Abstract
Although accumulating evidence has demonstrated the key roles of enhancers in gene expression regulation, the contribution of genome-wide enhancer-enhancer interactions to developmental decisions remains unclear. Here we explored the cooperative regulation patterns among enhancers to understand their regulatory mechanism. We first filtered robust enhancers in embryonic stem cells (ESCs) through integrating bidirectional transcription, genomic location, and epigenetic modification. Genome-wide enhancer-enhancer interactions were then identified based on enhancer-promoter relationships that were derived from Hi-C data. We further explored the interacting principles of the identified enhancer-enhancer interactions. The results revealed that the observed cooperativity occurred mainly between enhancers distributed within a 1-kb to 10-Mb distance across the genome. In addition, enhancer-enhancer pairs had higher expression correlations than non-interacting pairs. Finally, we identified robust enhancers during human cardiac commitment, and we found that enhancers exhibited strong stage-specific expression patterns. We further inferred the enhancer-enhancer interactions based on RNA sequencing (RNA-seq) data in heart development, according to the regulatory principles characterized from Hi-C data. The identified enhancer-enhancer interaction networks (EEINs) presented highly dynamic linkages. Moreover, enhancers cooperatively targeted many marker genes in each developmental stage to regulate stage-specific functions, which contribute to the organization of cell identity in heart development. Our work will increase the understanding of enhancer regulation in human heart development.
Keywords: enhancer, eRNA, histone modification, cooperative regulation, heart development, Hi-C, RNA-seq
Introduction
Precise temporal regulation of gene expression patterns is essential for heart development, and disruption of transcriptional networks in heart development may lead to congenital heart disease.1, 2, 3 Studies have investigated the dynamic changes in the transcriptome during different stages of cardiac differentiation.4, 5 To better understand development-related changes in gene expression, it is important to consider changes in mechanisms that regulate gene expression, such as enhancer regulation.
We have learned that the genome is folded in three-dimensional (3D) space inside the cell nucleus, thereby allowing long-range chromatin interactions and exhibiting more complex gene regulation.6, 7, 8 Specially, distal enhancers physically loop to their target gene promoters and play an important role in the transcriptional control of genes. Enhancers can be separated from promoters over extensive and highly variable genomic distances.9 Wamstad et al.5 previously reported that enhancers play a critical role in the temporal and cell type-specific activation of gene expression and stage-specific distal enhancer elements orchestrate cardiac differentiation. However, the contribution of genome-wide enhancer-anchored interactions to human developmental decisions remains poorly understood.
A very recent study discovered that enhancer-promoter contacts are highly dynamic and cell type specific during mouse neural differentiation,10 indicating that the 3D genome was globally reorganized during development. In addition, studies showed that several enhancers were predicted to converge on the regulation of a single gene.11, 12, 13 Beyond this, local and global chromosome conformation capture analysis demonstrated that enhancers associate not just in linear sequence in the form of super-enhancers but also in 3D space and these enhancer-enhancer interactions contribute to the regulation of gene expression.14 Although individual enhancer-enhancer interactions already have been uncovered,11, 12, 13, 14, 15 genome-wide analysis of cooperations among enhancers has not yet been conducted. Moreover, the role of enhancer-enhancer cooperative regulations during cardiac differentiation remains unclear.
On the other hand, linking enhancers to their target genes remains a major challenge. It is a commonly used method to identify enhancer regulations based on the correlation between chromatin signals at enhancers and transcriptional activity of genes. For example, Shen et al.13 predicted enhancer-promoter pairs based on the correlation of the chromatin state at enhancer and the RNA polymerase II (Pol II) intensity at promoter for each possible pair of elements along a chromosome. Moreover, Lin et al.12 identified putative enhancer-gene interactions that are contained within the same topologically associated domain and exhibit significant positive correlations between enhancer H3K27ac signal and gene expression.
Bidirectional transcription is a marker of active enhancers,16, 17 and the induced bidirectional enhancer RNA (eRNA) as functional transcript is required for enhancer-dependent target gene activation events.18 As the abundance of eRNA is strongly related to enhancer activity, identifying enhancer regulations based on expression correlation is a relatively effective approach in this research area. The functional annotation of the mammalian genome 5 (FANTOM5) project cap analysis of gene expression (CAGE) atlas provides enhancer expression across the majority of human cell types and tissues. Active enhancers were de novo identified based on the bidirectional transcription signature from CAGE data.17 Yao et al.19 identified robust enhancer regions expressed in the human brain and constructed eRNA-gene coexpression networks based on FANTOM5 data to identify brain region-specific or developmental stage-specific coexpression modules. Moreover, Andersson and colleagues17 associated enhancer-promoter pairs based on pairwise expression correlation, and they found that transcription is a better predictor of regulatory targets than chromatin accessibility.
