SUMMARY
Proper transcriptome reprogramming is critical for hosts to launch an effective defense response upon pathogen attack. How immune-related genes are regulated at the posttranscriptional level remains elusive. We demonstrate here that P-bodies, the non-membranous cytoplasmic ribonucleoprotein foci related to 5′-to-3′ mRNA decay, are dynamically modulated in plant immunity triggered by microbe-associated molecular patterns (MAMPs). The DCP1-DCP2 mRNA decapping complex, a hallmark of P-bodies, positively regulates plant MAMP-triggered responses and immunity against pathogenic bacteria. MAMP-activated MAP kinases directly phosphorylate DCP1 at the serine237 residue, which further stimulates its interaction with XRN4, an exonuclease executing 5′-to-3′ degradation of de-capped mRNA. Consequently, MAMP treatment potentiates DCP1-dependent mRNA decay on a specific group of MAMP-downregulated genes. Thus, the conserved 5′-to-3′ mRNA decay elicited by the MAMP-activated MAP kinase cascade is an integral part of plant immunity. This mechanism ensures a rapid posttranscriptional downregulation of certain immune-related genes that may otherwise negatively impact immunity.
Graphical Abstract
In Brief
Yu et al. show that P-body core components DCP1 and DCP2 positively regulate plant pattern-triggered immunity. DCP1 phosphorylation by immune-activated MAPKs contributes to P-body disassembly and mRNA decay on a subset of immune-regulated genes, revealing mRNA-decay-mediated posttranscriptional regulation is an integral part of plant immunity.
INTRODUCTION
Detecting the presence of microbial components is crucial for hosts to launch an effective defense response against pathogen invasion. Plant pattern-triggered immunity (PTI) is initiated by the recognition of microbe-associated molecular patterns (MAMPs) by cell-surface-resident pattern recognition receptors (PRRs) (Couto and Zipfel, 2016; Yu et al., 2017). Flagellin-sensing 2 (FLS2) and elongation factor EF-TU receptor (EFR), two PRRs from Arabidopsis, recognize flg22 and elf18, the synthetic peptides derived from bacterial MAMP flagellin and EF-Tu, respectively (Gómez-Gómez and Boller, 2000; Zipfel et al., 2006). Both FLS2 and EFR are leucine-rich repeat-receptor-like kinases (LRR-RLKs) and heterodimerize with the shared co-receptor brassinosteroid insensitive 1-associated kinase 1 (BAK1) family LRR-RLKs to form an active PRR complex (Chinchilla et al., 2007; Heese et al., 2007; Sun et al., 2013). Members of the Botrytis-induced kinase 1 (BIK1) family receptor-like cytoplasmic kinases (RLCKs) associate with multiple PRR complexes and bifurcate the signaling to downstream components (Kadota et al., 2014; Li et al., 2014c; Lu et al., 2010; Zhang et al., 2010). The PRR complex undergoes layered positive and negative regulations (Böhm et al., 2014; Couto and Zipfel, 2016), and activates two major intracellular signaling modules, namely, mitogen-activated protein kinases (MAPKs) and calcium-dependent protein kinases (CDPKs) that play a primary role in transcriptional control of immune-related gene expression (Asai et al., 2002; Boudsocq et al., 2010).
In addition to the transcriptional regulation, the posttranscriptional control adds another crucial layer of regulation of immune gene expression (Carpenter et al., 2014; Staiger et al., 2013). As an important mechanism in the posttranscriptional regulation, mRNA decay could degrade both aberrant and normal transcripts (Decker and Parker, 2012). Degradation of mRNA is typically initiated with deadenylation, followed by exosome-mediated 3′-to-5′ exonucleolytic decay. More often, the deadenylated mRNAs are decapped by the decapping enzyme decapping 2 (DCP2) along with its co-activator DCP1 and other cofactors and then degraded by 5′-to-3′ exonuclease exoribonuclease (XRN) (Decker and Parker, 2012; Schoenberg and Maquat, 2012). The decapping machinery, together with associated non-translated mRNAs, are conditionally assembled into RNA Processing bodies (P-bodies). P-bodies, a type of ribonucleoprotein particle (RNP) granules, are microscopically visible and non-membranous cytoplasmic foci consisting of aggregations of translationally repressed mRNAs bound by various catalytic and regulatory proteins and frequently non-coding RNAs (Mitchell and Parker, 2014). Increasing evidence indicates that P-bodies act as a storage compartment for translationally repressed mRNAs, including in the plant system (Brengues et al., 2005; Hubstenberger et al., 2017; Jang et al., 2019).
P-bodies have been observed in a wide range of organisms, including yeast, metazoans, and plants (Decker and Parker, 2012; Schoenberg and Maquat, 2012; Xu and Chua, 2011). Mutations in DCP1 and DCP2 in Arabidopsis cause growth defects, suggesting the importance of P-body core components in regulating gene expression essential for plant development (Xu et al., 2006). In addition, P-body components are suggested to be involved in abiotic stresses, such as drought, cold, and salinity (Perea-Resa et al., 2016; Xu and Chua, 2012). How P-body assembly is initiated and participates in a specific physiological response remain elusive. In this study, we observed rapid P-body disassembly and re-assembly upon MAMP elicitation, indicating the dynamic fate changes of mRNAs stored in P-bodies as part of immune responses. PRR-activated MAPKs directly phosphorylate DCP1 and stimulate DCP1 dissociation from DCP2 and association with exoribonuclease 4 (XRN4). Consistent with the P-body dynamics as an integral part of plant immunity, MAMP treatment potentiates DCP1-dependent mRNA decay on a subset of MAMP-downregulated genes. Such mRNA-decay-mediated posttranscriptional regulation likely underpins the ability of hosts to launch an expeditious and effective immune response by rapid reprogramming of primary immune genes.
RESULTS
Pathogen-Induced P-body Dynamics in Arabidopsis
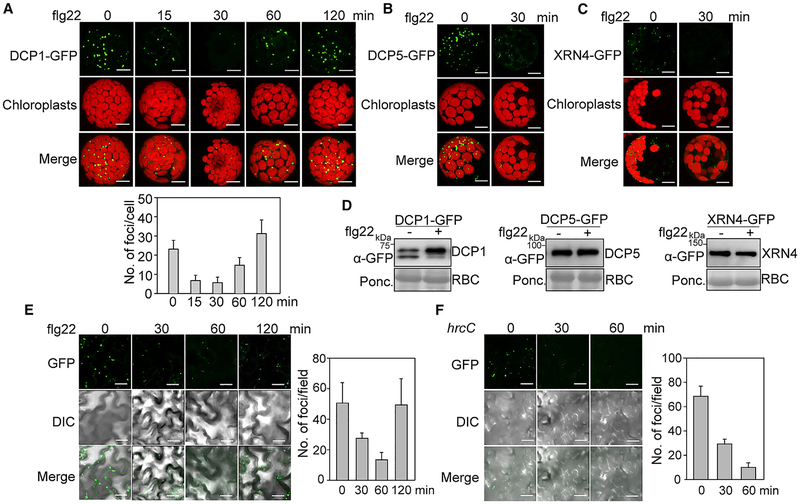
We have deployed a series of random mutagenesis screens for components involved in plant PTI based on the transcriptional response of an immune reporter gene (Feng et al., 2015; Li et al., 2014a). A candidate gene identified from the screen encodes a protein localizing to P-bodies (study in progress). To reveal the potential link between P-bodies and plant immunity, we examined the dynamics of P-body assembly upon PTI elicitation. Interestingly, we observed a significant reduction of DCP1-GFP-labeled P-bodies at 15 and 30 min after flg22 treatment in Arabidopsis Col-0 protoplasts. The DCP1-GFP-labeled P-bodies re-appeared at 60 min after flg22 treatment (Figure 1A). The flg22-triggered P-body disassembly was not observed in the fls2 mutant, indicating that this dynamics is specifically triggered by the flg22 recognition (Figure S1A). Similarly, flg22 treatment for 30 min led to the disassembly of P-bodies labeled with DCP5-GFP (Figure 1B) or XRN4-GFP (Figure 1C). Apparently, the reduction of P-bodies was not due to the reduced expression of DCP1-GFP, DCP5-GFP, or XRN4-GFP proteins (Figure 1D), suggesting that the disappearance of P-bodies was caused by the P-body disassembly. Furthermore, elf18 treatment also induced the disappearance of DCP1-GFP-, DCP5-GFP-, or XRN4-GFP-associated P-bodies (Figure S1B). We further generated transgenic plants expressing DCP1-GFP under the control of the 35S promoter. The number of DCP1-GFP-labeled P-bodies in transgenic plants was reduced upon flg22 treatment within 30 to 60 min (Figure 1E). In addition, infiltration with a non-pathogenic bacterium Pseudomonas syringae pv. tomato DC3000 (Pst) type III secretion mutant, hrcC, which carries multiple MAMPs but does not secrete type III effectors, also triggered the disappearance of P-bodies in DCP1-GFP transgenic plants (Figure 1F). However, flg22-induced P-body disappearance was not observed with PAT1-GFP-labeled P-bodies, likely due to the enhanced accumulation of PAT1 proteins and/or differential response of different P-body species (e.g., PAT1 versus DCP1/DCP5/XRN4) to flg22 treatment (Figures S1C and S1D) (Roux et al., 2015). Together, the results indicate that the presence of P-bodies is dynamically modulated in response to MAMP perception.
Figure 1. Pathogen-Induced P-Body Dynamics in Arabidopsis.
(A) DCP1-GFP-labeled P-body dynamics upon flg22 treatment in Arabidopsis protoplasts. Protoplasts from wild-type (WT) Col-0 were expressed with DCP1-GFP, and treated with 100 nM flg22 for indicated times. Images were taken with a confocal microscope. Autofluorescence of chloroplasts is shown in the middle panels. Bar, 10 μm. Quantification of DCP1-GFP-labeled cytoplasmic foci with ImageJ 1.49v software is shown on the bottom.
(B and C) The flg22-triggered P-body disassembly as indicated by DCP5-GFP (B)- or exoribonuclease 4 (XRN4)-GFP (C)-labeled cytoplasmic foci. Protoplasts expressing DCP5-GFP or XRN4-GFP were treated with 100 nM flg22 for 30 min. Bar, 10 μm.
(D) Expression of DCP1-GFP, DCP5-GFP, and XRN4-GFP proteins without and with flg22 treatment. Protoplasts expressing DCP1-GFP, DCP5-GFP, or XRN4-GFP were treated without or with 100 nM flg22 for 30 min, and resulting protein extracts were subjected to immunoblotting (IB) with an α-GFP antibody. Protein loading is shown by Ponceau S staining for RuBisCo (RBC).
(E) The flg22-induced P-body dynamics in 35S::DCP1-GFP transgenic plants. One-week-old seedlings were treated with 100 nM flg22 for indicated times. Bar, 20 μm. DIC is differential interference contrast microscopy. Quantification of DCP1-GFP-labeled fluorescence foci is shown on the right.
(F) P-body dynamics induced by Pst hrcC infection. Leaves of 4-week-old 35S::DCP1-GFP transgenic plants were hand-inoculated with Pst hrcC at OD600 = 0.1, and imaged at the indicated time points. Quantification is shown on the right.
