Abstract
Background:
Circular RNAs (circRNAs) have displayed dysregulated expression in several types of cancer. Nevertheless, their function and underlying mechanisms in cervical cancer remains largely unknown. This study aimed to describe the regulatory mechanisms in cervical cancer.
Methods:
We downloaded the circRNAs expression profiles from Gene Expression Omnibus database, and RNAs expression profiles from The Cancer Genome Atlas database. We established a circRNA-miRNA-mRNA and circRNA-miRNA-hubgene network. The interactions between proteins were analyzed using the STRING database and hubgenes were identified using MCODE plugin. Then, we conducted a circRNA-miRNA-hubgenes regulatory module. Functional and pathway enrichment analyses were conducted using R packages “Clusterprofile”.
Results:
Six circRNAs, 15 miRNAs, and 158 mRNAs were identified to construct the ceRNA network of cervical cancer. PPI (protein-protein interaction) network and module analysis identified 7 hubgenes. Then, a circRNA-miRNA-hubgene subnetwork was constructed based on the 1 DEcircRNAs, 3 DEmiRNAs, and 3 DEmRNAs. The KEGG pathway analysis indicated DEmRNAs are involved in progesterone-mediated oocyte maturation, cell cycle, and oocyte meiosis.
Conclusion:
These ceRNAs are critical in the pathogenesis of cervical and may serve as future therapeutic biomarkers.
Keywords: cervical cancer, circRNA, competitive endogenous RNA, microRNA
1. Introduction
Cervical cancer (CC) is one of the most common female genital malignant tumors, with high incidence and high mortality. There were approximately 500,000 new-diagnosed cases and 250,000 deaths.[1,2] The primary treatment of early-stage cervical cancer is surgery followed by chemotherapy or chemoradiation. However, in the event of tumor recurrence, patients with cervical cancer have a poor prognosis because of limited clinical strategies. Moreover, nearly one-third of patients die from recurrence or progression of the disease. Therefore, it is critical to investigate the molecular mechanisms involved in the development and progression of cervical cancer.[3,4]
Circular RNAs (circRNAs) are a class of noncoding RNAs with continuous, covalently closed circular structures. Owing to its absence of 3’, 5’ end and poly A tail, circRNAs can avoid the degradative effect of exonuclease and RNase R.[5,6] Compared with linear RNA, circRNAs are more conservative and stable and show cell-specific, tissue-specific and stage-specific.[7,8] With the development of high-throughput sequencing technologies and analysing, scientists gradually realize that circRNAs play important regulatory roles in the development of tumor, such as non-small cell lung cancer, colorectal cancer, and esophageal squamous cell carcinoma.[9–11] It has been demonstrated that circRNAs act as competitive endogenous RNAs (ceRNAs), namely microRNA sponges, regulating target gene expression. They could also serve as transcriptional regulator or protein-binding RNA, or even be directly translated into proteins under certain circumstances.[12–14] However, the expression and function of circRNAs in CC have rarely been explored.
In this study, circRNA microarray and RNA-Seq was used to screen CC tissues and pair-matched adjacent normal tissues to find the differential expressed circRNAs (DEcircRNAs) and their targets, by the aim to investigate potential markers for the progression of CC and elucidate the possible mechanism involved for new insights of molecular therapy.
2. Materials and methods
2.1. Data acquisition and processing
The circRNA expression profile of GSE102686 were downloaded from the GEO database, including 5 cervical tissues and 5 normal tissues. The mRNA expression profile (306 cervical tissues and 3 normal tissues) and miRNA expression profiles (309 cervical tissues and 3 normal tissues) were obtained from The Cancer Genome Atlas (TCGA) database. Neither ethical approval nor informed consent was required in this study because of the public-available data.
2.2. Identification of DEGs
We applied Limma package to identify differentially expressed circRNAs (DEcircRNAs) with thresholds of |log2 fold change (FC)| >2 and adjusted P value <.01. Additionally, the edgeR package was used to screen differentially expressed miRNA (DEmiRNA) and mRNA (DEmRNA) with thresholds of |log 2 (FC)| >2 and adjusted P value <.05.
