Abstract
Background:
The inherited predisposition to development of specific histologic subtypes of invasive breast carcinoma has been incompletely investigated. Using a large, population-based database, we sought to investigate familial clustering of breast cancer by histologic subtype.
Methods:
Using the Utah Population Database (UPDB), which links genealogy records to the state-wide National Cancer Institute Surveillance, Epidemiology, and End-Results cancer registry, we identified patients with breast cancer by histology and tested for evidence of shared genetic predisposition to histologic specific subtypes by examining pairwise relatedness and estimating relative risk (RR) among first-, second-, and third-degree relatives.
Results:
We identified 23,629 individuals in the UPDB with at least 3 generations of genealogy and at least one primary breast cancer, 2883 (12.2%) of which were specific histologic subtypes other than invasive ductal carcinoma (including inflammatory [n=178], lobular [n=1688], and mucinous [n=542]). Statistically significant excess distant relatedness was identified for the mucinous subtype (p=0.011) as well as for inflammatory breast cancers (p=0.024). The RR for breast cancer of any histology in second-degree relatives was significantly increased for patients with inflammatory (RR 1.32 [1.02, 1.68]; p=0.03), lobular (RR 1.36 [1.25, 1.47]; p<0.001), and mucinous (RR 1.27 [1.12, 1.44]; p=0.00021) subtypes.
Conclusions:
These findings provide evidence for significant familial clustering within histological subtypes for lobular, mucinous and inflammatory breast carcinomas. Further research is required to identify the underlying genetic variants responsible for the increased risk. Studies of high risk pedigrees segregating a specific histologic subtype could be a powerful design for predisposition gene identification.
Keywords: breast cancer, inflammatory, mucinous, lobular, familiality, UPDB
Precis:
Using the Utah Population Data Base, we found evidence of histology-specific familial clustering for mucinous, lobular, and inflammatory breast carcinomas. Breast cancer cases with specific histologies appear to cluster more in pedigrees than expected, and the homogeneous pedigrees observed may be informative for identification of the predisposition genes responsible.
Introduction
Breast cancer is a heterogeneous disease. According to the World Health Organization, breast cancer can be classified based on histopathologic characteristics including cell morphology, architecture, and growth patterns into 21 distinct subtypes.1, 2 While the majority of invasive breast cancers are ductal carcinomas, there are a number of more uncommon histologic subtypes that are associated with either a more aggressive or a more indolent disease course.
Pure mucinous and tubular breast cancers are examples of more indolent tumor subtypes. Mucinous tumors comprise 1–2% of breast cancers, and are typically hormone receptor positive and HER2 negative.3 They are frequently diagnosed at an older age and are associated with a better than average breast cancer-specific survival. In addition to being histologically distinct, mucinous cancers also differ genomically from more common breast cancer subtypes, including lower genomic instability and a lower frequency of mutations in genes in the PI3K pathway.3, 4
Inflammatory breast cancer, in contrast, is an aggressive type of breast cancer. It is not a specific histology per se, but rather is diagnosed based on characteristic clinical changes including rapid development of diffuse erythema and edema of the breast.5–8 About 2.5% of patients diagnosed with breast cancer have inflammatory breast cancer, and it is often at advanced stage at the time of diagnosis.6 Dermal lymphatic invasion can be noted on pathology, although this finding is not required for the diagnosis. Prior reports have shown an increased risk of inflammatory breast cancer in patients with younger age and higher body mass index.9
About 5–10% of breast cancers are inherited. Patients with mutations in specific genes, including BRCA1, BRCA2, TP53, and PTEN, have a high risk of developing breast cancer.10 More recently, it has become apparent that mutations in numerous additional genes, including CHEK2 and PALB2, are associated with a more moderate risk of breast cancer.11 However, the only histologic subtypes that have been associated with mutations in specific genes are lobular carcinoma with CDH1, which encodes e-cadherin,12 and medullary carcinoma with BRCA1.13 Other histologies, including mucinous carcinoma, have not previously been found to be clearly associated with a family history of breast cancer. Recently, inflammatory breast cancer has been reported to be associated with a first-degree family history of breast cancer in some but not all studies; mutations in specific genes have not yet been identified.9, 14, 15
The possibility of additional genetic contributions to breast cancer exists. Identification of excess familial clustering using the genealogical index of familiality (GIF) is a method that can be used to provide evidence that risk of a disease is mediated by inherited genetic factors. The GIF has previously been described in studies evaluating the familiality of cancer.16–19 Determining whether a disease exhibits familial clustering can be used to calculate disease risk in relatives, to increase the understanding of the disease pathogenesis, and ultimately can permit identification of underlying causative mutations. In the context of the excess familial clustering that is generally recognized for breast cancers of any histology, we used this established methodology to investigate hypotheses of additional genetic contributions to predisposition to specific breast cancer histologic subtypes and inflammatory breast cancer using a large population-based database.
