THE CURRENT CLINICAL STATUS OF NEUROENDOCRINE TUMOR DISEASE
Neuroendocrine neoplasms (NENs), also called neuroendocrine tumors (NETs), and generically referred to as “carcinoids,” represent a spectrum of tumors with a diverse range of molecular abnormalities that share a common neuroendocrine cell origin (Table 1).1–9 Anatomically, lesions arise from the diffuse neuroendocrine system of the lungs, gastrointestinal tract, and pancreas as well as discrete organs sites, such as the thymus, pituitary, and adrenal. Functionally, they produce a wide variety of biologically active amines and peptides. As might be predicted, given the diverse cell and tumor types involved, their 5-year survival rates diverge as widely (15%–95%) as their clinical presentations. Overall, this reflects the biological heterogeneity (diverse cell types, disparate molecular regulatory mechanisms, and ill-understood oncogenic drivers) of the tumors and, in reality, suggests that these tumors often bear little relation to each other than their putative common cell of origin.10,11
Table 1.
Biological and clinical utility of neuroendocrine tumor biomarkers
| Detection Indices | Monoanalyte | Circulating Tumor Cells | MicroRNA | mRNA |
|---|---|---|---|---|
| Pathobiology | ||||
| Mutations | No | No | No | Yes |
| Proliferation | No | No | No | Yes |
| Secretion | Yes | No | No | Yes |
| Metabolism | No | No | No | Yes |
| Epigenetic remodeling | No | No | No | Yes |
| Apoptosis | No | No | No | Yes |
| Signaling pathway activity | No | No | No | Yes |
| Cell of origin | Yes | Noc | No | Yes |
| Clinical utility | ||||
| Diagnosis | Yes | No | No | Yes |
| NET disease identification | Yes | No | No | Yes |
| Somatostatin receptor expression quantification | No | No | No | Yes |
| Prediction of therapy efficacy | No | Minimal data | No | Yes |
| Measurement of treatment response | Noa | Minimal data | No | Yes |
| Identification of a residual disease | Nob | Minimal data | No | Yes |
Only symptomatic therapy.
Only in specific cases, for example, gastrinoma/insulinoma.
Detection technique identifies EPCAM (Epithelial Cell Adhesion Molecule).
Their management reflects varied approaches often based on local practical experience, eminence-based medicine, or the availability of certain therapies or drug studies. Despite the repetitive development of classification systems and wearisome guidelines (eg, World Health Organization12 and European Neuroendocrine Tumor Society),13 there are few evidence-based standardized approaches, particularly for indolent disease or for appropriate sequencing of therapy. Most studies are retrospective, are underpowered, and exhibit significant design flaws. Apart from early identified (usually serendipitous) appendiceal, rectal, or gastric NETs, cure is uncommon and the overwhelming majority of management approaches reflect diverse combinations of strategies in an attempt to delay local or metastatic disease progression and subdue clinical symptomatology.14 In those with indolent tumor behavior or evidence of stable disease, a watch-and-wait-strategy is considered appropriate by some physicians.15 Current therapeutic strategies include somatostatin receptor agonists and antagonists, targeted agents (mammalian target of rapamycin inhibitors and vascular endothelial growth factor antagonists), immunotherapy (interferon), cytotoxic chemo-therapy, peptide receptor radionuclide therapy (PRRT), external radiation, and interventional radiological or probe-directed ablation.16 Management choice is often based on local experience, current ongoing pharmaceutical trials, and the composition of the multidisciplinary tumor board rather than a delineation of the molecular biology of the tumor. Relatively simplistic grading and staging classifications tend to drive most decision making in the absence of state-of-the-art assessment of the genomic basis of the individual tumor and the application of system biology techniques to advancing knowledge of the disease.17
LIMITATIONS IN THE DELINEATION OF DISEASE STATUS
The continual assessment by imaging, biomarker levels, symptomatology, and evaluation of progression-free survival (PFS) represents the fundamental basis on which NEN management strategies are based. Typically, disease recurrence, progression, or deficits in therapeutic efficacy are defined using an amalgamation of anatomic/morphologic and functional imaging interpolated with alterations in symptomatology and perturbations in biomarkers. Anatomic imaging using the Response Evaluation Criteria in Solid Tumors (RECIST) has well-documented limitations that include suboptimal reproducibility, insensitivity in the interpretation of disease responsiveness to targeted therapies, and relatively low discriminant indices in the identification of metastatic disease.18–20 Functional imaging with somatostatin receptor–based strategies, for example, 68Ga–somatostatin analog (SSA) PET/CT, has considerable value21 but limited spatial resolution (several millimeters for PET scanners), and partial volume effects constrain the ability to delineate small lesions. Although the development of new lesions is probably the most powerful indicator of disease progression, the monitoring of therapeutic efficacy and the early detection of residual or progressive disease using imaging remain challenging and are suboptimal.22–24 Current biomarkers in use are secretory monoanalytes (gastrin and insulin), protein cosecretory products (chromogranin A [CgA]), and urinary degradatory amines (5-hydroxyindoleacetic acid [5-HIAA]) which, in general, have limited predictive or prognostic value.25
LIMITATIONS OF CURRENT BIOMARKERS
Biomarkers are tools that diagnose a disease and monitor or predict the outcome of treatment of disease. They are cellular, biochemical, or molecular alterations measurable in biological media, such as tissues, cells, or fluids.26 NENs secrete bioactive products, including amines and peptides, into the circulation, which are detectable and quantifiable. These include analytes specific to an individual cell type, for example, gastrin (gastrinomas), as well as cosecreted products common to all NENs, for example, CgA or neuron-specific enolase.
CgA is a constitutive product of the neuroendocrine cell secretory granule and is measurable in serum or plasma. It may correlate with tumor mass and seems to function as a prognostic agent.27,28 Small tumors, however, may be hypersecretory whereas large tumors can exhibit low secretion. Specific receptor targeting agents, for example, SSAs, decrease CgA secretion through inhibition of synthesis and the secretory machinery. The sensitivity of CgA ranges from 60% to 90% with a specificity less than 50% (depending on the population studied).29 CgA does not correlate with imaging, in particular 68Ga-SSA and fludeoxyglucose F 18 imaging, and its utility with CT or MRI remains to be determined. Biochemical responses to therapy, as measured by changes in circulating CgA levels also generally are nonconcordant with image-based assessments.25 Poor laboratory metrics, nonspecificity, and diagnostic inaccuracy further contribute to the low enthusiasm for its clinical utility.25
Such limitations emphasize the need for alternative tools, such as microRNA (miRNA) or circulating tumor cells (CTCs), or informative molecular tools, such as multianalyte biomarkers that delineate significant biological characteristics of the disease state. Currently, the measurement of miRNA remains complex and is not adequately standardized for clinical usage.30 CTCs, although intuitively attractive as a direct measurement of tumor cell-related events, have to date failed to provide evidence of broad clinical utility.25 Dynamic characterization of tumor behavior based on blood-derived genomic information can only be derived from assessment of circulating real-time multianalyte genetic information (NETest, Clifton Life Sciences, Nevis) (see Table 1). Blood-based transcriptome analysis and interrogation of the specific genomic drivers of a tumor provide a liquid biopsy, which is an optimal platform to assess tumor status on a real-time basis.31
RATIONALE FOR MOLECULAR BIOMARKERS
The complexity and diversity in cancer biological behavior and responses to therapy cannot be adequately defined through measurements of secretory products.32,33 Measurements of exocytotic and secreted proteins do not adequately capture the biological activity of an active tumor cell, which includes proliferation, metabolic activity, growth factor signaling, and so forth.33 Clinical scientists in diverse oncological disciplines have, therefore, concluded that a dynamic and panoramic delineation of the molecular biological topography of an evolving neoplasm, that is, the hallmarks of cancer, is best assessed through a multidimensional appraisal of the molecular genomic machinery of the tumor cell. This includes the measurement of mRNA and DNA as well as the delineation of mutational status, and the application of systems biology to the identification of master regulators and oncogenic checkpoints.10 There is, thus, a focus on the application of molecular technologies to better define a cancer cell state focusing both on detection of mutations (typically in circulating tumor DNA [ctDNA] or transcriptional profiles, including mRNA and signal pathway analyses).34,35
Examples of the utility of tumor tissue–based mRNA approaches include MammaPrint (Agendia, Irvine, California), which is a 70-gene assay for predicting the risk of recurrence in an early stage breast cancer while considering adjuvant treatment.36 Other genomic tests to stratify the risk of cancer recurrence while considering adjuvant treatment are for example, Oncotype DX (Genomic Health, Redwood City, California), used in breast, colon, prostate cancer, or MammoStrat (Clarient Diagnostic Services, Aliso Viejo, California), used in early-stage, hormone receptor–positive breast cancer. The clinical utility of these approaches is highlighted by the recent endorsement of MammaPrint by the American Society for Clinical Oncology, to guide treatment decisions on adjuvant systemic therapy in women with early-stage invasive breast cancer.37 Although this information underscores the importance and usefulness of gene expression tests, such technology is currently limited to tissue-based testing and would require repetitive biopsy to provide real-time clinical information.