Data in (A), (E), and (F) are shown as mean ± SD (n = 10). The above experiments were repeated three times with similar results.
See also Figure S1.
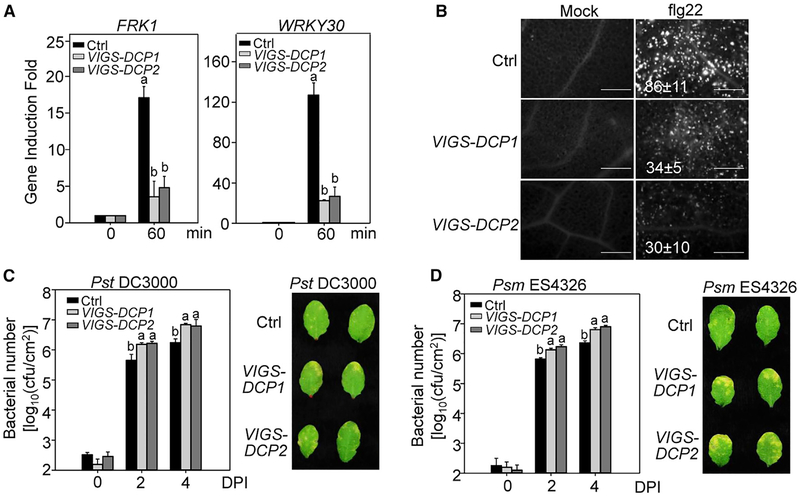
DCP1 and DCP2 Positively Regulate Plant Immunity
The null mutants of dcp1 or dcp2 are post-embryonic lethal (Xu et al., 2006). To elucidate the functional relevance of P-body components in plant immunity, we silenced DCP1 or DCP2 in wild-type (WT) Col-0 plants with virus-induced gene silencing (VIGS) after germination at the 2-week-old seedling stage. Silencing of DCP1 or DCP2 did not cause detectable growth defects (Figures S1E and S1F), which is consistent with the hypothesis that the seedling lethality of dcp1 and dcp2 is due to the accumulation of certain proteins whose mRNAs are normally degraded after germination (Xu and Chua, 2012). Significantly, the flg22-induced expression of FRK1 and WRKY30, two PTI marker genes, was reduced in DCP1- and DCP2-silenced plants compared to plants inoculated with a GFP vector control (Figure 2A). In addition, DCP1- or DCP2-silenced plants showed compromised flg22-induced callose deposition, a late PTI response, compared to control plants (Figure 2B). Furthermore, both DCP1- and DCP2-silenced plants were more susceptible to the infection by the virulent bacteria Pst (Figure 2C) and P. syringae pv. maculicola ES4326 (Psm) (Figure 2D) than control plants at 2 and 4 days post-inoculation (dpi). The inoculated leaves of DCP1-and DCP2-silenced plants developed more severe chlorosis than control plants at 4 dpi (Figures 2C and 2D). Thus, DCP1 and DCP2, the integral components of P-bodies, positively contribute to plant immunity.
Figure 2. DCP1 and DCP2 Positively Regulate Plant Immunity.
(A) Reduced immune gene expression in DCP1- and DCP2-silenced plants. Leaves from VIGS-silenced DCP1 and DCP2 plants at the 4-week-old stage were hand-infiltrated with 100 nM flg22 for 60 min. The VIGS vector containing a GFP fragment was used as control (Ctrl). Data were normalized to the expression of ACTIN2 in qRT-PCR analysis.
(B) Compromised flg22-triggered callose deposition in DCP1- and DCP2-silenced plants. Callose deposits were stained with aniline blue 24 h after treatment with 100 nM flg22 or H2O. The number of callose deposits per mm2 is shown as mean ± SD from three biological repeats. Bar, 0.5mm.
(C) Enhanced susceptibility to Pst infection in DCP1- and DCP2-silenced plants. Leaves from 4-week-old plants were hand-inoculated with Pst at OD600 = 5 × 10−4, and bacterial counting was performed at 0, 2, and 4 days post-inoculation (dpi) (left). Pictures of inoculated leaves were taken at 4 dpi (right).
(D) Enhanced susceptibility to Psm infection in DCP1- and DCP2-silenced plants.
Data in (A)–(D) are shown as mean ± SD from three independent repeats. Different letters indicate significant differences according to one-way ANOVA test (p < 0.01). The above experiments were repeated three times with similar results.
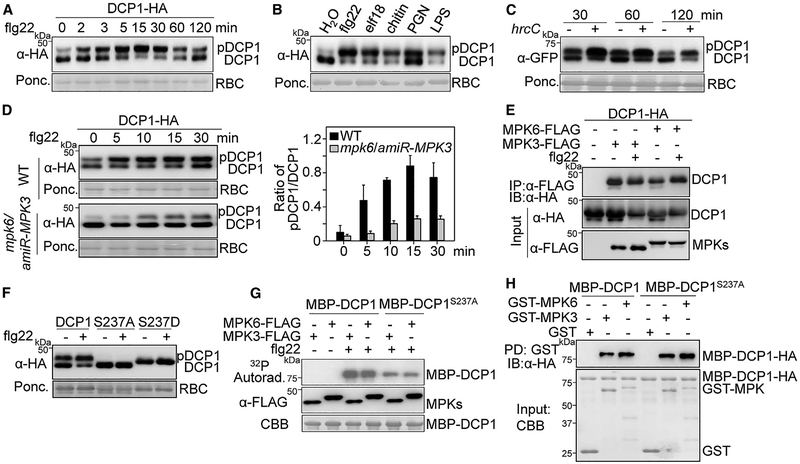
MAMP-Activated MPK3/MPK6 Phosphorylate DCP1 on Ser237
Notably, DCP1-GFP proteins were detected as two major poly-peptide bands in immunoblots with a shift from low to high molecular weight after flg22 treatment (Figure 1D). The shifted band of DCP1-hemagglutinin (HA) was observed as early as in 2 min, peaking at 15 min, and gradually decreasing at 60 min after flg22 treatment (Figure 3A). The flg22-induced mobility shift of DCP1-GFP proteins was also detected in the 35S::DCP1-GFP transgenic plants (Figure S2A). In addition, other MAMPs including bacterial elf18, lipopolysaccharide (LPS) and peptidoglycan (PGN), and fungal chitin also induced mobility shift of DCP1 in both transgenic plants and protoplasts (Figures 3B and S2B). Furthermore, infiltration of Pst hrcC in 35S::DCP1-GFP transgenic plants induced DCP1-GFP mobility shift (Figure 3C). Compared to individual MAMPs, Pst hrcC induced a long-lasting mobility shift of DCP1-GFP, which might be attributed to the presence of multiple MAMPs in the nonpathogenic bacterium. The flg22-induced mobility shift of DCP1-HA was largely restored to the unmodified form after the treatment of lambda protein phosphatase (λPP) or calf intestinal alkaline phosphatase (CIP) (Figure S2C), implying that DCP1 undergoes phosphorylation modification upon flg22 perception. MAPK cascade is a key signaling module in plant PTI responses (Meng and Zhang, 2013; Tena et al., 2011). The flg22-induced phosphorylation of DCP1-HA was blocked by either an MAPK kinase (MEK) inhibitor PD184161 or co-expression of the MAPK-specific phosphatase MKP (Figure S2D). Furthermore, expression of the full-length MEKK1, a top tier kinase in the flg22-activated MAPK cascade, triggered constitutive DCP1-HA phosphorylation without flg22 treatment (Figure S2D). Together, these data suggest that the flg22-induced MAPK cascade is required for DCP1 phosphorylation.
Figure 3. MAMP-Activated MPK3/MPK6 Phosphorylate DCP1 on Ser237.
(A) Mobility shift of DCP1-HA upon flg22 treatment. Protoplasts expressing DCP1-HA were treated with 100 nM flg22 for indicated times. Protein expression was analyzed with an α-HA immunoblot. The intensity of shifted and unshifted polypeptide bands was quantified by ImageJ software, and the percentage of shifted to total proteins (pDCP1/DCP1) is shown on the bottom as a bar graph.
(B) DCP1-HA mobility shift induced by multiple MAMPs. Protoplasts expressing DCP1-HA were treated with 100 nM flg22, 100 nM elf18, 50 μg/mL chitin, 50 μg/mL PGN, or 50 μg/mL LPS for 15 min. The quantification is shown on the bottom.
(C) DCP1 mobility shift induced by Pst hrcC in 35S::DCP1-GFP transgenic plants. Leaves from 4-week-old plants were hand-inoculated with H2O (−) or hrcC (+) at OD600 = 0.1 for α-GFP immunoblot. The quantification is shown on the bottom.
(D) The flg22-induced DCP1 phosphorylation depends on MPK3 and MPK6. Protoplasts from WT Col-0 and the mpk6/amiR-MPK3 plants were transfected with DCP1-HA and treated with 100 nM flg22 for the indicated times. The quantification is shown on the right. Data in (A)–(D) are shown as mean ± SD (n = 3).
(E) DCP1 associates with MPK3 and MPK6 in N. benthamiana. Agrobacterium carrying 35S::DCP1-HA was co-infiltrated with Agrobacterium carrying 35S::MPK3-FLAG, 35S::MPK6-FLAG, or an empty vector (Ctrl.) into leaves of N. benthamiana. Leaf samples were collected 2 dpi for co-immunoprecipitation (coIP) assay with α-FLAG (IP: α-FLAG), and the proteins were analyzed by IB with α-HA (top panel). The input control is shown on the bottom two panels.
(F) Ser237 is important for flg22-induced DCP1 phosphorylation in vivo. Protoplasts expressing HA-tagged DCP1, DCP1S237A (phospho-inactive), or DCP1S237D (phospho-mimetic) were treated without or with 100 nM flg22 for 15 min.
(G) Ser237 is important for MPK3/MPK6-mediated DCP1 phosphorylation. Protoplasts expressing MPK3-FLAG or MPK6-FLAG were treated with 100 nM flg22 for 15 min. MAPK proteins were immunoprecipitated with a-FLAG antibody and subjected to an in vitro kinase assay by using MBP-DCP1 or MBP-DCP1S237A as a substrate in the presence of [γ-32P]-ATP. Proteins were analyzed by SDS-PAGE autoradiography (upper panel), and MAPK expression is shown by immunoblot (middle panel). MBP-DCP1 proteins are shown by Coomassie brilliant blue (CBB) staining.
(H) Both MBP-DCP1 and MBP-DCP1S237A interact with GST-MPK3 and GST-MPK6 in vitro. GST, GST-MPK3, or GST-MPK6 immobilized on glutathione Sepharose beads was incubated with MBP-DCP1 or MBP-DCP1S237A proteins. Beads were washed and pelleted for immunoblot analysis with α-HA antibody. PD, pull-down. The above experiments were repeated three times with similar results.
See also Figure S2.