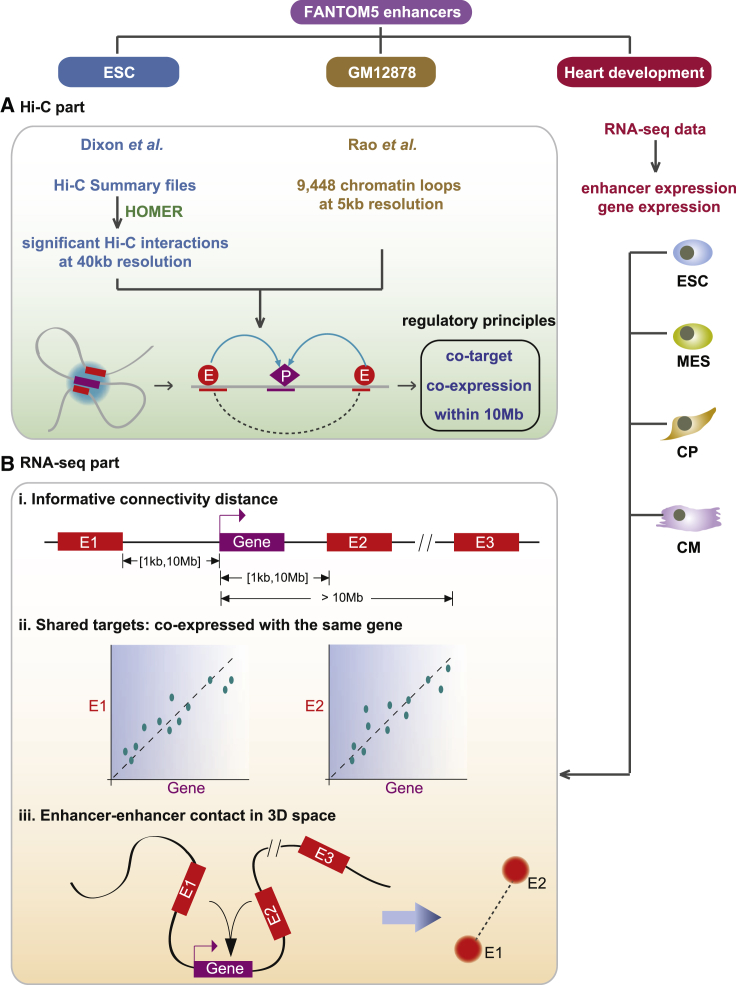
Motivated by the aforementioned observations, we aimed to identify genome-wide cooperative interactions among enhancers based on enhancer-target relationships. The workflow mainly included two modules (Figure 1). First, we filtered robust enhancers in embryonic stem cell through integrating multi-omics information. We subsequently identified genome-wide enhancer-enhancer interactions based on enhancer-promoter associations that were derived from Hi-C data, and we further explored the cooperative interacting principles of the interactions, which were confirmed by Hi-C data with high resolution of GM12878. Second, based on the identified robust enhancers in the cardiac commitment system, a sequential differentiation system including human embryonic stem cells (ESCs), mesoderm (MES), cardiac precursors (CPs), and cardiomyocytes (CMs), we predicted enhancer-promoter regulatory associations and enhancer-enhancer cooperative interactions from RNA sequencing (RNA-seq) data, according to the interacting principles characterized from Hi-C data. The assembled enhancer-enhancer interaction networks (EEINs) showed dynamic linkages during cardiac development. This work increases the understanding of the effect of non-coding regulatory elements on human heart development, and it highlights novel mechanisms underlying enhancer regulation.
Figure 1.
Workflow for Inferring Enhancer-Enhancer Interactions
(A) Genome-wide enhancer-enhancer interactions were identified based on the enhancer-promoter relationships that were derived from Hi-C data. (B) Enhancer-promoter and enhancer-enhancer interactions were predicted in each developmental stage based on the RNA-seq data according to the interacting principles from Hi-C data.
Results
Identifying Robust Enhancers in ESCs
Currently, genomic locations of enhancers can be detected by the mapping of chromatin marks20 or based on bidirectional transcriptional activity.17 As transcription is a marker of active enhancers, we first identified candidate active enhancers in ESCs based on the FANTOM5 CAGE atlas, which encompassed ∼1,000 human primary cell, tissue, and cell line samples. More than 2,700 enhancers were expressed in at least one sample above 0.5 TPM (tags per million) (Figure 2A). As the bidirectional expression of enhancers in the close vicinity of transcriptional start sites (TSSs) may be difficult to distinguish from expression at promoter regions,19 we limited our analyses to intronic and intergenic enhancers. Among these, 92.4% were intergenic or intronic, consistent with previous studies.19, 21
Figure 2.
Enhancer-Enhancer Interactions Identified from Hi-C Data of ESCs
(A) The pipeline for robust enhancer identification of ESCs. (B) Bar graph detailing the number of enhancers assigned to N promoters. (C) Bar graph detailing the number of genes assigned to N enhancers. (D) Schematic diagrams indicating how enhancer-enhancer interactions can be extracted from enhancer-promoter associations. (E) Hi-C interacting enhancer-promoter pairs and (F) Hi-C interacting enhancer-enhancer pairs show significant expression correlation compared with random interacting pairs. The density plot is for 1,000 times of random procedures, and the arrow points to the average correlation value for true pairs. Both Pearson (blue) and Spearman (green) methods were used. (G) Distance distribution of the identified enhancer-promoter and enhancer-enhancer pairs. (H) Tracks showing multi-omics signals around the enhancer-enhancer interacting region. RNA-seq track is shown in blue, ChIP-seq H3K4me1 profiles are shown in green, and H3K27ac is in purple. Magenta boxes at the bottom correspond to candidate enhancer regions. The line connecting them indicates enhancer-enhancer interaction. The visualization was presented by WashU EpiGenome Browser.
Given that enhancers tend to be expressed at lower levels22 and that the FANTOM5 sample size is relatively small, we therefore replicated enhancer expression from independent total RNA-seq datasets from GEO, which included 25 ESC samples. The expression of 1,467 enhancer loci was replicated in the independent dataset, with the detection threshold of 0.5 TPM in at least two distinct samples. In addition, recent studies have shown that chromatin profiling provides a powerful means for the characterization of enhancer portions of the genome.14, 20, 23 Therefore, in order to further refine our confident enhancer list, we used histone modification data from the Roadmap Epigenomics Project24 to assess the overlap of enhancer-specific chromatin marks (H3K4me1 or H3K27ac) with the 1,467 enhancer regions. Ultimately, a total of 973 robust enhancers overlapped with Roadmap H3K27ac or H3K4me1 peaks of ESCs were obtained for subsequent analysis (Figure 2A).
Characterizing Interacting Principles of Enhancer-Enhancer Interactions from Hi-C Data
The relevance of 3D physical interactions within chromosomes for transcriptional regulation and, thereby, for cellular fate at large is now widely accepted.25 More and more studies have demonstrated that the relationship between eRNA and mRNA is associated with physical contact between enhancers and promoters.16, 19, 25 Moreover, recent study has already uncovered individual enhancer-enhancer interactions, implying the cooperative regulations between enhancers. Specifically, Hi-C technology26 renders whole-genome contact maps by offering the advantage of interrogating all-to-all interactions, including relatively long-range enhancer-enhancer and enhancer-promoter interactions, providing opportunities to understand the structural and functional genome organization.