LIQUID BIOPSY STRATEGIES IN OTHER NEOPLASIA
The evolution of strategies to evaluate circulating molecular information from neoplasia, however, has advanced to the point that blood sampling can provide considerable oncological information.34,35 There is significant interest in the identification and application of such strategies, or liquid biopsies, in the field of oncology (Fig. 1).38,39 In this respect, a circulating neoplastic molecular signature, that is, a circulating Mammaprint-like signature, would have clinical utility to limit invasive biopsies, define therapeutic targets, and provide a real-time monitoring tool to evaluate disease status.40
Fig. 1.
Numbers of publications (PubMed) or Web focus (Google Trending) relating to “liquid biopsy”. Significant public interest was initially noted in 2004 and has escalated since 2012. Academic interest initially lagged but has subsequently escalated from 2012. There has been an exponential explosion in interest in both domains though the medical scientific community appear to be less receptive than the public.
Given the invasive nature and the technical limitations of tissue biopsy, there is added enthusiasm for the development of surrogate markers that can be quantified in blood on a real-time basis.31 Research has focused on measurement of circulating genetic information, for example, ctDNA, RNA, or tumor cells, or the identification of actionable mutation events, for example, BRAF mutations in ctDNA. As such, the use of liquid biopsy allows for patient stratification (eg, as a companion diagnostic) for screening and for monitoring treatment responses. Examples include identification of mutation T790M in ctDNA, which allows for monitoring of treatment responses to EFGR inhibitors in lung cancer.41,42 Measurements of ctDNA levels have been used for the detection of minimal residual disease after surgery/recurrence for example, in colon cancer.43
Unlike a majority of cancers, however, activating mutations are infrequent if not largely unknown in NENs,10 and most tumors exhibit somatic mutations (when identified) in tumor suppressor genes, for example, MEN-1, the predominant pancreatic mutation.44 The clinical usefulness of other alterations for example, in ATRX, DAXX,45 or YY146 (all identified as sporadic mutations in pancreatic NEN) remains to be proved. In addition, the clinical usefulness of copy number and chromosomal imbalances as well as chemical-based DNA modifications, for example, methylation, requires elucidation.10 To date, the clinical utility of the measurement of molecular signals, such as ctDNA, methylated gene targets, or CTCs have, therefore, been limited in NENs.44
The focus in NENs has thus switched to mRNA-based liquid biopsy approaches, which have been demonstrated to have utility in other diseases. For example, FibroSure/FibroTest (BioPredictive S.A.S., Paris, France) is a blood-based, biochemical, algorithmic test for liver damage used to detect hepatitis C.47 The FibroSure is a repeatable, noninvasive test that is considered to have high accuracy with the added value of reducing the discomfort or complications related to a liver biopsy.
SCIENTIFIC BASIS OF THE NETest
Transcriptome-based evaluations have proved useful in identifying and differentiating the different subtypes of NEN (based on origin [eg, pancreatic vs small intestinal] and aggressiveness [eg, nonprogressive vs malignant/metastatic]).48,49 These mRNA-based evaluations also have demonstrable predictive utility at a tissue level.50
Transcriptional profiling of tumor tissue has identified a series of neuroendocrine transcripts that are detectable in the circulation51 and can be used clinically to evaluate gastrointestinal, pancreatic, and bronchopulmonary (BP) NENs.52–59 This strategy has also been used to define tumors originating in the nervous system, including paragangliomas and adrenal glands, that is, pheochromocytomas.60 This blood-based multianalyte transcript analysis is the most extensively investigated liquid biopsy tool for this class of tumor.
Individual genes were selected by analyzing microarray data sets of cellular profiles from fresh frozen tumors as well as from whole blood (3 microarray data sets: tumor tissue [n = 15], peripheral blood [n = 7], and adenocarcinoma [n = 363 tumors]) to identify similarities in expression patterns (Fig. 2).51 Once defined, coexpression network inference against normal tissue gene expression was undertaken to eliminate genes that were unlikely to be neoplasia relevant.51 Tumor-associated genes from other tumor types for example, breast, colon, and so forth, were similarly excluded; 51 candidate marker genes, detectable in the peripheral circulation, were identified to encompass the hallmarks of a NEN. The candidate gene signature was then examined in a training set of 130 blood samples (NEN: n = 63) and validated in 2 independent sets (set 1 [NENs: n = 72] and set 2 [NENs: n = 58]). Correlation analysis of matched blood/tumor identified this as highly significant (R2 = 0.62–0.91; P<.0001), indicating the blood-based measurements were directly attributable to a tumor gene expression signature.
Fig. 2.
Computational pipeline used to derive a set of marker genes, the NET Marker Panel that identifies GEP-NEN/NET disease in the blood. Step 1: gene coexpression networks inferred from 2 independent data sets (GEP-NEN-A and GEP-NEN-B) are intersected to produce the GEP-NEN network. Step 2: coexpression networks from neoplastic and normal tissue microarray data sets are combined to produce the normal and neoplastic networks. Step 3: links present in normal and neoplastic networks are subtracted from the GEP-NEN network. Step 4: up-regulated genes in both the GEP-NEN-A and GEP-NEN-B data sets (n = 21) are mapped to the consensus GEP-NEN network. Step 5: identification of consistently up-regulated genes in GEP-NEN blood transcriptome and GEP-NEN-A and GEP-NENB data sets, provided 32 putative genes. Step 6: literature curation and cancer mutation database search yielded an additional panel of 22 putative marker genes. A total of 75 marker genes was analyzed prior to delineation of the final NET marker panel. Step 7: the final NETest liquid biopsy includes 51 marker genes that were validated in 3 independent cohorts totaling 193 NETs and 172 controls. RT-PCR, reverse transcription PCR. (Modified from Modlin I, Drozdov I, Kidd M. The identification of gut neuroendocrine tumor disease by multiple synchronous transcript analysis in blood. PLoS One 2013;8:e63364; with permission.)
