Abstract
Agrobacterium mediated genetic transformation has become an important tool in crops for molecular breeding. Am-SOD quality containing transgenic plants were created from embryogenic calli of Sambha mahsuri and cotton sannalu by Agrobacterium tumifaciens co-development. The superoxide desmutase quality was housed responsible for CaMV 355 promoter and Nos polyadenylation motion in double vector pCAMBIA 1301. Good change productivity was gotten. Mix of quality at genome level in the plants was exhibited by PCR examination and Southern smear, and furthermore delineated by a few physiological studies.
Keywords: Agrobacterium, SOD quality, β-Glucouronidase, PCR
1. Introduction
Oryza sativa is the staple nourishment in India with an aggregate generation of 548 million tons developed in 20.0 million ha arrive, (Deepa et al., 2011). More than one million hectare of rice land including 68% of the waterfront territories is salt influenced because of an unnatural weather change and late unfriendly environmental change (Ingram et al., 2001). In the natural elements soil saltiness is one of the central point affecting from seedling to reaping stage severe (Gupta and Bingru, 2014). In spite of the fact that some saltiness tolerant assortments are as of now grew yet these assortments are not reasonable to beat higher worries of saltiness in rice generation to battle environmental change. Close to traditional rearing, hereditary change assumes an imperative part under existing climatic worry to bring novel qualities into product, for resistance and better yield (Dong et al., 2001).
New rearing lines ought to produce for better adjusted assortments for saltiness inclined zone. With in rice species there is fantastic variety for resistance of salt which gives a chance to create salt tolerant assortments by hereditary change (Hiei et al., 1997). In this manner, it is most imperative to help the creation of rice to experience the issue of increment in populace which is relied upon to achieve 9.1 billion by 2050. A few strategies for conventional rearing have been attempted to conquer the issue however with little advance accomplished. In the rearing projects hereditary change can be a noticeable apparatus to manage the issues. It opens path to a massive quality pool which takes into consideration exchange of coveted qualities (Tyagi et al., 1999). In the previous decades the advancement of plant change methods has made conceivable to present cloned qualities and enhance the harvest plants. Exchange of remote DNA into the plant cell and recovery of plants from the changed cells are the two essential and basic strides in fruitful change of rice (Taylor and Fauquet, 2002).
In tissue culture of rice, callus acceptance and recovery of plants rely on upon number of elements like media supplementation, for example, basal salts, natural segments, development controllers, kind of the explants and the genotype utilized (Gao et al., 2007). Rice is known as the model framework in grain genomics, and since it is the most imperative product on the planet, consequently creating plants with biotic and abiotic stretch resilience by hereditary building is a test in rice biotechnology examine (Lafitte et al., 2004).
In rice hereditary designing is utilized as a notable device for development. In Japonica rice a few research gatherings are performing change examines routinely, yet indica rice is thought to be a hard and convoluted framework. The quantity of duplicates of qualities and chromosomal area of those qualities is not controllable yet, and furthermore among the transformants the statement of the presented qualities contrast. In this way to choose attractive transformants and to concentrate the expression, a generally huge number of transgenic plants must be produced. Agrobacterium based change and biolistic approach are the normally utilized instruments in rice hereditary building.
Addition of unrearranged portions of DNA in the beneficiary plant genome at low duplicate number is normally come about by agrobacterium-interceded change (Li et al., 2007). In addition, nearly vast DNA pieces with unmistakable closures (i.e. left and right T-DNA fringes) can be incorporated at high rate into beneficiary plant genome (Sarangi et al., 2011). The perfect technique for quality move into rice plants is Agrobacterium-interceded change, since this procedure has various focal points in examination with direct DNA take-up methods (Yu et al., 2007) and (Corrado and Karali, 2009). For effective hereditary change of rice a skillful and reproducible change convention is required (Zhang et al., 2012, Zhang et al., 2013).
Diverse anxieties like osmotic, temperature, overwhelming metals, and herbicides causes over generation of receptive oxygen species (ROS), bringing on cell harm. Plants have their own particular guard framework to conquer oxidative worry with a variety of qualities, similar to superoxide dismutase (SOD). They evacuate O2-in the cells when they are produced. The super oxide radicals O2-, hydrogen peroxide (H2O2) and hydroxyl radicals (OH-) are created in plants at the season of stress which causes harm to the cell layer and other vital particles like DNA, lipids, proteins and photosynthetic shades. Thus these ROS shaped because of abiotic stresses are should have been rummaged for insurance and typical development of the plants and furthermore profitability of the plants. With this foundation we have attempted to utilize Am-SOD quality which is as of late been appeared to oppose abundance salt. Oxidative anxiety is overseen by the quality through rummaging superoxides, peroxidase and singlet oxygen to give saltiness resistance in rice.
