Mid-aged leaves of rice upregulate OsSGR expression to accelerate chlorophyll degradation for Mg remobilization and photooxidative protection under Mg-limited conditions.
Abstract
Magnesium (Mg) is a relatively mobile element that is remobilized in plants under Mg-limited conditions through transport from old to young tissues. However, the physiological and molecular mechanisms underlying Mg remobilization in plants remain poorly understood. In this study, we investigated Mg remobilization in rice (Oryza sativa) as facilitated through a Mg dechelatase gene involved in chlorophyll degradation, STAY-GREEN (OsSGR). We first observed that mid-aged leaves of rice are more susceptible to Mg deficiency. Expression of OsSGR was specifically upregulated by Mg deficiency, and the response was more pronounced in mid-aged leaves. Knockout of OsSGR exhibited the stay-green phenotype, which hindered the mobility of Mg from mid-aged leaves to young developing leaves. This decline in Mg mobility was associated with inhibited growth of developing leaves in mutants under Mg-limited conditions. Furthermore, Mg deficiency enhanced reactive oxygen species (ROS) generation in mid-aged leaves. ROS levels, particularly hydrogen peroxide, in turn, positively regulated OsSGR expression, probably through chloroplast-to-nucleus signaling, which triggers chlorophyll degradation to protect mid-aged leaves from photodamage. Taken together, these results show that OsSGR-mediated chlorophyll degradation contributes to not only internal remobilization of Mg from mid-aged leaves to developing leaves, but also photooxidative protection of mid-aged leaves under Mg-limited conditions. ROS appear to act as feedback regulators of OsSGR expression to precisely govern chlorophyll degradation in mid-aged leaves where Mg and photosynthetic capacities are relatively high.
Magnesium (Mg) is the second most abundant cation in plants. Optimal plant growth requires 1.5–3.5 g of Mg per kilogram of dry matter for many physiological and biochemical processes (Marschner, 2012; Verbruggen and Hermans, 2013). The most widely studied aspect of Mg activity in plants is the role it plays in photosynthesis. One-fifth of total Mg in plants is bound in chloroplasts, mainly as a key component of chlorophyll molecules participating in light harvesting in PSI and PSII (Rissler et al., 2002; Karley and White, 2009; Cakmak and Yazi̇ci̇, 2010). In addition, many photosynthetic enzymes involved in carbon fixation in chloroplasts are activated by Mg2+ (Sugiyama et al., 1968; Pierce, 1986; Lundqvist and Schneider, 1991). Mg also plays a crucial role in carbohydrate partitioning. In the very early stage of Mg deficiency, phloem export of Suc is remarkably blocked, resulting in accumulation of carbohydrates in source leaves and consequently disruption of photosynthetic carbon metabolism and restriction on CO2 fixation (Fischer and Bremer, 1993; Cakmak et al., 1994a, 1994b; Hermans and Verbruggen, 2005). Under such conditions, photosynthetic electrons not utilized in CO2 fixation are transferred to molecular O2, leading to the generation of highly reactive oxygen species (ROS) and subsequent cell damage (Cakmak and Kirkby, 2008). Besides, Mg is essential for energy metabolism in plants and mainly acts as a cofactor of enzyme activity with ATP (Marschner, 2012). In general, up to 50% of total cellular Mg2+ binds with ATP, specifically used for ATP hydrolysis and synthesis (Maguire and Cowan, 2002; Gout et al., 2014).
Plant growth is greatly inhibited under Mg deficiency stress (−Mg), which leads to detrimental effects on crop productivity and quality in agricultural systems (Aitken et al., 1999). Given that Mg is relatively mobile in plants, Mg in older tissues is preferentially transported to young tissues to ensure continued growth and development under Mg-limited conditions. Thus, the first visual symptoms of –Mg are typically observed in older leaves (Bergmann, 1994; Hermans and Verbruggen, 2005; Cakmak and Yazi̇ci̇, 2010). However, hydroponic studies on sugar beet (Beta vulgaris), Arabidopsis (Arabidopsis thaliana), and rice (Oryza sativa) reveal young mature leaves are more susceptible to −Mg stress, exhibiting much quicker declines of chlorophyll and Mg concentrations than the oldest leaves (Hermans et al., 2005; Hermans and Verbruggen, 2005; Kobayashi et al., 2013; Chen et al., 2018). This indicates that Mg recycling probably is more vigorous in young mature leaves through much of the plant kingdom. Moreover, −Mg leads to simultaneous high accumulations of Suc, starch, and anthocyanins in plants (Hermans et al., 2004; Kobayashi et al., 2013), which is different from other mineral deficiencies that only accumulate one of them. This suggests that Mg remobilization and plant responses to −Mg are backed by a distinct set of physiological mechanisms that can only be fully understood through specific investigation focused on Mg effects in various plant tissues.
Leaf senescence is a highly programmed process that facilitates plant growth through recycling of nutrients from senescing leaves to developing tissues and organs (Hörtensteiner and Feller, 2002; Park et al., 2007; Liu et al., 2008; Hörtensteiner and Kräutler, 2011). A key process in leaf senescence is the degradation of chlorophyll, which includes the following steps. First, chlorophyll b is reduced to chlorophyll a by chlorophyll b reductase (Scheumann et al., 1996) and 7-hydroxymethyl chlorophyll a reductase (Meguro et al., 2011). Second, the central Mg in chlorophyll a is removed by Mg dechelatase (STAY-GREEN, SGR; Shimoda et al., 2016), and a phytol side chain is cut by pheophytinase (Schelbert et al., 2009). Third, pheophorbide a is further catabolized into primary fluorescent chlorophyll catabolite by pheophorbide a oxygenase (Pruzinská et al., 2003) and red chlorophyll catabolite reductase (Rodoni et al., 1998; Schelbert et al., 2009). Finally, primary fluorescent chlorophyll catabolite is converted into nonfluorescent chlorophyll catabolites or dioxobilin-type nonfluorescent chlorophyll catabolites under acidic conditions within vacuoles (Hörtensteiner and Kräutler, 2011; Chen et al., 2016). Among chlorophyll catabolic enzymes (CCEs), SGR catalyzes the first step of chlorophyll degradation and thereby plays an important role in the regulation of chlorophyll degradation (Sato et al., 2007; Shimoda et al., 2016). AtSGR1 in Arabidopsis physically interacts with not only all CCEs, but also light-harvesting complex subunits of PSII (Park et al., 2007; Sakuraba et al., 2012). Thus, SGR-CCEs-light-harvesting complex subunits of PSII complexes are key components of chlorophyll breakdown and PS degradation during senescence (Shimoda et al., 2016; Sato et al., 2018).
Besides normal senescence during fruit ripening, a variety of stimuli can promote chlorophyll degradation and leaf senescence during vegetative growth (Hörtensteiner and Feller, 2002; Lim and Nam, 2005). Although −Mg is known to trigger obvious leaf chlorosis symptoms (Bennett, 1993), it remains unknown whether this −Mg-induced leaf chlorosis is accompanied by other typical leaf senescence processes. It might be, as previously hypothesized, leaf chlorosis and declines in photosynthesis associated with −Mg are due to hindered chlorophyll synthesis (Hermans et al., 2004). However, considering that leaf chlorosis always appears during the later stages of −Mg, it has been proposed that ROS generated by impaired photosynthetic systems cause oxidative damage to chloroplasts, rather than the lack of Mg atoms for chlorophyll chelation (Cakmak and Kirkby, 2008; Verbruggen and Hermans, 2013; Kobayashi and Tanoi, 2015).
In short, the mechanisms underlying −Mg-triggered leaf chlorosis and Mg remobilization are not well understood (Chen et al., 2018). In this study, we first observed that −Mg triggers chlorosis and internal Mg remobilization in mid-aged leaves of rice. Through identifying a Mg dechelatase gene that is induced specifically by Mg deficiency, we found that −Mg stimulates chlorophyll degradation for subsequent Mg remobilization and photooxidative protection in mid-aged leaves, in which Mg pool and photosynthetic capacities are always highest. We further determined that −Mg enhances generation of ROS, which act as a feedback signal to positively regulate OsSGR expression.
RESULTS
Mid-aged Leaves Are More Susceptible to Mg Deficiency
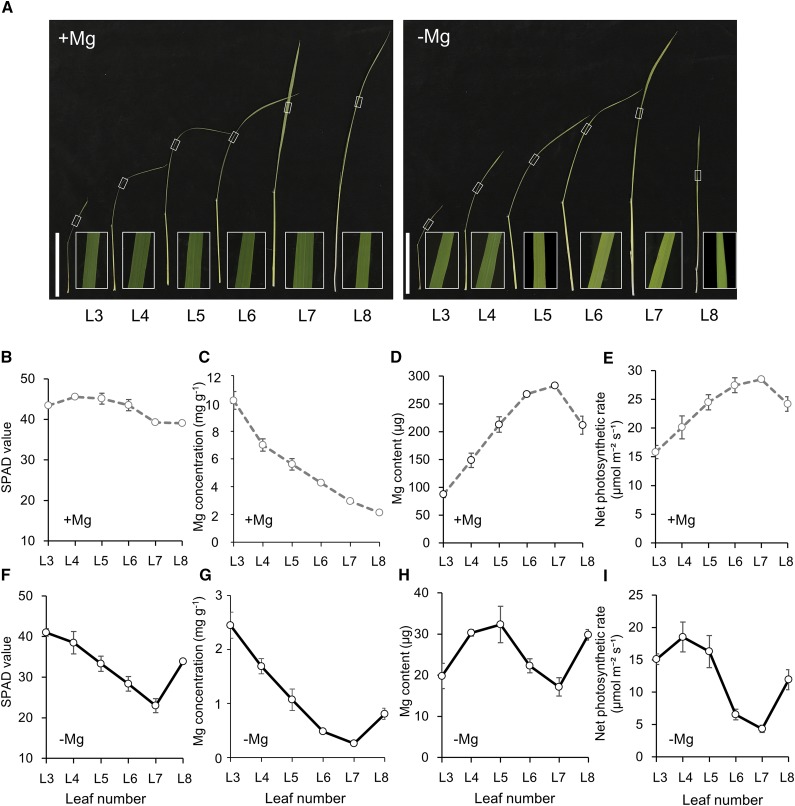
To investigate variation in the physiological responses to −Mg stress among rice leaves, we determined chlorophyll concentration (measured nondestructively as spectral plant analysis diagnostic [SPAD] values; Fig. 1, B and F), Mg concentration (Fig. 1, C and G), Mg content (Fig. 1, D and H), and net photosynthetic rate (Fig. 1, E and I) under both +Mg and −Mg conditions. Phenotypic observation showed that leaf chlorosis in response to Mg depletion was most pronounced in mid-aged leaves (L5, L6, and L7), particularly L6 and L7, whereas −Mg only mildly affected the old (L3 and L4) and the new leaves (L8; Fig. 1, A, B, and F). Consistent with the observed phenotypes, Mg concentration, Mg content and net photosynthetic rate in mid-aged leaves were the lowest among rice leaves grown in −Mg conditions (Fig. 1, G–I). These results confirm that Mg deprivation leads to chlorosis and initiates Mg recycling in mid-aged leaves of rice.