Sequential phosphorylation of two canonical MAPK cascades upon MAMP perception leads to the activation of MPK3/MPK6 and MPK4, respectively (Bi et al., 2018; Meng and Zhang, 2013; Sun et al., 2018; Tena et al., 2011). The flg22-induced DCP1 phosphorylation was unaffected in the mpk3, mpk6, or mpk4 single mutants (Figures S2E and S2F). MPK3 and MPK6 play redundant roles in plant growth and immunity, and the null mutant of mpk3mpk6 is embryonic lethal (Meng and Zhang, 2013). The flg22-induced DCP1 phosphorylation was largely reduced in the mpk6/amiR-MPK3 plants in which MPK3 was silenced by an artificial microRNA (amiR-MPK3) in the mpk6 background (Figure 3D), suggesting that flg22-induced DCP1 phosphorylation depends on MPK3 and MPK6. We also observed that DCP1-HA was co-immunoprecipitated with MPK3-FLAG and MPK6-FLAG when co-expressed in Nicotiana benthamiana (Figure 3E). Apparently, flg22 treatment did not affect DCP1 association with MPK3 or MPK6 (Figure 3E). It has been reported that MPK6 phosphorylates DCP1 at the serine 237 (Ser237) residue in response to dehydration stress (Xu and Chua, 2012). In line with this observation, we found that DCP1S237A, the phospho-inactive mutant, displayed no mobility shift upon flg22 treatment, whereas DCP1S237D, the phospho-mimetic mutant, showed a constitutive mobility shift in the absence of flg22 treatment (Figure 3F). In addition, flg22-activated MPK3 or MPK6 strongly phosphorylated DCP1 proteins fused to maltose-binding protein (MBP) in vitro but to a less extent toward MBP-DCP1S237A proteins (Figure 3G). Both MBP-DCP1 and MBP-DCP1S237A could pull down MPK3 and MPK6 proteins (Figure 3H), suggesting a direct interaction between DCP1 and MPK3 or MPK6. These results indicate that Ser237 is a major DCP1 phosphorylation site by flg22-activated MPK3 and MPK6.
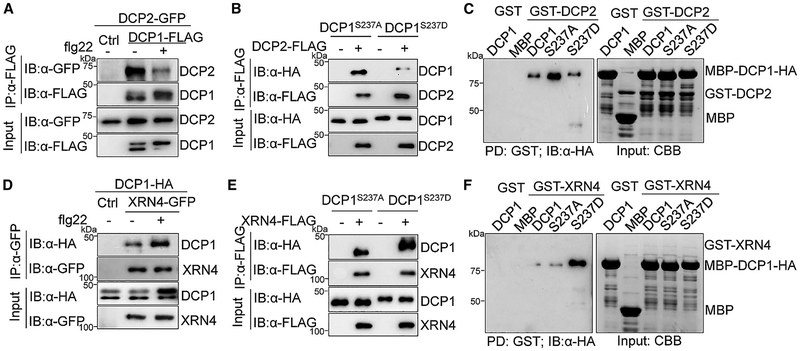
MAMP-Induced DCP1 Association Dynamics with DCP2 and XRN4
DCP1 functions as a co-activator for DCP2, the major decapping enzyme, to regulate its catalytic activity. In addition, DCP1 interacts with itself and other P-body components, such as the translation repressor DCP5 (Borja et al., 2011; She et al., 2008; Tritschler et al., 2009; Xu and Chua, 2009; Xu et al., 2006). We found here that DCP1 interacted with itself and DCP5 in a flg22-independent manner in Arabidopsis (Figures S3A and S3B). DCP1 also co-immunoprecipitated with DCP2 when co-expressed in Arabidopsis protoplasts (Figure 4A) or N. benthamiana (Figure S3C). Interestingly, flg22 treatment reduced DCP1-DCP2 interaction (Figures 4A and S3C). In line with the observation that flg22 treatment induced DCP1 phosphorylation at Ser237, DCP1S237A exhibited a stronger association with DCP2 than DCP1S237D in Arabidopsis (Figure 4B) and N. benthamiana (Figure S3C). An in vitro pull-down assay indicated that MBP-DCP1 proteins directly interacted with DCP2 proteins fused with glutathione S-transferase (GST). Similar to in vivo results, DCP2 interacted with DCP1S237A stronger than that with DCP1S237D (Figure 4C). The data indicate that DCP1 interacts with DCP2 at the steady state and disassociates from DCP2 upon flg22-activated MAPK-mediated phosphorylation. Remarkably, we consistently observed that flg22 treatment enhanced DCP1 interaction with XRN4 (Figures 4D and S3D), a 5′–3′ exoribonuclease that degrades the decapped mRNAs (Souret et al., 2004). In addition, XRN4 associated with DCP1S237D stronger than that with DCP1S237A in Arabidopsis and N. benthamiana (Figures 4E and S3D). Similarly, in vitro pull-down assays showed that DCP1S237D had a stronger affinity with XRN4 than DCP1S237A (Figure 4F). Collectively, the results suggest that PRR activation upon MAMP perception triggers rapid phosphorylation of the decapping co-activator DCP1 by MAPKs, which subsequently leads to DCP1 dissociation from the decapping enzyme DCP2 and association with the exonuclease XRN4. MAMP-induced DCP1 dissociation from DCP2 and association with XRN4 might facilitate the engagement of XRN4 in mRNA degradation once mRNAs are decapped by DCP2. It is likely that mRNAs are decapped before the DCP1-DCP2 dissociation upon flg22 treatment.
Figure 4. MAMP-Induced DCP1 Association Dynamics with DCP2 and XRN4.
(A) Treatment with flg22 induces DCP1-DCP2 dissociation. Protoplasts co-expressing DCP1-FLAG and DCP2-GFP or a vector control were treated with 100 nM flg22 for 15 min. CoIP was performed with α-FLAG (IP: α-FLAG), and the proteins were analyzed by IB with α-GFP or α-FLAG (top two panels). The input control is shown on the bottom two panels.
(B) DCP1S237A has a higher affinity with DCP2 than DCP1S237D in Arabidopsis protoplast coIP assay.
(C) DCP1S237A shows higher affinity with DCP2 than DCP1S237D in vitro. GST or GST-DCP2 immobilized on glutathione Sepharose beads was incubated with MBP, MBP-DCP1, MBP-DCP1S237A, or MBP-DCP1S237D proteins. Beads were washed and pelleted for IB with a-HA (left). Input proteins are shown by CBB staining (right).
(D) Treatment with flg22 enhances DCP1-XRN4 association in Arabidopsis protoplast coIP assay.
(E) DCP1S237D has a higher affinity with XRN4 than DCP1S237A in Arabidopsis protoplast coIP assay.
(F) DCP1S237D shows higher affinity with XRN4 than DCP1S237A in vitro. The experiment was carried out as described in (C). The above experiments were repeated three times with similar results.
See also Figure S3.
The flg22-Induced DCP1 Phosphorylation Is Required for Its Function in Plant Immunity and P-Body Disassembly
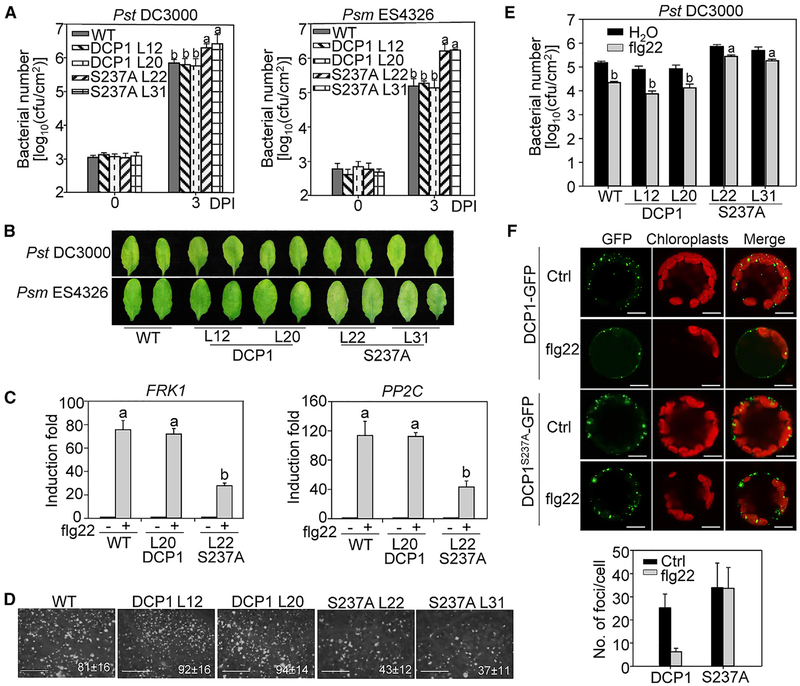
To determine the biological significance of MAMP-induced DCP1 phosphorylation in plant immunity, we complemented the dcp1–1 mutant with GFP-tagged WT DCP1 or DCP1S237A under the control of its native promoter. Both pDCP1::DCP1-GFP and pDCP1::DCP1S237A-GFP complementation lines rescued the seedling lethality of the dcp1–1 mutant and were indistinguishable from WT Col-0 plants at mature stage (Figures S4A, S4B, and S4C), suggesting that phosphorylation at Ser237 is not required for DCP1 function in plant growth. When challenged with bacterial pathogen Pst or Psm, the population of bacterial growth in pDCP1::DCP1-GFP transgenic lines (L12 and L20) was similar with that in WT plants (Figure 5A). Notably, the pDCP1::DCP1S237A-GFP complementation lines (L22 and L31) supported more bacterial growth of Pst or Psm at 3 dpi than WT and pDCP1::DCP1-GFP transgenic lines (Figure 5A). The disease symptom also appeared more severe in the pDCP1::DCP1S237A-GFP lines than that in WT and pDCP1::DCP1-GFP lines (Figure 5B). In addition, the pDCP1::DCP1S237A-GFP transgenic lines displayed compromised induction of immune-related genes FRK1 and PP2C upon flg22 treatment (Figure 5C). The pDCP1::DCP1S237A-GFP transgenic lines deposited less callose upon flg22 elicitation than WT and pDCP1::DCP1-GFP transgenic lines (Figure 5D). The number of callose deposits in the pDCP1::DCP1S237A-GFP lines was about half of what was observed in WT and pDCP1::DCP1-GFP transgenic lines. Furthermore, the flg22 treatment primed plant resistance against Pst infection in both WT and pDCP1::DCP1-GFP transgenic lines. However, this flg22-induced resistance was compromised in pDCP1::DCP1S237A-GFP lines (Figure 5E). These data suggest that phosphorylation of DCP1 at Ser237 by flg22-activated MPK3/MPK6 is essential for its function in plant immunity.
Figure 5. The flg22-Induced DCP1 Phosphorylation Is Required for Its Function in Plant Immunity.
(A) DCP1S237 is critical for DCP1 function in bacterial disease resistance. Leaves of 4-week-old WT Col-0, pDCP1::DCP1-GFP/dcp1–1 and pDCP1::DCP1S237AGFP/dcp1–1 plants were hand-inoculated with Pst (left) or Psm (right) at OD600 = 5 × 10−4 for bacterial growth assay at 0 and 3 dpi.
(B) Disease symptom of infected leaves. Pictures of inoculated leaves from (A) were taken at 3 dpi.
