Abstract
Cardiovascular diseases (CVDs) cause high mortality throughout the world. Existing fibrinolytic agents are highly expensive and have many side effects. Microbial fibrinolytic enzymes are very much considered as novel therapeutic candidate for the treatment of CVDs. Reports on fibrinolytic enzyme from Xanthomonas sp. is lacking. This study reports fibrinolytic enzymes from Xanthomonas oryzae IND3 as it shows hyperactivity on fibrin-agarose plates. This organism utilized various agro-industrial wastes for enzymes production. Among all, cow dung enhanced more enzyme production, hence it was used as the low-cost substrate for statistical optimization of fibrinolytic protease in Solid state fermentation. Response surface methodology was employed to optimize the factors and enhanced yield by 4-fold. The interactions among the variables, viz, sucrose, yeast extract, and pH of the medium were investigated using Central Composite Design (CCD). The predicted fibrinolytic enzyme activity was 2340 U/g, and the observed fibrinolytic enzyme activity was 2294 ± 12.8 U/g. The fibrinolytic enzyme degraded blood clot in vitro completely. This study is the first report on statistical optimization of fibrinolytic enzyme production in SSF from Xanthomonas sp. The crude extract has immense activity on proteinaceous wastes. The production of fibrinolytic protease using the low-cost substrate could reduce the production cost of enzyme.
Keywords: Cardiovascular diseases, Xanthomonas oryzae, Solid-state fermentation, Fibrinolytic enzyme, Response surface methodology, Blood clot lysis
1. Introduction
Thrombosis is one of the important Cardiovascular diseases cause more deaths throughout the world. Thrombolytic agents such as streptokinase, tissue plasminogen activator (tPA) and urokinase plasminogen activator (u-PA) are commonly used to treat thrombosis. Although t-PA and u-PA are sate to treat thrombosis but every expensive. In the other hand, streptokinase cause serious side effects (Collen and Gold, 1990). In recent years, microbial fibrinolytic enzymes have been reported to treat and prevent CVDs. These enzymes have various therapeutic applications, including, oncolyticus, anticoagulants, thrombolytics and anti-inflammatories. There are various reports on fibrinolytic enzymes with thrombolytic applications from many sources, such as earth worm, snake venom, and fermented foods (Peng et al., 2003). Many studies involved the characterization of fibrinolytic enzyme, however, very few studies were carried on culture media optimization using statistical approach (Mukherjee and Rai, 2011). The fermentation media are the major cost determining factors for bacterial enzyme production, however the application of agro-residues could minimize the enzyme production cost and also valorization of agro-wastes to decrease environmental pollution (Bajaj et al., 2014). Many studies were conducted on agro-residues, such as pigeon pea (Johnvesly et al., 2002), potato peel (Mukherjee et al., 2008), apple pomace (Dhillon et al., 2012b), rice chaff (Tao et al., 1997), fish waste (Ramkumar et al., 2016), wheat bran (Almalki, 2018) and agro-industrial wastes (Sadh et al., 2018) for the production of enzymes. These reported substrates were highly useful for the production of enzymes in solid-state fermentation (SSF).