Identification of Enhancer-Enhancer Interactions
Here, to identify genome-wide enhancer-enhancer interactions and further reveal the underlying regulatory principles of the interactions, we first used 3D chromatin interaction data assayed by Hi-C technology in ESCs to identify enhancer-promoter associations. As a result, a total of 749 Hi-C-defined enhancer-promoter interactions were identified. Among them, we observed that 49.7% of enhancers were assigned to only a single gene promoter. On average, an enhancer was associated with 2.12 gene promoters (Figure 2B), confirmed by previous studies.12, 17, 27 Moreover, 17.1% of promoters were associated with 2 or more enhancers (Figure 2C), indicating that multiple enhancers may work in concert to regulate the expression of a gene.17 We assumed that two enhancers would loop to a same protein-coding gene simultaneously if both of them interact with the gene in 3D space, implying that these two enhancers would, therefore, have contact with each other. Based on this hypothesis, we finally identified 92 enhancer-enhancer interactions from enhancer-promoter relationships in ESCs (Figures 1 and 2D).
Higher Expression Correlation between Enhancer-Enhancer Interacting Pairs
Although genome-wide chromatin interactions have been mapped, it remains to be determined whether the interactions are bona fide regulatory interactions to be predictive of expression levels. To this end, we explored the expression correlations of the identified enhancer-promoter and enhancer-enhancer interacting pairs. Starting from the premise that enhancers should be coexpressed with their target genes,19 we indeed found that enhancer-promoter interacting pairs show significant coexpression compared with randomly selected pairs (p for Pearson = 0, p for Spearman = 0.001, permutation test; Figure 2E).
To further analyze the association between enhancer regulation and gene expression, we divided the ESC samples into three groups according to enhancer activity. As a result, we found that the expression of the target gene was higher in samples where the two regulating enhancers were both active (i.e., TPM > 0.5) than in samples where only one regulating enhancer was active. Moreover, the target gene expression was the lowest in samples where the two regulating enhancers were both inactive (Figures S1A–S1F). These results indicated that the expression level of target genes is correlated with the number of regulating enhancers. We also observed that the expression levels between enhancer-enhancer interacting pairs were more highly correlated (p for Pearson = 0, p for Spearman = 0, permutation test; Figure 2F), indicating that the two enhancers targeting the same protein-coding gene were both in active state and transcribed eRNAs simultaneously.
Genomic Distance Distribution of Enhancer-Enhancer Interactions
We next analyzed the genomic distance distribution of enhancer-anchored interactions, and we found that the vast majority of the interactions were located within 10 Mb (Figure 2G), consistent with previous study that 3D interactions with informative connectivity information occur within a 1-kb to 10-Mb distance.28 This analysis indicated that cooperativity occurred mainly between enhancers distributed within a 1-kb to 10-Mb distance across the genome. As an example, one of our identified enhancer-enhancer interactions is shown in Figure 2H. Both en_chr6:14211644-14212034 and en_chr6:14381333-14381628 are intronic enhancers and they interact with each other in 3D space. Their genomic distance is 169,299 bp. We could observe RNA-seq reads and histone modification signals over the enhancer regions (Figure 2H; Figure S1G).
Robustness of the Inferred Enhancer Interacting Principles
Additionally, considering that Hi-C data with high resolution could give more exact characterization of the distance distribution of enhancer cooperativity, we therefore analyzed a total of 9,448 chromatin loops at 5-kb resolution in GM1287829 to identify enhancer-promoter relationships. This resulted in the identification of 320 interactions involving 15,803 promoters and 3,123 intergenic or intronic enhancers (Figure 3A). Likewise, 77.2% of enhancers were assigned to only a single gene target. On average, 1.33 gene promoters were connected to a same enhancer in GM12878 (Figure 3B).
Figure 3.
Enhancer-Enhancer Interactions Identified from Hi-C Data of GM12878
(A) Distribution of the identified enhancers across the genome. (B) Bar graph detailing the number of enhancers assigned to N promoters. (C) Bar graph detailing the number of genes assigned to N enhancers. (D) Distance distribution of the identified enhancer-promoter and enhancer-enhancer pairs.
Compared with enhancer-promoter interactions detected in ESCs, we found that 34.7% of promoters were regulated by multiple enhancers (Figure 3C), indicating the more globally cooperative regulatory pattern of enhancers. Importantly, only 2.19% of the enhancer-promoter interactions were located beyond 10 Mb across the genome (Figure 3D). Moreover, all inferred enhancer-enhancer interactions were located within a 10-Mb distance, suggesting that the enhancer-enhancer cooperative interaction occurs within informative connectivity distance. In addition, a recent study showed that enhancers can be separated from promoters by distances ranging from a few kilobases to a little over 1 Mb.30 Actually, in our analysis, we found that 63.2% and 83.8% of the enhancer-promoter interactions identified from Dixon et al.31 and Rao et al.,29 respectively, were distributed within 1 kb to 1 Mb across the genome, consistent with the previous study.
Here, we evaluated the effect of the local sequence region of each promoter on the identification of enhancer-promoter interactions. We selected regions surrounding ±1 kb of TSSs as promoters12 to identify enhancer-promoter associations from Hi-C data. The results revealed that the identified enhancer-promoter associations were reproducible between different promoter definitions (Table S4).
Collectively, these regulatory principles of enhancer-enhancer interactions elucidated that the inferred relationship between enhancer and enhancer is associated with physical contact between them.
Stage-Specific Expression Pattern of Enhancers during Cardiac Differentiation
As enhancer activity exhibits a correlation with heart-specific programs,5 we thus identified robust enhancers integrating FANTOM5 data, RNA-seq data, and histone modification data to systematically elucidate the role of enhancers during cardiac commitment.
More than 14,000 enhancers were expressed in at least two distinct samples above 0.5 TPM. Among these, 94.53% were intergenic or intronic (Figure 4A). It has been shown that enhancers are significantly cell type specific, with a majority being specific to a single cell type.20 Here we also observed that these intergenic or intronic enhancers showed cell type-specific expression (top 500 enhancers with most variable expression, Figure 4B), demonstrating the temporally related expression of enhancers.
