SUMMARY
The Retinoid inducible nuclear factor (Rinf), also known as CXXC5, is a nuclear protein, but its functions in the context of the chromatin are poorly defined. We find that in mouse embryonic stem cells (mESCs), Rinf binds to the chromatin and is enriched at promoters and enhancers of Tet1, Tet2, and pluripotency genes. The Rinf-bound regions show significant overlapping occupancy of pluripotency factors Nanog, Oct4, and Sox2, as well as Tet1 and Tet2. We found that Rinf forms a complex with Nanog, Oct4, Tet1, and Tet2 and facilitates their proper recruitment to regulatory regions of pluripotency and Tet genes in ESCs to positively regulate their transcription. Rinf deficiency in ESCs reduces expression of Rinf target genes, including several pluripotency factors and Tet enzymes, and causes aberrant differentiation. Together, our findings establish Rinf as a regulator of the pluripotency network genes and Tet enzymes in ESCs.
In Brief
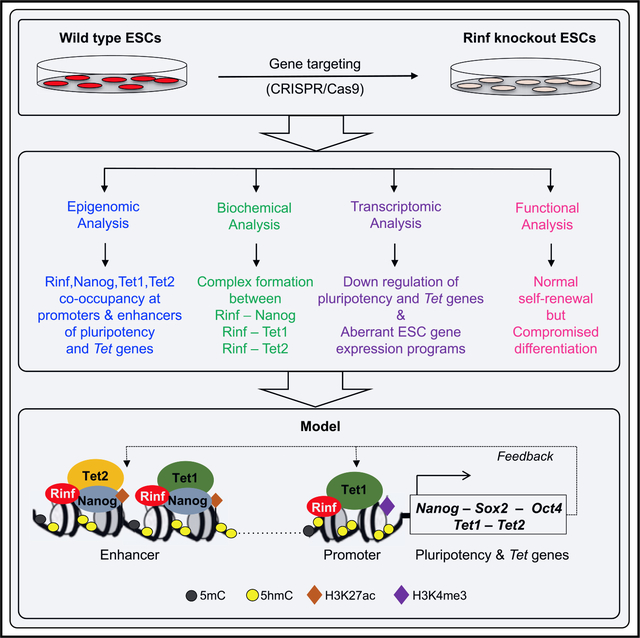
In embryonic stem cells, pluripotency factors and Tet enzymes regulate gene expression and differentiation programs. Ravichandran et al. find that Rinf facilitates their recruitment to promoters and enhancers of stemness genes, including several pluripotency and Tet genes. Rinf positively regulates their transcription and ensures proper differentiation of embryonic stem cells.
Graphical Abstract
INTRODUCTION
Embryonic stem cells (ESCs) are pluripotent cells that give rise to all cell types of the three embryonic germ layers (Hanna et al., 2010). ESC pluripotency is tightly regulated by the expression of core pluripotency genes, including the transcription factors Oct3/4, Sox2, and Nanog (Jaenisch and Young, 2008). These factors regulate themselves and each other and constitute the core pluripotency circuitry in ESCs. Epigenetic mechanisms involving DNA methylation and histone modifications are also involved in proper expression of pluripotency factors and maintaining ESC state gene expression programs (Pastor et al., 2013; Smith and Meissner, 2013). The Ten eleven translocation (Tet) family of dioxygenases (Tet1/2/3), which promote DNA demethylation by converting 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) and further derivatives, are involved in gene regulation in ESCs (Pastor et al., 2013). Tet1 and Tet2 are expressed in ESCs and enriched at gene regulatory regions to facilitate gene expression. Through a feedback mechanism, they are also targets of pluripotency factors (Koh et al., 2011). Proper expression and recruitment of Tets and pluripotency factors to their targets are essential for maintaining the pluripotent state (Jaenisch and Young, 2008; Wu and Zhang, 2010).
Pluripotency transcription factors bind to specific DNA motifs (Jaenisch and Young, 2008), whereas Tet enzymes are recruited to CpG-containing regions (Pastor et al., 2013). Tet1 and Tet3 have distinct CXXC domains in their N-terminal regions that bind to CpGs and, in part, facilitate their targeting to chromatin. In contrast, Tet2 does not contain a CXXC domain (Ko et al., 2013; Liu et al., 2013). It is believed that its CXXC domain has undergone an evolutionary chromosomal gene inversion and is separated from the Tet2 genomic sequence to become an independent gene called CXXC4 or Idax (Inhibitor of disheveled and axin). Idax has similarity not only to the N-terminal region of Tet1 and Tet3 but also to the related proteins CXXC5 or Rinf (Retinoid inducible nuclear factor) (Ko et al., 2013). This similarity suggests that Rinf may also have evolved from ancestral Tet genes or by duplication and/or translocation of Idax. Rinf and Idax are ~30-kDa proteins that are expressed in various cell types and present in the cytoplasm and nucleus. They possess both CXXC-DNA binding and Dishevelled (Dvl) binding domains. Their binding to Dvl negatively regulate Wnt signaling with implications in hematopoiesis, neurogenesis, wound healing, and cancer (Kim et al., 2010; Knappskog et al., 2011; Lee et al., 2015; Pendino et al., 2009).
Although the cytoplasmic functions of Rinf and Idax involve regulation of Wnt signaling, their nuclear functions remain poorly investigated. It is likely that their CXXC domain, which recognizes and binds to CpGs, is involved in targeting them to gene regulatory regions and is responsible for their nuclear functions. There is some evidence in support of Rinf facilitating transcription in selected somatic cell types (Kim et al., 2014, 2016; Li et al., 2014; Ma et al., 2017). Rinf and Idax are also implicated in negatively regulating Tet2 through Parp-mediated degradation of the Tet2 protein (Ko et al., 2013). Although these studies allude to some nuclear functions of Rinf in various cell types, the role of Rinf in regulation of ESC biology has not been investigated. In this study, we have established how Rinf interacts with the chromatin and regulates gene expression in mouse ESCs. We find that Rinf, but not Idax, is expressed in ESCs and is mainly present in the nucleus where it occupies gene regulatory regions along with core pluripotency factors (Nanog, Oct3/4, and Sox2) and Tet1/2 enzymes. We show that Rinf facilitates recruitment of pluripotency factors and Tet enzymes to promoters and enhancers of pluripotency and Tet genes to regulate their expression. Our findings identify Rinf as a regulator of gene expression in ESCs and propose a mechanism for its involvement in modulating the pluripotency network.
RESULTS
Rinf Is Expressed in ESCs and Binds to the Chromatin
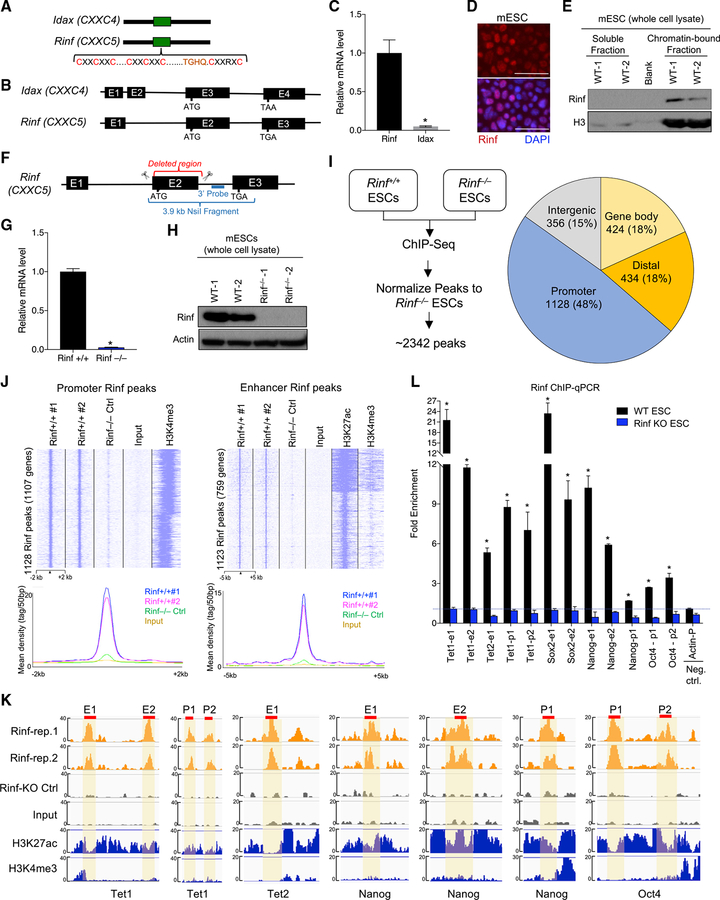
Rinf and Idax are CXXC-domain-containing proteins (Figure 1A) and have a similar gene structure (Figure 1B). We find that Rinf, but not Idax, is expressed in mouse ESCs (Figure 1C; Figure S1A) and is mainly present in the nucleus (Figure 1D). To examine if Rinf is targeted to the chromatin, we analyzed Rinf protein levels in soluble and chromatin-bound fractions of ESC lysate. We found that Rinf is primarily present in the chromatin-bound fraction (Figure 1E), suggesting that it may play a role in regulation of gene expression in ESCs. To establish the molecular and biological significance of Rinf in ESCs, we generated Rinf knockout (Rinf−/−) ESC by CRISPR/Cas9 by using two guide RNAs (gRNAs) flanking the Rinf exon 2 (Figure 1F). This exon encodes a major portion of the protein, and its deletion completely abolished Rinf expression. We confirmed genotypes of properly targeted Rinf−/− ESC lines by PCR and Southern blot (Figures S1B and S1C) and validated the complete loss of Rinf mRNA and protein by qRT-PCR and western blot, respectively (Figures 1G and 1H). We also noted that the loss of Rinf did not lead to an induction of Idax in Rinf−/− ESCs (Figures S1D and S1E).
