Abstract
Thrombin, derived from zymogen prothrombin (ProT), is a serine protease involved in procoagulation, anticoagulation, and platelet activation. Thrombin’s actions are regulated through Anion Binding Exosites I and II (ABE I and ABE II) that undergo maturation during activation. Mature ABEs can utilize exosite-based communication to fulfill thrombin functions. However, the conformational basis behind such long-range communication and the resultant ligand binding affinities are not well understood. Protease Activated Receptors (PARs), involved in platelet activation and aggregation, are known to target thrombin ABE I. Unexpectedly, PAR3 (44–56) can already bind to pro-ABE I of ProT. Nuclear Magnetic Resonance (NMR) ligand-enzyme titrations were used to characterize how individual PAR1 (49–62) residues interact with pro-ABE I and mature ABE I. 1D proton line broadening studies demonstrated that binding affinities for native PAR1P (49–62, P54) and for the weak binding variant PAR1G (49–62, P54G) increased as ProT was converted to mature thrombin. 1H,15N-HSQC titrations revealed that PAR1G residues K51, E53, F55, D58, and E60 exhibited less affinity to pro-ABE I than comparable residues in PAR3G (44–56, P51G). Individual PAR1G residues then displayed tighter binding upon exosite maturation. Long range communication between thrombin exosites was examined by saturating ABE II with phosphorylated GpIbα (269–282, 3Yp) and monitoring binding of PAR1 and PAR3 peptides to ABE I. Individual PAR residues exhibited increased affinities in this dual ligand environment supporting the presence of inter-exosite allostery. Exosite maturation and beneficial long range allostery are proposed to help stabilize an ABE I conformation that can effectively bind PAR ligands.
INTRODUCTION
Thrombin (factor IIa) is originally expressed as the zymogen prothrombin (ProT, UniprotKB P00734).1, 2 Following a vascular injury, the prothrombinase complex consisting of Factor Xa, cofactor Factor Va, Ca2+ ion, and a phospholipid membrane proteolytically converts ProT to the serine protease thrombin.3, 4 Besides the active site region, thrombin functions are controlled by regulatory surface loops and two anion binding exosites (ABE I and ABE II) that are each positioned at opposite sides of the active site region.5, 6
Engagement with the anion binding exosites helps dictate the fate of thrombin as a procoagulant or an anticoagulant.5, 6 Procoagulant functions include activating FV, FVIII, and FXIII, converting fibrinogen into a fibrin clot, and activating platelets. Protein C activation in the presence of thrombomodulin is a critical anticoagulant thrombin function.3, 4 Physiological peptide ligands targeting thrombin ABE-I include fibrinogen,7 thrombomodulin,8 Protease Activated Receptors PAR19 and PAR3,10 and leech-derived inhibitor hirudin.11, 12 ABE II directed ligands include heparin,13 heparin analogs,14 FVIII,15 fibrinogen γ՛16, 17, and platelet receptor protein GpIbα.18, 19 Reports indicate that allosteric communication is possible between ABE I and II, and effects may be ligand dependent.18, 20–23
Zymogen prothrombin contains immature pro-exosites that mature into active exosites without any major structural changes to the ABE I regions of these two protein forms.5, 24 We propose that a conformational maturation process occurs in which the dynamic pro-exosite/exosite populations progress to states that are fully competent to bind a ligand. Interestingly, ABE I ligands derived from hirudin, DNA/RNA aptamers, and PAR3 already have the ability to interact with immature pro-ABE I on ProT.25–28 Recent published data with PAR3 (44–56) based peptide ligands demonstrated that solution NMR methods could be used to detect conformational environments that accumulate during exosite maturation.28 Using individual 15N-labeled peptides of PAR3G (44–56, P51G), both electrostatic (E48, D54) and hydrophobic PAR3 residues (F47, L52) made unique contributions toward interacting with pro-ABE I and ABE I.28
PAR3 is just one member of the protease activated receptor family. PARs are membrane bound proteins containing seven transmembrane domains,29 and thrombin exhibits the greatest substrate specificity towards PAR130. Thrombin cleaves PAR1 at the R41-S42 peptide bond, and a portion of the new N-terminus then serves as a tethered ligand to help activate PAR1.31 PAR1 is well known for its roles in platelet activation, aggregation, and for helping to trigger inflammatory processes.32, 33 PAR1 has gained attention for its role in controlling the metastasis of cancer cells.34 Interestingly, PAR3 and PAR1 both assist in cleavage of PAR4 which lacks an ABE I binding segment.35 Following their activations, PAR1 and PAR4 can illicit transmembrane signaling events like coupling to heterotrimeric G-proteins, regulating kinase signaling cascades, and promoting receptor phosphorylation and internalization.33, 36
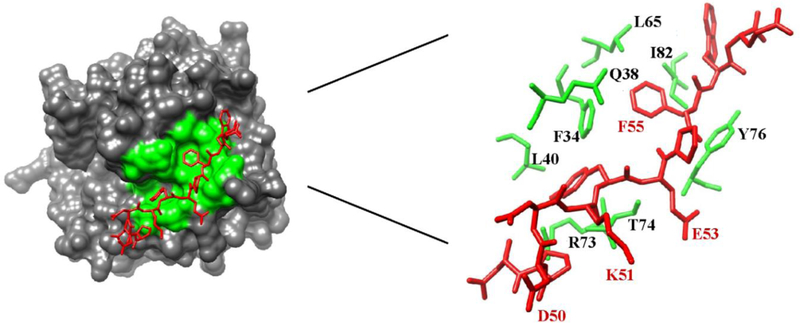
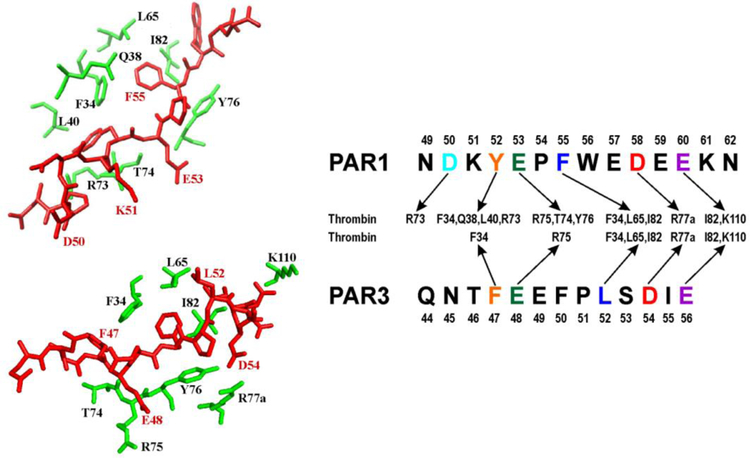
Gandhi and coworkers reported the X-ray crystal structure of PAR1 (33–62, 33ATNATLDPRSFLLRNPNDKYEPFWEDEEKN62) bound to catalytically inactive thrombin S195A.9, 37 The 38LDPRSFLLRNP48 segment spans the thrombin active site region. Similar to the 56FEEI59 sequence of hirudin, the PAR1 segment 50DKYEPF55 is hypothesized to be important for targeting ABE I and utilizes both electrostatic and hydrophobic PAR1 residue contributions.9, 11, 37, 38 An evaluation of the X-ray crystal structure indicated that PAR1 residues F34, Q38, L40, L65, R73, T74, R75, Y76, I82, and K149 are positioned less than 4 Å away from that thrombin ABE I (Figure 1).9 Molecular modeling studies have suggested that the C-terminal PAR1 58DEEKN62 binds to ABE I,39 but this flexible PAR1 segment is not observed in the Gandhi X-ray crystal structure.9, 37
Figure 1:
An X-ray crystal structure of human PAR1 fragment (49–57) bound to thrombin (PDB code 3LU9). The thrombin surface is rendered in gray. The PAR1 residues are labeled as red sticks whereas ABE I residues situated < 4 Å from PAR1 are shown as green sticks or green surfaces. Residues later selected for 1H,15N-HSQC NMR titration studies are highlighted in red.
Previously published NMR studies have shown that thrombin is highly dynamic and achieves its various functions by adopting distinct states in the presence of ligands.40–44 When thrombin is active-site inhibited with PPACK, the dynamic ABE I residues make up one of the few regions whose NMR peaks are difficult to detect.40, 41 Importantly, NMR relaxation rate studies have been carried out on the active site mutant thrombin S195M versus PPACK-thrombin.44 These results demonstrated that thrombin ABE I and the surface loop regions maintained flexibility even with an active site inhibited serine protease. Furthermore, no major structural changes were observed.41–44 Our current NMR studies then employ isotopically labeled peptides to characterize binding of individual PAR ligand residues to the (pro)-ABE I regions of ProT versus PPACK-thrombin.
