rearrangement, we further examined the single filament dynamics in root cells following TIBA treatment. In mock-treated cells, growing actin filaments elongated at a mean rate of 1.5 µm s−1 to an average maximum filament length of 7 µm (Table 1). Filaments were present for 15 s before disassembling through severing activity (Table 1). Most severed ends did not regrow (2.4%) or anneal to other filaments (11.7%). Actin bundle formation occurred at a frequency of 6.1 × 10−6 events/µm2/filament/s (Table 1). The filament-debundling events, however, were less frequent (1.7 × 10−6 events/µm2/filament/s; Table 1). After treatment of TIBA, a subset of these parameters changed significantly (Table 1). Notably, the filament elongation rate increased significantly. TIBA also elicited an increase in the average maximum length and lifetime of growing filaments, as well as the frequency of bundle formation. A 2-fold reduction in severing frequency was detected in TIBA-treated cells when compared with the mock control (Table 1). However, filament–filament annealing, regrowth, and debundling frequency were not significantly altered following TIBA treatment. In summary, TIBA treatment reduces actin filament turnover and increases actin filament bundle formation.
Table 1. Single filament dynamics in wild-type treated with mock or 10 µm TIBA.
Values given are the mean ± se, with n > 50 filaments from n > 10 root epidermal cells and at least 10 seedlings per treatment. Significantly different from mock control value by t test; **P < 0.01, ***P < 0.001. nd, Not significantly different from mock control value by t test; P > 0.05.
| Stochastic Dynamics Parameters | Mock | TIBA |
|---|---|---|
| Elongation rate (µm/s) | 1.5 ± 0.1 | 1.8 ± 0.1** |
| Max. filament length (µm) | 7.1 ± 0.3 | 9.6 ± 0.5*** |
| Max. filament lifetime (s) | 14.7 ± 1.3 | 23.1 ± 1.7*** |
| Severing frequency (breaks/µm/s; 10−2) | 3.1 ± 0.2 | 1.8 ± 0.2*** |
| Regrowth frequency (%) | 2.4 ± 0.1 | 2.2 ± 1.1 (nd) |
| Annealing frequency (%) | 11.7 ± 3.9 | 16.4 ± 3.6 (nd) |
| Bundling frequency (events/µm2/filament/s; 10−6) | 6.1 ± 1.1 | 17.4 ± 2.3*** |
| Debundling frequency (events/µm2/filament/s; 10−6) | 1.7 ± 0.6 | 2.3 ± 0.9 (nd) |
Altered Sensitivity of vln4 Mutants to TIBA
To identify the response regulators involved in TIBA-induced actin rearrangement, we tested the sensitivity of different Arabidopsis mutants for key actin-binding proteins to TIBA by measuring their root gravitropic response. Among all tested lines, the VLN4 knockout mutants (Zhang et al., 2011) exhibited significantly reduced sensitivity to TIBA (Fig. 2, A and B; Supplemental Fig. S3), as did vln2vln3, the double mutant of two other villin isovariants (Supplemental Fig. S4A). However, mutations of Capping protein (Li et al., 2012), class XI myosins (XI-1, XI-2, XI-k; Cai et al., 2014), or actin-related protein 2 and 3 complex (Li et al., 2014b) showed wild-type responses to this drug (Supplemental Fig. S4, B–D).
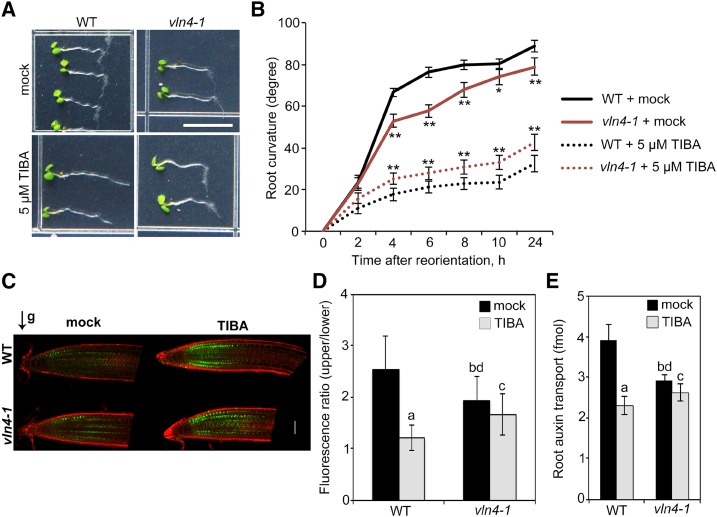
Figure 2.
TIBA sensitivity is altered in vln4-1 mutant. A, Gravitropic response of 4-d-old wild-type (WT) and vln4-1 seedlings grown on medium supplemented with mock or 5 µm TIBA for 24 h. Bar = 0.5 cm. B, Root tropic bending curvatures were measured at the indicated time points. Values given are means ± se (n = 50 seedlings for each genotype and treatment; *P < 0.05; **P < 0.01 between wild-type and vln4-1 mutant; t test). C, Representative images of DII-VENUS in drug-treated (5 µm) wild-type and vln4-1 roots after 1-h gravity stimulation. Green channel, DII-VENUS; red channel, cell walls stained with propidium iodide. Bar = 50 µm. D, The ratio of DII-VENUS fluorescence between the upper and lower sides of wild-type and vln4-1 roots. E, Basipetal auxin transport was detected in the roots treated with 30 µm TIBA for 15 h. Values given are means ± se (n = 5 [D] and 15 [E] roots for each genotype and treatment; a, significantly different from mock control within the same genotype; b, significantly different from wild-type treated with mock; c, no significant difference from mock control within the same genotype; d, significantly different from wild-type treated with TIBA; t test).
The action of ATIs is known to interfere with auxin transport. TIBA acts on root gravitropic response by disrupting asymmetric auxin redistribution between the upper and lower sides of root tips (Dhonukshe et al., 2008). To test the effect of VLN4 in this process, a sensitive auxin input reporter, DII-VENUS (Brunoud et al., 2012), was introduced into both wild-type and vln4-1 mutant. After gravistimulation for 1 h, DII-VENUS signal was reduced on the lower side of the wild-type root tip (Fig. 2, C and D), suggesting that auxin was redistributed to this side (Brunoud et al., 2012). However, TIBA treatment led to no establishment of the asymmetric signal in wild-type seedlings (Fig. 2, C and D). In the case of the vln4-1 mutant, DII-VENUS–monitored asymmetric auxin distribution was affected, suggesting the requirement of VLN4 for the establishment of differential auxin distribution (Fig. 2, C and D). Moreover, TIBA had a weaker effect on vln4-1 than it did on wild-type at various concentrations (Fig. 2, C and D; Supplemental Fig. S5, A and B). We further compared the effect of TIBA on PAT between wild-type and the vln4-1 mutant. As shown in Figure 2E, our measurements of basipetal auxin transport in root using 3H-indole-3-acetic acid showed that the rate of PAT was slower in the vln4-1 mutant than in wild-type. Additionally, the inhibition of PAT by TIBA was attenuated in the mutant (Fig. 2E; Supplemental Fig. S5, C and D). Taken together, these data suggest a link between VLN4 and TIBA activity in plants.
TIBA-Induced Actin Rearrangement Is Impaired by Loss of VLN4
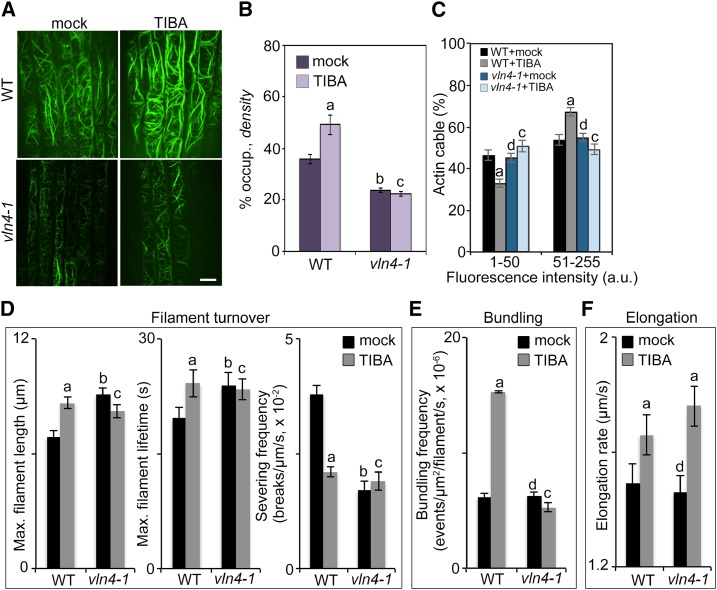
To test whether VLN4 mediates the action of TIBA on actin reorganization, we applied TIBA to vln4 mutants and compared actin architecture with wild-type quantitatively. Cortical actin arrays in vln4 mutants were less dense than in wild-type (Fig. 3, A and B; Supplemental Figs. S6 and S7A). However, the frequency distribution across the intensity classes was not significantly different between wild-type and vln4 mutants (Fig. 3, A and C; Supplemental Fig. S7B). To investigate whether actin arrays show different responses to TIBA depending on the developmental stage of the root cells, we further quantified the actin organization in cells from transition zone (TZ) and elongation zone (EZ; Supplemental Fig. S8). Consistent with previous data (Akkerman et al., 2011; Takatsuka et al., 2018), two actin configurations were observed in TZ and EZ: diffusive actin filaments in TZ cells, and thick actin filament bundles interspersed with fine actin filament and areas devoid of F-actin in EZ cells. Here, we confirmed this observation by actin architecture analyses (Supplemental Fig. S8). TZ cells had higher actin density than EZ cells (Supplemental Fig. S8, A and B). Additionally, the frequency distribution across the intensity classes was different between the two zones. In TZ cells, peaks with low intensity were more abundant, whereas EZ cells had a higher population of thick bundles (Supplemental Fig. S8, C and D), further confirming that actin reorganization is involved in root cell elongation (Takatsuka et al., 2018). In the vln4-1 mutant, actin arrays in both zones were less dense compared to wild-type (Supplemental Fig. S8, A and B). Moreover, the differences in bundle distribution between the two zones were impaired in the mutant (Supplemental Fig. S8, C and D). These data suggest that loss of VLN4 in root epidermal cells results in a less abundant actin array, and that VLN4 regulates the actin bundle formation associated with root cell elongation.
Figure 3.
Loss of VLN4 in cells leads to impaired actin responses to TIBA treatment. A, Representative images of root epidermal cells from wild-type (WT) and vln4-1 mutant expressing GFP-fABD2 after treatment with mock or 10 µm TIBA for 5 min. Bar = 10 µm. B and C, Actin architecture analyses were performed on images shown in A. D to F, Individual filament dynamics were measured on wild-type and vln4-1 mutant seedlings treated with mock or 10 µm TIBA for 5 min. See also Supplemental Table S1 for full analyses of single-filament parameters. Values given are means ± se (for actin architecture analysis, n > 300 images from 25 seedlings for each genotype and treatment; for single-filament analysis, n > 50 filaments from 10 seedlings for each genotype and treatment; a, significantly different from mock control within the same genotype; b, significantly different from wild-type treated with mock; c, no significant difference from mock control within the same genotype; d, no significant difference from wild-type treated with mock; t test; Pearson’s χ2 test was applied to evaluate significant differences in the frequency distribution across intensity classes between genotypes and treatments).
