Abstract
Background
The development of nanocarrier technology has attracted great interest in the last decade. Biodegradable spheres made of functionalized silk have considerable potential to be used as drug delivery systems for cancer treatment. A targeting ligand displayed at the surface of a carrier, with a specific affinity towards a particular receptor, can further enhance the accumulation and uptake of nanoparticles at the site of a tumor.
Materials and methods
The hybrid constructs were obtained by adding a Her2-binding peptide (H2.1) to MS1 and MS2 bioengineered silks based on the MaSp1 and MaSp2 proteins from N. clavipes, respectively. The H2.1MS1 and H2.1MS2 proteins were blended at a weight ratio of 8:2. Stable silk particles were formed by mixing a soluble protein with potassium phosphate using a micromixing technique. We used specific inhibitors of endocytosis to determine the cellular uptake pathway of the silk nanoparticles in human Her2-positive breast cancer cells. The subcellular distribution of silk particles was investigated by evaluating the signal colocalization with organelle-specific tracker. Moreover, lysosomal and exosomal inhibitors were implemented to evaluate their impact on the silk spheres behavior and degradation.
Results
The functionalized spheres were specifically taken up by Her2-positive cancer cells. Silk particles facilitated the entry into cells through both the clathrin- and caveola-dependent pathways of endocytosis. Upon entering the cells, the particles accumulated in the lysosomes, where intracellular degradation occurred.
Conclusions
The present study demonstrated directly that the lysosomal function was essential for silk-based carrier elimination. The degradation of the carrier is of great importance to develop an optimal drug delivery system.
Keywords: silk spheres, endocytosis, nanoparticle trafficking, lysosomal degradation, targeted drug delivery, cancer therapy
Introduction
Multidisciplinary research in the field of nanomedicine has led to the development of a variety of nanoparticle-based carrier systems. The main goal of nanomedicine is to design a formulation that i) would target a drug to the desired site of action with simultaneous reduction of the access to sites of toxicity, ii) control the location and rate of drug release, and iii) in certain cases, transport a drug across a biological barrier.1 The discovery of the enhanced permeability and retention (EPR) effect2,3 associated with tumors has emerged as the foundation for the passively targeted nanomedicines that preferentially accumulate in tumors.4 However, the therapeutic outcome based on exploiting the EPR effect can be inconsistent, mainly due to the heterogeneity associated with tumor tissues.5 Inclusion of targeting ligands displayed at the surface of a carrier, with a specific affinity towards a particular molecule expressed at the target site, can further enhance the accumulation and uptake of nanoparticles at the site of action. The enhanced cellular uptake of nanoparticles is of great importance because targets of many theranostics agents are localized in subcellular compartments.6 Knowledge about the mechanism of cellular uptake, intracellular routing and fate of nanoparticles, is essential.
The nanomedicines routinely used in the clinic and the growing number entering preclinical and clinical trials, such as liposomes, polymeric nanoparticles, dendrimers, and gold nanoparticles, have demonstrated remarkable potential as carrier systems for drugs.7 Some polymeric materials are often not advantageous for drug delivery due to the required use of organic solvents or harsh formulation conditions during their processing. Therefore, the polymers of natural origin, which can be processed in an aqueous environment, have received growing attention. Among natural polymers, spider silk proteins are promising candidates. Their noncytotoxicity, biotechnological accessibility and simple processability into various morphologies, make them suitable material for biomedical applications.8,9 Moreover, bioengineered spider silk can be functionalized via genetic engineering.10,11 Nanoparticles made of bioengineered silk that are functionalized with tumor-homing peptides have shown enhanced cellular uptake.12–16
Previously, we designed a system for targeted cancer therapy that was composed of blended bioengineered silks functionalized with a Her2-binding peptide, ie, H2.1.12,13,17 The specificity of the H2.1 peptide was previously analyzed.18 Her2-binding ligand enabled the internalization of blended spheres into target cells upon binding with the Her2 receptor.12 The encapsulated drug (doxorubicin) was released from silk nanoparticles and localized in the nucleus, which caused the death of cells.12 Furthermore, we developed the automated sphere production process under controllable and repeatable conditions by using micromixing in combination with ultrafiltration.17 The shear stress generated during micromixing and ultrafiltration did not impair the binding properties of the H2.1-functionalized spheres.17 However, a more advanced studies are required to indicate the mechanism involved in the uptake, intracellular trafficking and the final fate of silk spheres.
The potential application of particles made of silk of various origin as drug carriers has been evaluated in vitro.12,13,17,19–23 Several studies to characterize the silk particles degradation have been conducted. Lysosomal inhibitors were used to study their effect on drug release from silkworm silk spheres and thus as an indirect indication of sphere degradation.24,25 Upon their application, reduced drug release was observed.24 Moreover, silk particles were exposed ex vivo to lysosomal enzyme preparations, and after 5 days the morphological changes to silk nanoparticles following enzymatic treatment were indicated.25 Additionally, enzymatic degradation studies of eADF4(C16) particles based on Araneus diadematus spider silk using trypsin and elastase showed that the silk spheres upon enzyme treatment decreased in size.22 Moreover, Lammel et al performed a drug release study employing a model drug loaded into eADF(C16) particles.22 An accelerated drug release upon enzymatic treatment of the silk spheres was observed. However, none of those studies directly examined the fate of the silk particles in the live cell.
Our aim was to analyze the cellular uptake, intracellular distribution and fate of Her2-targeting silk nanospheres inside cells. We used inhibitors of the clathrin- and caveola-mediated endocytosis to determine the cellular uptake pathway of the silk nanoparticles. Moreover, inhibitors of the lysosomal acidification and enzymatic activity, as well as inhibitors of the exosomal release, were implemented to evaluate their impact on the silk spheres behavior. This study provides evidence of the subsequent accumulation of silk spheres in lysosomes upon entering the cell and the importance of lysosomal activity for enzymatic degradation of the silk nanoparticles in live cells.
Materials and methods
Expression and purification of bioengineered spider silks
The proteins H2.1MS1 and H2.1MS2 based on the Nephila clavipes MaSp1 and MaSp2 spidroins were designed, expressed and purified as indicated previously.12,13,26,27
Cell culture
Human Her2-overexpressing breast cancer cells SKBR3 (ATCC, Manassas, VA) were used in the study. Cells were maintained in Dulbecco’s Modified Eagle Medium (Biowest, Nuaillé, France) supplemented with 10% fetal bovine serum (Biowest, Nuaillé, France) and 80 µg/mL gentamycin (KRKA, Novo Mesto, Slovenia). Cells were grown at 37 °C in a humidified atmosphere containing 5% CO2.