Presently, Bacillus sp. and fungal isolates were very much exploited for fibrinolytic enzyme production. However, very fewer reports available on other bacterial species. The search for novel bacteria for bioprocess optimization for enzyme production is a continuous process. Xanthomonas sp. has the ability to produce various enzymes and was reported by various research groups. For example, Xanthomonas axonopodis pv. punicae strain has the ability to produce xylanolytic and cellulolytic enzymes (Amat et al., 2014). Lytic enzyme was produced from Xanthomonas campestris using low cost substrates (Da Silva et al., 2014). Kalashnikova et al. (2003) used three Xanthomonas campestris pv. campestris strains for their ability to produce proteolytic enzymes. However, the fibrinolytic enzyme production from the genus is highly limited. For industrial production of enzymes, a suitable culture medium is critical to enhance the yield for any selected isolates. Also, important prerequisites for commercial production are to reduce the cost of enzymes. This can be achieved by optimizing culture medium using agro-industrial residues. The conventional method of optimization is One-Factor-At-A-Time (OFAT) experiment, however this procedure is laborious and time consuming. Also, this method frequently fails to provide the interactive effect among the selected process parameters on product yield (Singh and Bajaj, 2016). Statistical optimization methods have several advantages over OVAT approach. A response surface methodology (RSM) has been widely used for the production of various enzymes, including nattokinase (Liu et al., 2005). Proteolytic enzymes have many applications in dairy-, meat-industries, and to hydrolyse proteinaceous discharges in aquatic environment. In municipal wastewater treatment, proteases hydrolyse proteins into smaller units (Cadoret et al., 2002). The therapeutic application of fibrinolytic protease is to dissolve the blood clot directly. The use of fibrinolytic proteases could avoid the use of highly toxic chemicals. The production of low-cost fibrinolytic enzyme is possible only by applying cheap agro-residues in SSF. The report on firinolytic protease production using low cost substrate by Xanthomonas oryzae is not available. The main objective of the present study is to optimize fibrinolytic enzyme from Xathomonas oryzae for biomedical applications.
2. Materials and methods
2.1. Screening and isolation of fibrinolytic protease producing bacteria
Rice was boiled for 60 min and allowed for microbial fermentation (32 ± 2 °C) for 48 h. It was further used for the screening of potent fibrinolytic protease producing bacteria. About one gram of rice was mixed with 100 ml sterile distilled water and plated on skimmed milk agar (Himedia, Mumbai, India) plates. Ten protease producing bacterial isolates were purified by standard method and further subjected for secondary screening. For secondary screening the bacterial isolates were cultured in submerged fermentation in nutrient broth and cell free extract was used for screening.
The protease producing bacteria were cultured in nutrient broth (Himedia, Mumbai, India) for 48 h at 37 ± 2 °C. After 48 h, fermented medium was centrifuged (5000g at 4 °C) for 15 min, and the supernatant was used for screening. The fibrinolytic protease activity was determined on fibrin-agarose plate (Astrup and Mullertz, 1952). In this plate, 2.5 ml fibrinogen (0.5%, w/v) and 100 µl thrombin (100 NIH U/ml) were incorporated. It was allowed to stand at 30 ± 2 °C for 1 h and 15-µl sample was dropped into wells. The plates were incubated at 30 ± 2 °C, and the fibrinolytic protease activity exhibited zone of hydrolysis around the well.
2.2. Molecular identification
The hyperactive fibrinolytic enzyme producing X. oryzae IND3 was cultured in the nutrient broth medium for 18 h at 37 ± 2 °C. The genomic DNA was extracted using a commercial kit (QIAGEN genomic DNA purification kit) according to the manufacturer’s instructions. The16S rDNA gene was amplified using a PCR—Peltier Thermal Cycler Machine (USA). The amplified 16S rDNA was sequenced, and 857-bp16S rDNA gene sequences of the bacterial isolate were submitted to GenBank under the accession number: KF250419.
2.3. Solid state fermentation
In this study, banana peel, green gram husk, rice bran, wheat bran and cow dung was used as the substrate. About 5.0 g of these substrates were individually transferred in a 100-ml Erlenmeyer flask and moisture content was maintained at 90% (v/w) level using Tris-buffer (100 mM, pH 8.0). All Erlenmeyer flasks were sterilized and cooled. These flasks were inoculated individually with 0.5 ml of 18-h grown bacterial culture (OD 600 nm = 1.98 ± 0.240) and incubated at 37 ± 2 °C for 48 h in an incubator. After incubation, enzyme was extracted and the supernatant was used to screen the efficacy of substrate on enzyme production.