Materials and methods
Utah Population Database
A unique Utah resource consisting of the genealogy of the Utah Northern European founders from the mid 1800s and their descendants to modern day linked to a statewide National Cancer Institute Surveillance, Epidemiology, and End-Results (SEER) Cancer Registry from 1973 was used to describe the familial clustering of breast cancer cases by histologic subtypes and of inflammatory breast cancer. This resource, the Utah Population DataBase (UPDB),20, 21 includes data on over 11 million individuals. The approximately 3 million of these individuals who have at least 3, and up to 16, generations of genealogy data connecting to Utah founders were analyzed here. The Utah Cancer Registry was established statewide in 1966 and became one of the original SEER Registries in 1973. All independent primary cancers diagnosed or treated in Utah are recorded. Study approval was obtained from the University of Utah Institutional Review Board and the Utah Resource for Genetic and Epidemiology Research Review Board. Statistical analyses were performed using tools created specifically for the UPDB.
Breast Cancer Cases
Approximately 150,000 of the 3 million individuals with genealogy in UPDB have a linked Utah Cancer Registry record; 23,629 of these linked cancer records are for individuals diagnosed with breast cancer, and 155 of these breast cancers occurred in males. Breast cancer cases were identified using the International Classification of Diseases for Oncology (ICD-Oncology) Revision 3 definition of the primary site 500–509 and including histology codes 8000 to 9589 (leukemias and lymphomas excluded). In particular, this classification was used to assign specific histologic subtypes as follows: angiosarcoma (9120); apocrine (8401); inflammatory (8530); lobular (8520); medullary (8510, 8512, 8513); metaplastic (8570–8572, 8575); mucinous (8480); phyllodes (9020); and tubular (8211).
Genealogical Index of Familiality Method
The genealogical index of familiality (GIF) method was developed for use with the UPDB and allows a test of the hypothesis of excess relatedness among individuals with a phenotype of interest. The GIF compares the average pairwise relatedness of a set of individuals (e.g. all breast cancer cases) to the expected pairwise relatedness for a similar set of individuals in the UPDB. The coefficient of kinship is used to measure relatedness,22 and pairs can be defined based on their genetic distance. The expected pairwise relatedness for a group of individuals is estimated in a set of randomly selected matched controls. Randomly selected controls from the UPDB were matched to cases by sex, 5-year birth year cohort, and birth place (Utah or not Utah). For each GIF test of a set of cases, the expected pairwise relatedness was estimated as the average pairwise relatedness computed for 1,000 sets of matched controls. The significance of the case GIF was assessed empirically by its position within the 1,000 control GIF statistic values. Breast cancer histologic subsets with sample sizes less than 100 were not analyzed with the GIF method.
The GIF method tests for excess relatedness or familiality, but does not distinguish between relatedness due to genetics versus common environment. The distant GIF (dGIF) test was therefore created as an extension of the GIF statistic; it is performed while ignoring all relationships closer than third-degree, to test for an excess of distant relationships, which is unlikely in the absence of a heritable contribution.
Estimation of Relative Risk in Relatives
Evidence for a familial or genetic contribution to disease is commonly considered using estimates of relative risk in relatives. Published risks for cancer in relatives are typically limited to close relationships (first-degree). Relative risks (RR) for breast cancer were estimated in both close and distant relatives in the UPDB utilizing birth- and sex-specific cohort rates of breast cancer estimated internally from the UPDB as follows.
All individuals in the UPDB genealogy with at least 3 generations of genealogy were assigned membership to a birth year- (5-year groups), sex-, and birthplace-specific (Utah or not Utah) cohort. Internal, cohort-specific rates of breast cancer (and histologic subtypes) were calculated for all cohorts separately, by summing the number of individuals with the selected breast cancer histology in each cohort, and dividing by the total number of individuals in the cohort. Expected numbers of cancer cases for a set of individuals (e.g. first-degree relatives of mucinous breast cancer cases) were estimated by counting all first-degree relatives of the set of mucinous breast cancer cases by cohort, multiplying the number of relatives per cohort by the cohort-specific cancer rate, and then summing over all cohorts. The observed numbers of cancer cases were counted, without duplication, in the set of relatives being considered. RR = (observed number of cases)/(expected number of cases) is an unbiased estimator of RR. Exact two-sided Poisson probabilities were calculated under the null hypothesis that the RR = 1.0, and 95% confidence intervals were calculated based on the assumption that the number of observed cases follows a Poisson distribution, with mean equal to the expected number of cancers.