Figure 1.
Physiological responses to −Mg stress among rice leaves. A, Growth and chlorosis phenotypes in different leaves. White frame photo is magnified section of each leaf blade. Scale bars = 10 cm. B to I, SPAD values (B and F), Mg concentration (C and G), Mg content (D and H), and net photosynthetic rate (E and I) under +Mg (B−E) or −Mg (F–I) conditions. Rice seedlings were grown in nutrient solution containing 0- or 250-μm Mg for 8 d. The SPAD value of each leaf was determined using a chlorophyll meter. Mg was determined by inductively coupled plasma-mass spectrometry (ICP-MS). Net photosynthetic rates were measured using a portable photosynthesis analysis system. L3–L8 are labels for leaves ordered from old to young.
Mg Deficiency Enhances the Expression of OsSGR in Mid-aged Leaves
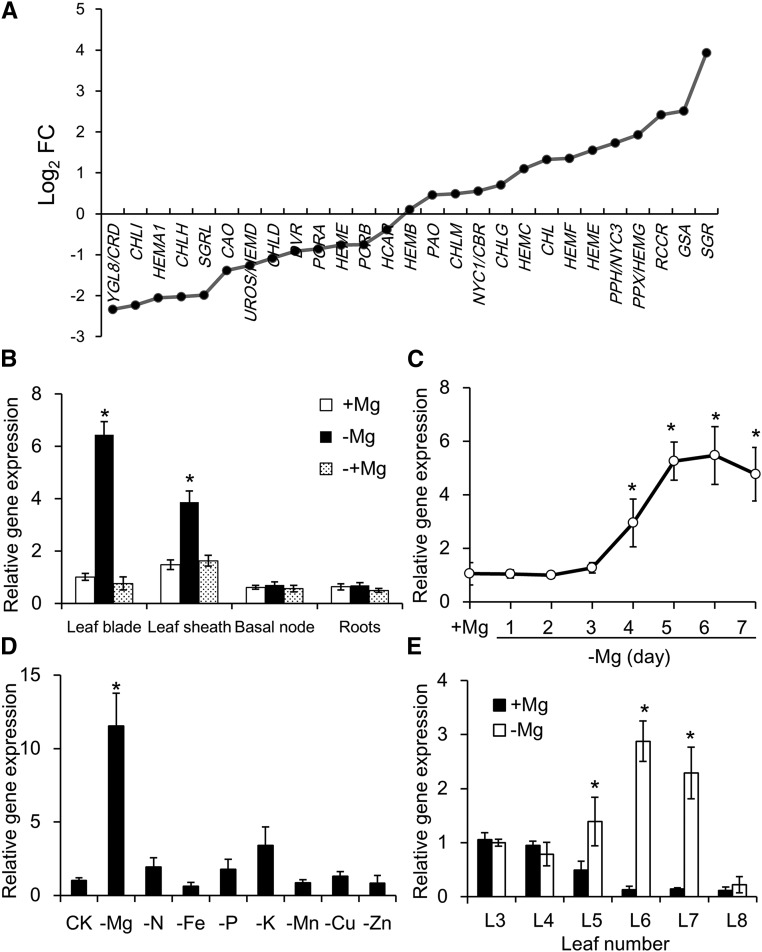
To investigate the molecular mechanisms underlying −Mg responses in rice, we conducted RNA sequencing (RNA-seq) transcriptomic analysis with plants grown in +Mg and −Mg nutrient solutions. In this analysis, many metabolic processes and signaling pathways were affected by −Mg stress. Notably, transcription of genes involved in chlorophyll biosynthesis (e.g. Glutamyl-tRNA reductase1, Mg-chelatase H, D, and I subunits) was slightly repressed, while transcription of genes involved in chlorophyll degradation (e.g. pheophytinase, red chlorophyll catabolite reductase, and SGR) was significantly enhanced in response to −Mg stress (Fig. 2A). These results suggest that Mg deficiency initiates leaf senescence processes. Among the senescence-related differentially expressed genes, OsSGR was the most highly upregulated. Considering the RNA-seq results, along with the key role filled by SGRs in chlorophyll degradation, we examined gene expression patterns of OsSGR in response to −Mg treatments in more detail. Expression of OsSGR increased in response to –Mg in shoots (both leaf blades and sheaths) but not in roots or the basal node (Fig. 2B). Furthermore, this upregulation was entirely eliminated by the addition of Mg to the growth medium for 24 h (Fig. 2B). In a time-course experiment, OsSGR expression in shoots increased gradually upon transfer to the −Mg treatment and remained high after 5 d of −Mg treatment (Fig. 2C). None of the other tested nutrient stresses (−N, −Fe, −P, −K, −Mn, −Cu, and −Zn) significantly affected OsSGR expression (Fig. 2D), indicating that the OsSGR expression response is specific to −Mg conditions. Furthermore, OsSGR was only responsive to −Mg in mid-aged leaves (L5, L6, and L7; Fig. 2E), which is consistent with the leaf chlorosis responses shown in Figure 1, A and B.
Figure 2.
Gene expression patterns in response to Mg deprivation. A, Transcriptomic analysis of genes involved in chlorophyll biosynthesis and degradation of rice shoots. Fold change (FC) of fragments per kilobase million value in −Mg-treated plants relative to +Mg-treated plants is shown. B, Tissue-specific expression of OsSGR. Rice seedlings were grown in nutrient solution containing 0- or 250-μ m Mg for 7 d. Afterward, Mg-deficient seedlings were resupplied with 250-μ m Mg for another day. Expression is shown relative to expression in +Mg leaf blades. C, Time-dependent expression of OsSGR. Three rice seedlings were put into −Mg nutrient solution each day and all were harvested 7 d after transferring the first plants. Expression is shown relative to expression in +Mg plants. D, Mg-specific response of OsSGR expression. Rice seedlings were grown in normal nutrient solution (CK) or in the nutrient solution without Mg, N, Fe, P, K, Mn, Cu, or Zn for 7 d. Expression is shown relative to expression in CK plants. E, Expression of OsSGR in different leaves. L3–L8 leaves were separated and harvested after growing plants in nutrient solution containing 0- or 250-μ m Mg for 7 d. Expression is shown relative to expression in +Mg L3 leaves. The expression level was determined by quantitative reverse transcription PCR. Data are means ± sd (n = 3). The asterisk shows a significant difference compared to +Mg treatment (P < 0.05 by Tukey’s test).
Subcellular Localization of OsSGR
We investigated the subcellular localization of OsSGR by transiently expressing OsSGR-GFP in rice protoplasts. The green signal observed inside protoplasts completely overlapped with the pink autofluorescence signal from chloroplasts (Supplemental Fig. S1, A–D), indicating that OsSGR is a chloroplast-localized protein. Rice protoplasts expressing 35S:GFP, as a control, exhibited fluorescence signals in the cytosol and nucleus (Supplemental Fig. S1, E–H).
Chlorophyll-Degrading Activity of OsSGR
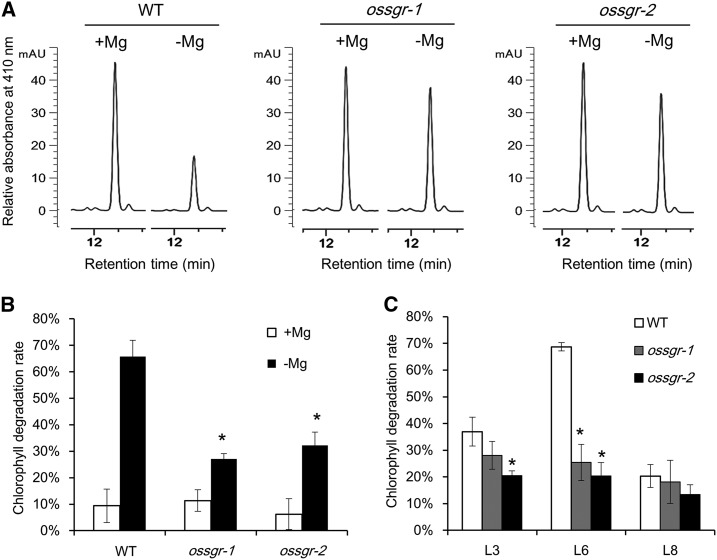
To investigate the physiological roles filled by OsSGR in plants under −Mg stress, we conducted further experiments with two independent null mutants (ossgr-1 and ossgr-2) containing 8- and 1-bp deletions in the first and second exons of OsSGR, respectively (Supplemental Fig. S2A). Chlorophyll degradation rates were first investigated by incubation of chlorophyll a standard substance with total protein extract from the wild type and mutants, respectively. Mg deficiency resulted in chlorophyll degradation of up to 70% in wild type, whereas only 30% degradation occurred in two null mutants (Fig. 3, A and B). Furthermore, mid-aged leaf (L6) of wild type showed much greater degradation than old (L3) and young (L8) leaves; however, there was no apparent difference observed among these leaves in the two null mutants (Fig. 3C). These results reveal that OsSGR activity makes a major contribution to –Mg-induced chlorophyll degradation.
Figure 3.
Chlorophyll-degrading activity of OsSGR. A, Chlorophyll a analysis by high performance liquid chromatography (HPLC) after incubation with total protein extract. Chlorophyll a was incubated with total protein extract from wild type (WT) and two null mutants after 0- or 250-μ m Mg treatment for 8 d. After incubation for 90 min, chlorophyll a was analyzed by HPLC at 410 nm. B, Comparison of chlorophyll degradation rate between +Mg and −Mg treatments. C, Comparison of chlorophyll degradation rate among different leaves under −Mg conditions. L3, old; L6, mid-aged; L8, young. Chlorophyll degradation rate = (chlorophyll a without protein incubation – chlorophyll a with protein incubation)/chlorophyll a without protein incubation. Data are means ± sd (n = 3). The asterisk shows a significant difference compared to wild type (P < 0.05 by Tukey’s test).