(C) Reduced immune marker gene expression in pDCP1::DCP1S237A-GFP/dcp1–1 plants. Leaves of 4-week-old plants were infiltrated with 100 nM flg22 or H2O for 60 min for qRT-PCR analysis. Data in (A) and (C) are shown as mean ± SD from three independent repeats. The different letters indicate significant differences according to one-way ANOVA test (p < 0.01).
(D) Reduced callose deposition in pDCP1::DCP1S237A-GFP/dcp1–1 plants. Callose deposits were stained with aniline blue at 24 h after 100 nM flg22 or H2O treatment in 4-week-old plants. The number of callose deposits per mm2 is shown as mean ± SD from three biological repeats. Bar, 0.5mm. The above experiments were repeated three times with similar results.
(E) Compromised flg22-mediated resistance to Pst infection in pDCP1::DCP1S237A-GFP/dcp1–1 plants. Leaves from 4-week-old plants were hand-inoculated with H2O or 100 nM flg22 and 24 h later hand-inoculated with Pst at OD600 = 5 × 104. Bacterial counting was performed at 3 dpi.
(F) flg22-induced P-body disassembly is blocked in p35S::DCP1S237A-GFP/Col-0 plants. Protoplasts isolated from p35S::DCP1-GFP/Col-0 and p35S::DCP1S237A-GFP/Col-0 plants were treated without or with 100 nM flg22 for 30 min. Bar, 10 μm. Quantification is shown on the bottom as mean ± SD (n = 8).
See also Figure S4.
To determine whether flg22-induced P-body disassembly is dependent on DCP1 phosphorylation at Ser237, we compared the dynamics of DCP1-GFP- and DCP1S237A-GFP-labeled P-bodies before and after flg22 treatment. Consistently, flg22 treatment reduced both size and number of DCP1-GFP-labeled P-bodies (Figure 5F). However, the reduction was not observed for DCP1S237A-GFP-labeled P-bodies after flg22 treatment (Figure 5F). This result indicates that MAPK-mediated DCP1 phosphorylation is required for flg22-induced P-body disassembly. To gain insights into the possible consequence of MAMP-induced DCP1 association with XRN4, we evaluated the mRNA decay of ERF1, AT2G25250, AT1G10120, and AT1G30210, which have been shown to be the targets of XRN4 (Nguyen et al., 2015; Rymarquis et al., 2011), in pDCP1::DCP1-GFP and pDCP1::DCP1S237A-GFP complementation lines. Interestingly, the mRNA stability of these genes tended to be higher in pDCP1::DCP1S237A-GFP than that in pDCP1::DCP1-GFP complementation lines (Figure S4D). This result suggests that the interaction between phosphorylated DCP1 and XRN4 may promote XRN4 to degrade its substates.
DCP1 Phosphorylation-Dependent mRNA Decay Contributes to flg22-Triggered Gene Downregulation
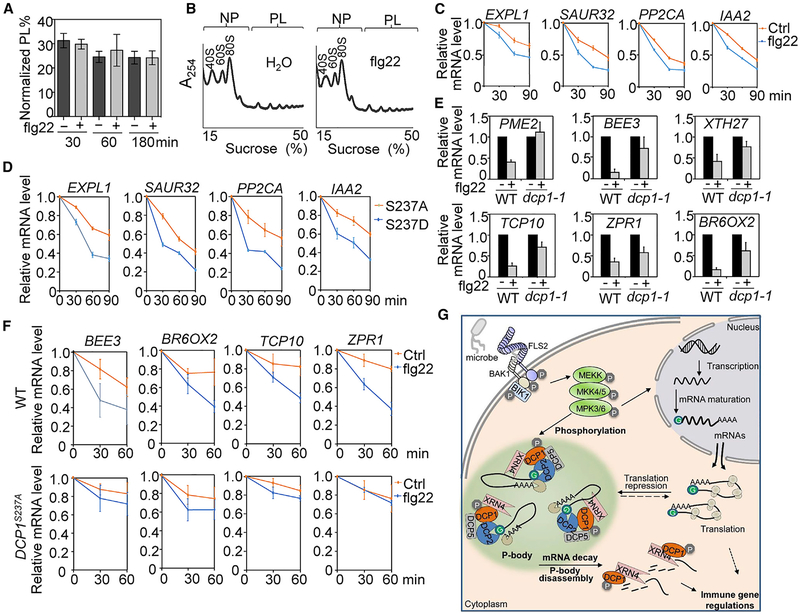
MAMP-induced P-body disassembly may be due to a reduced mRNA pool stored in P-bodies after PTI elicitation. These P-body-associated and translationally suppressed mRNAs are, therefore, either fated for degradation or re-entry to the translation cycle (Decker and Parker, 2012). This P-body-mediated posttranscriptional regulation may allow hosts to launch a rapid and effective immune response by degrading the transcripts of negative regulators and/or timely translation of positive regulators. We first examined whether flg22 treatment could induce a global translational change. The degree of active translation was estimated by calculating the percentage of cellular RNAs in the polysome fraction (PL%) resolved by sucrose gradient. Results showed no significant difference of PL% between samples treated without or with flg22 for 30, 60, or 180 min (Figures 6A and 6B). Thus, flg22 treatment does not seem to trigger a detectable remodeling of global protein translation. However, we cannot exclude the possibility that specific immune-related transcripts originally stored in P-bodies may be reengaged with polysomes for translation upon MAMP treatment. A recent study shows that elf18 induces translational reprogramming of a suite of Arabidopsis genes (Xu et al., 2017).
Figure 6. DCP1 Phosphorylation-Dependent mRNA Decay Contributes to flg22-Triggered Gene Downregulation.
(A) No detectable global mRNA translational changes upon flg22 treatment. The bar graph shows the RNA loading percentage in polysome fraction (PL%) in WT Col-0 seedlings treated without or with 100 nM flg22 for different times. The data are shown as mean percentage ± SE from three biological replicates.
(B) A representative polysome profile of WT seedlings treated without or with 100 nM flg22 for 60 min. Ribosome subunits (40S and 60S), mono-ribosome (80S), non-polysomal (NP), and polysomal (PL) fractions are marked. The data are shown with a representative absorbance profile of fractionated ribosomes from three independent biological replicates. A254 indicates absorbance at 254 nm.
(C) Enhanced mRNA decay upon flg22 treatment. Ten-day-old WT seedlings were pre-treated with transcriptional inhibitor cordycepin at 150 μg/ml for 30 min and then treated without or with 100 nM flg22 for 0, 30, 60, and 90 min. The relative mRNA level at different time points is shown as the ratio to the mRNA level at 0 min (right before flg22 treatment), which is set as 1. ACTIN2 was used as an internal control.
(D) pDCP1::DCP1S237D-GFP/dcp1–1 transgenic plants have a faster mRNA decay rate than pDCP1::DCP1S237A-GFP/dcp1–1. Ten-day-old seedlings were treated with cordycepin for indicated times for qRT-PCR analysis.
(E) DCP1 is required for flg22-induced gene downregulation. Five-day-old WT and dcp1–1 seedlings were treated with 100 nM flg22 for 60 min for qRT-PCR analysis. Gene expression level was normalized with ACTIN2.
(F) DCP1 phosphorylation-dependent mRNA decay is essential for downregulation of flg22-repressed genes. Ten-day-old seedlings from WT and pDCP1::DCP1S237A-GFP/dcp1–1 plants were pretreated with cordycepin for 30 min and then treated without or with 100 nM flg22 for indicated times. Data from (C)–(F) are shown as means ± SD from three independent biological replicates.
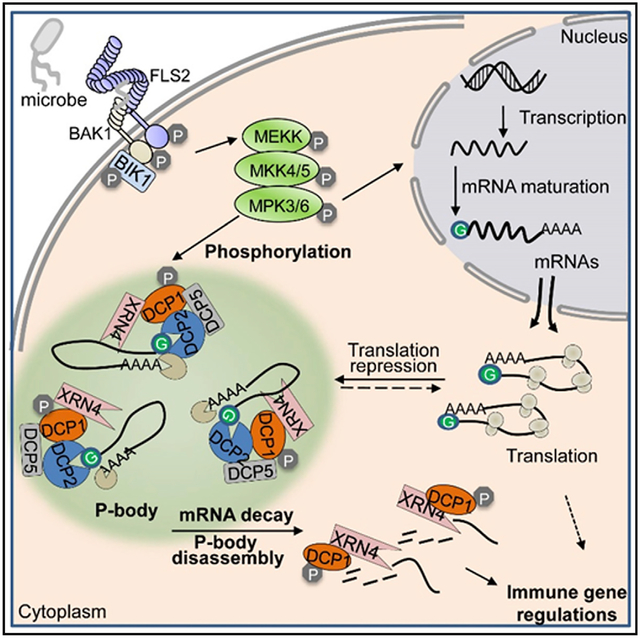
(G) A model of P-body dynamics in plant immunity. Plant immunity is initiated by the recognition of MAMP by cell-surface-resident PRR complex, which activates the convergent MAPK cascade. The activated MPK3/MPK6 directly phosphorylate DCP1, the co-activator of decapping enzyme DCP2 in P-bodies, and leads to DCP1 disassociation from DCP2, and association with XRN4, an exonuclease degrading decapped mRNAs. Degradation of mRNAs stored in P-bodies results in P-body disassembly and downregulation of some immune-related genes. Certain translationally repressed mRNAs in P-bodies may also move to polysomes for translation. This P-body-mediated posttranscriptional regulation may provide hosts a means to launch a rapid and effective immune response by degrading the transcripts of negative regulators and/or timely translation of positive regulators.
See also Figures S5 and S6.
We then determined whether flg22 perception might regulate the mRNA decay. According to the previous reports (Gutierrez et al., 2002; Narsai et al., 2007), we identified seven genes (AT3G45970/EXPL1, AT2G46690/SAUR32, AT2G24570/WRKY17, AT3G11410/PP2CA, AT3G23030/IAA2, AT3G26810/AFB2, and AT1G78080/RAP2.4) that possess highly unstable mRNAs by monitoring the transcript level in the presence of transcriptional inhibitor cordycepin (Figure S5). Significantly, the mRNA decay rates of four genes (EXPL1, SAUR32, PP2CA, and IAA2) were accelerated upon flg22 treatment within the 90-min period examined (Figure 6C), whereas the mRNA decay of the other three genes (WRKY17, AFB2, and RAP2.4) was not changed upon flg22 treatment. To determine whether the observed flg22-induced mRNA decay is linked with MAPK-mediated DCP1 phosphorylation, we compared the mRNA decay rates of these genes in transgenic plants expressing DCP1 phospho-inactive mutant (DCP1S237A) or phospho-mimetic mutant (DCP1S237D). Consistently, the flg22-induced mRNA decay rates of EXPL1, SAUR32, PP2CA, and IAA2 were higher in pDCP1::DCP1S237D-GFP/dcp1–1 than those in pDCP1::DCP1S237A-GFP/dcp1–1 lines (Figure 6D). These results indicate that MAMP treatment induces DCP1 phosphorylation-dependent decay of specific mRNAs.