In vitro, recombinant VLN4 protein exhibits actin filament bundling, Ca2+-dependent severing, and barbed-end-capping activities (Zhang et al., 2011). To dissect VLN4’s contribution to actin dynamics in vivo, we examined the dynamic behavior of actin filaments in the vln4-1 mutant (Fig. 3, D–F; Supplemental Table S1). When compared with wild-type, actin filaments in the vln4-1 mutant had significantly increased length and lifetime. The severing frequency was decreased 2-fold in the mutant compared to the wild-type control (Fig. 3D; Supplemental Table S1). However, loss of VLN4 did not affect other dynamic parameters tested. In particular, the bundling and debundling frequencies did not change in vln4-1 compared with wild-type (Fig. 3E; Supplemental Table S1). The filament elongation rate was not altered in the mutant (Fig. 3F; Supplemental Table S1). The regrowth and annealing frequencies, which were used to describe the behavior of filament ends (Li et al., 2012), showed no significant differences between wild-type and the mutant (Supplemental Table S1). Collectively, analyses of actin organization and dynamics suggest that VLN4 regulates actin filament bundling during root cell elongation, and it also contributes to filament severing and facilitates actin turnover.
Following TIBA treatment, increased filament abundance and bundling was shown in wild-type. Moreover, wild-type cells from TZ and EZ showed similar responses to this inhibitor. TIBA-triggered actin reorganization was disrupted in vln4 mutants (Fig. 3, A–C; Supplemental Figs. S6–S8). In addition, a subset of actin filament parameters did not change following treatment of the mutant with TIBA. For example, the length and lifetime of actin filaments in vln4-1 mutant did not differ significantly from the mock control (Fig. 3D; Supplemental Table S1). Moreover, TIBA treatment did not decrease severing frequency in the vln4-1 mutant as it did in wild-type (Fig. 3D; Supplemental Table S1). Surprisingly, the filament-bundling frequency in mutant cells was unresponsive to TIBA treatment (Fig. 3E; Supplemental Table S1). Collectively, these findings suggest that VLN4 is required for actin reorganization following TIBA treatment. The dynamics of single actin filaments indicate the involvement of VLN4 in actin filament turnover and filament bundling during TIBA-induced actin remodeling. In addition, our data implied additional actin-binding proteins in TIBA-induced actin dynamics, since the filament elongation rate still increased in the vln4-1 mutant to a level comparable to wild-type after TIBA treatment (Fig. 3F; Supplemental Table S1).
VLN4 Is Involved in the Action of TIBA on Actin-Based PIN2 Dynamics
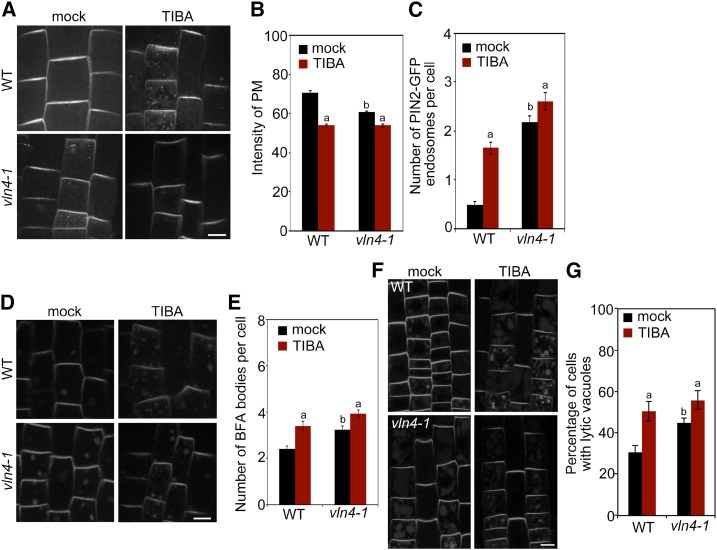
The role of actin cytoskeleton in auxin transporter trafficking has been long recognized (Geldner et al., 2001). The action of TIBA on actin-based PIN trafficking has been suggested to underlie its effect on auxin transport. To test whether this requires VLN4, we first examined the subcellular distribution of PIN2 in vln4-1 mutant cells in the presence or absence of TIBA. Neither TIBA nor loss of VLN4 had a noticeable impact on the polar distribution of PIN2 to the PM (Fig. 4A), suggesting that the PIN2 polar-targeting machinery remains intact. However, the level of PIN2 at the PM was significantly reduced, and more PIN2 was detected in the endosomal compartment in vln4-1 (Fig. 4, A–C), which explains the decreased PAT in the mutant (Fig. 2E). To test whether the reduced PM localization and increased intercellular accumulation of PIN2 result from enhanced endocytosis, we analyzed the uptake of the endocytic tracer N-(3-Triethylammoniumpropyl)-4-(6-(4-(Diethylamino) Phenyl) Hexatrienyl) Pyridinium Dibromide (FM4-64). In the vln4-1 mutant, FM4-64 uptake was significantly increased compared to wild-type (Supplemental Fig. S9), suggesting that VLN4 plays a negative role in endocytosis. When treated with the recycling inhibitor BFA, internalized PIN2 proteins accumulate in compartments termed BFA bodies (Grebe et al., 2002; Paciorek et al., 2005). Loss of VLN4 resulted in increased PIN2 accumulation in BFA bodies (Fig. 4, D and E), suggesting more active internalization of PIN2 in the mutant. Following BFA washout, the PIN2-containing BFA bodies mostly disappeared in both wild-type and vln4-1 cells, and no significant differences were found between wild-type and the mutant (Supplemental Fig. S10), suggesting that VLN4 is not involved in PIN2 recycling back to the PM. Targeting PIN2 to the lytic vacuole for degradation is a well-characterized mechanism of regulating PM-localized PIN2 protein levels (Kleine-Vehn et al., 2008). We also found that the vln4-1 mutant accumulated more PIN2 in the lytic vacuole (Fig. 4, F and G), indicating that the trafficking of PIN2 to the vacuole was enhanced in the mutant. TIBA treatment in wild-type cells led to defects in PIN2 dynamics similar to those caused by loss of VLN4. Furthermore, PIN2 dynamics in the vln4-1 mutant were less responsive to TIBA (Fig. 4). PIN2 trafficking was also separately examined in TZ and EZ (Supplemental Fig. S11). PIN2 localization and dynamics showed no obvious differences between TZ and EZ in wild-type; the defects in PIN2 dynamics caused by VLN4 deletion were consistent across TZ and EZ cells (Supplemental Fig. S11). TIBA treatment disrupted PIN2-GFP behavior similarly in both TZ and EZ cells. Moreover, PIN2 trafficking in mutant cells from both zones was less responsive to the inhibitor (Supplemental Fig. S11). Collectively, our findings suggest that VLN4 plays a role in regulating PIN2 dynamics in response to TIBA.
Figure 4.
PIN2 dynamics in wild-type (WT) and vln4-1 show different sensitivity to TIBA. A, Subcellular distribution of PIN2 in wild-type and the vln4-1 mutant following 5- to 15-min treatment with 10 µm TIBA. B and C, PIN2 signal intensity at the PM (B) or the number of PIN2-containing endosomes (C) was measured in wild-type and mutant. D and E, BFA-induced PIN2 internalization under TIBA treatment. Seedlings were treated with mock or 10 µm TIBA for 5 min, followed by treatment with 25 µm BFA and 10 µm TIBA for 30 min. F and G, Vacuolar trafficking of PIN2. Wild-type and mutant seedlings were pretreated with mock or 5 µm TIBA for 2 h, followed by a 3-h dark treatment on growth medium supplemented with mock or 5 µm TIBA. Bars = 10 µm (A, D, and F). Values given are means ± se (n > 400 cells for each genotype and treatment; a, significantly different from mock control within the same genotype; b, significantly different from wild-type treated with mock; c, no significant difference from mock control within the same genotype; t test).
TIBA Binds to the C-terminal Headpiece Domain of VLN4
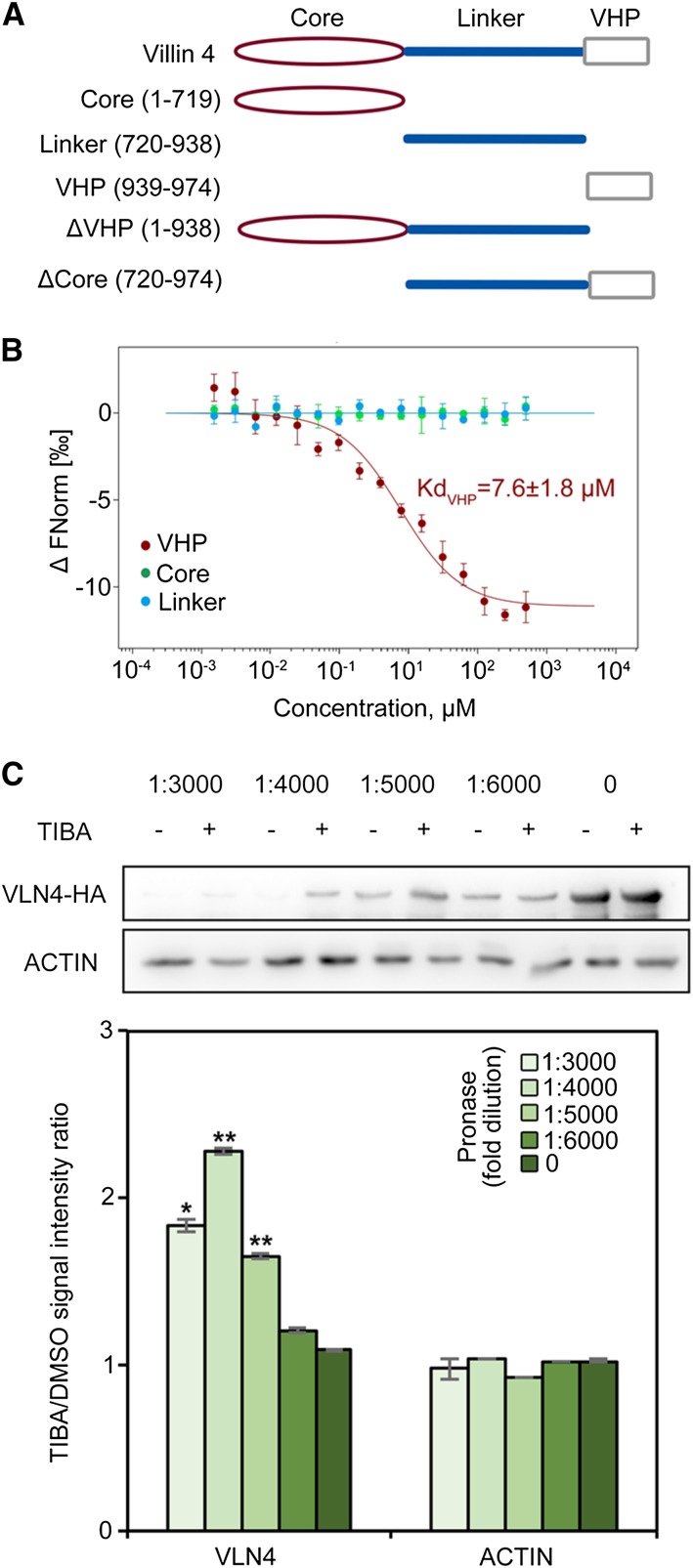
To test whether TIBA interacts with VLN4 directly, a microscale thermophoresis (MST) assay was performed (Wienken et al., 2010). Full-length VLN4 protein easily aggregates and adsorbs to capillary wall under MST conditions. To avoid experimental artifacts, VLN4 truncated proteins were used. Arabidopsis VLNs contain an N-terminal gelsolin core (core), an VHP, and a long linker between the core and the VHP (Fig. 5A; Khurana et al., 2010). VLN4 fragments were generated based on these three predicted domains (Fig. 5A) and subjected to MST experiments. As shown in Figure 5B, VHP domain directly interacts with TIBA, and the Kd for this interaction is 7.6 ± 1.8 µm. However, we failed to detect binding between TIBA and the other two domains (Fig. 5B). We further determined the binding affinity of the inactive analog, BA, for VHP. The Kd for the interaction of VHP with BA (80 ± 2.8 µm) is 10 times higher than the Kd for VHP with TIBA, suggesting a much weaker interaction (Supplemental Fig. S12A).