Knockout of OsSGR Yields Stay-Green Leaves and Growth Retardation under −Mg Stress
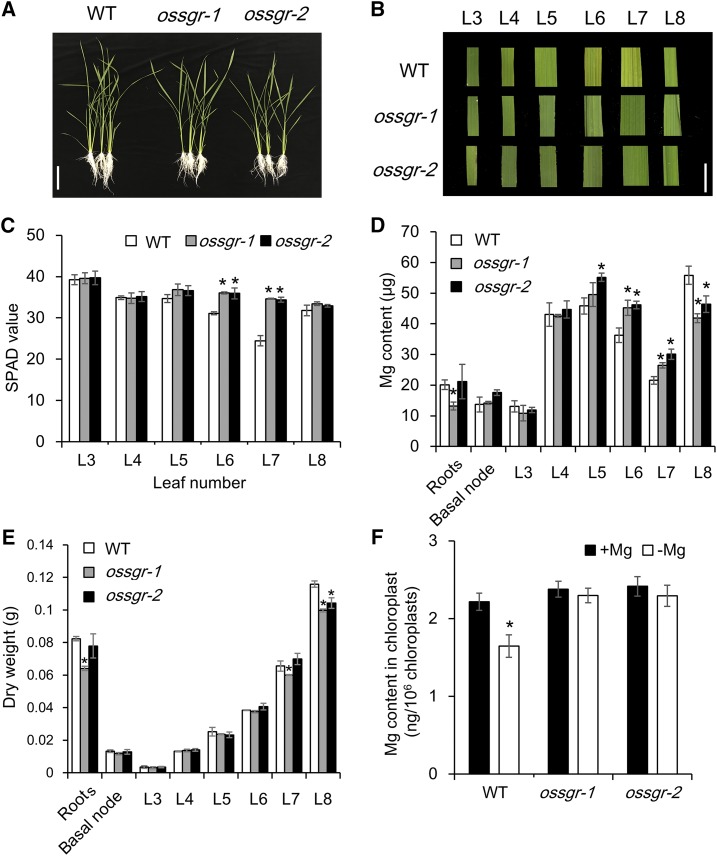
There were no significant phenotypic differences between wild type and two ossgr mutants during the vegetative growth period in +Mg hydroponics cultures (Supplemental Fig. S3). However, after transferring to the −Mg treatment for 8 d, mid-aged leaves (L6 and L7) of wild-type plants became chlorotic, while those of the mutants remained green (Fig. 4, A and B). Consistent with this difference in leaf phenotypes, the chlorophyll concentration of mid-aged leaves was significantly lower in wild type than that in ossgr mutants (Fig. 4C; Supplemental Fig. S4B), but was not changed in old (L3) and young (L8) leaves (Fig. 4C; Supplemental Fig. S4, A and C). Notably, knockout of OsSGR did not alter Mg uptake, translocation, or distribution under −Mg conditions as observed after feeding plants the stable isotope 25Mg for 24 h (Supplemental Fig. S5). Yet, a time-course experiment showed that two null mutants had higher Mg concentration in mid-aged leaves but lower in young leaves than wild type after 5 d of −Mg treatment (Supplemental Fig. S4, E and F). Over 8 d of −Mg, the two mutants accumulated more Mg in mid-aged leaves (L6 and L7) and less Mg in newly developing leaves (L8) than wild-type plants (Fig. 4D). In addition, the two mutant lines displayed an increased sensitivity to −Mg stress, as indicated by lower biomass in newly developing leaves than expected solely from reduced Mg import (Fig. 4E). To further confirm that the transferred Mg is derived from chlorophyll breakdown, Mg content was measured for chloroplasts isolated from mid-aged leaves. As expected, Mg content in wild-type chloroplasts declined significantly in response to −Mg stress, whereas −Mg had no effect on chloroplast Mg content in the two mutant lines (Fig. 4F), which suggests that −Mg accelerates OsSGR-regulated chlorophyll degradation for internal Mg remobilization in mid-aged leaves. Because the expression of SGR-like (SGRL) is also altered by −Mg (Fig. 2A), we further investigated the physiological roles of OsSGRL using two independent clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated system 9 (Cas 9) mutants (ossgrl-1 and ossgrl-2; Supplemental Fig. S2B). However, unlike ossgr mutants, ossgrl-1 and ossgrl-2 did not exhibit the stay-green phenotype under low-Mg conditions (Supplemental Fig. S6), suggesting that the SGRL-dependent chlorophyll degradation pathway is not initiated by −Mg.
Figure 4.
Phenotypes and Mg accumulation in ossgr mutants under −Mg stress. A and B, Growth (A) and chlorosis phenotypes (B) in different leaves of wild-type (WT) and mutant plants. Images in (B) are digitally abstracted and shown as a composite for comparison. Scale bars = 10 cm (A) and 1 cm(B). C, SPAD value. D, Mg content. E, Dry weight. F, Mg content in chloroplasts. Both wild-type and mutant rice seedlings were grown in a nutrient solution containing 0- or 250-μm Mg for 8 d. The SPAD value of each leaf was measured using a chlorophyll meter. Intact chloroplasts from wild-type and mutant plants were obtained using the Percoll gradient method. Mg content was determined by ICP-MS. L3–L8 are labels for leaves ordered from old to young. Data are means ± sd (n = 3). The asterisk shows a significant difference compared to wild type (P < 0.05 by Tukey’s test).
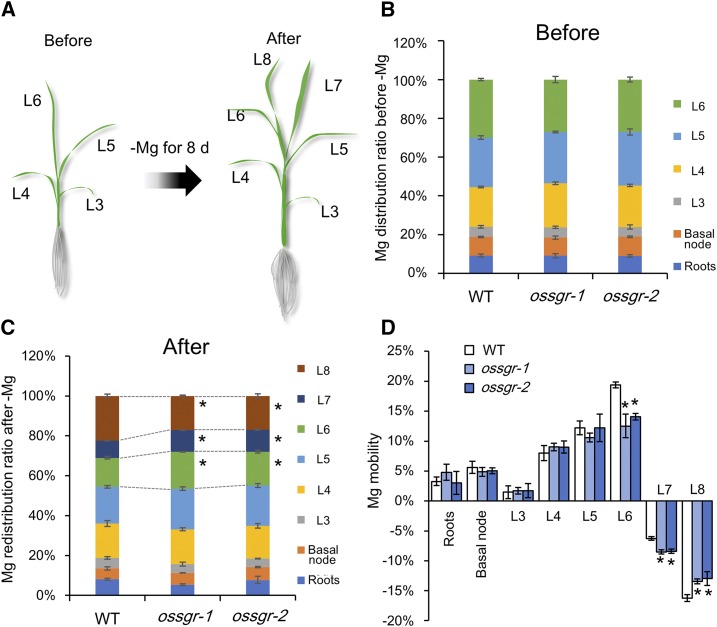
Knockout of OsSGR Decreases Mg Mobility in Mid-aged Leaves under −Mg Stress
To further examine the involvement of OsSGR in the remobilization of Mg from mid-aged leaves to newly developing tissues, we determined Mg distributions among leaves. Both wild-type and mutant plants were grown in +Mg nutrient solution until the L6 leaf was fully expanded and then transferred to −Mg nutrient solutions for 8 d (Fig. 5A). Before the initiation of −Mg treatments, there were no significant differences observed in Mg distributions among leaves between wild-type and mutant plants (Fig. 5B). Then, after 8 d of −Mg treatment, Mg distribution ratios were lower in mid-aged leaves (L6 and L7) and higher in newly developing leaves (L8) for wild-type plants than for the two knockout lines (Fig. 5C). Mg mobility was calculated by dividing the decrease in Mg content in response to −Mg treatment for each part by total plant Mg content. In this way, it was determined that mid-aged leaves are the predominant contributors to Mg recycling in both wild-type and mutant plants (Fig. 5D). Even so, Mg mobility from L6 and L7 leaves was much lower in mutant plants than in wild-type plants (Fig. 5D). Because L7 leaves were developing during the −Mg treatment period, net mobilization from these leaves was negative on balance. These results further reinforce the conclusion that OsSGR is required for remobilization of Mg from mid-aged leaves.
Figure 5.
Mg redistribution and mobility in ossgr mutants under −Mg stress. A, Scheme for Mg redistribution. Rice seedlings of both wild-type (WT) and mutant plants were first grown in nutrient solution containing 250-μm Mg until the L6 leaves were fully expanded. Then plants were transferred to −Mg nutrient solution for 8 d. Plant parts (roots, basal node, and the six labeled leaves) were separately harvested before and after exposure to the −Mg treatment. B, Mg distribution ratio before transfer to −Mg media. C, Mg redistribution ratio after 8 d in −Mg nutrient solution. Ratios were calculated by dividing Mg content in each fraction by total plant Mg content. D, Mg mobility. The mobility of Mg was calculated by dividing the decrease of Mg content in each fraction after 8 d in −Mg media by total plant Mg contents. Data are means ± sd (n = 3). The asterisk shows a significant difference compared to wild type (P < 0.05 by Tukey’s test).