Silk sphere preparation
The H2.1MS1 and H2.1MS2 silks were labeled with fluorophore ATTO647N according to the manufacturer’s instructions (Sigma, St. Louis, MO). Next, the ATTO647N conjugated proteins H2.1MS1 and H2.1MS2 were blended at a weight ratio of 8:2, respectively, and then used at concentration of 0.5 mg/mL to form H2.1MS1:H2.1MS2 spheres.13 Mixing of 2 M potassium phosphate (pH 8) with silk solution using a syringe pump system (neMESYS 2600N, Cetoni GmbH, Korbuβen, Germany) was implemented for the sphere preparation as described previously.17 Briefly, the micromixing was conducted at a silk:phosphate volumetric ratio of 1:10 and a flow rate of 10:100 μL/s. A T-shaped mixing element with a circular mixing zone having an inner diameter of 150 µm (P-890, PEEK tee, Upchurch Scientific, Oak Harbor, USA), the feed tubes of 250 µm (Upchurch Scientific, Oak Harbor, USA) and the outlet tube of 500 µm in diameter (Upchurch Scientific, Oak Harbor, USA) were applied.
Cellular uptake of silk spheres – confocal laser scanning microscopy (CLSM) study
To study cellular uptake, the SKBR3 cells were seeded onto 8-well Lab-Tek chambered cover glasses (Nunc, Naperville, IL, USA) at a density of 1×105 cells/well and then cultured for 24 h. Next, the cells were incubated with ATTO647N-labeled H2.1MS1:H2.1MS2 particles (10 µg/mL) for 4 h at 37 °C or 2 h at 4 °C. After washing with PBS, the cells were fixed with 4% paraformaldehyde (PFA; Sigma, St. Louis, MO, USA), and then the cell membranes were stained with ConA-FITC (Sigma, St. Louis, MO, USA) at a concentration of 50 μg/mL for 30 min. Subsequently, the cells were washed with PBS, immersed in Fluoroshield mounting medium with DAPI (Sigma, St. Louis, MO), and then analyzed under an Olympus FV1000 Confocal Laser Scanning Microscope (CLSM; Shinjuku, Tokyo, Japan) connected to a blue laser diode and an argon laser. Image acquisition and analysis were performed with a 60× objective, a 1.4 N.A. oil immersion lens and FLUOVIEW Viewer software, ver. 4.1. The DAPI-stained nuclei were visualized using 405 nm excitation and 440–480 nm emission wavelengths. Images of the silk spheres labeled with ATTO647N were visualized using 635 nm excitation and 575–675 nm emission wavelengths. To visualize cell membranes, 488 nm excitation and 495–525 nm emission wavelengths (FITC) were applied.
Cellular uptake of silk spheres – flow cytometry (FCM) study
The SKBR3 cells were seeded onto 12-well plates at a density of 2.5×105 cells per well. Next, the cells were preincubated separately for 30 min with endocytosis inhibitors: 10 µg/mL chlorpromazine (CPM; Sigma, St. Louis, MO) or 5 µg/mL filipin (FLP; Sigma, St. Louis, MO, USA). Then, the ATTO647N-labeled H2.1MS1:H2.1MS2 spheres were added to the cell culture at a final concentration of 10 µg/mL and incubated for an additional 4 h. The cells receiving silk spheres but without inhibitor treatment were used as a positive control (w/o inhibitor); the cells without any treatment were used as a negative control (control). Next, the cells were washed with PBS and detached with nonenzymatic cell dissociation solution (Sigma, St. Louis, MO, USA). Fluorescence data were collected in FL4 channel of BD FACSAria flow cytometer (BD Pharmingen, San Jose, CA, USA), and analyzed using FlowJo software (Tree Star, Ashland, OR, USA). Three independent experiments were performed.
The intracellular distribution of silk spheres – CLSM study
The SKBR3 cells were seeded onto 8-well Lab-Tek chambered cover glasses (Nunc, Naperville, IL) at a density of 1×105 cells/well and then cultured for 24 h. To determine the intracellular localization of the silk spheres, the cells were incubated with ATTO647N-labeled H2.1MS1:H2.1MS2 particles (10 µg/mL) for 4 h at 37 °C. After washing with PBS, the endosomes and lysosomes were stained with 50 nM LysoTracker Green (Molecular Probes, Eugene, OR, USA) for 30 min at 37 °C. Subsequently, the cells were washed with PBS, fixed with 4% PFA and immersed in the Fluoroshield mounting medium with DAPI. The intracellular distribution of silk spheres was observed using an Olympus FV1000 CLSM as described above. The endo-/lysosome fluorescence was analyzed using a 488 nm excitation and 495–525 nm emission wavelengths. Three independent experiments were performed.
The intracellular degradation and exosomal release of silk spheres – CLSM study
The SKBR3 cells were plated onto 8-well Lab-Tek chambered cover glasses (Nunc, Naperville, IL) at a density of 5×104 cells/well and then cultured for 24 h before the experiment. For the degradation studies of silk spheres, the ATTO647N-conjugated H2.1MS1:H2.1MS2 spheres at a final concentration of 10 µg/mL and one of three lysosomal inhibitors, i) chloroquine (50 µM; Santa Cruz Biotechnology, Dallas, TX, USA), ii) cathepsin G inhibitor I (100 µM; Santa Cruz Biotechnology, Dallas, TX, USA) or iii) chymostatin (50 µM; Sigma, St. Louis, MO, USA), were added. Cells were incubated at 37 °C for additional 4 h. To analyze the exosome-mediated elimination of silk spheres, the cells were incubated at 37 °C for 4 h with H2.1MS1:H2.1MS2 spheres at a final concentration of 10 µg/mL in the presence of an inhibitor of exosomal release: i) dimethyl amiloride (DMA, 10 µg/mL; Sigma, St. Louis, MO, USA), ii) omeprazole (OMP, 50 µg/mL; Sigma, St. Louis, MO, USA) or iii) brefeldin A (BFA, 10 µg/mL, Sigma, St. Louis, MO, USA). After washing with PBS to remove unbound particles, the cells were immersed in a fresh medium containing appropriate inhibitors and further cultured for the indicated periods of time. The cells incubated with silk spheres but without any inhibitor treatment was used as a control. Next, the cells were washed, fixed and observed using a CLSM applying the settings described above. The intensity of the fluorescence signal from silk spheres was assessed with FLUOVIEW Viewer software Ver. 4.1. At least 10 individual regions of interest (ROIs) were analyzed on three randomly selected CLSM images to calculate an average fluorescence intensity of the silk spheres. The experiments were repeated three times.
Statistics
The statistical significance of the differences between groups was calculated using analysis of variance (ANOVA). In case of significance, post hoc tests with Bonferroni correction were performed. The differences were considered significant when p<0.001 (***), p<0.01 (**), p<0.05 (*). The data are expressed as the means ± SEM. The significant differences were calculated with GraphPad Prism 5 software.
