Abstract
Nonstructural protein 1 (NS1) is a multifunctional virulence factor of influenza virus. The effector domain (ED) of influenza viruses is capable of binding to a variety of host factors, however, the molecular basis of the interactions remains to be investigated. The isolated NS1-ED exists in equilibrium between the monomer and homodimer. Although the structural diversity of the dimer interface has been well-characterized, limited information is available regarding the internal conformational heterogeneity of the monomeric NS1-ED. Here, we present the solution NMR structure of the NS1-ED W187R of the 1918 influenza A virus, which caused the “Spanish flu.” Structural plasticity is an essential property to understand the molecular mechanism by which NS1-ED interacts with multiple host proteins. Structural comparison with the NS1-ED from influenza A/Udorn/1972 (Ud) strain revealed a similar overall structure but a distinct conformational variation and flexibility. Our results suggest that conformational flexibility of the NS1-ED might differ depending on the influenza strain.
Keywords: 1918 Influenza A virus, nonstructural protein 1, structural plasticity, conformational dynamics, NMR, virulence factor
Introduction
Influenza virus is responsible for seasonal flus, resulting in 30,000 deaths every year in the United States alone [1]. The 1918 influenza A virus (1918 IAV) caused the worst flu pandemic by killing more than 50 million people worldwide [2]. The genome of the 1918 IAV was determined in 2005 [3,4], however, the molecular determinants of its high virulence remain unclear.
Nonstructural protein 1 (NS1) is a multifunctional virulence factor of influenza viruses [5]. The NS1 protein plays diverse roles in suppressing anti-viral immune responses of host cells during the infection cycle [6]. NS1 consists of three domains [6,7], an RNA-binding domain (RBD), an effector domain (ED) and a disordered C-terminal tail (Figure 1A). The NS1-ED interacts with a number of host factors, including cellular 30 kDa subunit of the cleavage and polyadenylation specificity factor (CPSF 30) [8,9], phosphoinositide 3 kinase (PI3K) [10], tripartite motif-containing protein 25 (TRIM25) [11,12], and CT-10 regulatory of kinase (CRK) [13,14,15]. These interactions contribute to the suppression of innate immune responses [6] to promote viral replication. Thus, the NS1-ED is considered a potential target for the development of anti-influenza reagents [16,17].
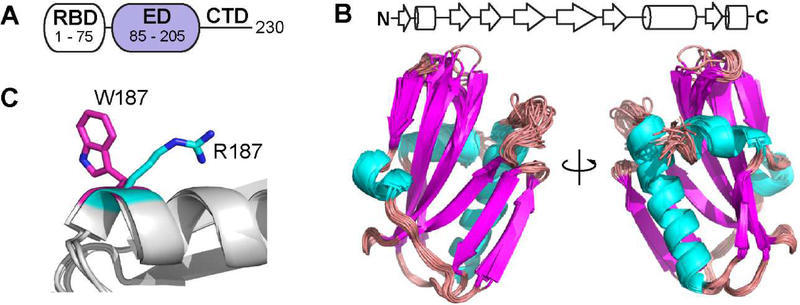
Figure 1.
Structure of the 1918 NS1-ED. (A) Domain organization of NS1. (B) Ensemble view of the 20 lowest energy structure of 1918 NS1-ED. (C) Conformations of W187 and R187 in Ud and 1918 NS1-ED are shown in magenta and cyan, respectively.
Structural plasticity was suggested to be essential for NS1 to sample a conformational space, that is necessary to bind multiple host factors [18,19]. Moreover, the variability in the interactions mediated by the NS1-ED was suggested to contribute to strain-specific pathogenesis of influenza viruses [9,20,21]. Therefore, studies on the conformational flexibility of NS1-ED proteins are important to understand the molecular mechanism by which NS1 binds to diverse host factors.
The isolated NS1-ED forms a homodimer through either strand-strand [22] or helix-helix [23,24] dimer interfaces. The homodimerization plays a role in enhancing the affinity of the RBD to dsRNA in full-length NS1 [25,26]. However, recent structural studies showed that the monomeric NS1-ED is important for binding to host proteins such as CPSF30 [9], PI3K [27], and TRIM25 [28]. Therefore, studies on the structure and dynamics of the NS1-ED monomer are important for elucidating the underlying molecular mechanisms by which NS1 binds host proteins.