Besides local enzyme-ligand interactions, long range communication can also occur across the thrombin surface. Events at thrombin ABE I have been influenced by occupancy at ABE II.16, 18, 21, 22 Similar to PAR1 and PAR3, glycoprotein GpIbα is also found on the surface of platelets.45 GpIbα recruits thrombin to the platelet environment by binding an anionic cluster (269–286) to thrombin ABE II (R93, R101, R126, K235, K236, and K240).18, 19, 46 The GpIbα-thrombin complex enhances PAR1 hydrolysis,23 hinders release of FpA,47 and decreases affinity for FVIII.48
Studies have shown that Protease Activated Receptors (PARs) play critical roles in platelet activation and other physiological processes.33, 37 PARs 1 and 3 target thrombin but less is known about binding interactions with ProT. Important goals of the current project were thus 1) to characterize the ligand binding properties that occur as PAR1P (49–62, P54) and weaker binding PAR1G (49–62, P54G) interact with immature ProT compared to mature active thrombin and 2) to further explore long-range communication between exosites. We predict that a PAR binding competent state is already present in ProT. As the exosite matures, we propose that the population of binding competent states increases and is encountered more frequently. The current NMR 1H-15N-HSQC titrations revealed that individual PAR1G residues could indeed already bind to pro-ABE I. Affinities were, however, weaker than comparable residues in PAR3G (44–56, P51G). Individual PAR1G (49–62, P54G) residues then displayed tighter binding upon exosite maturation and could help probe the new chemical environment. Since GpIbα can function as a cofactor for PAR1 hydrolysis, we explored the binding affinities and chemical environments encountered as 1H-15N-labeled PAR1 and PAR3 bind to an unlabeled GpIbα:thrombin complex. Both PAR1G (49–62, P54G) and PAR3G (44–56, P51G) responded to the presence of the ABE II ligand GpIbα (269–286). The increased affinities detected for individual PAR residues in this dual ligand environment support the presence of beneficial inter-exosite allostery.
MATERIALS AND METHODS
Materials
Prothrombin (ProT) and thrombin (IIa) derived from human plasma were purchased from Haematologic Technologies, Inc (Essex Junction, VT). The serine protease inhibitor PPACK (D-phenylalanyl-L-prolyl-L-arginine chloromethyl ketone) (Calbiochem (San Diego, CA) was added to block the thrombin active site thus protecting the enzyme from autolysis. The D2O (99.96%) employed in this project was from Cambridge Isotope Laboratories (Andover, MA).
Synthetic Peptides
Custom synthesis of PAR1-derived peptides was performed by New England Peptide (Gardner, MA). The peptides are fully defined as PAR1P (49–62, P54), the variant PAR1G (49–62, P54G), and the variant PAR3G (44–56, P51G). 15N-labeled peptides were prepared as follows. PAR1 (49N 15D K Y E 54P 15F W E D E E K N62), abbreviated as PAR1PFD (49–62) is the native PAR1 sequence, and D50 and F55 were 15N-labeled at their amide nitrogens. PAR1 (49N 15D K Y E 54G 15F W E D E E K N62) abbreviated as PAR1GFD (49–62), has a Pro54 to Gly54 (54G) and furthermore, the D50 and F55 were 15N-labeled. PAR1 (49N D K Y 15E G F W E 15D E E K N62), abbreviated as PAR1GED (49–62), also has the Pro54 to Gly54 (54G) but now E53 and D58 are 15N-labeled. PAR1 (49N D 15K Y E G F W E D E 15E K N62), abbreviated as PAR1GKE (49–62), also has the Pro54 to Gly54 (54G), where K51 and E60 are 15N-labeled. PAR3 (44Q N T F 15E E F G 15L S D I E56), abbreviated as PAR3GEL (44–56), has the P51G (G), where E48 and L52 are 15N-labeled. GpIbα (269–286, 3YP) (269D E G D T D L YP P D YP P YP P P E E D T E G289) with the three tyrosines phosphorylated was synthesized by Bachem Bioscience Inc (Torrance, CA). The synthetic peptide purity was checked by both HPLC and MALD-TOF MS. Similar to previous studies with PAR3G (44–56, P51G), peptide stock solutions were dissolved in deionized water, and amino acid analysis (AAA Service Laboratory, Inc., Damascus, OR) was used to obtain accurate concentrations.28 NMR buffer composed of 25 mM H3PO4, 150 mM NaCl, and 0.2 mM EDTA (pH 6.5) was used to dilute the peptide for future NMR studies.28
1D Proton Line Broadening and 2D Transferred NOESY Experiments
Sample preparations and NMR methods similar to those of Billur et. al. were employed.28 To minimize serine protease autolysis, a 4:1 complex of PPACK to thrombin was incubated at 37 °C for 30 min. Employing a Vivaspin 2 ultrafiltration unit (5000 Da) from Sartorius (Göttingen, Germany), ProT and PPACK-thrombin samples were exchanged into NMR buffer (pH6.5). Extinction coefficients (E1%280 nm) of 13.8 for ProT and 18.3 for thrombin were used to determine protein concentrations.
As is typical for 1D proton line broadening studies, 2D TOCSY and 2D transferred-NOESY (tr-NOESY) experiments, a 10-fold excess of PAR1P (49–62, P54) peptide was employed for the (peptide ligand)/protein complex. The 1H chemical shifts for the ligand correlate with the solution environment experienced by the ligand when in the presence of protein.49–51 The complexes included 780 μM PAR1P (49–62, P54) with 59 μM ProT, 900 μM PAR1P (49–62, P54) with 97 μM PPACK-thrombin, 830 μM PAR1G (49–62, P54G) with 83 μM ProT, and 870 μM PAR1G (49–62, P54G) with 87 μM PPACK-thrombin. At least 1 mM peptide was used as the free ligand sample. All NMR experiments were carried out at 25 °C on a Varian Inova 700 MHz spectrometer equipped with pulsed-field Z-axis gradients and a triple resonance cold probe.28 Mnova NMR (Mestrelab Research software) was used to process the 1D 1H NMR spectra. NMRPipe and nmrDraw were used to process 2D TOCSY, 2D tr-NOESY, and 2D 1H,13C-HSQC data. Sparky was used to visualize these 2D NMR results.28
The 1H chemical shift assignments for all PAR1 peptide residues were derived from 2D TOCSY and 2D tr-NOESY experiments. Standard pulse sequences were employed. Since the aromatic residues (F55 and W56) were challenging to assign for PAR1P and PAR1G (49–62), 2D 1H,13C-HSQC natural abundance experiments were also employed. Each aromatic carbon and hydrogen of F55, W56, and Y52 were first assigned in 2D 1H,13C-HSQC spectra. Later, aromatic hydrogens were matched with the fingerprint and amide-amide regions of 2D TOCSY and 2D tr-NOESY spectra.
1D and 2D 1H,15N-HSQC NMR Titration
In heteronuclear single quantum coherence (HSQC) NMR, all 1H that are covalently attached to a 15N are detected. 1D and 2D 1H,15N-HSQC NMR titrations were employed to determine if specific 15N-labeled peptide ligand residues exhibited fast, intermediate, or slow exchange on/off the surface of the enzyme.28, 52 For such conditions, kex is the exchange rate of the interaction and Δω is the difference in resonance frequency between the bound and free states. During the titrations, chemical shift changes are recorded as a function of protein-ligand ratios. For the weak binding, fast exchange regime (kex >> |Δω|), each 15N-labeled residue exhibits one signal which reflects the population-weighted averages of chemical shift, line width, and intensity. The resultant NMR chemical shift changes can be used to determine individual binding affinities for each 15N-labeled residue.28, 52–54 For the intermediate exchange regime (kex ≈ |Δω|), extensive line broadening may cause the 15N peak to disappear during the titration and later reappear upon increasing the bound population. Such line broadening is often due to intermediate, interconversion rates occurring during the NMR detection period. Finally, for the tight binding, slow exchange regime (kex << |Δω|), both free and bound signal states are observed reflecting their distinct chemical shifts, line widths, and intensities.52–54
To deal with solubility issues at higher protein concentrations, we employed an NMR titration strategy where the ligand was 15N-labeled and protein was unlabeled.28 Protein concentrations (100–200 μM) were diluted serially while at the same time keeping a constant concentration of 15N-labeled ligand. For each titration point, a defined volume of the protein-ligand was taken out and replaced with an equal ligand volume. NMR titrations were therefore started with a high protein:ligand ratio and were stopped upon reaching low protein:ligand ratios. Throughout this process, a constant peptide ligand concentration was maintained. The ability of ProT or PPACK-inhibited thrombin to interact with the 15N-labeled PAR1 or PAR3 residue was thus being monitored by our NMR titration strategy.28
In preparation for such NMR titrations, Vivaspin 2 ultrafiltration units (5000 Da MW cutoff) were used to buffer exchange ProT and active site inhibited PPACK-thrombin into NMR buffer. The starting complexes for the initial PAR1 binding studies contained 50 μM PAR1PFD (49–62) with 115 μM ProT or 150 μM PPACK-thrombin. The serial dilutions for both ProT:PAR1PFD and PPACK-thrombin:PAR1PFD resulted in ratios that spanned at least from 2:1 to 0.1:1.