2.3. Construction of the ceRNA network
We used the Cancer-specific CircRNA (http://gb.whu.edu.cn/CSCD/) database to predict the miRNA binding sites (MREs). Only overlapping genes were selected as candidate target miRNAs. These target miRNAs were further screened by the DEmiRNAs. Then, we used miRTarBase and TargetScan databases to identify miRNA-targeted mRNAs.[15,16] Only mRNAs recognized by all 2 databases were considered as candidate mRNAs and intersected with the DEmRNAs to determine the DEmRNAs that were targeted by the DEmiRNAs. Basing on the DEmiRNA-DEcircRNA and DEmiRNA–DEmRNA interactions, we constructed circRNA-miRNA-mRNA regulatory network, which was visualized using Cytoscape 3.7.0 software.
2.4. Construction of PPI network
To assess the interactions between DEmRNAs, we established a protein–protein interaction (PPI) network by the Search Tool for the Retrieval of Interacting Genes (STRING). The cut-off criteria were a combined score of > 0.9 for a PPI network and a node degree of ≥6 for screening hub genes. Cytoscape 3.7.0 was used for visualization. We used the MCODE app to extract hub genes from the PPI network.[17]
2.5. Functional enrichment analysis
To assess the main function of the DEmRNAs in the ceRNA network in tumorigenesis, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were conducted using the clusterProfiler package of R software.[18]P value <.05 was set as the cutoff criterion.
3. Results
3.1. Identification of differentially expressed circRNA, mRNA, and miRNA
A total of 7 the differentially expressed circRNAs (DEcircRNAs) were screened from GSE102686 dataset, including of 2 upregulated and 5 downregulated circRNAs (Fig. 1). In addition, 124 DEmiRNAs (67 upregulated and 57 downregulated miRNAs) and 2368 mRNAs (1049 upregulated and 1319 downregulated mRNAs) from RNA-Seq between CC tissues and normal cervival tissues were screened (Fig. 2A and B). The basic information of the 7 circRNAs is shown in Table 1. The basic structural patterns of the 7 circRNAs are listed in Figure 3.
Figure 1.
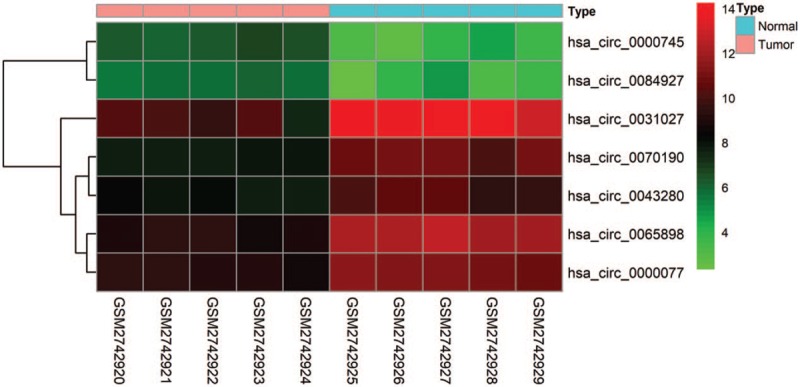
Heatmap of the differentially expressed circular RNAs.
Figure 2.
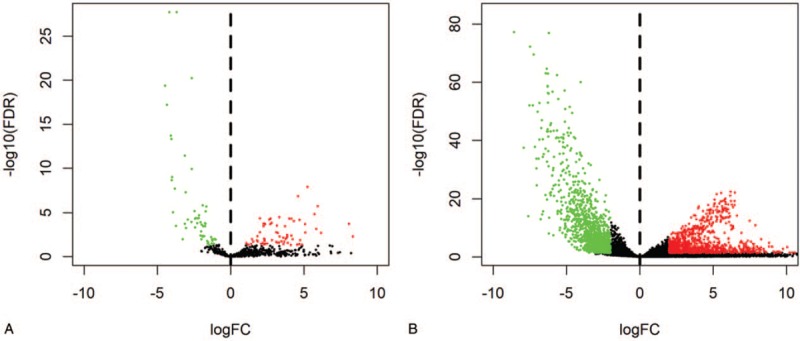
Volcano plot of differentially expressed miRNAs (A) and mRNAs (B).