The signature—the NETest—can identify all types NEN, including small nonmetastatic tumors. The sensitivity is such that image-negative lesions in the liver can be identified and subsequently confirmed by histologic demonstration of microscopic tumor deposits. Comparison assessment with other NET biomarkers identifies it to significantly out-perform single analyte-based assays for detection.51,57 In addition, levels correlate with clinical status, for example, stable or progressive disease.61 Mathematical analyses have demonstrated that this technique is superior to single analyte assays in the diagnosis of NEN.11 For example, the area under the curve (AUC) for the NEN gene-based classifier was 0.95 to 0.98 compared with 0.64 for CgA (Z-statistic 6.97–11.42; P<.0001).51 In comparisons with other commonly used biomarkers, like pancreastatin and neurokinin A (AUC: 0.58–0.63), the NETest AUC was 0.98 (area differences: 0.284–0.403, Z statistic 4.85–5.9; P<.0001).57 The utility of the NETest was mathematically confirmed by the use of predictive feature analysis, which established that measuring multiple genes in the circulation exhibited the highest value (69%) for NEN diagnosis compared with measurement of single secreted products (CgA: 13%; pancreastatin: 9%; and neurokinin A: 9%).57
MATHEMATICAL BASIS OF THE NETest
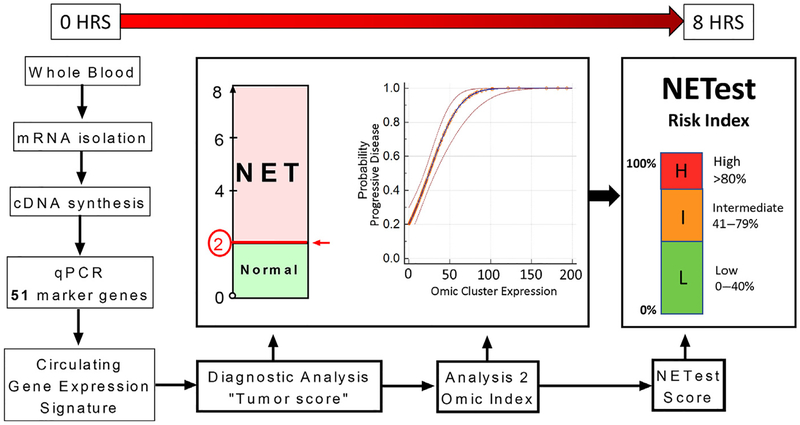
The test uses a 2-step protocol (mRNA isolation, cDNA production, and polymerase chain reaction [PCR])51,62 from EDTA-collected whole blood (Fig. 3).51,62 Blood gene expression of the 51 markers is normalized to housekeepers and quantified versus a population control.51 Gene expression levels are related to an outcome using supervised machine learning algorithms, including support vector machines and linear discriminant analyses, both of which have been applied extensively in clinical medicine.63–68 These algorithms use gene expression levels to learn whether a sample should be categorized as either, for example, tumor or control. Algorithm performance is subsequently validated using an independent test set.
Fig. 3.
The multistep protocol used to provide a multianalyte gene expression panel for GEP-NETs. A 2-step protocol (mRNA isolation and cDNA synthesis) is undertaken prior to quantitative PCR gene expression. mRNA levels are normalized using house-keeping gene expression. The normalized 51-marker signature is then interrogated using 2 separate mathematical algorithmic analyses. This provides two readouts. The first generates a score that identifies whether the sample is a NET or non-NET (score 0–8). Samples scored 0 to 2 are classified as normals and levels of 3 to 8 are categorized as NETs. The second analysis evaluates expression of defined clusters of genes involved in the biologically relevant NET pathways. Omic values greater than or equal to 50 have a greater than 75% probability of identifying progressive disease. These 2 information sets are condensed to a single score, which is scaled 0% to 100% (the NETest score). Scaling is undertaken based on weighting the classification score (analysis 1), with the biological gene expression linked to disease status (analysis 2). The NETest delineates in a specific patient whether the tumor falls into a category of low risk (<40%), moderate risk (40%–79%), or high risk (≥80%) for disease activity. HRS, hours; qPCR, quantitative PCR.
The NETest uses 4 different mathematical tools: support vector machine, linear discriminant analysis, k-nearest neighbors, and the naïve Bayes algorithm. These were taught (or trained) using a training set of 130 blood samples (NENs: n = 63 and controls: n = 67) and validated (or tested) in 2 independent sets (set 1 [NENs: n = 72, controls: n = 43] and set 2 [NENs: n = 58; controls: n = 62]). All algorithms were designed to differentiate controls from tumors and stable disease from progressive disease. First, each algorithm labels an unknown sample as either 0 or 1, which corresponds to a prediction of control or tumor, respectively. Then, prediction is applied to tumor samples, labeling them as either 0 (stable) or 1 (progressive). This categorization results in a 0 to 8 score.51,62 Scores greater than 2 are considered tumor. In the test sets, the AUC values were 0.98 and 0.95, respectively. The test exhibited a high sensitivity (85%–98%), specificity (93%–97%), positive predictive value (95%–96%), and negative predictive value (87%–98%).51,62 These data confirm that learning algorithms can successfully classify (and, therefore, diagnose) NETs in blood.
To expand the utility of the test from a pure diagnostic to a tool that could capture the biology of neuroendocrine neoplasia, the authors subsequently undertook regulatory network analysis. Briefly, this approach involves mapping NET-specific genes to a database of human protein-protein interactions, thus visualizing marker genes in the context of their respective biological function. This strategy identified 8 biologically relevant gene “omic” clusters (SSTRome, proliferome, signalome, metabolome, secretome, epigenome, plurome, and apoptome), which define the NEN fingerprint and constitute the oncobiome of the NET cell.31 Differential analysis of gene expression in 6 of the clusters (SSTRome, proliferome, metabolome, secretome, epigenome, and plurome) can then be mathematically analyzed to deduce stable from progressive disease.31 These constitute a molecular representation of the biological signature of an individual tumor. Overall, this strategy captures the biology of a specific NEN and defines the molecular status of an individual tumor.31
To facilitate the clinical interpretation of this information, the diagnostic score is represented as a clinical activity score ranging from 0% (low activity) to 100% (high activity).31 Thus, a high score, for example, 8, with elevated expression of genes in omic clusters is scaled to 100% (high activity). In contrast, the same score (8) in which a low expression of omic gene clusters is identified is weighted to 53% (see Fig. 3). In both examples, the samples are tumor (a score of 8 is equivalent to all 4 algorithms [discussed previously], classifying the sample as a tumor). The difference between the 2 samples reflects differential tumor biology, as captured by omic gene clusters. For example, a score of 100% identifies a more aggressive tumor phenotype than a score of 53%. Using Kaplan-Meier analyses (n = 63, time period 60 months), the authors correlated clinical determinants with gene expression levels. These are low biological activity, less than or equal to 40%; intermediate biological activity, 41% to 79%; and high (biologically aggressive) activity, 80% to 100%.31,55,56 A similar spectrum of ranges has been identified in BP tumors.59
LABORATORY METRICS OF THE NETest
The multianalyte algorithmic analysis procedure has been validated62 and is undertaken in a Clinical Laboratory Improvement Amendments–certified clinical laboratory (State of Connecticut: 07D2081388). The interassay variability is 2.14% ± 1.14% and the intra-assay variability is 1.02% ± 0.74%.62 In clinical studies, the intra-assay reproducibility ranges 0.4% to 1% for individual gene expression. Assessment of PCR cycle times, normalized gene expression, and scoring demonstrates high correlation levels (Spearman >0.90). Consecutive daily analysis of NEN samples had a Spearman correlation for scored expression of 0.96 (P<.001; coefficient of variation <5%). The summated NETest data assessed in approximately 5500 patient samples from approximately 100 different institutions and approximately 150 physicians indicate that the day-to-day variability for the test is extremely low (<2%) and the test is highly reproducible (sample concordance >95%).