To weaken the affinity of the PAR1P peptide for PPACK-thrombin, P54 was replaced with the flexible glycine. For such PAR1GFD binding studies, the starting complexes included 50 μM PAR1GFD (49–62) with 150 μM ProT or with 200 μM PPACK-thrombin. Serial dilutions for both ProT:PAR1GFD and PPACK-thrombin:PAR1GFD resulted in protein-peptide ratios that at least covered the range 3:1 to 0.1:1. For the PAR1GKE titrations, initial complexes contained 50 μM PAR1GKE (49–62) with 150 μM ProT or with 150 μM PPACK-thrombin. The serial dilutions for both ProT:PAR1GKE and PPACK-thrombin:PAR1GKE resulted in ratios that at least covered the range 3:1 to 0.1:1. For the PAR1GED titrations, the initial complexes contained 50 μM PAR1GED (49–62) with 150 μM ProT or with 200 μM PPACK-thrombin. The protein:ligand ratio of ProT:PAR1GED and PPACK-thrombin:PAR1GED complexes that at least covered the range 3:1 to 0.1:1
For the 1D 1H,15N-HSQC titrations of PAR1P and PAR1G complexes, the NMR parameters included 512 transients, 9000 Hz sweep width, and 4096 complex points in the direct dimension. For the 2D 1H,15N-HSQC titrations, the parameters included 16 transients, 64 complex points in the indirect dimension, 1242 complex points collected in the direct dimension, and 9000 Hz sweep widths for both direct and indirect dimensions. As an NMR control sample, 50 μM of 15N-labeled free peptide was employed. Mnova NMR was used to stack the 1D 1H,15N-HSQC data. NMRPipe, nmrDraw, and Sparky were used to process and then visualize the 2D 1H,15N-HSQC data.28
In-house scripts developed in Python were used to obtain quantitative binding estimates for individual 15N-labeled peptide ligand residues interacting with specific proteins.55 The script includes total enzyme concentrations[P0], total peptide concentrations[L0], and the NMR chemical shift difference (Δobs) between each set of free and bound conditions employed in different titration points. The current NMR titrations measured protein binding to a specific peptide ligand concentration. Thus, the equation was adjusted so that the usual [P0] in the denominator is replaced with [L0]. All the titrations were repeated at least twice. A Monte-Carlo approach was employed to calculate the errors in KD values. An error of 10% was applied to address the use of serially diluted thrombin concentrations.28, 55
1D and 2D 1H,15N-HSQC NMR Titration for Long-Range Communication Studies
To better investigate long-range communication between thrombin ABE II and ABE I, an unlabeled GpIbα peptide was bound to ABE II and 15N-labeled PAR peptides were targeted to ABE I. Triply phosphorylated GpIbα (269–286, 3Yp) has been reported to bind to thrombin ABE II with a KD of 5.9 nM.56 The two PAR based 15N-labeled peptide ligands chosen for this project included PAR1GED (49–62) and PAR3GEL (44–56). Sample conditions were maintained such that GpIbα (269–286, 3Yp) was at least 99.7 % bound during the entire course of the titration. The following equation was used to determine the % GpIbα ligand (L) bound to protein/receptor (R).
The experimental strategies employed for these NMR titrations were similar to the previously described titrations in this project. The only difference was adding the GpIbα to the thrombin prior to starting the PAR titration. For both PAR1GED and PAR3GEL NMR studies, the initial complexes included 150 μM PPACK-thrombin, 200 μM GpIbα, and 50 μM of 15N-labeled PAR peptide. The protein:ligand ratio of PPACK-thrombin:PAR1GED/PAR3GEL complexes covered the range 3:1 to 0.1:1. PPACK-thrombin concentration was diluted from 150 μM to 6 μM and GpIbα concentration was diluted from 200 μM to 8 μM. At each titration point, the concentration of 15N-labeled PAR ligands was kept constant. The same instrument, parameters, and processing software were used to investigate long-range communication in the serine protease thrombin as used for the single ligand binding sites.
RESULTS
1D Proton Line Broadening Experiments with PAR1P (49–62, P54)
1D proton line broadening NMR experiments were carried out for PAR1P (49–62, P54) in the presence and absence of enzyme. The appearance of peak broadening can be correlated with particular peptide protons that directly contact the enzyme surface. Substantial amide proton peak broadening was detected for PAR1P (49–62, P54) in the presence of ProT (Figure S1). Similar to previous work with PAR3 (44–56, P51),28 the pro-ABE I site on ProT can already accommodate PAR1P (49–62, P54). PAR1 residues displaying effective line broadening included D50, K51, E53, F55, D58, and E60. The increase in line broadening observed with PPACK-thrombin supports a proposal for improved ligand contact upon exosite maturation (Figure S1). Later, the residues exhibiting substantial line broadening were selected for 15N labeling.
1H,15N-HSQC Titration Studies with PAR1PFD (49–62) Labeled at D50 and F55
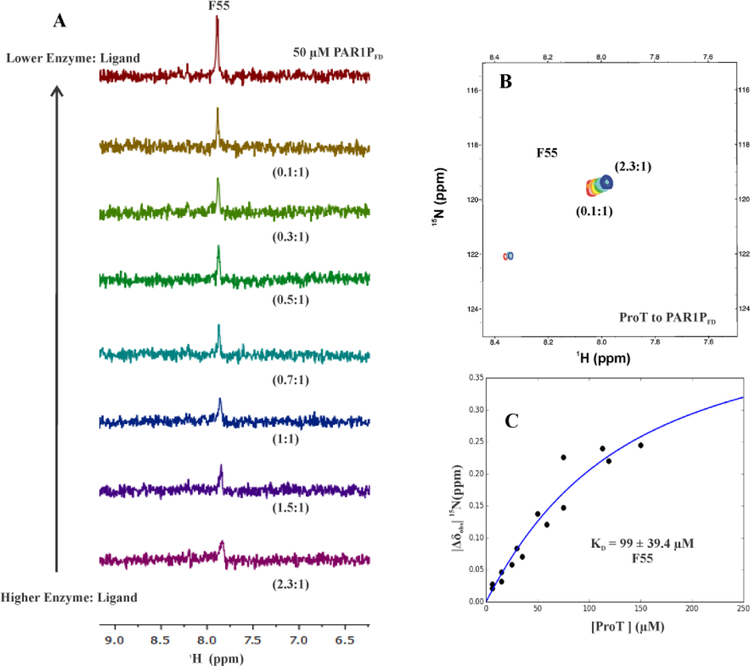
To characterize individual binding properties of ABE I directed ligands and to probe for evidence of exosite maturation, 1H,15N-HSQC based protein-ligand titrations were first carried out with PAR1PFD (49–62) containing 15N-labeled D50 and F55. As shown in Figure 1, PAR1 D50 is in ionic interaction with R73 of thrombin and F55 is surrounded by the hydrophobic cluster (F34, L65 and I82) of ABE I.9 Thrombin ABE I is mainly composed of the 30 and 70 loops which are responsible for electrostatic and hydrophobic interactions with physiological ligands.5 1H,15N-HSQC titrations started with ProT:PAR1PFD ratios of 2.3:1. Unfortunately, the [15N]-D50 amide proton was not seen throughout the NMR titration series due to fast exchange with the solvent. This amide proton could be detected at pH 3.0; however, this environment is far more acidic than what ProT/thrombin encounters physiologically. By contrast, [15N]-F55 could be detected during the full NMR titration series at pH 6.5. 1D 1H,15N-HSQC titration revealed that the [15N]-F55 peak exhibited line broadening at high ProT:PAR1PFD ratios when a substantial percentage of the peptide could interact with ProT. Sharper peaks started to develop as the enzyme was diluted with a constant amount of 15N-labeled PAR1PFD (Figure 2A) and a greater amount of free PAR1PFD peptide was accumulating.
Figure 2:
1D, 2D 1H,15N-HSQC NMR titrations and KD value of PAR1PFD (49–62) in the presence of ProT. For both of these NMR studies, the initial ProT:PAR1PFD (49–62) ratio was 2.3:1 and then gradually diluted down to 0.1:1. A) 1D 1H,15N-HSQC NMR titrations for starting conditions of 50 μM PAR1PFD (49–62, [15N]-D50, [15N]-F55) in 115 μM ProT. B) 2D 1H,15N-HSQC NMR titrations for starting conditions of 50 μM PAR1PFD (49–62, [15N]-D50, [15N]-F55) in 115 μM ProT. Serially diluting the ProT:PAR1PFD complex led to ratios of 2.3:1(blue) to 0.1(red). Representative titration series are shown. C) Calculating affinity (KD) for 15N-labeled F55 of PAR1PFD binding to prothrombin. ProT and PAR1P [15N]-F55 binding exhibited a KD of 99 ± 39.4 μM. The titrations were carried out in duplicate. In-house scripts developed in Python were used to determine the KD values. The y-axis |Δδobs| 15N ppm = δ15Nbound – δ15N free corresponds to the absolute difference in chemical shift between bound and free states of an individual 15N-residue. A Monte-Carlo approach that assumes a 10% error in the serially diluted protein samples was employed for error analysis.
2D 1H,15N-HSQC cross peaks for [15N]-F55 could be monitored for the full titration series. Such results are consistent with this residue being in fast exchange on/off the ProT surface (Figure 2B). Moreover, there was a clear change in F55 chemical shift position (and thus chemical environment) resulting in a binding affinity (KD) of 99 ± 39.4 μM (Figure 2C). These results indicated that pro-ABE I is already available for hydrophobic interactions with the PAR1PFD peptide. Upon exosite maturation, [15N]-F55 showed extensive line broadening even at the very lowest PPACK-IIa:PAR1PFD (0.1:1) ratios. Such line broadening was due to extensive contacts with the mature ABE I surface of thrombin and is consistent with intermediate enzyme-ligand exchange properties. As a result, KD values for this ligand-protein interaction could not be determined.
1H,15N-HSQC Titration Studies with PAR1GFD (49–62) Labeled at D50 and F55
A potentially weaker binding version of PAR1P (49–62), containing a P54G substitution, was used next to probe the availability of (pro)-ABE I for binding. A similar P to G substitution had been utilized successfully in our previous PAR3 peptide project.28 In the current project, 1D proton line broadening could still be observed for PAR1G (49–62, P54G) in the presence of ProT and increased further with the mature thrombin (Figure S1). 1H,15N-HSQC NMR titrations with PAR1GFD (49–62) were thus initiated with a 3:1 (ProT:PAR1GFD) protein:ligand ratio where D50 and F55 were 15N labeled. Both 1D and 2D 1H,15N-HSQC titrations revealed that the [15N]-F55 peak showed no change in chemical shift position with ProT (Figure S2A and S2B). These results suggest that the new PAR1GFD [15FD N]-F55 containing sequence may exhibit less effective interactions with the hydrophobic cluster (F34, L65, I82) of zymogen ProT than PAR1PFD. Overall, the PAR1 P54G substitution had caused a weakening of binding affinity.