Figure 1. Expression and Chromatin Enrichment of Rinf in Mouse ESCs.
(A) Schematic of protein domains of Rinf and Idax.
(B) Schematic of exon and intron structures of Rinf and Idax.
(C) Rinf and Idax mRNA levels quantified by qRT-PCR in ESCs. Data normalized to glyceraldehyde 3-phosphate dehydrogenase (Gapdh).
(D) Expression of Rinf in mouse ESCs detected by immunofluorescence.
(E) Rinf protein level in soluble and chromatin-bound fractions of ESC lysate.
(F) Schematic of gene targeting strategy for generating Rinf knockout ESCs..
(G) Rinf mRNA level quantified by qRT-PCR in targeted ESC clones. Data normalized to Gapdh.
(H) Quantification of Rinf protein by western blot in targeted ESC clones.
(I) Schematic of ChIP-seq strategy (left), and distribution of Rinf peaks in the genome (right).
(J) ChIP-seq read densities at Rinf peaks presented as heatmaps (top) and line graphs (bottom). All rows are peaks and centers are the summits of Rinf peaks. The H3K4me3 and H3K27ac data were obtained from a previous study (see STAR Methods).
(K) Enrichment of Rinf ChIP-seq signals at regulatory regions of selected pluripotency and Tet genes. H3K27ac and H3K4me3 tracks are used to depict enhancers and promoters (±2 kb of TSS). Selected regions of these peaks (red line) are validated by ChIP-qPCR in (L).
(L) Quantification of enrichment of Rinf at regulatory elements of indicated genes by ChIP-qPCR in ESCs (data normalized to immunoglobulin G [IgG]). Rinf KO ESCs are used as control for antibody specificity. Actin is used as a negative control.
For all panels, data are presented as mean ± SD. *Statistically significant (p < 0.05). E, enhancer; p, promoter. Scale bars, 50 mm (see also Figure S1).
Rinf Is Enriched at Promoters and Enhancers in ESCs
Because we found that Rinf is a chromatin-bound protein, we mapped its genome-wide-binding distribution and enrichment at genes and regulatory regions by performing chromatin immunoprecipitation using two independent wild-type ESC clones with a specific antibody against Rinf, followed by DNA sequencing (chromatin immunoprecipitation sequencing [ChIP-seq]). To ensure the specificity of the antibody, we also performed ChIP-seq in a Rinf−/− ESC line as a negative control. The ChIP-seq analysis identified a total of 2,342 Rinf peaks that were mapped to promoters and gene bodies as well as distal regulatory elements and intergenic regions (Figure 1I). We found a strong enrichment of Rinf at “promoters” (±2 kb of transcriptional start sites [TSS], supported by high H3K4me3 signals), with a total of 1,128 peaks mapped to 1,107 genes (Figure 1J). Likewise, we observed a strong enrichment of Rinf at “enhancers” (±50 kb from genes, supported by high H3K27ac and low H3K4me3 signals) with a total of 1,123 peaks mapped to 759 genes (Figure 1J). The ChIP-seq data between the two wild-type ESC replicates were highly reproducible. In contrast, no or limited enrichment was seen in Rinf−/− ESCs (Figure 1J), ascertaining the high specificity of our antibody and Rinf peak calling. This result established that Rinf is primarily targeted to promoters and enhancers in ESCs. To examine if Rinf binds to any specific DNA sequence, we performed a motif enrichment analysis of the peaks. We found that the Rinf-bound DNA sequences at non-promoter regions, but not at promoters, were significantly enriched for the binding motifs of known pluripotency factors, including Oct4, Sox2, Nanog, and Esrrb (Figure S1F). Gene Ontology (GO) analysis on the Rinf-bound genes identified regulation of gene expression and transcription and stem cell maintenance and development as key enriched terms (Figure S1G). This suggests that Rinf target genes are involved in gene regulation and biological properties of ESCs. These genes include the core pluripotency factors Nanog, Oct4, and Sox2 as well as epigenetic modifiers Tet1 and Tet2 (Figure 1K). We validated Rinf occupancy at the promoters and enhancers of these genes by ChIP-qPCR (Figure 1L). These findings established that Rinf is present at the promoters and enhancers of pluripotency and Tet genes in ESCs and may regulate their expression.
Significant Co-occupancy of Rinf, Pluripotency Factors, and Tet1/2 Enzymes at Chromatin
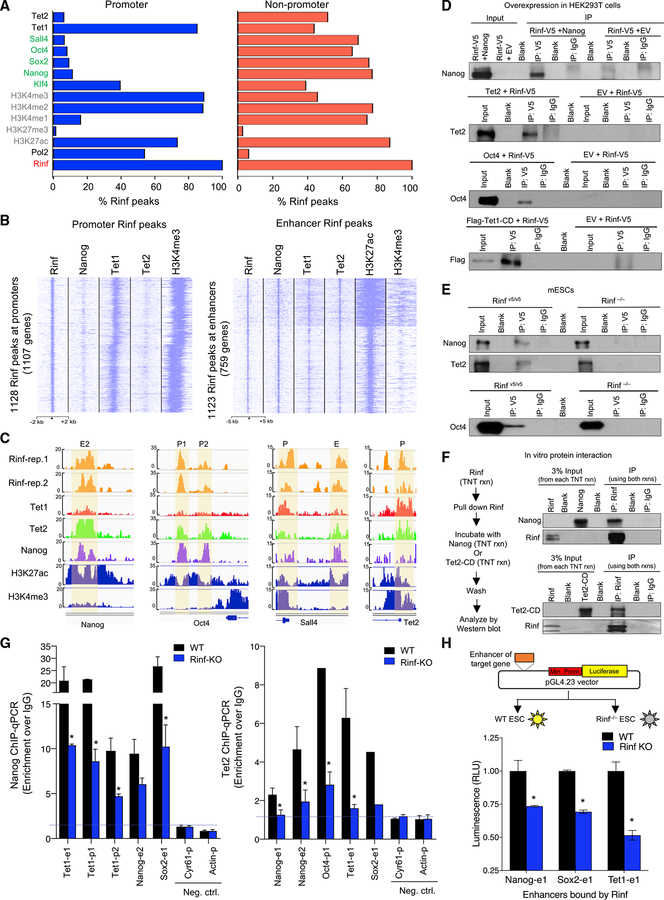
Because Rinf-bound regions are enriched for binding motifs of pluripotency factors, we examined how Rinf occupancy compares to those of pluripotency factors and associated chromatin modifiers and activating or repressing histone marks. We compared the Rinf ChIP-seq peaks to the peaks of pluripotency factors (Nanog, Sall4, Oct4, Sox2, and Klf4), epigenetic modifiers Tet1 and Tet2, RNA polymerase II, and activating or repressive histone marks from published datasets (Figures 2A–2C; Figures S2A and S2B). We found that at promoters ~90% of Rinf peaks overlapped with active histone marks and Tet1 peaks (p < 0.001), while ~10% of Rinf peaks overlapped with peaks of Tet2 and pluripotency factors. In contrast, at non-promoter regions ~80% of Rinf peaks overlapped with those of pluripotency factors and active histone marks, and ~50% of Rinf peaks overlapped with those of Tet1 and Tet2 (p < 0.001). Conversely, between 10%–25% of the total ChIP-seq peaks of each Tet1, Tet2, Nanog, Oct4, Sox2, Klf4, and Sall4 overlapped with Rinf peaks at promoter and non-promoter regions (Figure S2B). These findings suggest that Rinf is associated with active chromatin and its occupancy overlaps with that of Tet1 at promoters but with those of Tet1, Tet2, and pluripotency factors at enhancers. The data also suggest that Rinf likely functions cooperatively with pluripotency factors on a subset of their targets.
Figure 2. Co-occupancy of Rinf with Pluripotency Factors and Tet Enzymes at Gene Regulatory Regions.
(A) Overlapping analysis of Rinf peaks (from this study) with those of pluripotency factors, Tet enzymes, and activating/repressive histone marks (from previous studies, see STAR Methods). Data presented as % of Rinf peaks overlapping with each of the factors.
(B) Enrichment of Rinf peaks at ChIP-seq signals of Nanog, Tet1, and Tet2 at promoters (±2 kb of TSS and H3K4me3 positive, left) and enhancers (H3K27ac positive and H3K4me3 low, right).