Measurement of the NETest signature in blood is robust and not affected by food intake.62 Assessment of the gene expression measurements using unsupervised hierarchical clustering failed to identify intrinsic relationships between feeding and gene expression, and the NETest was not altered over a 4-hour period after a test meal.62 No relationship between age, gender, ethnicity, and proton pump inhibitor (PPI) usage was identifiable.62 The latter is a particular issue in the measurement of CgA because PPIs significantly increase CgA levels.69 NETest scores are not affected by long-term PPI treatment (>1 year). Overall, the test has been demonstrated to exhibit a reliably high level of sensitivity and specificity (both >95%),51 to be standardized and reproducible (interassay and intra-assay coefficient of variation <2%)62 and not to be affected by age, gender, ethnicity, fasting, or PPI medication.52,62
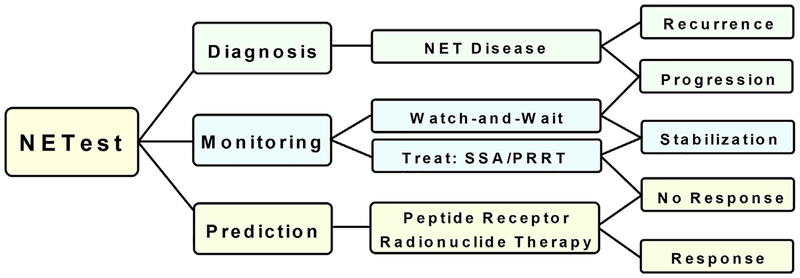
CLINICAL APPLICATIONS OF THE NETest
Diagnosis
Small bowel neuroendocrine neoplasm
The NETest accurately identifies small bowel NENs and differentiates these from other small and large bowel cancers (Fig. 4). In 1 prospective study, the accuracy for detecting small bowel NEN was 93% (all NETs positive and 3 [12%] colorectal tumors were positive).54 CgA was positive in 80%, but 29% (n = 7) of colorectal cancers also exhibited elevated circulating CgA levels. Gene expression scores were elevated (P<.05) in subjects with metastatic disease and were more accurate (76%–80%) than CgA levels (20%–32%) for detecting NEN disease.54 Overall, as a diagnostic test, the NETest was significantly more sensitive than CgA for small bowel NEN.
Fig. 4.
Clinical utility of a multianalyte assay (NETest) for NET diagnosis and management. The NETest identifies disease status, detects disease progression, is prognostic, and can be used to predict PRRT efficacy. Diagnosis: the NETest can detect BP NET, pancreatic, and gastrointestinal tract NET with greater than or equal to 95% accuracy. In addition, it is effective in the diagnosis of PPGLs (≥95%). Management: NETest has clinical utility in 3 areas: (1) evaluate the effectiveness of a surgical procedure; this allows for a prediction/identification of disease recurrence; (2) evaluate treatment response to SSA or PRRT; and (3) predict treatment failure/disease progression; response to PRRT can be predicted using the NETest and subsequent measurement of transcript levels over time monitor treatment response.
Pancreatic neuroendocrine neoplasm
The NETest is also useful for accurately confirming neuroendocrine disease (compared with other cancers and non-neoplastic diseases [eg, chronic pancreatitis]) in blood samples from pancreatic disease patients.54 In 1 study, the accuracy was 94%54; 6% (2/31) of intraductal papillary mucinous neoplasms were positive, consistent with the reported coexistence of NEN and these lesions.70 Only 29% of pancreatic NETs were CgA positive in blood; the overall accuracy of CgA was 56%54 (Fig. 5).
Fig. 5.
Comparison of the accuracy of circulating NET transcript measurement (NETest) to CgA. The MAAA (multianalyte algorithm analysis) (NETest) is positive in 96% to 100% of bronchopulmonary, pancreatic, and small bowel NET. CgA in contrast is significantly less accurate. It is positive (elevated) in only approximately 30% to 60%. In pNETs, CgA is elevated in only 30% of tumors. Overall, CgA levels are normal in 40% to 70%, significantly limiting its clinical utility as a biomarker.
Bronchopulmonary neuroendocrine neoplasm
Detectable mRNA was identified in blood from lung tumors with a neuroendocrine phenotype in greater than 90%.59 A receiver operating characteristic (ROC) analysis AUC was 0.99 for differentiating lung carcinoids (typical or atypical) versus controls. The sensitivity and specificity ranged from 93% to 95% and from 82% to 93%, respectively.58 In individuals with RECIST-defined progressive disease, NETest levels were significantly increased (72% ± 23%) irrespective of histology compared with stable disease (33% ± 17%) or those considered surgical cures (10% ± 5%). Levels were greater in metastatic disease (63% ± 26%) in comparison to localized disease (45% ± 21%). As a comparator, blood CgA was elevated in less than 50%. Decision curve analysis demonstrated a greater than 75% standardized clinical net benefit up to a risk threshold of 90% for gene expression analysis compared with CgA. Thus, the use of CgA as a biomarker exhibited a net clinical benefit in less than 30% of patients.
Paragangliomas and pheochromocytomas
The neural-derived lesions, paragangliomas and pheochromocytomas (PPGLs), are NETest-positive in 100% of cases.60 An ROC analysis AUC was 0.98 for differentiating PPGLs versus controls. Although mutation status was not directly linked to blood gene expression levels, metastatic (80% ± 9%) and multicentric (64% ± 9%) disease had significantly (P<.04) higher scores than localized disease (43% ±7%). Progressive disease had the highest scores (86% ± 2% vs stable 41% ± 2%; P<.0001).60 Overall, the NETest was highly sensitive (>95%) as a diagnostic test for PPGLs.
Assessment of the Effectiveness of Surgery for Gastroenteropancreatic and Bronchopulmonary Neuroendocrine Neoplasia
Gastroenteropancreatic neuroendocrine neoplasia
In a prospective GEP-NEN study,53 the score was elevated in all 35 patients (100%) preoperatively. In comparison, only 14 (40%) had elevated CgA. Resection reduced the NETest from 80% ± 5% to 29% ± 5 (P<.0001). NETest decreases correlated with diminished tumor volume (R2 = 0.29; P = .03). CgA decrease was insignificant and did not correlate with the extent of tumor resection. The assessment of R0 resections was of particular interest in that 4 (36%) of 11 resections reported as complete had an elevated NETest at 1 month. All 4 subsequently developed positive tumor imaging within 6 months of surgery.
Bronchopulmonary neuroendocrine neoplasm
A prospective BP-NEN study was undertaken in 21 patients.58 At 6 months after surgery 9 (43%) had evidence of disease (residual/recurrence) and 12 (57%) were disease-free. In the recurrent group, levels were unchanged from before (71% ± 11%) to after surgery (66% ± 8%; P = not significant). In the disease-free group, presurgery gene expression levels (70% ± 7%) were significantly reduced by surgery to 23% ± 3% (P = .0005).
These results demonstrate that blood NET transcripts delineate surgical resection/cytoreduction and facilitate early identification of residual disease in GEP-NEN and BP-NEN.
Monitoring Therapeutic Efficacy
Somatostatin analog
Efficacy was evaluated in a prospective, blinded study.56 The utility of the NETest was evaluated compared with CgA for the ability to predict treatment failure. In 28 patients receiving SSAs (octreotide [n = 14] and lanreotide [n = 14]), univariate analysis identified that the NETest (P = .002) and tumor grade (P = .054) were associated with therapy responses. Multiple regression analysis identified that only the NETest predicted disease progression during SSA usage (P = .0002). NETest changes occurred significantly earlier than image changes (approximately 5 months prior to image-defined progression). It also was 100% effective in identifying patients who progressed. Multiple regression analysis did not identify CgA as predictive of SSA therapy. This study56 identified that the NETest exhibited utility in predicting SSA treatment response.