Upon thrombin exosite maturation, [15N]-F55 underwent a substantial increase in chemical shift during the titration series. The presence of a new binding nvironment around the hydrophobic cluster (F34, L65, I82) of thrombin was thus detected upon ABE I maturation (Figure S3A, S3B). Also, the chemical shift changes for [15N]-F55 of PAR1GFD titrations proceeded in the same direction across the panels as [15N]-F55 of PAR1PFD. This common pattern is consistent with the idea that both versions of PAR1 are encountering the same environment within the (pro)- ABE I. The estimated binding affinity (KD) of PAR1GFD [15N]-F55 for PPACK-IIa was 251μM (Figure S3C, Table 1).
Table 1:
Evaluation of KD and |Δωmax| values derived from 2D 1H,15N-HSQC titrations of 15N-labeled PAR1GFD, PAR1GKE, and PAR1GED bound to PPACK-Thrombina
| Peptide | Residue | KD for PPACK-IIa (μM) | |Δωmax| (ppm) | IIa residues in contact with PAR1 (49–62) |
|---|---|---|---|---|
| PAR1GFD | F55 | 251 | NA | F34, L65, 182 |
| PAR1GFD | D50 | Solvent exchange issue | NA | R73 |
| PAR1GKE | K51 | 167 ± 49 | 0.09 ± 0.02 | In vicinity of R73 |
| PAR1GKE | E60 | 280 | NA | Not seen in X-ray (might contact I82, K110) |
| PAR1GED | E53 | Too weak for KD calculation | NA | T74, Y76, and R75 |
| PAR1GED | D58 | 36 ± 6.6 | 0.41 ± 0.05 | Not seen in X-ray (might contact R77a) |
For these HSQC titrations, the PAR1 peptide concentrations were maintained at 50 μM while the PPACK-thrombin concentrations were diluted serially. KD values were determined from in-house scripts written in Python. Experimental data employed for these calculations included the various concentrations of protein and peptide. In addition, 15N chemical shift differences for sets of free and bound conditions were utilized for F55, E60, and D58. 1H chemical shift differences were employed for the K51. The HSQC titrations were carried out at least in duplicate. A Monte-Carlo approach that assumes a 10% error in the serially diluted protein samples was employed for error analysis.
1H,15N-HSQC Titration Studies with PAR1GKE (49–62) Labeled at K51 and E60
Basic PAR1 K51 is located just after D50 and acidic PAR1 E60 is located toward the C-terminal end of the PAR1GKE (49–62) peptide. The X-ray structure of PAR1 (49–62) – thrombin (Figure 1) showed that PAR1 K51 is in close vicinity of thrombin ABE I residues R73 and T74.9 By contrast, researchers were unable to detect the electron density for the acidic C-terminal tail of PAR1 (58DEEKN62).9 1H,15N-HSQC NMR studies on PAR1 E60 would provide an alternative strategy to probe this region. [15N]-K51 and [15N]-E60 did not show any change in chemical shift position in the presence of ProT indicating little if any new binding environment encountered by the peptide residues upon encountering pro-ABE I (Figure S4A and S5A). However upon exosite maturation, the 2D 1H,15N-HSQC studies revealed that PAR1 residue K51 showed modest changes in chemical shift in the hydrogen dimension with a KD of 167 ± 49.2 μM and for PAR1 E60 a KD of 280 μM in the nitrogen dimension (Figure S4B, S5B, S6A, S6B, Table 1). Overall, both [15N]-K51 and [15N]-E60 exhibited improved interactions with PPACK-thrombin.
1H,15N-HSQC Titration Studies with PAR1GED (49–62) Labeled at E53 and D58
Two additional acidic PAR1G residues were then probed. Gandhi et al., had shown that PAR1 E53 is involved in interactions with thrombin T74, R75, and Y76.9 Labeling at E53 would thus provide another opportunity to probe the 70 loop of (pro)-ABE I. To further characterize unresolved PAR1 (58DEEKN62), the residue D58 was 15N labeled.9 1D line broadening studies revealed that PAR1 E53 and D58 both showed modest proton line broadening with ProT. By contrast, both [15N]-E53 and [15N]-D58 within PAR1GED (49–62) exhibited no change in chemical shift position in the HSQC titration series with ProT. These results indicated that both PAR1 E53 and D58 have quite weak interactions with pro-ABE I of ProT and are not experiencing much alteration in chemical environment compared to free peptide (Figure S7).
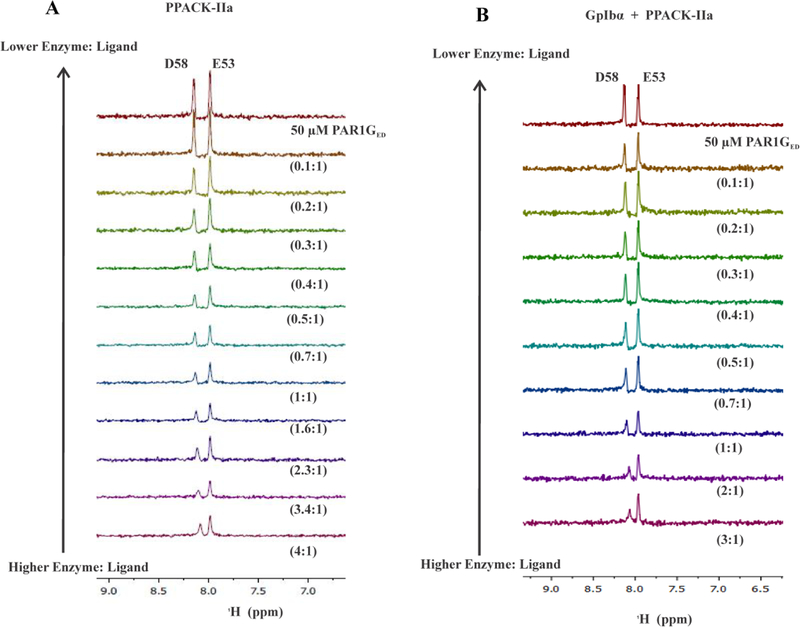
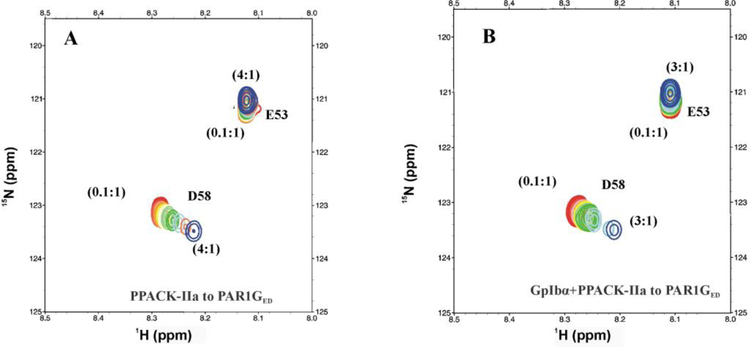
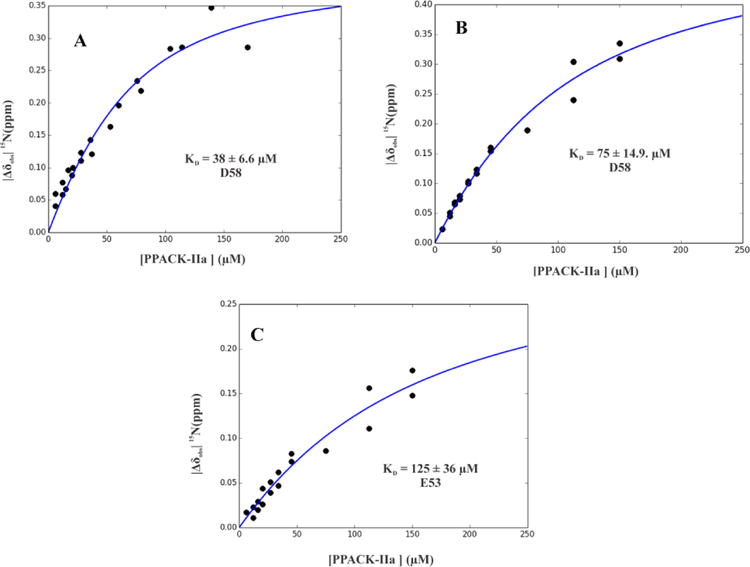
After ABE I maturation, PAR1GED [15N]-E53 still did not exhibit any substantial change in chemical environment with PPACK-IIa (Figure 3A and 4A). By contrast, the 1D 1H,15N-HSQC spectra of the PAR1GED [15N]-D58 peak started to show line broadening and chemical shifts already at high PPACK-IIa:ligand ratios consistent with effective thrombin binding (Figure 3A). This effect was confirmed with 2D 1H,15N-HSQC spectra, where chemical shift changes for PAR1GED [15N]-D58 were clearly observed for the full 4:1 to 0.1:1 PPACK-IIa to PAR1GED ratios (Figure 4A). The NMR titration series revealed that PAR1GED [15N]-D58 binds PPACK-IIa with KD of 38 ± 6.6 μM (Figure 5A, Table 1). This result provided valuable information about a flexible C-terminal region whose electron density could not be defined by X-ray crystallography.9
Figure 3:
1D 1H,15N-HSQC NMR titrations of PAR1GED (49–62) interacting with PPACK-IIa and GpIbα3P – PPACK-IIa. For these 1D NMR studies, the initial (PPACK-IIa or GpIbα3P – PPACK-IIa) to PAR1GED (49–62) ratio was at least 3:1 and then gradually diluted down to 0.1:1. A) 1D 1H,15N-HSQC NMR titrations for starting conditions of 50 μM PAR1GED (49–62, [15N]-E53, [15N]-D58) in 200 μM PPACK-IIa. B) 1D 1H,15N-HSQC NMR titrations for starting conditions of 50 μM PAR1GED (49–62, [15N]-E53, [15N]-D58) in 150 μM PPACK-IIa:GpIbα3P. NMR titrations were performed in duplicate.