Results
Cellular uptake of silk spheres
H2.1MS1:H2.1MS2 blended silk spheres were generated using a micromixing technique by salting out with 2 M potassium phosphate (pH 8). Scanning electron microscopy confirmed that the obtained particles were spherical with a uniform particle size of 251±16 nm (data not shown). The size, size distribution, zeta potential, colloidal stability, and production efficiency of the H2.1MS1:H2.1MS2 blended spheres were investigated in previous reports.13,17 Moreover, in our previous work, we showed that the flow cytometry analysis indicated a basal level of binding of the control spheres (without functionalization) to Her2-positive and -negative cells, as well as functionalized particles to control Her2-negative cells.12,13,17 However, the confocal microscopy revealed that in such cases spheres were on the surface of the cells and were not internalized.12,17 Therefore, to investigate the degradation of spheres within cells, we performed our analysis using Her2-positive cells and spheres targeting Her2 molecule.
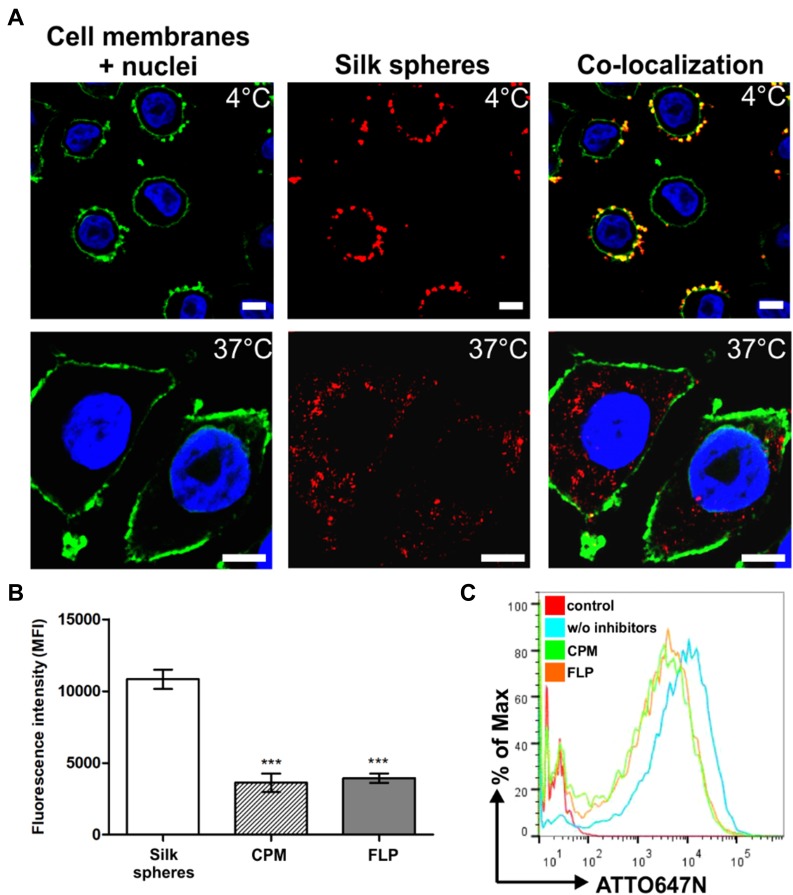
To analyze the general internalization pathway of the nanoparticles, we incubated the SKBR3 cells with the fluorescently labeled spheres at different temperatures. As shown in Figure 1A, after the incubation at 4 °C, the silk spheres did not enter the cells and were mostly observed at the surface of cell membranes. In contrast, after the incubation at 37 °C, the functionalized nanoparticles were internalized into cells and were detected in the cytoplasm (Figure 1A). To further elucidate pathways of the potential cellular uptake of the functionalized spheres, CPM (10 µg/mL; clathrin-mediated pathway inhibitor) or FLP (5 µg/mL; caveola-mediated pathway inhibitor) was used.28 The FCM studies indicated that the clathrin- and caveola-mediated pathways were both involved in the uptake of the functionalized spheres (Figure 1B, C). Compared with the cells not treated with inhibitors (w/o inhibitors), in the presence of CPM or FLP, the cellular uptake of the silk nanoparticles was significantly reduced to 33% or 36%, respectively.
Figure 1.
The cellular uptake of the functionalized silk spheres analyzed by confocal microscopy and flow cytometry. (A) SKBR3 cells were incubated with functionalized silk spheres at 4 °C (top) or 37 °C (bottom). Red, ATTO647N-conjugated spheres; green, cell membranes stained with ConA-FITC; blue, nuclei stained with DAPI. Scale bar: 10 µm. (B, C) Effect of endocytosis inhibitors on the cellular uptake of silk spheres. SKBR3 cells were preincubated with endocytosis inhibitor CPM or FLP, then incubated with functionalized silk spheres for 4 h at 37 °C. (B) The mean fluorescence intensity (MFI ± SEM) of three independent experiments is shown. (***) indicates statistical significance with p<0.001. (C) Representative graphs of the flow cytometry analysis of SKBR3 cells treated with ATTO647N-labeled H2.1MS1:H2.1MS2 blended spheres in the presence or w/o inhibitors. Control – nontreated cells.
The intracellular distribution of silk spheres
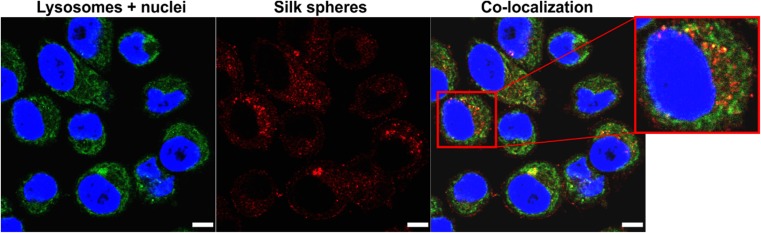
In Figure 2, the intracellular distribution of the functionalized silk nanoparticles is indicated. The functionalized spheres were internalized and mostly trafficked to the endo- and lysosomal compartments, which was indicated by observing the colocalization signals (yellow) of silk spheres (red) and endo/lysosomes (green) inside the cells (Figure 2).
Figure 2.
The intracellular localization of the silk particles by CLSM. SKBR3 cells were incubated with spheres at 37 °C for 4 h, followed by staining with LysoTracker Green for the endo-/lysosome visualization. Red, silk spheres labeled with ATTO647N; green, endo-/lysosomes stained with LysoTracker Green; blue, nuclei stained with DAPI. Scale bar: 10 μm.