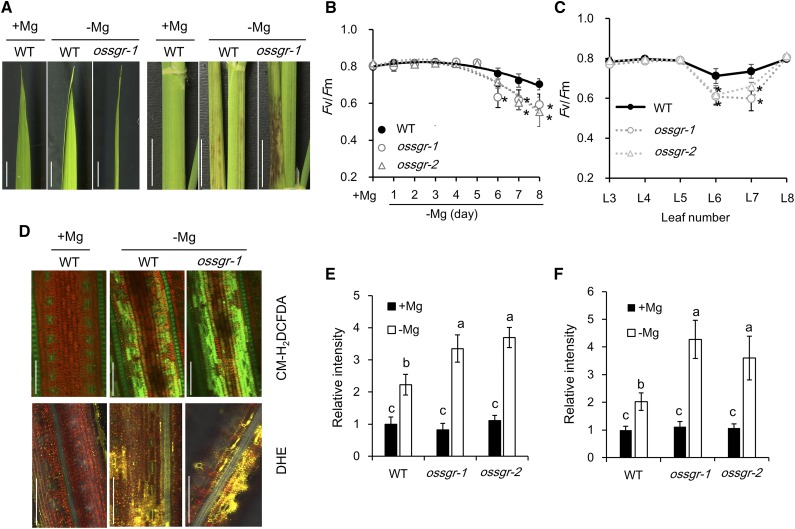
OsSGR Prevents Photodamage and ROS Generation by −Mg in Mid-aged Leaves
Although mutant leaves remained green under −Mg conditions, leaf blades exhibited early curling and leaf sheaths developed necrotic spots, suggesting that these stay-green leaves are more sensitive to −Mg stress than chlorotic wild-type leaves (Fig. 6A). To determine the involved mechanisms, we measured the chlorophyll fluorescence parameter Fv/Fm, the maximal quantum yield of PSII photochemistry, which is regarded as a sensitive indicator of photoinhibition (Barber and Andersson, 1992; Kasahara et al., 2002). Our results showed that Fv/Fm gradually decreased with exposure time to –Mg (Fig. 6B), and the decrease of Fv/Fm only occurred in the mid-aged leaves of wild-type plants (Fig. 6C). The ROS also were determined by using different fluorescence chemicals. Interestingly, −Mg stress significantly increased the production of hydrogen peroxide (H2O2, by 5-[and-6]-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate acetyl ester [CM-H2DCFDA]; Fig. 6, D and E) and superoxide (·O2−, by dihydroethidium [DHE]; Fig. 6, D and F) but did not alter the generation of singlet oxygen (1O2, by singlet oxygen sensor green [SOSG] reagent; Supplemental Fig. S7), suggesting that −Mg stress enhances the overflow of photosynthetic electrons to molecular O2. Furthermore, two null mutants exhibited lower values of Fv/Fm and higher levels of ROS (H2O2 and ·O2−) than wild type (Fig. 6, B–F), indicating that in the absence of SGR, more photodamage and ROS are produced in mid-aged leaf by –Mg stress.
Figure 6.
ROS generation in mid-aged leaves under −Mg stress. A, Phenotypic profile of leaf blades (left) and leaf sheaths (right) of mid-aged leaves. Both wild-type (WT) and mutant rice seedlings were grown in nutrient solution containing 0- or 250-μm Mg for 8 d. Scale bars = 1 cm. B and C, Comparison of Fv/Fm values in different Mg deficient times (B) and different leaves (C). Rice seedlings were dark-adapted overnight. The maximum efficiency of PSII photochemistry Fv/Fm = (Fm − F0)/Fm, was determined by a portable photosynthesis system (model no. LI-6800; Li-Cor). Data represent means ± sd (n = 3). D, CM-H2DCFDA and DHE staining of ROS intermediates in leaf blades. Mid-aged leaves were incubated in 10-μm CM-H2DCFDA or 40-μm DHE for 30 min and imaged by confocal microscopy. Scale bars = 100 μm. E and F, Relative intensity of CM-H2DCFDA (E) and DHE (F) staining. Data represent means ± sd (n = 6). The asterisk in (B) and (C) shows a significant difference compared to wild type (P < 0.05 by Tukey’s test). Means with different letters in (E) and (F) are significantly different (P < 0.05 by Tukey’s test).
H2O2 Regulates OsSGR Expression and Chlorophyll Degradation
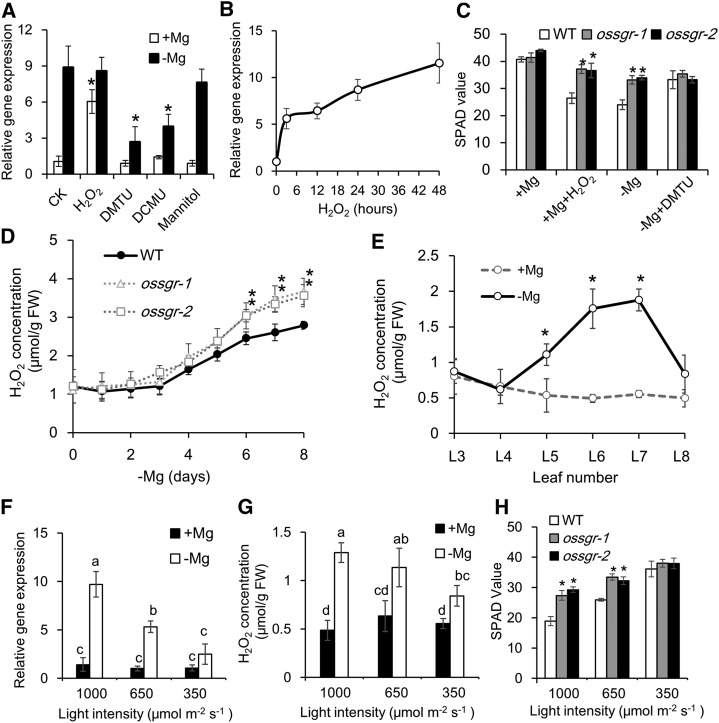
Accumulation of carbohydrates in source leaves and subsequent restriction on CO2 fixation by −Mg stress is thought to be the source of ROS generation (Cakmak et al., 1994a, 1994b; Hermans and Verbruggen, 2005; Cakmak and Kirkby, 2008). Thus, we firstly measured Suc concentration in each leaf of the wild type and mutants. Indeed, Suc accumulated quickly after exposure to −Mg, particularly in mid-aged leaves (Supplemental Fig. S8A), but there was no difference in Suc concentrations between wild type and mutants (Supplemental Fig. S8B), indicating that OsSGR-regulated photoprotection and endogenous ROS balance might not be associated with carbohydrate status in rice leaves.
Considering ROS burst (particularly H2O2 and ·O2−) by −Mg stress (Fig. 6), we conducted experiments to test whether H2O2 might act as a signal molecule in the regulation of chlorophyll degradation (Orozco-Cárdenas et al., 2001; Veal et al., 2007). As expected, the expression of OsSGR in mid-aged leaves was highly induced by exogenous addition of H2O2 (Fig. 7A), and this induction was very fast (within 3 h) and in a time-dependent manner (Fig. 7B). The –Mg-induced expression of OsSGR was suppressed by the H2O2 scavenger 1,3-dimethyl-2-thiourea (DMTU; Levine et al., 1994) and also by the photosynthetic electron transfer inhibitor 3-(3, 4-dichlorphenyl)-1,1-dimethylurea (DCMU; Exposito-Rodriguez et al., 2017) but not by hydroxyl radical (OH·) scavenger mannitol (Patel and Williamson, 2016; Fig. 7A). Accordingly, both addition of H2O2 and −Mg stress led to reductions in chlorophyll concentrations in wild-type plants, which were not matched in ossgr mutants (Fig. 7C). The reductions were prevented by addition of DMTU to −Mg treated wild-type plants (Fig. 7C). On the whole, these results suggest that −Mg stress induces OsSGR expression and chlorophyll degradation through upregulation of endogenous H2O2 levels.
Figure 7.
ROS-mediated OsSGR expression and chlorophyll degradation. A, The effects of exogenous addition of H2O2 and ROS scavengers on OsSGR expression in mid-aged leaves. Rice seedlings were grown in +Mg or −Mg nutrient solution for 8 d and treated with H2O2, DMTU, DCMU, or mannitol for 12 h. B, Time-dependent expression of OsSGR in response to H2O2. C, The effects of H2O2 and DMTU on leaf chlorosis. Rice seedlings were grown in +Mg solution with or without H2O2, or −Mg solution with or without DMTU addition. D and E, H2O2 concentrations in different Mg-deficient times (D) and in different leaves (E). F to H, Effects of light intensity on OsSGR expression (F), H2O2 concentration (G), and SPAD values (H) in mid-aged leaves. Rice seedlings were exposed to light intensities of 1,000, 650, or 350 μmol m−2 s−1 with or without Mg supplied. Gene expression is shown relative to expression of CK (+Mg) in (A), 0 h in (B), and 1,000 PPFD (+Mg) in (F). H2O2 concentration was quantified by the FOX method. SPAD values were measured using a chlorophyll meter. Data are means ± sd (n = 3). The asterisk in (A), (C), (D), (E), and (H) shows a significant difference compared to CK (A), wild type (WT; C), (D), and (H), and +Mg (E; P < 0.05 by Tukey’s test). Means with different letters in (F) and (G) are significantly different (P < 0.05 by Tukey’s test). FW, fresh weight.
Detection of plant internal H2O2 revealed that H2O2 consistently aligned with the OsSGR expression pattern, which started to increase after 4 d of –Mg treatment (Fig. 7D) and exhibited this increase only in mid-aged leaves (Fig. 7E). Moreover, the two ossgr mutants had much higher concentrations of H2O2 in mid-aged leaves than wild-type plants (Fig. 7D), confirming that OsSGR-mediated chlorophyll breakdown suppresses the generation of ROS in response to −Mg stress in mid-aged leaves. These observations support the conclusion that ROS appear to act as feedback regulators of OsSGR expression to precisely govern chlorophyll degradation in mid-aged leaves.
Because ROS generation in response to −Mg stress is known to result from photooxidation (Cakmak and Kirkby, 2008), we exposed rice seedlings to different light intensities (1,000, 650, and 350 μmol m−2 s−1 photosynthetic photon flux density [PPFD]) and subsequently assayed for variation in endogenous ROS levels. Decreased light intensity lowered both OsSGR expression and H2O2 levels (Fig. 7, F and G), while chlorophyll concentration increased significantly with decreasing light intensity in the absence of Mg (Fig. 7H). These results further indicate that photooxidation-derived ROS positively regulate leaf senescence through stimulation of OsSGR expression.