Figure 5.
TIBA binds to VLN4. A, Diagram of VLN4 truncated fragments. B, Thermophoresis binding curves of fragmented VLN4 proteins titrated with TIBA from experiments of three biological replicates. C, The drug affinity responsive target stability (DARTS) assay shows that TIBA protects VLN4, but not actin, from degradation. Top, Western blot of DARTS samples treated with different concentrations of pronase using anti-HA (Human influenza hemagglutinin) and antiactin antibodies. Bottom, Quantification of the protein band intensity between TIBA and dimethyl sulfoxide (DMSO) reveals that the presence of TIBA stabilizes VLN4 in cells. Values given are means ± se (n = 3 independent experiments; *P < 0.05; **P < 0.01 between TIBA and DMSO treatment; t test).
To further confirm the results from MST, we performed the DARTS assay (Lomenick et al., 2009). The DARTS approach was developed based on the observation that some proteins are protected from degradation by proteases when bound to the ligand (Lomenick et al., 2009). Pretreatment with TIBA resulted in protection from protease-induced degradation of VLN4 (Fig. 5C). The protection was specific because antibodies against actin in the same protein extraction showed no significant difference between TIBA and the DMSO control (Figs. 5C). Additionally, treatment with BA failed to protect VLN4 from protease-induced degradation (Supplemental Fig. S12B). These results indicate that VLN4 is stabilized specifically in the presence of TIBA, suggesting that TIBA directly targets VLN4 in cells.
Next, we asked if other Arabidopsis villin isovariants could also be targets of TIBA. We selected VLN2 and VLN3 because of their similar biochemical properties to VLN4 and abundant expression in roots (Zhang et al., 2011; Bao et al., 2012; van der Honing et al., 2012; Huang et al., 2015). The interaction of TIBA with these two isovariants was also detected in both MST and DARTS assays (Supplemental Fig. S13). Results from MST showed that the VHP domains of VLN2 and VLN3 bind to TIBA (Supplemental Fig. S13A). However, the VHP domain of VLN4 had the lowest Kd among these villins (Kd values for VLN2, VLN3, and VLN4 were 21.3 ± 8.5 µm, 16.2 ± 4.9 µm, and 7.4 ± 1.9 µm, respectively), thus indicating a stronger binding to TIBA (Supplemental Fig. S13A). We conclude from these assays that Arabidopsis VLNs are targeted by TIBA, and VHP domain contributes to this interaction.
The Bundling Activity of VLN4 Is Enhanced by TIBA
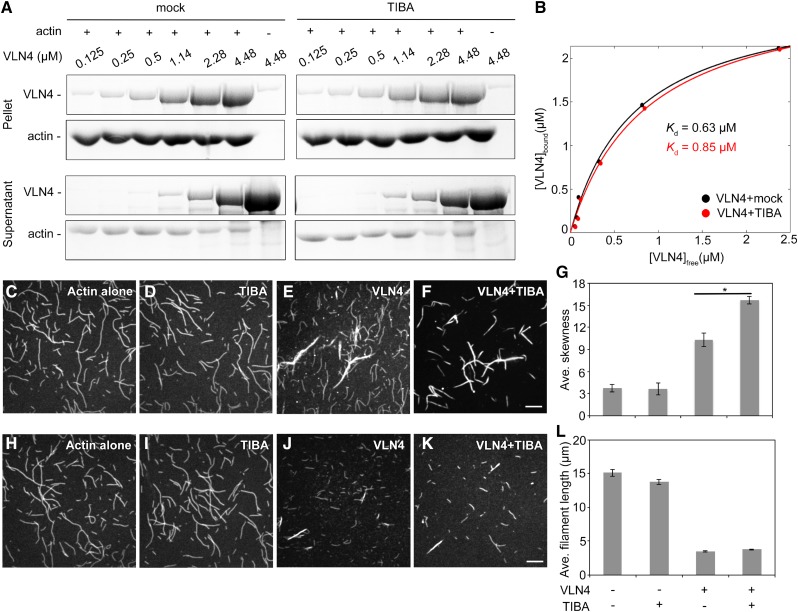
To test whether TIBA binding affects the biochemical properties of VLN4, high-speed cosedimentation assays were performed to determine the filament-binding ability of VLN4 in the absence or presence of the drug. Addition of TIBA had no obvious impact on the binding of VLN4 to actin filaments (Fig. 6, A and B). The filament-bundling and -severing activities of VLN4 were further assessed in the presence of TIBA by fluorescence microscopy. In control samples, individual actin filaments were visualized (Fig. 6, C and H). Actin bundles were induced by VLN4 (Fig. 6E; Zhang et al., 2011). In comparison, the extent of bundle formation was significantly increased when VLN4 was incubated with TIBA (Fig. 6F). The skewness parameter has been used to quantify the extent of actin filament bundling in a reconstituted system comprising muscle actin filaments and recombinant actin-bundling proteins (Khurana et al., 2010). Here, we used this analysis to compare single actin filaments and actin filament bundles in the presence of VLN4. Images of single actin filaments had average skewness values around 3. In the presence of VLN4, average skewness values increased significantly. This value was enhanced further when TIBA was present with VLN4 in the reaction (Fig. 6G). These results were confirmed by low-speed cosedimentation assays (Supplemental Fig. S14, A and B), in which addition of TIBA significantly increased the amount of sedimented actin by VLN4 (Supplemental Fig. S14, A and B).
Figure 6.
TIBA enhances the bundling activity of VLN4. A and B, A high-speed cosedimentation assay was used to examine the effect of TIBA on VLN4 binding to F-actin. Increasing concentrations of VLN4 were cosedimented with F-actin (3 µm). The supernatants and pellets obtained were subjected to SDS-PAGE and Coomassie stained. B, The experiments in A were repeated three times. After gel quantification, the concentration of bound VLN4 was plotted against the concentration of free VLN4 and fitted with a hyperbolic function. C to F, The effect of TIBA on VLN4-induced actin bundle formation. Prepolymerized actin (Oregon green-labeled, 3 µm) alone (C), with 500 µm TIBA (D), or 200 nm VLN4 in the absence (E) or presence (F) of 500 µm TIBA. EGTA (2 mm) was added in each reaction mix. Bar = 10 µm (F). G, Skewness analyses were performed on images shown in C to F. Values given are means ± se (n = 50 images for each treatment; *P < 0.05; t test). (H to K, TIBA does not affect the severing activity of VLN4. Prepolymerized actin (Oregon green-labeled, 3 µm) alone (H), with 500 µm TIBA (I), or 150 nm VLN4 in the absence (J) or presence (K) of 500 µm TIBA. All reactions were supplemented with 200 µm free Ca2+. Bar = 10 µm (K). L, Average filament lengths were measured on images shown in H to K. Values given are means ± se (n > 300 filaments for each treatment).
In the presence of 200 µm free Ca2+, actin filaments were severed by VLN4 and were significantly shorter than those in the control samples (Fig. 6, H and J; Zhang et al., 2011). The reduction in filament length caused by VLN4 did not show significant differences when TIBA was added to the reaction (Fig. 6, J and K). Similar results were obtained by high-speed cosedimentation assays (Supplemental Fig. S14, C and D). Consistent with a previous study (Dhonukshe et al., 2008), TIBA alone did not show any noticeable effect on actin dynamics (Fig. 6, D and I; Supplemental Fig. S14). These data suggest that TIBA regulates VLN4 by enhancing its filament-bundling activity, whereas the severing activity of VLN4 is not affected by this interaction. Surprisingly, these biochemical data do not support the in vivo mechanism of TIBA-induced decrease in actin turnover through the inhibition of VLN4. However, it is possible that TIBA treatment leads to more VLN4 involved in filament bundle formation in plants, consequently resulting in less VLN4 contributing to actin turnover.