2. Materials and methods
2.1. Plant material
High yielding assortment Sambha mahsuri, and Cotton dora sannalu cultivar acquired from ARS (Agricultural Research Station), Warangal, were utilized in this review.
2.2. Bacterial strains and plasmids
In this review, pCAMBIA 1305.2 vector was utilized. The vector comprise of GUS quality is having better strength at 60 °C and within the sight of formaldehyde and glutaraldehyde fixatives. It likewise has solid and in situ cell particular quality identification movement. It is additionally having rice glycine rich protein flag peptide at Ncol and BgIII locales for additional cell discharge that gives quick in vivo GUS examines without utilizing UV recognition. Furthermore, pCAMBIA 1305.2 is a packed twofold vector with a pBR322 ori for high duplicate replication in Escherichia coli and a broad host go ori for low duplicate stable replication in Agrobacterium. It comprise of kanamycin safe quality for bacterial determination and hygromycin at XhoI site for choice of plants. An anxiety tolerant quality pSFSOD1 was utilized as a vector with Am-SOD as quality of intrigue and hygromycin as plant choice marker for improvement of transgenic plants.
2.3. Acceptance of callus
Chosen rice varietiy seeds were dehusked and washed utilizing 5% watery Teepol arrangement (fluid cleanser) by consistent shaking on a revolving shaker at 80 rpm for 10 min (Remi, Model-CIS 24). Later the seeds were washed with refined water and surface disinfected with 0.1% HgCl2 with two drops of Tween 20 as wetting specialist in a laminar wind stream (LAF), and after that the seeds were washed trice with sterile refined water. Surface sanitized seeds were smear dried on sterile tissue paper and immunized onto callus enlistment medium (MS6 + 2 mg L−1 2,4-D)4 and hatched at 25 ± 2 °C in dull The essential callus (10-d-old) were further subcultured on callus keeping up MS medium having 1 mg L−1 2,4-D. Enthusiastically developing embryogenic calli were utilized further for Agrobacterium-intervened hereditary change..
2.4. Agrobacterium-interceded transformation
LBA 4404 (pCAMBIA 1305.2) Agrobacterium strain was utilized to assess hereditary change skill of indica rice assortment Sambha mahsuri and Cotton dora sannalu utilizing embryogenic calli. Culture was developed in a BOD hatchery shaker at 28 °C at 250 rpm. The bacterial suspension was utilized for change when the OD estimation of 1.0 at 600 nm was come to (UV–VIS spectrophotometer, Chemito 1200).Actively developing embryogenic calli were taken of all assortments and inundated in Agrobacterium suspension culture containing 200 µM acetosyringone (Aldrich Chemical Company, USA, Cat No.D13 440–6) for 10 min. At that point the Calli were expelled from the bacterial suspension and smear dried with sterile tissue paper to evacuate overabundance microscopic organisms. These calli were put on to co-development MS with 2 mg L−1 2,4-D medium comprising acetosyringone. The way of life were brooded at 25 ± 2oC for 72 h for co-culture. Later the co-developed calli were flushed thrice in channel sterilzed cefotaxime 250 mg L for 1 min each and took after by washing with sterile refined water four circumstances to totally expel Agrobacterium cells. Again the calli were dried on sterile tissue paper and arbitrarily chose for transient GUS expression and remaining calli were put on callus determination MS-AS + 250 mg L−1 cefotaxime + 50 mg L−1 hygromycin medium. This convention was enhanced by taking after this strategy in every one of the trials alongside changing a few parameters. (See Fig. 1, Fig. 2)
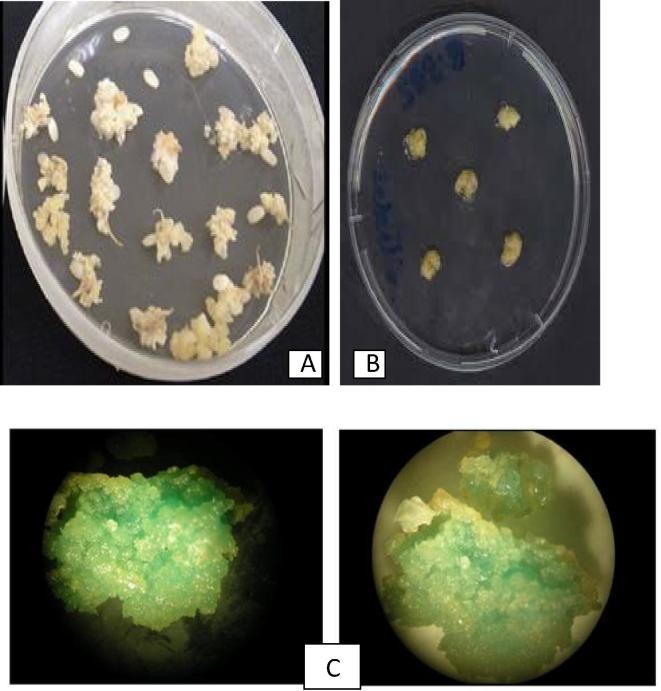
Fig. 1.