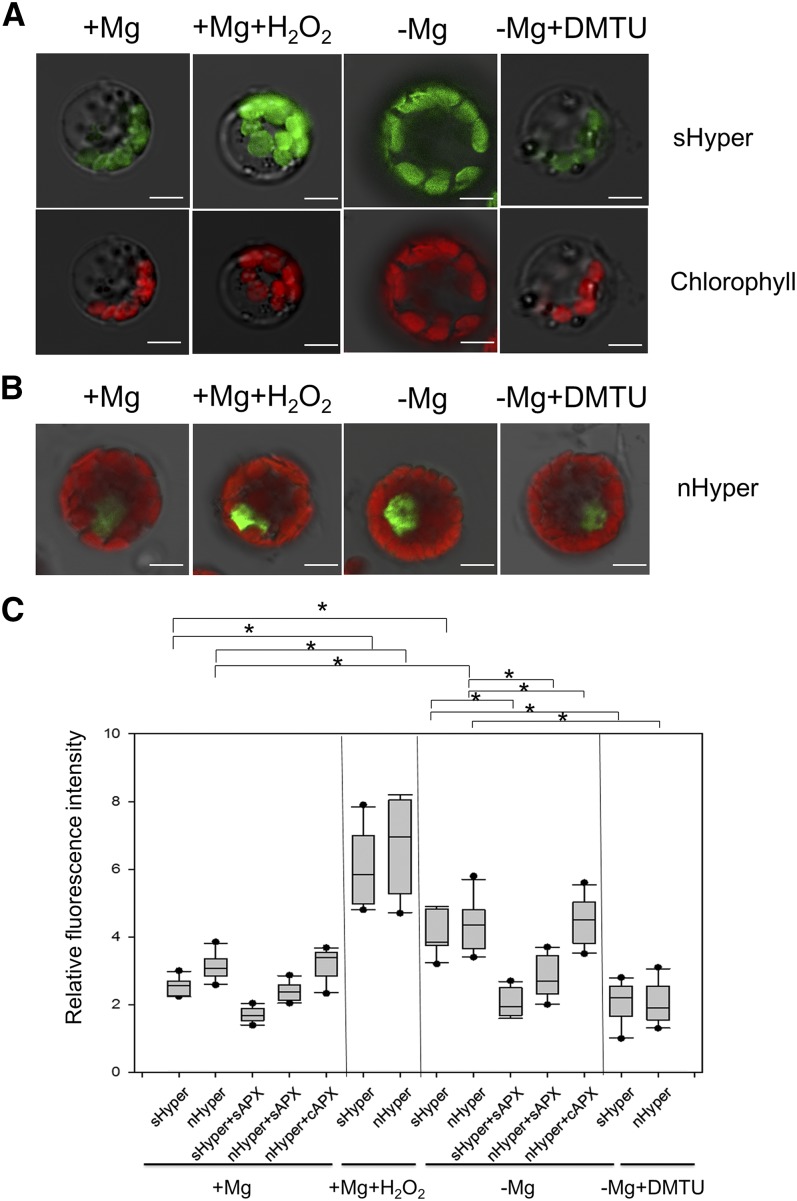
Mg Deficiency Increases the Production of Chloroplastic and Nuclear H2O2
Considering that OsSGR is localized at the chloroplast but its expression is activated by H2O2 at the nucleus, we investigated H2O2 production in the chloroplast and nucleus of rice protoplasts using a genetically encoded fluorescent H2O2 sensor, HyPer, which shows an increase in fluorescence emission at 530 nm when excited at 488 nm in the presence of H2O2 (Exposito-Rodriguez et al., 2017). Our results revealed that either −Mg stress or H2O2 treatment increased H2O2 production, whereas addition of DMTU to −Mg stressed protoplasts suppressed H2O2 production in both the chloroplast and nucleus by transformation with a stroma-localized Hyper (sHyper) or a nucleus-localized Hyper (nHyper; Fig. 8, A–C). Overexpressing a stromal ascorbate peroxidase (sAPX) to increase the capacity of chloroplastic H2O2 scavenging decreased H2O2 production by –Mg not only in the chloroplast (sHyper + sAPX; Fig. 8C), but also in the nucleus (nHyper + sAPX; Fig. 8C). However, overexpressing a cytosolic ascorbate peroxidase (cAPX) had no such alleviating effects on nuclear H2O2 production (nHyper + cAPX; Fig. 8C). Therefore, the close association of H2O2 production between the chloroplast and nucleus strongly suggests the important role of H2O2 in chloroplast-to-nucleus signaling.
Figure 8.
Production of chloroplastic and nuclear H2O2 by −Mg. A and B, Responses of the stroma or nuclear targeted Hyper in rice protoplasts to oxidized state (5-mm H2O2 for 10 min) or reduced state (100-μm DMTU for 20 min) under Mg-sufficient (+) and -deficient (−) conditions. Scale bars = 5 μm. C, Fluorescence intensity of Hyper relative to chlorophyll autofluorescence. Box-whisker plots show signal variations in protoplasts (n = 10). The asterisk shows a significant difference (P < 0.05 by Tukey’s test).
DISCUSSION
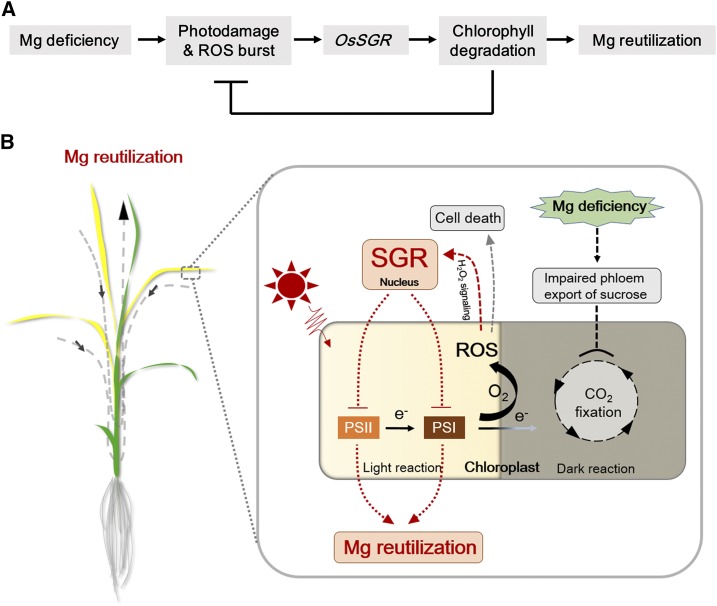
Many nutrient deficiency stresses trigger obvious leaf chlorosis symptoms in plants, but the underlying mechanisms are seldom studied. Through transcriptomic analysis of rice plants, Mg deficiency was observed to stimulate chlorophyll degradation in mid-aged leaves (Fig. 2A). The fact that substantial amounts of Mg are bound to chlorophyll led to testing of whether Mg freed from degrading chlorophyll can be reutilized across plant tissues in response to −Mg stress. Interestingly, expression of a key regulator gene for chlorophyll degradation, OsSGR, is highly and specifically induced by Mg depletion in mid-aged rice leaves (Fig. 2), which is consistent with the leaf chlorosis phenotype caused by −Mg stress (Fig. 1, A and B). So far, SGRs have been identified in several plant species as putative Mg dechelatases that remove Mg from chlorophyll a (Matsuda et al., 2016; Shimoda et al., 2016). Furthermore, the function of SGR proteins appears to be conserved among plant species, because knockout of SGRs lead to leaf stay-green phenotypes (Jiang et al., 2007; Park et al., 2007; Ren et al., 2007; Sato et al., 2007; Fang et al., 2014). Therefore, it is reasonable to hypothesize that OsSGR is involved in remobilization of chlorophyll Mg in rice under −Mg stress. However, plants have developed several strategies to overcome nutrient deficiencies, such as strengthening nutrient uptake, translocation, distribution, and redistribution. We found that OsSGR is not required for Mg uptake, root-to-shoot translocation, or distribution because knockout of OsSGR did not alter these processes (Supplemental Fig. S5). However, we found that OsSGR is required for remobilization of Mg from mid-aged leaf to young developing tissues. This is supported by results showing wild type significantly increased Mg mobility in mid-aged leaves through accelerating chlorophyll degradation and release of Mg (Fig. 4F), which resulted in more Mg being transferred to developing tissues in rice plants under −Mg stress (Figs. 4 and 5). The end result is that growth of young developing leaves is curtailed considerably in ossgr mutant lines (Fig. 4E). The released Mg from chlorophyll degradation by OsSGR accounts for one-quarter of the total amount of remobilized Mg, based on the difference in mobility between wild type and mutants (Fig. 5D). The other three quarters are probably from vacuolar and cytosolic Mg (weak binding with ATP and ribosomes), which account for 60% to 80% total Mg in cells. On the whole, these results strongly indicate that OsSGR-mediated chlorophyll degradation and Mg remobilization are stimulated by –Mg stress in mid-aged leaves of rice. Our study identifies a gene encoding a Mg dechelatase that is induced specifically by −Mg stress and in the leaves where Mg deficiency is most apparent. Thus, it provides evidence for a key biochemical step in the mechanism of –Mg stress, which to date was only known quite descriptively.
Our results further demonstrate that endogenous ROS levels, particularly H2O2, are the trigger for −Mg-induced OsSGR expression (Fig. 7, A and B). ROS are constantly generated at basal levels and keep balance between production and elimination, playing an important signaling role in plants. However, they also cause extensive cell damage once the endogenous ROS balance is disturbed by biotic and abiotic environmental stresses (Das and Roychoudhury, 2014). The major ROS species include 1O2, H2O2, ·O2−, and OH·. Among them, 1O2 and H2O2 are known as signal molecules regulating different cellular signaling pathways (Apel and Hirt, 2004; Reczek and Chandel, 2015). −Mg stress may enhance the overflow of photosynthetic electrons to O2, as H2O2 and ·O2− were largely generated in our study (Fig. 6, D–F). However, −Mg stress does not affect the reaction of chlorophyll triplet state in the antenna system with O2, as 1O2 was little affected (Supplemental Fig. S7). We hypothesize the H2O2 signaling pathway is initiated by −Mg stress because despite Mg supply, approaches by artificially altering endogenous H2O2 levels exclusively changed the expression level of OsSGR (Fig. 7A). In addition, DCMU stimulates the release of 1O2 (Wagner et al., 2004), further confirming that ROS signaling is not from 1O2. Considering the important regulatory roles of H2O2 in plant physiological processes (Hossain et al., 2015), H2O2 might not simply be passively damaging chloroplasts in plants under −Mg stress, but may be active feedback regulators participating in −Mg-induced chlorophyll degradation through governing of OsSGR expression. Strong evidence supporting this view is that two mutants’ lack of OsSGR for chlorophyll degradation led to much higher accumulation of ROS levels and more serious photodamage (Fig. 6). Thus, chlorophyll degradation by OsSGR not only provides Mg for remobilization, but also might slow down photosynthetic electron generation and thereby reduce photodamage in mid-aged leaves caused by −Mg stress (Fig. 9).
Figure 9.
Proposed model of −Mg-triggered chlorophyll degradation and Mg remobilization. A, Simplified model. B, Detailed model. Mg depletion impairs the phloem export of Suc in the early stages of Mg deprivation. This leads to high accumulations of carbohydrates in source leaves, which inhibits photosynthetic CO2 fixation. As a result, more photosynthetic electron transfer to O2 occurs, which increases the generation of ROS. ROS levels, particularly H2O2, positively regulate OsSGR expression through chloroplast-to-nucleus signaling and thereby accelerate chlorophyll degradation to release Mg for remobilization. Thus, chlorophyll breakdown not only facilitates Mg remobilization but also slows down photosynthetic electron generation, protecting leaves from further photodamage.