Table 1.
Basic characteristics of the 7 differently expressed circRNAs.
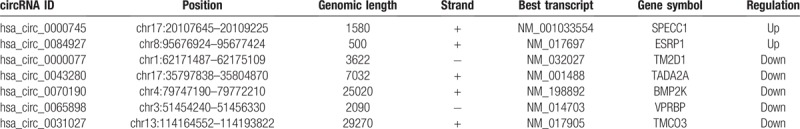
Figure 3.
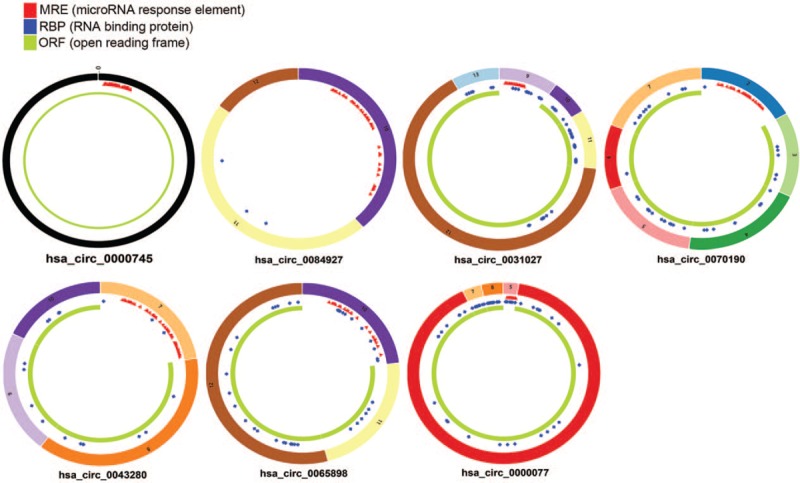
Structural patterns of the 7 circular RNAs: (A) hsa_circ_0000745, (B) hsa_circ_0084927, (C) hsa_circ_0031027, (D) hsa_circ_0070190, (E) hsa_circ_0043280, (F) hsa_circ_0065898, (G) hsa_circ_0000077.
3.2. Construction of the ceRNA network
The potential target miRNAs of 7 DEcircRNAs were retrieved from the CSCD online database. A total of 342 circRNA-miRNA pairs were identified. After intersecting with the DEmiRNAs, only 21 circRNA–miRNA pairs, including 6 circRNAs and 15 DEmiRNAs, remained. Furthermore, 1739 mRNAs predicted by 2 databases (miRTarBase and TargetScan) were identified. Then, the 1739 targeted mRNAs were compared to the 2368 DEmRNAs and only overlapping genes were selected as candidate genes. The results indicated that 158 DEmRNAs were involved in ceRNA network. Finally, we constructed a ceRNA network based on 6 circRNAs, 15 miRNAs, and 158 mRNAs (Fig. 4).
Figure 4.
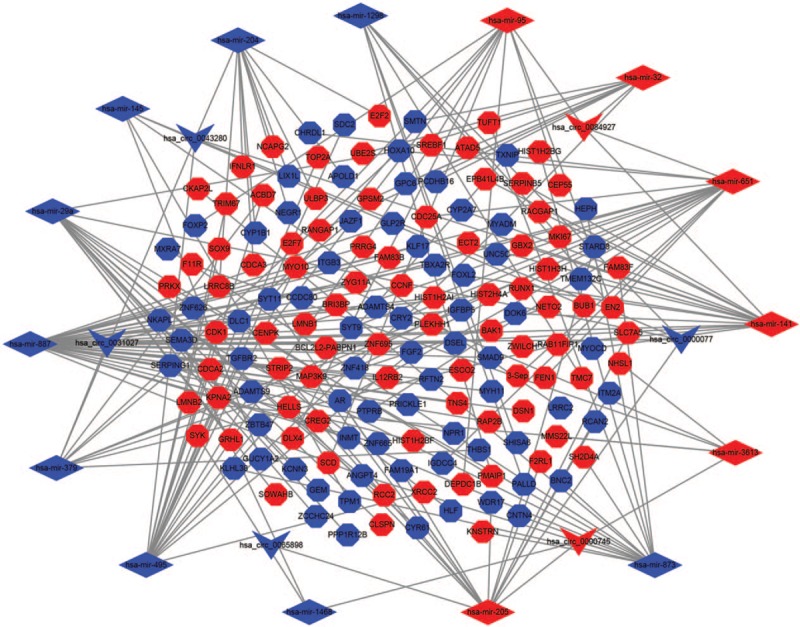
The ceRNA network of circRNA-miRNA-mRNA in CC. V indicate circRNA, diamond indicate miRNA, and octagon indicate mRNA. The nodes highlighted in red and blue represent upregulation and downregulation, respectively.