Figure 4.
Identification and Characterization of Cardiac Developmental Enhancers
(A) The detailed process for identifying robust enhancers in cardiac development. (B) Heatmap of the top 500 intronic and intergenic enhancers with most variable expression. Color represents the FANTOM5 expression level and the cell type is specified in the top bar. (C) Heatmap of the top 500 robust enhancers with most variable expression. Color represents the RNA-seq expression level and the cell type is specified in the top bar.
Given that enhancers tend to be expressed at lower levels (Figure S2A), we thus replicated enhancer expression from an independent total RNA-seq dataset, which included 76 samples consisting of four key differentiation stages from ESC to CM. The expression of 8,887 enhancer loci was replicated in our independent dataset, with the same detection threshold of 0.5 TPM in at least two distinct samples. Enhancer expression at a threshold of ≥10 uniquely mapped reads was also measured.32 A total of 6,516 enhancers were confirmed, the vast majority of which belong to the enhancer set filtered by TPM-normalized expression (Figure S2B).
In addition, we used histone modification data to assess the overlap of enhancer-specific chromatin marks with the 8,887 enhancer regions. Ultimately, a total of 7,195 robust enhancers overlapped with Roadmap H3K27ac or H3K4me1 peaks of ESCs, and heart tissues were obtained for subsequent analysis (Figure 4A). These enhancers showed stage-specific RNA-seq expression patterns (top 500 enhancers with most variable expression, Figure 4C), similar to protein-coding genes (15,803 protein-coding genes were filtered with the detection threshold of 1 FPKM [fragments per kilobase of transcript per million fragments mapped] in at least one sample; Figure S2C), thereby indicating that enhancers play important roles in heart development. Specifically, we collected the marker genes in each stage and discovered that marker genes were specifically expressed at corresponding stages (Figure S2D).
We also selected the top 25% of enhancers or protein-coding genes with the most variable expression to analyze the dynamic expression pattern. As a result, we also observed the stage-specific expression pattern of enhancers and genes during cardiac differentiation (Figure S3).
Enhancer-Enhancer Interaction Landscape during Cardiac Differentiation
Previous study has shown that transcription is a better predictor of enhancer targets than chromatin accessibility.17 Therefore, based on the above interacting features characterized from Hi-C data that enhancer-promoter pairs show significant coexpression and tend to distribute within 10 Mb, we further predicted genome-wide enhancer-promoter associations in heart development based on an expression correlation with the requirement of informative connectivity distance. Specifically, enhancers were connected to genes within 1 kb to 10 Mb28, 30 that exhibited concordant expression changes to identify specific enhancer-promoter regulations in each developmental stage (Figure 1).
Stage-Specific Enhancer-Enhancer Interactions during Cardiac Differentiation
Therefore, the EEIN was constructed in each developmental stage and visualized in a circular ideogram layout using the Circos tool33 (Figures 5A and 5B). These enhancer-enhancer interactions distributed across all chromosomes (Figure S4A). There were a hugely varying number of interactions identified, with ESCs having the most (27,378 interactions in ESCs, 1,497 in MES, 393 in CPs, and 746 in CMs; Figure 6A). This implies that ESCs require more complex enhancer coregulations to initiate development and the enhancer regulations become more specific with maturation of the human heart. Moreover, the Spearman’s rank correlation between the number of regulating enhancers and the expression level of the gene was 0.93 in ESCs (p < 3.14e−06), which was calculated on the average value. In addition, we calculated the associations between gene expression and the number of regulating enhancers based on the individual data points, and we observed that gene expression was also significantly correlated with the number of enhancers in ESCs (Spearman’s rank correlation rho = 0.28, p < 2.2e−16; Figure S4B).
Figure 5.
Enhancer-Enhancer Interactions during Cardiac Development
(A) Circos plot of the identified genome-wide intra-chromosomal enhancer-enhancer interactions in each developmental stage. The outermost layer ring labeled different colors represents different chromosomes of the human genome. Each link of the four layers inside denotes one enhancer-enhancer interaction of each developmental stage. The gray links represent background interactions, i.e., the union of interactions of all four stages except the interactions of the current stage. The bar plot in the inner area of the Circos plot is showing the total number of enhancer-enhancer interactions in each stage and the percentage of specific interactions of each developmental stage. (B) The representative chromosome 1 is enlarged to see more clearly.
Figure 6.
Cell Type-Specific Enhancer-Enhancer Interactions Contribute to the Organization of Cell Identity in Heart Development
(A) Number of enhancer-enhancer interactions in multiple developmental stages. For each developmental stage, the whole bar represents the union of interactions of all four stages, and the number labeled in the bar refers to the interaction number occurring in the current stage. (B) The overlapped interactions between each two stages. (C) Interaction between en_chr19:1213864-1214370 and en_chr19:7482509-7482624 is one of the ESC-specific enhancer-enhancer interactions. Tracks show RNA-seq signals around the enhancer-enhancer interacting region across the four developmental stages. Magenta boxes at the bottom correspond to candidate enhancer regions. The line connecting them indicates enhancer-enhancer interaction. The visualization was presented by WashU EpiGenome Browser. (D) Enhancer-enhancer pairs from ESCs cooperatively regulate pluripotency genes NANOG, SOX2, and POU5F1, which can also bind and regulate downstream enhancers to form the core pluripotency circuitry. (E–H) Enriched Gene Ontology (GO) terms for the target genes associated with the enhancers from networks identified in ESCs (E), MESs (F), CPs (G), and CMs (H), respectively.
The Circos plots of these intra-chromosomal interactions showed that interactions tended to occur within a relatively short distance, which was confirmed by a previous study.34 Our analysis also revealed that there are distinct enhancer-enhancer linkages across the four stages. The shared interactions between each two stages were very small (Figure 6B) and the vast majority of the interactions were stage specific (Tables S5 and S6). As an example, en_chr19:1213864-1214370 interacts with en_chr19:7482509-7482624 only in ESCs (Figure 6C). All these results indicate that these significant stage-specific enhancer-enhancer interactions might contribute to stage-specific enhancer-enhancer cooperative regulations.