This regulatory role for H2O2 might have arisen from −Mg stress leading to the generation of ROS through photooxidation. This is due to the fact that phloem export of Suc is severely impaired during early stages of Mg depletion, which results in the inhibition of CO2 fixation and subsequent excessive flows of absorbed light energy to molecular oxygen to generate ROS (Cakmak and Kirkby, 2008). In this study, support for photooxidation effects on H2O2 regulation of OsSGR expression and chlorophyll degradation was produced in light intensity experiments. Endogenous H2O2 levels (Fig. 7G) and expression of OsSGR (Fig. 7F) both decreased with decreases in light intensity. As a consequence, chlorophyll degradation slowed in wild-type plants growing in −Mg conditions (Fig. 7H). These results suggest that photooxidation-derived H2O2 is important for regulating OsSGR expression and chlorophyll degradation in response to Mg availability. On the other hand, H2O2 is able to move out of chloroplasts (Mubarakshina et al., 2010), and chloroplast-derived H2O2 under light conditions has been proposed to participate in chloroplast-to-nucleus signaling (Bobik and Burch-Smith, 2015; Gollan et al., 2015; Exposito-Rodriguez et al., 2017). Our results showed nuclear H2O2 is closely associated with chloroplastic H2O2 generation under −Mg conditions (Fig. 8), supporting the possibility that −Mg-induced photooxidative stress triggers chloroplast-to-nucleus H2O2 signaling.
In general, nutrient deficiency stresses lead to plant leaf chlorosis phenotypes either starting from the oldest (e.g. N, K) or the youngest leaves (e.g. Fe, Ca). However, Mg deficiency first triggers chlorosis in mid-aged leaves and simultaneously triggers Mg remobilization from mid-aged to youngest leaves (Figs. 1, 4, and 5). This symptom of −Mg stress differentiates this deficiency from others, even though the underlying mechanisms were still unclear (Chen et al., 2018). In this study, we observed that mid-aged leaves of rice have relatively high photosynthetic capacities under +Mg conditions (Fig. 1E), which suggest relatively high rates of carbohydrate production. Once carbohydrate export is blocked in the early stages of −Mg stress, more carbohydrates accumulate in mid-aged leaves (Supplemental Fig. S8A). This impairs CO2 fixation (Fig. 1I), which results in more photosynthetic electron transfer to O2 to generate ROS in −Mg stressed mid-aged leaves (Figs. 6 and 7D). We speculate here that rice plants counter oxidative stress by accelerating OsSGR-mediated chlorophyll breakdown, which leads to the chlorosis phenotypes observed first in mid-aged leaves of plants subjected to Mg deprivation. On the other hand, we also observed that mid-aged leaves have relatively large Mg pools under +Mg condition (Fig. 1C). It is reasonable for plants to develop such a strategy to preferentially reutilize Mg from the regions with high Mg content once Mg is deficient.
In conclusion, the experiments herein demonstrate that Mg deficiency initiates OsSGR-mediated chlorophyll degradation in mid-aged leaves of rice. Upregulation of OsSGR is an adaptive strategy to accelerate Mg remobilization and protect mid-aged leaves from photodamage under −Mg stress.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The rice (Oryza sativa) ossgr-1 mutant used in this study was donated for this work by Prof. Makoto Kusaba (Sato et al., 2007). The other mutant, ossgr-2, which is a CRISPR/Cas9 knockout line of OsSGR in the cv Nipponbare background, was generated according to the protocol described by Ma et al. (2015). The target site primers for OsSGR (F, 5′-GCCGGGTGTCGCACACCATCAACC-3′ and R, 5′-AAACGGTTGATGGTGTGCGACACC-3′) were introduced into the pYLsgRNA-U6a vector using BsaI. The integrated single guide RNA was then amplified in two rounds of PCR using the primers U-F/gR-R and Pps-GGL/Pgs-GGR, respectively. Amplified fragments were then introduced into the pYLCRISPR/Cas9 Pubi-H vector using BsaI. The CRISPR/Cas9 transgenic seedlings were then obtained through Agrobacterium tumefaciens (EHA105)-mediated transformation. Cultivator Nipponbare was used as the wild-type rice in this work.
Seeds of wild-type and mutant rice were soaked in deionized water at 30°C in the dark for 2 d. Subsequently, seedlings were transferred to a net floating on 0.5-mm CaCl2 solution for 7 d. The seedlings were then grown in half-strength Kimura B nutrient solution (pH 5.6) in a growth chamber (12-h light at 30°C/12-h dark at 25°C, 60% humidity) for 12 d. This was followed by 8 d of growth in a Mg-sufficient (+Mg, 250 μm) treatment or a Mg-deficient (−Mg, 0 μm) treatment. For oxidative stress treatment, 10-mm H2O2 was added to the +Mg nutrient solution. For H2O2 trap treatment, 0.2-mm DMTU was added into –Mg nutrient solution. Dynamics of Mg turnover were also assessed by adding 200-μm 25Mg (Taiyo Nippon Sansho) into –Mg nutrient solution. Effects of light intensity were also studied by exposing plants in both Mg treatments to a PPFD of 1,000, 650, or 350 μmol m−2 s−1. All experiments were repeated at least twice with three replicates each.
Phenotypic Analysis
Seedlings of both wild type and ossgr mutants reared in hydroponics were photographed upon harvesting. Seedlings were separated into roots, nodes, and each leaf for further analysis. The SPAD value for chlorophyll concentration of each leaf was measured using a chlorophyll meter (SPAD-502 Plus; Konica Minolta). The dry weight of each part was measured after drying in a 65°C oven for 3 d.
The net photosynthetic rate was measured with a portable photosynthesis system (model no. LI-6800; Li-Cor). Measurements were taken during the daytime in the growth chamber with real-time illumination, leaf temperatures of 30°C, and a relative humidity of 60%. The CO2 level in the chamber was 400 μmol mol−1, and the PPFD was 650 μmol m−2 s−1. For chlorophyll fluorescence analysis, rice seedlings were dark-adapted overnight. The maximum efficiency of PSII photochemistry Fv/Fm = (Fm − F0)/Fm, was determined as described in Andrews et al. (1993). F0 and Fm are the minimum and maximum fluorescence in the dark-adapted state, respectively.
Transcriptomic and Gene Expression Analysis
Rice seedlings (20-d–old) were grown in +Mg or −Mg hydroponic cultures for 7 d before sampling the shoots for RNA-seq analysis. Total RNA was isolated and reverse-transcribed into cDNA. The appropriate cDNA fragments from agarose gel electrophoresis were amplified by PCR and then sequenced using the Illumina HiSeq 2500 (Novogene Biotechnology).
To investigate the response of OsSGR expression to −Mg stress, rice seedlings (20-d–old) were grown in +Mg or −Mg nutrient solution for 7 d. Roots, basal nodes, leaf blades and leaf sheaths were then separately harvested from a portion of the plants. The remainder of the Mg deficient seedlings were resupplied with 250-μm Mg for 1 d before harvesting. For −Mg time-course experiments, a portion of rice seedlings were transferred into –Mg nutrient solution daily. Seven d after the first transfer, the shoots of all samples were harvested. To differentiate expression among rice leaves, L3–L8 (old to young) leaves were separately harvested after 7 d in +Mg or −Mg nutrient solution. To investigate OsSGR expression responses to other nutrient stresses, rice seedlings (20-d–old) were grown in normal nutrient solution (half-strength Kimura B nutrient solution) or in nutrient solution lacking Mg, N, Fe, P, K, Mn, Cu, or Zn for 7 d before harvesting the shoots for RNA extraction. For oxidative stress and ROS trap treatments, 10-mm H2O2, 30-mm mannitol, and 0.2-mm DMTU or 10-μm DCMU was added into the +Mg and –Mg nutrient solution for 12 h. For H2O2 time-course experiments, a portion of rice seedlings was transferred into 10-mm H2O2 for 0, 3, 12, 24, and 48 h, and the mid-aged leaves were harvested.
Total RNA was extracted using the TransZol Up Plus RNA Kit (TransGen Biotech). cDNA synthesis was performed using the ReverTra Ace qPCR RT Master Mix with genomic DNA Remover (TOYOBO) following the manufacturer’s instructions. Gene expression levels were analyzed by quantitative reverse transcription PCR on a LightCycler 96 (Roche) using the TranStart Top Green qPCR SuperMix (TransGen Biotech). OsActin was selected as an internal standard. Expression was normalized by the ΔΔCt method. The specific primers used were F, 5′-CTGCAGGGGTGGTACAACAA-3′ and R, 5′-TGGACGAACGCCTTCAGAAC-3′ for OsSGR and F, 5′-GACTCTGGTGATGGTGTCAGC-3′ and R, 5′-GGCTGGAAGAGGACCTCAGG-3′ for OsActin.
OsSGR Activity Assay
To verify the activity of OsSGR, total proteins from mid-aged leaves were extracted by extraction buffer containing 25-mm Tris-HCl at pH 7.5, 10-mm NaCl, 10-mm MgCl2, 1 × ProteinSafe Protease Inhibitor Cocktail (TransGen Biotech), and 5-mm dithiothreitol and incubated with chlorophyll a standard substance (275 pmol; Santa Cruz Biotechnology) for 90 min at room temperature in a 30-μL reactive system containing 50-mm Tris-HCl at pH 7.5, 100-mm NaCl, and 0.05% (v/v) polysorbate 20. Samples were added with 60-μL acetone and kept on ice and in the dark for 10 min. After centrifugation at 21,000g for 15 min at 4°C, the pigments extracted from the reaction mixture were analyzed by HPLC according to Das et al. (2018). A model no. C18 Hypersil ODS column (125 × 4.0 mm, 5 μm; Thermo Fisher Scientific) was used for analysis. The solvents (solvent A, ammonium acetate:MeOH = 20:80; solvent B: acetone:MeOH = 20:80 [v/v]) at a flow rate of 1.0 mL/min were run by following the HPLC program: solvent A for 4 min, A to B for 5 min, solvent B for 9 min, and A for 2 min. We set column temperature to 28°C and injection volume to 20 μL and monitored the elution profiles with a diode array detector at 410-nm excitation (1260 Infinity; Agilent).
Mg Determination
Upon conclusion of exposure to the various treatments, harvested samples were separated into root, basal node, and each leaf. Roots were washed three times with ice-cold 0.5-mm CaCl2 to remove apoplastic-bound Mg. After drying at 65°C for 3 d, samples were weighed and digested in concentrated HNO3. The concentrations of 24Mg and 25Mg were determined by ICP-MS using a model no. 7900 Mass Spectrometer (Agilent).