Agrobacterium-mediated genetic transformation in rice; (A) Mature seed derived embroygenic calli of rice var.Sambha mahsuri used in transformation: (B) Co-cultivation of embryogenic calli with Agrobacterium:(C) Difference in intensities of strained callus,
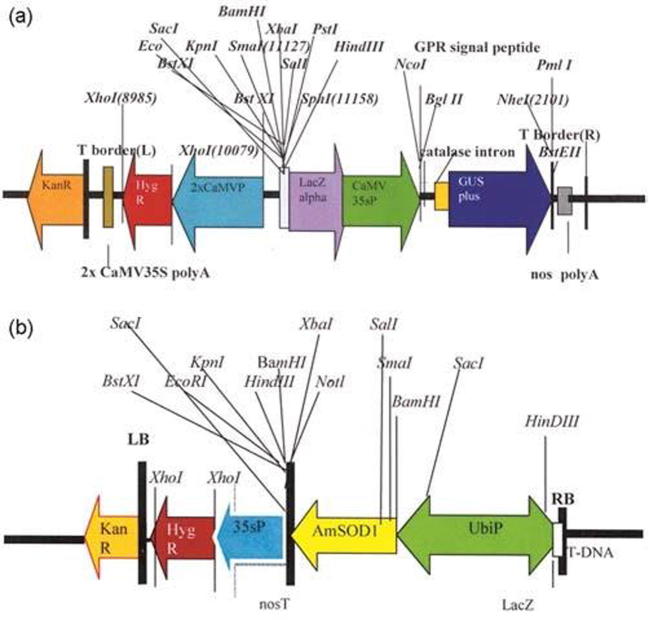
Fig. 2.
T-DNA maps: (a) Diagrammatic representation of T-DNA of the binary vector pCAMBIA 1305.2 with GUS as gene of interest and hygromycin as plant selection marker; (b) Diagrammatic representation of vector pSFSOD1 with Am-SOD as gene of interest and hygromycin as plant selection marker.
2.5. Histochemical Assay of β-Glucouronidase (GUS)
Jefferson5 technique was taken after for examine of GUS expression in putative changed calli and plantlets.. Changed calli were brooded with 25 mM phosphate support and 1% Triton-X for 1 h at 37 °C. To the calli (under LAF) measure arrangement was included, and hatched overnight at 37 °C after vacuum invasion for 10 min. The tested calli were checked under stereomicroscope for blue spots, their number and force of shading following day. Dousing time of calli cells Agrobacterium bears key significance as the bacterial development on the explants assumes imperative part in hereditary change by Agrobacterium. Change may increment by stress too. Consequently for improvement these parameters were thought about. (See Table 1, Table 2, Table 3, Table 4)
Table 1.
Agrobacterium strains in influencing transformation efficiency in rice varities.
| Genotype | Agrobacterium strain | No. of calli plated | No. of GUS stained calli | Staining pattern (No. of calli) |
Stain intensity** | |||
|---|---|---|---|---|---|---|---|---|
| Full | 1/3 | 1/2 | 1/4 | |||||
| Sambha mahuri | EHA105 | 40 | 10 | 5 | 2 | 00 | 3 | ± |
| (pCAMBIA 1305.2)* | (25.0) | (50.0) | (20.0) | (0.00) | (30.0) | |||
| LB4404 | 40 | 20 | 2 | 5 | 5 | 2 | + | |
| (pCAMBIA 1305.2) | (50.0) | (10.0) | (25.0) | (25.0) | (10.0) | |||
Figures within parentheses indicate percent values.
Moderate (±); deep (+).
Table 2.