The mRNA decay constitutes an important step in the regulation of gene expression by maintaining proper mRNA levels both in the steady-state and under perturbations. We determined whether DCP1 and mRNA decay were involved in flg22-triggered downregulation of immune genes. Among flg22-downregulated genes (Li et al., 2015), 6 of 15 examined genes, including AT1G53830/PME2, AT1G73830/BEE3, AT2G01850/XTH27, AT2G31070/TCP10, AT2G45450/ZPR1, and AT3G30180/BR6OX2, showed de-repression in the dcp1–1 mutant compared to WT plants (Figures 6E and S6A), indicating that flg22-triggered downregulation of these genes depends on DCP1. We further determined whether flg22 treatment promoted mRNA decay of these genes. Among them, BEE3, BR6OX2, XTH27, and ZPR1 showed accelerated mRNA decay upon flg22 treatment (Figures 6F and S6B). Notably, flg22-induced mRNA decay of these genes was considerably blocked in the pDCP1::DCP1S237A-GFP/dcp1–1 plants compared to WT plants (Figure 6F). Together, the results indicate that DCP1 phosphorylation-dependent mRNA decay in P-bodies attributes to flg22-triggered gene downregulation (Figure 6G).
DISCUSSION
Activation of PRRs induces a large-scale and dynamic transcriptome reprogramming, culminating in immunity to invading pathogens. The multi-layered transcriptional and posttranscriptional regulation of immune responsive genes is central for launching an effective defense response in hosts. We provide evidence that the components of P-bodies, the cytoplasmic granules that serve as locales for translationally repressed mRNA turnover and storage, play an important role in plant PRR-mediated immunity. P-body assembly is modulated in a dynamic manner during plant immune activation, and P-body components DCP1 and DCP2 are essential in plant immunity. PRR-activated MPK3 and MPK6 directly phosphorylate DCP1, a co-activator for decapping enzyme DCP2, and stimulate DCP1 association with XRN4, an exonuclease executing 5′-to-3′ degradation of de-capped mRNA (Figure 6G). Complementation analysis indicates that MAMP-induced DCP1 phosphorylation by the MAPK cascade is essential for its function in plant immunity. PRR-activated DCP1 phosphorylation also regulates flg22-induced mRNA decay. Thus, our studies provide a mechanism of immune gene downregulation upon PRR activation and reveal that DCP1 phosphorylation-dependent mRNA decay is an essential host immune response.
Pathogen-induced transcriptome reprogramming leads to dynamic up- and downregulation of a large number of immune-related genes (Li et al., 2016). Downregulation of negative regulators is as crucial as upregulation of positive regulators for hosts to launch an effective defense response. We hypothesize that DCP1 phosphorylation-dependent mRNA decay may facilitate the degradation of the transcripts of negative regulators. The homologs of several DCP1-dependent and flg22 downregulated genes have been shown to negatively regulate plant immunity. AT1G73830/BEE3 encodes a basic helix-loop-helix (bHLH) transcription factor, which is closely related to BEE2 and HBI1. Both BEE2 and HBI1 negatively regulate plant PTI and mediate tradeoff between PTI and hormone brassinosteroid (BR)-mediated growth (Fan et al., 2014; Malinovsky et al., 2014). BR perception and signaling inhibit PTI responses (Albrecht et al., 2012; Belkhadir et al., 2012). AT3G30180/BR6OX2, a key BR biosynthetic gene, is downregulated by multiple MAMPs (Jiménez-Góngora et al., 2015). Apparently, suppression of BR biosynthesis by turnover of BR biosynthetic genes is associated with PRR activation. The transient degradation of mRNA upon flg22 treatment may include both positive and negative regulators in the immune signaling, which, in turn, regulates some other immune responses, including gene expression, callose deposition, and immunity.
Nonsense-mediated mRNA decay (NMD) is part of mRNA quality control that prevents translation of aberrant proteins by degrading their mRNAs. It has been shown that NMD is involved in another branch of plant immunity triggered by pathogen effectors by intracellular nucleotide-binding domain leucine-rich repeat (NLR) proteins. NMD controls the constitutive turnover of the transcripts of several Toll/interleukin-1 receptor (TIR) domain-containing NLRs, the overexpression of which often leads to plant autoimmunity (Gloggnitzer et al., 2014). Thus, impairment of NMD leads to a constitutive activation of plant immune responses. NMD also serves as a defense system to viral replication by eliminating (+) strand RNAs (Garcia et al., 2014). We show here that the 5′-to-3′ exonucleolytic decay, a major mRNA decay pathway, plays an important role in plant cell surface-resident PRR-mediated immunity. Unlike NMD, P-body components positively modulate PTI responses. In addition, the P-body-localized Arabidopsis tandem zinc finger 9 (TZF9) is required for PTI responses (Maldonado-Bonilla et al., 2014). Interestingly, TZF9 is also a substrate of MAMP-activated MPK3 and MPK6 (Maldonado-Bonilla et al., 2014). In contrast to the positive role of MPK3 and MPK6, MPK4 negatively regulates plant defense (Petersen et al., 2000). MPK4 phosphorylates protein associated with topoisomerase II 1 (PAT1), a decapping enhancer in RNA decay (Roux et al., 2015). Similar to mpk4, the pat1 mutant displays constitutive defense and autoimmunity in an NLR protein SUMM2-dependent manner (Roux et al., 2015). Flg22-treatment induced PAT1-GFP-labeled P-bodies, likely due to the increased abundance of PAT1 proteins after MAMP treatment (Figures S1C and S1D) (Roux et al., 2015). Apparently, differential phosphorylation of mRNA decay components by MAPKs may specifically affect the fates of targeted immune-related genes upon MAMP treatments.
P-body components also play important roles in plant abiotic stress responses (Perea-Resa et al., 2016; Xu and Chua, 2012). Osmotic-stress-activated subclass I SnRK2s phosphorylate varicose (VCS), an mRNA decapping activator, to regulate mRNA decay under osmotic stress conditions (Soma et al., 2017). Upon dehydration stress, activated MPK6 phosphorylates DCP1 at Ser237, leading to an enhanced association with DCP5, and likely DCP2, to promote mRNA decapping (Xu and Chua, 2012). Here, we observed that MAMP-activated MPK3 and MPK6 phosphorylate DCP1, leading to its disassociation from DCP2 and association with XRN4. It is intriguing that flg22- and dehydration-stress-activated MPK3/MPK6 phosphorylate DCP1 at the same site but could lead to different interaction dynamics with DCP2 and DCP5. As both elicitations could induce global changes of numerous proteins, it is possible that flg22 or dehydration stress could induce specific proteins or their modifications to differentially affect P-body composition, assembly, and function. In the mRNA decay process, DCP-mediated decapping is followed by exoribonuclease-mediated 5′-to-3′ mRNA degradation. In Drosophila, DCP1 directly interacts with exoribonuclease 1 and functions as a molecular link between DCP2 activation and the subsequent mRNA turnover (Braun et al., 2012). Our data suggest that upon MAMP perception, DCP1 is rapidly phosphorylated by MAPKs and disengaged with DCP2, then interacts with XRN4, likely to promote the degradation of a subset of mRNAs, and, hence, leads to the disassembly of P-bodies and plant immune activation (Figure 6G). DCP1 and XRN4 association could promote exonuclease activity of XRN4. Alternatively, DCP1 and XRN4 association could increase the accessibility of the decapped mRNAs to XRN4. The interaction between DCP1 and DCP5 (an RNA binding protein) (Xu and Chua, 2009) (Figure S3B) could hold XRN4 in close vicinity to the decapped mRNAs and make the decapped mRNA substrates more accessible to XRN4.
The mechanism underlying P-body assembly and disassembly is still poorly understood. It has been suggested that the dynamics of assembly results from either protein-protein interaction dynamics or the availability of mRNAs as a platform for protein binding (Standart and Weil, 2018). Accordantly, we observed flg22-induced DCP1 phosphorylation at Ser237 contributes to the degradation of a subset of mRNAs, which may explain the disassembly of P-bodies upon MAMP treatment. Consistently, flg22-induced P-body disassembly was not observed with DCP1S237A-GFP. Furthermore, a subset of P-body-stored translationally repressed mRNAs could reengaged with polysomes for translation in response to MAMP elicitation. Taken together, our data uncover that modulation of P-body assembly dynamics and DCP1 phosphorylation-dependent mRNA decay activated by a canonical MAPK cascade downstream of multiple PRRs provide a means for hosts to launch a prompt defense response by posttranscriptional reprogramming of primary immune genes.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Ping He (pinghe@tamu.edu). We will distribute the plasmids and transgenic plants freely to the scientific community upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Arabidopsis thaliana and growth conditions
All Arabidopsis plants used in this study were in the Columbia-0 (Col-0) background, except mpk4 mutant which was in the Ler background. The various mutants and transgenic lines used in this study were described in the Key Resources Table. Arabidopsis lines were grown in soil (Metro Mix 366) in a growth room at 23°C, 45% humidity and 85 μE m−2s−1 light with a 12-hr light/12-hr dark photo-period. Four-week-old plants were used for protoplast isolation and defense-related assays. Seedlings were germinated on 1/2 Murashige and Skoog (MS) plates with 1% sucrose and 0.8% agar, and grown under the same growth condition as above for 10 days. The seedlings were transferred to a 6-well tissue culture plate with 2 mL H2O for overnight, and then used for indicated assays.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-HA-Peroxidase | Roche | Cat # 12013819001; RRID:AB_439705 |
| Anti-FLAG-Peroxidase | Sigma-Aldrich | Cat # A8592; RRID:AB_259529 |
| Anti-GFP | Roche | Cat # 11814460001; RRID:AB_390913 |
| Anti-Mouse IgG HRP-linked antibody | Cell Signaling | Cat # 7076; RRID:AB_330924 |
| Anti-FLAG M2 Affinity gel | Sigma-Aldrich | Cat # 2220; RRID:AB_10063035 |
| Protein G Agarose | Roche | Cat #5015952001 |
| Bacterial and Virus Strains | ||
| E.coli MC1061 | Li et al., 2015 | N/A |