Figure 4:
2D 1H,15N-HSQC NMR titrations of PAR1GED (49–62) in the presence of PPACK-IIa and GpIbα3P – PPACK-IIa. For these 2D NMR studies, the initial (PPACK-IIa or GpIbα3P – PPACK-IIa) to PAR1GED (49–62) ratio was 4:1 or 3:1 (blue) and then gradually diluted down to 0.1:1 (red). A) 2D 1H,15N-HSQC NMR titrations for 50 μM PAR1GED (49–62, [15N]-E53, [15N]-D58) in 200μM PPACK-IIa. B) 1H,15N-2D HSQC of 50 μM PAR1GED (49–62, [15N]-E53, [15N]-D58) in 150 μM GpIbα3P – PPACK-IIa.
Figure 5:
Calculating affinity (KD) for 15N-labeled E53 and D58 of PAR1GED (49–62) binding to PPACK-IIa and GpIbα3P – PPACK-IIa. KD values for PAR1G D58 went from A) 38 ± 6.6 μM with PPACK-IIa to B) 75 ± 14.9 μM with GpIbα3P – PPACK-IIa. KD values for PAR1G E53 went from too weak to calculate by NMR to C) 125 ± 36 μM GpIbα3P – PPACK-IIa. NMR titrations were carried out in duplicate. The KD values were assessed from scripts written in-house through Python. A Monte-Carlo approach that assumes a 10% error in the serially diluted protein samples was employed for error analysis.
1D and 2D 1H,15N-HSQC NMR Titration for Long-Range Communication Studies
Previously published HDX-MS (hydrogen-deuterium exchange mass spectrometry) studies demonstrated that when a triply phosphorylated GpIbα (269–282, 3YP) peptide bound to thrombin ABE II, there was a long-range influence over to the ABE I region.18, 22 To further characterize this dual ligand binding system, NMR titration studies were carried out with complexes consisting of unlabeled GpIbα (269–282, 3YP), PPACK-thrombin, and two distinct 15N-labeled PAR based peptides: PAR1GED (49–62) and PAR3GEL (44–56).28
To monitor such dual ligand allostery, a saturated amount of triply phosphorylated GpIbα (269–282, 3YP) was first added to PPACK-thrombin followed by the desired 15N-labeled PAR peptide. Both GpIbα and PPACK-IIa were then serially diluted with constant amounts of 15N-labeled peptide. Interestingly, titrations with PAR1GED revealed that [15N]-E53 went from an affinity for thrombin ABE I that was too weak to detect by NMR to one with an improved KD of 125 ± 36 μM in the presence of GpIbα peptide (Figure 3B, 4B, 5C, Table 2). By contrast, PAR1 [15N]-D58 did not respond much to the presence of GpIbα binding at ABE II. The KD for D58 went from 36 ± 6.6 μM to 75 ± 14.9 μM (Figure 3B, 4B, 5A, 5B, and Table 2).
Table 2:
Evaluation of KD and |Δωmax| values derived from 2D 1H,15N-HSQC titrations of 15N-PAR1GED when bound to GpIbα+PPACK-IIaa
| Peptide | Residue | KD for PPACK-IIa (μM) | |Δωmax| (ppm) | IIa residues in contact with PAR1 (49–62)9 |
|---|---|---|---|---|
| PAR1GED | E53 | KD too weak for NMR analysis | NA | In vicinity of T74, R75, and Y76 |
| PAR1GED+GpIbα | E53 | 126±36 | 0.32 ± 0.05 | In vicinity of T74, R75, and Y76 |
| PARIGED | D58 | 36±6.6 | 0.41 ± 0.05 | Not seen in X-ray (might contact R77a) |
| PAR1GED+GpIbα | D58 | 75±15 | 0.52 ± 0.05 | Not seen in X-ray (might contact R77a) |
For these 1H-15N-HSQC titrations, the PAR1 peptide concentrations were maintained at 50 μM and the PPACK-thrombin concentrations were diluted serially. KD values were determined from in-house scripts written in Python. Experimental data employed for these calculations included the various concentrations of protein and peptide. In addition, 15N NMR chemical shift differences for sets of free and bound conditions were utilized These HSQC titration series were performed at least in duplicate. A Monte-Carlo approach that assumes a 10% error in the serially diluted protein samples was employed for error analysis.
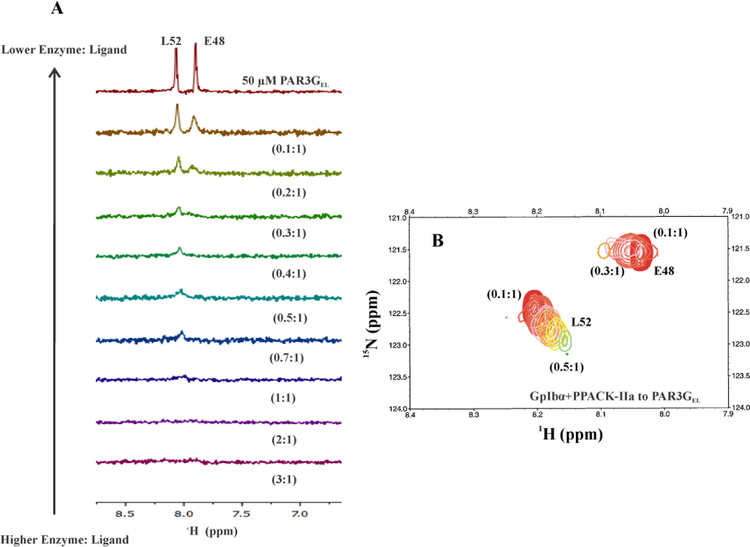
The PAR3GEL peptide displayed a more dramatic response to GpIbα binding at thrombin ABE II than PAR1GED (Figure 6A, 6B). Prior studies had shown that [15N]-L52 bound to ABE I with a KD of 47 ± 6 μM and [15N]-E48 was in higher affinity, intermediate exchange already at a 0.8:1 enzyme to ligand ratio.10, 28 With GpIbα binding to ABE II, the PAR3 peaks broadened further, and the effect started to occur at lower thrombin concentrations, consistent with increased affinity. [15N]-L52 of PAR3 could now only be quantitated from 0.5:1 to 0.1:1 (PAR3GEL:PPACK-IIa). As result, there were an insufficient number of data points for [15N]-L52 to calculate the KD values. Originally, PAR3GEL [15N]-E48 had exhibited intermediate exchange properties at a PPACK-IIa to ligand ratio of 0.8:1.28 In the presence of GpIbα, such extreme broadening occurred already at 0.3:1 protein to ligand ratio. The binding affinity of PAR3GEL to thrombin ABE I had clearly increased in the presence of the ABE II ligand. These studies demonstrate that NMR could document long range influence from thrombin ABE II over to ABE I (Figure 6, Table 3). Both PAR1 and PAR3 binding affinities at ABE I had been affected by binding GpIbα at ABE II.
Figure 6:
1D, and 2D 1H,15N-HSQC NMR titrations of PAR3GEL (44–56) with GpIbα3P – PPACK-IIa. For these NMR studies, the initial GpIbα3P – PPACK-IIa to PAR3GEL (44–56) ratio was 3:1 and then gradually diluted down to 0.1:1. A) 1D 1H,15N-HSQC NMR titrations for starting concentrations of 50 μM PAR3GEL (44–56, [15N]-E48, [15N]-L52) in at least 150 μM GpIbα3P – PPACK-IIa. B) 2D 1H,15N-HSQC NMR titrations of 50 μM PAR3GEL (44–56, [15N]-E48, [15N]-L52) in at least 150 μM GpIbα3P – PPACK-IIa.