The intracellular degradation of silk spheres
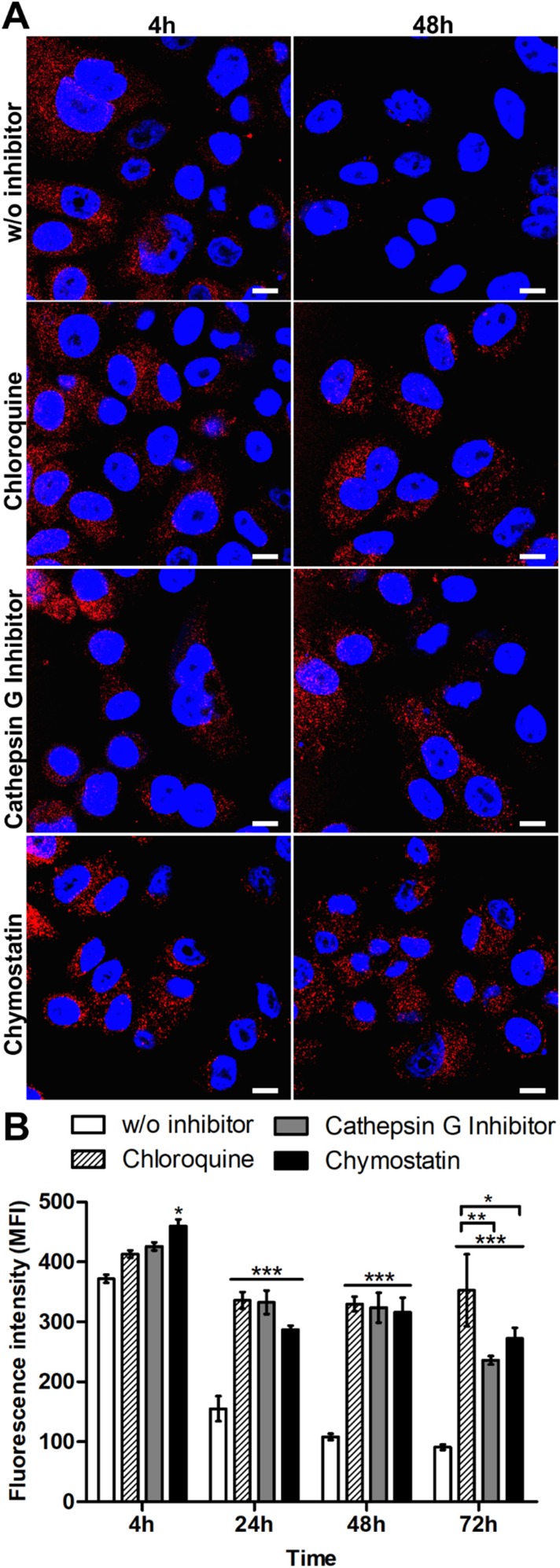
The impact of lysosomal acidification and proteolytic lysosomal enzymes in silk spheres processing was assessed. The SKBR3 cells were incubated with functionalized silk spheres alone or in the presence of chloroquine, cathepsin G inhibitor I or chymostatin for the indicated periods of time at 37 °C and subsequently analyzed using CLSM. As shown in Figure 3, in the control cells, the signal from internalized spheres decreased with time, and the spheres were hardly visible after 48 h of incubation (Figure 3A). In the presence of agent that suppressed the lysosomal acidification or inhibitors of lysosomal enzymes, the strong signal of the internalized spheres was detected after 48 h (Figure 3A). Moreover, to quantify the obtained results, the fluorescence intensity of the spheres in the individual cells was measured, and the mean value of spheres’ fluorescence per cell was calculated (Figure 3B). As the incubation time increased, the intensity of the fluorescence signal remained at the same level inside the cells after chloroquine and inhibitor treatment, and it was maintained throughout the experiment. Moreover, in the presence of chloroquine, the cell-associated fluorescence after 72 h was similar to that observed after 24 and 48 h treatment. However, for samples treated with lysosomal enzymes inhibitors, a further signal decrease was observed (Figure 3B).
Figure 3.
Analysis of the intracellular processing of the silk spheres in SKBR3 cells. (A) The cells were incubated with ATTO647N-labeled H2.1MS1:H2.1MS2 spheres in the presence of lysosomal inhibitor chloroquine, cathepsin G inhibitor I or chymostatin for the indicated time periods at 37 °C. The silk particles were observed using CLSM. Red, spheres conjugated with ATTO647N; blue, nuclei stained with DAPI; scale bar: 10 μm. (B) The fluorescence intensity of the silk spheres was measured by the mean fluorescence value (±SEM) per cell (n=30). Statistical significance with p<0.001 (***), p<0.01 (**), p<0.05 (*).
The exosomal release of silk spheres
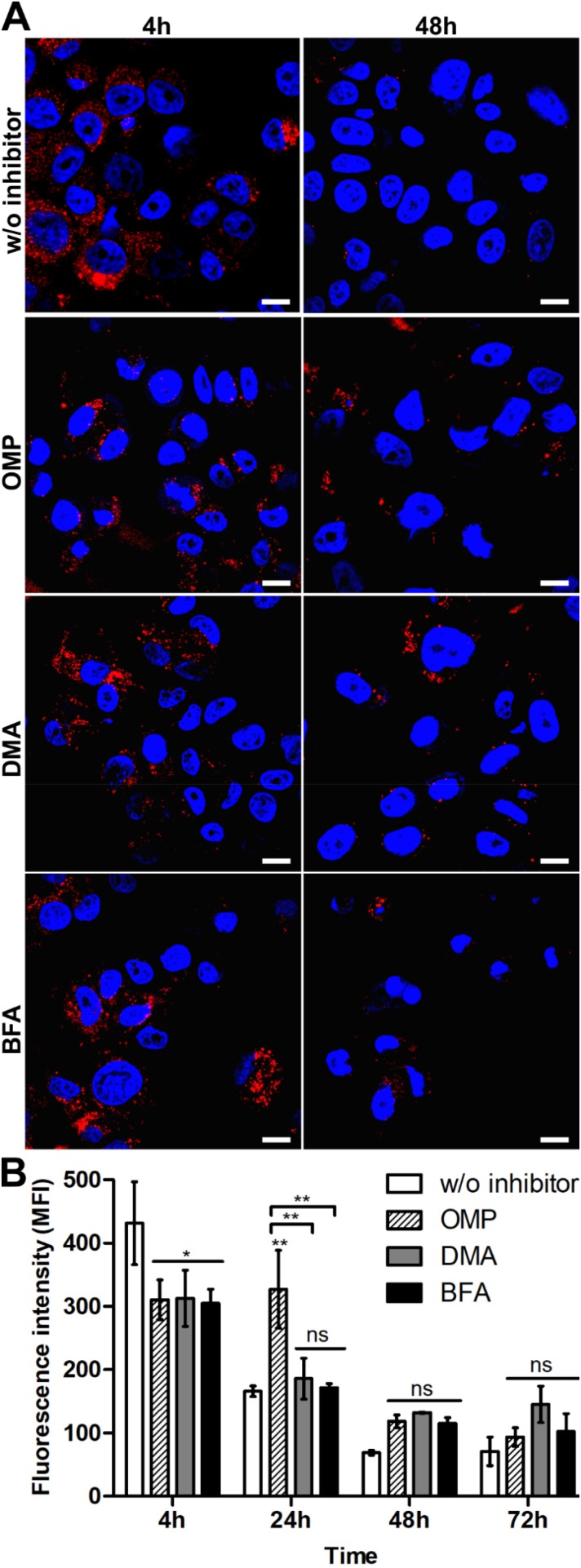
To determine the functional relevance of exosomes in the elimination of silk particles from cells, SKBR3 cells were incubated with functionalized spheres alone or in the presence of exosome inhibitors for the indicated periods of time at 37 °C (Figure 4). When SKBR3 cells were treated with OMP, DMA or BFA, a significantly reduced signal of functionalized spheres after 4 h incubation was observed compared with cells not treated with inhibitors (Figure 4B). The DMA and BFA treatments did not contribute to the spheres’ processing since the intensity of the fluorescence signal of spheres decreased with the time of incubation, similarly to the control cells (Figure 4B). The functionalized spheres were more abundant in the cells after 24 h of OMP treatment compared with control cells (Figure 4B). After 48 and 72 h of incubation, OMP did not affect the secretion of the functionalized spheres, similarly to DMA and BFA (Figure 4).