| Agrobacterium tumefaciens GV3101 | Li et al., 2015 | N/A |
| E. coli BL21 | Li et al., 2015 | N/A |
| Pseudomonas syringae pv. tomato DC3000 (Pst) | He et al., 2006 | N/A |
| P. syringae pv. maculicola ES4326 (Psm) | Li et al., 2014a | N/A |
| Pseudomonas syringae pv. tomato DC3000 (Pst) type III secretion mutant hrcC | He et al., 2006 | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| PD184161 | A.G. Scientific | Cat # P-1713 |
| Calf intestinal alkaline phosphatase (CIP) | New England BioLabs | Cat # M0290S |
| Lambda protein phosphatase (λPP) | New England BioLabs | Cat # P0753S |
| Cordycepin | Sigma-Aldrich | Cat # C9137 |
| RiboZol RNA Extraction Reagent | AMRESCO | Cat # N580 |
| RNase-free DNase I | New England BioLabs | Cat # M0303L |
| flg22 | Genscript | N/A |
| elf18 | Genscript | N/A |
| chitin | Sigma-Aldrich | Cat # C9752 |
| peptidoglycan | Sigma-Aldrich | Cat #77140 |
| lipopolysaccharide | Sigma-Aldrich | Cat # L2012 |
| IPTG | Sigma-Aldrich | Cat# I6758 |
| Pierce glutathione agarose | Thermo Scientific | Cat# 16101 |
| amylose resin | New England BioLabs | Cat# E8021L |
| Critical Commercial Assays | ||
| M-MuLV Reverse Transcriptase | New England BioLabs | Cat # M0253L |
| iTaq SYBR green Supermix | Bio-Rad | Cat # 1725124 |
| Experimental Models: Organisms/Strains | ||
| Arabidopsis thaliana Col-0 wild-type | Li et al., 2015 | N/A |
| Arabidopsis thaliana Ler ecotype | Li et al., 2015 | N/A |
| mpk3 | ABRC | SALK_151594 |
| mpk6 | ABRC | Salk_073907 |
| mpk4 | ABRC | CS5205, Ler background |
| fls2 | ABRC | Salk_141277 |
| dcp1–1 | ABRC | GABI-844B03 |
| p35S::DCP1-GFP/Col-0 | This paper | N/A |
| p35S::DCP1S237A-GFP/Col-0 | This paper | N/A |
| pDCP1::DCP1-GFP/dcp1–1 | This paper | N/A |
| pDCP1::DCP1S237A-GFP/dcp1–1 | This paper | N/A |
| pDCP1::DCP1S237D-GFP/dcp1–1 | This paper | N/A |
| mpk6/amiR-MPK3 | This paper | N/A |
| Oligonucleotides | ||
| Primers for cloning, point mutation and VIGS, see Table S1 | This paper | N/A |
| Primers for genotyping and RT-PCR, see Table S1 | This paper | N/A |
| Primers for qRT-PCR, see Table S1 | This paper | N/A |
| Recombinant DNA | ||
| pYL156 (pTRV-RNA2) | Li et al., 2014a | N/A |
| pTRV-RNA1 | Li et al., 2014a | N/A |
| pYL156-GFP | Li et al., 2014a | N/A |
| pHBT | He et al., 2006 | N/A |
| pGST | Lu et al., 2010 | N/A |
| pMAL-c2 | Lu et al., 2010 | N/A |
| pCB302 | Li et al., 2014a | N/A |
| pHBT-amiRNA-ICE1 | Li et al., 2014b | N/A |
| pTA7002-AvrPto | He et al., 2006 | N/A |
| pYL156-DCP1 | This paper | N/A |
| pYL156-DCP2 | This paper | N/A |
| pHBT-DCP1-GFP | This paper | N/A |
| pHBT-DCP1-HA | This paper | N/A |
| pHBT-DCP1-FLAG | This paper | N/A |
| pHBT-DCP2-GFP | This paper | N/A |
| pHBT-DCP2-FLAG | This paper | N/A |
| pHBT-XRN4-GFP | This paper | N/A |
| pHBT-XRN4-HA | This paper | N/A |
| pHBT-XRN4-FLAG | This paper | N/A |
| pHBT-DCP5-GFP | This paper | N/A |
| pHBT-DCP5-HA | This paper | N/A |
| pHBT-PAT1-GFP | This paper | N/A |
| pHBT-DCP1S237A-HA | This paper | N/A |
| pHBT-DCP1S237D-HA | This paper | N/A |
| pHBT-MEKK1-FLAG | Li et al., 2015 | N/A |
| pHBT-MPK3-FLAG | Li et al., 2014a | N/A |
| pHBT-MPK6-FLAG | Li et al., 2014a | N/A |
| pCB302–35S::DCP1-GFP | This paper | N/A |
| pCB302–35S::DCP1 S237A-GFP | This paper | N/A |
| pCB302–35S::DCP1 S237D-GFP | This paper | N/A |
| pCB302–35S::DCP2-FLAG | This paper | N/A |
| pCB302–35S::XRN4-HA | This paper | N/A |
| pCB302–35S::MPK3-FLAG | This paper | N/A |
| pCB302–35S::MPK6-FLAG | This paper | N/A |
| pCB302-pDCP1::DCP1-GFP | This paper | N/A |
| pCB302-pDCP1::DCP1 S237A-GFP | This paper | N/A |
| pCB302-pDCP1::DCP1 S237D-GFP | This paper | N/A |
| pMAL-DCP1 | This paper | N/A |
| pMAL-DCP1 S237A | This paper | N/A |
| pMAL-DCP1 S237D | This paper | N/A |
| pGST-DCP2 | This paper | N/A |
| pGST-XRN4 | This paper | N/A |
| pGST-MPK3 | This paper | N/A |
| pGST-MPK6 | This paper | N/A |
| pTA7002-amiR-MPK3 | This paper | N/A |
| Software and Algorithms | ||
| Olympus Fluoview Viewer | Olympus | Version 3.0 |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
Nicotiana benthamiana and growth conditions
Nicotiana benthamiana was grown in greenhouses in soil under a 12-hr light/12-hr dark photoperiod at 23°C.
Bacterial strains
The various bacteria strains used in this study were described in the Key Resources Table. Pseudomonas syringae pv. tomato (Pst) DC3000 and Pst DC3000 hrcC were grown on the King’s B medium plates with 50 μg/ml rifampicin. P. syringae pv. maculicola ES4326 (Psm) was grown on the King’s B medium plates with 50 μg/ml Streptomycin. All the Pseudomonas strains were grown on plates at 28°C for 2 days for further inoculum preparation.
METHOD DETAILS
Constructs and transgenic plant generation
To generate VIGS DCP1 and DCP2 constructs, fragments of DCP1 (~450bp) and DCP2 (~430bp) were PCR-amplified from Arabidopsis Col-0 cDNA, digested with EcoRI and KpnI, and ligated into the pYL156 (pTRV-RNA2) vector. The DCP1, DCP2, DCP5, XRN4 and PAT1 genes were amplified from Col-0 cDNA and introduced into the plant gene expression vector pHBT with an HA, FLAG or GFP epitope-tag at the C terminus. The point mutations of DCP1S237A and DCP1S237D were generated by site-directed mutagenesis. To generate constructs for E. coli. fusion protein isolation, DCP1, DCP1S237A, DCP1S237D, DCP2, and XRN4 were released from pHBT vector and subcloned into a modified pGST or pMAL-c2 vector. To generate the 35S promoter-driven genes in the binary vector, DCP1, DCP1S237A, DCP1S237D, DCP2, and XRN4 were sub-cloned into the pCB302 binary vector with a 35S promotor and an HA, FLAG or GFP epitope-tag at the C terminus. To construct the native promoter-driven genes in the binary vector, the DCP1-GFP fragment and its mutants were released from pHBT-35S::DCP1-GFP, and ligated into the pCB302 vector. The native promoter of DCP1 (~660 bp upstream of the start codon) was amplified from Col-0 genomic DNA and ligated into the abovementioned pCB302–35S::DCP1-GFP construct. The DNA fragments amplified from PCR were fully sequenced to validate the sequences after cloning into the perspective vectors. The MEKK1, MKP, MPK3, MPK4 and MPK6 clones in the pHBT vector were reported previously (Li et al., 2014a, Li et al., 2015). To generate transgenic plants, the standard protocol for Agrobacterium tumefaciens-mediated floral dip was used. The transgenic plants were selected by glufosinate-ammonium (50 μg/mL). Multiple transgenic lines were analyzed by immunoblotting for protein expression and two lines with a single T-DNA insertion and similar protein expression levels were chosen for further phenotypic and molecular characterization.
Agrobacterium-mediated floral dipping
Agrobacterium tumefaciens GV3101 containing the binary vector was cultured at 28°C in LB liquid medium with 50 μg/ml Kanamycin and 25 mg/ml Gentamicin. Bacteria were harvested by centrifugation at 3000 rpm for 15 min and the pellet was suspended with buffer containing 50 mM MES (pH 5.5–5.7), 5% sucrose and 200 μl/L silwetL-77 at the density of OD600 = 0.8. Arabidopsis flower buds were dipped thoroughly to the bacteria solution and then the dipped plants were covered with a dome for 24 hr to maintain high humidity. After that, plants were placed in the greenhouses under 12-hr light/12-hr dark light period and seeds were harvested for transgenic plants selection.
Elicitor and chemical inhibitor treatments
MAMPs were used in a final concentration of 100 nM for flg22, 100 nM for elf18, 50 μg/mL for chitin, 50 μg/mL for PGN and 50 μg/mL for LPS. 5 μM of the MEK inhibitor PD184161 was added to protoplasts 1 hr before flg22 treatment. Calf intestinal alkaline phosphatase (CIP) and Lambda protein phosphatase (λPP) treatments were performed following the instruction. The working concentration of transcriptional inhibitor cordycepin was 150 μg/mL for RNA decay assay.
Generation of mpk6/amiR-MPK3 plants and treatment
Artificial mircoRNA (amiRNA) was constructed as previously described (Li et al., 2014b). In brief, primers for cloning amiRNAs were generated according to the website (http://wmd3.weigelworld.org/cgi-bin/webapp.cgi). The cognate fragments were cloned into the modified pHBT-amiRNA-ICE1 vector (Li et al., 2014b), and then subcloned into pTA7002-AvrPto vector (He et al., 2006) with XhoI and StuI digestion to generate pTA7002-amiR-MPK3 construct. The pTA7002-amiR-MPK3 construct was transformed into the mpk6 mutant (SALK_062471) and the transgenic plants were selected with hygromycin resistance. To silence MPK3 expression, the transgenic plants were sprayed with 30uM dexamethasone supplemented with 0.01% Silwet L-77 at 6 days and 3 days before protoplast isolation.
Callose deposition
Leaves of four-week-old plants were hand-infiltrated with 500 nM flg22 or ddH2O with a needleless syringe. The treated leaves were harvest 24 h later and then fixed with FAA solution (10% formaldehyde, 5% acetic acid and 50% ethanol), cleared in 95% ethanol, rinsed twice with 75% ethanol and ddH2O, and stained for 30 min in 0.01% aniline blue solution (150 mM KH2PO4, pH 9.5). The leaf samples were mounted in 50% glycerol and callose deposits were visualized under a UV filter with a fluorescence microscope. The number of callose deposits was counted using ImageJ software and expressed as number/mm2 with the mean ± SD from at least three different leaves.
Bacterial infection assay
Pseudomonas syringae pv. tomato DC3000 (Pst) and P. syringae pv. maculicola ES4326 (Psm) strains were cultured for overnight at 28°C in the King’s B medium with appropriate antibiotics. Bacteria were harvested by centrifugation at 3500 rpm for 5 min and washed twice with ddH2O. The pellet was resuspended with 10 mM MgCl2 and adjusted to the density of OD600 = 5 × 10−4. Leaves of four-week-old plants were hand-infiltrated with bacterial suspension using a needleless syringe. For flg22 protection assay, leaves were pre-infiltrated with 100nM flg22 or water as a control for 24 h before bacterial pathogen infiltration. To measure bacterial growth, six leaf discs separated as three repeats were ground and serial dilutions were plated on Petri dish plates with medium (1% tryptone, 1%sucrose, 0.1% glutamatic acid and 1.5% agar) containing corresponding antibiotics. The plates were cultured in a 28°C incubator for two days and bacterial colony forming units (cfu) were counted.