Here we present the solution NMR structure of the 1918 NS1-ED W187R. We find that the 1918 NS1-ED W187R exists as a monomer in solution. We also characterize the intrinsic conformational plasticity in the 1918 NS1-ED. By comparing the solution NMR structures of the 1918 and Ud (A/Udorn/307/1972) NS1-ED [25,29] proteins, we reveal that the spatial distribution of conformationally flexible regions are different between the two proteins. Our results indicate that the difference in the conformational flexibility of the NS1-ED should be considered to fully understand strain-specific functions of NS1.
Materials and methods
Protein sample preparation
Genes encoding the 1918 NS1-ED was prepared by gene-synthesis service from Genscript. NS1-ED W187R was expressed in BL21 (DE3) E. coli cells with a His6 and SUMO tags, and purified by Ni2+ NTA column and gel-filtration chromatography. Purity of protein samples was confirmed using SDS-PAGE. For stable isotope labeling, 2 L bacterial cultures were grown in LB media to an OD600 of ~0.6 and then, bacteria were spun-down to transfer to 1 L of M9 minimal media supplemented with 1 g/L of 15N NH4Cl and 2g/L 13C D-glucose. Protein expression was then induced with 0.5 mM IPTG for 4 h at 37 °C.
NMR sample preparation
The NMR sample of the 1918 NS1-ED was prepared in an NMR buffer consisting of 20 mM sodium phosphate (pH 7.0), 80mM NaCl, 1mM EDTA, 10% D2O, 10 mM DSS (4,4-dimethyl-4-silapentane-sulfonate). For resonance assignment and structural determination 13C, 15N-labled NS1-ED was concentrated to 150 μM in NMR buffer.
NMR resonance assignment and structure determination of 1918 NS1-ED
NMR data were collected at 298 K on Bruker Avance III 600 MHz and 800 MHz spectrometers equipped with a cryogenic probe at Biomolecular NMR facility (Texas A&M University). The data were processed using NMRPipe [30] and resonance assignment was conducted using NMRFAM-Sparky [31]. Backbone and sidechain resonance assignments were performed using a standard backbone-directed triple resonance experiments in combination with NOE spectroscopy (NOESY) [32]. Backbone and sidechain chemical shift assignments have been deposited into the Biological Magnetic Resonance Databank under the BMRB accession code 12032. The structure of free 1918 NS1-ED was calculated using simulated annealing methods based on NOESY-derived distance restraints and torsion angle restraints calculated by TALOS+ [33]. Distance restraints were derived from NOE cross peaks in 13C-NOESY-HSQC and 15N-NOESY-HSQC spectra with mixing times of 80 and 100 ms. The automated NOESY peak assignment was performed via the CYANA Noeassign macro [34]. Cross-peak intensities were converted into distances using CYANA. 100 structures were analyzed and the 40 structure with the lowest residual CYANA target function values were subjected to further energy minimization with R2/R1 ratio and explicit water refinement protocol of Xplor-NIH [35,36]. The resulting 20 lowest structures were further refined using YASARA webserver [37]. Atomic coordinates have been deposited in the Protein Data Bank (PDB ID: 6NU0).
NMR 15N relaxation measurements
Measurements for 15N R1 and R2 rate constants were performed at 800 MHz magnetic field strength as described elsewhere [38]. Briefly, for R1, five relaxation time points were taken between 100 ms and 1 s. For R2, five relaxation time points were taken between 4 and 120 ms. For R1 and R2 measurements, a recycle delay of 2 s was used between transitions. Calculation of 15N R1 and R2 was conducted using HYDRONMR [39] with the atomic element radius of 3.1 Å.