Agrobacterium-mediated genetic transformation efficiency in different rice varieties.
| Genotype | No. of calli selected for staining | No. of GUS stained calli | Staining pattern (No. of calli) |
Stain | |||
|---|---|---|---|---|---|---|---|
| intensity** | |||||||
| Full | 2/3rd | 1/2 | 1/4th | ||||
| Cotton dora sannalu | 40 | 10 | 5 | 1 | 3 | 1 | ± |
| (25.0) | (50.0) | (10.0) | (30.0) | (10.0) | |||
| Sambha mahsuri | 40 | 12 | 6 | 3 | 4 | 2 | + |
| (28.0) | (50.0) | (25.0) | (30.0) | (20.0) | |||
* Figures within parentheses indicate per cent value.
Moderate (±); deep (+).
Table 3.
Acetosyringone concentration in influencing genetic transformation involving LBA4404 (pCAMBIA 1305.2).
| Genotype | Acetosyringone concentration (μM/mL) | No. of calli selected for staining | No of GUS stained calli | Staining pattern (No. of calli) |
Stain intensity** | |||
|---|---|---|---|---|---|---|---|---|
| Full | 2/3rd | ½ | 1/4th | |||||
| Sambha | 100 | 30 | 4 | 2 | 0 | 2 | 0 | (–) |
| mahsuri | (13.3) | (10.0) | (00.0) | (10.0) | (00.0) | |||
| 150 | 30 | 6 | 1 | 2 | 1 | 2 | (±) | |
| (20.0) | (16.6) | (33.3) | (16.6) | (33.3) | ||||
| 200 | 30 | 12 | 3 | 4 | 5 | 0 | (+) | |
| (40.0) | (15.3) | (38.4) | (46.1) | (00.0) | ||||
| 250 | 30 | 15 | 0 | 6 | 3 | 3 | (+) | |
| (50.0) | (00.0) | (46.6) | (26.6) | (26.6) | ||||
* Figures within parentheses indicate percent values.
Faint (–); moderate (±); deep (+).
Table 4.
Co-cultivation duration in influencing genetic transformation involving LBA4404 (pCAMBIA 1305.2).
| Genotype | Co-cultivation intensity** | No. of calli selected for staining | No. of GUS stained calli | Staining pattern (No. of calli) |
Stain intensity** | |||
|---|---|---|---|---|---|---|---|---|
| Full | 2/3rd | 1/2 | 1/4th | |||||
| 24 | 40 | 2 | 2 | 1 | 2 | 00 | – | |
| (05.0)* | (100.0) | (50.00) | (100.0) | (0.00) | ||||
| Sambha mahsuri | 48 | 40 | 10 | 00 | 5 | 1 | 5 | ± |
| (25.0) | (0.00) | (50.0) | (10.00) | (50.0) | ||||
| 72 | 40 | 15 | 4 | 8 | 2 | 4 | ± | |
| (37.5) | (25.0) | (50.00) | (12.5) | (25.0) | ||||
| 96 | 40 | 1 | 1 | 0 | 0 | 1 | _ | |
| (02.5) | (100.0) | (0.00) | (0.00) | (100.0) | ||||
Figures within parentheses indicate percent values.
Faint (-), moderate (±), and deep (+).
2.6. Methyl viologen test of putative transgenic plants
One tiller from each of 8 recovered putative plantlets and a non-transgenic control plantlet were dunked in 10 mL of 20, 100 and 150 µM methyl viologen (MV) in 50 mL limit culture tubes. After 24 and 48 h of introduction to MV those were exchanged to faucet water. As apparent from Table 5, among 8 putative transgenic plantlets, one (TB10) reliably demonstrated no indications of harm at even 100 µM of MV (Fig.3a and b). Transgenic rice plantlets with constitutively hoisted levels of SOD demonstrate an expanded level of resilience to MV poisonous quality. A considerable contrast was perceptible among control plantlets and MV tolerant transgenic plantlets. After 24 h of MV treatment, suseceptible plants began demonstrating leaf rolling. Second day onwards these plantlets began indicating drying side effects lastly kicked the bucket (Table 6). TB3, TB4, TB6, TB8 and TB10 putative transgenics demonstrated relatively preferable execution over the control plant (Table 5). This improved level of resistance in putative transgenic lines might be because of coordination of Am-SOD in the genome (18). ROS delivered in non-changed plants can't detoxify and endure because of oxidative anxiety. These middle people influence layer trustworthiness and cause extreme impedance of a few physiological procedures and biochemical responses, at last bringing about death of plant. A few reports demonstrate that transgenic plant can search ROS and secure them. Chloroplast was recommended to be the principle focus of MV activity; be that as it may, it was likewise found to deliver superoxide radicals in other cell parts as well.