TIBA-Enhanced Bundling Requires C-terminal Headpiece Domain of VLN4
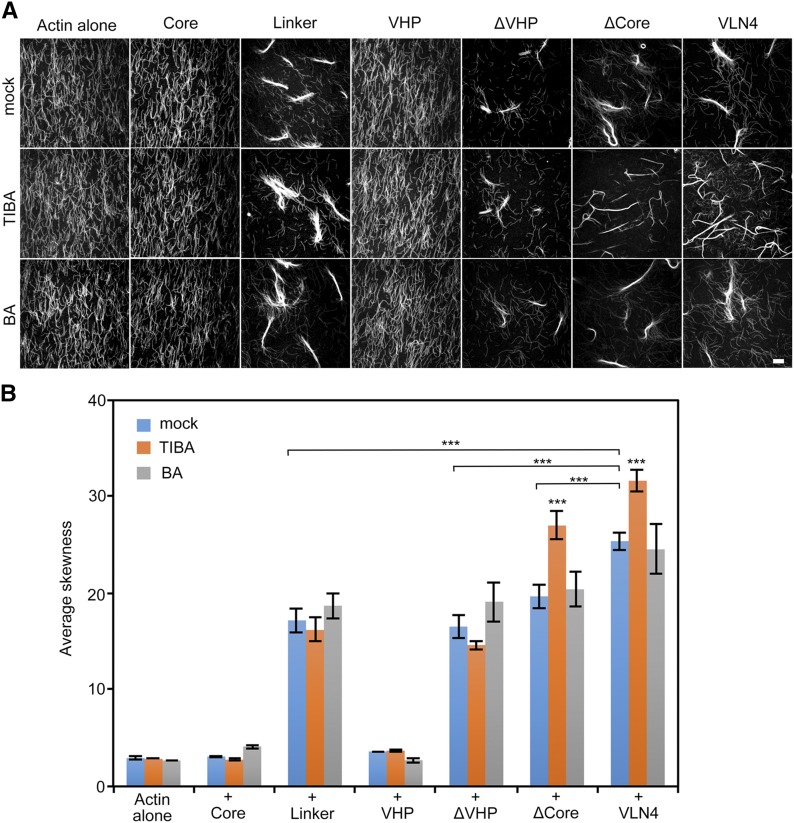
Based on the data above, we sought to dissect how TIBA enhances the bundling activity of VLN4. However, the molecular mechanism by which Arabidopsis villins bundle actin filaments remains unclear. Data from vertebrate villins have suggested that both the core and the headpiece contain an actin filament-binding domain. This led to the hypothesis that villin bundles actin as a monomer (Glenney and Weber, 1981; Friederich et al., 1999; Hampton et al., 2008). George et al. (2007) suggested that villin forms dimers, which allows each VHP domain to bind an actin filament. To understand the exact mechanism of bundling by Arabidopsis villins, we took advantage of truncated VLN4 fragments and studied their biochemical activities. First, the actin-binding and -severing domain of VLN4 was determined by high-speed cosedimentation assays (Supplemental Fig. S15). In the presence of EGTA, the core domain is not required for filament binding. Instead, the linker region of VLN4 displays strong actin-binding ability. VHP also binds to actin filament, but the interaction was much weaker than the linker region (Supplemental Fig. S15A). In the presence of Ca2+, the amount of actin filament in the pellets was decreased in the presence of full-length VLN4 and VLN4 mutants that contain the core region, whereas VHP and linker did not show similar effects on actin filaments (Supplemental Fig. S15B). Their effects on bundling were further assessed by fluorescence microscopy (Fig. 7A). We found that VLN4 fragments that contain the linker region all led to actin bundle formation, whereas the core and VHP domains alone failed to bundle actin filaments (Fig. 7A). Collectively, these data suggest that the core domain of Arabidopsis villins is not necessary for the actin-bundling function but plays an essential role in the severing activity. Linker region alone is sufficient to bind and cross link actin filaments in vitro. Neither of these properties requires the C-terminal headpiece domain of VLN4.
Figure 7.
Effects of VLN4 fragments on actin bundle formation under TIBA treatment. A, Micrographs of actin bundles formed in the presence of VLN4 fragments with or without 500 µm TIBA or BA. EGTA (2 mm) was added in each reaction mix. Bar = 10 µm. B, Skewness analyses were performed on images shown in A. Values given are means ± se (n = 50 images for each treatment; ***P < 0.001; t test).
In the presence of TIBA, both full-length VLN4 and deletion of the core domain (ΔCore) displayed stronger bundling activity (Fig. 7A), whereas the activities of core, linker, and VHP fragments were not altered by drug (Fig. 7A). Additionally, TIBA failed to increase actin bundles when VHP-deleted VLN4 protein was added. Applying inactive analog, BA, did not show a noticeable impact on the activity of these fragments (Fig. 7A). These images were further quantified by a skewness analysis (Fig. 7B). The skewness value increased when linker region was present in the reaction. Higher values were obtained from bundles formed by VLN4 or ΔCore after incubation with TIBA (Fig. 7B). However, no significant difference in the extent of bundling was caused by TIBA when incubated with VLN4 deletion mutant lacking VHP (Fig. 7B). Taken together, out data suggest that TIBA-enhanced bundling requires the C-terminal VHP domain of VLN4.
TIBA Binding Promotes VLN4 Oligomerization
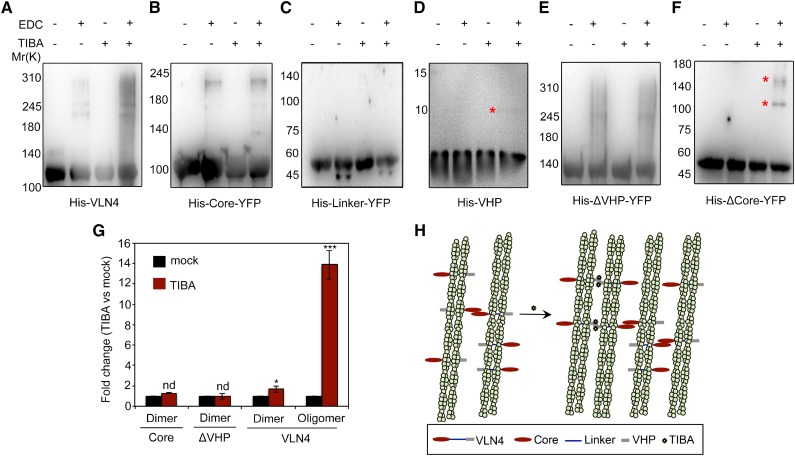
We hypothesized that bundling function of VLN4 in the presence of a single actin-binding site could be determined by villin self-association. Additionally, TIBA-enhanced bundling may result from increased villin oligomerization via TIBA-VHP interaction. To test these hypotheses, both full-length and truncated VLN4 proteins were cross linked by the zero-length cross linker [3-(dimethylamino)propyl]carbodiimide (EDC; Fig. 8, A–F). When full-length VLN4 was cross linked, a band with higher molecular mass was detected (Fig. 8A), indicating that VLN4 dimerizes by itself. However, this self-association was suppressed by deletion of the core domain (Fig. 8, C, D, and F), suggesting that the core domain is required for VLN4 self-interaction. Addition of TIBA induced oligomerization of the ΔCore and VHP fragments (Fig. 8, D and F) but failed to do so when VHP was further deleted (Fig. 8E). Moreover, TIBA led to an increase in the higher oligomeric population of full-length VLN4 (Fig. 8, A and G), whereas the oligomeric states of core and ΔVHP mutant fragments were not altered (Fig. 8, B, E, and G). Collectively, our data suggest two alternative mechanisms for regulating actin bundling by VLN4 (Fig. 8H). VLN4 bundles actin with the linker region as a monomer; an alternative is that villin forms dimers via the core domain, which allows each linker region to bind an actin filament (Fig. 8H). TIBA binding induces additional interaction between VHP domains, which further promotes the formation of VLN4 oligomers (Fig. 8), offering a third mechanism for VLN4 to bundle filaments (Fig. 8H).
Figure 8.
TIBA binding induces the oligomerization of VLN4 through its VHP domain. A to F, Oligomerization of VLN4 truncated variants. Each VLN4 variant (1.2, 8 µm for the VHP domain) was incubated with or without 500 µm TIBA and cross linked with 2 mm EDC for 1 h (30 min for VHP) at room temperature. Stars indicate oligomeric states that appeared when VHP (D) and ΔCore (F) were incubated with TIBA. G, Oligomeric populations of core, ΔVHP, and full-length VLN4 after TIBA treatment were measured quantitatively by densitometric analysis and plotted as fold change compared with mock control. Values given are means ± se from five replicates (nd, no significant difference from mock control; *P < 0.05; ***P < 0.001; t test). H, Schematic illustration of the positive regulatory mechanisms of TIBA in VLN4-mediated bundling.
VHP Domain Contributes to TIBA-Induced Actin Reorganization in Plants
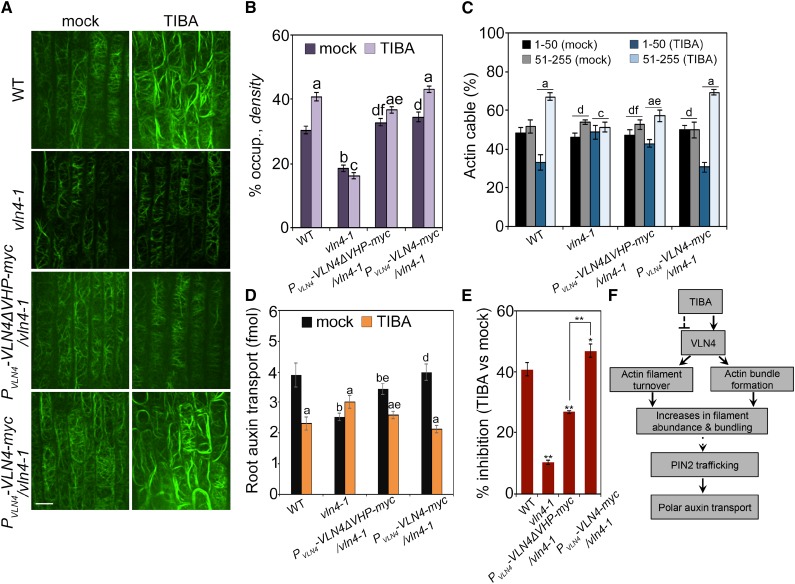
The data above suggested that TIBA binds to VHP domains to regulate bundling activity of VLN4 in vitro. We hypothesized that the C-terminal headpiece domain is also required for TIBA-induced actin reorganization in vivo. To test this hypothesis, we transformed full-length VLN4 and VLN4 mutant lacking VHP domain into vln4-1 plant, all driven by the endogenous VLN4 promoter. These constructs were able to rescue the short root hair phenotype in vln4-1 mutant (Supplemental Fig. S16). Additionally, wild-type and the complemented lines showed no significant differences in actin organization (Fig. 9, A–C). The reduced rate of PAT in vln4-1 mutant was recovered by expressing VLN4. Expressing VLN4ΔVHP restored the PAT in mutant, but to a lesser extent than did wild-type and full-length VLN4 transgenes (Fig. 9D). These results suggest that the VHP domain is not essential for VLN4 to regulate actin dynamics, PAT, and root hair growth under normal conditions. Following treatment with TIBA, VLN4ΔVHP failed to restore actin response to the wild-type level as full-length VLN4 did in cells (Fig. 9, A–C). The rate of auxin transport was further measured in wild-type, vln4-1 mutant, and the complemented lines. As shown in Figure 9, D and E, the inhibitory effect of TIBA on auxin transport in mutant transformed with full-length VLN4 was similar to that in wild-type. In both vln4-1 mutant and VLN4ΔVHP-complemented lines, however, the extent of inhibition was attenuated (Fig. 9, D and E). These data suggest that the villin headpiece region contributes to the action of TIBA on actin remodeling and PAT.
Figure 9.