3.3. Construction of PPI network and module analysis
Based on the DEmRNAs, PPI network was conducted, involving 57 nodes and 90 edges (Fig. 5A). To explore the hubgenes in the process of PDAC carcinogenesis, the degree, betweenness centrality, key circRNA-miRNA-mRNA regulatory module was extracted using MCODE approach from the PPI network. The significant module contains 7 nodes and 21 edges. These hubgenes were CDK1, BUB1, CENPK, ZWILCH, DSN1, RCC2, and RANGAP1 (Fig. 5B). Then, we established a circRNA-miRNA-hugene subnetwork (Fig. 6), including 3 subnetwork regulatory modules (hsa_circ_0084927-hsa-mir-141-ZWILCH, hsa_circ_0084927-hsa-mir-32-RANGAP1, and hsa_circ_0084927-hsa-mir-95-CENPK).
Figure 5.
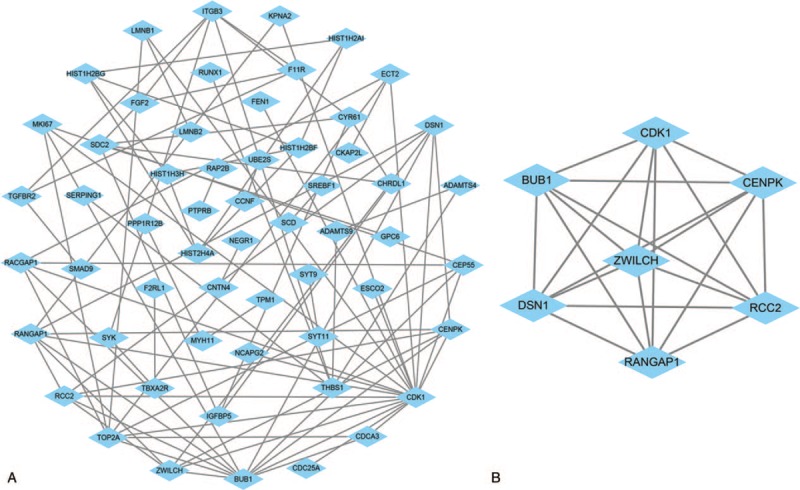
Identification of hubgenes from the protein–protein interaction (PPI) network with the MCODE algorithm. (A) PPI network of 158 genes. (B) PPI network of 7 hubgenes that extracted from (A).
Figure 6.
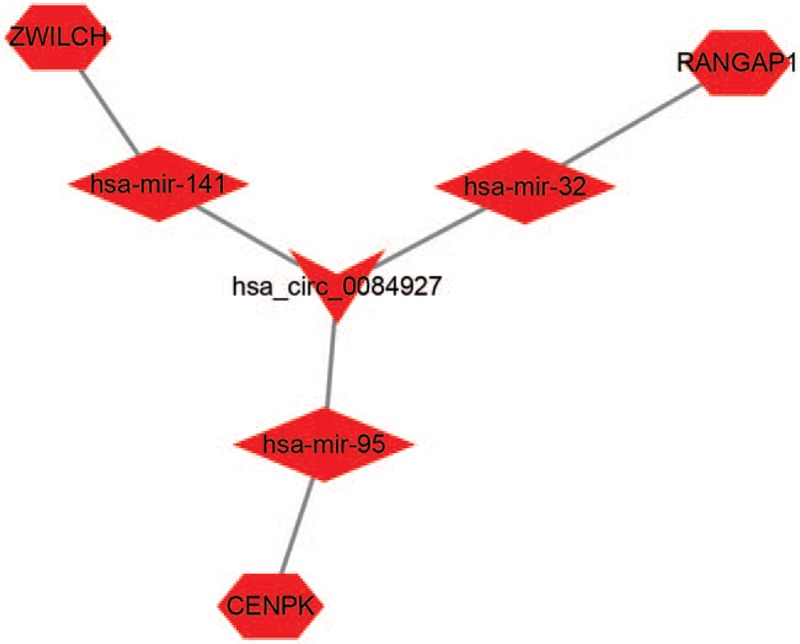
CircRNA-miRNA-hubgene network consisting of 1 cricRNA, 3 miRNAs, and 3 hubgenes.