(C) Enrichment of ChIP-seq signals showing co-occupancy of Rinf, pluripotency factors, and Tet enzymes at selected pluripotency and Tet genes. H3K27ac and H3K4me3 tracks are used as reference to depict enhancers and promoters (±2 kb of TSS).
(D) Co-immunoprecipitation of exogenously expressed V5-tagged Rinf with Nanog, Oct4, Tet1 catalytic domain (Tet1-CD), and Tet2 in HEK293T cells using anti-V5 antibody.
(E) Co-immunoprecipitation of endogenous Rinf with Nanog, Oct4, and Tet2 in mouse ESCs using anti-V5 antibody. Rinf−/− ESCs are used as negative control.
(F) Co-immunoprecipitation of in vitro transcribed and translated Rinf, Nanog, and Tet2 catalytic domain (Tet2-CD) from rabbit reticulocyte lysate.
(G) Enrichment of Nanog and Tet2 at regulatory elements of indicated genes quantified by ChIP-qPCR in wild-type and Rinf−/− ESCs (data normalized to IgG). Actin and Cyr61 are used as negative controls.
(H) Schematic of dual luciferase reporter assay applied to test enhancers targeted by Rinf (top). Transcriptional activity is presented as relative luminescence unit (RLU), which is normalized to the luminescence from cells that express the empty vector only (bottom).
Data presented as mean ± SEM. *Statistically significant (p < 0.05). E, enhancer; p, promoter (see also Figure S2).
Rinf Forms a Complex with Nanog and Tet2 and Mediates Their Recruitment to Regulatory Regions of Pluripotency and Tet Genes
Given the co-occupancy of Rinf with pluripotency factors and Tet enzymes at gene regulatory regions in ESCs, we examined whether Rinf forms a complex with them. To this end, we performed co-immunoprecipitation in HEK293T cells that overexpress V5-tagged Rinf along with Nanog, Oct4, Tet1 catalytic domain (Tet1-CD), or Tet2, and found that Rinf forms a complex with each of these proteins (Figure 2D). Next, to establish if these complexes are formed in ESCs, we immunoprecipitated endogenous Rinf from nuclear lysates of endogenously V5-tagged Rinf ESCs (Rinf v5/v5) or Rinf−/− ESCs and probed for Nanog, Oct4, and Tet2. We found that Rinf specifically immunoprecipitated with these proteins in Rinf v5/v5 lysates (Figure 2E). These interactions are likely direct, as in-vitro-coupled transcription and translation of Rinf, Nanog, and Tet2 catalytic domain (Tet2-CD) in rabbit reticulocyte lysate, a system devoid of any chromatin regulatory complexes, followed by co-immunoprecipitation confirmed complex formation between Rinf and Nanog or Rinf and Tet2-CD (Figure 2F). This also suggests that the catalytic domain of Tet2 is sufficient for complex formation with Rinf. To establish the significance of this complex formation in ESCs, we tested the hypothesis that Rinf facilitates the recruitment of Nanog and Tet enzymes to gene regulatory regions of pluripotency and Tet genes. Using wild-type and Rinf−/− ESCs, we examined the enrichment of Nanog and Tet2 at the promoters and enhancers of several pluripotency and Tet genes that are bound by Rinf. We found that the loss of Rinf led to ~50% reduction in the enrichment of Nanog and Tet2 at promoters and enhancers of their target genes (Figure 2G). This suggests that Rinf facilitates recruitment of Nanog and Tet2 to gene regulatory regions in ESCs.
Rinf Facilitates Transcription from Enhancers of Pluripotency Genes and Tet Enzymes
Next, we examined whether the regulatory regions of the pluripotency and Tet genes that are bound by Rinf can indeed influence transcription. We used a dual luciferase system and cloned the enhancers of Nanog, Sox2, and Tet1 upstream of the minimal promoter that drives luciferase expression (Figure2H). We transfected these constructs into wild-type and Rinf−/− ESCs and quantified the luminescence normalized to empty vector as a measure of transcriptional activity. We observed 25%–50% reduced transcriptional activity from these enhancers in Rinf−/− ESCs compared to wild-type ESCs. We conclude that Rinf modulates transcription from specific enhancers of pluripotency and Tet genes.
Loss of Rinf Compromises Proper Expression of Pluripotency Genes and Epigenetic Regulators in ESCs
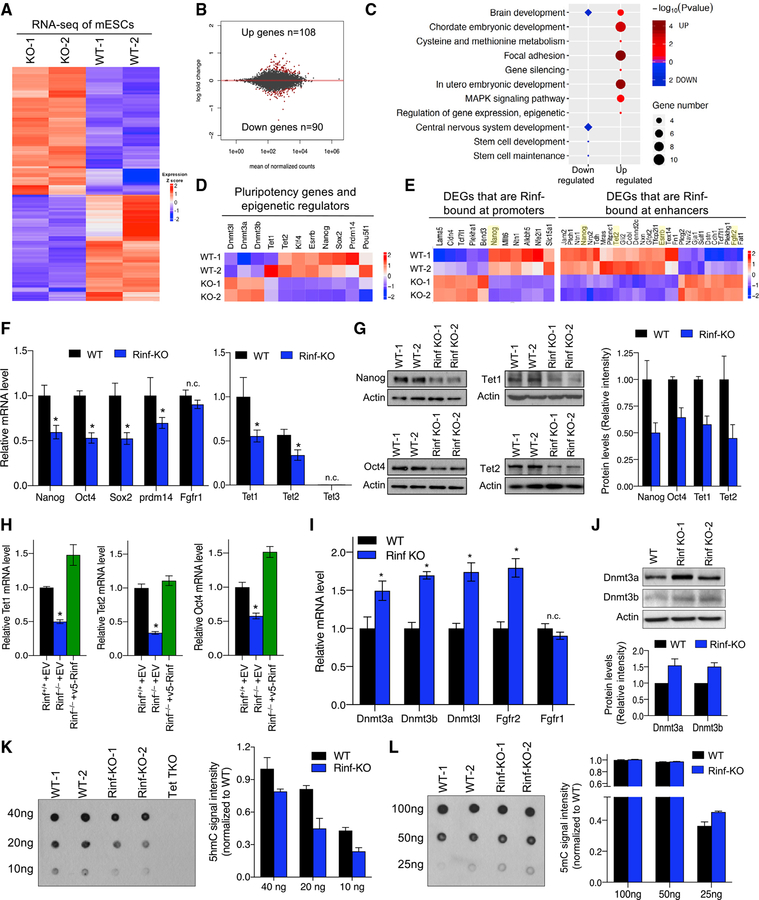
To further examine the role of Rinf in regulation of ESC gene expression programs, we compared the transcriptome of Rinf−/− and wild-type ESCs by RNA sequencing (RNA-seq) (Figure 3A; Figure S3A). This analysis identified ~200 genes that were significantly deregulated (108 up and 90 down) in Rinf−/− ESCs (Figure 3B), but the extent of changes was subtle. GO analysis found that the differentially expressed genes (DEGs) were enriched in various biological processes, including stem cell maintenance, embryonic development, gene regulation, and nervous system development (Figure 3C). The DEGs included several pluripotency factors (Nanog, Esrrb, Prdm14, and Klf4) and Tet enzymes (Tet1 and Tet2) that were downregulated (Figure 3D). This indicates that Rinf facilitates the expression of pluripotency genes and Tet enzymes in ESCs. A comparison of DEGs to Rinf-bound genes identified 44 deregulated genes as direct targets of Rinf (11 bound by Rinf at promoters and 33 bound by Rinf at enhancers, p < 0.01), including Tet enzymes and pluripotency factors (Figure 3E; Figure S3B). This further supports our earlier observations that Rinf is involved in transcriptional regulation of pluripotency and Tet genes. We validated the downregulation of several pluripotency factors and Tet enzymes in Rinf−/− ESCs both at mRNA (Figure 3F) and protein levels (Figure 3G). Furthermore, we showed that re-expression of Rinf in Rinf−/− ESCs restored their normal expression (Figure 3H). This confirmed that the loss of Rinf leads to downregulation of pluripotency factors and Tet enzymes in ESCs. We also identified de novo DNA methyltransferases (Dnmt3a, Dnmt3b, and Dnmt3L) and Fgf receptor 2 (Fgfr2) to be upregulated in Rinf−/− ESCs (Figure 3D) and validated these findings by qRT-PCR (Figure 3I) and western blot (Figure 3J). Dnmts and Tet enzymes regulate DNA methylation (5mC) and hydroxymethylation (5hmC) in ESCs, respectively (Pastor et al., 2013). Consistently, we observed a significant reduction in 5hmC levels in Rinf−/− ESCs (Figure 3K) concomitant with a subtle increase in 5mC levels (Figure 3L) in these cells. This suggests that Rinf can also influence the establishment and maintenance of DNA methylation and hydroxymethylation landscapes of ESCs.
Figure 3. Loss of Rinf Reduces the Expression of Pluripotency Factors and Tet Enzymes in ESCs and Perturbs Gene Expression Programs.
(A) Heatmap of differentially expressed genes identified. Colors indicating relative expression.
(B) MA plot of the average gene expression level and change, with DEGs in red.
(C) GO enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of the differentially expressed genes.