Results and discussion
Overall structure of the 1918 NS1-ED
The structure of the 1918 NS1-ED W187R (hereinafter 1918 NS1-ED unless otherwise defined) was determined using solution NMR spectroscopy as described in the Methods. The ensemble of the 20 lowest energy structures is shown in Figure 1B. The full structural statistic for the ensemble is shown in Supplementary Table 1
Overall, the solution NMR structure showed an α-helix β-crescent fold that is common to all known structures of the NS1-ED. In this study, we incorporated the W187R mutation into 1918 NS1-ED to abolish homodimerization. Both W187R and W187A mutations can abolish NS1-ED homodimerization; however, the W187R mutation is the only amino acid substitution allowed for position 187 of NS1 that does not affect the overlapping open reading frame of NS2, which is an alternatively spliced protein of the NS1 gene [40]. The side chain of W187 is exposed to solvent in its monomeric form (Figure 1C). Our NMR structure also showed that the side chain of R187 is fully exposed to solvent and does not interact with other residues of the protein (Figure 1C). The side chain of R187 was also fully exposed to solvent in the NMR structure of the Ud NS1-ED W187R [25]. These results indicate that the W187R mutation does not affect the overall structure of the 1918 NS1-ED.
The 1918 NS1-ED W187R exists as a monomer in solution
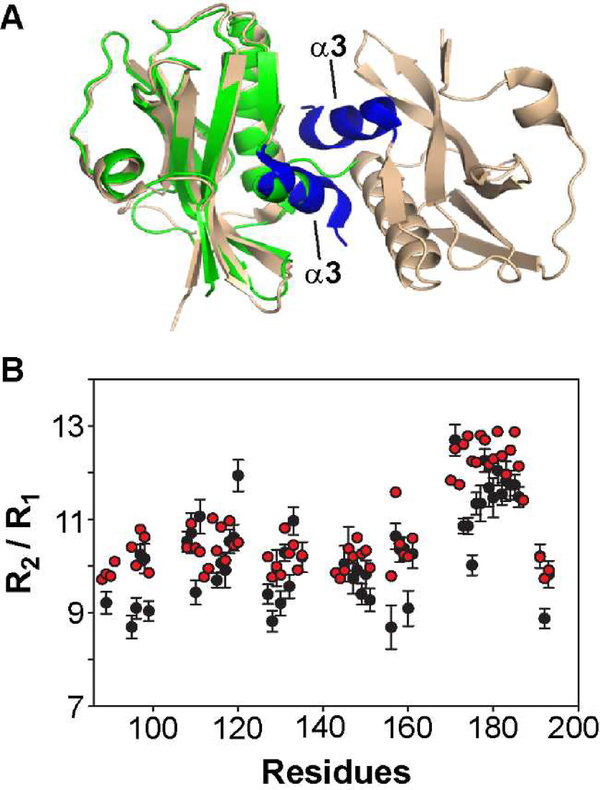
The overall structure of the 1918 NS1-ED W187R was highly similar to the recently reported crystal structure of the 1918 NS1-ED W187A [41] (PDB ID: 6DGK) (Figure 2A). However, while the NMR structure is a monomer, the crystal structure of the 1918 NS1-ED W187A showed a dimeric form [41]. The crystal structure contained two NS1-ED molecules in an asymmetric unit, although the W187A mutation is supposed to abolish the dimeric form. Intriguingly, the crystal structure revealed a new dimer interface, the α3-α3 interface, mediated by W203. However, it remained unclear whether the 1918 NS1-ED W187A or W187R forms a dimer in solution. This is an important question to address because the interactions with host factors such as CPSF30, PI3K, and TRIM25 were suggested to be mediated by the NS1-ED monomer [9,27].
Figure 2.
1918 NS1-ED exists as a monomer in solution. (A) Crystal structure of 1918 NS1-ED W187A (PDB ID: 6DGK). The α3-α3 dimeric interface is shown in blue. For comparison, NMR structure of 1918 NS1-ED W187R (shown in green) is superimposed to chain A of the crystal structure. (B) The NMR R2 / R1 ratio as a function of primary structure of 1918 NS1-ED. The experimental and HYDRONMR-calculated values of conformationally rigid residues are shown in black and red circles, respectively.