VHP domain of VLN4 contributes to the effect of TIBA on actin dynamics and PAT in Arabidopsis. A, Representative images of root epidermal cells from wild-type (WT), vln4-1, and vln4-1–complemented lines. Seedlings were treated with mock or 10 µm TIBA for 5 min. Bar = 10 µm. B and C, Actin architecture analyses were performed on images shown in A. D, Basipetal auxin transport was detected in the roots treated with 30 µm TIBA for 15 h. E, The percentage inhibition of PAT in seedlings treated with TIBA relative to the mock control was calculated. Values given are means ± se (for actin architecture analysis, n > 300 images from 25 seedlings for each genotype and treatment; for PAT analysis, n = 15 roots for each genotype and treatment; for calculating the percentage inhibition, data were from five replicates; a, significantly different from mock control within the same genotype; b, significantly different from wild-type treated with mock; c, no significant difference from mock control within the same genotype; d, no significant difference from wild-type treated with mock; e, significantly different from vln4-1 complemented with full-length VLN4 within the same treatment; f, no significant difference from vln4-1 complemented with full-length VLN4 within the same treatment; * P < 0.05; **P < 0.01; t test; Pearson’s χ2 test was applied to evaluate significant differences in the frequency distribution across intensity classes between treatments). F, Schematic illustration summarizing VLN4 as a direct-binding target of TIBA to regulate actin dynamics and PAT. TIBA stimulates actin bundling and suppresses filament turnover in a VLN4-dependent fashion, leading to a more bundled cortical actin array in cells. The changes in actin organization and dynamics alter PIN2 localization and trafficking, which are critical for the maintenance of PAT. Solid and dashed lines indicate verified and unclear interactions, respectively.
DISCUSSION
In this study, we utilize TIBA to investigate the mechanisms of ATI-induced actin organization and dynamics. TIBA treatment results in high abundance of thick bundles in the cortical actin array. Genetic dissection revealed that Arabidopsis VLN4 mediates the action of TIBA on actin cytoskeleton and auxin transport. VLN4-dependent actin dynamics are involved in the action of TIBA to increase PIN2 internalization and sorting to the vacuole for degradation, resulting in reduced PIN2 level at the PM. TIBA enhanced the bundling activity of VLN4 in vitro but had no apparent impact on other biochemical properties of this protein. Furthermore, TIBA directly interacts with the C-terminal headpiece domain of VLN4. The VHP–TIBA interaction induces additional oligomeric VLN4 to cross link actin filament via intermolecular association between VHP domains, which normally does not occur in the mechanism of VLN4-mediated bundling. VLN4 mutant lacking the VHP domain has impaired actin and auxin transport responses to TIBA in plants, suggesting that the villin C-terminal headpiece contributes to in vivo TIBA sensitivity. Our study provides mechanistic insight into the link between actin cytoskeleton, vesicle trafficking, and auxin transport (Fig. 9F).
It has been reported that VLN-dependent actin remodeling is involved in various auxin transport-dependent physiological processes. For example, the vln2 single mutant in rice and the vln2vln3 double mutant in Arabidopsis both display hypergravitropism (Wu et al., 2015; this study) and twisted organ growth (van der Honing et al., 2012; Wu et al., 2015), a phenotype that is associated with altered PIN2 polar distribution (Lanza et al., 2012). Moreover, mutation of rice VLN2 disrupts cycling of PIN2 and PAT (Wu et al., 2015). Our study suggests that loss of VLN4 leads to reduced root gravitropic response, less PM presence of PIN2, and a slower rate of auxin transport, further demonstrating that VLN-mediated actin reorganization plays a role in coordinating trafficking of auxin transporters, PAT, and auxin-dependent growth. Villins are widely expressed in most eukaryotic cells. We show that, in addition to VLN4, other villin isovariants, including VLN2 and VLN3, are also targeted by TIBA, suggesting that actin dynamics modulated by villins might be a conserved mechanism for the action of several ATIs among plant and mammalian cells (Dhonukshe et al., 2008; Zhu et al., 2016). However, the extent of involvement of these VLNs in the action may depend on the dosage of drug and/or duration of treatment. For example, when plants are treated with TIBA at low concentration or for short periods of time, this inhibitor may preferentially bind to VLN4 but not to other VLNs, because VLN4 has the highest binding affinity to TIBA among the VLNs tested. This may explain why actin filament bundling and turnover in the mutant were totally unresponsive to 5-min treatment of TIBA (Fig. 3, E and F). However, a higher dose or longer-term treatment of TIBA requires more VLNs to be involved, as suggested by PAT assays, in which the rate of auxin transport in the vln4 mutant was less sensitive to but still inhibited by TIBA (Supplemental Fig. S5).
Our data uncovered the mechanism by which TIBA regulates bundling activity of VLNs, whereas how severing function is involved in TIBA action remains unclear. In vivo analyses showed that actin turnover in the vln4 mutant was insensitive to TIBA, suggesting the requirement of severing activity (Fig. 3F), but in vitro biochemical data suggested otherwise (Fig. 6, H–L). How to explain this contradiction is worthy of future investigation. Nevertheless, our data demonstrate that VLN-dependent actin dynamics contribute to TIBA action. How ATI-induced actin dynamics associate with PAT inhibition remains unclear. It has been hypothesized that auxin regulates its own transport by unbundling actin filaments (Holweg et al., 2004; Paciorek et al., 2005; Zhu and Geisler, 2015), and that ATIs may counteract the effect of auxin by promoting actin bundling and subsequent auxin exporter vesicle trafficking defects and thus altered PAT (Kleine-Vehn et al., 2006; Dhonukshe et al., 2008). However, this study and others suggest that there is no direct correlation between a specific alteration of actin architecture and the status of PIN2 localization and trafficking (Dhonukshe et al., 2008; Lanza et al., 2012; Li et al., 2014a; Wu et al., 2015; Zhu et al., 2016). Instead, our results showed that TIBA not only stimulates actin bundling but also suppresses actin filament turnover. The resulting reduction of actin dynamics may predominantly lead to the trafficking defects (Dhonukshe et al., 2008; this study).
Using advanced live-cell imaging and quantitative analyses of actin organization, we find that actin rearrangement induced by TIBA occurs much faster than what was shown previously (Rahman et al., 2007; Dhonukshe et al., 2008); it can be detected within minutes after application. TIBA treatment results in an increased optical density in the cortical array. The optical density measures the occupancy of GFP signals; it does not discriminate well between single actin filaments above threshold and filaments that are bundled over a spatial scale below optical resolution. Therefore, this value does not reflect the actual density of assembled actin filaments. To solve this, we estimated actin filament density by analyzing the total intensity in filamentous structures. The total intensities were then normalized to the intensity of single actin filaments to account for variance in actin reporter expression or optical efficiency during imaging. The data suggest that the relative amount of actin filaments was significantly increased after TIBA treatment, which is consistent with the results from optical density analysis. Collectively, data from different analyses demonstrate that TIBA treatment enhances the density of thick actin bundles in the cortical array, which is consistent with previous studies (Rahman et al., 2007; Dhonukshe et al., 2008). Quantitative assessment of the dynamics of single actin filaments further suggests that the increased filament abundance and bundling result from increased filament assembly and reduced filament disassembly, namely, a reduced severing frequency. We also detected an increase in the bundling frequency in TIBA-treated cells, suggesting an enhancement of filament bundle formation. Similar to TIBA, another auxin transport inhibitor, NPA, also induces excessively bundled actin arrays in Arabidopsis (Zhu et al., 2016). However, the underlying single filament dynamics are distinct between TIBA and NPA. For example, the bundling frequency decreases after NPA treatment (Zhu et al., 2016). NPA also reduces the debundling frequency (Zhu et al., 2016), whereas TIBA has no apparent effect on this parameter. These data suggest that actin bundle formation induced by ATIs may require different classes of actin-bundling proteins.
The Arabidopsis genome encodes five isovariants of villin, VLN1 to VLN5 (Klahre et al., 2000; Huang et al., 2005). They are abundantly expressed in a wide range of tissues, with elevated expression levels in certain types of cells (Klahre et al., 2000). Except for VLN1, which is a simple filament-bundling protein that lacks severing ability (Huang et al., 2005; Khurana et al., 2010), the other four VLNs contain the full suite of actin-modifying activities, including bundling, capping, and filament severing (Khurana et al., 2010; Zhang et al., 2010, 2011; Bao et al., 2012). The function of VLNs in actin organization and dynamics has been studied in different tissues and cell types. Loss of VLN2 and VLN3 results in fewer thick bundles and more thin bundles in the cortical actin array, leading to impaired directional organ growth and sclerenchyma development (Bao et al., 2012; van der Honing et al., 2012). VLN2 and VLN5 are responsible for filament severing at pollen tube tips, which facilitates the construction of actin collars (Qu et al., 2013). Moreover, the decreased skewness value in the subapical region of vln2vln5 mutant pollen tubes implies an involvement of villin-mediated bundling activity (Qu et al., 2013). A previous study by Zhang et al. (2011) shows that loss of VLN4 inhibits root hair growth and causes thinning of actin bundles, suggesting that VLN4 contributes to root hair growth by bundling actin filaments. Consistently, we further demonstrate that deletion of VLN4 reduces the actin array abundance and impairs the enhanced actin bundling associated with root cell elongation, resulting in fewer thick bundles in the cortical actin array. VLN4 also facilitates filament turnover by severing actin filaments. Its loss leads to decreased severing frequency and longer actin filament length and lifetime. Compared to the filament bundling and severing activity, the barbed-end-capping activity of VLNs seems less important in cells, because the dynamic behaviors of actin filament ends are not altered by loss of VLNs (Wu et al., 2015; this study). This study and others suggest that members of the Arabidopsis villin family primarily function in actin filament bundling and turnover in different tissues and cell types.
Unlike their vertebrate homologs, plant villins have a much longer linker polypeptide connecting the core and headpiece. In addition, the linker–headpiece connection region in plant villins lacks sequence homology to the vertebrate villin sequences. It is unknown to what extent the function of these structural regions in plant villins resembles those of the well-studied vertebrate counterparts. Here, the biochemical properties of each domain were characterized in vitro. In the presence of EGTA, the core domain of VLN4 does not bind and bundle actin filaments. The actin-binding and -bundling activities are predominantly contributed by the linker region. In the presence of high calcium (200 µm), the core domain, but not the linker, displays strong severing activity. This is consistent with earlier studies in which high calcium releases an autoinhibited conformation of vertebrate villins, thus exposing the actin-binding site in the core and regulating the actin-severing activity (Kumar et al., 2004). Our in vitro cross-linking studies with villin truncation mutants suggest that villin dimerizes between core domains, which facilitates bundle formation. Based on these data, we proposed two alternative mechanisms for VLN4-induced bundling. First, VLN4 bundles actin with the linker region as a monomer; an alternative is that villin forms dimers via the core domain, which allows each linker region to bind an actin filament. Thus, our detailed functional analyses provide insight into the actin-severing and -bundling activities of plant villins.