High-risk Pedigrees
Given a set of individuals selected from the UPDB, all related clusters of these individuals who descend from a common ancestor can be identified. All such clusters, or pedigrees, were identified for each subset of breast cancer cases with a specific histology; pedigrees were never completely overlapping, but cases could appear in more than 1 pedigree. To determine whether a pedigree is high-risk, the observed number of cases among the descendants is compared to the expected number of cases. The expected number of, for example, breast cancer cases with mucinous histology, among the descendants of a set of pedigree founders is calculated by counting all descendants by cohort, multiplying the number of descendants in each cohort by the cohort-specific rate of mucinous type breast cancer (estimated as described above), and summing over all cohorts. An excess of observed to expected affected descendants of p<0.05 was used to classify a descending pedigree as high-risk for a specific subtype.
Results
We identified 23,629 individuals with at least 3 generations of genealogy connecting to Utah founders who had at least one primary breast cancer recorded in the Utah Cancer Registry. The majority of these cases were ductal histology or mammary carcinoma not otherwise specified; 53 apocrine carcinomas, 178 inflammatory breast cancers, 1688 lobular carcinomas, 542 mucinous carcinomas, 134 tubular carcinomas, 38 metaplastic carcinomas, 32 phyllodes tumors, 341 medullary carcinomas, and 12 angiosarcomas were also identified.
Since inflammatory breast cancer does not represent a specific histology but rather is a clinical diagnosis, the clinical and pathologic characteristics can be heterogeneous. In this analysis, the cohort of 178 patients with inflammatory breast cancers had median age 58 and median survival of 31 months (Supplemental Table 1). Just over half of cancers were poorly differentiated. Almost 15% had metastatic disease at the time of diagnosis. In contrast, the invasive lobular and mucinous cancers were diagnosed in women at an older age, at an earlier stage, with a lower grade, and had a longer overall survival.
Genealogical Index of Familiality
Table 1 shows the results of the GIF test for excess relatedness for all breast cancers and for the 5 histologic subtypes with at least 100 cases observed. Shown for each subtype is the number of cases (N), the average pairwise relatedness (Case GIF), the mean GIF statistic for the 1,000 sets of matched controls (Mean Control GIF), the empirical significance for the overall GIF test (GIF p value), the average pairwise relatedness ignoring first- and second-degree relationships (Case dGIF), the mean dGIF statistic for the 1,000 sets of matched controls (Mean Control dGIF), and the empirical significance for the distant GIF test (dGIF p value).
Table 1. Genealogical Index of Familiality (GIF) Relatedness Analysis.
The number (N) of cases of all breast cancer and each subtype of breast cancer included in the analysis, the GIF for cases and controls and the p value for the comparison, as well as the distant GIF (dGIF) for cases and controls and the p value for the comparison, are provided.
| Subtype | N | Case GIF | Mean Control GIF | GIF P value |
Case dGIF | Mean Control dGIF | dGIF P value |
|---|---|---|---|---|---|---|---|
| Breast | 23,629 | 2.93 | 2.72 | <0.001 | 2.37 | 2.38 | 0.676 |
| Inflammatory | 178 | 3.54 | 2.68 | 0.147 | 3.54 | 2.36 | 0.024 |
| Lobular | 1688 | 3.07 | 2.72 | 0.007 | 2.36 | 2.40 | 0.619 |
| Mucinous | 542 | 3.65 | 2.71 | 0.003 | 2.88 | 2.34 | 0.011 |
| Tubular | 134 | 2.52 | 2.86 | 0.529 | 2.52 | 2.47 | 0.426 |
| Medullary | 341 | 2.12 | 2.69 | 0.897 | 2.12 | 2.33 | 0.723 |
Statistically significant excess relatedness was observed for all breast cancer cases (p<0.001), and for the lobular (p=0.007) and mucinous subtypes (p=0.003) (Table 1). When close relationships were ignored, the dGIF test identified significant excess distant relatedness for the mucinous subtype (p=0.011) as well as for inflammatory breast cancers (p=0.024), but not for all breast cancers considered together or for the other tumor subtypes.