Stage-Specific Function Regulation by Enhancer-Enhancer Interactions
Importantly, marker genes of the four stages of differentiation are frequently regulated by enhancers participating in their corresponding interaction network. Enhancers of undifferentiated ESCs cooperatively regulate pluripotency genes, such as POU5F1/OCT4, NANOG, and SOX2 (Table 1; Figure 6D), which are pluripotent master transcription factors that can bind enhancer elements and recruit Mediator to activate much of the gene expression program of pluripotent ESCs.35, 36, 37, 38 Indeed, we found that enhancers collaborate to form core transcriptional regulatory circuitry consisting of autoregulatory and feedforward loops (Figure 6D),39, 40, 41 suggesting the important roles of enhancer interactions in mediating the maintenance of ESC pluripotency. In addition, mesodermal markers TWIST1 and EOMES, cardiac precursor marker MEF2C, and CM-specific gene MYH11 are cooperatively regulated by enhancers from the corresponding developmental stage. These imply that the stage-specific expression of marker genes can be due to the stage-specific regulations of enhancer-enhancer interactions.
Table 1.
Marker Genes Cooperatively Regulated by Enhancer-Enhancer Pairs of Each Developmental Stage
| Developmental Stage | ESC | MES | CP | CM |
|---|---|---|---|---|
| Marker genes | CD9, DNMT3B, NANOG, PODXL, DPPA2, SOX2, POU5F1, LEFTY1, GAL, TCL1A, DPPA4, GRB7, ZFP42, GDF3, TDGF1 | TWIST1, EOMES, T | MEF2C | MYH11 |
Finally, functional analysis of enhancer target genes was further performed and revealed that the enriched categories become CM specific progressively (Figures 6E–6H). Each stage enriched the expected functional categories, such as developmental growth, positive regulation of cell cycle, and embryo development for ESCs (Figure 6E); stem cell differentiation and positive regulation of cell growth involved in cardiac muscle cell development for MES (Figure 6F); primary heart field specification, secondary heart field specification, and ventricular cardiac muscle tissue morphogenesis for CPs (Figure 6G); and cardiac conduction, heart contraction, and regulation of heart contraction for CMs (Figure 6H). Various general functions related to development and metabolism were also identified, such as regulation of signal transduction, cell proliferation, cell cycle, and negative regulation of Wnt-signaling pathway.
Together, these findings formed a landscape to understand developmental enhancer-enhancer cooperative regulations during lineage commitment, and they increased our understanding of spatial principles underlying the chromosomal organization and the effect of enhancer regulation on human heart development.
Discussion
Enhancers are a new class of gene-regulatory elements controlling gene expression and cell identity. Gene regulation by cell type-specific enhancers is fundamental to the organization of cell identity in development.42 In the present study, we first identified robust enhancers integrating widely used features of enhancers, including bidirectional transcription, genomic location, and epigenetic modification. Furthermore, we revealed several regulatory principles of enhancer-enhancer interactions based on Hi-C data to further predict genome-wide enhancer-enhancer interactions in different developmental stages, based on generally used RNA-seq data. We finally found that enhancer-enhancer linkages were dramatically dynamic during human cardiac commitment. On the other hand, to measure nonlinearly direct dependencies between variables,43 we have applied the Spearman’s rank correlation to further measure the enhancer-promoter regulatory associations during heart development. At the same coexpression threshold, the results revealed that 54.15%, 81.24%, 80.68%, and 83.66% of the enhancer-promoter relationships based on Spearman correlation in ESCs, MES, CPs, and CMs, respectively, were captured by Pearson correlation.
Enhancers exhibited strong stage-specific expression patterns, indicating that enhancers are essential for precise heart development. In addition to the stage-specific expression of enhancers, the protein-coding genes also exhibited strong stage-specific expression patterns during cardiac differentiation. However, the CTCF protein (CCCTC-binding factor) was ubiquitously expressed to form loops between binding sites flanking the globin locus.44 As a looping partner of CTCF,45, 46, 47 cohesin protein complex is also stage invariant, consistent with previous studies that the domain boundaries are largely invariant between cell types.31
In this study, we just focused on intra-chromosomal Hi-C interactions and ignored inter-chromosomal interactions. In fact, Lan et al.34 demonstrated that inter-chromosomal interactions represent only 5% of all interactions. Moreover in our analysis, we first explored the regulatory principles of the cooperative interactions based on Dixon’s Hi-C data of ESCs, which provides a picture of genomic architecture with a resolution of 40 kb. Therefore, the analysis of chromatin loops at 5-kb resolution of GM12878 in our study is a good complement to identify more comprehensive interactions to analyze the interacting principles.
It was recently found that the Linc1405/Eomes complex can promote cardiac mesoderm specification and further cardiogenesis, highlighting the critical long intergenic noncoding RNA (lincRNA)-mediated mechanism in the regulation of cardiogenesis in vitro and in vivo.48 In addition to lincRNA, our work illustrated that enhancer regulation is an additional layer of epigenetic regulation of cardiogenesis, which contributes to a better understanding of the cardiac regulation. Another recent study found that cardiac reprogramming factors synergistically activate enhancers highlighted by Mef2c-binding sites and that Hand2 and Akt1 coordinately recruit other reprogramming transcription factors (TFs) to enhancer elements.49 The scenario looks very intriguing when combining our enhancer-enhancer cooperative regulation network. It forms a complicated cardiac reprogramming regulatory network centered at MEF2C involving complex crosstalk between TFs and enhancers, where TFs synergistically activate enhancers and enhancers in turn synergistically activate target genes.
In summary, we identified the dynamic enhancer-enhancer interactions during cardiac commitment based on the generally used high-throughput dataset with relatively low cost by integrating the FANTOM5 data and RNA-seq data. We demonstrated that cell type-specific enhancer-enhancer interactions contribute to the organization of cell identity in heart development. Our work highlights the power of systems biology approaches to represent a complement for the current Hi-C technique on the characterization of coordination among enhancers and delineate the enhancer-enhancer interaction landscape for heart development. There are both opportunities and challenges for developing more computational models to identify enhancer-promoter and enhancer-enhancer interactions in future studies.