Because biomarker assays that exhibit utility in clinical academic trials do not always effectively translate to real world settings,71,72 a US registry study () was undertaken to confirm the clinical utility of the assay in a prospective observational investigation. A study of 51 SSA-treated patients in the United States identified that all patients (n = 37) with a low score (NETest ≤40%) were able to continue without any modification in therapy (type or dose). In contrast, all those with a high score (NETest ≥80%) (n = 24) either underwent a treatment modification (86%) or remained on the current treatment regimen. All (n = 24) exhibited disease progression consistent with failure to respond to SSA. In 21 of these patients, appropriate treatment modifications (dose increases, changing type of SSA, or introduction of selective internal radiation therapy or PRRT) were undertaken. All (n = 21) exhibited disease stabilization at follow-up (6 months: image-based confirmation). The median PFS (mPFS) was not reached for those with low scores. A high score was associated with a mPFS of 5 months (χ2 = 27.7; hazard ratio [HR] 60.2 (18–201); P<.0001) (Fig. 6).
Fig. 6.
Relationship between NETest score and PFS in a prospective observational registry cohort. (A) Watch-and-wait cohort: a low NETest score was associated with mPFS of 12 months, and a high score was associated with an mPFS of 3 months. This difference was significant (HR 30.4; P<.0001). (B) Treatment cohort: a low score was associated with an mPFS that was not reached at 12-months, and a high score was associated with an mPFS of 5 months; this difference was significant (HR 60.2; P<.0001).
Peptide radioreceptor therapy
Although previously regarded as an experimental or Hail Mary therapeutic strategy, PRRT is an effective and well-established NET therapy.73 In principle, selection is based on image-based assessment of somatostatin receptor uptake, although not all patients respond.74 To better predict efficacy, a combination of tumor grade and a specific omic analysis variant of the NETest gene signature (encompassing growth factor signaling and metabolomic gene expression) was developed as a predictive quotient.75 This was evaluated in 3 prospective studies (n = 158) prior to PRRT therapy.76 This predictor signature has 2 outputs—positive (predicts response to therapy) and negative (nonresponders). Mathematical assessment using decision curve analysis exhibited greater than 90% standardized predictive benefit up to a risk threshold of 80% for the predictor biomarker analysis. The benefit of a CgA value or grade stratification was equivalent to not using a biomarker (<10% across comparable risk thresholds). Overall, the biomarker was 94% to 97% accurate for predicting a tumor response to PRRT. The mPFS was never reached in those predicted to respond to PRRT (up to 31 months after treatment initiation). For nonresponders, mPFS ranged from 8 months to 14 months. The overall HR for the PRRT predictive biomarker was 47.
As a monitor, measurement of all 51 marker genes identified that the NETest correlated accurately (94%) with PRRT responders (97%) versus nonresponders (91%). Moreover, during therapy, changes in gene expression scores accurately (89%, P<10−6) correlated with treatment response assessment (RECIST). In contrast, changes in CgA were only 24% accurate. Overall, the NETest and the predictor analysis (PRRT predictive quotient) identified that PRRT efficacy could be accurately predicted and monitored in greater than 90% of individuals.75
Assessment of Long-term Management
Retrospective cohort analysis
A long-term (5-year) study in 34 patients identified that the NETest has predictive and prognostic utility for GEP-NETs. Blood measurement of transcript levels identified clinically actionable alterations approximately 1 year before image-based evidence of progression.55 Cox modeling identified that the only factor associated with PFS was the NETest. A baseline NETest greater than 80% was significantly associated with disease progression (mPFS: 0.68 years vs 2.78 years, with <40% levels). In contrast, baseline NETest levels greater than 40% in those defined as clinically stable were 100% prognostic of disease progression. Baseline NETest values less than 40% accurately (100%) predicted stability over 5 years. A χ2 analysis that compared alterations in NETest values to CgA levels demonstrated the NETest 96% more informative than CgA (P<.001) in predicting disease status alteration.55
Prospective observational study
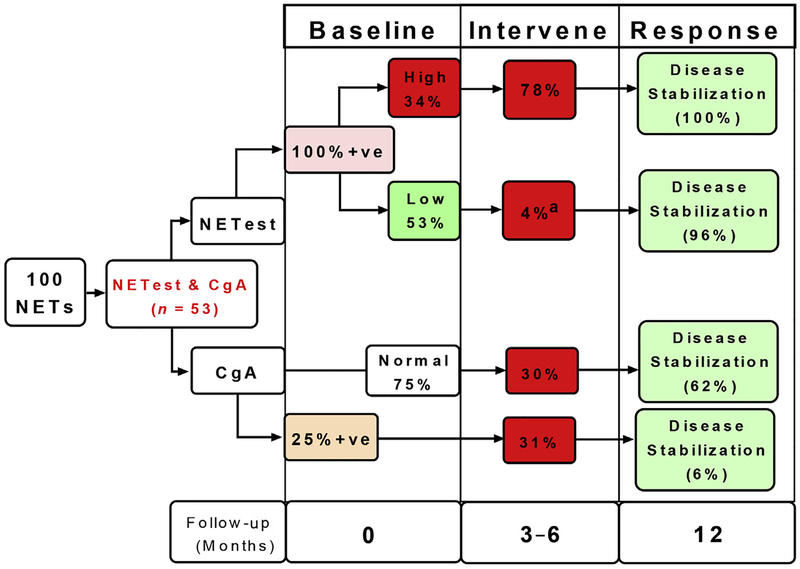
In a registry study () (n = 100), a low NETest score was associated with conservative management and maintenance in a watch-and-wait program (n = 28). At follow-up (12 months), all remained stable. All patients (n = 12) with a high score (NETest ≥80%) required treatment interventions and at follow-up (12 months) had disease stabilization (imaging or symptom diminution). A low score was associated with mPFS of 12 months. A high score was associated with an mPFS of 3 months. This difference was significant (χ2 = 27.7; HR 30.4 [95% CI, 8.5–108]; P<.0001). Comparison of the NETest with CgA using the McNemar test (evaluates 2 biomarkers in paired sample sets) demonstrated CgA of no clinical value in decision making. Overall, a low NETest score reduced imaging approximately 40% (Fig. 7).
Fig. 7.
Comparative clinical utility for CgA and NETest; 100 patients were studied, of whom 53 had both a NETest and CgA. NETest was positive in all 53 samples. CgA levels were elevated in 13 (25%) and were normal in 40 (75%). High NETest scores were noted in 18 (34%) of the 53 patients. Alterations in clinical management (intervene) were made in 78%. All demonstrated disease stabilization at subsequent follow-up (12 months). Low scores were associated with a management change in 1 patient (4%). This patient progressed on everolimus. All other patients (96%) exhibited disease stabilization. CgA was associated with alterations in clinical management in approximately 30% of patients, irrespective of whether the CgA level was elevated. Disease stabilization ranged from 6% to 62% based on intervention and score. CgA levels, therefore, are unable to effectively guide disease management. a P<.0001 versus high score. F/Up, Follow-up; Mo, months; 1ve, positive.