(D) Expression patterns of deregulated pluripotency genes, Tets, and Dnmts.
(E) Expression patterns of differentially expressed genes that are bound by Rinf.
(F) mRNA levels of pluripotency and Tet genes quantified by qRT-PCR. Three ESC lines of each genotype are used. Fgfr1 is used as a negative control (unchanged gene in RNA-seq). Expression of Tet enzymes in the left plot is normalized to the wild-type levels of Tet1.
(G) Protein levels of indicated genes quantified by western blot. Quantification of signal intensity of bands by ImageJ is plotted on the right.
(H) Restoration of the expression of pluripotency and Tet genes in Rinf−/− ESCs upon expression of exogenous Rinf. Data represent mRNA levels quantified by qRT-PCR.
(I) mRNA levels of indicated genes quantified by qRT-PCR. Three independent ESC lines of each genotype are used. Fgfr1 is used as a negative control (unchanged gene in RNA-seq).
(J) Protein levels of Dnmt3a and Dnmt3b quantified by western blot in ESCs. Quantification of signal intensity of bands by ImageJ is plotted on the right.
(K) Global 5hmC levels in ESC DNA quantified by dot blot. Tet1/2/3 triple knockout (TKO) ESC DNA is used as negative control. Average 5hmC signal intensity was plotted.
(L) Global 5mC levels in ESC DNA quantified by dot blot. Average 5mC signal intensity was plotted.
Data presented as mean ± SD. *Statistically significant (p < 0.05). n.c., no change (see also Figure S3).
Rinf Deficiency in ESCs Does Not Affect Self-Renewal but Compromises Differentiation
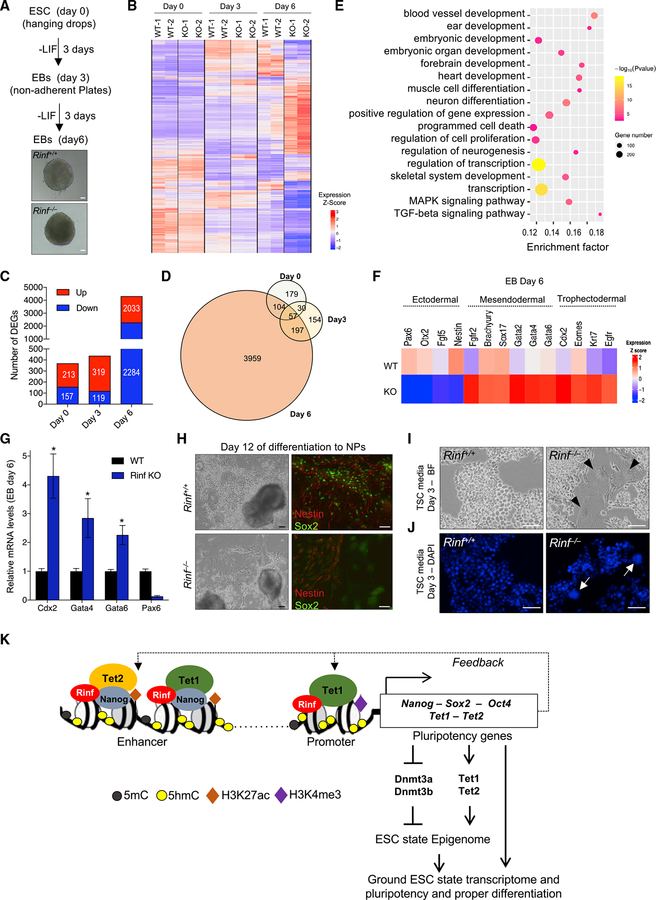
We examined whether Rinf deficiency and its associated gene expression changes affect ESC maintenance and differentiation. We found that Rinf−/− ESCs were morphologically indistinguishable from wild-type ESCs in culture (Figure S4A) and had comparable proliferation rates (Figure S4B). This suggests that the loss of Rinf does not affect ESC self-renewal and maintenance. To test if Rinf is important for differentiation and lineage specification of ESCs, we differentiated wild-type and Rinf−/− ESCs to embryoid bodies (EBs) and analyzed their transcriptomic differences by RNA-seq at three time points (day 0, 3, and 6) during differentiation (Figures 4A–4D). We found that the loss of Rinf led to distinct gene expression changes not only in ESC state but also during differentiation. Notably, at day 6 of differentiation to EBs, we found ~4,000 genes aberrantly expressed and enriched for various differentiation and developmental GO terms. These terms included ectodermal lineage development (neural differentiation and brain formation) and mesodermal lineage development (heart and muscle formation) as well as signaling pathways critical for mesendoderm and trophectoderm differentiation (mitogen-activated protein kinase [MAPK] and transforming growth factor beta [TGF-beta]) (Figure 4E). We noted downregulation of several neuroectodermal markers (such as Pax6) as well as upregulation of several mesendodermal markers (such as Gata4 and Gata6) and trophectodermal markers (such as Cdx2), findings that were validated by qRTPCR (Figures 4F and 4G). This suggests that deficiency of Rinf in ESCs compromises the normal differentiation programs by skewing differentiation toward mesendoderm and trophectoderm at the expense of neuroectoderm. Consistent with these observations, we found that Rinf−/− ESCs, when differentiated to neural progenitors (NPs), formed fewer Nestin-Sox2-positive NPs (Figure 4H). Likewise, Rinf−/− ESCs, when cultured in trophoblast stem cell (TSC) media, exhibited increased propensity to form cell types of trophoblast lineage, including giant cells marked by large nuclei (Figures 4I and 4J). Nonetheless, the loss of Rinf did not block the ability of ESCs to form cell types of the three germ layers. Teratomas derived from Rinf−/− ESCs contained ectodermal, mesodermal, and endodermal cell types (Figure S4C). Because Rinf regulates Tet enzymes in ESCs and its loss leads to their downregulation, we tested if overexpression of Tet1-CD in Rinf−/− ESCs can restore some of the pheno-types observed in these cells. We found that it restored proper expression of pluripotency genes in Rinf−/− ESCs and of lineage markers in Rinf−/− EBs (Figure S4D). It also improved the differentiation potential of Rinf−/− ESCs toward NPs (Figure S4E) and reduced the aberrant or skewed differentiation capacity of Rinf−/− ESCs toward trophectoderm lineage (Figure S4F). This suggests that Rinf−/− ESCs, although pluripotent in a teratoma assay, have aberrant differentiation potential, which can be in part rescued by Tet catalytic activity. Taken together, our findings support a model for the nuclear functions of Rinf in ESCs whereby Rinf facilitates transcription of pluripotency genes and Tet enzymes to regulate gene expression and differentiation programs in ESCs (Figure 4K).
Figure 4. Rinf-Deficient ESCs Have Compromised Differentiation.
(A) Schematic of differentiation of ESCs to EBs over 6 days and bright-field images of EBs.
(B) Heatmap of DEGs identified at the indicated time points during differentiation to EBs. Colors represent relative expression. Two independent ESC lines were used in the analysis.
(C) Number of DEGs between wild-type and Rinf−/− EBs plotted for the indicated time points during differentiation.
(D) Venn diagram illustrating overlap of DEGs between wild-type and Rinf−/− EBs for each time point during differentiation.
(E) GO enrichment and KEGG pathway analysis of the DEGs at day 6 of differentiation to EBs.
(F) Expression patterns of deregulated lineage marker genes at day 6 of differentiation.
(G) Quantification of mRNA levels of indicated lineage markers by qRT-PCR in day 6 EBs. Three independent ESC lines of each genotype were used.
(H) Bright-field images of culture of ESCs at day 12 of differentiation to neural progenitors (NPs) (left). Immunofluorescence images of NPs derived from ESCs and stained for Nestin and Sox2 at day 12 of differentiation (right).
(I) Bright-field images of ESCs cultured in TSC media for 3 days. Arrowheads indicate trophoblast giant cells.
(J) 4′,6-Diamidino-2-phenylindole (DAPI) staining of ESCs cultured in TSC media for 3 days. Arrowheads indicate large nuclei of trophoblast giant cells.
(K) Model for the role of Rinf in transcriptional regulation of pluripotency and Tet genes in ESCs.
Data presented as mean ± SD. *Statistically significant (p < 0.05). All scale bars, 50 mm (see also Figure S4).