2.4. Fibrinolytic protease assay
The crude enzyme was mixed with Tris-HCl buffer (2.5 ml, 0.1 M, pH 7.8, 0.01 M calcium chloride) and fibrin (2.5 ml, 1.2%, w/v) and incubated at 37 °C for 30 min. The reaction was terminated by adding 5.0 ml of trichloroaceticacid (0.11 M), containing sodium acetate (0.22 M) and acetic acid (0.33 M). The absorbance of the sample was measured at 275 nm against blank (Ansen, 1938). Total protein estimation was carried out by the method of Lowry et al. (1951).
2.5. Screening of variables by one-variable-at-a-time approach
SSF was carried out as described previously. Cow dung which showed maximum enzyme production among the selected substrates was used as the substrate for optimization studies, until otherwise stated. The important nutrient factors, such as carbon source ([1%, w/w], glucose, sucrose, maltose, starch, and xylose), nitrogen source ([1%, w/w]; peptone, casein, yeast extract, gelatine, and urea), and inorganic ions ([1%, w/w], calcium chloride, magnesium chloride, sodium di-hydrogen phosphate, zinc sulphate, and mercury chloride), were screened.
2.6. Elucidation of process variables by a two-level full factorial design
A two-level full factorial design (25) was employed to screen the important factors affecting fibrinolytic protease production. The five selected factors were moisture, pH, sucrose (carbon source), yeast extract (nitrogen source), and sodium di-hydrogen phosphate (mineral salt). Each factor was analyzed at two different levels (Table 1a). The two-level full factorial design experiment was based on the first-order polynomial equation described below:
where Y = fibrinolytic protease activity, αij = ijth interaction coefficient, αijk = ijkth interaction coefficient, and α0 = intercept
Table 1a.
Factors involved according to a two-level full factorial design for optimization of fibrinolytic protease production.
| Symbol | Variables | Units | Coded levels |
|
|---|---|---|---|---|
| −1 | 1 | |||
| A | Moisture | % | 80 | 100 |
| B | pH | 7 | 9 | |
| C | Sucrose | % | 0.1 | 1 |
| D | Yeast extract | % | 0.1 | 1 |
| E | NaH2PO4 | % | 0.01 | 1 |
Fibrinolytic protease assay was carried out in duplicate experimental runs, and the average value was taken as response “Y.” Analysis of variance (ANOVA) was used to test the statistical significance and p-value < 0.05 was considered as statistically significant. The entire experimental setup for a two-level full factorial design for five variables is described in Table 1b. The most significant process parameters (p < 0.05) which influence on enzyme production were further optimized by RSM.
Table 1b.
Statistical analysis of 25 full factorial design showing values for each variables for fibrinolytic enzyme activity.
| Run | Sucrose | Peptone | NaH2PO4 | pH | Moisture | Enzyme |
|---|---|---|---|---|---|---|
| A | B | C | D | E | activity (U/g) | |
| 1 | −1 | 1 | −1 | 1 | −1 | 275 |
| 2 | −1 | 1 | 1 | 1 | 1 | 540 |
| 3 | 1 | 1 | 1 | 1 | −1 | 850 |
| 4 | −1 | −1 | 1 | 1 | −1 | 385 |
| 5 | −1 | −1 | −1 | 1 | 1 | 230 |
| 6 | 1 | −1 | 1 | −1 | 1 | 460 |