Table 5.
methyl viologen effect on Sambha mahsuri transgenic lines harbouring Am-SOD gene.
| Putative transgenic line | Treatment of methyl viologen (µM) | No. of leaves | Observation |
|||
|---|---|---|---|---|---|---|
| 24 h Symptom | Scale (0–9)* | 48 h Symptom | Scale (0–9) | |||
| TBc | 20 | 4 | The entire leaves rolled, LG** | 8 | Remained rolled, dried | 7 |
| 100 | 3 | The entire leaves rolled, LG | 7 | To some extaet dried | 8 | |
| 150 | 4 | The entire leaves rolled, LG | 7 | To some extent dried | 9 | |
| TBd | 20 | 4 | 3 leaves opened, G● | 8 | Dried, rolled | 8 |
| 100 | 4 | The entire leaves rolled, LG | 7 | Dried, rolled | 8 | |
| 150 | 3 | The entire leaves rolled, G | 9 | Dried, rolled | 9 | |
| TBe | 20 | 4 | Partly dried, G | 7 | To some extent dried, rolled | 9 |
| 100 | 3 | Entirely rolled, LG | 8 | Dried, rolled | 8 | |
| TBf | 150 | 4 | All leaves completely rolled, LG | 7 | Dried, rolled | 8 |
| 20 | 3 | All leaves completely rolled, G | 7 | Dried, rolled | 9 | |
| TBh | 100 | 4 | 2 open 2 completely rolled, LG | 6 | Dried, rolled | 7 |
| 150 | 3 | Fully rolled, LG | 8 | To some extent dried, rolled | 9 | |
| 2 | Fully rolled, G | 9 | Dried, rolled | 9 | ||
| TBi | 20 | 4 | Partly rolled, LG | 8 | To some extent dried, rolled | 8 |
| 100 | 4 | Total leaves rolled, dried, LG | 7 | To some extent dried, rolled | 8 | |
| 150 | 3 | Total leaves dried rolled, Y | 8 | Dried, rolled | 8 | |
| TBj | 20 | 3 | 1 leaves open, G | 3 | To some extent dried, rolled | 9 |
| 100 | 2 | 1 leaves fully open, LG | 4 | Were fresh, hardly rolled | 7 | |
| TBn | 20 | 4 | Total leaves rolled, LG | 7 | Were rolled, dried | 8 |
| 100 | 3 | Total leaves rolled, G | 9 | Partly dried | 8 | |
| 150 | 2 | Total leaves rolled, LG | 8 | Partly dried | 9 | |
| Control | 20 | 4 | All leaves rolled except one, G | 7 | Dried, rolled | 8 |
| (Non transgenic plant) | 100 | 3 | Yellow patches in leaves and yellowish stem totally rolled Yellowish tinge on stem and leaves, | 9 | Dried, rolled Dried, rolled | 8 |
| 150 | 9 | 9 | ||||
| totally rolled | ||||||
0 = No effect, 1–3: Highly tolerant to leaf rolling and tip burning, 4–6: Moderate, 7–9: Highly susceptible.
G, LG & Y denote to green, light green and yellow leaf colour.
Fig. 3.
(A) Tillers of putative transgenic plants showing tolerance to methyl viologen; (B) Tillers of non-transgenic plants showing susceptibility to methyl viologen.
Table 6.
Stress recovery in Sambha Mahsuri transgenics harbouring Am-SOD gene after methyl viologen treatment.