Virus-induced gene silencing assay
Plasmids containing binary TRV vectors pTRV-RNA1, pTRV-RNA2 (pYL156), pYL156-GFP, pYL156-DCP1, or pYL156-DCP2 were introduced into Agrobacterium tumefaciens strain GV3101 by electroporation. Agrobacterium culture was grown at 28°C in LB medium containing 50 μg/ml Kanamycin and 25 μg/ml Gentamicin. Bacteria were pelleted by centrifugation, and re-suspended in the induction medium (10 mM MgCl2, 10 mM MES and 200 μM acetosyringone). Cell suspensions were adjusted to a final concentration OD600 = 1.5 and then incubated at room temperature for at 3 hr. Agrobacteria containing pTRV-RNA1 and derivatives of pTRV-RNA2 were mixed at a 1:1 ratio, infiltrated into the first pair of true leaves of two-week-old soil-grown plants with a needleless syringe. Two weeks after infiltration, pYL156-DCP1, pYL156-DCP2 and pYL156-GFP (control) inoculated plants were used for PTI response as-says and pathogen infection.
RNA isolation, qRT-PCR analysis, and mRNA half-life determination
To determine flg22-induced immune gene expression, leaves of four-week-old plants were hand-infiltrated with 100 nM flg22 or ddH2O with a needleless syringe and samples were harvested at 1 hour post infiltration (hpi). To measure the unstable mRNA decay rate, ten-day-old seedlings from 1/2 MS plates were transferred to ddH2O for overnight. Before the transcriptional inhibitor cordycepin (150 μg/ml) treatment, seedlings were pre-soaked in the incubation buffer (1 mM PIPES, pH = 6.25, 1 mM sodium citrate, 1 mM KCl, 15 mM sucrose, and 0.08% Silwet L-77) for 30 min. Total RNAs from plant samples were extracted by RiboZol RNA Extraction Reagent and quantified with NanoDrop (Thermo Scientific). RNA was treated with RNase-free DNase I to remove DNA contamination. The first strand cDNA was synthesized by reverse transcription with M-MuLV reverse transcriptase and oligo (dT) primer. qRT-PCR analysis was performed using iTaq SYBR green Supermix with an ABI GeneAmp® PCR System 9700 following a standard protocol. The expression of each gene was normalized to the expression of internal control ACTIN2. The percentages of mRNAs at each time point calculated relative to the zero time point value were plotted against time and a regression curve was obtained.
Confocal microscopy and image analysis
For transient protoplast expression, protoplasts were transfected with GFP-tagged DCP1, DCP5, XRN4 or PAT1 vectors and were incubated for 12 hr in a 12-well tissue culture plate followed by flg22 treatment. For pCB302–35S::DCP1-GFP/Col-0 and pCB302–35S::DCP1S237A-GFP/Col-0 transgenic plants, five-day-old seedlings or protoplasts isolated from four-week-old plants were used for flg22 treatment, and leaves of four-week-old plants were used for Pst hrcC infiltration. The fluorescence images were taken by the FLUOVIEW FV1200 (Olympus) confocal system. The excitation wavelength is 488 nm for GFP, and 635 nm for Chlorophyll, which serves as the internal control. Emission is detected at 505–525 nm for GFP, and 660–680 nm for Chlorophyll. Images were converted to 8-bit grayscale with ImageJ software. To quantify the number of P-bodies, the threshold was adjusted to allow the cytoplasmic foci above a certain intensity to be detected. The same threshold was applied to all samples in one experiment. The number of P-bodies was counted using the “analyses particles” function and particle size was limited to those greater than 0.2 μm2 to get rid of noises. Quantification data are shown as mean ± SD from at least 10 cells or images.
In vivo co-immunoprecipitation (Co-IP) assay
Arabidopsis protoplasts were transfected with a pair of constructs tested (the empty vector as the negative control) and incubated for 12 hr. Samples were collected by centrifugation and lysed with Co-IP buffer (20 mM Tris-HCl, pH7.5, 100 mM NaCl, 1 mM EDTA, 10% Glycerol, 0.5% Triton X-100 and protease inhibitor cocktail from Roche) by vortexing. For Co-IP in Nicotiana benthamiana, leaves of three-week-old soil-grown plants were hand-infiltrated with different pairs of Agrobacterium tumefaciens carrying indicated vectors. Overnight cultured bacteria were harvested by centrifugation and re-suspended in buffer (10 mM MES, pH5.7, 10 mM MgCl2, 200 μM acetosyringone) at OD600 = 1.5. Leaf samples were harvested two days post inoculation and subjected to homogenization with Co-IP buffer. Protein extract was pre-incubated with protein-G-agarose beads for 1 hr at 4°C with gentle shaking on a rocker. Immunoprecipitation was carried out with an α-GFP or α-FLAG antibody for 2 hr and then with protein-G-agarose beads for another 2 hr at 4°C. The beads were collected and washed three times with washing buffer (20 mM Tris-HCl, pH7.5, 100 mM NaCl, 1 mM EDTA, 0.1% Triton X-100). The immunoprecipitated proteins and input proteins were analyzed by immunoblotting with indicated antibodies.
In vitro pull-down and immunocomplex kinase assays
Fusion proteins were expressed in E. coli BL21 strain using LB medium supplemented with 0.25 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG). Glutatione-S-transferase (GST), GST-MPK3, GST-MPK4 and GST-MPK6 were purified with Pierce glutathione agarose, and maltose binding protein (MBP), MBP-DCP1, MBP-DCP1S237A and MBP-DCP1S237D proteins were purified using amylose resin according to standard protocols. MBP fusion proteins (tagged with HA) were pre-incubated with prewashed glutathione agarose in 300 mL incubation buffer (20 mM Tris-HCl, pH7.5, 100 mM NaCl, 0.1mM EDTA and 0.5% Triton X-100) for 0.5 hr at 4°C. After centrifugation, the supernatant was collected and incubated with prewashed GST, GST-MPK3 or GST-MPK6 beads for another 1 hr. The beads were collected and washed three times with washing buffer (20 mM Tris-HCl, pH7.5, 300 mM NaCl, 0.1mM EDTA and 0.1% Triton X-100). Proteins were detected with an a-HA antibody by immunoblotting. For immunocomplex kinase assay, FLAG-tagged MPK3, MPK4, and MPK6 were expressed in Col-0 protoplasts for 12 hr and then treated with 100 nM flg22 for 15 min. The activated MPK proteins were immunoprecipitated using α-FLAG agarose following Co-IP procedure. The beads were spun down and re-suspended in 50 mM Tris-HCl, pH7.5. The MPK-bounded beads were incubated with MBP-DCP1 or MBPDCP1S237A fusion proteins in the kinase reaction buffer (20 mM Tris-HCl, pH7.5, 20 mM MgCl2, 5 mM EDTA, 1 mM DTT and 100 μM ATP) in the presence of 5 αCi [32P]-γ-ATP for 2 hr at room temperature. The reactions were stopped by adding SDS sample buffer, and protein phosphorylation was visualized by autoradiography in 10% SDS-PAGE.
Analysis of translation efficiency
Ribosome profiles were performed as described previously (Liu et al., 2012). In brief, polysomal RNA was extracted with polysome extraction buffer. The resuspended mixture was incubated on ice for 5 min, and then spun at 15000 g for 5 min at 4°C. Supernatant was loaded on a continuous sucrose gradient (15%–50%) and spun at 210000 g for 3.5 hr at 4°C. The distribution of the nucleic acids was examined by a UV254 absorbance profile (model #UV-6, ISCO). Total RNAs from non-polysome fractions (NP) or polysome fractions (PL) were used for the calculation of PL% (PL/NP+PL) after normalization by spike-in RNA DAP (Affymetrix, GeneChip Poly-A RNA Control Kit).
QUANTIFICATION AND STATISTICAL ANALYSIS
In general, data are represented by mean ± SD. The numbers of biological repeats (n) are provided in each figure legend. One-way analysis of variance (ANOVA) was performed to check for statistically significant differences. For qRT-PCR analysis, each biological repeat is based on an RNA sample extracted from four seedlings.
DATA AND CODE AVAILABILITY
This manuscript did not generate new datasets or code.
Supplementary Material
Highlights.