Relative risks
RRs were estimated for first- (FDR; Table 2), second- (SDR; Table 3), and third-degree relatives (TDR; Table 4) for all breast cancers, and for each of the histologic subgroups. Each table shows the subgroup, number of relatives (# FDRs, # SDRs, # TDRs, respectively), observed number of breast cancer cases of the same histologic subgroup (Obs), expected number of breast cancer cases of the same histologic subgroup (Exp), significance of the test of RR = 1.0 (p value), and estimated relative risk and 95% confidence interval (RR [CI]); the observed and expected numbers of cases and the significance and RR with CI for breast cancer of any histologic type are also shown for each histologic subgroup.
Table 2. Estimated relative risks (RR) for breast cancer of the same histologic subgroup, and for breast cancer of any histology in first-degree relatives (FDR) of proband breast cancer cases by histologic subgroup.
CI: 95% confidence interval; Exp: expected; Obs: observed
| Same histology subgroup | Any breast cancer histology | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Subgroup | #FDRs | Obs | Exp | P value | RR (CI) | Obs | Exp | P value | RR (CI) |
| Breast cancer | 190,576 | -- | -- | -- | -- | 5,846 | 3,308.2 | <0.001 | 1.77 (1.72,1.81) |
| Angiosarcoma | 96 | 0 | 0 | 6 | 2.3 | 0.03 | 2.59 (1.13, 5.63) | ||
| Apocrine | 436 | 0 | 0.02 | 20 | 7.84 | 0.0002 | 2.55 (1.56, 3.94) | ||
| Inflammatory | 1493 | 0 | 0.2 | 42 | 26.4 | 0.0046 | 1.59 (1.14, 2.15) | ||
| Lobular | 14,097 | 48 | 18.2 | <0.001 | 2.64 (1.95, 3.50) | 551 | 248.4 | <0.001 | 2.22 (2.04, 2.41) |
| Mucinous | 5,056 | 6 | 2.2 | 0.025 | 2.73 (1.19, 5.95) | 168 | 90.4 | <0.001 | 1.86 (1.59, 2.16) |
| Phyllodes | 251 | 0 | 0.01 | 5 | 3.97 | 0.61 | 1.26 (0.41, 2.94) | ||
| Tubular | 1264 | 0 | 0.15 | 47 | 23.6 | 1.8e-5 | 2.00 (1.47, 2.65) | ||
| Medullary | 3,058 | 0 | 0.79 | 102 | 54.1 | <0.001 | 1.88 (1.54, 2.29) | ||
| Metaplastic | 313 | 0 | 0.01 | 9 | 5.43 | 0.099 | 1.66 (0.76, 3.15) | ||
Table 3. Estimated relative risks (RR) for breast cancer of the same histologic subgroup, and for breast cancer of any histology in second-degree relatives (SDR) of proband breast cancer cases by histologic subgroup.
CI: 95% confidence interval; Exp: expected; Obs: observed
| Same subgroup type | Any breast cancer | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Subgroup | #SDRs | Obs | Exp | P value | RR (CI) | Obs | Exp | P value | RR (CI) |
| Breast cancer | 559,835 | 7,395 | 5,916 | <0.001 | 1.25 (1.22, 1.28) | ||||
| Angiosarcoma | 294 | 0 | 0 | † | † | 0.56 | 1.29 (0.35, 3.32) | ||
| Apocrine | 1518 | 0 | 0.03 | 15 | 17.4 | 0.72 | 0.87 (0.48, 1.43) | ||
| Inflammatory | 4,983 | 0 | 0.33 | 66 | 49.9 | 0.03 | 1.32 (1.02, 1.68) | ||
| Lobular | 46,875 | 49 | 31.3 | 0.003 | 1.56 (1.16, 2.07) | 642 | 473.3 | <0.001 | 1.36 (1.25, 1.47) |
| Mucinous | 17,889 | 6 | 4.6 | 0.47 | 1.32 (0.48, 2.86) | 251 | 197.2 | 2.1e-4 | 1.27 (1.12, 1.44) |
| Phyllodes | 913 | 0 | 0.01 | 8 | 10.3 | 0.64 | 0.77 (0.33, 1.53) | ||
| Tubular | 4,252 | 0 | 0.21 | 52 | 42.9 | 0.17 | 1.21 (0.91, 1.59) | ||
| Medullary | 10,316 | 0 | 1.49 | 115 | 106.3 | 0.21 | 1.08 (0.89, 1.30) | ||
| Metaplastic | 981 | 0 | 0.01 | 13 | 10.4 | 0.25 | 1.24 (0.66, 2.13) | ||
Observed counts < 5 are censored
Table 4. Estimated relative risks (RR) for breast cancer of the same histologic subgroup, and for breast cancer of any histology in third-degree relatives (TDR) of proband breast cancer cases by histologic subgroup.