| Observation |
||||||
|---|---|---|---|---|---|---|
| Putative transgenic line | Treatment of methyl viologen (µM) | No.of leaves | 24 h Symptom | Scale (0–9)* | 48 h Symptom | Scale (0–9)* |
| TBc | 20 | 4 | 3 leaves open, LG** | 7 | Dried, rolled | 7 |
| 100 | 4 | Total leaves rolled, G | 8 | Dried, rolled | 8 | |
| 150 | 4 | Total leaves rolled, G | 8 | Dried, rolled | 7 | |
| TBd | 20 | 4 | Partly dried, G | 7 | Partly dried, rolled | 8 |
| 100 | 4 | Completely rolled, LG | 8 | Dried, rolled | 9 | |
| 150 | 4 | Completely rolled, G | 7 | Dried, rolled | 8 | |
| TBe | 20 | 3 | Partly rolled, LG | 7 | Partly dried, rolled | 8 |
| 100 | 4 | Total leaves rolled, dried, G | 7 | Partly dried, rolled | 9 | |
| 150 | 3 | Total leaves dried rolled, Y | 8 | Dried, rolled | 9 | |
| TBf | 20 | 4 | 1 leaves open, G | 6 | Partly dried, rolled | 8 |
| 100 | 2 | 3 leaves fully open, LG | 4 | Were fresh, hardly rolled | 7 | |
| 150 | 3 | Entire leaves rolled, LG | 8 | Dried, rolled | 9 | |
| TBh | 20 | 4 | Entire leaves rolled, G | 7 | Were rolled, dried | 9 |
| 100 | 2 | Entire leaves rolled, LG | 8 | Partly dried | 8 | |
| 150 | 3 | Entire leaves rolled, LG | 9 | Partly dried | 9 | |
| TBi | 20 | 4 | Entire leaves rolled, G | 8 | Dried, rolled | 8 |
| 100 | 4 | Entire leaves rolled, G | 8 | Dried, rolled | 9 | |
| 150 | 2 | Entire leaves rolled, LG | 9 | Dried, rolled | 8 | |
| TBj | 20 | 4 | Every single leaf totally rolled, G | 4 | Dried, rolled | 8 |
| 100 | 3 | Every leaf totally rolled, LG | 2 | Dried, rolled | 7 | |
| 150 | 4 | Every leaf totally rolled, G | 8 | Dried, rolled | 9 | |
| TBn | 20 | 3 | 2 open 1 totally rolled, LG | 5 | Dried, rolled | 9 |
| 100 | 2 | Completely rolled, LG | 7 | Party dried, rolled | 8 | |
| 150 | 3 | Completely rolled, G | 8 | Dried, rolled | 8 | |
| Control (Non | 20 | 3 | Total leaves rolled except one, G | 9 | Dried, rolled | 8 |
| transgenic plant) | 100 | 2 | Y patches in leaves and Y stem totally rolled Y tinge on stem and leaves, totally | 9 | Dried, rolled Dried, rolled | 9 |
| 150 | Rolled | 9 | 9 | |||
0 = No effect, 1–3: Highly tolerant to leaf rolling and tip burning, 4–6: Moderate, 7–9: Highly susceptible.
G, LG & Y denote to green, light green and yellow colour of leaf/stem.
2.7. Seed germination test for Inheritance of transgene
Germination test was performed including seeds from transgenic plants of T0 era in hygromycin-supplemented media. Safe seeds developed, while the non changed (helpless) seeds did not grow or kicked the bucket after germination as the vector utilized for transmitting the quality of intrigue had hygromycin as selectable marker. In dominant part of the cases 3:1 proportion was discernable, which demonstrates the nearness of single duplicate of the transgene. Out of the tried seeds from transgenic plants including T0 seeds, TB3 offspring indicated > 95% likelihood of 3:1 proportion, while a couple, viz., TB10 and TB9, demonstrated likelihood in the range 0.05–0.09 and few like TB8, TB5 and TB14 appeared in the range 0.02–0.05, and others not exactly these too. Diverse proportions seen might be because of little size of the specimens utilized as a part of the review.
2.8. NaCl tolerance
Potential part of SOD against saltiness worry in transgenic plants is accounted for (19). Transgenic lines of T0 and T1 eras were inspected for resistance to NaCl-incited salt anxiety and were discovered tolerant up to 150 mM fixation (Table 7). In the present review, when plant tillers were presented to hoisted levels of salt anxiety, few plants like TB3, TB10 and TB8 were more endure to saltiness.
Table 7.
Variation in enhanced salinity tolerance of tillers of putative transgeniccs after 24 and 48 h in 150 mM NaCl.