The headpiece domain of vertebrate villin contains an additional actin filament-binding domain that is crucial for its bundling capacity (Friederich et al., 1992, 1999; George et al., 2007). This often leads to the question of whether the properties of the VHP domain from plant villins are similar to those of vertebrate villins. Previous studies from Klahre et al. (2000) report that the GFP-tagged headpieces from Arabidopsis VLNs (VLN1, VLN2, and VLN3) localize to actin filaments in Bright Yellow-2 cells (Klahre et al., 2000). This is supported by the results in which the VHP domain of VLN4 also shows weak interaction with actin filaments in vitro. Our study suggests that the C-terminal headpiece domain of VLN4 is not critical for actin-severing and -bundling activities. In addition, expressing VLN4 in the mutant lacking the VHP domain is sufficient to recover the actin organization defects in vln4-1, suggesting that the VHP domain is not critical for VLN4 function both in vitro and in vivo. This is in contrast to the conclusion made by van der Honing et al. (2012). The authors report that deletion of the headpiece domain disrupts localization of VLN3 to thick actin bundles, and conclude that VLN3 requires the headpiece region for a correct localization to actin filament bundles and for actin filament bundling (van der Honing et al., 2012). However, a functional analysis of VLN3ΔVHP is lacking. Additionally, the actin network in the vln2vln3 double mutant may fail to support the correct localization of VLN3ΔVHP. We also cannot rule out the possibility that the VHP domains from different VLNs have distinct functions.
It has been shown that the bundling activity of villins is modulated by a variety of factors, such as phosphorylation and phosphoinositide lipids (Zhai et al., 2001; Kumar et al., 2004). In human villin, the interaction with phosphatidylinositol biphosphate (PIP2) at the VHP enhances villin bundling (Kumar et al., 2004). It is hypothesized that PIP2 interactions with VHP could result in dimerization or oligomerization of villin proteins, thus enhancing actin cross linking by villin in the presence of PIP2 (Kumar et al., 2004; George et al., 2007). Furthermore, circular dichroism studies suggest that there is a conformational change in VHP induced by PIP2, and this facilitates the actin cross-linking activity (Kumar et al., 2004). Similarly, our results show that TIBA interacts with the VHP domain of Arabidopsis villins. The VHP–TIBA interaction induces the formation of additional oligomeric VLN4, leading to enhanced bundling activity of VLN4. In cells with deletion of VHP, the actin array is less responsive to TIBA. These data suggest that the C-terminal headpiece domain might respond to environmental and developmental signals and regulate villin activity. However, the mechanism of TIBA-induced VLN4 oligomerization between VHP domains requires future investigation.
In addition to villins, it is possible that there are additional actin-binding proteins targeted by TIBA. Our analysis of single filament dynamics showed that the filament elongation rate still increases following TIBA treatment in the vln4-1 mutant as it does in wild-type cells. Potential candidates could be formin proteins. Formins directly stimulate both the nucleation and elongation of actin filament assembly (Goode and Eck, 2007; Chesarone et al., 2010). The involvement of formins in plant auxin transport has been demonstrated by Li et al. (2014a), who reported that a type II formin in rice, rice morphology determinant, modulates actin organization during auxin signaling and is required for cycling of PIN2.
CONCLUSIONS
The use of ATIs provides insights into the importance of actin cytoskeleton for auxin transport. In this study, using high-performance, live-cell imaging approaches, the effect of TIBA on actin cytoskeleton was characterized in detail. We found that TIBA induces high abundance of thick bundles in the cortical array. A genetic screen identified Arabidopsis Villin4 as a cytoskeletal target of TIBA. VLN4 contributes to the plant sensitivity to TIBA, including actin remodeling and PAT inhibition. In addition, TIBA interacts with the VLN4 C-terminal headpiece domain to modulate the bundling activity of VLN4, which is involved in the proper subcellular localization of PIN2. Finally, we provide genetic evidence that the C-terminal headpiece domain is important for exerting the inhibitory effect of TIBA on actin cytoskeleton and auxin transport in plants.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The homozygous vln4-1 (SALK_049058) and vln4-2 (SAIL_517_A03) single mutants were crossed to wild-type Arabidopsis (Arabidopsis thaliana) Columbia-0 expressing GFP-fABD2; DII-VENUS or PIN2ρPIN2-GFP reporter (Sheahan et al., 2004; Staiger et al., 2009; Zhang et al., 2011) and homozygotes were recovered from F2 populations. To generate the VLN4-complemented plants, a native promoter of 2,000 bp in length and cDNA were PCR amplified (Supplemental Table S2) and cloned into pMDC32-DEST; the resulting constructs were introduced into vln4-1 plants by Agrobacterium-mediated transformation. Surface-sterilized seeds were plated onto 0.5× Murashige and Skoog medium supplemented with 1% (w/v) Suc and 0.8% (w/v) agar. Plants were grown vertically in an environmental chamber under long-day lighting conditions (16 h light, 8 h dark) at 22°C for 4–5 d.
Plant Treatments
Before use, TIBA or BA (Sigma-Aldrich) was dissolved in DMSO and diluted to various concentrations. For fluorescence imaging, 5-d-old seedlings were treated with 10 µm TIBA for 5 min or the indicated time within figure legends. For gravitropic response assays, 4-d-old seedlings were transferred to 0.5× Murashige and Skoog solid medium supplemented with 5 µm TIBA and recovered for 6 h before 90° rotation. An equivalent amount of solvent was used as a negative control.
Protein Purification
To generate full-length and truncated villin constructs, the corresponding fragments were PCR amplified (Supplemental Table S2) from cDNA and cloned into pET-30a(+). The constructs were expressed in strain BL21 (DE3) of Escherichia coli. Cells were grown in Luria-Bertani medium at 37°C until optical density600 = 0.6, then induced for 12 h at 16°C with the addition of 0.5 mm isopropyl β-d-thiogalactopyranoside. Cells were collected and resuspended in binding buffer (400 mm NaCl, 40 mm phosphate-buffered saline, pH 8.0) supplemented with 0.1 mm phenylmethylsulfonyl fluoride and sonicated. The sonicate was clarified at 12,000g for 10 min. This was followed by purification using Ni-NTA His Bind Resin (GE Healthcare). The purified protein was dialyzed overnight against buffer Tris and KCl (5 mm Tris, 50 mm KCl, 0.5 mm dithiothreitol [DTT], 0.5 mm EDTA, pH 7.5). The purified proteins were separated into aliquots, frozen in liquid nitrogen, and stored at −80°C. Actin was purified from rabbit skeletal muscle acetone powder as described by Pardee and Spudich (1982). Protein concentrations were determined with the Bradford reagent (Bio-Rad) using bovine serum albumin as a standard.
MST
MST experiments were carried out using a Monolith NT.115 (NanoTemper Technologies GmbH). The core and linker domains of VLN4 were tagged with GFP by PCR amplification and purified from E. coli. His-VHP fragments were fluorescently labeled using MO-L008 Monolith His-tag labeling Kit RED-Tris-NTA (available from NanoTemper Technologies GmbH). Increasing concentrations of titrant (either TIBA or BA) were titrated against constant concentrations (50 nm) of target proteins in a standard MST buffer (50 mm Tris, pH 7.5; 150 mm NaCl; 10 mm MgCl2; 0.05% [v/v] Tween 20). MST premium-coated capillaries (Monolith NT.115 MO-K005) were used to load the samples into the MST instrument. Triplicate time traces were acquired for each test system. The binding curves were fit with Kd mode in MO analysis software (NanoTemper Technologies GmbH). Data analyses were performed with NTAnalysis (NanoTemper Technologies GmbH).
Measurements of Basipetal Auxin Transport in Roots
Auxin transport assay was performed according to previous description (Lewis and Muday, 2009). Six-day-old wild-type, vln4-1, and vln4-1–complemented seedlings were transferred to medium supplemented with 30 µm TIBA or solvent for a 10-h pretreatment. To test the basipetal auxin transport, small 7-µl droplets of agar containing 100 nm 3H`-indole-3-acetic acid (25 Ci mmol−1; Amersham) were applied at the root tip. After incubation in darkness for 5 h, the apical 2 mm of root was excised and discarded, and a 5-mm segment basal to that was collected and placed into scintillation vials containing 1 ml of scintillation fluid to examine the radioactivity with a liquid scintillation counter (HIDEX 300SL; Hidex).
DARTS Assay
DARTS assay was performed as previously described by Lomenick et al. (2011) with minor changes. In brief, wild-type protoplasts were transfected with HA-tagged villins. Total protein was extracted after transfection for 24 h with extraction buffer (50 mm Tris, 150 mm NaCl, 10% [v/v] glycerol, 0.1% [v/v] Nonidet P-40, 5 mm DTT, 2 mm MgCl2, 0.1 mm phenylmethylsulfonyl fluoride, pH7.5). The supernatant from 16,000g was used as the input. Protein extract (300 µL) was incubated with 500 μm TIBA, BA, or an equal volume of DMSO for 1 h at room temperature. This mixture was divided into six aliquots of 50 μl, to which different concentrations of pronase (Sigma-Aldrich) were added, and digested for 15 min at room temperature. Samples were loaded onto SDS-PAGE gels, and immunoblotting was performed. Membranes were probed with either anti-HA (1:2,000) or antiactin (1:10,000; Sigma-Aldrich).
Fluorescence Microscopy and Image Analysis
Images were acquired with a PerkinElmer UltraView Vox spinning disk microscope equipped with 60×/1.42 Numerical Aperture objective. Different genotypes and treatments were imaged with their respective controls side by side with fixed microscope settings on a given day. A fixed specimen exposure time and gain setting were used such that individual actin filament could be seen, but actin bundles were not saturated. Images were processed with ImageJ software (National Institutes of Health). For actin architecture analysis in vivo, the percentage of occupancy (density) was measured as previously reported (Higaki et al., 2010; Henty et al., 2011). In brief, filament density was calculated as the percentage occupancy of GFP-fABD2 signal separated from background by setting a minimal threshold to include all actin filaments, and then images were converted to binary black and white images. Because there were no z-series projections, and spinning disk confocal microscopy generates relatively high-contrast, low-background images, we did not apply Gaussian blur, high band-pass filter, or skeletonization processing steps. Quantification of the fluorescence intensity of actin cables was performed as previously described (Martin et al., 2007; van der Honing et al., 2012). Profiles of GFP fluorescence intensities were created parallel to the longitudinal cell axis. We selected an area in which no actin filaments were visible and subtracted the mean fluorescence intensity of this region from the fluorescence intensities of the intensity profile. This resulted in a new plot profile, which was used to distribute the peaks into two classes. Only peaks that were at least 10 units higher in fluorescence intensity than the intensities of the left and right bases of the peaks were included. Each experiment was repeated three times independently. All image collection and data analyses were performed as double-blind experiments. For time-lapse imaging of actin filament dynamics, the cortical actin array in root epidermal cells was recorded at 1-s intervals, and a series of 100 images were collected. Quantitative analyses of the dynamic behavior of individual actin filaments were performed as described previously (Staiger et al., 2009; Henty et al., 2011; Li et al., 2012). To minimize the effects of filament abundance or the density of the filament array, we normalized the bundling and debundling frequency to average filament number within the region of interest (Staiger et al., 2009; Zheng et al., 2013; Hoffmann et al., 2014; Li et al., 2015).