To address whether the 1918 NS1-ED-W187R exists as a monomer or dimer in solution, we measured the correlation time (τc) for the rotational diffusion process of the 1918 NS1-ED using the NMR 15N R2/R1 ratio, which is a highly sensitive probe of molecular size and shape [39,42]. By only including the residues located in stable secondary structures without a chemical exchange effect, we estimated that the τc is 7.2 ns [43], which is consistent with the value (τc = 7.4 ns) calculated using the software HYDRONMR with a monomeric NMR structure [39]. Thus, the HYDRONMR-calculated R2/R1 ratio is consistent with the experimentally measured value (Figure 2B). This shows that the 1918 NS1-ED W187R exists as a monomer in solution, although more thorough examination might be needed.
However, we note that long-term solubility of 1918 NS1-ED W187R was limited. For example, the protein was precipitated by ~20% over a week in the NMR condition, in which protein concentration was about 150 μM. The limited solubility suggests that even NS1-ED W187R undergoes oligomerization in slower timescale despite that its mechanism and relationship to biological function remain to be determined. We speculate that the slow-timescale precipitation could be induced by α3-α3 or strand-strand dimeric interfaces, which are not suppressed by W187R mutation.
Intrinsic conformational plasticity of the 1918 NS1-ED
Conformational plasticity might be important for the NS1-ED to adopt slightly different conformations to bind to diverse host factors. However, most NS1-ED structures have been determined by crystallography, and thus, it has remained unclear whether the conformational variations were due to crystal contacts or the intrinsic structural plasticity of the protein.
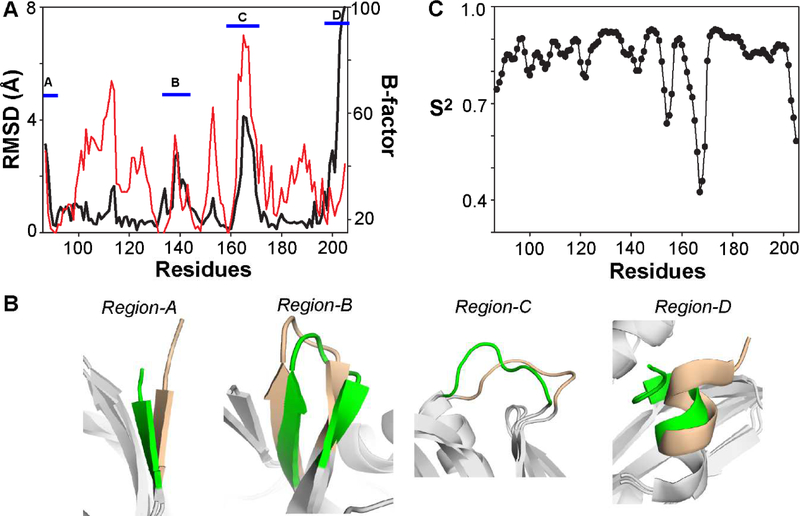
Therefore, we first examined the conformational plasticity of the 1918 NS1-ED. For this purpose, we compared the lowest energy NMR and crystal structures (PDB ID: 6DGK). The root-mean-square-deviation (RMSD) between the two structures was considerably large (> 2 Å) in the following four regions (Figure 3A and 3B). Region-A (residues 87 – 88) corresponds to the short N-terminal β1 strand. Region-B (residues 134 – 142) includes parts of the β4 and β5 strands and a flexible loop connecting the two β strands (β4-β5 loop). We also noticed that the region has relatively high B-factors in the crystal structure (Figure 3A). Region-C (residues 164 – 169) is a Pro-rich loop connecting the β6 strand and x2 helix (β6-α2 loop). This region showed the highest B-factors in the crystal structure (Figure 3A). Region-D (residues 199 – 205) corresponds to the C-terminal α3 helix. The RMSD in this region is due to the difference in the orientation of the α3 helix between the two structures. Although the B-factors of this region are lower than the other three regions (Figure 3A), this may be because the region is stabilized by the crystallographic dimer interface (i.e., α3-α3 interface). Therefore, the α3 helix would have more motional freedom in the solution state as reflected in our NMR structure.
Figure 3.