DISCUSSION
Rinf is expressed in ESCs, but its roles in regulation of gene expression and ESC biology are not defined. We find a nuclear role for Rinf in transcription of pluripotency and Tet genes and modulating ESC gene expression and differentiation programs. Rinf is mainly bound at promoters and enhancers in ESCs, where it shows strong co-occupancy with pluripotency factors and Tet enzymes. In contrast to ~2,300 Rinf peaks in ESCs, there are >8,000 peaks for each Tet1, Tet2, and Nanog (Whyte et al., 2013; Wu et al., 2011; Xiong et al., 2016). Thus, not all Nanog, Tet1, and Tet2 peaks overlap with Rinf. Rather, we find a majority of Rinf peaks overlap with Tet1 at promoters and with Tet2 and Nanog at enhancers. As such, Rinf does not seem responsible for the recruitment of Nanog and Tets to all of their genomic targets but instead only at selected target genes. We identify these targets to be regulatory regions of pluripotency genes and epigenetic modifiers, including Nanog and Tet1/2 enzymes themselves. This places Rinf as an upstream modulator of pluripotency and Tet genes in ESCs. The loss of Rinf reduces but does not abolish recruitment of Nanog and Tet2, just as it reduces but does not block their expression. This indicates that Rinf mainly acts as a facilitator of transcription. It is also possible that the loss of Rinf was partially compensated by other parallel mechanisms in ESCs. We can at least rule out Idax because it is not expressed in ESCs and is not induced in Rinf−/− ESCs. On a different note, Tet1, in contrast to Tet2, has a CXXC domain. This raises a question about justifying Tet1 dependency on Rinf. It is possible that the Tet1 CXXC domain is not equivalent to Rinf and is not as effective in recruiting Tet1 to chromatin or specifically to gene regulatory regions. This would warrant a role for Rinf in facilitating this process and perhaps providing the specificity. Moreover, variants of Tet1 that lack the CXXC domain and are expressed in some cell types may justifiably rely on Rinf.
Downregulation of pluripotency factors and Tet enzymes in Rinf−/− ESCs to near half of normal levels can lead to aberrant expression of their target genes, compromising ESC and differentiation gene expression programs. Transcriptomic analysis of Rinf−/− ESCs during differentiation to EBs reveals downregulation of gene expression programs involved in ectodermal and upregulation of genes involved in mesendodermal and trophectodermal differentiation. Consistently, these cells show reduced neural and enhanced mesendoderm and trophoblast differentiation. Nonetheless, this does not completely block ESC maintenance and pluripotency. Rinf−/− ESCs can form tissues of the three germ layers in a teratoma assay, albeit a qualitative assay that does not account for quantitative changes in differentiation. This observation is in agreement with the biology of Nanog haplo-insufficient ESCs (Mitsui et al., 2003) or Tet1/2/3 double and triple heterozygous ESCs (Dawlaty et al., 2013, 2014) where reduction of these proteins to half of normal levels does not block pluripotency and differentiation completely in a teratoma assay. We propose that the loss of Rinf deregulates proper expression of pluripotency and Tet genes impacting ESC and differentiation gene expression programs. This compromises, but does not block, differentiation along the three germ layer lineages. It is also possible that Rinf, in addition to facilitating transcription of pluripotency and Tet genes in ESCs, has a role during differentiation to directly regulate Tet proteins and lineage markers. This will be of interest to explore in the future by mapping Rinf genomic occupancy during differentiation and identifying its target genes.
Deregulation of pluripotency factors and Tet enzymes in Rinf−/− ESCs impacts their downstream effectors. For example, Prdm14 represses de novo DNA methyltransferases and Fgf signaling (Grabole et al., 2013; Yamaji et al., 2013). Together with Nanog-Oct3/4-Sox2, it helps maintain the naive pluripotent states in ESCs. The loss of Rinf and subsequent downregulation of Prdm14 lead to upregulation of Dnmt3a, Dnmt3b, and Fgfr2. Upregulation of de novo methyltransferases and Fgfr2 and downregulation of pluripotency factors and Tet enzymes, as observed in Rinf−/− ESCs, are features of epiblast stem cells (EpiSC) or primed pluripotency (Grabole et al., 2013; Hanna et al., 2010; Yamaji et al., 2013). Thus, our findings also implicate Rinf in promoting the ground state and preventing the primed state pluripotency gene expression programs in ESCs. We also note that the loss of Rinf, by downregulating Tet enzymes and upregulating Dnmt3a/3b, reduces 5hmC and increases 5mC levels in ESCs. This suggests that Rinf can influence gene expression indirectly by modulating DNA methylation and hydroxylation levels. Although the decrease in global 5hmC levels (~50%) leads to a subtle increase in global 5mC levels, it may have a prominent impact on gene expression in a locus- and target-specific fashion. Future studies involving genome-wide mapping of 5hmC and 5mC distribution in Rinf−/− ESCs can elaborate on this.
Although Rinf and Idax are implicated in the negative regulation of Wnt signaling in the cytoplasm (Kim et al., 2010; Kojima et al., 2009), we find that in ESCs Idax is not expressed and Rinf is only present in the nucleus. This suggests that the main functions of Rinf in ESCs are nuclear involving the chromatin and gene expression. Another study implicates Rinf and Idax in regulating Tet2 protein levels, where their overexpression promotes Parp-mediated degradation of exogenous Tet2 in HEK293T cells, while Idax knockdown during ESC differentiation increases Tet2 protein levels (Ko et al., 2013). As such, the loss of Rinf is expected to increase Tet2 protein levels. However, we find that Tet2 protein levels are not increased in Rinf−/− ESCs. Rather, both Tet1 and Tet2 are decreased at mRNA and protein levels. This shows that Rinf is a positive transcriptional regulator of Tet2 in ESCs and does not negatively affect its protein levels. Therefore, a role for Rinf in regulating Tet2 protein levels is likely cell type (HEK293T versus ESC), dosage (overexpression versus endogenous), and context specific (pluripotent versus differentiated state). It is also possible that Idax and Rinf have unique functions or distinct effects on Tet enzymes in ESC versus during differentiation.
This study also has implications beyond ESCs. Nanog and Tet1/2 are expressed in germ cells, which are also in a pluripotent state (Hackett et al., 2012; Hayashi et al., 2007), so it will be of interest to investigate if Rinf plays similar roles in germ cells. Tet enzymes and Rinf are also expressed in various somatic cells. It will be worthy to explore if Rinf regulates recruitment of Tet proteins in those contexts, as is suggested in one study (Ma et al., 2017). Moreover, Rinf is deregulated in several malignancies (Astori et al., 2013; Knappskog et al., 2011; Pendino et al., 2009), and its transcriptional roles, as defined by this study, may have implications in the oncogenic gene expression programs and etiology of cancers. Finally, Rinf and Idax, by virtue of their genomic architecture and protein domain similarities, may functionally compensate for each other in cell types that express both. Although ESCs only express Rinf, studying the combined functions of Rinf and Idax in other cell types will elaborate more on their biological requirements.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, M.M.D. (meelad.dawlaty@einstein.yu.edu)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Rinf knockout ESCs
Wild-type mouse ESCs (Line: V6.5, Background: mixed 129/B6, Sex: male) were genetically manipulated by CRISPR/Cas9 to generate Rinf knockout (Rinf−/−) ESCs. Two pX330 vectors expressing Cas9 and gRNAs flanking exon 2 of Rinf were used for gene editing as described before (Wang et al., 2013). Targeted clones were screened by PCR or Southern blot using NsiI digest and a 3′ probe. Cycling conditions for PCR were 95°C 5min, (95°C 45sec, 58°C 45sec, 72°C 1min 30 s) X 35, 72°C 10min, 12°C Hold. Loss of Rinf was confirmed at mRNA and protein levels by RT-qPCR and western blot, respectively. All oligo sequences are provided in Supplemental Information. The Rinfv5/v5 ESC line was generated by targeting a V5 tag sequence after the start codon of Rinf in wild-type mouse ESCs (Line: V6.5, Background: mixed 129/B6, Sex: male), using a donor vector carrying a V5 tag sequence and 500bp flanking sequences of Rinf start codon as well as a gRNA directed against the start codon sequence. Properly targeted homozygous clones were screened by PCR and confirmed by sanger sequencing. The resulting ESC lines used in the study were Rinf−/− (Line: V6.5, Background: mixed 129/B6, Sex: male) and Rinfv5/v5 ESC (Line: V6.5, Background: mixed 129/B6, Sex: male).
SCID Mice
Severe Combined Immuno Deficient (SCID) mice (Strain: IcrTac:ICR-Prkdcscid, Genotype: sp/sp, Age: 8-weeks-old, Sex: Male) were purchased from Taconic (Cat# ICRSC-M) and used for teratoma formation assay as described in STAR Methods. Mice were housed in SPF barrier facility and used in experiments in accordance with our Institutional Animal Care and Use Committee (IACUC) approved protocols overseen by the Institute for Animal Studies at Albert Einstein College of Medicine.
METHOD DETAILS
Culture of mouse ESCs
All ESC lines were cultured on irradiated feeders or on gelatin (0.2%) coated plates in media containing serum/LIF. ESCs stably expressing Rinf or Tet1-CD were generated by transfecting Rinf−/− ESCs with PiggyBac-hygro-mRinf-V5 or PiggyBac-hygro-Flag-mTet1-CD or empty vector using Xfect mESC transfection reagent (Clontech) and selecting with hygromycin (125ug/mL) for 10 days. For RNA and DNA extraction, ESCs were pre-plated to remove feeders and then seeded on gelatin overnight before harvest. For embryoid body (EB) formation assays, pre-plated ESCs were seeded in media without LIF in hanging drops for 3 days followed by culturing on non-adherent plastic surface for 3 days. EBs were harvested on day 6 for analyses.