Materials and Methods
FANTOM5 Enhancers
Both the genome coordinates and TPM-normalized CAGE expression data of human enhancers during cardiac commitment (Table S1) were downloaded from the FANTOM5 data portal.
RNA-Seq Data for Heart Development
Publicly available heart development-related datasets (GEO: GSE64417 and GSE54969) were downloaded and profiled by high-throughput transcriptome sequencing (Table S2). The Sequence Read Archive (SRA) files were converted to FASTQ format using fastq-dump–split-3 for paired-end reads. Then reads were aligned to the human reference genome GRCh38/hg38 (University of California, Santa Cruz [UCSC] gene transfer format [GTF] annotation) by using TopHat50 version (v.)2.1.0 to generate the Binary Alignment Map (BAM) files.
To compute the expression level of an enhancer for each sample, we calculated the count coverage within the enhancer region using the BEDTools software51 based on the mapped BAM file, and we measured the expression value as the total number of RNA-seq reads overlapping the enhancer region, normalized to library size, i.e., TPM. On the other hand, the gene FPKM values were called by using Cufflinks52 software 2.2.1 release with default parameters.
Histone Modification Data from Roadmap
H3K27ac and H3K4me1 peaks for ESCs and heart tissues were obtained from https://egg2.wustl.edu/roadmap/web_portal/processed_data.html (Table S3). We downloaded the narrowPeaks and converted their genomic locations to the hg38 build.
3D Chromatin Interaction Data
Calling of Interactions from Hi-C Data of ESCs
The mapped Hi-C summary files of the two replicates of H1 cell line based on hg18 were downloaded from Dixon et al.31 (GEO: GSE35156). Hypergeometric Optimization of Motif Enrichment (HOMER)53 was subsequently used to call significant Hi-C interactions from the mapped reads (Figure 1A). It mainly involves two analysis steps. First we used the makeTagDirectory program to process paired-end sequencing into a tag directory. Then the analyzeHiC command with the option “-interactions” was used to search for significant interactions in the genome at 40-kb resolution. We combined the results from the two replicates for further analysis.
Chromatin Loops of GM12878
9,448 chromatin loops detected in the in situ Hi-C map for GM12878 at 5-kb matrix resolution were downloaded from Rao et al.29 and converted to the hg38 build (Figure 1A).
Identification of Robust Enhancers in ESCs
Robust active enhancers in ESCs were identified based on three steps of filtration, mainly integrating bidirectional transcription, genomic location, and epigenetic modification of enhancers. As transcription is a marker of active enhancers,17 we first identified candidate active enhancers expressed above 0.5 TPM in at least one sample, based on the FANTOM5 CAGE atlas and at least two samples in the replicated RNA-seq datasets, respectively. Therefore, enhancers analyzed in our study were all representative of active enhancers. Furthermore, enhancers were annotated based on GENCODE release 24 to limit those either intergenic (located more than 2 kb away from a GENCODE gene) or overlapped with introns, but not overlapped with promoter regions (TSS ± 2 kb) or exon regions. Finally, robust enhancers were identified if enhancer regions were marked by epigenetic modification signals (H3K27ac or H3K4me1 peaks).
Identification of Robust Enhancers during Human Cardiac Commitment
On the other hand, we identified robust enhancers during human cardiac commitment based on the similar workflow. Considering the larger sample size here, the expression values of enhancer candidates must be above 0.5 TPM in at least two samples in both FANTOM5 and the replicated RNA-seq datasets, respectively.
Identification of Enhancer-Enhancer Interactions Based on Hi-C Data
Based on the significant intra-chromosomal interactions detected by HOMER from Hi-C data, the enhancer-promoter interactions were detected only if they were separately overlapped with two different bins that significantly contacted each other in 3D space. The overlaps between enhancers-promoters and bins were searched by using the BEDTools software. Enhancer-enhancer interactions were identified if two enhancers both interacted with a same protein-coding gene in 3D space (Figure 1A).
Construction of the EEIN Based on RNA-Seq Data in Each Differentiation Stage
The construction of the EEIN in each developmental stage of cardiac commitment was implemented as the following two steps (Figure 1B):
Identifying Enhancer-Promoter Regulatory Associations
All genes within 1-kb to 10-Mb distance of an enhancer were selected as candidate targets, and the Pearson correlation coefficient between each candidate target gene and the enhancer was further computed across RNA-seq samples of each differentiation stage. If the coexpression correlation was at a false discovery rate (FDR) value less than 0.05, we considered the candidate target gene as the confident target.
Constructing the EEIN
On the basis of enhancer-promoter regulatory relationships, we constructed an EEIN for each developmental stage. Two enhancers were defined as association only if they shared at least one target gene. At last, all enhancer-enhancer interactions were assembled into the EEIN, where nodes correspond to enhancers, and two enhancers were connected to each other if they had an association.
Hierarchical Clustering Analysis
Hierarchical clustering analysis was applied to expression of the top 500 enhancers or genes with the most expression variations across all samples, which was realized by the R package pheatmap. The dendrogram was constructed based on the average distance algorithm.
Identification of TF-Enhancer Regulations in ESCs
To identify TF-enhancer regulations, we first downloaded the enhancer sequences in FASTA format from the UCSC table browser in the hg38 build. Then, we searched potential binding sites for transcription factors using the Match tool that is integrated in TRANSFAC Professional (release 2013.6). The pre-calculated minFP cutoff was applied for searching the enhancer sequences, and a high-quality matrix was created. TFs belonging only to the human genome were retained. We required that there must exist binding sites of TFs in enhancers and that the TF-enhancer pairs should be coexpressed at the FDR value less than 0.05 in RNA-seq samples of ESCs.
Network Visualization
Network visualization was carried out using Cytoscape 3.3.0.54
The Functional Annotation of Enhancer Targets
The gene2go table was downloaded from the NCBI (2016/7/12), and we extracted human-related biological process (BP) ontology. A cumulative hypergeometric distribution test was used to identify the significantly overrepresented biological function categories for enhancer targeted protein-coding genes. All human genes were defined as a background gene set. Functional categories with the enriched p values < 0.05 were considered in our analyses.