Conformational plasticity of 1918 NS1-ED. (A) Black line corresponds to the backbone (Cα) RMSD between the NMR and crystal structures of 1918 NS1-ED. Red line corresponds to the Cα B-factor of the crystal structure. The four regions that have a large RMSD between the two structures are marked using blue bars. (B) Superimposed structures of four conformationally plastic regions in the 1918 NS1-ED. NMR and crystal structures are shown in green and wheat, respectively. Other regions in both structures are shown in gray. (C) RCI-derived order parameters of the 1918 NS1-ED.
To further examine the intrinsic conformational flexibility of the 1918 NS1-ED in the sub-ns timescale, we calculated the random-coil index (RCI)-derived backbone order parameters [44]. Three regions were identified to be conformationally flexible (S2 < 0.7), including residues 154 – 155, 165 – 169 (region-C), and 203 – 205 (region-D) (Figure 3C). This result showed that the conformational variation in regions-C and -D is mainly due to the intrinsic flexibility of the 1918 NS1-ED in the ps – ns timescale. Residues 154 – 155 correspond to a loop connecting the β3 and β4 strands. Although the region did not show a noticeable RMSD between the NMR and crystal structures, it showed slightly elevated B-factors in the crystal structure, indicating reasonably high conformational flexibility of the region. This loop is involved in the binding site for CPSF30 [9]. Intriguingly, regions-C and -D are also parts of the binding interface for TRIM25 [28]. Thus, our result suggests that the flexibility might have a role in binding to host proteins.
Comparison of conformational plasticity between the 1918 and Ud NS1-ED proteins
Detailed understanding of the conformational plasticity of NS1-ED proteins from diverse influenza viruses is important because it may provide structural insights into the strain-specific interactions of NS1 with host factors [21,29,45,46]. First, we sought to test whether there is a difference in conformationally plastic regions among NS1-ED proteins from different influenza viruses. To this end, we compared the conformational heterogeneity in the following two NMR ensemble structures: the 1918 and Ud NS1-EDs (PDB ID: 2KKZ) (Figure 4A). We chose NMR structures to avoid effects from crystal contacts. Moreover, structures of both free and CPSF30-bound forms are available for the Ud NS1-ED [9], and this allowed us to examine the conformational plasticity of the CPSF30-binding site in the NS1-ED.
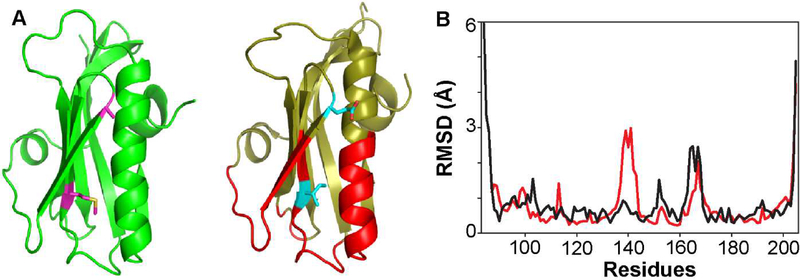
Figure 4.
Structural comparison of 1918 and Ud NS1-ED. (A) Solution NMR structures of 1918 (left panel) and Ud (right panel) NS1-ED. The CPSF30-binding sites in Ud NS1-ED are shown in red. Residues 112 and 119 are shown as a stick model. (B) The RMSD plots of the 20 lowest energy structures of 1918 and Ud NS1-ED are shown in red and black, respectively.
Figure 4B shows the average RMSD plots of the 20 lowest energy structures of each protein. Although the two proteins have virtually identical overall structure, they showed stark differences in the spatial distribution of the conformationally plastic regions. The Ud NS1-ED showed significantly higher conformational heterogeneity in the N-terminal β1 strand (region A) than the 1918 NS1-ED. In contrast, the 1918 NS1-ED showed higher RMSD in region-B than the Ud NS1-ED. Both proteins showed a high RMSD in region-C; however, conformational variation in the Ud NS1-ED was extended to a larger extent in the primary sequence than in the 1918 NS1-ED. This difference might be due to a single amino acid change in this region; residue 166 is Leu in the 1918 and Phe in the Ud strain. While the side chain of F166 in the Ud NS1-ED is exposed to solvent, the side chain of L166 is partially exposed to solvent in the 1918 NS1-ED; partially buried L166 might stabilize the loop conformation.