THE FUTURE
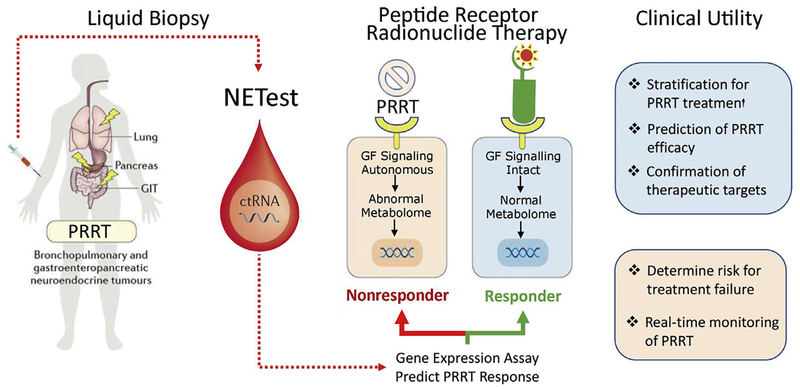
An optimal biomarker needs to have 3 capabilities, namely diagnostic, predictive, and prognostic. Thus, a disease can be identified early, the effect of therapy predicted, and the status of disease monitored. A liquid biopsy fulfills the criteria for real-time disease management and avoids the negative invasive implications and single time-point limitations of tissue biopsy. In addition, it provides information that is of adjunctive value to imaging but is easier to repeat and does not have any of the radiation-related concerns. More recently it has become apparent that a critical requirement is the development of a liquid biopsy that can better identify an appropriate drug target in a particular NEN and, thereafter, define treatment response. This is critical because many therapeutic strategies have limited efficacy, exhibit significant adverse events, and are expensive. In this respect, the PRRT predictive signature demonstrates the utility of circulating RNA as a biomarker (Fig. 8). Both the growth factor and the metabolomic genes captured by the signature are specifically related to oxidative stress, metabolism, and hypoxic signaling.77–79 It is likely that elevated expression of these genes in blood identifies tumors that are more radiosensitive given the role of hypoxia, oxidative stress, and loss of DNA repair associated with radiation responsivity.80 The specificity of PRRT efficacy prediction, therefore, reflects the identification of molecular mechanisms related to radiation response–associated genes, which modulate tumor response to PRRT.81
Fig. 8.
Cartoon of tumor cell response to 177Lu-octreotate therapy. Tumors (blue) responsive to PRRT exhibit a circulating gene fingerprint that has intact, regulated growth factor signaling pathways and normal metabolic pathways. These tumors are predicted to undergo significant DNA damage and tumor apoptosis leading to regression or disease stabilization. Tumors (orange) that are autonomous of growth factor modulation and exhibit abnormal metabolome (highly metabolically active) have variable responses to PRRT. Clinical progression is identified after PRRT in the majority (85%–100%) of tumors with predicted nonresponse gene signature. Evaluating blood NET gene expression prior to PRRT facilitates the precise identification of PRRT-responsive tumors. (Modified from Kidd M, Modlin IM. Therapy: the role of liquid biopsies to manage and predict PRRT for NETs. Nat Rev Gastroenterol Hepatol 2017;14:6:331–2; with permission.)
Given the recent demonstration of the efficacy of PRRT, a predictive strategy is of special relevance. Although PRRT is extremely well tolerated, there is evidence of modest toxicity to the kidney and bone marrow. These are largely unpredictable, because their pathogenesis is poorly understood and, in some cases, apparently idiosyncratic.16 Irrespective of the etiology, the need to predict or accurately assess the risk of such events is crucial. Blood-based gene expression measurements, therefore, provide an opportunity to identify transcripts relevant to either nephron or myelotoxicity. The development of such a test would provide a complement to the current PRRT predictive quotient and provide a molecular assessment of the risk-benefit ratio for a specific patient.
CODA
The NETest has been evaluated in more than 5500 NEN patients and identified to exhibit clinical utility in several different areas. These include the assessment of the effectiveness of curative surgery, assessment of the efficacy of SSA therapy, prediction of disease stability/progression, and identification of response to PRRT. The signature was decreased by surgery and values corresponded to the completeness of tumor removal.53,58 In addition, elevated levels afer R0 resection accurately predicted subsequent disease recurrence. In a separate study, elevated transcript levels were prognostic of SSA failure/disease progression.56 Alterations in transcript levels occurred significantly earlier than RECIST-based or somatostatin receptor imaging-based measures of disease progression.56 Finally, levels were prognostic for PRRT efficacy and could be used to evaluate therapeutic efficacy and correlated with image-based assessment.75
Current data identify the value of transcript analysis in the monitoring of a variety of therapeutic modalities, particularly in conjunction with other clinical and imaging parameters to monitor disease progression. The authors predict that future strategies for refining and improving the evaluation of therapy will be provided by incorporating imaging modalities and the blood-based molecular information provided by tumor transcriptome analysis.
KEY POINTS.
The NETest is a blood biomarker test for diagnosis and management of gastroentero-pancreatic and bronchopulmonary neuroendocrine neoplasia.
The test measures 51 individual circulating genes in 1 mL of blood and algorithmic analysis provides a numeric score of disease status.
The sensitivity and specificity of the test are respectively >95% and >90%.
In head-to-head comparisons, the test is ~4-fold more precise than CgA and for monitoring disease progress, it is ~10-fold more accurate.
Clinically, the test can define the completeness of surgical resection, identify residual disease, monitor disease progression and determine efficacy of treatment.
PRRT efficacy can be accurately (~95%) predicted using a Predictor Quotient gene set and Ki67 (PPQ).
Neuroendocrine disease status (stable/progressive) can be assessed by regular monitoring of blood NETest levels.
Footnotes
Disclosure: The authors have nothing to disclose.
REFERENCES
- 1.de Mestier L, Dromain C, d’Assignies G, et al. Evaluating digestive neuroendocrine tumor progression and therapeutic responses in the era of targeted therapies: state of the art. Endocr Relat Cancer 2014;21(3):R105–20. Print 2014. [DOI] [PubMed] [Google Scholar]
- 2.Bergsland EK. The evolving landscape of neuroendocrine tumors. Semin Oncol 2013;40(1):4–22. [DOI] [PubMed] [Google Scholar]
- 3.Wang H, Chen Y, Fernandez-Del Castillo C, et al. Heterogeneity in signaling pathways of gastroenteropancreatic neuroendocrine tumors: a critical look at notch signaling pathway. Mod Pathol 2012;24(10):143. [DOI] [PubMed] [Google Scholar]
- 4.Sundin A, Rockall A. Therapeutic monitoring of gastroenteropancreatic neuroendocrine tumors: the challenges ahead. Neuroendocrinology 2012;96(4):261–71. [DOI] [PubMed] [Google Scholar]
- 5.Kidd M, Schimmack S, Lawrence B, et al. EGFR/TGFalpha and TGFbeta/CTGF signaling in neuroendocrine neoplasia: theoretical therapeutic targets. Neuroendocrinology 2012;15:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan JA, Kulke MH. New treatment options for patients with advanced neuroendocrine tumors. Curr Treat Options Oncol 2011;12(2):136–48. [DOI] [PubMed] [Google Scholar]
- 7.Oberg K Pancreatic endocrine tumors. Semin Oncol 2010;37(6):594–618. [DOI] [PubMed] [Google Scholar]
- 8.Garcia-Carbonero R, Capdevila J, Crespo-Herrero G, et al. Incidence, patterns of care and prognostic factors for outcome of gastroenteropancreatic neuroendocrine tumors (GEP-NETs): results from the National Cancer Registry of Spain (RGETNE). Ann Oncol 2010;21(9):1794–803. [DOI] [PubMed] [Google Scholar]
- 9.Strosberg J, Gardner N, Kvols L. Survival and prognostic factor analysis of 146 metastatic neuroendocrine tumors of the mid-gut. Neuroendocrinology 2009; 89(4):471–6. [DOI] [PubMed] [Google Scholar]