Converting Genome Coordinates
All genomic coordinates were based on the human reference genome hg38. Genome locations originally provided in hg18 or hg19 were all converted to the hg38 build using the LiftOver tool in the UCSC genome browser.55
Randomization Test
To determine the significance of the coexpression of enhancer-promoter and enhancer-enhancer interacting pairs, we performed the randomization test by randomly selecting enhancer-promoter and enhancer-enhancer pairs from all possible pairs with the same number as the true pairs, respectively. We repeated the procedure 1,000 times, and the significance was defined as the proportion of times in which, in random conditions, the average correlation values were higher than the real average correlation.
Author Contributions
H.C., Hui Liu, and J. Xu designed the project. Hongyu Liu and X. Li supervised the project. H.C., J. Xiao, and T.S. collected all the data and designed the algorithm. H.C., J. Xiao, and J. Xu performed data analysis and wrote the manuscript. L.W., J.B., X.L., and N.D. reviewed the results and interpreted the enhancer regulation in the heart development. H.C. and J. Xiao were the major contributors in writing the manuscript. All authors read and approved the manuscript and its contents.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Key R&D Program of China (2018YFC2000100), the National Natural Science Foundation of China (31571331, 61873075, 31871338 and 31970646), the Weihan Yu Youth Science Fund Project of Harbin Medical University, the University Nursing Program for Young Scholars with Creative Talents in Heilongjiang Province (UNPYSCT-2016189, UNPYSCT-2017060), the Natural Science Foundation for Distinguished Young Scholars of Heilongjiang Province (JQ2019C004), Postdoctoral Scientific Research Developmental Fund of Heilongjiang Province (LBH-Q17110) and the Health and Family Planning Commission Scientific Research Subject of Heilongjiang Province (2017-168).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.07.015.
Contributor Information
Hui Liu, Email: liuhui870320@gmail.com.
Hongyu Liu, Email: hyliu1963@163.com.
Juan Xu, Email: xujuanbiocc@ems.hrbmu.edu.cn.
Xia Li, Email: lixia@hrbmu.edu.cn.
Supplemental Information
References
- 1.Bruneau B.G. The developmental genetics of congenital heart disease. Nature. 2008;451:943–948. doi: 10.1038/nature06801. [DOI] [PubMed] [Google Scholar]
- 2.Evans S.M., Yelon D., Conlon F.L., Kirby M.L. Myocardial lineage development. Circ. Res. 2010;107:1428–1444. doi: 10.1161/CIRCRESAHA.110.227405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Srivastava D. Making or breaking the heart: from lineage determination to morphogenesis. Cell. 2006;126:1037–1048. doi: 10.1016/j.cell.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Liu Q., Jiang C., Xu J., Zhao M.T., Van Bortle K., Cheng X., Wang G., Chang H.Y., Wu J.C., Snyder M.P. Genome-Wide Temporal Profiling of Transcriptome and Open Chromatin of Early Cardiomyocyte Differentiation Derived From hiPSCs and hESCs. Circ. Res. 2017;121:376–391. doi: 10.1161/CIRCRESAHA.116.310456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wamstad J.A., Alexander J.M., Truty R.M., Shrikumar A., Li F., Eilertson K.E., Ding H., Wylie J.N., Pico A.R., Capra J.A. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151:206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavalli G., Misteli T. Functional implications of genome topology. Nat. Struct. Mol. Biol. 2013;20:290–299. doi: 10.1038/nsmb.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dekker J., Rippe K., Dekker M., Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 8.Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 9.Jin F., Li Y., Dixon J.R., Selvaraj S., Ye Z., Lee A.Y., Yen C.A., Schmitt A.D., Espinoza C.A., Ren B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonev B., Mendelson Cohen N., Szabo Q., Fritsch L., Papadopoulos G.L., Lubling Y., Xu X., Lv X., Hugnot J.P., Tanay A., Cavalli G. Multiscale 3D Genome Rewiring during Mouse Neural Development. Cell. 2017;171:557–572.e24. doi: 10.1016/j.cell.2017.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chepelev I., Wei G., Wangsa D., Tang Q., Zhao K. Characterization of genome-wide enhancer-promoter interactions reveals co-expression of interacting genes and modes of higher order chromatin organization. Cell Res. 2012;22:490–503. doi: 10.1038/cr.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin C.Y., Erkek S., Tong Y., Yin L., Federation A.J., Zapatka M., Haldipur P., Kawauchi D., Risch T., Warnatz H.J. Active medulloblastoma enhancers reveal subgroup-specific cellular origins. Nature. 2016;530:57–62. doi: 10.1038/nature16546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen Y., Yue F., McCleary D.F., Ye Z., Edsall L., Kuan S., Wagner U., Dixon J., Lee L., Lobanenkov V.V., Ren B. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ing-Simmons E., Seitan V.C., Faure A.J., Flicek P., Carroll T., Dekker J., Fisher A.G., Lenhard B., Merkenschlager M. Spatial enhancer clustering and regulation of enhancer-proximal genes by cohesin. Genome Res. 2015;25:504–513. doi: 10.1101/gr.184986.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perry M.W., Boettiger A.N., Levine M. Multiple enhancers ensure precision of gap gene-expression patterns in the Drosophila embryo. Proc. Natl. Acad. Sci. USA. 2011;108:13570–13575. doi: 10.1073/pnas.1109873108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim T.K., Hemberg M., Gray J.M., Costa A.M., Bear D.M., Wu J., Harmin D.A., Laptewicz M., Barbara-Haley K., Kuersten S. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., Boyd M., Chen Y., Zhao X., Schmidl C., Suzuki T. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li W., Notani D., Ma Q., Tanasa B., Nunez E., Chen A.Y., Merkurjev D., Zhang J., Ohgi K., Song X. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao P., Lin P., Gokoolparsadh A., Assareh A., Thang M.W., Voineagu I. Coexpression networks identify brain region-specific enhancer RNAs in the human brain. Nat. Neurosci. 2015;18:1168–1174. doi: 10.1038/nn.4063. [DOI] [PubMed] [Google Scholar]