Table 3:
Evaluation of KD and |Δωmax| values derived from 2D 1H,15N-HSQC titrations for 15N labeled PAR3GEL when bound to GpIbα+PPACK-IIaa
| Peptide | Residue | KD for PPACK-IIa (μM) | |Δωmax| (ppm)28 | IIa residues in contact with PAR3 (44–56)10 |
|---|---|---|---|---|
| PAR3GEL28 | L52 | 47 ± 6 | 1.84± 0.08 | F34, L65, I82 |
| PAR3GEL+GpIbα | L52 | KD too tight for NMR analysis | NA | F34, L65, I82 |
| PAR3GEL28 | E48 | KD too tight for NMR analysis | Insufficient data points | R75 |
| PAR3GEL+GpIbα | E48 | KD too tight for NMR analysis | NA | R75 |
For these 1H-15N-HSQC titrations, the PAR1 peptide concentrations were maintained at 50 μM and the PPACK-thrombin concentrations were diluted serially. KD values were determined from in-house scripts written in Python. Experimental data employed for these calculations included the various concentrations of protein and peptide. In addition, 15N NMR chemical shift differences for sets of free and bound conditions were utilized These HSQC titration series were performed at least in duplicate. A Monte-Carlo approach that assumes a 10% error in the serially diluted protein samples was employed for error analysis
DISCUSSION
Thrombin is a multifunctional serine protease that is originally derived from the zymogen Prothrombin. Direct thrombin inhibitors typically bind to the thrombin active site.57, 58 However, these inhibitors may lead to bleeding issues, and regulating/reversing such effects can become a major medical challenge.59, 60 A promising alternative is to exploit the concept of allostery and develop therapeutics that target distant thrombin exosites.6, 61, 62 As a part of this process, it is also critical to know whether such drug candidates target or avoid specific regions of immature pro-ABE I or mature ABE I. Protease Activated Receptors PAR1 and PAR3 have segments that target ABE I but whether they can both target pro-ABE I was less clear.9, 10 A key strength of the current project was the ability to use NMR to measure binding affinities for individual [15N]-labeled PAR1 residues interacting with ProT versus thrombin. Moreover, any changes in chemical environments could be examined. Results were compared with previously published [15N]-labeled PAR3 titrations.28
1D 1H line broadening studies revealed that PAR1P (49–62, P54) and PAR1G (49–62, P54G) peptides showed moderate interactions with ProT, and the interactions increased upon formation of mature thrombin. PAR1 thus joins PAR3 as being two examples of Protease Activated Receptors that can already interact with the pro-ABE I site of ProT.28 Ligand binding properties and exosite maturation were then probed using PAR1 peptides custom synthesized with [15N]-amide labeling at K51, E53, F55, D58, and E60.
The [15N]-F55 of PAR1PFD (49–62) is involved in hydrophobic interactions with thrombin F34, L65, and I82 (Figure 7).9 The NMR titration results indicated that pro-ABE I is already available for hydrophobic interactions with this aromatic PAR1P residue (Figure 2A–C, Table 1). When [15N]-F55 PAR1PFD encountered PPACK-thrombin, affinities increased to a level where KD values could no longer be determined by NMR. Similar to our earlier PAR3 studies, ligand affinities could be reduced when a Pro to Gly substitution was introduced to generate PAR1G (49–62, P54G).28 Unexpectedly, peptides that were [15N]-labeled at the PAR1G F55, K51, E53, D58, and E60 positions all exhibited very weak binding with pro-ABE I on ProT. In contrast, PAR3G (44–56, P51G) with [15N]-labeled residues F47, E48, L52, and D54 could still bind to ProT and report on the structural features and binding affinities encountered (Figure 7).10, 28 Therapeutic agents mimicking PAR3G peptides would thus be more likely to target pro-ABE I than agents based on PAR1G peptides.
Figure 7:
Comparing interactions between thrombin ABE I (30 and 70 loop), PAR1 (49–62), and PAR3 (44–56). Crystal structures of thrombin in complex with human PAR1 (49–57) [PDB 3LU9] and murine PAR3 (44–56) [PDB 2PUX] were employed as structural guides. Since PAR1 (58–62) was unresolved by crystallography, proposed interactions with thrombin residues are provided for PAR1 D58 and E60. The PAR fragments target similar thrombin ABE I regions but to varying extents.
Following thrombin maturation, [15N]-F55 of PAR1GFD (49–62) exhibited greater interactions with the ABE I surface of PPACK-thrombin. Both PAR1G F55 and PAR3G L52 share common hydrophobic interactions with thrombin residues (F34, L65, and I82) (Figure 7).9,10 Interestingly, [15N]-L52 in PAR3GEL had a KD value that was 5-fold tighter than the [15N]-F55 in PAR1GFD.28 Interactions with this hydrophobic cluster are thus more effective in promoting binding affinity of L52 in PAR3GEL than F55 in PAR1GFD.
Acidic PAR1 E53 is in the vicinity of thrombin residues T74, R75, and Y76.9 The NMR titrations studies revealed that [15N]-E53 of PAR1GED (49–62) showed no substantial interaction with either ProT or PPACK-IIa (Figures S7, 3A, and 4A). E53 is located just N-terminal to the PAR1G P54G substitution. As a result, E53 may be hindered from adopting an effective orientation to promote beneficial interactions with (pro)-ABE I. By contrast, PAR3G [15N]-E48 is known to participate in a neighboring electrostatic interaction with ABE I R75 and exhibit a KD value too tight to calculate by NMR.10, 28 Basic residue PAR1 K51 is also located in the vicinity of thrombin R73 and T74.9 In the presence of PPACK-thrombin, the [15N]-K51 amide nitrogen did not exhibit chemical shift changes, but the amide hydrogen did undergo movement in chemical shift (Figure S4A, S5A, S6A, Table 1). These observations highlight NMR as a sensitive technique to explore changing chemical environments around different atoms such as hydrogen and nitrogen.
PAR1 D58 is the first residue of a flexible acidic C-terminal tail of PAR1 (58DEEKN62). Our NMR titration results thus provided the first solution-based evidence that acidic PAR1G residues in the (58DEEKN62) segment could make direct contact with ABE I, and a new chemical environment was encountered. A review of affinities later revealed that PAR1G [15N]-D58 is better anchored to ABE I than PAR1G [15N]-E60. An alignment of PAR3 and PAR1 residues suggests that PAR1 D58 may be near thrombin R77a (Figure 7).37 Thrombin R77a could thus help to stabilize PAR1 D58 similar to the strong electrostatics between PAR3G D54 and thrombin R77a monitored by both X-ray crystallography and NMR.10, 28 As with PAR3 E56, the unresolved PAR1 E60 may be directed toward thrombin I82 and K110.9, 10, 37
After characterizing PAR1 and PAR3 binding properties to ProT versus thrombin, our studies investigated whether binding of GpIbα at ABE II would influence the affinities of ABE I-directed PAR peptides. Allosteric communication between exosites and the active site is known to play a regulatory role in thrombin substrate specificity and catalysis.4, 6 Moreover, long-range communication between the thrombin exosites is possible even when the active site is covalently inhibited.18, 22, 43, 44
A triply phosphorylated GpIbα (269–282, 3YP) served as the ABE II ligand and PAR peptides targeted ABE I. With the GpIbα3Yp:PPACK-IIa complex, the weak affinity of PAR1 [15N]-E53 from PAR1GED (49–62) improved significantly for ABE I (Figure 3B, 4B, 5B, 5C, Table 1, and Table 2). The previous, non-optimal orientation of E53 may have been redirected by the introduction of GpIbα. By contrast, GpIbα did not have much of an effect on the binding affinity of the C-terminal PAR1 residue D58 (Figure 3B, 4B, 5B, Table 1, and Table 2). Putative interactions with thrombin R77a may have already been fairly well optimized. (Figure 7)
NMR titrations involving 15N-labeled PAR3GEL (44–56) and GpIbα3Yp:PPACK-IIa revealed that peaks for PAR3G [15N]-E48 and [15N]-L52 both exhibited extensive interactions within the dual ligand thrombin complex (Figure 6A, 6B, Table 3). The affinity of PAR3 [15N]-L52 for ABE I improved to a KD value too tight to calculate in the presence of the ABE II ligand GpIbα.28 PAR3 [15N]-E48 already exhibited strong affinity toward ABE I on PPACK-IIa. The presence of GpIbα caused further enhancements in PAR3 binding making it even harder to assess a KD value. (Figure 6A, 6B, Table 3).
When aligning the PAR3 and PAR1 sequences (Figure 7), it is interesting to note that PAR3 E48, PAR3 L52, and PAR1 E53 all experienced increased affinity in the presence of GpIbα-thrombin. The corresponding thrombin residues that may interact with these PAR residues can be clustered to (F34, L65, I82) and (T74, R75, Y76).9, 10 In support of the current NMR titrations, HDX-MS studies previously reported that GpIbα binding exerts a long-range solvent accessibility effect over to the thrombin 65–84 ABE I region.18, 22
In a physiological platelet environment, binding of GpIbα to ABE II helps promote thrombin catalyzed cleavage and subsequent activation of PAR1.23 Introducing a Fbg γ’ peptide has been shown to hinder platelet PAR1 activation by competing with GpIbα for the ABE II binding site.63 Thus, our dual ligand NMR titration results with GpIbα and PAR peptides support the presence of beneficial inter-exosite communication. By contrast, ABE II ligands can also exert a negative influence over to thrombin ABE I. When GpIbα, the DNA aptamer HD22, or Fbg γ’ bind to ABE II, the affinity of thrombomodulin TM456 for ABE I decreases.64 As a result, Protein C activation at the thrombin catalytic site is hindered. The ABE II directed ligands aptamer HD22, Fbg γ’, and prothrombin F2 have all been shown to inhibit thrombin binding to fibrin γAγ21. The current results suggest that the effect of interexosite communication is ligand dependent. PAR1 and PAR3 are important examples of ABE I ligands that benefit from such long range interactions.28
Unlike our NMR studies, there are no striking differences in the X-ray crystal structures of the pro-ABE I or ABE I regions of the following protein systems: prothrombin, active site inhibited PPACK-thrombin, ABE I-directed PAR3-thrombin, ABE-I directed PAR1-thrombin, and ABE II-directed GpIbα-thrombin (Figure S8).5, 9, 10, 24, 65–67 These protein systems likely have similar, transient structural states. We predicted that a PAR binding competent state is already present in ProT. Our NMR results then revealed that ProT does have a population of binding sites that can interact with PAR3 (44–56, P51) and PAR3G (44–56, P51G), and we can now include PAR1 (49–62, P54) and weaker binding PAR1G (49–62, P54G).28 The PAR1 and PAR3 receptors can thus join hirudin and DNA/RNA aptamers in being ligands that can target immature Pro-ABE I on ProT. As the exosite matures, the population of binding competent states is predicted to increase.68–70 Mature thrombin then promotes higher affinities for ABE I ligands by encountering binding competent states more frequently.