- 10.Kidd M, Modlin I, Bodei L, et al. Decoding the molecular and mutational ambiguities of gastroenteropancreatic neuroendocrine neoplasm pathobiology. Cell Mol Gastroenterol Hepatol 2015;1:131–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis MA, Yao JC. Molecular pathology and genetics of gastrointestinal neuroendocrine tumours. Curr Opin Endocrinol Diabetes Obes 2013;4:4. [DOI] [PubMed] [Google Scholar]
- 12.Bosman FT. 4th edition WHO classification of tumours of the digestive system, volume 3 Lyon (France): World Health Organization; International Agency for Research on Cancer; 2010. [Google Scholar]
- 13.Salazar R, Wiedenmann B, Rindi G, et al. ENETS 2011 consensus guidelines for the management of patients with digestive neuroendocrine tumors: an update. Neuroendocrinology 2012;95(2):71–3. [DOI] [PubMed] [Google Scholar]
- 14.Pavel M Translation of molecular pathways into clinical trials of neuroendocrine tumors. Neuroendocrinology 2013;97(1):99–112. [DOI] [PubMed] [Google Scholar]
- 15.Alexandraki KI, Kaltsas GA, Grozinsky-Glasberg S, et al. Appendiceal neuroendocrine neoplasms: diagnosis and management. Endocr Relat Cancer 2016; 23(1):R27–41. [DOI] [PubMed] [Google Scholar]
- 16.Frilling A, Modlin I, Kidd M, et al. Recommendations for management of patients with neuroendocrine liver metastases. Lancet Oncol 2014;15(1):e8–21. [DOI] [PubMed] [Google Scholar]
- 17.Alvarez MA, Subramaniam PS, Grunn A, et al. Pharmacological targeting of master regulator proteins in neuroendocrine tumors: a novel strategy for precision cancer medicine applications. Nat Genet, in press. [Google Scholar]
- 18.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45(2): 228–47. [DOI] [PubMed] [Google Scholar]
- 19.Neperud J, Mahvash A, Garg N, et al. Can imaging patterns of neuroendocrine hepatic metastases predict response yttruim-90 radioembolotherapy? World J Radiol 2013;5(6):241–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Denecke T, Baur AD, Ihm C, et al. Evaluation of radiological prognostic factors of hepatic metastases in patients with non-functional pancreatic neuroendocrine tumors. Eur J Radiol 2013;82(10):e550–5. [DOI] [PubMed] [Google Scholar]
- 21.Toumpanakis C, Kim MK, Rinke A, et al. Combination of cross-sectional and molecular imaging studies in the localization of gastroenteropancreatic neuroendocrine tumors. Neuroendocrinology 2014;21:21. [DOI] [PubMed] [Google Scholar]
- 22.Binderup T, Knigge U, Loft A, et al. 18F-fluorodeoxyglucose positron emission tomography predicts survival of patients with neuroendocrine tumors. Clin Cancer Res 2010;16(3):978–85. [DOI] [PubMed] [Google Scholar]
- 23.Castano JP, Sundin A, Maecke HR, et al. Gastrointestinal neuroendocrine tumors (NETs): new diagnostic and therapeutic challenges. Cancer Metastasis Rev 2014;5:5. [DOI] [PubMed] [Google Scholar]
- 24.Faivre S, Ronot M, Dreyer C, et al. Imaging response in neuroendocrine tumors treated with targeted therapies: the experience of sunitinib. Target Oncol 2012; 7(2):127–33. [DOI] [PubMed] [Google Scholar]
- 25.Oberg K, Modlin I, DeHerder W, et al. Biomarkers for neuroendocrine tumor disease: a delphic consensus assessment of multianalytes, genomics, circulating cells and monoanalytes. Lancet Oncol 2015;16:e435046. [Google Scholar]
- 26.Hulka B Overview of biological markers In: Hulka B, Griffith J, Wilcosky T, editors. Biological markers in epidemiology. New York: Oxford University Press; 1990. p. 3–15. [Google Scholar]
- 27.Modlin IM, Gustafsson BI, Moss SF, et al. Chromogranin A–biological function and clinical utility in neuro endocrine tumor disease. Ann Surg Oncol 2010; 17(9):2427–43. [DOI] [PubMed] [Google Scholar]
- 28.Yao JC, Pavel M, Phan AT, et al. Chromogranin A and neuron-specific enolase as prognostic markers in patients with advanced pNET treated with everolimus. J Clin Endocrinol Metab 2011;96(12):3741–9. [DOI] [PubMed] [Google Scholar]
- 29.Lawrence B, Gustafsson BI, Kidd M, et al. The clinical relevance of chromogranin A as a biomarker for gastroenteropancreatic neuroendocrine tumors. Endocrinol Metab Clin North Am 2011;40(1):111–34, viii. [DOI] [PubMed] [Google Scholar]
- 30.Malczewska A, Kidd M, Matar S, et al. A comprehensive assessment of the role of miRNAs as biomarkers in gastroenteropancreatic neuroendocrine tumors. Neuroendocrinology 2018;107(1):73–90. [DOI] [PubMed] [Google Scholar]
- 31.Kidd M, Drozdov I, Modlin I. Blood and tissue neuroendocrine tumor gene cluster analysis correlate, define hallmarks and predict disease status. Endocr Relat Cancer 2015;22(4):561–75. [DOI] [PubMed] [Google Scholar]
- 32.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000;100(1):57–70. [DOI] [PubMed] [Google Scholar]
- 33.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144(5):646–74. [DOI] [PubMed] [Google Scholar]
- 34.Walenkamp A, Crespo G, Fierro Maya F, et al. Hallmarks of gastrointestinal neuroendocrine tumours: implications for treatment. Endocr Relat Cancer 2014;21(6): R445–60. [DOI] [PubMed] [Google Scholar]
- 35.Wang E, Zaman N, McGee S, et al. Predictive genomics: a cancer hallmark network framework for predicting tumor clinical phenotypes using genome sequencing data. Semin Cancer Biol 2014;18(14):00050–9. [DOI] [PubMed] [Google Scholar]
- 36.Cardoso F, van’t Veer LJ, Bogaerts J, et al. 70-gene signature as an aid to treatment decisions in early-stage breast cancer. N Engl J Med 2016;375(8):717–29. [DOI] [PubMed] [Google Scholar]
- 37.Krop I, Ismaila N, Andre F, et al. Use of biomarkers to guide decisions on adjuvant systemic therapy for women with early-stage invasive breast cancer: american society of clinical oncology clinical practice guideline focused update. J Clin Oncol 2017;35(24):2838–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Curtis C Genomic profiling of breast cancers. Curr Opin Obstet Gynecol 2015; 27(1):34–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014;32(6):579–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siravegna G, Marsoni S, Siena S, et al. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol 2017;2(10):14. [DOI] [PubMed] [Google Scholar]
- 41.Oxnard GR, Paweletz CP, Kuang Y, et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res 2014;20(6):1698–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Remon J, Caramella C, Jovelet C, et al. Osimertinib benefit in EGFR-mutant NSCLC patients with T790M-mutation detected by circulating tumour DNA. Ann Oncol 2017;28(4):784–90. [DOI] [PubMed] [Google Scholar]
- 43.Reinert T, Scholer LV, Thomsen R, et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut 2015; 4(308859):625–34. [DOI] [PubMed] [Google Scholar]
- 44.Kidd M, Modlin I, Oberg K. Towards a new classification of gastroenteropancreatic neuroendocrine neoplasms. Nat Rev Clin Oncol 2016;13(11):691–705. [DOI] [PubMed] [Google Scholar]
- 45.Jiao Y, Shi C, Edil BH, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science 2011; 331(6021):1199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shay JW, Reddel RR, Wright WE. Cancer. Cancer and telomeres–an ALTernative to telomerase. Science 2012;336(6087):1388–90. [DOI] [PubMed] [Google Scholar]
- 47.Patel K, Friedrich-Rust M, Lurie Y, et al. FibroSURE and FibroScan in relation to treatment response in chronic hepatitis C virus. World J Gastroenterol 2011; 17(41):4581–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kidd M, Modlin IM, Drozdov I. Gene network-based analysis identifies two potential subtypes of small intestinal neuroendocrine tumors. BMC Genomics 2014;15: 595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duerr EM, Mizukami Y, Ng A, et al. Defining molecular classifications and targets in gastroenteropancreatic neuroendocrine tumors through DNA microarray analysis. Endocr Relat Cancer 2008;15(1):243–56. [DOI] [PubMed] [Google Scholar]