CI: 95% confidence interval; Exp: expected; Obs: observed
| Same subgroup type | Any breast cancer | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Subgroup | #TDRs | Obs | Exp | P value | RR (CI) | Obs | Exp | P value | RR (CI) |
| Breast cancer | 1,140,944 | 12,138 | 11,260 | <0.001 | 1.08 (1.06, 1.10) | ||||
| Angiosarcoma | 872 | 0 | 0 | 6 | 9.3 | 0.41 | 0.65 (0.24, 1.41) | ||
| Apocrine | 4,188 | 0 | 0.10 | 64 | 46.9 | 0.02 | 1.36 (1.05, 1.74) | ||
| Inflammatory | 13,624 | † | † | 0.27 | 2.00 (0.24, 7.21) | 139 | 143.3 | 0.77 | 0.97 (0.82, 1.15) |
| Lobular | 123,396 | 118 | 87.3 | 0.001 | 1.35 (1.12, 1.62) | 1527 | 1267 | <0.001 | 1.21 (1.15, 1.27) |
| Mucinous | 51,280 | 23 | 12.5 | 0.007 | 1.84 (1.16, 2.76) | 560 | 512.9 | 0.04 | 1.09 (1.02, 1.19) |
| Phyllodes | 2,696 | 0 | 0.03 | 24 | 27.9 | 0.57 | 0.86 (0.55, 1.28) | ||
| Tubular | 11,228 | † | † | 0.17 | 2.73 (0.33, 9.85) | 129 | 114.2 | 0.17 | 1.13 (0.94, 1.34) |
| Medullary | 28,053 | † | † | 0.23 | 0.49 (0.06, 1.77) | 319 | 278.4 | 0.02 | 1.15 (1.02, 1.28) |
| Metaplastic | 2,527 | 0 | 0.05 | 26 | 25.6 | 0.49 | 1.02(0.66, 1.49) | ||
Observed counts < 5 are censored
FDRs with the same histologic type as the proband were only observed for the 2 largest sets of probands, lobular and mucinous histologies, and significantly increased RR was observed for each (Table 2). The RR for breast cancer of any histology in FDRs was significantly increased for patients with all examined histologic subtypes; and was increased, but not significantly, for 2 of the smallest subgroups, phyllodes and metaplastic tumors.
Similarly for SDRs, affected relatives were only observed for the 2 largest subgroups, lobular and mucinous; significantly increased risk in second degree relatives was only observed for the lobular subgroup (RR = 1.56, p=0.003) (Table 3). The RR for breast cancer of any histology in SDRs was significantly increased for patients with inflammatory (RR 1.32 [1.02, 1.68]; p=0.03), lobular (RR 1.36 [1.25, 1.47]; p=<0.001), and mucinous (RR 1.27 [1.12, 1.44]; p=0.00021) subtypes.
For TDRs, significantly elevated risk for breast cancer of the same tumor subtype was observed for lobular (RR 1.35 [1.12, 1.62], p=0.001) and mucinous (RR 1.84 [1.16, 2.76]; p=0.007) cancers (Table 4). The RR for breast cancer of any histology in TDRs was significantly elevated for patients with lobular (RR 1.21 [1.15, 1.27]; p=<0.001), mucinous (RR 1.09 [1.02, 1.19]; p=0.04), medullary (RR 1.15 [1.02, 1.28]; p=0.02), and apocrine (RR 1.36 [1.05, 1.74]; p=0.02) subtypes.