3.4. Functional enrichment analysis of DEmRNA
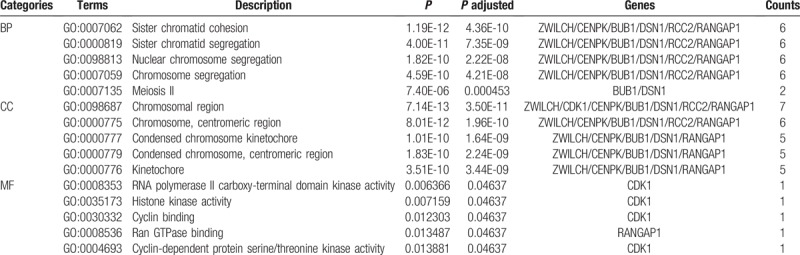
According to GO analysis, we found that the most enriched GO terms in biological process were sister chromatid cohesion, sister chromatid segregation, nuclear chromosome segregation, and chromosome segregation; in terms of cellular components, DEmRNAs were mostly enriched in chromosomal region. Among the 7 molecular function terms, the most enriched GO term was RNA polymerase II carboxy-terminal domain kinase activity (P < .05). The top 5 GO terms are indicated in Table 2. Moreover, KEGG pathway analysis indicated that most of DEmRNAs are involved in progesterone-mediated oocyte maturation, cell cycle, and oocyte meiosis.
Table 2.
The top 5 GO terms enriched by DEmRNA involved in the ceRNA network.
4. Discussion
With the continuous maturity and development of bioinformatics, scientists gradually realize that circRNA is a novel noncoding RNA which have conserved sequence and can stably express in mammals. CircRNAs can regulate the expression of gene at transcriptional or posttranscriptional.[19,20] Many studies have revealed the abnormal expression and regulation of circRNAs can affect the occurrence and course of cancers,[21,22] and thus have the potential to serve as biomarkers of malignancies.[23–25] However, the exact role of circRNAs in CC remains largely unclear. In this study, we firstly integrated circRNA, miRNA, and mRNA data between CC tissues and nontumor tissues from GEO and TCGA database and constructed the circRNA-miRNA-mRNA regulatory network.
Several studies have indicated that circRNA have displayed dysregulated expression in CC and is linked to the pathogenesis and prognosis, and is considered to be a tumor-related biomarker.[26–28] Zhang et al[26] showed that hsa_circ_0023404 was significantly upregulated in CC tissues compared to cervical normal tissues. Elevated hsa_circ_0023404 expression was linked to worse survival, and hsa_circ_0023404 may serve as an independent prognostic biomarker. Silence of hsa_circ_0023404 reduced cell proliferation, induced cell cycle arrest, and inhibited cell migration and invasion. Furthermore, they found that hsa_circ_0023404 can regulate the expression of TFCP2 by sponging miR-136, leading to CC development and progression. Similarly, elevated circRNA8924 was observed in CC tissues and was associated with tumor size, FIGO staging, and myometrial invasion. The knockdown of circRNA8924 significantly inhibited the proliferation, migration, and invasion of CC cells.[27] In our study, a total of 6 circRNAs were identified involved in the ceRNA network. Hsa_circ_0000745 may serve as a diagnostic marker for gastric cancer.[29] Hsa_circ_0084927 has been found to be significantly upregulated in malignant pleural effusion by qRT-PCR.[30] However, none of the other 6 circRNAs have been reported in CC.