| Putative Transgenic Line | No. of leaves | Observation |
|||
|---|---|---|---|---|---|
| 24 h |
48 h |
||||
| Symptom | Scale (0–9)* | Symptom | Scale (0–9)* | ||
| TBd | 4 | 2fully rolled, 2 entirely open, LG | 7 | 1 fully rolled, 3 fully open, Y | 8 |
| TBe | 3 3 fully open, LG | 4 | 1 fully open, 2 partly open, LG | 4 | |
| TBi | 4 | 2 fully rolled, 2 entirely open, G | 5 | 2 partly rolled, 2 fully open, G | 5 |
| TBj | 3 | 3 fully open, LG | 2 | 2 partly open, Y | 3 |
| TBf | 2 | 2 leaves fully rolled, G | 6 | 1 totally rolled, 1 fully open, LG | 6 |
| TBh | 3 | 2 leaves tip rolled, 1 fully open, LG | 7 | All fully open, LG | 8 |
| Control (Non transgenic plant) | 3 | 3 leaves tip Rolled, G | 8 | 3 totally rolled, y | 8 |
2.9. PCR analysis
Nanogram amounts of genomic DNA were observed to be sufficient for PCRintensification. For intensification reason, grouping (i) a forward groundwork:
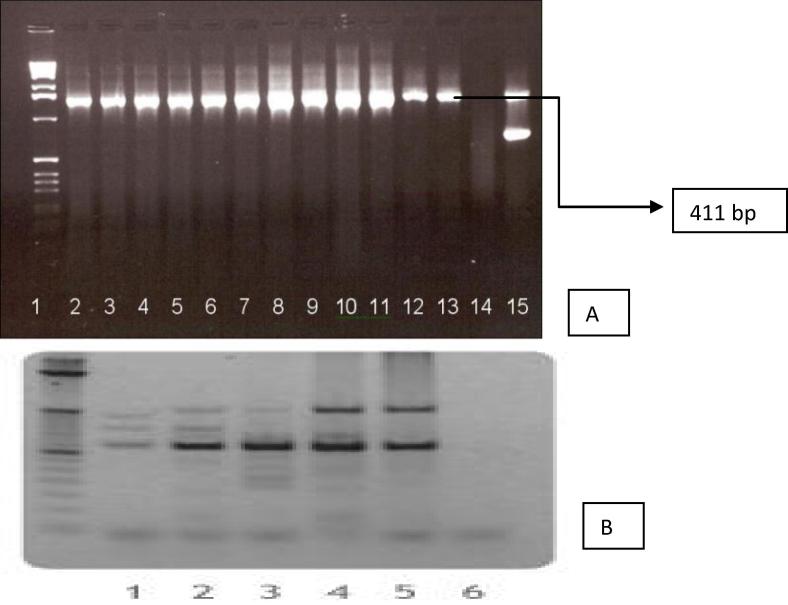
5΄ATGCCGAAGGCTGTCGCCGTACTT3΄ and (ii) a switch preliminary 5΄ TCAGCCCTGAAG ACCAATGATACC3΄ those are particular to the coding area of Am-SOD quality were utilized and were relied upon to open up a 411 bp section of Am-SOD quality succession. PCR amplicon profile indicated event of expected band of 411 bp in eight plantlets (Fig.4a). Of 17 plantlets inspected, some demonstrated clear opened up groups, while control plantlets did not show any sections enhanced, proposing reconciliation of Am-SOD quality in a portion of the putative transgenic plants.
Fig. 4.
(A) PCR amplification of Am-SOD gene involving genomic DNA extracted from putative transgenic plants of Sambha mahsuri [1, Molecularruler(1 kb ladder); 15, Positivecontrol (plasmidDNA); 14, Wildtype (nontransformedSambhamahsuri); & Lanes 2–13, Representputativetransgenicplants]. (B) Southern analysis showing the integration of the transgene in putative transgenic plants of Sambha mahsuri Lanes 1,2,3, 4 5 & 6 transgenic plants showing integration of Am-SOD gene in rice var. Sambha mahsuri.
2.10. Southern blot analysis
PCR positive and hygromycin safe plantlets derived from transformed call showed their genomic DNA from transgenic plantlets was utilized for Southern smear investigation to identify the nearness of Am-SOD quality. Southern smear uncovered the nearness of Am-SOD quality in the transformants. Genomic DNA was confined with Sac1 to discharge the quality in place as one section of a size of 550 bp and the test utilized was a limitation piece produced by utilizing BamH1, which gives a part of the quality of enthusiasm of a size ∼ 430 bp. On the off chance that the quality is incorporated, flag is required to be seen around a size of 550 bp, which should be 150 bp more than the extent of the test. The outcome as found in Fig.4b exhibits the nearness of the band of expected size for both the positive transgenic plants (550 bp) and for the test (435 bp). This outcome plainly shows genuine transgenic nature of the putative transgenic plants tried. Southern smear additionally uncovered incorporated outsider Am-SOD quality in every single positive plant and no improvements were seen in any of the transgenic plants considered.
3. Conclusion
In the present situation, populace development is expanding step by step alongside interest for sustenance supply. Hereditary change must be actualized to conquer any hindrance amongst generation and human need. Thus in the present we have communicated Am-SOD quality in our rice plants which have effectively demonstrated saltiness and dry spell resistance.