High-risk Pedigrees
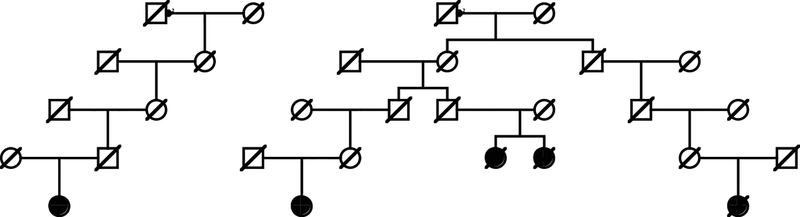
Analysis of all relationships among breast cancer cases for the inflammatory and mucinous subtypes (the 2 subtypes with significant excess relatedness observed) identified 48 high risk inflammatory breast cancer pedigrees including between 2 and 4 inflammatory breast cancer cases, and 110 high-risk mucinous breast cancer pedigrees including between 2 and 13 mucinous breast cancer cases. Figure 1 shows an example high-risk mucinous breast cancer pedigree. The top generation shows a male with 2 spouses. Of the total of over 10,630 descendants of this male founder in UPDB, 5 have been diagnosed with mucinous breast cancer (0.8 expected; p = 0.0018). Overall there are 65 breast cancer cases among the descendants with 43.9 expected (p=0.0017); the other breast cancers observed include 2 inflammatory, 2 lobular and 1 medullary breast cancer. Analysis of all relationships among lobular breast cancer cases (the subtype with significant excess risks for first-, second-, and third-degree relatives) identified 273 high-risk lobular breast cancer pedigrees including between 2 and 14 related lobular breast cancer cases.
Figure 1. Example of a high-risk mucinous breast cancer pedigree from the Utah Population DataBase.
Females are designated by circles and males by squares. Individuals with mucinous breast cancer are designated by a solid symbol. Deceased individuals are shown with a hash-mark through the symbol.
Discussion
Using a unique population-based resource linking decades of statewide cancer data to over 150 years of genealogy data, the hypothesis of histology-specific clustering of breast cancer cases has been investigated. Previously published findings from this resource provided evidence for clustering of lobular breast cancers and these results have been confirmed here.16 In addition, evidence was observed for significant familial clustering for mucinous and inflammatory breast carcinomas. No significant evidence for increased relatedness or risk for the same histologic subtype of breast cancer was observed for the other breast cancer histologies considered; however small sample sizes for many of these uncommon subgroups limited power to identify clustering. Most histological subtypes were associated with increased RR for breast cancer of any histologic type in first-, second-, and third-degree relatives, which is unsurprising given that it is well recognized that a diagnosis of breast cancer is associated with increased risk to relatives.
The GIF analysis for excess familiality was limited to the larger histologic subgroups, and identified significant excess relatedness for distant relationships for both inflammatory and mucinous subgroups, providing strong evidence for a heritable, rather than just a shared environment, contribution for these specific types of breast cancer. The RR analysis considered even the smaller histologic subgroups, but limited numbers of affected relatives with breast cancer of the same histology were observed. Nevertheless, a significantly increased risk for lobular breast cancers in the first-, second-, and third-degree relatives of lobular cancer cases was observed, again providing strong evidence for a heritable contribution to predisposition. Elevated risk for mucinous breast cancer was also observed in first-, second-, and third-degree relatives, and the elevation was statistically significant in the first-, and third-degree relatives. These findings suggest genetic contribution to risk, although additional environmental contribution to risk cannot be excluded.
In a nested case control study from the Breast Cancer Surveillance Consortium that included 617 inflammatory breast cancer cases, first-degree family history of breast cancer was associated with increased risk of inflammatory breast cancer.9 The multivariable rate ratio was 1.52 (95% CI 1.15, 2.01). In a second study, patients with inflammatory breast cancer had a higher likelihood of family history of breast cancer compared to unaffected controls, although a lower likelihood compared to patients with non-inflammatory breast cancer.14 Interestingly, in the latter cohort they identified several inflammatory breast cancer patients with multiple first- and second-degree family members with non-inflammatory breast cancer. Our findings both confirm and extend this finding, as we were able to examine relationships out to third-degree (first cousins) and identified a significant excess of distant relatedness. The RR for third-degree relatives for inflammatory breast cancer was elevated, but not significantly (RR=2.00; 95%CI 0.24, 7.21); sample sizes are small, but this indicates that the evidence for the significant excess relatedness observed in the dGIF test came from an excess of even more distant relationships.
It was somewhat surprising that we did not identify increased relatedness among patients with medullary carcinoma given its known association with tumors with pathogenic variants in BRCA1. However, according to the literature only 10–15% of BRCA1 mutated tumors have pure medullary histology.13 In addition, only about 11% of medullary tumors, regardless of family history, had identified pathogenic variants in BRCA1.23 Therefore, our results support prior findings that only a minority of medullary breast cancer cases are likely due to an inherited predisposition.