ChIP-seq and data analysis
ChIP-seq was performed on two independent wild-type V6.5 mESC lines and one Rinf−/− ESC line (negative control) as previously described (Johnson et al., 2007). Briefly, ESCs were cultured on gelatin, harvested, crosslinked, lysed, sonicated and subjected to ChIP using an anti-Rinf antibody (84546S, CST). Sequencing was performed at the Einstein Epigenomics core following their established protocols using Illumina HiSeq 2500 platform. Reads were mapped to the mouse genome (mm10) using the software Bowtie2 (VN: 2.2.3) with default (Langmead and Salzberg, 2012). The Rinf binding peaks were called with the software MACS2 using the input as controls and default parameters (Zhang et al., 2008), with the final peaks called from the merged reads of the two biological replicates. Application of the same pipeline generated < 300 peaks in the Rinf−/− ESC samples, only ~80 were also present in the WT samples. The final Rinf peaks were associated to genes and separated into promoter peaks (< +/− 2kb of transcription start sites; TSSs), gene body peaks and distal regulatory peaks (< +/−50 kb of genes). The gene body and distal regulatory peaks were considered as “enhancer” peaks. Motif analyses was performed by the HOMER (v 4.7) software (Heinz et al., 2010). Rinf bound genes were subjected to Gene Ontology analysis using DAVID software (Jiao et al., 2012). Rinf peaks were compared to those of pluripotency factors, Tets and histone marks using published datasets (Ma et al., 2011; Rockowitz and Zheng, 2015; Wu et al., 2011; Xiong et al., 2016). In comparison of peaks, overlapping peaks were defined as those sharing at least one base pair. The ChIP-seq read density heatmaps were generated by the software seqMINER (Ye et al., 2011) by sampling same number of total reads for each sample to account for different sequencing depths.
Gene expression profiling by RNA-seq and data analysis
Total RNA was extracted from two independent ESC of each genotype (Omega E.Z.N.A Total RNA kit), barcoded and used to prepare libraries. ERCC spike in controls were included. The libraries were subjected to 150 bp paired-end sequencing using an Illumina Next-Seq 500 platform at the Einstein Epigenomics core following their protocols. We generated ~25 million reads per sample. The reads were trimmed using trim galore (v 0.4.1, https://github.com/FelixKrueger/TrimGalore) to remove adapters and then mapped to mouse genome (mm10) by tophat software (v 2.0.13) with default parameters (Kim et al., 2013). The read pair numbers mapped to each gene in the Refseq gene annotation (downloaded from UCSC genome browser in 03/2017) were calculated with HTseq (v 0.6.1; Anders et al., 2015) using “–s reverse” parameter to obtain read counts for each gene. The Fragments Per Kilobase of transcript per Million (FPKMs) for each gene were calculated using cufflinks package (v 2.2.1; Trapnell et al., 2010). Read counts at the genes with FPKM > 1 were analyzed by the DESeq2 software (Love et al., 2014) for differential expression. We used False Discovery Rate (FDR) < 0.05 as criteria to identify differentially expressed genes (DEGs) between Rinf−/− and Rinf+/+ samples. Functional enrichment of DEGs was performed via DAVID (https://david.ncifcrf.gov/tools.jsp). Bubble plots were used to show enriched GO terms and KEGG pathways. In the bubble plots, enrichment factors were calculated as the ratio of gene counts that mapped to a certain pathway versus the total gene number of that pathway. Gene expression patterns were identified by hierarchical clustering and displayed as heatmap using R.
For transcriptomic analysis of ESCs during differentiation to EBs, RNA was isolated at three time points (day 0, 3, 6) of differentiation to EBs and subjected to RNA-seq (75bp single-end sequencing, Illumina Next-Seq 500 platform) as described above. We generated ~30 million reads per sample. Data analysis and identification of differentially expressed genes between the two genotypes for each time point was performed using similar pipelines and approaches as described for ESCs above.
RT-qPCR
1.5 μg of RNA extracted from feeder free ESCs or day 6 EBs (Omega E.Z.N.A Total RNA kit) was used to synthesize cDNA (Superscript III First-Strand synthesis system, Invitrogen). Real time quantitative PCR was performed using SYBR green master mix (Applied Biosystems) in QuantStudio 6 Flex Real-Time PCR system following standard protocols. Relative gene expression level was analyzed by comparative Ct method and was normalized to Gapdh. Sequences of primers used are listed in Supplemental Information (Table S1).
ChIP-qPCR
ChIP experiments were performed on ESCs cultured on gelatin following published protocols (Johnson et al., 2007) using antibodies against Rinf (84546S, CST), Tet2 (ab124297, abcam) and Nanog (A300–397A, Bethyl Laboratories). DNA concentration was measured using Qubit 2.0 Fluorometer (Invitrogen). Enrichment at specific loci was quantified by qPCR as mentioned above. ChIP-qPCR signals were calculated as fold enrichment using IgG as control. Primer sequences and their genomic location are listed in Supplemental Information (Tables S1 and S2).
Immunoprecipitation
For immunoprecipitation (IP) of endogenous proteins, nuclear extracts were isolated from Rinfv5/v5 ESCs and treated with benzonase nuclease (Millipore E1014, conc. 75 units/IP) as described before (Chrysanthou et al., 2018) and incubated with 3 μg of antibody (anti-V5, 13202S, CST; Rabbit-IgG, 3900S, CST) crosslinked to Protein G-conjugated magnetic beads (Dynabeads protein G, Invitrogen) overnight at 4°C. IgG was used as control. Immunocomplexes were washed with buffer containing 20mM HEPES, pH 7.6, 10% glycerol, 100 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA. The proteins were eluted in 2X Laemelli buffer at 95°C and analyzed by western blot as above. For IP of exogenous proteins, HEK293T cells were transfected with plasmids expressing V5-Rinf, Nanog, Tet1-CD or Tet2 using X-tremegene GENE DNA transfection reagent (Roche). IP was performed on benzonase nuclease (Millipore E1014, conc. 75 units/IP) -treated total cell lysate and analyzed by western blot as described before (Ko et al., 2013). Briefly, cells were lysed in lysis buffer (50 mM Tris- HCl, pH 7.4, 150 mM NaCl, 1mM EDTA and 1% Triton X-100) supplemented with PIC and subjected to IP as described above using anti-V5 antibody. IgG was used as control. The protein-bead complexes were washed five times with wash buffer (50 mM Tris- HCl, pH 7.4, 150 mM NaCl and 0.05% Triton X-100) and eluted in 2X Laemelli buffer. The eluted proteins were analyzed by western blot as above. For in vitro protein interactions, mouse Rinf, Nanog, and Tet2-CD transgenes were cloned into pRUTH5 vector and transcribed and translated by TNT Quick Coupled Transcription/Translation System (Promega L1170). Reactions were mixed and subjected to co-IP using anti-Rinf and analyzed by western blot as above.
Western blot and cell fractionation
For western blots, cells were lysed in RIPA buffer (50 mM Tris-HCl, pH 7.4, 250 mM NaCl, 2% Nonidet-P40, 2.5 mM EDTA, 0.1% SDS,0.5% DOC) supplemented with PIC and PMSF, resolved on 6%–10% SDS-PAGE (Mini-PROTEAN electrophoresis chamber, Bio-Rad), and transferred on PVDF membranes (Mini Trans-Blot apparatus, Bio-Rad) following manufacturer’s protocols. Membranes were blocked in 5% milk in PBS with 0.1% tween (PBST) and incubated overnight at 4°C, or for 1hr at room temperature with primary antibodies (Rinf 84546S, CST 1:1000; Nanog A300–397A, Bethyl Laboratories 1:2000; Oct4 SC-5279, SantaCruz 1:500; H3 ab1791, abcam 1:15000; Tet2 ab124297, abcam 1:1000; Flag 14739S, CST 1:1000; Actin AC-15, abcam 1:40000). Secondary antibody incubations (HRP-anti mouse, 401253, or anti rabbit, 401393, 1:5000, CalBiochem) were carried out for 1hr at room temperature. For quantifying Rinf protein levels in chromatin bound and soluble fractions of the cell, ESC lysate was fractionated as described before (Zhang et al., 2016). Each fraction was analyzed by western blot using anti-Rinf antibody. In all experiments Actin or H3 were used as loading controls.
Immunofluorescence
Mouse ESCs cultured on coverslips were washed twice with PBS and fixed with 4% paraformaldehyde for 15 mins at room temperature. Cells were permeabilized with 0.1% Triton X-100 in PBS for 15 mins and blocked in 0.1% Tween, 5% BSA in PBS for 30 min at room temperature. Primary antibody (anti-Rinf, 84546S, CST, 1:1000) and secondary antibody (Alexa Flour 488 -anti-rabbit, A21206, Life Technologies, 1:1000) incubations were carried out at room temperature for an hour. Nuclei were stained with DAPI (1:1000, 5ug/ml stock). Likewise, neural progenitors were stained with Nestin (MAB533, EMD Millipore Corp., 1:200) and Sox2 (AB5603, EMD Millipore Corp., 1:200) as described above. Similarly, ESCs cultured in TSC media were stained with anti-E-cadherin (610182, BD Bioscience, 1:200) and DAPI (1:1000, 5ug/ml stock). Microscopy was performed after final washes using Zeiss Axio Observer.A1 inverted microscope.