Figure 4.
Analysis of the exosomal release of the silk spheres in SKBR3 cells. (A) The cells were incubated with ATTO647N-labeled H2.1MS1:H2.1MS2 spheres in the presence of an inhibitor of exosomal release, DMA, OMP, or BFA, for the indicated time periods at 37 °C. The silk particles were observed using CLSM. Red, spheres conjugated with ATTO647N; blue, nuclei stained with DAPI; scale bar: 10 μm. (B) The fluorescence intensity of the spheres was measured by the mean fluorescence value (±SEM) per cell (n=30). Statistical significance with p<0.01 (**), p<0.05 (*), p≥0.05 (ns).
Discussion
Silk nanoparticles are a promising approach for the intracellular delivery of therapeutic agents, but the knowledge regarding their intracellular fate is negligible. Accordingly, one of the most important aspects of the application of nanoparticles is the answer to the questions i) whether the particles are degradable and ii) how their degradation is followed.
In a previous study, we indicated that the Her2-binding domain was exposed on the surface of the silk spheres and preserved its binding capacity.12 The functionalized nanoparticles were internalized into cells overexpressing Her2, and the silk spheres were observed in the cytoplasm. However, the route of uptake utilized by nanoparticles to enter cells needed to be determined. The present study revealed that the clathrin- and caveola-mediated pathways were both involved in the uptake of the functionalized spheres. Generally, the major uptake routes for macromolecules are clathrin-mediated endocytosis, caveola-mediated endocytosis and micropinocytosis.29 Larger particles are predominantly nonselectively taken up by macropinocytosis.29 However, this mechanism is unlikely to be involved in the cellular uptake of the silk nanospheres functionalized with a peptide that binds to a specific receptor. Our results are consistent with research by Chen et al, which used specific inhibitors of endocytosis to assess the cellular uptake of micelles decorated with the same Her2-binding peptide.30 The receptor-mediated endocytosis, both clathrin- and caveola-mediated, is the predominant pathway of micelle entry into cells.30 Moreover, Chen et al indicated that macropinocytosis was not closely involved in the internalization of these particles.30 EGF receptors utilize multiple endocytotic pathways, possibly depending upon the ligand type and its concentration in the extracellular milieu.31 At a low concentration of ligand, the EGF-type receptors are internalized through clathrin-dependent endocytosis, and at high ligand concentration, a caveola-dependent endocytotic pathway is also used.31
A few preliminary studies tracing the intracellular fate of nanoparticles formed of silk of different origin were recently performed.15,16,32,33 The uptake route of the silkworm silk nanoparticles to MCF7 human breast cancer cells was an energy-dependent endocytosis process mediated by clathrin.33 Moreover, the application of the chemical inhibitors at the uptake studies of eADF4(C16) silk particles and eADF4(C16) particles functionalized with cell-homing peptide Tat, RGD or poly-lysine indicated a clathrin-mediated mechanism of the spheres’ internalization into HeLa cells.15,16 However, in general, the efficacy of endocytosis inhibitors is highly cell line dependent.34 Moreover, the particles presented above either did not have any functional domains or were functionalized with nonspecific cell-homing peptides; hence, they would interact with any cell type (due to the nonspecific interactions). Furthermore, they differed in size and silk origin. Therefore, the separately performed internalization study of each type of silk nanoparticle is essential. Our strategy implemented the silk spheres that were derived from Nephila clavipes MaSp1 and MaSp2 spidroins and were functionalized with Her2-binding peptide. The H2.1 functional domain presented on the surface of the silk carriers should guide the nanoparticle to a specific receptor, thus determining the predominant entryway into cells and providing their high selectivity.
In the present study, we indicated that the functionalized spheres upon their internalization were mostly trafficked to the endo-/lysosomal compartments. Maitz et al demonstrated localization of silk nanoparticles in lysosomes upon entering blood cells.32 Their findings concerned the influence of PEGylation on the cellular uptake of the silkworm silk particles.32 The observed localization of the particles could result from the surface modification of spheres and could be particle-type and/or cell-type specific. Therefore, similarly to the internalization aspect, the intracellular distribution study should be performed separately for each type of silk nanoparticle.
Previously, we indicated that the morphology of cells upon sphere internalization was different from nontreated cells; upon the massive uptake of silk spheres, the cell size increased and normalized with time.12 Moreover, the signal of the internalized spheres disappeared with time. Furthermore, we indicated in previous studies that a considerable amount of Dox molecules was accumulated in the nucleus.12 Because the silk spheres were observed mostly in the cytoplasm and did not penetrate the nucleus, the Dox deposited in the nucleus was released from the spheres before entering the nucleus. Also, our previous drug release studies showed the significantly increased Dox release from silk spheres at low pH.12,13 These observations might suggest that the lysosomal microenvironment could be responsible for Dox release. From one hand, the low pH in the lysosomes could increase the Dox release, and from the other hand, the proteolytic enzymes present in the lysosomes might degrade the spheres, which could further enhance the drug release. To answer the question whether the silk spheres were proteolytically degraded inside the cells, the effects of different compounds disrupting lysosomal proteolysis were evaluated in this study either by suppressing the lysosomal acidification or by inhibiting directly the cathepsins. Neutralization of the internal acidic environment by weak alkaline compounds, such as chloroquine, inhibits lysosomal function. Chloroquine-mediated lysosomal dysfunction leads to the increase of the intralysosomal pH, accumulation of ineffective autophagosomes and impairment of autophagic protein degradation.35,36 Moreover, the role of lysosomal enzymes in silk sphere degradation was investigated by using the lysosomal protease inhibitors chymostatin and cathepsin G inhibitor I. Chymostatin has been used extensively as an inhibitor of lysosomal function, and its primary mode of action is the inhibition of serine proteases, such as cathepsins A and G.37,38 Its inhibition of other cellular proteases has also been reported.39
In the present study, treatment with chloroquine, chymostatin or cathepsin G inhibitor I induced a significant enhancement of cell-associated silk sphere fluorescence compared with control cells. These results indicated that the silk sphere degradation was mediated by both the low pH and the proteolytic environment in the lysosomal compartment. We demonstrated a higher dependence of silk degradation on the lysosomal acidification than on the enzymatic activity, especially after 72 h of inhibitor application. One possible explanation of this observation might be the dependence of the activity of lysosomal enzymes on pH.24,40