For imaging actin filaments in vitro, actin (75% Oregon green-labeled; Kuhn and Pollard, 2005) was dialyzed against buffer F (5 mm Tris-HCl, 0.1 mm CaCl2, 1 mm NaN3, 0.2 mm ATP⋅2Na, 0.5 mm DTT, 50 mm KCl, 2.5 mm MgCl2, and 0.1 mm ATP, pH 7.0) and prepolymerized in 1× KMEI buffer (50 mm KCl, 1 mm MgCl2, 1 mm EGTA, 10 mm imidazole, pH 7.0) at room temperature for 2 h. To test the bundling activity of VLN4 or VLN4 fragments, 3 µm F-actin was incubated with different VLN4 proteins in the absence or presence of TIBA or BA at room temperature for 30 min. Reaction mixtures were supplemented with 2 mm EGTA. Skewness analysis was performed to compare single actin filaments and reconstituted actin filament bundles in vitro as described previously (Khurana et al., 2010). To test the severing activity of VLN4, 3 µm F-actin was incubated with VLN4 in the presence of 200 µm free Ca2+. Before imaging, all of the polymerized F-actin was diluted to 250 nm with fluorescence buffer containing 10 mm imidazole, pH 7.0; 50 mm KCl; 1 mm MgCl2; 1 mm DTT; 0.2 mm ATP; 15 mm Glc; 20 µg mL−1 catalase; 100 µg mL−1 Glc oxidase; and 0.5% (w/v) methylcellulose.
High-Speed Cosedimentation Assay
High-speed cosedimentation assays were used to determine the actin-binding activity of VLN4 and various VLN4 fragments (Zhang et al., 2011). Actin was dialyzed overnight against buffer G (5 mm Tris-HCl, 0.1 mm CaCl2, 1 mm NaN3, 0.2 mm ATP·2Na, 0.5 mm DTT, pH 7.0). Before use, proteins were clarified by centrifugation at 55,000g for 1 h. Actin was prepolymerized in 1× KMEI buffer (50 mm KCl, 1 mm MgCl2, 1 mm EGTA, 10 mm imidazole, pH 7.0) at room temperature for 2 h. In a 100-µL reaction volume, VLN4 fragments were incubated with 3.0 µm preformed actin filaments. Following incubation, the reaction mixture was centrifuged for 1 h at 55,000g. Supernatant (80 µL) was removed to a separate tube, and 16 µL of 6× protein-loading buffer was added to it. The pellet was suspended by 100 µL of buffer G, and 20 µL of 6× protein-loading buffer was added. The samples were then separated by SDS-PAGE and stained with Coomassie Brilliant Blue R (Sigma-Aldrich). To determine the effects of TIBA on binding, VLN4 was preincubated with 500 µm TIBA on ice for 30 min. Increasing amounts of VLN4 were incubated with 3.0 µm preformed actin filaments in the presence of 500 µm TIBA. The amount of VLN4 in the pellets or supernatants was quantified using ImageJ software. The Kd value for VLN4 bound to actin filaments was calculated by fitting the data of bound protein versus free protein to a hyperbolic function using PRISM 5 software (GraphPad Software).
Cross-Linking Assay
After centrifugation at 20,000g for 20 min, proteins were mixed with 500 µm TIBA or solvents in PEM buffer (1 mm MgCl2, 1 mm EGTA, and 100 mm PIPES-KOH, pH 6.9) for 30 min on ice. Zero-length cross-linker EDC (2 mm; Pierce Biotechnology) was then added, and the solution was kept at room temperature for 1 h. Since the VHP domain is easily degraded under standard assay conditions, to detect the cross linking between VHP domains, protein was freshly purified before use, and after a higher protein concentration (8 µm) and 1% (v/v) protease inhibitor cocktail (2 mm o-phenanthroline, 0.5 mg/mL leupeptin, 2 mg/mL aprotinin, 1 mg/mL pepstatin) were added into PEM buffer, the cross-linking reaction time was further reduced to 30 min. Proteins separated by SDS-PAGE were transferred to nitrocellulose membranes and probed with anti-His antibody. The blots were incubated in horseradish peroxidase–coupled secondary antibody (Sigma-Aldrich) at 1:100,000 dilution and developed with SuperSignal Pico West chemiluminescent substrate (Thermo Scientific) according to the manufacturer’s instructions. Densitometric analysis was performed for quantitative measurements of protein levels using ImageJ (National Institutes of Health). Fold change in oligomeric population, normalized to total protein loaded and relative to oligomeric protein levels in the control sample, was calculated.
Statistical Analyses
Mean values, se, and statistical tests were calculated with Microsoft Excel software. Statistical significance was assessed by one-tailed Student’s t test with unequal variance and between control and treatment. Pearson’s χ2 test was applied to evaluate significant differences in the frequency distribution across intensity classes between genotypes and treatments, as stated in the figure legends. Each experiment was repeated at least three times with similar results; results from one biological replicate were presented.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under accession numbers: At VLN2, At2g41740; At VLN3, At3g57410; and At VLN4, At4g30160.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The relative amount of actin filaments in the cortical array was increased following TIBA treatment.
Supplemental Figure S2. Actin response to TIBA treatments is dose-dependent.
Supplemental Figure S3. Reduced sensitivity to TIBA in the vln4-2 mutant.
Supplemental Figure S4. TIBA sensitivities of different actin-binding protein mutants.
Supplemental Figure S5. Reduced TIBA sensitivity in the vln4-1 mutant.
Supplemental Figure S6. TIBA treatment does not affect the amount of actin filaments in the vln4-1 mutant.
Supplemental Figure S7. Actin arrays in the vln4-2 mutant are unresponsive to TIBA treatment.
Supplemental Figure S8. Actin responses to TIBA in cells from transition zone and elongation zone in wild type and vln4-1 root.
Supplemental Figure S9. Enhanced FM4-64 uptake by TIBA is impaired in the vln4-1 mutant.
Supplemental Figure S10. VLN4 is not required for PIN2 recycling to the PM.
Supplemental Figure S11. PIN2 dynamics in cells from TZ and EZ in wild type and vln4-1 root.
Supplemental Figure S12. Analysis of the interaction between VLN4 and BA.
Supplemental Figure S13. TIBA also binds to other villin isovariants in Arabidopsis.
Supplemental Figure S14. Validation of the effect of TIBA on VLN4 activity by low-speed and high-speed cosedimentation assays.
Supplemental Figure S15. The actin binding and severing activity of different VLN4 fragments.
Supplemental Figure S16. Root hair phenotype of the vln4-1 mutant and vln4-1-complemented lines.
Supplemental Table S1. Single filament dynamics in wild type and the vln4-1 mutant following TIBA treatment.
Supplemental Table S2. Primers used in this study.
Acknowledgments
We thank Dr. Ying Fu (China Agriculture University) for generously providing PIN2ρPIN2-GFP lines. DII-VENUS lines and vln2vln3 double mutant are kindly shared by Dr. Jie Le (Institute of Botany, Chinese Academy of Sciences) and Dr. Yi Zhang (Beijing Normal University), respectively. We thank Dr. Jing Zhang and Yang Li (China Agriculture University) for help on the PAT assay. We thank Dr. Christopher J. Staiger (Purdue University) for valuable suggestions on this study.
Footnotes
This work was supported by the Fundamental Research Funds for the Central Universities (2016NT07 to J.L.).
Articles can be viewed without a subscription.
References
- Akkerman M, Overdijk EJR, Schel JHN, Emons AMC, Ketelaar T (2011) Golgi body motility in the plant cell cortex correlates with actin cytoskeleton organization. Plant Cell Physiol 52: 1844–1855 [DOI] [PubMed] [Google Scholar]
- Bao C, Wang J, Zhang R, Zhang B, Zhang H, Zhou Y, Huang S (2012) Arabidopsis VILLIN2 and VILLIN3 act redundantly in sclerenchyma development via bundling of actin filaments. Plant J 71: 962–975 [DOI] [PubMed] [Google Scholar]
- Benjamins R, Scheres B (2008) Auxin: The looping star in plant development. Annu Rev Plant Biol 59: 443–465 [DOI] [PubMed] [Google Scholar]
- Bennett T, Leyser O (2014) The auxin question: A philosophical overview. In Zažímalová E, Petrasek J, Benková E, eds, Auxin and Its Role in Plant Development. Springer, New York, pp 3–19 [Google Scholar]
- Blancaflor EB. (2013) Regulation of plant gravity sensing and signaling by the actin cytoskeleton. Am J Bot 100: 143–152 [DOI] [PubMed] [Google Scholar]
- Brunoud G, Wells DM, Oliva M, Larrieu A, Mirabet V, Burrow AH, Beeckman T, Kepinski S, Traas J, Bennett MJ, Vernoux T (2012) A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 482: 103–106 [DOI] [PubMed] [Google Scholar]
- Cai C, Henty-Ridilla JL, Szymanski DB, Staiger CJ (2014) Arabidopsis myosin XI: A motor rules the tracks. Plant Physiol 166: 1359–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarone MA, DuPage AG, Goode BL (2010) Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat Rev Mol Cell Biol 11: 62–74 [DOI] [PubMed] [Google Scholar]
- Dhonukshe P, Grigoriev I, Fischer R, Tominaga M, Robinson DG, Hasek J, Paciorek T, Petrásek J, Seifertová D, Tejos R, Meisel LA, Zazímalová E, et al. (2008) Auxin transport inhibitors impair vesicle motility and actin cytoskeleton dynamics in diverse eukaryotes. Proc Natl Acad Sci USA 105: 4489–4494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friederich E, Vancompernolle K, Huet C, Goethals M, Finidori J, Vandekerckhove J, Louvard D (1992) An actin-binding site containing a conserved motif of charged amino acid residues is essential for the morphogenic effect of villin. Cell 70: 81–92 [DOI] [PubMed] [Google Scholar]
- Friederich E, Vancompernolle K, Louvard D, Vandekerckhove J (1999) Villin function in the organization of the actin cytoskeleton. Correlation of in vivo effects to its biochemical activities in vitro. J Biol Chem 274: 26751–26760 [DOI] [PubMed] [Google Scholar]
- Friml J. (2010) Subcellular trafficking of PIN auxin efflux carriers in auxin transport. Eur J Cell Biol 89: 231–235 [DOI] [PubMed] [Google Scholar]
- Geldner N, Friml J, Stierhof Y-D, Jürgens G, Palme K (2001) Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413: 425–428 [DOI] [PubMed] [Google Scholar]
- George SP, Wang Y, Mathew S, Srinivasan K, Khurana S (2007) Dimerization and actin-bundling properties of villin and its role in the assembly of epithelial cell brush borders. J Biol Chem 282: 26528–26541 [DOI] [PubMed] [Google Scholar]