- 20.Ernst J., Kheradpour P., Mikkelsen T.S., Shoresh N., Ward L.D., Epstein C.B., Zhang X., Wang L., Issner R., Coyne M. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vermunt M.W., Reinink P., Korving J., de Bruijn E., Creyghton P.M., Basak O., Geeven G., Toonen P.W., Lansu N., Meunier C., Netherlands Brain Bank Large-scale identification of coregulated enhancer networks in the adult human brain. Cell Rep. 2014;9:767–779. doi: 10.1016/j.celrep.2014.09.023. [DOI] [PubMed] [Google Scholar]
- 22.Core L.J., Martins A.L., Danko C.G., Waters C.T., Siepel A., Lis J.T. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat. Genet. 2014;46:1311–1320. doi: 10.1038/ng.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farh K.K., Marson A., Zhu J., Kleinewietfeld M., Housley W.J., Beik S., Shoresh N., Whitton H., Ryan R.J., Shishkin A.A. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518:337–343. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu J., Adli M., Zou J.Y., Verstappen G., Coyne M., Zhang X., Durham T., Miri M., Deshpande V., De Jager P.L. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell. 2013;152:642–654. doi: 10.1016/j.cell.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Won H., de la Torre-Ubieta L., Stein J.L., Parikshak N.N., Huang J., Opland C.K., Gandal M.J., Sutton G.J., Hormozdiari F., Lu D. Chromosome conformation elucidates regulatory relationships in developing human brain. Nature. 2016;538:523–527. doi: 10.1038/nature19847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B.R., Sabo P.J., Dorschner M.O. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fukaya T., Lim B., Levine M. Enhancer Control of Transcriptional Bursting. Cell. 2016;166:358–368. doi: 10.1016/j.cell.2016.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sahlén P., Abdullayev I., Ramsköld D., Matskova L., Rilakovic N., Lötstedt B., Albert T.J., Lundeberg J., Sandberg R. Genome-wide mapping of promoter-anchored interactions with close to single-enhancer resolution. Genome Biol. 2015;16:156. doi: 10.1186/s13059-015-0727-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rao S.S., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S., Aiden E.L. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vernimmen D., Bickmore W.A. The Hierarchy of Transcriptional Activation: From Enhancer to Promoter. Trends Genet. 2015;31:696–708. doi: 10.1016/j.tig.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 31.Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu H., Nord A.S., Akiyama J.A., Shoukry M., Afzal V., Rubin E.M., Pennacchio L.A., Visel A. Tissue-specific RNA expression marks distant-acting developmental enhancers. PLoS Genet. 2014;10:e1004610. doi: 10.1371/journal.pgen.1004610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lan X., Witt H., Katsumura K., Ye Z., Wang Q., Bresnick E.H., Farnham P.J., Jin V.X. Integration of Hi-C and ChIP-seq data reveals distinct types of chromatin linkages. Nucleic Acids Res. 2012;40:7690–7704. doi: 10.1093/nar/gks501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ng H.H., Surani M.A. The transcriptional and signalling networks of pluripotency. Nat. Cell Biol. 2011;13:490–496. doi: 10.1038/ncb0511-490. [DOI] [PubMed] [Google Scholar]
- 36.Orkin S.H., Hochedlinger K. Chromatin connections to pluripotency and cellular reprogramming. Cell. 2011;145:835–850. doi: 10.1016/j.cell.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young R.A. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kagey M.H., Newman J.J., Bilodeau S., Zhan Y., Orlando D.A., van Berkum N.L., Ebmeier C.C., Goossens J., Rahl P.B., Levine S.S. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu Y., Loh Y.H., Li H., Cesana M., Ficarro S.B., Parikh J.R., Salomonis N., Toh C.X., Andreadis S.T., Luckey C.J. Alternative splicing of MBD2 supports self-renewal in human pluripotent stem cells. Cell Stem Cell. 2014;15:92–101. doi: 10.1016/j.stem.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buganim Y. Tex10: A New Player in the Core Pluripotency Circuitry. Cell Stem Cell. 2015;16:572–573. doi: 10.1016/j.stem.2015.04.019. [DOI] [PubMed] [Google Scholar]
- 42.Lee T.I., Young R.A. Transcriptional regulation and its misregulation in disease. Cell. 2013;152:1237–1251. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao J., Zhou Y., Zhang X., Chen L. Part mutual information for quantifying direct associations in networks. Proc. Natl. Acad. Sci. USA. 2016;113:5130–5135. doi: 10.1073/pnas.1522586113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Splinter E., Heath H., Kooren J., Palstra R.J., Klous P., Grosveld F., Galjart N., de Laat W. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parelho V., Hadjur S., Spivakov M., Leleu M., Sauer S., Gregson H.C., Jarmuz A., Canzonetta C., Webster Z., Nesterova T. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 46.Wendt K.S., Yoshida K., Itoh T., Bando M., Koch B., Schirghuber E., Tsutsumi S., Nagae G., Ishihara K., Mishiro T. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 47.Hadjur S., Williams L.M., Ryan N.K., Cobb B.S., Sexton T., Fraser P., Fisher A.G., Merkenschlager M. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo X., Xu Y., Wang Z., Wu Y., Chen J., Wang G., Lu C., Jia W., Xi J., Zhu S. A Linc1405/Eomes Complex Promotes Cardiac Mesoderm Specification and Cardiogenesis. Cell Stem Cell. 2018;22:893–908.e6. doi: 10.1016/j.stem.2018.04.013. [DOI] [PubMed] [Google Scholar]
- 49.Hashimoto H., Wang Z., Garry G.A., Malladi V.S., Botten G.A., Ye W., Zhou H., Osterwalder M., Dickel D.E., Visel A. Cardiac Reprogramming Factors Synergistically Activate Genome-wide Cardiogenic Stage-Specific Enhancers. Cell Stem Cell. 2019;25:69–86.e5. doi: 10.1016/j.stem.2019.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.