Subcellular Localization of OsSGR
Subcellular localization of OsSGR was investigated by introducing OsSGR-GFP into rice leaf protoplasts. The open reading frame of OsSGR was amplified by PCR using the primers 5′-TTTGGAGAGGACACGCTCGAGATGGCTGCTGCTACTTCGAC-3′ and 5′-GCCCTTGCTCACCATGGCGCGCCGCTGCTGCGGCTGGCCGTCGG-3′, and then inserted into the XhoI and AscI sites of the pFGC5941-GFP vector along with a CaMV35S promoter. The leaf protoplasts used for transient expression analysis were extracted from rice seedlings (14-d–old) by the polyethylene glycol method (Chen et al., 2006). GFP signals were observed by confocal laser-scanning microscopy (LSM880; Carl Zeiss).
Detection of ROS
H2O2 content was quantified by the ferrous oxidation-xylenol orange (FOX) method as described in Kaur et al. (2016) and Mátai and Hideg (2017). Briefly, 50-mg samples were ground in liquid nitrogen and homogenized in 0.5 mL of 5% (w/v) trichloroacetic acid. After centrifuging at 5,000 g at 4°C for 10 min, 25 μL of the supernatant was mixed with 475 μL of FOX reagent (Kaur et al., 2016). Samples were analyzed using a spectrophotometer with the wavelength set at 560 nm (UV-1780; Shimadzu).
For CM-H2DCFDA, DHE, and SOSG staining, mid-aged leaves were incubated in 50-mm phosphatic buffer solution (pH 7.4) containing 10-μm CM-H2DCFDA (Invitrogen), 40-μm DHE (Sigma-Aldrich), or 10-μm SOSG (Invitrogen) for 30 min at room temperature. These samples were then washed with phosphatic buffer solution three times and used for fluorescent imaging by confocal microscopy (model no. LSM880; Carl Zeiss). Excitation was detected at 488 nm and emission at 517–540 nm for CM-H2DCFDA. Excitation was detected at 514 nm and emission at 520–600 nm for DHE. Excitation was detected at 488 nm and emission at 520–550 nm for SOSG.
Suc Determination
Suc was determined by the resorcinol method according to Peng et al. (2018) using a Suc Assay Kit (Comin Biotechnology) as directed by the manufacturer. In brief, each sample was mixed with 1 mL of extract solution and heated at 80°C for 10 min. After centrifuging, the supernatant was mixed with activated carbon and heated to 80°C for 30 min before adding 1 mL of extract solution. After centrifuging, the supernatant was mixed with Reagent II at 95°C for 5 min and then mixed with Reagent III and Reagent IV and incubated at 95°C for 30 min. Samples were chilled before colorimetric analysis using a model no. UV-1780 spectrophotometer with the wavelength set at 480 nm (Shimadzu).
Isolation of Intact Chloroplasts
Intact chloroplasts were isolated using the Percoll gradient method as described in Kunst (1998) with slight modification. All operations were performed at 4°C. Approximately 7 g of fresh rice seedling leaf tissue was homogenized in a buffer containing 50-mm HEPES-KOH at pH 7.7, 331.2-mm sorbitol, 1-mm MgCl2, and 2-mm EDTA-KOH at pH 8.0. After filtering through two layers of Miracloth (Millipore), the homogenate was centrifuged at 3,200 g for 7 min with a swing-out rotor centrifuge (5810R; Eppendorf). Pellets were then gently resuspended in SH buffer (50-mm HEPES-KOH, at pH 8.0, 331.2-mm sorbitol, and 1-mm dithiothreitol) and carefully transferred onto a 35%/70% (v/v) Percoll gradient (GE Healthcare) for centrifugation at 3,200 g for 30 min to obtain intact chloroplasts at the interface of the two Percoll phases. After washing three times with SH buffer, chloroplasts were counted using a hemocytometer under a Primo Star light microscope (Carl Zeiss). Samples were then digested in concentrated HNO3 for measurement of Mg concentrations using ICP-MS, as described in "Mg Determination".
Transient Expression of Hyper System in Rice Protoplasts
The plasmids sHyper, nHyper, sHyper + sAPX, nHyper + sAPX, and nHyper + cAPX were purchased from Addgene (www.addgene.org) and were originally provided by Exposito-Rodriguez et al. (2017). Five-d–old rice seedlings (cv Nipponbare) were treated with 0- or 250-μm Mg for 7 d, and leaf protoplasts were extracted and used for transient expression by the polyethylene glycol method (Chen et al., 2006). For H2O2 treatment, Mg-sufficient protoplasts were treated with 5-mm H2O2 for 10 min before imaging. For DMTU treatment, Mg-deficient protoplasts were treated with 100-μm DMTU for 20 min before imaging. The signals of Hyper and chlorophyll were observed by a model no. LSM880 confocal laser-scanning microscope (Carl Zeiss) at 488 nm and 633 nm, respectively.
Accession Numbers
Sequence data from this article can be found in the GenBank/European Molecular Biology Laboratory data libraries under the following accession numbers: OsSGR (Os09g0532000) and OsSGRL (Os04g0692600).
Supplemental Data
The following materials are available.
Supplemental Figure S1. Subcellular localization of OsSGR.
Supplemental Figure S2. Gene structure of OsSGR and OsSGRL.
Supplemental Figure S3. Phenotype and Mg accumulation in ossgr mutants under +Mg conditions.
Supplemental Figure S4. Time-dependent decline of chlorophyll and Mg concentrations in different leaves by −Mg.
Supplemental Figure S5. 25Mg uptake, translocation, and distribution in ossgr mutants.
Supplemental Figure S6. Phenotype of ossgrl mutants in rice.
Supplemental Figure S7. SOSG staining of singlet oxygen in rice leaves.
Supplemental Figure S8. Suc accumulation by −Mg stress.
Acknowledgments
We are grateful to Prof. Makoto Kusaba for providing ossgr-1 mutant rice, and we thank Prof. Hong Liao, Prof. Leon Kochian, and Dr. Thomas Walk for critical comments and reading of the article, Prof. Deshu Lin for providing CM-H2DCFDA and DHE chemicals, and Prof. Philip Mullineaux for depositing these Hyper plasmids at Addgene.
Footnotes
This work was financially supported by the National Natural Science Foundation of China (No. 31872171 to Z.C.C.), the China National Key Program for Research and Development (2016YFD0100700 to Z.C.C.), and by Grant-in-Aid for Specially Promoted Research (Japan Society for the Promotion of Science KAKENHI grant no. 16H06296 to J.F.M.).
Articles can be viewed without a subscription.
References
- Aitken RL, Dickson T, Hailes KJ, Moody PW (1999) Response of field-grown maize to applied magnesium in acidic soils in north-eastern Australia. Aust J Agric Res 50: 191–198 [Google Scholar]
- Andrews JR, Bredenkamp GJ, Baker NR (1993) Evaluation of the role of state transitions in determining the efficiency of light utilisation for CO2 assimilation in leaves. Photosynth Res 38: 15–26 [DOI] [PubMed] [Google Scholar]
- Apel K, Hirt H (2004) Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Barber J, Andersson B (1992) Too much of a good thing: Light can be bad for photosynthesis. Trends Biochem Sci 17: 61–66 [DOI] [PubMed] [Google Scholar]
- Bennett WF. (1993) Nutrient Deficiencies and Toxicities in Crop Plants. American Phytopathological Society Press, Saint Paul, MN [Google Scholar]
- Bergmann W. (1994) Nutritional Disorders of Plants: Development, Visual and Analytical Diagnosis. Gustav Fischer, Jena, Germany [Google Scholar]
- Bobik K, Burch-Smith TM (2015) Chloroplast signaling within, between and beyond cells. Front Plant Sci 6: 781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cakmak I, Kirkby EA (2008) Role of magnesium in carbon partitioning and alleviating photooxidative damage. Physiol Plant 133: 692–704 [DOI] [PubMed] [Google Scholar]
- Cakmak I, Yazi̇ci̇ AM (2010) Magnesium: A forgotten element in crop production. Better Crops 94: 23–25 [Google Scholar]
- Cakmak I, Hengeler C, Marschner H (1994a) Changes in phloem export of sucrose in leaves in response to phosphorus, potassium and magnesium deficiency in bean plants. J Exp Bot 45: 1251–1257 [Google Scholar]
- Cakmak I, Hengeler C, Marschner H (1994b) Partitioning of shoot and root dry matter and carbohydrates in bean plants suffering from phosphorus, potassium and magnesium deficiency. J Exp Bot 45: 1245–1250 [Google Scholar]
- Chen J, Ren G, Kuai B (2016) The mystery of Mendel’s Stay-Green: Magnesium stays chelated in chlorophylls. Mol Plant 9: 1556–1558 [DOI] [PubMed] [Google Scholar]
- Chen S, Tao L, Zeng L, Vega-Sanchez ME, Umemura K, Wang GL (2006) A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol Plant Pathol 7: 417–427 [DOI] [PubMed] [Google Scholar]
- Chen ZC, Peng WT, Li J, Liao H (2018) Functional dissection and transport mechanism of magnesium in plants. Semin Cell Dev Biol 74: 142–152 [DOI] [PubMed] [Google Scholar]
- Das K, Roychoudhury A (2014) Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front Environ Sci 2: 53 [Google Scholar]
- Das A, Guyer L, Hörtensteiner S (2018) Chlorophyll and chlorophyll catabolite analysis by HPLC. Methods Mol Biol 1744: 223–235 [DOI] [PubMed] [Google Scholar]
- Exposito-Rodriguez M, Laissue PP, Yvon-Durocher G, Smirnoff N, Mullineaux PM (2017) Photosynthesis-dependent H2O2 transfer from chloroplasts to nuclei provides a high-light signalling mechanism. Nat Commun 8: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang C, Li C, Li W, Wang Z, Zhou Z, Shen Y, Wu M, Wu Y, Li G, Kong LA, et al. (2014) Concerted evolution of D1 and D2 to regulate chlorophyll degradation in soybean. Plant J 77: 700–712 [DOI] [PubMed] [Google Scholar]
- Fischer ES, Bremer E (1993) Influence of magnesium deficiency on rates of leaf expansion starch and sucrose accumulation, and net assimilation in Phaseolus vulgaris. Physiol Plant 89: 271–276 [Google Scholar]