miRNAs are a class of small (approximately 22 nucleotides in length) and highly conserved noncoding RNAs.[31] They can post-transcriptionally regulate gene expression by binding to 3’ untranslated regions, resulting in translation repression or mRNA degradation.[32] MiRNAs play key roles in many biological processes, including cell proliferation, differentiation, development, immune response.[32,33] In this study, we indentified 15 DEmiRNAs in the ceRNA network. Some researchers have studied the binding of circRNAs to miRNAs and their interactions in CC.[28,34] Cai et al[34] indicated that hsa_circ_0000263 promoted CC cell proliferation, apoptosis, and migration by sponging miR-150–5p. Ma et al[28] reported that circRNA-000284 promotes progression of CC by sponging miR-506. In present study, we predicted the correlation between 6 circRNAs and 15 miRNAs involved in ceRNA network. Of these 15 miRNAs, 7 have been reported in CC.[35–41]
To further identify the key circRNAs participating in the regulatory network, we estibalished the PPI network, screening 7 hubgenes. Then, we established the circRNA-miRNA-hubgene network, including 3 circRNA-miRNA-mRNA axes. To understand the underlying biological processes and pathways between DEmRNAs in the ceRNA network, we performed the GO and KEGG enrichment analyses. The result indicated that the DEmRNAs were involved in many important tumor-associated biological functions.[42,43] However, our study presents several limitations. First, the dataset is relatively small. Second, these DEcircRNAs and their predicted interactions are needed to be validated by experimental methods. In the future, we will perform substantial experiments in vitro and in vivo to verify our findings.
5. Conclusions
We have screened several dysregulated circRNAs and established a circRNA-associated ceRNA network by bioinformatics analysis. The result demonstrated that hsa_circ_0084927 may play important roles in CC, which provides new insight into the pathogenesis and may offer potential therapeutic targets for CC.
Author contributions
Conceptualization: Jun Gong, Yin-zhi Wei.
Data curation: Jun Gong, Hui Jiang, Mei-qin Hu, Rong-feng Li, Yin-zhi Wei.
Formal analysis: Jun Gong, Hui Jiang, Mei-qin Hu, Qin Liu, Rong-feng Li, Yin-zhi Wei.
Funding acquisition: Hui Jiang.
Investigation: Jun Gong, Chang Shu, Qin Liu, Yin-zhi Wei.
Methodology: Jun Gong, Chang Shu, Yan Huang, Qin Liu, Rong-feng Li, Yin-zhi Wei.
Project administration: Chang Shu, Mei-qin Hu, Yan Huang, Qin Liu.
Resources: Chang Shu, Mei-qin Hu, Yan Huang, Qin Liu, Rong-feng Li.
Software: Jun Gong, Yan Huang, Rong-feng Li.
Supervision: Jun Gong, Yin-zhi Wei.
Validation: Jun Gong, Yin-zhi Wei.
Visualization: Jun Gong, Yin-zhi Wei.
Writing – original draft: Jun Gong, Hui Jiang, Chang Shu, Mei-qin Hu, Yan Huang, Qin Liu, Rong-feng Li, Yin-zhi Wei.
Writing – review & editing: Jun Gong, Hui Jiang, Chang Shu, Mei-qin Hu, Yan Huang, Qin Liu, Rong-feng Li, Yin-zhi Wei.
Footnotes
Abbreviations: CC = cervical cancer, circRNAs = circular RNAs, DEmiRNAs = differentially expressed miRNAs, DEmRNAs = differentially expressed mRNAs, FC = fold change, GEO = Gene Expression Omnibus, GO = GO, MREs = miRNA binding sites, PPI = protein–protein interaction, TCGA = The Cancer Genome Atlas.
The authors report no conflicts of interest.