Exosomes are secreted membrane vesicles that share structural and biochemical characteristics with intraluminal vesicles of multivesicular endosomes (MVEs). Exosomes are distinguished from apoptotic bodies and microvesicles by their size, origin and composition. These vesicles do not originate by direct budding or shedding of plasma membrane.41 Instead, they are formed via inward budding of endosomal membranes, resulting in the formation of intracellular multivesicular bodies that later fuse with the plasma membrane and release exosomes to the exterior.42
In the present study, the effect of inhibitors of exosomal release, OMP, DMA and BFA, was examined. OMP, a K+/H+ ATPase inhibitor, is involved in exosome depletion and reduction of exosome release. DMA, an inhibitor of the H+/Na+ and Na+/Ca2+ channels, perturbs exosome secretion.43,44 BFA, an inhibitor of ER/Golgi-dependent protein secretion, blocks the transport of polypeptides from ER to the Golgi apparatus and dysregulates membrane traffic throughout the vacuolar system.45 Moreover, it inhibits exosome secretion from cells.46 Our analysis revealed that all tested exosomal inhibitors induced the significant reduction of the signal from functionalized spheres after 4 h of incubation compared with control cells. If inhibitors restrained the secretion of exosomes and thus the silk sphere removal, the signal deriving from ATTO647N-labeled spheres would be higher compared with control. We observed the opposite effect. In general, the tested inhibitors affect a wide range of processes related to intracellular trafficking, including endocytosis, exocytosis, and vacuolar targeting.45,47 We presume that we altered membrane secretory pathways triggered by the in vitro exposure to the proton pump inhibitors (PPIs) and BFA-induced disruption, and thus the endocytosis-dependent uptake of silk spheres was impaired. However, the functionalized spheres were more abundant in the cells after 24 h of OMP treatment compared with control and other samples (Figure 4B), indicating that OMP could inhibit exosome secretion and thus the elimination of silk spheres. OMP, which is a proton pump blocker, could potentially inhibit lysosomal acidification and subsequent proteolytic digestion of the silk spheres.48 Its role needs further investigation. In contrast, DMA and BFA did not inhibit the secretion of silk nanoparticles over time, indicating that the secretion from cells mediated by exosomes did not contribute to the elimination of the silk nanoparticles by cells.
The degradation of biomaterials is obviously an important feature for their medical application. Silk biomaterials are generally considered biodegradable, and numerous studies have described the mechanism of silk degradation as depending on proteases.49–51 However, these studies mostly analyzed the responses of various silk biomaterial structures to enzyme treatments, eg, films,52 hydrogels,52 fibers53 or scaffolds,54 while the assessment of the degradation behavior of the silk nanoparticles remained poorly characterized. According to the literature, the biodegradation rate largely depends on the morphology and structural characteristics of the biomaterials, such as size and porosity.49 For example, α-chymotrypsin degrades silkworm fibers;49 however, α-chymotrypsin does not have an appreciable effect on the degradation of the films made of regenerated silkworm silk.55
As mentioned above, studies regarding the degradation of the silkworm silk nanoparticles are relatively insufficient. The silk mass loss, particle swelling, surface charge reduction, and the aggregation of silkworm silk nanoparticles in response to protease XIV and papain were observed.56 Papain is an enzyme similar to mammalian lysosomal cathepsins B, H, L, and S. Moreover, following a 5-day exposure to a cocktail of lysosomal enzymes isolated from the rat liver, Wongpinyochit et al demonstrated a significant reduction of the silk nanoparticle size.56 The relatively slower and partial carrier degradation might have been due to the presence of PEG on the silk surface, which hindered access of the proteolytic enzymes, and the general ex vivo conditions of the experiment.56 In another study, Totten et al showed that doxorubicin was released more rapidly from silkworm silk nanoparticles in the presence of papain.24 This indicated that the activity of lysosomal enzymes was important for drug release. Moreover, the role of lysosomal enzyme activity and lysosomal acidification in triggering drug release was also indicated in cells. However, the applied examination strategy could be crucial, since pH played a major role in doxorubicin release, while the impact of the enzymatic activity on drug release was comparatively lower. Nevertheless, these studies provided evidence for lysosomal drug delivery in live cells.24
Studies by Wongpinyochit and Totten analyzed regenerated silkworm silk nanoparticles. Our spider silk sequences differed from the Bombyx mori sequence, and they lacked the amino acids that are recognized by cysteine proteases in the silkworm silk. Therefore, in our study, as a potential target for inhibitors, we selected serine proteases. Previously, Lammel et al employed an elastase and trypsin, ie, serine proteases, to study the degradation of eADF4(C16) silk spheres.22 Indeed, the degradation experiments showed that the silk particles decreased in size following these enzyme exposures. Moreover, a model drug release study upon sphere degradation indicated an accelerated drug release behavior.22 However, in contrast to the studies mentioned above, our approach determined directly the silk sphere degradation in the lysosomes of live cells. Moreover, we observed more rapid degradation that occurred over 48 h compared with a partial degradation over a 5-day ex vivo lysosomal enzyme preparation treatment.56 In contrast to the study by Totten et al that measured drug trafficking to the nucleus, providing indirect evidence of silk nanoparticle degradation in the lysosomes, we present direct evidence of the involvement of lysosomes in silk carrier elimination.
Conclusion
The degradation behavior of biomaterials made of different silk origin can vary essentially, depending on their amino acid sequence. Moreover, the complex in vivo environment specific to the cell/tissue type and location can determine the rate of the degradation of silk biomaterial. We analyzed the fate of functionalized spheres inside cancer cells. The spheres were made of bioengineered silks based on the Nephila clavipes MaSp1 and MaSp2 spidroins. The functionalized silk spheres were processed into cells by the clathrin- and caveola-mediated endocytosis pathways and were subsequently trafficked to lysosomes. Our results indicate that the exosome-mediated secretory pathway was not or was marginally involved in the removal of the functionalized silk spheres from cells. The lysosomal function was essential for silk-based carrier elimination. The degradation of the carrier is of great importance to develop a safe drug delivery system. We consider these findings important evidence supporting the application of functionalized silk nanoparticles as drug carriers in the in vivo treatment of tumors.
Acknowledgment
The project was supported by an internal grant of the Medical Faculty II of Poznan University of Medical Sciences (502-14-22333810-10604), Poland, and partially by a grant from The National Science Centre, Poland (2014/15/B/NZ7/00903).
Disclosure
The authors reports no conflicts of interest in this work.