| 7 | 1 | −1 | 1 | 1 | −1 | 875 |
| 8 | 1 | 1 | 1 | −1 | −1 | 455 |
| 9 | 1 | 1 | 1 | −1 | 1 | 1400 |
| 10 | 1 | 1 | −1 | 1 | −1 | 1000 |
| 11 | 1 | −1 | −1 | −1 | 1 | 1190 |
| 12 | −1 | −1 | −1 | 1 | −1 | 1590 |
| 13 | −1 | 1 | −1 | −1 | 1 | 495 |
| 14 | −1 | −1 | 1 | 1 | 1 | 700 |
| 15 | −1 | 1 | −1 | −1 | −1 | 2245 |
| 16 | −1 | 1 | 1 | −1 | −1 | 260 |
| 17 | 1 | −1 | 1 | −1 | −1 | 545 |
| 18 | 1 | 1 | 1 | 1 | 1 | 1220 |
| 19 | 1 | −1 | −1 | 1 | −1 | 1525 |
| 20 | −1 | −1 | −1 | −1 | −1 | 900 |
| 21 | −1 | 1 | −1 | 1 | 1 | 1325 |
| 22 | 1 | −1 | 1 | 1 | 1 | 120 |
| 23 | −1 | −1 | 1 | −1 | −1 | 375 |
| 24 | −1 | −1 | −1 | −1 | 1 | 475 |
| 25 | 1 | 1 | −1 | −1 | 1 | 790 |
| 26 | −1 | 1 | 1 | −1 | 1 | 780 |
| 27 | 1 | −1 | −1 | 1 | 1 | 925 |
| 28 | 1 | 1 | −1 | −1 | −1 | 505 |
| 29 | −1 | 1 | 1 | 1 | −1 | 510 |
| 30 | −1 | −1 | 1 | −1 | 1 | 670 |
| 31 | 1 | −1 | −1 | −1 | −1 | 325 |
| 32 | 1 | 1 | −1 | 1 | 1 | 680 |
2.7. Central composite design
Central composite design (CCD) was frequently used to find the optimum concentration of variables. In our study, the selected factors, including, pH, sucrose, and yeast extract were employed at five levels (−α, −1, 0, +1, and +α) (Table 2a). For three variables, a total of 20 experiments run, including six center, six axial, and eight factorial points. SSF was carried out in duplicates as described earlier with the specific experimental matrix described in Table 2b. The results obtained were analyzed by ANOVA and the second-order polynomial equation is as follows.
Table 2a.
Range of media components for the production of fibrinolytic protease.
| Variables | Symbol | Coded values |
||||
|---|---|---|---|---|---|---|
| −α | −1 | 0 | 1 | +α | ||
| pH | A | 6.32 | 7 | 8 | 9 | 9.68 |
| Sucrose | B | −0.21 | 0.1 | 0.55 | 1 | 1.31 |
| Yeast extract | C | −0.21 | 0.1 | 0.55 | 1 | 1.31 |
Table 2b.
Central composite design matrix for the production of fibrinolytic protease from X. oryzae.
| Std | A:pH | B:Sucrose | C:Yeast extract | Enzyme activity (U/g) |
|---|---|---|---|---|
| 1 | −1 | 1 | 1 | 622 |
| 2 | 1 | 1 | 1 | 1590 |
| 3 | 0 | 0 | 0 | 1269 |
| 4 | 1 | 1 | −1 | 2068 |
| 5 | 1.682 | 0 | 0 | 1050 |
| 6 | 0 | 0 | 0 | 1231 |
| 7 | 0 | 0 | 0 | 1290 |
| 8 | 0 | 0 | 0 | 1249 |
| 9 | 0 | 1.682 | 1 | 1500 |
| 10 | 0 | 0 | 1.682 | 1508 |
| 11 | −1 | 1 | −1 | 841 |
| 12 | −1 | −1 | 1 | 2213 |
| 13 | 1 | −1 | −1 | 873 |
| 14 | 0 | 0 | 0 | 1348 |
| 15 | −1 | −1 | −1 | 1372 |
| 16 | 0 | −1.682 | 0 | 2110 |
| 17 | 0 | 0 | −1.682 | 1200 |
| 18 | −1.682 | 0 | 0 | 810 |
| 19 | 0 | 0 | 0 | 1308 |
| 20 | 1 | −1 | 1 | 1480 |
2.8. Application of fibrinolytic enzyme
2.8.1. Activity of fibrinolytic enzyme on blood clot in vitro
The Goat blood was collected from a slaughter house. Furthermore, the clot was washed several times with phosphate buffered saline (PBS) (pH 7.2) and cut into small pieces aseptically. The enzyme was diluted appropriately (200 U/ml) and incubated with blood clot (500 ± 50 mg) at 30 ± 2 °C. To the control vial, enzyme was not added with PBS. The tubes were incubated for 24 h and blood clot lysis was observed (Najafi et al., 2005).