A similar conformational property has been reported for active site directed FPR binding to thrombin versus ProT. FPR binding occurs by selection of an optimal conformation from a pre-existing equilibrium of E* (closed) and E (open) states.71, 72 ProT and Prethrombin-2 have greater populations of E*. The ABE I ligands thrombomodulin and hirugen have also been shown to probe ensembles of rapidly interconverting conformations for binding states.71, 72 Our NMR studies further reveal that PAR1 and PAR3 can detect the changing chemical environments of ProT and PPACK-thrombin and then report on KD values.
In conclusion, our NMR studies have successfully examined the binding of PAR1 and PAR3 based peptide ligands to ProT versus thrombin. The NMR titration methods have the advantage of monitoring unique chemical environments and binding affinities at the single residue level. Our results indicate that ProT has a population of pro-ABE I binding sites that can interact with PAR1P (49–62, P54) and PAR1G (49–62, P54G). After exosite maturation, PAR1 and PAR3 peptide ligands both target similar areas of the thrombin ABE-I 30 and 70 loop regions with increased affinities. The individual PAR amino acids, however, make unique contributions to the overall binding affinities. The current project also demonstrates that binding of phosphorylated GpIbα (269–282, 3Yp) to ABE II leads to increased ABE I affinities for selected residues of PAR1G (49–62, P54G) and PAR3G (44–56, P51G). The increased affinities observed in this dual ligand environment support the presence of inter-exosite allostery. Exosite maturation and beneficial long range allostery are proposed to help stabilize an ABE I conformation that can effectively bind PAR ligands. Therapeutic agents that target distant thrombin exosites may be promising alternatives to direct thrombin inhibitors whose actions can be difficult to control.
Supplementary Material
ACKNOWLEDGEMENTS
N. Stolowich is thanked for guidance with NMR instrumentation. Research discussions with B.A. Anokhin and F.D.O. Ablan were helpful and appreciated. This research was supported by National Institutes of Health grants R01HL6844 (M.C.M) and R15HL120068 (M.C.M), a University of Louisville Research Initiation Grant (M.C.M), and by start-up funds provided by the James Graham Brown Cancer Foundation (T.M.S).
Footnotes
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest with the contents of this article.
REFERENCES
- [1].Krishnaswamy S (2013) The transition of prothrombin to thrombin, J Thromb Haemost 11 Suppl 1, 265–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Pozzi N, Chen Z, Zapata F, Niu W, Barranco-Medina S, Pelc LA, and Di Cera E (2013) Autoactivation of thrombin precursors, J Biol Chem 288, 11601–11610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Di Cera E, Page MJ, Bah A, Bush-Pelc LA, and Garvey LC (2007) Thrombin allostery, Phys Chem Chem Phys 9, 1291–1306. [DOI] [PubMed] [Google Scholar]
- [4].Lane DA, Philippou H, and Huntington JA (2005) Directing thrombin, Blood 106, 2605–2612. [DOI] [PubMed] [Google Scholar]
- [5].Bode W, Turk D, and Karshikov A (1992) The refined 1.9-A X-ray crystal structure of D-Phe-Pro-Arg chloromethylketone-inhibited human alpha-thrombin: structure analysis, overall structure, electrostatic properties, detailed active-site geometry, and structure-function relationships, Protein Sci 1, 426–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bock PE, Panizzi P, and Verhamme IM (2007) Exosites in the substrate specificity of blood coagulation reactions, J Thromb Haemost 5 Suppl 1, 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Pechik I, Madrazo J, Mosesson MW, Hernandez I, Gilliland GL, and Medved L (2004) Crystal structure of the complex between thrombin and the central “E” region of fibrin, Proc Natl Acad Sci U S A 101, 2718–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Fuentes-Prior P, Iwanaga Y, Huber R, Pagila R, Rumennik G, Seto M, Morser J, Light DR, and Bode W (2000) Structural basis for the anticoagulant activity of the thrombin-thrombomodulin complex, Nature 404, 518–525. [DOI] [PubMed] [Google Scholar]
- [9].Gandhi PS, Chen Z, and Di Cera E (2010) Crystal structure of thrombin bound to the uncleaved extracellular fragment of PAR1, J Biol Chem 285, 15393–15398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bah A, Chen Z, Bush-Pelc LA, Mathews FS, and Di Cera E (2007) Crystal structures of murine thrombin in complex with the extracellular fragments of murine protease-activated receptors PAR3 and PAR4, Proc Natl Acad Sci U S A 104, 11603–11608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Rydel TJ, Ravichandran KG, Tulinsky A, Bode W, Huber R, Roitsch C, and Fenton JW 2nd. (1990) The structure of a complex of recombinant hirudin and human alpha-thrombin, Science 249, 277–280. [DOI] [PubMed] [Google Scholar]
- [12].Rydel TJ, Tulinsky A, Bode W, and Huber R (1991) Refined structure of the hirudin-thrombin complex, J Mol Biol 221, 583–601. [DOI] [PubMed] [Google Scholar]
- [13].Carter WJ, Cama E, and Huntington JA (2005) Crystal structure of thrombin bound to heparin, J Biol Chem 280, 2745–2749. [DOI] [PubMed] [Google Scholar]
- [14].Sidhu PS, Liang A, Mehta AY, Abdel Aziz MH, Zhou Q, and Desai UR (2011) Rational design of potent, small, synthetic allosteric inhibitors of thrombin, Journal of medicinal chemistry 54, 5522–5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Esmon CT, and Lollar P (1996) Involvement of thrombin anion-binding exosites 1 and 2 in the activation of factor V and factor VIII, J Biol Chem 271, 13882–13887. [DOI] [PubMed] [Google Scholar]
- [16].Sabo TM, Farrell DH, and Maurer MC (2006) Conformational analysis of gamma’ peptide (410–427) interactions with thrombin anion binding exosite II, Biochemistry 45, 7434–7445. [DOI] [PubMed] [Google Scholar]
- [17].Lovely RS, Moaddel M, and Farrell DH (2003) Fibrinogen gamma’ chain binds thrombin exosite II, Journal of thrombosis and haemostasis : JTH 1, 124–131. [DOI] [PubMed] [Google Scholar]
- [18].Sabo TM, and Maurer MC (2009) Biophysical investigation of GpIbalpha binding to thrombin anion binding exosite II, Biochemistry 48, 7110–7122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lechtenberg BC, Freund SM, and Huntington JA (2014) GpIbalpha interacts exclusively with exosite II of thrombin, Journal of molecular biology 426, 881–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Fredenburgh JC, Stafford AR, and Weitz JI (1997) Evidence for allosteric linkage between exosites 1 and 2 of thrombin, J Biol Chem 272, 25493–25499. [DOI] [PubMed] [Google Scholar]
- [21].Petrera NS, Stafford AR, Leslie BA, Kretz CA, Fredenburgh JC, and Weitz JI (2009) Long range communication between exosites 1 and 2 modulates thrombin function, J Biol Chem 284, 25620–25629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Malovichko MV, Sabo TM, and Maurer MC (2013) Ligand binding to anion-binding exosites regulates conformational properties of thrombin, J Biol Chem 288, 8667–8678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].De Candia E, Hall SW, Rutella S, Landolfi R, Andrews RK, and De Cristofaro R (2001) Binding of thrombin to glycoprotein Ib accelerates the hydrolysis of Par-1 on intact platelets, J Biol Chem 276, 4692–4698. [DOI] [PubMed] [Google Scholar]
- [24].Pozzi N, Chen Z, Gohara DW, Niu W, Heyduk T, and Di Cera E (2013) Crystal structure of prothrombin reveals conformational flexibility and mechanism of activation, J Biol Chem 288, 22734–22744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ni F, Ning Q, Jackson CM, and Fenton JW 2nd. (1993) Thrombin exosite for fibrinogen recognition is partially accessible in prothrombin, The Journal of biological chemistry 268, 16899–16902. [PubMed] [Google Scholar]
- [26].Anderson PJ, Nesset A, Dharmawardana KR, and Bock PE (2000) Characterization of proexosite I on prothrombin, J Biol Chem 275, 16428–16434. [DOI] [PubMed] [Google Scholar]
- [27].Bompiani KM, Monroe DM, Church FC, and Sullenger BA (2012) A high affinity, antidote-controllable prothrombin and thrombin-binding RNA aptamer inhibits thrombin generation and thrombin activity, Journal of thrombosis and haemostasis : JTH 10, 870–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Billur R, Ban D, Sabo TM, and Maurer MC (2017) Deciphering Conformational Changes Associated with the Maturation of Thrombin Anion Binding Exosite I, Biochemistry 56, 6343–6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kahn ML, Zheng YW, Huang W, Bigornia V, Zeng D, Moff S, Farese RV Jr., Tam C, and Coughlin SR (1998) A dual thrombin receptor system for platelet activation, Nature 394, 690–694. [DOI] [PubMed] [Google Scholar]
- [30].Ayala YM, Cantwell AM, Rose T, Bush LA, Arosio D, and Di Cera E (2001) Molecular mapping of thrombin-receptor interactions, Proteins 45, 107–116. [DOI] [PubMed] [Google Scholar]