References
- [1].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7–30. [DOI] [PubMed] [Google Scholar]
- [2].Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69–90. [DOI] [PubMed] [Google Scholar]
- [3].Niu C, Sun X, Zhang W, et al. NR2F6 expression correlates with pelvic lymph node metastasis and poor prognosis in early-stage cervical cancer. Int J Mol Sci 2016;17:E1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kim MK, Jo H, Kong HJ, et al. Postoperative nomogram predicting risk of recurrence after radical hysterectomy for early-stage cervical cancer. Int J Gynecol Cancer 2010;20:1581–6. [PubMed] [Google Scholar]
- [5].Liu J, Liu T, Wang X, et al. Circles reshaping the RNA world: from waste to treasure. Mol Cancer 2017;16:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Li J, Yang J, Zhou P, et al. Circular RNAs in cancer: novel insights into origins, properties, functions and implications. Am J Cancer Res 2015;5:472–80. [PMC free article] [PubMed] [Google Scholar]
- [7].Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013;19:141–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Rybak-Wolf A, Stottmeister C, Glazar P, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell 2015;58:870–85. [DOI] [PubMed] [Google Scholar]
- [9].Qin S, Zhao Y, Lim G, et al. Circular RNA PVT1 acts as a competing endogenous RNA for miR-497 in promoting non-small cell lung cancer progression. Biomed Pharmacother 2018;111:244–50. [DOI] [PubMed] [Google Scholar]
- [10].Li XN, Wang ZJ, Ye CX, et al. RNA sequencing reveals the expression profiles of circRNA and indicates that circDDX17 acts as a tumor suppressor in colorectal cancer. J Exp Clin Cancer Res 2018;37:325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zhang Y, Wang Q, Zhu D, et al. Up-regulation of circ-SMAD7 inhibits tumor proliferation and migration in esophageal squamous cell carcinoma. Biomed Pharmacother 2019;111:596–601. [DOI] [PubMed] [Google Scholar]
- [12].Liu G, Huang K, Jie Z, et al. CircFAT1 sponges miR-375 to promote the expression of Yes-associated protein 1 in osteosarcoma cells. Mol Cancer 2018;17:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Li X, Zhang Z, Jiang H, et al. Circular RNA circPVT1 promotes proliferation and invasion through sponging miR-125b and activating E2F2 signaling in non-small cell lung cancer. J Cell Biochem 2018;51:2324–40. [DOI] [PubMed] [Google Scholar]
- [14].Wang R, Zhang S, Chen X, et al. EIF4A3-induced circular RNA MMP9 (circMMP9) acts as a sponge of miR-124 and promotes glioblastoma multiforme cell tumorigenesis 2018;17:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res 2015;43:D146–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Fromm B, Billipp T, Peck LE, et al. A uniform system for the annotation of vertebrate microRNA genes and the evolution of the human microRNAome. Annu Rev Genet 2015;49:213–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bandettini WP, Kellman P, Mancini C, et al. MultiContrast Delayed Enhancement (MCODE) improves detection of subendocardial myocardial infarction by late gadolinium enhancement cardiovascular magnetic resonance: a clinical validation study. J Cardiovasc Magn Reson 2012;14:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lux S, Bullinger L. Circular RNAs in cancer. Adv Exp Med Biol 2018;1087:215–30. [DOI] [PubMed] [Google Scholar]
- [20].Chen S, Zhao Y. Circular RNAs: Characteristics, function, and role in human cancer. Histol Histopathol 2018;33:887–93. [DOI] [PubMed] [Google Scholar]
- [21].Kristensen LS, Hansen TB, Veno MT, et al. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene 2018;37:555–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zhong Y, Du Y, Yang X, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer 2018;17:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Xu Y, Yao Y, Gao P, et al. Upregulated circular RNA circ_0030235 predicts unfavorable prognosis in pancreatic ductal adenocarcinoma and facilitates cell progression by sponging miR-1253 and miR-1294. Biochem Biophys Res Commun 2019;509:138–42. [DOI] [PubMed] [Google Scholar]