Interestingly, we noticed that parts of the CPSF30-binding site in the NS1-ED showed the opposite pattern of the conformational heterogeneity between the two proteins. For example, while the backbone of F103 showed considerable RMSD among the ensemble of Ud NS1-ED, the same position was well-converged in the 1918 NS1-ED, resulting in low RMSD. F103 interacts with hydrophobic residues of CPSF30 [9], stabilizing the tetrameric complex with CPSF30. Moreover, the β3-β4 loop (residues 150 – 155) is also involved in the CPSF30-binding site and showed large RMSD in the Ud-NS1, while it was well-converged in the 1918 NS1-ED. This result suggests that the 1918 and Ud NS1-ED proteins may have different binding properties, such as Kd, to CPSF30.
The CPSF30-binding interface consists of 27 residues in the Ud NS1-ED (Figure 4A). These residues are conserved in the 1918 NS1-ED except for two residues; E112 and I119 in the Ud NS1-ED are replaced by A112 and M119 in the 1918 NS1-ED, respectively. Interestingly, these two positions were well-converged in both ensemble structures despite the sequence variation. However, the effects of these mutations on the binding affinity to CPSF30 remain to be investigated.
These results indicate that the spatial distribution of conformationally plastic regions varies among NS1-ED proteins from different influenza viruses. A caveat in this analysis should be noted. The conformational heterogeneity of the NMR structure could be affected by nonstructural reasons such as lack of enough distance-constraints during structural calculations. Although it is beyond the scope of this report, a systematic NMR dynamics study on NS1-ED proteins from various influenza viruses would be highly informative to reveal the strain-specific conformational flexibility of NS1-ED proteins. Moreover, it would be important to examine whether the conformational heterogeneity and dynamics of NS1-ED proteins would affect differential recognition of host factors. The contribution of conformational plasticity and dynamics to molecular recognition was highlighted in recent studies [47,48]. Thus, the binding thermodynamics and kinetics between the NS1-ED and host factors might be different for different influenza viruses, although further studies will be needed to test this hypothesis.
Supplementary Material
Highlights.
Solution NMR structure of the NS1-ED of the 1918 influenza A virus
NS1-ED W187R of the 1918 influenza A virus exists as a monomer in solution
The 1918 NS1-ED is structurally plastic in the binding sites for host proteins
Spatial distribution of structurally plastic regions might vary among NS1-ED proteins
Acknowledgements
This was supported by the National Institutes of Health to J.H.C (1R01GM 127723-01A1).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Flannery B, Chung JR, Belongia EA, et al. , Interim Estimates of 2017–18 Seasonal Influenza Vaccine Effectiveness — United States, MMWR Morb Mortal Wkly Rep 67 (2018) 180–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Taubenberger JK, Reid AH, Janczewski TA, et al. , Integrating historical, clinical and molecular genetic data in order to explain the origin and virulence of the 1918 Spanish influenza virus, Philos Trans R Soc Lond B Biol Sci 356 (2001) 1829–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Tumpey TM, Basler CF, Aguilar PV, et al. , Characterization of the reconstructed 1918 Spanish influenza pandemic virus, Science 310 (2005) 77–80. [DOI] [PubMed] [Google Scholar]
- [4].Taubenberger JK, Reid AH, Lourens RM, et al. , Characterization of the 1918 influenza virus polymerase genes, Nature 437 (2005) 889–893. [DOI] [PubMed] [Google Scholar]
- [5].Lamb RA, Choppin PW, Segment 8 of the influenza virus genome is unique in coding for two polypeptides, Proc Natl Acad Sci USA 76 (1979) 4908–4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hale BG, Randall RE, Ortin J, et al. , The multifunctional NS1 protein of influenza A viruses, J Gen Virol 89(Pt 10) (2008) 2359–2376. [DOI] [PubMed] [Google Scholar]