P-body dynamics is regulated in plant pattern-triggered immunity (PTI)
P-body components DCP1 and DCP2 positively regulate plant PTI
Microbe-associated molecular pattern-activated MAP kinases phosphorylate DCP1
DCP1 phosphorylation contributes to mRNA decay of certain immune-related genes
ACKNOWLEDGMENTS
We thank Drs. Jianfeng Li (Sun Yat-Sen University, China) and Jen Sheen (Harvard Medical School, USA) for amiRNA construct and members in He and Shan laboratories for comments. The work was supported by National Science Foundation (NSF) (IOS-1252539) and NIH (R01GM092893) to P.H.; NIH (1R01GM097247) and the Robert A. Welch foundation (A-1795) to L.S.; NSF (MCB-1906060) to J.-C.J., P.H., and L.S.; the Frontier Science Research Program from the Ministry of Science and Technology of Taiwan (MOST105–2321-B-001–017) to S.-H.W.; and Ohio Agricultural Research and Development Center SEEDS (2018–007) to J.-C.J.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.07.054.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Albrecht C, Boutrot F, Segonzac C, Schwessinger B, Gimenez-Ibanez S, Chinchilla D, Rathjen JP, de Vries SC, and Zipfel C (2012). Brassinosteroids inhibit pathogen-associated molecular pattern-triggered immune signaling independent of the receptor kinase BAK1. Proc. Natl. Acad. Sci. USA 109, 303–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, and Sheen J (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977–983. [DOI] [PubMed] [Google Scholar]
- Belkhadir Y, Jaillais Y, Epple P, Balsemão-Pires E, Dangl JL, and Chory J (2012). Brassinosteroids modulate the efficiency of plant immune responses to microbe-associated molecular patterns. Proc. Natl. Acad. Sci. USA 109, 297–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi G, Zhou Z, Wang W, Li L, Rao S, Wu Y, Zhang X, Menke FLH, Chen S, and Zhou JM (2018). Receptor-Like Cytoplasmic Kinases Directly Link Diverse Pattern Recognition Receptors to the Activation of Mitogen-Activated Protein Kinase Cascades in Arabidopsis. Plant Cell 30, 1543–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhm H, Albert I, Fan L, Reinhard A, and Nürnberger T (2014). Immune receptor complexes at the plant cell surface. Curr. Opin. Plant Biol 20, 47–54. [DOI] [PubMed] [Google Scholar]
- Borja MS, Piotukh K, Freund C, and Gross JD (2011). Dcp1 links coactivators of mRNA decapping to Dcp2 by proline recognition. RNA 17, 278–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq M, Willmann MR, McCormack M, Lee H, Shan L, He P, Bush J, Cheng SH, and Sheen J (2010). Differential innate immune signalling via Ca(2+) sensor protein kinases. Nature 464, 418–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun JE, Truffault V, Boland A, Huntzinger E, Chang CT, Haas G, Weichenrieder O, Coles M, and Izaurralde E (2012). A direct interaction between DCP1 and XRN1 couples mRNA decapping to 5′ exonucleolytic degradation. Nat. Struct. Mol. Biol 19, 1324–1331. [DOI] [PubMed] [Google Scholar]
- Brengues M, Teixeira D, and Parker R (2005). Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310, 486–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter S, Ricci EP, Mercier BC, Moore MJ, and Fitzgerald KA (2014). Post-transcriptional regulation of gene expression in innate immunity. Nat. Rev. Immunol 14, 361–376. [DOI] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nürnberger T, Jones JD, Felix G, and Boller T (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448, 497–500. [DOI] [PubMed] [Google Scholar]
- Couto D, and Zipfel C (2016). Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol 16, 537–552. [DOI] [PubMed] [Google Scholar]
- Decker CJ, and Parker R (2012). P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb. Perspect. Biol 4, a012286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan M, Bai MY, Kim JG, Wang T, Oh E, Chen L, Park CH, Son SH, Kim SK, Mudgett MB, and Wang ZY (2014). The bHLH transcription factor HBI1 mediates the trade-off between growth and pathogen-associated molecular pattern-triggered immunity in Arabidopsis. Plant Cell 26, 828–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B, Liu C, de Oliveira MV, Intorne AC, Li B, Babilonia K, de Souza Filho GA, Shan L, and He P (2015). Protein poly(ADP-ribosyl)ation regulates arabidopsis immune gene expression and defense responses. PLoS Genet 11, e1004936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D, Garcia S, and Voinnet O (2014). Nonsense-mediated decay serves as a general viral restriction mechanism in plants. Cell Host Microbe 16, 391–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloggnitzer J, Akimcheva S, Srinivasan A, Kusenda B, Riehs N, Stampfl H, Bautor J, Dekrout B, Jonak C, Jiménez-Gómez JM, et al. (2014). Nonsense-mediated mRNA decay modulates immune receptor levels to regulate plant antibacterial defense. Cell Host Microbe 16, 376–390. [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L, and Boller T (2000). FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5, 1003–1011. [DOI] [PubMed] [Google Scholar]
- Gutierrez RA, Ewing RM, Cherry JM, and Green PJ (2002). Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: rapid decay is associated with a group of touch- and specific clock-controlled genes. Proc. Natl. Acad. Sci. USA 99, 11513–11518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He P, Shan L, Lin NC, Martin GB, Kemmerling B, Nürnberger T, and Sheen J (2006). Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell 125, 563–575. [DOI] [PubMed] [Google Scholar]
- Heese A, Hann DR, Gimenez-Ibanez S, Jones AM, He K, Li J, Schroeder JI, Peck SC, and Rathjen JP (2007). The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. USA 104, 12217–12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubstenberger A, Courel M, Bénard M, Souquere S, Ernoult-Lange M, Chouaib R, Yi Z, Morlot JB, Munier A, Fradet M, et al. (2017). P-Body Purification Reveals the Condensation of Repressed mRNA Regulons. Mol. Cell 68, 144–157.e5. [DOI] [PubMed] [Google Scholar]
- Jang GJ, Yang JY, Hsieh HL, and Wu SH (2019). Processing bodies control the selective translation for optimal development of Arabidopsis young seedlings. Proc. Natl. Acad. Sci. USA 116, 6451–6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Góngora T, Kim SK, Lozano-Durán R, and Zipfel C (2015). Flg22-Triggered Immunity Negatively Regulates Key BR Biosynthetic Genes. Front. Plant Sci 6, 981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota Y, Sklenar J, Derbyshire P, Stransfeld L, Asai S, Ntoukakis V, Jones JD, Shirasu K, Menke F, Jones A, and Zipfel C (2014). Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell 54, 43–55. [DOI] [PubMed] [Google Scholar]
- Li F, Cheng C, Cui F, de Oliveira MV, Yu X, Meng X, Intorne AC, Babilonia K, Li M, Li B, et al. (2014a). Modulation of RNA polymerase II phosphorylation downstream of pathogen perception orchestrates plant immunity. Cell Host Microbe 16, 748–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JF, Zhang D, and Sheen J (2014b). Epitope-tagged protein-based artificial miRNA screens for optimized gene silencing in plants. Nat. Protoc 9, 939–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Li M, Yu L, Zhou Z, Liang X, Liu Z, Cai G, Gao L, Zhang X, Wang Y, et al. (2014c). The FLS2-associated kinase BIK1 directly phosphor-ylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15, 329–338. [DOI] [PubMed] [Google Scholar]
- Li B, Jiang S, Yu X, Cheng C, Chen S, Cheng Y, Yuan JS, Jiang D, He P, and Shan L (2015). Phosphorylation of trihelix transcriptional repressor ASR3 by MAP KINASE4 negatively regulates Arabidopsis immunity. Plant Cell 27, 839–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Meng X, Shan L, and He P (2016). Transcriptional Regulation of Pattern-Triggered Immunity in Plants. Cell Host Microbe 19, 641–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MJ, Wu SH, Chen HM, and Wu SH (2012). Widespread translational control contributes to the regulation of Arabidopsis photomorphogenesis. Mol. Syst. Biol 8, 566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Wu S, Gao X, Zhang Y, Shan L, and He P (2010). A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. USA 107, 496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado-Bonilla LD, Eschen-Lippold L, Gago-Zachert S, Tabassum N, Bauer N, Scheel D, and Lee J (2014). The Arabidopsis tandem zinc finger 9 protein binds RNA and mediates pathogen-associated molecular pattern-triggered immune responses. Plant Cell Physiol 55, 412–425. [DOI] [PubMed] [Google Scholar]
- Malinovsky FG, Batoux M, Schwessinger B, Youn JH, Stransfeld L, Win J, Kim SK, and Zipfel C (2014). Antagonistic regulation of growth and immunity by the Arabidopsis basic helix-loop-helix transcription factor homolog of brassinosteroid enhanced expression2 interacting with increased leaf inclination1 binding bHLH1. Plant Physiol 164, 1443–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, and Zhang S (2013). MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol 51, 245–266. [DOI] [PubMed] [Google Scholar]
- Mitchell SF, and Parker R (2014). Principles and properties of eukaryotic mRNPs. Mol. Cell 54, 547–558. [DOI] [PubMed] [Google Scholar]
- Narsai R, Howell KA, Millar AH, O’Toole N, Small I, and Whelan J (2007). Genome-wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana. Plant Cell 19, 3418–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AH, Matsui A, Tanaka M, Mizunashi K, Nakaminami K, Hayashi M, Iida K, Toyoda T, Nguyen DV, and Seki M (2015). Loss of Arabidopsis 5′-3′ Exoribonuclease AtXRN4 Function Enhances Heat Stress Tolerance of Plants Subjected to Severe Heat Stress. Plant Cell Physiol 56, 1762–1772. [DOI] [PubMed] [Google Scholar]
- Perea-Resa C, Carrasco-López C, Catalá R, Turečková V, Novak O, Zhang W, Sieburth L, Jiménez-Gómez JM, and Salinas J (2016). The LSM1–7 Complex Differentially Regulates Arabidopsis Tolerance to Abiotic Stress Conditions by Promoting Selective mRNA Decapping. Plant Cell 28, 505–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen M, Brodersen P, Naested H, Andreasson E, Lindhart U, Johansen B, Nielsen HB, Lacy M, Austin MJ, Parker JE, et al. (2000). Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103, 1111–1120. [DOI] [PubMed] [Google Scholar]
- Roux ME, Rasmussen MW, Palma K, Lolle S, Regué AM, Bethke G, Glazebrook J, Zhang W, Sieburth L, Larsen MR, et al. (2015). The mRNA decay factor PAT1 functions in a pathway including MAP kinase 4 and immune receptor SUMM2. EMBO J 34, 593–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymarquis LA, Souret FF, and Green PJ (2011). Evidence that XRN4, an Arabidopsis homolog of exoribonuclease XRN1, preferentially impacts transcripts with certain sequences or in particular functional categories. RNA 17, 501–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenberg DR, and Maquat LE (2012). Regulation of cytoplasmic mRNA decay. Nat. Rev. Genet 13, 246–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She M, Decker CJ, Svergun DI, Round A, Chen N, Muhlrad D, Parker R, and Song H (2008). Structural basis of dcp2 recognition and activation by dcp1. Mol. Cell 29, 337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soma F, Mogami J, Yoshida T, Abekura M, Takahashi F, Kidokoro S, Mizoi J, Shinozaki K, and Yamaguchi-Shinozaki K (2017). ABA-unresponsive SnRK2 protein kinases regulate mRNA decay under osmotic stress in plants. Nat. Plants 3, 16204. [DOI] [PubMed] [Google Scholar]
- Souret FF, Kastenmayer JP, and Green PJ (2004). AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol. Cell 15, 173–183. [DOI] [PubMed] [Google Scholar]
- Staiger D, Korneli C, Lummer M, and Navarro L (2013). Emerging role for RNA-based regulation in plant immunity. New Phytol 197, 394–404. [DOI] [PubMed] [Google Scholar]
- Standart N, and Weil D (2018). P-Bodies: Cytosolic Droplets for Coordinated mRNA Storage. Trends Genet 34, 612–626. [DOI] [PubMed] [Google Scholar]
- Sun Y, Li L, Macho AP, Han Z, Hu Z, Zipfel C, Zhou JM, and Chai J (2013). Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 342, 624–628. [DOI] [PubMed] [Google Scholar]
- Sun T, Nitta Y, Zhang Q, Wu D, Tian H, Lee JS, and Zhang Y (2018). Antagonistic interactions between two MAP kinase cascades in plant development and immune signaling. EMBO Rep 19, e45324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tena G, Boudsocq M, and Sheen J (2011). Protein kinase signaling networks in plant innate immunity. Curr. Opin. Plant Biol 14, 519–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritschler F, Braun JE, Motz C, Igreja C, Haas G, Truffault V, Izaurralde E, and Weichenrieder O (2009). DCP1 forms asymmetric trimers to assemble into active mRNA decapping complexes in metazoa. Proc. Natl. Acad. Sci. USA 106, 21591–21596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, and Chua NH (2009). Arabidopsis decapping 5 is required for mRNA decapping, P-body formation, and translational repression during postembryonic development. Plant Cell 21, 3270–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, and Chua NH (2011). Processing bodies and plant development. Curr. Opin. Plant Biol 14, 88–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, and Chua NH (2012). Dehydration stress activates Arabidopsis MPK6 to signal DCP1 phosphorylation. EMBO J 31, 1975–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Yang JY, Niu QW, and Chua NH (2006). Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. Plant Cell 18, 3386–3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Greene GH, Yoo H, Liu L, Marqués J, Motley J, and Dong X (2017). Global translational reprogramming is a fundamental layer of immune regulation in plants. Nature 545, 487–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Feng B, He P, and Shan L (2017). From Chaos to Harmony: Responses and Signaling upon Microbial Pattern Recognition. Annu. Rev. Phytopathol 55, 109–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li W, Xiang T, Liu Z, Laluk K, Ding X, Zou Y, Gao M, Zhang X, Chen S, et al. (2010). Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudo-monas syringae effector. Cell Host Microbe 7, 290–301. [DOI] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, and Felix G (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125, 749–760. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This manuscript did not generate new datasets or code.