References
- 1.Duncan R, Richardson SC. Endocytosis and intracellular trafficking as gateways for nanomedicine delivery: opportunities and challenges. Mol Pharm. 2012;9(9):2380–2402. [DOI] [PubMed] [Google Scholar]
- 2.Maeda H. Tumor-selective delivery of macromolecular drugs via the EPR effect: background and future prospects. Bioconj Chem. 2010;21(5):797–802. [DOI] [PubMed] [Google Scholar]
- 3.Maeda H. The link between infection and cancer: tumor vasculature, free radicals, and drug delivery to tumors via the EPR effect. Cancer Sci. 2013;104(7):779–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prabhakar U, Maeda H, Jain RK, et al. Challenges and key considerations of the enhanced permeability and retention effect for nanomedicine drug delivery in oncology. Cancer Res. 2013;73(8):2412–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yameen B, Choi WI, Vilos C, Swami A, Shi J, Farokhzad OC. Insight into nanoparticle cellular uptake and intracellular targeting. J Control Release. 2014;190:485–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rajendran L, Knolker HJ, Simons K. Subcellular targeting strategies for drug design and delivery. Nat Rev Drug Discov. 2010;9(1):29–42. doi: 10.1038/nrd2897 [DOI] [PubMed] [Google Scholar]
- 7.Shi J, Kantoff PW, Wooster R, Farokhzad OC. Cancer nanomedicine: progress, challenges and opportunities. Nat Rev Cancer. 2017;17(1):20–37. doi: 10.1038/nrc.2016.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seib FP. Silk nanoparticles - an emerging anticancer nanomedicine. AIMS Bioeng. 2017;4(2):239–258. doi: 10.3934/bioeng.2017.2.239 [DOI] [Google Scholar]
- 9.Aigner TB, DeSimone E, Scheibel T. Biomedical applications of recombinant silk-based materials. Adv Mater. 2018;30(19):e1704636. doi: 10.1002/adma.v30.19 [DOI] [PubMed] [Google Scholar]
- 10.Jastrzebska K, Kucharczyk K, Florczak A, Dondajewska E, Mackiewicz A, Dams-Kozlowska H. Silk as an innovative biomaterial for cancer therapy. Rep Pract Oncol Radiother. 2015;20(2):87–98. doi: 10.1016/j.rpor.2014.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deptuch T, Dams-Kozlowska H. Silk materials functionalized via genetic engineering for biomedical applications. Materials. 2018;10(12):1417–1438. doi: 10.3390/ma10121417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Florczak A, Mackiewicz A, Dams-Kozlowska H. Functionalized spider silk spheres as drug carriers for targeted cancer therapy. Biomacromolecules. 2014;15(8):2971–2981. doi: 10.1021/bm500591p [DOI] [PubMed] [Google Scholar]
- 13.Florczak A, Jastrzebska K, Mackiewicz A, Dams-Kozlowska H. Blending two bioengineered spider silks to develop cancer targeting spheres. J Mater Chem B. 2017;5:3000–3011. doi: 10.1039/C7TB00233E [DOI] [PubMed] [Google Scholar]
- 14.Numata K, Mieszawska-Czajkowska AJ, Kvenvold LA, Kaplan DL. Silk-based nanocomplexes with tumor-homing peptides for tumor-specific gene delivery. Macromol Biosci. 2012;12(1):75–82. doi: 10.1002/mabi.201100274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elsner MB, Herold HM, Muller-Herrmann S, Bargel H, Scheibel T. Enhanced cellular uptake of engineered spider silk particles. Biomater Sci. 2015;3(3):543–551. doi: 10.1039/c4bm00401a [DOI] [PubMed] [Google Scholar]
- 16.Schierling MB, Doblhofer E, Scheibel T. Cellular uptake of drug loaded spider silk particles. Biomater Sci. 2016;4(10):1515–1523. doi: 10.1039/c6bm00435k [DOI] [PubMed] [Google Scholar]
- 17.Florczak A, Jastrzebska K, Bialas W, Mackiewicz A, Dams-Kozlowska H. Optimization of spider silk sphere formation processing conditions to obtain carriers with controlled characteristics. J Biomed Mater Res A. 2018;106(12):3211–3221. doi: 10.1002/jbm.a.36516 [DOI] [PubMed] [Google Scholar]
- 18.Urbanelli L, Ronchini C, Fontana L, Menard S, Orlandi R, Monaci P. Targeted gene transduction of mammalian cells expressing the HER2/neu receptor by filamentous phage. J Mol Biol. 2001;313(5):965–976. doi: 10.1006/jmbi.2001.5111 [DOI] [PubMed] [Google Scholar]
- 19.Jastrzebska K, Felcyn E, Kozak M, et al. The method of purifying bioengineered spider silk determines the silk sphere properties. Sci Rep. 2016;6:28106. doi: 10.1038/srep28106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jastrzebska K, Florczak A, Kucharczyk K, et al. Delivery of chemotherapeutics using spheres made of bioengineered spider silks derived from MaSp1 and MaSp2 proteins. Nanomedicine (Lond). 2018;13(4):439–454. doi: 10.2217/nnm-2017-0276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozlowska AK, Florczak A, Smialek M, et al. Functionalized bioengineered spider silk spheres improve nuclease resistance and activity of oligonucleotide therapeutics providing a strategy for cancer treatment. Acta Biomater. 2017;59:221–233. doi: 10.1016/j.actbio.2017.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lammel A, Schwab M, Hofer M, Winter G, Scheibel T. Recombinant spider silk particles as drug delivery vehicles. Biomaterials. 2011;32(8):2233–2240. doi: 10.1016/j.biomaterials.2010.11.060 [DOI] [PubMed] [Google Scholar]
- 23.Hofer M, Winter G, Myschik J. Recombinant spider silk particles for controlled delivery of protein drugs. Biomaterials. 2012;33(5):1554–1562. doi: 10.1016/j.biomaterials.2011.10.053 [DOI] [PubMed] [Google Scholar]
- 24.Totten JD, Wongpinyochit T, Seib FP. Silk nanoparticles: proof of lysosomotropic anticancer drug delivery at single-cell resolution. J Drug Target. 2017;25(9–10):865–872. doi: 10.1080/1061186X.2017.1363212 [DOI] [PubMed] [Google Scholar]
- 25.Wongpinyochit T, Uhlmann P, Urquhart AJ, Seib FP. PEGylated silk nanoparticles for anticancer drug delivery. Biomacromolecules. 2015;16(11):3712–3722. doi: 10.1021/acs.biomac.5b01003 [DOI] [PubMed] [Google Scholar]
- 26.Dams-Kozlowska H, Majer A, Tomasiewicz P, Lozinska J, Kaplan DL, Mackiewicz A. Purification and cytotoxicity of tag-free bioengineered spider silk proteins. J Biomed Mater Res A. 2013;101(2):456–464. doi: 10.1002/jbm.a.34353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huemmerich D, Helsen CW, Quedzuweit S, Oschmann J, Rudolph R, Scheibel T. Primary structure elements of spider dragline silks and their contribution to protein solubility. Biochemistry. 2004;43(42):13604–13612. doi: 10.1021/bi048983q [DOI] [PubMed] [Google Scholar]