- 50.Drozdov I, Kidd M, Nadler B, et al. Predicting neuroendocrine tumor (carcinoid) neoplasia using gene expression profiling and supervised machine learning. Cancer 2009;115(8):1638–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Modlin I, Drozdov I, Kidd M. The Identification of gut neuroendocrine tumor disease by multiple synchronous transcript analysis in blood. PLoS One 2013;8: e63364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Modlin IM, Aslanian H, Bodei L, et al. A PCR blood test outperforms chromogranin A in carcinoid detection and is unaffected by PPIs. Endocr Connect 2014;14: 14–0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Modlin IM, Frilling A, Salem RR, et al. Blood measurement of neuroendocrine gene transcripts defines the effectiveness of operative resection and ablation strategies. Surgery 2016;159(1):336–47. [DOI] [PubMed] [Google Scholar]
- 54.Modlin IM, Kidd M, Bodei L, et al. The clinical utility of a novel blood-based multitranscriptome assay for the diagnosis of neuroendocrine tumors of the gastrointestinal tract. Am J Gastroenterol 2015;110(8):1223–32. [DOI] [PubMed] [Google Scholar]
- 55.Pavel M, Jann H, Prasad V, et al. NET blood transcript analysis defines the crossing of the clinical rubicon: when stable disease becomes progressive. Neuroendocrinology 2017;104(2):170–82. [DOI] [PubMed] [Google Scholar]
- 56.Cwikla JB, Bodei L, Kolasinska-Cwikla A, et al. Circulating transcript analysis (NETest) in GEP-NETs treated with somatostatin analogs defines therapy. J Clin Endocrinol Metab 2015;100(11):E1437–45. [DOI] [PubMed] [Google Scholar]
- 57.Modlin IM, Drozdov I, Alaimo D, et al. A multianalyte PCR blood test outperforms single analyte ELISAs (chromogranin A, pancreastatin, neurokinin A) for neuroendocrine tumor detection. Endocr Relat Cancer 2014;21(4):615–28. [DOI] [PubMed] [Google Scholar]
- 58.Filosso PL, Kidd M, Roffinella M, et al. The utility of blood neuroendocrine gene transcript measurement in the diagnosis of bronchopulmonary neuroendocrine tumors (BPNET) and as a tool to evaluate surgical resection and disease progression. Eur J Cardiothorac Surg 2017;53(3):631–9. [DOI] [PubMed] [Google Scholar]
- 59.Kidd M, Modlin I, Drozdov I, et al. A liquid biopsy for bronchopulmonary/lung carcinoid diagnosis. Oncotarget 2017;9(6):7182–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peczkowska M, Cwikla J, Kidd M, et al. The clinical utility of circulating neuroendocrine gene transcript analysis in well-differentiated paragangliomas and pheochromocytomas. Eur J Endocrinol 2017;176(2):143–57. [DOI] [PubMed] [Google Scholar]
- 61.Modlin I, Drozdov I, Kidd M. A multitranscript blood neuroendocrine tumor molecular signature to identify treatment efficacy and disease progress. J Clin Oncol 2013;31(Suppl):A4137. [Google Scholar]
- 62.Modlin I, Drozdov I, Kidd M. Gut neuroendocrine Tumor Blood qPCR fingerprint assay: characteristics and reproducibility. Clin Chem 2014;52(3):419–29. [DOI] [PubMed] [Google Scholar]
- 63.Kononenko I Machine learning for medical diagnosis: history, state of the art and perspective. Artif Intell Med 2001;23(1):89–109. [DOI] [PubMed] [Google Scholar]
- 64.Darcy AM, Louie AK, Roberts LW. Machine learning and the profession of medicine. JAMA 2016;315(6):551–2. [DOI] [PubMed] [Google Scholar]
- 65.Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol 2000;25(2):169–93. [DOI] [PubMed] [Google Scholar]
- 66.Jia X, Ju H, Yang L, et al. A novel multiplex polymerase chain reaction assay for profile analyses of gene expression in peripheral blood. BMC Cardiovasc Disord 2012;12:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schmittgen TD, Zakrajsek BA, Mills AG, et al. Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: comparison of endpoint and real-time methods. Anal Biochem 2000;285(2):194–204. [DOI] [PubMed] [Google Scholar]
- 68.Ding C, Cantor CR. A high-throughput gene expression analysis technique using competitive PCR and matrix-assisted laser desorption ionization time-of-flight MS. Proc Natl Acad Sci U S A 2003;100(6):3059–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Giusti M, Sidoti M, Augeri C, et al. Effect of short-term treatment with low dosages of the proton-pump inhibitor omeprazole on serum chromogranin A levels in man. Eur J Endocrinol 2004;150(3):299–303. [DOI] [PubMed] [Google Scholar]
- 70.Ishida M, Shiomi H, Naka S, et al. Concomitant intraductal papillary mucinous neoplasm and neuroendocrine tumor of the pancreas. Oncol Lett 2013;5(1):63–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ginsburg GS, Kuderer NM. Comparative effectiveness research, genomics-enabled personalized medicine, and rapid learning health care: a common bond. J Clin Oncol 2012;30(34):4233–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Malik SM, Pazdur R, Abrams JS, et al. Consensus report of a joint NCI thoracic malignancies steering committee: FDA workshop on strategies for integrating biomarkers into clinical development of new therapies for lung cancer leading to the inception of “master protocols” in lung cancer. J Thorac Oncol 2014; 9(10):1443–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Strosberg J, El-Haddad G, Wolin E, et al. Phase 3 Trial of 177Lu-Dotatate for midgut neuroendocrine tumors. N Engl J Med 2017;376(2):125–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bodei L, Kidd M, Paganelli G, et al. Clinical features are not reliable in predicting long-term toxicity after PRRT - Evidence from >800 patients to support genetic screen development. Paper presented at: ENETS Conference Barcelona (Spain), March 6–8, 2014. [Google Scholar]
- 75.Bodei L, Kidd M, Modlin IM, et al. Measurement of circulating transcripts and gene cluster analysis predicts and defines therapeutic efficacy of peptide receptor radionuclide therapy (PRRT) in neuroendocrine tumors. Eur J Nucl Med Mol Imaging 2016;43(5):839–51. [DOI] [PubMed] [Google Scholar]
- 76.Bodei L, Kidd MS, Singh A, et al. PRRT genomic signature in blood for prediction of 177Lu-octreotate efficacy. Eur J Nucl Med Mol Imaging 2018;45(7):1155–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Valli A, Rodriguez M, Moutsianas L, et al. Hypoxia induces a lipogenic cancer cell phenotype via HIF1alpha-dependent and -independent pathways. Oncotarget 2015;6(4):1920–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Olsson AH, Yang BT, Hall E, et al. Decreased expression of genes involved in oxidative phosphorylation in human pancreatic islets from patients with type 2 diabetes. Eur J Endocrinol 2011;165(4):589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Day TF, Mewani RR, Starr J, et al. Transcriptome and proteome analyses of TNFAIP8 knockdown cancer cells reveal new insights into molecular determinants of cell survival and tumor progression. Methods Mol Biol 2017;1513: 83–100. [DOI] [PubMed] [Google Scholar]
- 80.Hill RP. The changing paradigm of tumour response to irradiation. Br J Radiol 2017;90(1069):20160474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kidd M, Modlin IM. Therapy: the role of liquid biopsies to manage and predict PRRT for NETs. Nat Rev Gastroenterol Hepatol 2017;14(6):331–2. [DOI] [PubMed] [Google Scholar]