Strengths of this analysis are the large number of primary cancers in the database, histopathological confirmation of all cases in the Utah Cancer Registry, and comprehensive genealogy information including large numbers of first-, second-, and third-degree relatives with known cancer status. The UPDB database includes genealogy from the mid-1800s and statewide cancer data from 1966; nevertheless, some breast cancer histologic subgroups were rarely observed, and significant conclusions for some hypotheses will require larger data sets. The total sample size for primary inflammatory breast cancers was limited, and no FDRs or SDRs diagnosed with inflammatory breast cancer were observed. However, it is possible that inflammatory breast cancer cases may be incompletely represented in the database since it is a clinical, rather than histologic, diagnosis, and cases without evidence of dermal lymphatic invasion in a pathology specimen may not have been identified.
Even among the histologic subgroups with small sample sizes some pedigrees including related breast cancer cases with the same histology were identified to have a significant excess of breast cancer. These rare pedigrees may provide a powerful resource to identify new breast cancer predisposition genes or to enhance our understanding of known predisposition genes.
The analysis has some limitations. Data for some individuals could have been censored due to diagnosis of cancer outside Utah or before 1966. Pathology reports and slides were not reviewed and some records date back decades. In addition, changes in histologic classification have occurred over time, and these updates or subclassifications may not be reflected in the available patient-level data. Although it would have been interesting to examine associations between familiality and both clinicopathologic characteristics and disease outcomes, especially for the inflammatory breast cancer cohort, unfortunately neither receptor status nor treatment information is available at this time.
Censoring also occurred for individuals who do not have genealogy data in the UPDB or who did not appropriately link to their genealogy data. Approximately 60% of Utah Cancer Registry records link to an individual with Utah genealogy data; females have lower record linking rates than males due to name changes. In addition, genealogy does not always represent biological relationships. These censorship issues can be assumed to occur uniformly across both cases and controls in UPDB and should not bias analyses, although they can lower power.
The Utah population represented in the UPDB largely consists of individuals of Northern European ancestry. The population has been shown to be genetically identical to other Northern European populations. The original Utah pioneers, who began arriving in Utah in 1847, were largely unrelated; the Utah population continues to have low or normal inbreeding compared to the United States.24 In addition, the Utah population has a high proportion of women with higher average number of pregnancies, younger age at first childbirth, lower alcohol and tobacco use, and lower rates of post-menopausal obesity, all of which could impact the rates of breast cancer incidence. Therefore the results of this study are likely applicable to populations of females similar to the Utah population, but should not be extrapolated to other populations without validation.
In summary, using genealogy data from a large population-based database, this study provides additional evidence supporting a genetic predisposition to inflammatory breast cancer as well as lobular and mucinous breast cancer histologies. While estimated RRs are modest, the important point is that breast cancer cases with specific histologies appear to cluster more in pedigrees than expected, and these homogeneous pedigrees may be informative for identification of the predisposition genes responsible. Subsequent identification of inherited genetic variants should be performed in order to identify potential etiologies of specific breast cancer subtypes.
Supplementary Material
Funding Sources:
This work was supported by: the Huntsman Cancer Institute to NLH. The work was also supported by the Utah Cancer Registry, which is funded by the National Cancer Institute’s SEER Program, Contract Number HHSN261201800016I, the US Center for Disease Control and Prevention’s National Program of Cancer Registries, Cooperative Agreement No. NU58DP0063200–01, with additional support from the University of Utah and Huntsman Cancer Foundation. Partial support for all datasets within the Utah Population Database is provided by the University of Utah, Huntsman Cancer Institute and the Huntsman Cancer Institute Cancer Center Support grant, P30 CA42014 from the National Cancer Institute.
Footnotes
Conflict of Interest: Neither author reports relevant conflicts of interests. NLH reports funding to her institution for the conduct of pharmaceutical-sponsored clinical trials by AstraZeneca, AbbVie, H3 Biosciences, Innocrin Pharmaceuticals, and Pfizer.