- [7].Bornholdt ZA, Prasad BV, X-ray structure of NS1 from a highly pathogenic H5N1 influenza virus, Nature 456 (2008) 985–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Nemeroff ME, Barabino SM, Li Y, et al. , Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3’end formation of cellular pre-mRNAs, Mol Cell 1 (1998) 991–1000. [DOI] [PubMed] [Google Scholar]
- [9].Das K, Ma LC, Xiao R, et al. , Structural basis for suppression of a host antiviral response by influenza A virus, Proc Natl Acad Sci USA 105 (2008) 13093–13098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ehrhardt C, Marjuki H, Wolff T, et al. , Bivalent role of the phosphatidylinositol-3-kinase (PI3K) during influenza virus infection and host cell defence, Cell Microbiol 8 (2006) 1336–1348. [DOI] [PubMed] [Google Scholar]
- [11].Gack MU, Albrecht RA, Urano T, et al. , Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I, Cell Host Microbe 5 (2009) 439–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Meyerson NR, Zhou L, Guo YR, et al. , Nuclear TRIM25 Specifically Targets Influenza Virus Ribonucleoproteins to Block the Onset of RNA Chain Elongation, Cell Host Microbe 22 (2017) 627–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Heikkinen LS, Kazlauskas A, Melen K, et al. , Avian and 1918 Spanish influenza a virus NS1 proteins bind to Crk/CrkL Src homology 3 domains to activate host cell signaling, J. Biol. Chem 283 (2008) 5719–5727. [DOI] [PubMed] [Google Scholar]
- [14].Shen Q, Zeng D, Zhao B, et al. , The molecular mechanisms underlying the hijack of host proteins by the 1918 Spanish influenza virus, ACS Chem Biol 12 (2017) 1199–1203. [DOI] [PubMed] [Google Scholar]
- [15].Shen Q, Shi J, Zeng D, et al. , Molecular mechanisms of tight binding through fuzzy interactions, Biophys J 114 (2018) 1313–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Min JY, Krug RM, The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: Inhibiting the 2’−5’ oligo (A) synthetase/RNase L pathway, Proc Natl Acad Sci USA 103 (2006) 7100–7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Twu KY, Noah DL, Rao P, et al. , The CPSF30 binding site on the NS1A protein of influenza A virus is a potential antiviral target, J Virol 80 (2006) 3957–3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hale BG, Conformational plasticity of the influenza A virus NS1 protein, J Gen Virol 95 (2014) 2099–2105. [DOI] [PubMed] [Google Scholar]
- [19].Carrillo B, Choi JM, Bornholdt ZA, et al. , The influenza A virus protein NS1 displays structural polymorphism, J Virol 88 (2014) 4113–4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Tscherne DM, García-Sastre A, Virulence determinants of pandemic influenza viruses., J Clin Invest 121 (2011) 6–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ayllon J, Hale BG, García-Sastre A, Strain-specific contribution of NS1-activated phosphoinositide 3-kinase signaling to influenza A virus replication and virulence, J Virol 86 (2012) 5366–5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bornholdt ZA, Prasad BV, X-ray structure of influenza virus NS1 effector domain, Nat Struct Mol Biol 13 (2006) 559–560. [DOI] [PubMed] [Google Scholar]
- [23].Hale BG, Barclay WS, Randall RE, et al. , Structure of an avian influenza A virus NS1 protein effector domain, Virology 378 (2008) 1–5. [DOI] [PubMed] [Google Scholar]
- [24].Xia S, Robertus JD, X-ray structures of NS1 effector domain mutants, Arch Biochem Biophys 494 (2010) 198–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Aramini JM, Ma LC, Zhou L, et al. , Dimer interface of the effector domain of nonstructural protein 1 from influenza A virus: an interface with multiple functions, J Biol Chem 286 (2011) 26050–26060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Cheng A, Wong SM, Yuan YA, Structural basis for dsRNA recognition by NS1 protein of influenza A virus, Cell Res 19 (2009) 187–195. [DOI] [PubMed] [Google Scholar]