Dual luciferase reporter assay
Enhancers of Nanog (chr6:122662781–122663192), Sox2 (chr3:34646228–34646529) or Tet1(chr10:62895488–62895876) were cloned upstream of the minimal promoter of PGL4.23 vector. In separate experiments ~200,000 ESCs were transfected with each vector or empty vector, along with pRL-CMV-Renilla using Xfect transfection reagent (Clonetech). The luciferase activity of firefly and Renilla was measured after 48 hours using Dual-luciferase reporter assay kit (Promega) following the manufacturer’s protocol using BioTek Synergy B multimode plate reader. As a transfection efficiency control, the firefly luciferase activity was divided by Renilla luciferase activity and the data were represented as relative luminescence unit (RLU) by normalizing the luciferase activity with that of empty vector.
Dot Blot for 5mC and 5hmC
Genomic DNA was isolated from ESCs, purified by phenol:chloroform:isoamyl alcohol and then analyzed by dot blot using anti 5hmC antibody (Active Motif 1:10,000) or anti 5mC (CST, 1:1000) following manufacturers’ protocols. Signal intensity was quantified by ImageJ software (Schneider et al., 2012) and average values of replicates were plotted.
Teratoma formation assay
1.5 x106 ESCs were injected subcutaneously into the flank of a SCID mouse (Taconic). Three mice per each ESC line were used. 4 weeks after injection mice were euthanized and tumors were removed and fixed in formalin for two days. They were imbedded in paraffin, sectioned and stained with hematoxylin and eosin for histological analysis.
In vitro differentiation to neural progenitors (NPs) and trophoblast stem cells (TSCs)
ESCs were differentiated to EBs by hanging drop for 4 days. EBs were transferred to tissue culture plates, allowed to attach for a day and then cultured in ITSFn media for 8 days. Upon passaging, neural precursors were propagated on poly-D-ornithine and laminin coated plates in N2 media containing bFGF (5 ng/ml), EGF (20 ng/ml) and Laminin (1 μg/ml). For differentiation to trophoblast cells, ESCs were cultured for 3 days on gelatin in TSC media (70% pre-conditioned media on MEFs, 30% TS base media {20% FBS, 1mM sodium pyruvate, 50uM β-mercaptoethanol, 1x PenStrep in RPMI 1640}, 1 ug/ml heparin, 25 ng/ml FGF) as described (Chrysanthou et al., 2018).
QUANTIFICATION AND STATISTICAL ANALYSIS
One-way-Anova test and GraphPad Prism 7 software was used for calculating statistical significance in RT-qPCR and ChIP-qPCR analyses and luciferase assays. Statistical methods for analysis of genome wide datasets involving RNA-seq and ChIP-seq are explained in detail under the respective sections as part of the detailed methods.
DATA AND CODE AVAILABILITY
The ChIP-seq and RNA-seq datasets have been deposited in the Gene Expression Omnibus (GEO) database (Accession number GSE132025).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Rinf | Cell signaling Technology | Cat# 84546; RRID:AB_2800040 |
| Anti-V5 | Cell signaling Technology | Cat# 13202; RRID:AB_2687461 |
| Anti-Flag | Cell signaling Technology | Cat# 14793; RRID:AB_2572291 |
| Anti-Tet1 | GenTex | Cat# GTX125888; RRID:AB_11164485 |
| Anti-Tet2 | Abcam | Cat# ab124297; RRID:AB_2722695 |
| Anti-Nanog | Bethyl Laboratories | Cat# A300-397A; RRID:AB_386108 |
| Anti-Oct4 | SantaCruz | Cat# SC-5279; RRID:AB_628051 |
| Anti-H3 | Abcam | Cat# ab1791; RRID:AB_302613 |
| Anti-Actin | Abcam | Cat# ab82618; RRID:AB_1658432 |
| Anti-Nestin | R&D | Cat# MAB533; RRID:AB_2070659 |
| Anti-Sox2 | EMD Millipore Corp. | Cat# AB5603; RRID:AB_2286686 |
| Anti-E-cadherin | BD Biosciences | Cat# 610182; RRID:AB_397581 |
| Anti-Dnmt3a | Novus Biologicals | Cat# NB120-13888; RRID:AB_789607 |
| Anti-Dnmt3b | Santa Cruz | Cat# SC-376043; RRID:AB_10988201 |
| Anti-5hmC | Active Motif | Cat# 39769; RRID:AB_10013602 |
| Anti-5mC | Cell Signaling Technology | Cat# 28692; RRID:AB_2798962 |
| Goat Anti-Rabbit IgG-HRP | Millipore | Cat# 401393-2ML; RRID:AB_10683386 |
| Goat Anti-Mouse IgG-HRP | Millipore | Cat# 401253; RRID:AB_437779 |
| Alexa Flour 488 -anti-rabbit | Life Technologies | Cat# A-21206; RRID:AB_2535792 |
| Alexa Flour 594 -anti-mouse | Life Technologies | Cat# A-11005; RRID:AB_2534073 |
| Critical Commercial Assays | ||
| Dual Luciferase reporter assay kit | Promega | E1910 |
| E.Z.N.A. Total RNA kit | Omega | R6834-02 |
| Superscript III first strand | Invitrogen | 18080-400 |
| Qubit dsDNA HS assay kit | Invitrogen | Q32851 |
| Xfect mESC polymer | Clonetech | 631320 |
| XtremeGene 9 DNA transfection reagent | Roche | 06365787001 |
| TNT Coupled transcription and translation kit | Promega | L1170 |
| Deposited Data | ||
| ChIP-seq data | This paper | GEO: GSE132025 |
| RNA-seq data | This paper | GEO: GSE132025 |
| Experimental Models: Cell Lines | ||
| Rinf−/− ESC | This paper | N/A |
| RinfV5/V5 ESC | This paper | N/A |
| Experimental Models: Organisms/Strains | ||
| SCID mice | Taconic | Model#ICRSC-M |
| Oligonucleotides | ||
| RT-qPCR primers, see Table S1 | This paper | N/A |
| ChIP-qPCR Primers, see Tables S1 and S2 | This paper | N/A |
| Genotyping Primers, see Table S1 | This paper | N/A |
| gRNA oligos for gene targeting, see Table S1 | This paper | N/A |
| Recombinant DNA | ||
| PiggyBac-hygro | This paper | N/A |
| PiggyBac-V5-mRinf-hygro | This paper | N/A |
| PiggyBac-mTet1-CD-hygro | This paper | N/A |
| PiggyBac-mTet2-hygro | This paper | N/A |
| FUW-Nanog | This paper | N/A |
| FUW-Oct4 | This paper | N/A |
| pGL4.23-empty vector | Promega | E8411 |
| pGL4.75-empty vector | Promega | E6931 |
| pGL4.23-Tet1 Enhancer | This paper | N/A |
| pGL4.23-Nanog Enhancer | This paper | N/A |
| pGL4.23-Sox2 Enhancer | This paper | N/A |
| pRUTH5-mRinf | This paper | N/A |
| pRUTH5-mNanog | This paper | N/A |
| pRUTH5-mTet2-CD (aa1040-1910) | This paper | N/A |
| Software and Algorithms | ||
| Trim galore v0.4.1 | Github | https://github.com/FelixKrueger/TrimGalore |
| MACS2 v2.1.0 | Zhang et al., 2008 | https://github.com/taoliu/MACS/ |
| seqMINER v1.2.1 | Ye etal., 2011 | http://bips.u-strasbg.fr/ |
| HOMER v4.7 | Heinz et al., 2010 | http://homer.ucsd.edu/homer/ |
| Bowtie v2.2.3 | John Hopkins University | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Tophat v2.0.13 | John Hopkins University | https://ccb.jhu.edu/software/tophat/index.shtml |
| HTSeq v0.6.1 | Anders et al., 2015 | https://github.com/simon-anders/htseq |
| Cufflinks v2.2 | Trapnell et al., 2010 | http://cole-trapnell-lab.github.io/cufflinks/ |
| DESeq2 v1.20.0 | Love et al., 2014 | http://www.bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Integrative Genomics Viewer (IGV) v2.5.0 | Broad Institute | http://software.broadinstitute.org/software/igv/ |
| DAVID 6.8 | Jiao et al., 2012 | https://david.ncifcrf.gov/ |
| GraphPad Prism 7 | GraphPad | https://www.graphpad.com/ |
| ImageJ | Schneider et al., 2012 | https://imagei.nih.gov/ii/ |
Highlights.