References
- 1.Kleihues P, Sobin LH. World Health Organization classification of tumors. Cancer. 2000;88: 2887. [DOI] [PubMed] [Google Scholar]
- 2.Tavassoli FA, Devilee P. Pathology and Genetics of Tumours of the Breast and Female Genital Organs. . Lyon, France: IARC Press, 2003. [Google Scholar]
- 3.Nguyen B, Veys I, Leduc S, et al. Genomic, transcriptomic, epigenetic, and immune profiling of mucinous breast cancer. J Natl Cancer Inst. 2019. [DOI] [PubMed] [Google Scholar]
- 4.Kehr EL, Jorns JM, Ang D, et al. Mucinous breast carcinomas lack PIK3CA and AKT1 mutations. Hum Pathol. 2012;43: 2207–2212. [DOI] [PubMed] [Google Scholar]
- 5.Dawood S, Merajver SD, Viens P, et al. International expert panel on inflammatory breast cancer: consensus statement for standardized diagnosis and treatment. Ann Oncol. 2011;22: 515–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hance KW, Anderson WF, Devesa SS, Young HA, Levine PH. Trends in inflammatory breast carcinoma incidence and survival: the surveillance, epidemiology, and end results program at the National Cancer Institute. J Natl Cancer Inst. 2005;97: 966–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Uden DJ, van Laarhoven HW, Westenberg AH, de Wilt JH, Blanken-Peeters CF. Inflammatory breast cancer: an overview. Crit Rev Oncol Hematol. 2015;93: 116–126. [DOI] [PubMed] [Google Scholar]
- 8.Woodward WA. Inflammatory breast cancer: unique biological and therapeutic considerations. Lancet Oncol. 2015;16: e568–576. [DOI] [PubMed] [Google Scholar]
- 9.Schairer C, Li Y, Frawley P, et al. Risk factors for inflammatory breast cancer and other invasive breast cancers. J Natl Cancer Inst. 2013;105: 1373–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walsh T, King MC. Ten genes for inherited breast cancer. Cancer Cell. 2007;11: 103–105. [DOI] [PubMed] [Google Scholar]
- 11.Stadler ZK, Thom P, Robson ME, et al. Genome-wide association studies of cancer. J Clin Oncol. 2010;28: 4255–4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pharoah PD, Guilford P, Caldas C, International Gastric Cancer Linkage C. Incidence of gastric cancer and breast cancer in CDH1 (E-cadherin) mutation carriers from hereditary diffuse gastric cancer families. Gastroenterology. 2001;121: 1348–1353. [DOI] [PubMed] [Google Scholar]
- 13.Mavaddat N, Barrowdale D, Andrulis IL, et al. Pathology of breast and ovarian cancers among BRCA1 and BRCA2 mutation carriers: results from the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA). Cancer Epidemiol Biomarkers Prev. 2012;21: 134–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moslehi R, Freedman E, Zeinomar N, Veneroso C, Levine PH. Importance of hereditary and selected environmental risk factors in the etiology of inflammatory breast cancer: a case-comparison study. BMC Cancer. 2016;16: 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gutierrez Barrera AM, Fouad TM, Song J, et al. BRCA mutations in women with inflammatory breast cancer. Cancer. 2018;124: 466–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allen-Brady K, Camp NJ, Ward JH, Cannon-Albright LA. Lobular breast cancer: excess familiality observed in the Utah Population Database. Int J Cancer. 2005;117: 655–661. [DOI] [PubMed] [Google Scholar]
- 17.Cannon-Albright LA, Thomas A, Goldgar DE, et al. Familiality of cancer in Utah. Cancer Res. 1994;54: 2378–2385. [PubMed] [Google Scholar]
- 18.Albright F, Teerlink C, Werner TL, Cannon-Albright LA. Significant evidence for a heritable contribution to cancer predisposition: a review of cancer familiality by site. BMC Cancer. 2012;12: 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cannon L, Bishop D, Skolnick M, S H, L L, CR S. Genetic epidemiology of prostate cancer in the Utah Mormon genealogy. Cancer Surv. 1982;1: 47–70. [Google Scholar]
- 20.Cannon Albright LA. Utah family-based analysis: past, present and future. Hum Hered. 2008;65: 209–220. [DOI] [PubMed] [Google Scholar]
- 21.Skolnick M The Utah genealogical database: a resource for genetic epidemiology In: Cairns J, Lyon JL, Skolnick M, editors. Cancer Incidence in Defined Populations. New York: Cold Spring Harbor Laboratory Press, 1980:285–297. [Google Scholar]
- 22.Malécot G Les mathématiques de l’hérédité. Paris: Masson & Cie, 1948. [Google Scholar]
- 23.Eisinger F, Jacquemier J, Charpin C, et al. Mutations at BRCA1: the medullary breast carcinoma revisited. Cancer Res. 1998;58: 1588–1592. [PubMed] [Google Scholar]
- 24.Jorde LB. Inbreeding in the Utah Mormons: an evaluation of estimates based on pedigrees, isonymy, and migration matrices. Ann Hum Genet. 1989;53: 339–355. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.