- [27].Hale BG, Kerry PS, Jackson D, et al. , Structural insights into phosphoinositide 3-kinase activation by the influenza A virus NS1 protein, Proc Natl Acad Sci USA 107 (2010) 1954–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Koliopoulos MG, Lethier M, van der Veen AG, et al. , Molecular mechanism of influenza A NS1-mediated TRIM25 recognition and inhibition, Nat Commun 9 (2018) 1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Xia S, Monzingo AF, Robertus JD, Structure of NS1A effector domain from the influenza A/Udorn/72 virus, Acta Crystallogr D D65 (2009) 11–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Delaglio F, Grzesiek S, Vuister GW, et al. , NMRPipe: a multidimensional spectral processing system based on UNIX pipes, J. Biomol. NMR 6 (1995) 277–293. [DOI] [PubMed] [Google Scholar]
- [31].Lee W, Tonelli M, Markley JL, NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy, Bioinformatics 31 (2015) 1325–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sattler M, Schleucher J, Griesinger C, Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients, Prog. Nucl. Magn. Reson. Spectrosc 34 (1999) 93–158. [Google Scholar]
- [33].Shen Y, Delaglio F, Cornilescu G, et al. , TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts, J Biomol NMR 44 (2009) 213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Güntert P, Mumenthaler C, Wuthrich K, Torsion angle dynamics for NMR structure calculation with the new program DYANA, J Mol Biol 273 (1997) 283–298. [DOI] [PubMed] [Google Scholar]
- [35].Schwiesters CD, Kuszewski JJ, Clore GM, Using Xplor-NIH for NMR molecular structure determination, Progr. NMR Spectroscopy 48 (2006) 47–62. [Google Scholar]
- [36].Schwieters CD, Kuszewski JJ, Tjandra N, et al. , The Xplor-NIH NMR Molecular Structure Determination Package, J Magn Res 160 (2003) 66–74. [DOI] [PubMed] [Google Scholar]
- [37].Krieger E, Koraimann G, Vriend G, Increasing the precision of comparative models with YASARA NOVA--a self-parameterizing force field, Proteins 47 (2002) 393–402. [DOI] [PubMed] [Google Scholar]
- [38].Bhatt VS, Zeng D, Krieger I, et al. , Binding mechanism of the N-terminal SH3 domain of CrkII and proline-rich motifs in cAbl Biophys. J 110 (2016) 2630–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Bernado P, Garcia J de la Torre, M. Pons, Interpretation of 15N NMR relxation data of globular proteins using hydrodynamic calculatioin with HYDRONMR, J. Biomol. NMR 23 (2002) 139–150. [DOI] [PubMed] [Google Scholar]
- [40].Ayllon J, Russell RJ, García-Sastre A, et al. , Contribution of NS1 effector domain dimerization to influenza A virus replication and virulence, J Virol 86 (2012) 13095–13098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kleinpeter AB, Jureka AS, Falahat SM, et al. , Structural analyses reveal the mechanism of inhibition of influenza virus NS1 by two antiviral compounds, J Biol Chem 293 (2018) 14659–14668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lee LK, Rance M, Chazin WJ, et al. , Rotational diffusion anisotropy of proteins from simultaneous analysis of 15N and 13C alpha nuclear spin relaxation, J. Biomol. NMR 9 (1997) 287–298. [DOI] [PubMed] [Google Scholar]
- [43].Fushman D, Weisemann R, Thuring H, et al. , Backbone dynamics of ribonuclease T1 and its complex with 2’GMP studied by two-dimensional heteronuclear NMR spectroscopy, J Biomol NMR 4 (1994) 61–78. [DOI] [PubMed] [Google Scholar]
- [44].Berjanskii MV, Wishart DS, A simple method to predict protein flexibility using secondary chemical shifts, J. Am. Chem. Soc 127 (2005) 14970–14971. [DOI] [PubMed] [Google Scholar]
- [45].Kerry PS, Long E, Taylor MA, et al. , Conservation of a crystallographic interface suggests a role for β-sheet augmentation in influenza virus NS1 multifunctionality, Acta Crystallogr F F67 (2011) 858–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Lopes AM, Domingues P, Zell R, et al. , Structure-Guided Functional Annotation of the Influenza A Virus NS1 Protein Reveals Dynamic Evolution of the p85β-Binding Site during Circulation in Humans, J Virol 91 (2017) e01081–01017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Frederick KK, Marlow MS, Valentine KG, et al. , Conformational entropy in molecular recognition by proteins, Nature 448 (2007) 325–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Tzeng SR, Kalodimos CG, Protein activity regulation by conformational entropy, Nature 488 (2012) 236–240. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.