Acknowledgements
Authors are grateful to CII (confederation of Indian industry), DST (Department of Science and Technology, Ministry of Science & Technology, New Delhi, India), Varsha Bioscience and Technology Private Limited for providing the Prime Minister’s Fellowship (Samar Shekar Reddy.S). Similarly, Shri Neeraj Verma, Director In-charge (State Farms Corporation of India Limited) for authenticating and providing Indica rice cultivars and Dr. C. Cheralu, Associate Director Agricultural Research Station, Warangal, India for authenticating and providing Indica rice cultivars for our present study. MTCC, Chandigarh, India for providing Agrobacterium strains and plasmids for our present study. Gitam university, Department of biotechnology, Vishakapatnam for providing lab and equipments for this work.
Footnotes
Peer review under responsibility of King Saud University.
References
- Deepa S.P., Saleh M.A., Selvaraj C. Rice breeding for salt tolerance. Res. Biotechnol. 2011;2(2):1–10. [Google Scholar]
- Ingram H.M., Livesey N.L., Power J.B., Davey M.R. Genetic transformation of wheat: progress during the 1990s into the millennium. Acta Physiol. Plant. 2001;23(2):221–239. [Google Scholar]
- Gupta B., Bingru H. Mechanism of Salinity Tolerance in Plants: Physiological, Biochemical and Molecular Characterization. Int. J. Genomics. 2014;14:1–18. doi: 10.1155/2014/701596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J.J., Kharb P., Teng W.M., Hall T.C. Characterization of rice transformed via an Agrobacterium-mediated inflorescence approach. Mol. Breed. 2001;7:187–194. [Google Scholar]
- Hiei Y., Ohta S., Komari T., Kubo T. Transformation of rice mediated by Agrobacterium tumefacien. Plant Mol. Biol. 1997;35:205–218. [PubMed] [Google Scholar]
- Tyagi A.K., Mohanty A., Bajaj S., Chaudhury A., Maheshwari S.C. Transgenic rice: a valuable monocot system for crop improvement and gene research. Critical Rev. Biotechnol. 1999;19:41–79. [Google Scholar]
- Taylor N.J., Fauquet C.M. Microparticle bombardment as a tool in plant science and agricultural biotechnology. DNA Cell Biol. 2002;21:963–977. doi: 10.1089/104454902762053891. [DOI] [PubMed] [Google Scholar]
- Gao J.-P., Chao D.-Y., Lin H.-X. Understanding abiotic stress tolerance mechanisms: recent studies on stress response in rice. J. Integr. Plant Biol. 2007;49(6):742–750. [Google Scholar]
- Lafitte H.R., Ismail A., Bennet J. Abiotic stress tolerance in rice for Asia: progress and the future, in new directions for a diverse planet. Plant Cell. 2004;14(3):537–545. [Google Scholar]
- Li S.H., Tian X.H., Huang Y.P., Liu A.Y. Preliminary analysis of high temperature harm in middle-season rice at flowering stage in Jianghan Plain in the latest 50 years. Chinese J. Agrometeorol. 2007;28(1):5–8. [Google Scholar]
- Sarangi S., Ghosh J., Bora A., Das S., Mandal A.B. Agrobacterium-mediated genetic transformation of indica rice varieties involving Am-SOD gene. Indian J. Biotechnol. 2011;10(1):9–18. [Google Scholar]
- Yu S.-M., Ko S.-S., Hong C.-Y. Global functional analyses of rice promoters by genomics approaches. Plant Mol. Biol. 2007;65(4):417–425. doi: 10.1007/s11103-007-9232-1. [DOI] [PubMed] [Google Scholar]
- Corrado G., Karali M. Inducible gene expression systems and plant biotechnology. Biotechnol. Adv. 2009;27(6):733–743. doi: 10.1016/j.biotechadv.2009.05.006. [DOI] [PubMed] [Google Scholar]
- Zhang X.W., Li J.P., Liu A.L. Expression profile in rice panicle: insights into heat response mechanism at reproductive stage. PLoS ONE. 2012;7(11):1–17. doi: 10.1371/journal.pone.0049652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.W., Rerksiri W., Liu A.L. Transcriptome profile reveals heat response mechanism at molecular and metabolic levels in rice flag leaf. Gene. 2013;530(2):185–192. doi: 10.1016/j.gene.2013.08.048. [DOI] [PubMed] [Google Scholar]