- [24].Arnaiz E, Sole C, Manterola L, et al. CircRNAs and cancer: biomarkers and master regulators. Semin Cancer Biol 2018. [DOI] [PubMed] [Google Scholar]
- [25].Yan B, Zhang W, Mao XW, et al. Circular RNA ciRS-7 correlates with advance disease and poor prognosis, and its down-regulation inhibits cells proliferation while induces cells apoptosis in non-small cell lung cancer. Eur Rev Med Pharmacol Sci 2018;22:8712–21. [DOI] [PubMed] [Google Scholar]
- [26].Zhang J, Zhao X, Zhang J, et al. Circular RNA hsa_circ_0023404 exerts an oncogenic role in cervical cancer through regulating miR-136/TFCP2/YAP pathway. Biochem Biophys Res Commun 2018;501:428–33. [DOI] [PubMed] [Google Scholar]
- [27].Liu J, Wang D, Long Z, et al. CircRNA8924 promotes cervical cancer cell proliferation, migration and invasion by competitively binding to MiR-518d-5p /519-5p family and modulating the expression of CBX8. Cell Physiol Biochem 2018;48:173–84. [DOI] [PubMed] [Google Scholar]
- [28].Ma HB, Yao YN, Yu JJ, et al. Extensive profiling of circular RNAs and the potential regulatory role of circRNA-000284 in cell proliferation and invasion of cervical cancer via sponging miR-506. Am J Transl Res 2018;10:592–604. [PMC free article] [PubMed] [Google Scholar]
- [29].Huang M, He YR, Liang LC, et al. Circular RNA hsa_circ_0000745 may serve as a diagnostic marker for gastric cancer. World J Gastroenterol 2017;23:6330–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hou Z, Wen Y, Wang Y, et al. Microarray expression profile and analysis of circular RNA regulatory network in malignant pleural effusion. Cell Cycle 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bartel DP. Metazoan MicroRNAs. Cell 2018;173:20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Liu C, Liu R, Zhang D, et al. MicroRNA-141 suppresses prostate cancer stem cells and metastasis by targeting a cohort of pro-metastasis genes. Nat Commun 2017;8:14270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Cai H, Zhang P, Xu M, et al. Circular RNA hsa_circ_0000263 participates in cervical cancer development by regulating target gene of miR-150-5p. J Cell Physiol 2019;234:11391–400. [DOI] [PubMed] [Google Scholar]
- [35].Pang H, Yue X. MiR-205 serves as a prognostic factor and suppresses proliferation and invasion by targeting insulin-like growth factor receptor 1 in human cervical cancer. Tumour Biol 2017;39:1010428317701308. [DOI] [PubMed] [Google Scholar]
- [36].Li JH, Zhang Z, Du MZ, et al. microRNA-141-3p fosters the growth, invasion, and tumorigenesis of cervical cancer cells by targeting FOXA2. Arch Biochem Biophys 2018;657:23–30. [DOI] [PubMed] [Google Scholar]
- [37].Liu YJ, Zhou HG, Chen LH, et al. MiR-32-5p regulates the proliferation and metastasis of cervical cancer cells by targeting HOXB8. Eur Rev Med Pharmacol Sci 2019;23:87–95. [DOI] [PubMed] [Google Scholar]
- [38].Sathyanarayanan A, Chandrasekaran KS, Karunagaran D. microRNA-145 modulates epithelial-mesenchymal transition and suppresses proliferation, migration and invasion by targeting SIP1 in human cervical cancer cells. Cell Oncol (Dordr) 2017;40:119–31. [DOI] [PubMed] [Google Scholar]
- [39].Zamani S, Sohrabi A, Hosseini SM, et al. Deregulation of miR-21 and miR-29a in cervical cancer related to HPV infection. MicroRNA 2019;8:110–5. [DOI] [PubMed] [Google Scholar]
- [40].Duan S, Wu A, Chen Z, et al. miR-204 regulates cell proliferation and invasion by targeting EphB2 in human cervical cancer. Oncol Res 2018;26:713–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Shi X, Xiao X, Yuan N, et al. MicroRNA-379 suppresses cervical cancer cell proliferation and invasion by directly targeting V-crk avian sarcoma virus CT10 Oncogene Homolog-Like (CRKL). Oncol Res 2018;26:987–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Luo Y, Wu Y, Peng Y, et al. Systematic analysis to identify a key role of CDK1 in mediating gene interaction networks in cervical cancer development. Ir J Med Sci 2016;185:231–9. [DOI] [PubMed] [Google Scholar]
- [43].Su Z, Yang H, Zhao M, et al. MicroRNA-92a promotes cell proliferation in cervical cancer via inhibiting p21 expression and promoting cell cycle progression. Oncol Res 2017;25:137–45. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]