Rinf is enriched at promoters and enhancers of pluripotency and Tet genes
Rinf forms a complex with Nanog, Oct4, Tet1, and Tet2
Rinf positively regulates transcription of pluripotency genes and Tet enzymes
Rinf loss compromises gene expression and differentiation programs in ESCs
ACKNOWLEDGMENTS
We thank the Einstein Epigenomics and Histopathology cores for NextGen sequencing and sectioning tissues, respectively. We are grateful to H. Su, H. Belalcazar, and B. Cattau for help with gene targeting and cloning. We also thank M. Suzuki (Greally lab), Y. Zhu (Suh lab), and F. Soldner for helpful discussions. M.M.D. is supported by Sidney Kimmel Foundation, Leukemia Research Foundation, R01GM122839, R01HL148852, NYSDOH/NYSTEM contract C32589GG, and funds from Albert Einstein College of Medicine Stem Cell Institute and Genetics Department. R.L. and S.C. are supported by The Einstein Training Program in Stem Cell Research from the Empire State Stem Cell Fund NYSDOH Contract C30292GG. D.S. is supported by R01GM108646. A.C. is supported by NIH/NEI EY014237 and EY012200.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.07.080.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Anders S, Pyl PT, and Huber W (2015). HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astori A, Fredly H, Aloysius TA, Bullinger L, Mansat-De Mas V, de la Grange P, Delhommeau F, Hagen KM, Récher C, Dusanter-Fourt I, et al. (2013). CXXC5 (retinoid-inducible nuclear factor, RINF) is a potential therapeutic target in high-risk human acute myeloid leukemia. Oncotarget 4, 1438–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrysanthou S, Senner CE, Woods L, Fineberg E, Okkenhaug H, Burge S, Perez-Garcia V, and Hemberger M (2018). A Critical Role of TET1/2 Proteins in Cell-Cycle Progression of Trophoblast Stem Cells. Stem Cell Reports 10, 1355–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty MM, Breiling A, Le T, Raddatz G, Barrasa MI, Cheng AW, Gao Q, Powell BE, Li Z, Xu M, et al. (2013). Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with post-natal development. Dev. Cell 24, 310–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty MM, Breiling A, Le T, Barrasa MI, Raddatz G, Gao Q, Powell BE, Cheng AW, Faull KF, Lyko F, and Jaenisch R (2014). Loss of Tet enzymes compromises proper differentiation of embryonic stem cells. Dev. Cell 29, 102–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabole N, Tischler J, Hackett JA, Kim S, Tang F, Leitch HG, Magnúsdóttir E, and Surani MA (2013). Prdm14 promotes germline fate and naive pluripotency by repressing FGF signalling and DNA methylation. EMBO Rep. 14, 629–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett JA, Zylicz JJ, and Surani MA (2012). Parallel mechanisms of epigenetic reprogramming in the germline. Trends Genet. 28, 164–174. [DOI] [PubMed] [Google Scholar]
- Hanna JH, Saha K, and Jaenisch R (2010). Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell 143, 508–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, de Sousa Lopes SMC, and Surani MA (2007). Germ cell specification in mice. Science 316, 394–396. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, and Young R (2008). Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell 132, 567–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X, Sherman BT, Huang W, Stephens R, Baseler MW, Lane HC, and Lempicki RA (2012). DAVID-WS: a stateful web service to facilitate gene/protein list analysis. Bioinformatics 28, 1805–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, and Wold B (2007). Genome-wide mapping of in vivo protein-DNA interactions. Science 316, 1497–1502. [DOI] [PubMed] [Google Scholar]
- Kim MS, Yoon SK, Bollig F, Kitagaki J, Hur W, Whye NJ, Wu Y-P, Rivera MN, Park JY, Kim H-S, et al. (2010). A novel Wilms tumor 1 (WT1) target gene negatively regulates the WNT signaling pathway. J. Biol. Chem 285, 14585–14593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, and Salzberg SL (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HY, Yang DH, Shin SW, Kim MY, Yoon JH, Kim S, Park HC, Kang DW, Min D, Hur MW, and Choi KY (2014). CXXC5 is a transcriptional activator of Flk-1 and mediates bone morphogenic protein-induced endothelial cell differentiation and vessel formation. FASEB J. 28, 615–626. [DOI] [PubMed] [Google Scholar]
- Kim M-Y, Kim H-Y, Hong J, Kim D, Lee H, Cheong E, Lee Y, Roth J, Kim DG, Min S, and Choi K-Y (2016). CXXC5 plays a role as a transcription activator for myelin genes on oligodendrocyte differentiation. Glia 64, 350–362. [DOI] [PubMed] [Google Scholar]
- Knappskog S, Myklebust LM, Busch C, Aloysius T, Varhaug JE, Lønning PE, Lillehaug JR, and Pendino F (2011). RINF (CXXC5) is overexpressed in solid tumors and is an unfavorable prognostic factor in breast cancer. Ann. Oncol 22, 2208–2215. [DOI] [PubMed] [Google Scholar]
- Ko M, An J, Bandukwala HS, Chavez L, Aijö T, Pastor WA, Segal MF, Li H, Koh KP, Lähdesmäki H, et al. (2013). Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature 497, 122–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, et al. (2011). Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell 8, 200–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima T, Shimazui T, Hinotsu S, Joraku A, Oikawa T, Kawai K, Horie R, Suzuki H, Nagashima R, Yoshikawa K, et al. (2009). Decreased expression of CXXC4 promotes a malignant phenotype in renal cell carcinoma by activating Wnt signaling. Oncogene 28, 297–305. [DOI] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-H, Kim M-Y, Kim H-Y, Lee Y-M, Kim H, Nam KA, Roh MR, Min S, Chung KY, and Choi K-Y (2015). The Dishevelled-binding protein CXXC5 negatively regulates cutaneous wound healing. J. Exp. Med 212, 1061–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Ye X, Peng X, Deng Y, Yuan W, Li Y, Mo X, Wang X, Wan Y, Liu X, et al. (2014). CXXC5 regulates differentiation of C2C12 myoblasts into myocytes. J. Muscle Res. Cell Motil. 35, 259–265. [DOI] [PubMed] [Google Scholar]
- Liu N, Wang M, Deng W, Schmidt CS, Qin W, Leonhardt H, and Spada F (2013). Intrinsic and extrinsic connections of Tet3 dioxygenase with CXXC zinc finger modules. PLoS One 8, e62755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Swigut T, Valouev A, Rada-Iglesias A, and Wysocka J (2011). Sequence-specific regulator Prdm14 safeguards mouse ESCs from entering extraembryonic endoderm fates. Nat. Struct. Mol. Biol 18, 120–127. [DOI] [PubMed] [Google Scholar]
- Ma S, Wan X, Deng Z, Shi L, Hao C, Zhou Z, Zhou C, Fang Y, Liu J, Yang J, et al. (2017). Epigenetic regulator CXXC5 recruits DNA demethylase Tet2 to regulate TLR7/9-elicited IFN response in pDCs. J. Exp. Med 163, 1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, and Yamanaka S (2003). The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113, 631–642. [DOI] [PubMed] [Google Scholar]
- Pastor WA, Aravind L, and Rao A (2013). TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat. Rev. Mol. Cell Biol. 14, 341–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendino F, Nguyen E, Jonassen I, Dysvik B, Azouz A, Lanotte M, Ségal-Bendirdjian E, and Lillehaug JR (2009). Functional involvement of RINF, retinoid-inducible nuclear factor (CXXC5), in normal and tumoral human myelopoiesis. Blood 113, 3172–3181. [DOI] [PubMed] [Google Scholar]
- Rockowitz S, and Zheng D (2015). Significant expansion of the REST/NRSF cistrome in human versus mouse embryonic stem cells: potential implications for neural development. Nucleic Acids Res. 43, 5730–5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, and Meissner A (2013). DNA methylation: roles in mammalian development. Nat. Rev. Genet 14, 204–220. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, and Pachter L (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, and Jaenisch R (2013). One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, and Young RA (2013). Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SC, and Zhang Y (2010). Active DNA demethylation: many roads lead to Rome. Nat. Rev. Mol. Cell Biol. 11, 607–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, D’Alessio AC, Ito S, Xia K, Wang Z, Cui K, Zhao K, Sun YE, and Zhang Y (2011). Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature 473, 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J, Zhang Z, Chen J, Huang H, Xu Y, Ding X, Zheng Y, Nishinakamura R, Xu G-L, Wang H, et al. (2016). Cooperative Action between SALL4A and TET Proteins in Stepwise Oxidation of 5-Methylcytosine. Mol. Cell 64, 913–925. [DOI] [PubMed] [Google Scholar]
- Yamaji M, Ueda J, Hayashi K, Ohta H, Yabuta Y, Kurimoto K, Nakato R, Yamada Y, Shirahige K, and Saitou M (2013). PRDM14 ensures naive pluripotency through dual regulation of signaling and epigenetic pathways in mouse embryonic stem cells. Cell Stem Cell 12, 368–382. [DOI] [PubMed] [Google Scholar]
- Ye T, Krebs AR, Choukrallah M-A, Keime C, Plewniak F, Davidson I, and Tora L (2011). seqMINER: an integrated ChIP-seq data interpretation platform. Nucleic Acids Res. 39, e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, and Liu XS (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Xia W, Wang Q, Towers AJ, Chen J, Gao R, Zhang Y, Yen CA, Lee AY, Li Y, et al. (2016). Isoform Switch of TET1 Regulates DNA Demethylation and Mouse Development. Mol. Cell 64, 1062–1073. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The ChIP-seq and RNA-seq datasets have been deposited in the Gene Expression Omnibus (GEO) database (Accession number GSE132025).