- Gollan PJ, Tikkanen M, Aro EM (2015) Photosynthetic light reactions: Integral to chloroplast retrograde signalling. Curr Opin Plant Biol 27: 180–191 [DOI] [PubMed] [Google Scholar]
- Gout E, Rébeillé F, Douce R, Bligny R (2014) Interplay of Mg2+, ADP, and ATP in the cytosol and mitochondria: Unravelling the role of Mg2+ in cell respiration. Proc Natl Acad Sci USA 111: E4560–E4567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermans C, Verbruggen N (2005) Physiological characterization of Mg deficiency in Arabidopsis thaliana. J Exp Bot 56: 2153–2161 [DOI] [PubMed] [Google Scholar]
- Hermans C, Johnson GN, Strasser RJ, Verbruggen N (2004) Physiological characterisation of magnesium deficiency in sugar beet: Acclimation to low magnesium differentially affects photosystems I and II. Planta 220: 344–355 [DOI] [PubMed] [Google Scholar]
- Hermans C, Bourgis F, Faucher M, Strasser RJ, Delrot S, Verbruggen N (2005) Magnesium deficiency in sugar beets alters sugar partitioning and phloem loading in young mature leaves. Planta 220: 541–549 [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S, Feller U (2002) Nitrogen metabolism and remobilization during senescence. J Exp Bot 53: 927–937 [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S, Kräutler B (2011) Chlorophyll breakdown in higher plants. Biochim Biophys Acta 1807: 977–988 [DOI] [PubMed] [Google Scholar]
- Hossain MA, Bhattacharjee S, Armin SM, Qian P, Xin W, Li HY, Burritt DJ, Fujita M, Tran LS (2015) Hydrogen peroxide priming modulates abiotic oxidative stress tolerance: Insights from ROS detoxification and scavenging. Front Plant Sci 6: 420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Li M, Liang N, Yan H, Wei Y, Xu X, Liu J, Xu Z, Chen F, Wu G (2007) Molecular cloning and function analysis of the STAY GREEN gene in rice. Plant J 52: 197–209 [DOI] [PubMed] [Google Scholar]
- Karley AJ, White PJ (2009) Moving cationic minerals to edible tissues: Potassium, magnesium, calcium. Curr Opin Plant Biol 12: 291–298 [DOI] [PubMed] [Google Scholar]
- Kasahara M, Kagawa T, Oikawa K, Suetsugu N, Miyao M, Wada M (2002) Chloroplast avoidance movement reduces photodamage in plants. Nature 420: 829–832 [DOI] [PubMed] [Google Scholar]
- Kaur N, Sharma I, Kirat K, Pati PK (2016) Detection of reactive oxygen species in Oryza sativa L. (rice). Bio Protoc 6: e2061 [Google Scholar]
- Kobayashi NI, Tanoi K (2015) Critical issues in the study of magnesium transport systems and magnesium deficiency symptoms in plants. Int J Mol Sci 16: 23076–23093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi NI, Saito T, Iwata N, Ohmae Y, Iwata R, Tanoi K, Nakanishi TM (2013) Leaf senescence in rice due to magnesium deficiency mediated defect in transpiration rate before sugar accumulation and chlorosis. Physiol Plant 148: 490–501 [DOI] [PubMed] [Google Scholar]
- Kunst L. (1998) Preparation of physiologically active chloroplasts from Arabidopsis. Methods Mol Biol 82: 43–48 [DOI] [PubMed] [Google Scholar]
- Levine A, Tenhaken R, Dixon R, Lamb C (1994) H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 79: 583–593 [DOI] [PubMed] [Google Scholar]
- Lim PO, Nam HG (2005) The molecular and genetic control of leaf senescence and longevity in Arabidopsis. Curr Top Dev Biol 67: 49–83 [DOI] [PubMed] [Google Scholar]
- Liu J, Yun HW, Yang JJ, Yu DL, Shen FF (2008) Protein degradation and nitrogen remobilization during leaf senescence. J Plant Biol 51: 11–19 [Google Scholar]
- Lundqvist T, Schneider G (1991) Crystal structure of activated ribulose-1,5-bisphosphate carboxylase complexed with its substrate, ribulose-1,5-bisphosphate. J Biol Chem 266: 12604–12611 [PubMed] [Google Scholar]
- Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, et al. (2015) A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8: 1274–1284 [DOI] [PubMed] [Google Scholar]
- Maguire ME, Cowan JA (2002) Magnesium chemistry and biochemistry. Biometals 15: 203–210 [DOI] [PubMed] [Google Scholar]
- Marschner H. (2012) Marschner’s Mineral Nutrition of Higher Plants. Academic Press, New York [Google Scholar]
- Mátai A, Hideg É (2017) A comparison of colorimetric assays detecting hydrogen peroxide in leaf extracts. Anal Methods 9: 2357–2360 [Google Scholar]
- Matsuda K, Shimoda Y, Tanaka A, Ito H (2016) Chlorophyll a is a favorable substrate for Chlamydomonas Mg-dechelatase encoded by STAY-GREEN. Plant Physiol Biochem 109: 365–373 [DOI] [PubMed] [Google Scholar]
- Meguro M, Ito H, Takabayashi A, Tanaka R, Tanaka A (2011) Identification of the 7-hydroxymethyl chlorophyll a reductase of the chlorophyll cycle in Arabidopsis. Plant Cell 23: 3442–3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mubarakshina MM, Ivanov BN, Naydov IA, Hillier W, Badger MR, Krieger-Liszkay A (2010) Production and diffusion of chloroplastic H2O2 and its implication to signalling. J Exp Bot 61: 3577–3587 [DOI] [PubMed] [Google Scholar]
- Orozco-Cárdenas ML, Narváez-Vásquez J, Ryan CA (2001) Hydrogen peroxide acts as a second messenger for the induction of defense genes in tomato plants in response to wounding, systemin, and methyl jasmonate. Plant Cell 13: 179–191 [PMC free article] [PubMed] [Google Scholar]
- Park SY, Yu JW, Park JS, Li J, Yoo SC, Lee NY, Lee SK, Jeong SW, Seo HS, Koh HJ, et al. (2007) The senescence-induced stay-green protein regulates chlorophyll degradation. Plant Cell 19: 1649–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel TK, Williamson JD (2016) Mannitol in plants, fungi, and plant-fungal interactions. Trends Plant Sci 21: 486–497 [DOI] [PubMed] [Google Scholar]
- Peng WT, Zhang LD, Zhou Z, Fu C, Chen ZC, Liao H (2018) Magnesium promotes root nodulation through facilitation of carbohydrate allocation in soybean. Physiol Plant 163: 372–385 [DOI] [PubMed] [Google Scholar]
- Pierce J. (1986) Determinants of substrate specificity and the role of metal in the reactions of ribulosebisphosphate carboxylase/oxygenase. Plant Physiol 81: 943–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruzinská A, Tanner G, Anders I, Roca M, Hörtensteiner S (2003) Chlorophyll breakdown: Pheophorbide a oxygenase is a Rieske-type iron-sulfur protein, encoded by the accelerated cell death 1 gene. Proc Natl Acad Sci USA 100: 15259–15264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reczek CR, Chandel NS (2015) ROS-dependent signal transduction. Curr Opin Cell Biol 33: 8–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G, An K, Liao Y, Zhou X, Cao Y, Zhao H, Ge X, Kuai B (2007) Identification of a novel chloroplast protein AtNYE1 regulating chlorophyll degradation during leaf senescence in Arabidopsis. Plant Physiol 144: 1429–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissler HM, Collakova E, DellaPenna D, Whelan J, Pogson BJ (2002) Chlorophyll biosynthesis. Expression of a second chl I gene of magnesium chelatase in Arabidopsis supports only limited chlorophyll synthesis. Plant Physiol 128: 770–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodoni S, Schellenberg M, Matile P (1998) Chlorophyll breakdown in senescing barley leaves as correlated with phaeophorbidea oxygenase activity. J Plant Physiol 152: 139–144 [Google Scholar]
- Sakuraba Y, Schelbert S, Park SY, Han SH, Lee BD, Andrès CB, Kessler F, Hörtensteiner S, Paek NC (2012) STAY-GREEN and chlorophyll catabolic enzymes interact at light-harvesting complex II for chlorophyll detoxification during leaf senescence in Arabidopsis. Plant Cell 24: 507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Shimoda Y, Matsuda K, Tanaka A, Ito H (2018) Mg-dechelation of chlorophyll a by Stay-Green activates chlorophyll b degradation through expressing Non-Yellow Coloring 1 in Arabidopsis thaliana. J Plant Physiol 222: 94–102 [DOI] [PubMed] [Google Scholar]
- Sato Y, Morita R, Nishimura M, Yamaguchi H, Kusaba M (2007) Mendel’s green cotyledon gene encodes a positive regulator of the chlorophyll-degrading pathway. Proc Natl Acad Sci USA 104: 14169–14174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelbert S, Aubry S, Burla B, Agne B, Kessler F, Krupinska K, Hörtensteiner S (2009) Pheophytin pheophorbide hydrolase (pheophytinase) is involved in chlorophyll breakdown during leaf senescence in Arabidopsis. Plant Cell 21: 767–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheumann V, Ito H, Tanaka A, Schoch S, Rüdiger W (1996) Substrate specificity of chlorophyll(ide) b reductase in etioplasts of barley (Hordeum vulgare L.). Eur J Biochem 242: 163–170 [DOI] [PubMed] [Google Scholar]
- Shimoda Y, Ito H, Tanaka A (2016) Arabidopsis STAY-GREEN, Mendel’s green cotyledon gene, encodes magnesium-dechelatase. Plant Cell 28: 2147–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Nakayama N, Akazawa T (1968) Structure and function of chloroplast proteins. V. Homotropic effect of bicarbonate in RuDP carboxylase reaction and the mechanism of activation by magnesium ions. Arch Biochem Biophys 126: 737–745 [DOI] [PubMed] [Google Scholar]
- Veal EA, Day AM, Morgan BA (2007) Hydrogen peroxide sensing and signaling. Mol Cell 26: 1–14 [DOI] [PubMed] [Google Scholar]
- Verbruggen N, Hermans C (2013) Physiological and molecular responses to magnesium nutritional imbalance in plants. Plant Soil 368: 87–99 [Google Scholar]
- Wagner D, Przybyla D, Op den Camp R, Kim C, Landgraf F, Lee KP, Würsch M, Laloi C, Nater M, Hideg E, et al. (2004) The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science 306: 1183–1185 [DOI] [PubMed] [Google Scholar]