- [31].Coughlin SR (2000) Thrombin signalling and protease-activated receptors, Nature 407, 258–264. [DOI] [PubMed] [Google Scholar]
- [32].Coughlin SR (1999) Protease-activated receptors and platelet function, Thromb Haemost 82, 353–356. [PubMed] [Google Scholar]
- [33].Nieman MT (2016) Protease-activated receptors in hemostasis, Blood 128, 169–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kamath L, Meydani A, Foss F, and Kuliopulos A (2001) Signaling from protease-activated receptor-1 inhibits migration and invasion of breast cancer cells, Cancer Res 61, 5933–5940. [PubMed] [Google Scholar]
- [35].Nakanishi-Matsui M, Zheng YW, Sulciner DJ, Weiss EJ, Ludeman MJ, and Coughlin SR (2000) PAR3 is a cofactor for PAR4 activation by thrombin, Nature 404, 609–613. [DOI] [PubMed] [Google Scholar]
- [36].Macfarlane SR, Seatter MJ, Kanke T, Hunter GD, and Plevin R (2001) Proteinase-activated receptors, Pharmacol Rev 53, 245–282. [PubMed] [Google Scholar]
- [37].Gandhi PS, Chen Z, Appelbaum E, Zapata F, and Di Cera E (2011) Structural basis of thrombin-protease-activated receptor interactions, IUBMB Life 63, 375–382. [DOI] [PubMed] [Google Scholar]
- [38].Jacques SL, LeMasurier M, Sheridan PJ, Seeley SK, and Kuliopulos A (2000) Substrate-assisted catalysis of the PAR1 thrombin receptor. Enhancement of macromolecular association and cleavage, J Biol Chem 275, 40671–40678. [DOI] [PubMed] [Google Scholar]
- [39].Seeley S, Covic L, Jacques SL, Sudmeier J, Baleja JD, and Kuliopulos A (2003) Structural basis for thrombin activation of a protease-activated receptor: inhibition of intramolecular liganding, Chem Biol 10, 1033–1041. [DOI] [PubMed] [Google Scholar]
- [40].Lechtenberg BC, Johnson DJ, Freund SM, and Huntington JA (2010) NMR resonance assignments of thrombin reveal the conformational and dynamic effects of ligation, Proc Natl Acad Sci U S A 107, 14087–14092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Fuglestad B, Gasper PM, Tonelli M, McCammon JA, Markwick PR, and Komives EA (2012) The dynamic structure of thrombin in solution, Biophys J 103, 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Fuglestad B, Gasper PM, McCammon JA, Markwick PR, and Komives EA (2013) Correlated motions and residual frustration in thrombin, J Phys Chem B 117, 12857–12863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Lechtenberg BC, Freund SM, and Huntington JA (2012) An ensemble view of thrombin allostery, Biol Chem 393, 889–898. [DOI] [PubMed] [Google Scholar]
- [44].Handley LD, Fuglestad B, Stearns K, Tonelli M, Fenwick RB, Markwick PR, and Komives EA (2017) NMR reveals a dynamic allosteric pathway in thrombin, Sci Rep 7, 39575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Clemetson KJ (2007) A short history of platelet glycoprotein Ib complex, Thrombosis and haemostasis 98, 63–68. [PubMed] [Google Scholar]
- [46].De Cristofaro R, De Candia E, Landolfi R, Rutella S, and Hall SW (2001) Structural and functional mapping of the thrombin domain involved in the binding to the platelet glycoprotein Ib, Biochemistry 40, 13268–13273. [DOI] [PubMed] [Google Scholar]
- [47].Li CQ, Vindigni A, Sadler JE, and Wardell MR (2001) Platelet glycoprotein Ib alpha binds to thrombin anion-binding exosite II inducing allosteric changes in the activity of thrombin, J Biol Chem 276, 6161–6168. [DOI] [PubMed] [Google Scholar]
- [48].De Cristofaro R, and De Filippis V (2003) Interaction of the 268–282 region of glycoprotein Ibalpha with the heparin-binding site of thrombin inhibits the enzyme activation of factor VIII, Biochem J 373, 593–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Cleary DB, Trumbo TA, and Maurer MC (2002) Protease-activated receptor 4-like peptides bind to thrombin through an optimized interaction with the enzyme active site surface, Archives of biochemistry and biophysics 403, 179–188. [DOI] [PubMed] [Google Scholar]
- [50].Campbell AP, and Sykes BD (1993) The two-dimensional transferred nuclear Overhauser effect: theory and practice, Annu Rev Biophys Biomol Struct 22, 99–122. [DOI] [PubMed] [Google Scholar]
- [51].Ni F, and Scheraga HA (1994) Use of the Transferred Nuclear Overhauser Effect to Determine the Conformations of Ligands Bound to Proteins, Accounts Chem Res 27, 257–264. [Google Scholar]
- [52].Teilum K, Kunze MB, Erlendsson S, and Kragelund BB (2017) (S)Pinning down protein interactions by NMR, Protein Sci 26, 436–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Fielding L (2003) NMR methods for the determination of protein-ligand dissociation constants, Curr Top Med Chem 3, 39–53. [DOI] [PubMed] [Google Scholar]
- [54].Kleckner IR, and Foster MP (2011) An introduction to NMR-based approaches for measuring protein dynamics, Biochimica et biophysica acta 1814, 942–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Iconaru LI, Ban D, Bharatham K, Ramanathan A, Zhang W, Shelat AA, Zuo J, and Kriwacki RW (2015) Discovery of Small Molecules that Inhibit the Disordered Protein, p27(Kip1), Sci Rep 5, 15686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].De Marco L, Mazzucato M, Masotti A, and Ruggeri ZM (1994) Localization and characterization of an alpha-thrombin-binding site on platelet glycoprotein Ib alpha, J Biol Chem 269, 6478–6484. [PubMed] [Google Scholar]
- [57].Connolly SJ, Ezekowitz MD, Yusuf S, Eikelboom J, Oldgren J, Parekh A, Pogue J, Reilly PA, Themeles E, Varrone J, Wang S, Alings M, Xavier D, Zhu J, Diaz R, Lewis BS, Darius H, Diener HC, Joyner CD, and Wallentin L (2009) Dabigatran versus warfarin in patients with atrial fibrillation, N Engl J Med 361, 1139–1151. [DOI] [PubMed] [Google Scholar]
- [58].Salmeron Febres LM, and Cuenca Manteca J (2017) Direct Oral Anticoagulants in the Treatment of Venous Thromboembolic Disease, Ann Vasc Surg. [DOI] [PubMed] [Google Scholar]
- [59].Dager WE, and Banares L (2017) Reversing the anticoagulation effects of dabigatran, Hosp Pract (1995) 45, 29–38. [DOI] [PubMed] [Google Scholar]
- [60].Weitz JI (2017) Reversal of Direct Oral Anticoagulants: Current Status and Future Directions, Semin Respir Crit Care Med 38, 40–50. [DOI] [PubMed] [Google Scholar]
- [61].Mehta AY, Thakkar JN, Mohammed BM, Martin EJ, Brophy DF, Kishimoto T, and Desai UR (2014) Targeting the GPIbalpha binding site of thrombin to simultaneously induce dual anticoagulant and antiplatelet effects, J Med Chem 57, 3030–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Mehta AY, Mohammed BM, Martin EJ, Brophy DF, Gailani D, and Desai UR (2016) Allosterism-based simultaneous, dual anticoagulant and antiplatelet action: allosteric inhibitor targeting the glycoprotein Ibalpha-binding and heparin-binding site of thrombin, J Thromb Haemost 14, 828–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Lancellotti S, Rutella S, De Filippis V, Pozzi N, Rocca B, and De Cristofaro R (2008) Fibrinogen-elongated gamma chain inhibits thrombin-induced platelet response, hindering the interaction with different receptors, The Journal of biological chemistry 283, 30193–30204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Chen K, Stafford AR, Wu C, Yeh CH, Kim PY, Fredenburgh JC, and Weitz JI (2017) Exosite 2-Directed Ligands Attenuate Protein C Activation by the Thrombin-Thrombomodulin Complex, Biochemistry 56, 3119–3128. [DOI] [PubMed] [Google Scholar]
- [65].Celikel R, McClintock RA, Roberts JR, Mendolicchio GL, Ware J, Varughese KI, and Ruggeri ZM (2003) Modulation of alpha-thrombin function by distinct interactions with platelet glycoprotein Ibalpha, Science 301, 218–221. [DOI] [PubMed] [Google Scholar]
- [66].Dumas JJ, Kumar R, Seehra J, Somers WS, and Mosyak L (2003) Crystal structure of the GpIbalpha-thrombin complex essential for platelet aggregation, Science 301, 222–226. [DOI] [PubMed] [Google Scholar]
- [67].Koradi R, Billeter M, and Wuthrich K (1996) MOLMOL: a program for display and analysis of macromolecular structures, J Mol Graph 14, 51–55, 29–32. [DOI] [PubMed] [Google Scholar]
- [68].Boehr DD, Nussinov R, and Wright PE (2009) The role of dynamic conformational ensembles in biomolecular recognition, Nat Chem Biol 5, 789–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Tzeng SR, and Kalodimos CG (2012) Protein activity regulation by conformational entropy, Nature 488, 236–240. [DOI] [PubMed] [Google Scholar]
- [70].Wei G, Xi W, Nussinov R, and Ma B (2016) Protein Ensembles: How Does Nature Harness Thermodynamic Fluctuations for Life? The Diverse Functional Roles of Conformational Ensembles in the Cell, Chem Rev 116, 6516–6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Vogt AD, Chakraborty P, and Di Cera E (2015) Kinetic dissection of the pre-existing conformational equilibrium in the trypsin fold, J Biol Chem 290, 22435–22445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Chakraborty P, Acquasaliente L, Pelc LA, and Di Cera E (2018) Interplay between conformational selection and zymogen activation, Sci Rep 8, 4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.