- 28.Akagi T, Shima F, Akashi M. Intracellular degradation and distribution of protein-encapsulated amphiphilic poly(amino acid) nanoparticles. Biomaterials. 2011;32(21):4959–4967. doi: 10.1016/j.biomaterials.2011.03.049 [DOI] [PubMed] [Google Scholar]
- 29.Conner SD, Schmid SL. Regulated portals of entry into the cell. Nature. 2003;422(6927):37–44. doi: 10.1038/nature01451 [DOI] [PubMed] [Google Scholar]
- 30.Chen Q, Long M, Qiu L, et al. Decoration of pH-sensitive copolymer micelles with tumor-specific peptide for enhanced cellular uptake of doxorubicin. Int J Nanomedicine. 2016;11:5415–5427. doi: 10.2147/IJN.S111950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu S, Olenyuk BZ, Okamoto CT, Hamm-Alvarez SF. Targeting receptor-mediated endocytotic pathways with nanoparticles: rationale and advances. Adv Drug Deliv Rev. 2013;65(1):121–138. doi: 10.1016/j.addr.2012.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maitz MF, Sperling C, Wongpinyochit T, Herklotz M, Werner C, Seib FP. Biocompatibility assessment of silk nanoparticles: hemocompatibility and internalization by human blood cells. Nanomedicine. 2017;13(8):2633–2642. doi: 10.1016/j.nano.2017.07.012 [DOI] [PubMed] [Google Scholar]
- 33.Tian Y, Jiang X, Chen X, Shao Z, Yang W. Doxorubicin-loaded magnetic silk fibroin nanoparticles for targeted therapy of multidrug-resistant cancer. Adv Mater. 2014;26(43):7393–7398. doi: 10.1002/adma.201403562 [DOI] [PubMed] [Google Scholar]
- 34.Vercauteren D, Vandenbroucke RE, Jones AT, et al. The use of inhibitors to study endocytic pathways of gene carriers: optimization and pitfalls. Mol Ther. 2010;18(3):561–569. doi: 10.1038/mt.2009.281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harhaji-Trajkovic L, Arsikin K, Kravic-Stevovic T, et al. Chloroquine-mediated lysosomal dysfunction enhances the anticancer effect of nutrient deprivation. Pharm Res. 2012;29(8):2249–2263. doi: 10.1007/s11095-012-0753-1 [DOI] [PubMed] [Google Scholar]
- 36.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140(3):313–326. doi: 10.1016/j.cell.2010.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seglen O. Inhibitors of lysosomal function In: Methods in Enzymology. Vol. 96 Elsevier Inc; 1983. [DOI] [PubMed] [Google Scholar]
- 38.Zou F, Schafer N, Palesch D, et al. Regulation of cathepsin G reduces the activation of proinsulin-reactive T cells from type 1 diabetes patients. PLoS One. 2011;6(8):e22815. doi: 10.1371/journal.pone.0022815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jung M, Lee J, Seo HY, Lim JS, Kim EK. Cathepsin inhibition-induced lysosomal dysfunction enhances pancreatic beta-cell apoptosis in high glucose. PLoS One. 2015;10(1):e0116972. doi: 10.1371/journal.pone.0116972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohkuma S, Poole B. Fluorescence probe measurement of the intralysosomal pH in living cells and the perturbation of pH by various agents. Proc Nat Acad Sci USA. 1978;75(7):3327–3331. doi: 10.1073/pnas.75.7.3327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9(8):581–593. doi: 10.1038/nri2567 [DOI] [PubMed] [Google Scholar]
- 42.Sun Y, Liu J. Potential of cancer cell-derived exosomes in clinical application: a review of recent research advances. Clin Ther. 2014;36(6):863–872. doi: 10.1016/j.clinthera.2014.04.018 [DOI] [PubMed] [Google Scholar]
- 43.Chalmin F, Ladoire S, Mignot G, et al. Membrane-associated Hsp72 from tumor-derived exosomes mediates STAT3-dependent immunosuppressive function of mouse and human myeloid-derived suppressor cells. J Clin Invest. 2010;120(2):457–471. doi: 10.1172/JCI40483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Savina A, Furlan M, Vidal M, Colombo MI. Exosome release is regulated by a calcium-dependent mechanism in K562 cells. J Biol Chem. 2003;278(22):20083–20090. doi: 10.1074/jbc.M301642200 [DOI] [PubMed] [Google Scholar]
- 45.Orci L, Tagaya M, Amherdt M, et al. Brefeldin A, a drug that blocks secretion, prevents the assembly of non-clathrin-coated buds on Golgi cisternae. Cell. 1991;64(6):1183–1195. doi: 10.1016/0092-8674(91)90273-2 [DOI] [PubMed] [Google Scholar]
- 46.Misumi Y, Misumi Y, Miki K, Takatsuki A, Tamura G, Ikehara Y. Novel blockade by brefeldin A of intracellular transport of secretory proteins in cultured rat hepatocytes. J Biol Chem. 1986;261(24):11398–11403. [PubMed] [Google Scholar]
- 47.Liu W, Baker SS, Trinidad J, et al. Inhibition of lysosomal enzyme activities by proton pump inhibitors. J Gastroenterol. 2013;48(12):1343–1352. doi: 10.1007/s00535-013-0774-5 [DOI] [PubMed] [Google Scholar]
- 48.Lee CM, Tannock IF. Inhibition of endosomal sequestration of basic anticancer drugs: influence on cytotoxicity and tissue penetration. Br J Cancer. 2006;94(6):863–869. doi: 10.1038/sj.bjc.6603010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cao Y, Wang B. Biodegradation of silk biomaterials. Int J Mol Sci. 2009;10(4):1514–1524. doi: 10.3390/ijms10041514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thurber AE, Omenetto FG, Kaplan DL. In vivo bioresponses to silk proteins. Biomaterials. 2015;71:145–157. doi: 10.1016/j.biomaterials.2015.08.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu B, Song YW, Jin L, et al. Silk structure and degradation. Colloids Surf B Biointerfaces. 2015;131:122–128. doi: 10.1016/j.colsurfb.2015.04.040 [DOI] [PubMed] [Google Scholar]
- 52.Brown J, Lu CL, Coburn J, Kaplan DL. Impact of silk biomaterial structure on proteolysis. Acta Biomater. 2015;11:212–221. doi: 10.1016/j.actbio.2014.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Horan RL, Antle K, Collette AL, et al. In vitro degradation of silk fibroin. Biomaterials. 2005;26(17):3385–3393. doi: 10.1016/j.biomaterials.2004.09.020 [DOI] [PubMed] [Google Scholar]
- 54.Wang Y, Rudym DD, Walsh A, et al. In vivo degradation of three-dimensional silk fibroin scaffolds. Biomaterials. 2008;29(24–25):3415–3428. doi: 10.1016/j.biomaterials.2008.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li M, Ogiso M, Minoura N. Enzymatic degradation behavior of porous silk fibroin sheets. Biomaterials. 2003;24(2):357–365. [DOI] [PubMed] [Google Scholar]
- 56.Wongpinyochit T, Johnston BF, Seib FP. Degradation behavior of silk nanoparticles - enzyme responsiveness. ACS Biomater Sci Eng. 2018;4:942–951. doi: 10.1021/acsbiomaterials.7b01021 [DOI] [PubMed] [Google Scholar]