- Glenney JRJ Jr., Weber K (1981) Calcium control of microfilaments: Uncoupling of the F-actin-severing and -bundling activity of villin by limited proteolysis in vitro. Proc Natl Acad Sci USA 78: 2810–2814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode BL, Eck MJ (2007) Mechanism and function of formins in the control of actin assembly. Annu Rev Biochem 76: 593–627 [DOI] [PubMed] [Google Scholar]
- Grebe M, Friml J, Swarup R, Ljung K, Sandberg G, Terlou M, Palme K, Bennett MJ, Scheres B (2002) Cell polarity signaling in Arabidopsis involves a BFA-sensitive auxin influx pathway. Curr Biol 12: 329–334 [DOI] [PubMed] [Google Scholar]
- Hampton CM, Liu J, Taylor DW, DeRosier DJ, Taylor KA (2008) The 3D structure of villin as an unusual F-Actin crosslinker. Structure 16: 1882–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henty JL, Bledsoe SW, Khurana P, Meagher RB, Day B, Blanchoin L, Staiger CJ (2011) Arabidopsis actin depolymerizing factor4 modulates the stochastic dynamic behavior of actin filaments in the cortical array of epidermal cells. Plant Cell 23: 3711–3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higaki T, Kutsuna N, Sano T, Kondo N, Hasezawa S (2010) Quantification and cluster analysis of actin cytoskeletal structures in plant cells: Role of actin bundling in stomatal movement during diurnal cycles in Arabidopsis guard cells. Plant J 61: 156–165 [DOI] [PubMed] [Google Scholar]
- Hoffmann C, Moes D, Dieterle M, Neumann K, Moreau F, Tavares Furtado A, Dumas D, Steinmetz A, Thomas C (2014) Live cell imaging reveals actin-cytoskeleton-induced self-association of the actin-bundling protein WLIM1. J Cell Sci 127: 583–598 [DOI] [PubMed] [Google Scholar]
- Holweg C, Süsslin C, Nick P (2004) Capturing in vivo dynamics of the actin cytoskeleton stimulated by auxin or light. Plant Cell Physiol 45: 855–863 [DOI] [PubMed] [Google Scholar]
- Huang S, Robinson RC, Gao LY, Matsumoto T, Brunet A, Blanchoin L, Staiger CJ (2005) Arabidopsis VILLIN1 generates actin filament cables that are resistant to depolymerization. Plant Cell 17: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Qu X, Zhang R (2015) Plant villins: Versatile actin regulatory proteins. J Integr Plant Biol 57: 40–49 [DOI] [PubMed] [Google Scholar]
- Khurana P, Henty JL, Huang S, Staiger AM, Blanchoin L, Staiger CJ (2010) Arabidopsis VILLIN1 and VILLIN3 have overlapping and distinct activities in actin bundle formation and turnover. Plant Cell 22: 2727–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klahre U, Friederich E, Kost B, Louvard D, Chua N-H (2000) Villin-like actin-binding proteins are expressed ubiquitously in Arabidopsis. Plant Physiol 122: 35–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J, Dhonukshe P, Swarup R, Bennett M, Friml J (2006) Subcellular trafficking of the Arabidopsis auxin influx carrier AUX1 uses a novel pathway distinct from PIN1. Plant Cell 18: 3171–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J, Leitner J, Zwiewka M, Sauer M, Abas L, Luschnig C, Friml J (2008) Differential degradation of PIN2 auxin efflux carrier by retromer-dependent vacuolar targeting. Proc Natl Acad Sci USA 105: 17812–17817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn JR, Pollard TD (2005) Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys J 88: 1387–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N, Zhao P, Tomar A, Galea CA, Khurana S (2004) Association of villin with phosphatidylinositol 4,5-bisphosphate regulates the actin cytoskeleton. J Biol Chem 279: 3096–3110 [DOI] [PubMed] [Google Scholar]
- Lanza M, Garcia-Ponce B, Castrillo G, Catarecha P, Sauer M, Rodriguez-Serrano M, Páez-García A, Sánchez-Bermejo E, T C M, Leo del Puerto Y, Sandalio LM, et al. (2012) Role of actin cytoskeleton in brassinosteroid signaling and in its integration with the auxin response in plants. Dev Cell 22: 1275–1285 [DOI] [PubMed] [Google Scholar]
- Laxmi A, Pan J, Morsy M, Chen R (2008) Light plays an essential role in intracellular distribution of auxin efflux carrier PIN2 in Arabidopsis thaliana. PLoS One 3: e1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DR, Muday GK (2009) Measurement of auxin transport in Arabidopsis thaliana. Nat Protoc 4: 437–451 [DOI] [PubMed] [Google Scholar]
- Li G, Liang W, Zhang X, Ren H, Hu J, Bennett MJ, Zhang D (2014a) Rice actin-binding protein RMD is a key link in the auxin-actin regulatory loop that controls cell growth. Proc Natl Acad Sci USA 111: 10377–10382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Henty-Ridilla JL, Huang S, Wang X, Blanchoin L, Staiger CJ (2012) Capping protein modulates the dynamic behavior of actin filaments in response to phosphatidic acid in Arabidopsis. Plant Cell 24: 3742–3754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Henty-Ridilla JL, Staiger BH, Day B, Staiger CJ (2015) Capping protein integrates multiple MAMP signalling pathways to modulate actin dynamics during plant innate immunity. Nat Commun 6: 7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Li J-H, Wang W, Chen N-Z, Ma T-S, Xi Y-N, Zhang X-L, Lin H-F, Bai Y, Huang S-J, Chen Y-L (2014b) ARP2/3 complex-mediated actin dynamics is required for hydrogen peroxide-induced stomatal closure in Arabidopsis. Plant Cell Environ 37: 1548–1560 [DOI] [PubMed] [Google Scholar]
- Lomenick B, Hao R, Jonai N, Chin RM, Aghajan M, Warburton S, Wang J, Wu RP, Gomez F, Loo JA, et al. (2009) Target identification using drug affinity responsive target stability (DARTS). Proc Natl Acad Sci USA 106: 21984–21989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomenick B, Jung G, Wohlschlegel JA, Huang J (2011) Target identification using drug affinity responsive target stability (DARTS). Curr Protoc Chem Biol 3: 163–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SG, Rincón SA, Basu R, Pérez P, Chang F (2007) Regulation of the formin for3p by cdc42p and bud6p. Mol Biol Cell 18: 4155–4167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedergang-Kamien E, Leopold AC (1957) Inhibitors of polar auxin transport. Physiol Plant 10: 29–38 [Google Scholar]
- Paciorek T, Zazímalová E, Ruthardt N, Petrásek J, Stierhof Y-D, Kleine-Vehn J, Morris DA, Emans N, Jürgens G, Geldner N, et al. (2005) Auxin inhibits endocytosis and promotes its own efflux from cells. Nature 435: 1251–1256 [DOI] [PubMed] [Google Scholar]
- Pardee JD, Spudich JA (1982) Purification of muscle actin. Methods Cell Biol 24: 271–289 [DOI] [PubMed] [Google Scholar]
- Petrásek J, Mravec J, Bouchard R, Blakeslee JJ, Abas M, Seifertová D, Wiśniewska J, Tadele Z, Kubeš M, Covanová M, et al. (2006) PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312: 914–918 [DOI] [PubMed] [Google Scholar]
- Qu X, Zhang H, Xie Y, Wang J, Chen N, Huang S (2013) Arabidopsis villins promote actin turnover at pollen tube tips and facilitate the construction of actin collars. Plant Cell 25: 1803–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A, Bannigan A, Sulaman W, Pechter P, Blancaflor EB, Baskin TI (2007) Auxin, actin and growth of the Arabidopsis thaliana primary root. Plant J 50: 514–528 [DOI] [PubMed] [Google Scholar]
- Sheahan MB, Staiger CJ, Rose RJ, McCurdy DW (2004) A green fluorescent protein fusion to actin-binding domain 2 of Arabidopsis fimbrin highlights new features of a dynamic actin cytoskeleton in live plant cells. Plant Physiol 136: 3968–3978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder WE. (1949) Some responses of plants to 2,3,5-triiodobenzoic acid. Plant Physiol 24: 195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger CJ, Sheahan MB, Khurana P, Wang X, McCurdy DW, Blanchoin L (2009) Actin filament dynamics are dominated by rapid growth and severing activity in the Arabidopsis cortical array. J Cell Biol 184: 269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuka H, Higaki T, Umeda M (2018) Actin reorganization triggers rapid cell elongation in roots. Plant Physiol 178: 1130–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Honing HS, Kieft H, Emons AMC, Ketelaar T (2012) Arabidopsis VILLIN2 and VILLIN3 are required for the generation of thick actin filament bundles and for directional organ growth. Plant Physiol 158: 1426–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienken CJ, Baaske P, Rothbauer U, Braun D, Duhr S (2010) Protein-binding assays in biological liquids using microscale thermophoresis. Nat Commun 1: 100. [DOI] [PubMed] [Google Scholar]
- Wu S, Xie Y, Zhang J, Ren Y, Zhang X, Wang J, Guo X, Wu F, Sheng P, Wang J, et al. (2015) VLN2 regulates plant architecture by affecting microfilament dynamics and polar auxin transport in rice. Plant Cell 27: 2829–2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai L, Zhao P, Panebra A, Guerrerio AL, Khurana S (2001) Tyrosine phosphorylation of villin regulates the organization of the actin cytoskeleton. J Biol Chem 276: 36163–36167 [DOI] [PubMed] [Google Scholar]
- Zhang H, Qu X, Bao C, Khurana P, Wang Q, Xie Y, Zheng Y, Chen N, Blanchoin L, Staiger CJ, Huang S (2010) Arabidopsis VILLIN5, an actin filament bundling and severing protein, is necessary for normal pollen tube growth. Plant Cell 22: 2749–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xiao Y, Du F, Cao L, Dong H, Ren H (2011) Arabidopsis VILLIN4 is involved in root hair growth through regulating actin organization in a Ca2+-dependent manner. New Phytol 190: 667–682 [DOI] [PubMed] [Google Scholar]
- Zheng Y, Xie Y, Jiang Y, Qu X, Huang S (2013) Arabidopsis actin-depolymerizing factor7 severs actin filaments and regulates actin cable turnover to promote normal pollen tube growth. Plant Cell 25: 3405–3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Geisler M (2015) Keeping it all together: Auxin-actin crosstalk in plant development. J Exp Bot 66: 4983–4998 [DOI] [PubMed] [Google Scholar]
- Zhu J, Bailly A, Zwiewka M, Sovero V, Di Donato M, Ge P, Oehri J, Aryal B, Hao P, Linnert M, et al. (2016) TWISTED DWARF1 mediates the action of auxin transport inhibitors on actin cytoskeleton dynamics. Plant Cell 28: 930–948 [DOI] [PMC free article] [PubMed] [Google Scholar]