2.8.2. Enzymatic hydrolysis of proteins from milk plant
Milk sewage hydrolysis experiment was carried out as described previously (Jung et al., 2002). Milk sewage was collected from Avin milk plant, Nagercoil, Tamilnadu, India. The protein hydrolysis was initiated by adding 200 U of crude fibrinolytic protease with milk sewage (100 ml) and incubated at room temperature (30 ± 2 °C) for 6 h. Total protein content was measured as described previously.
2.8.3. Hydrolysis of proteinaceous waste from sewage water
A liter of wastewater was collected at Nagercoil municipal area, Kanyakumari, Tamilnadu, India. The protein content of the wastewater was tested and concentrated by adding ammonium sulphate (70% saturation). The precipitated proteins were reconstituted at the concentration of 1 mg/ml and 0.1 ml enzyme solution was added to initiate the reaction. The reaction was terminated by 10% TCA (1 ml) solution after 0–60 min at a 15 min interval. Sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) (12%) was used to monitor the hydrolysis proteins (Laemmli, 1970).
3. Results and discussion
3.1. Screening of fibrinolytic enzyme producing X. oryzae IND3
In our study, a clear zone was observed on the fibrin-agarose plate showing fibrinolytic enzyme activity of X. oryzae IND3 than other isolates (Fig. 1). Many reports have demonstrated the production of proteases from various Xanthomonas sp., including, Xanthomonas maltophilia (Margesin and Schinner, 1991), Xanthomonas campestris pv. zinniae (Sun, 1991), Xanthomonas sp. (Silva et al., 2014), and Xanthomonas campestris (Dow et al., 1993). Recently, proteolytic enzyme was characterized from Xanthomonas campestris pv. vesicatoria by Sole et al. (2015). However, production of fibrinolytic enzyme from X. oryzae was not reported. Also, statistical optimization of fibrinolytic enzyme production other than Bacillus sp. is highly limited.
Fig. 1.
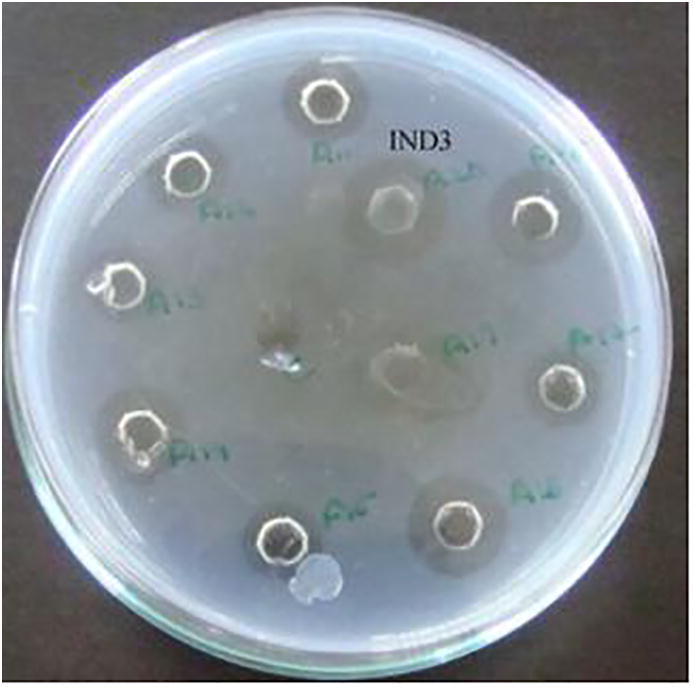
Fibrinolytic enzyme activity of the selected bacterial isolates.
3.2. Valorization of agro-residues for enzyme bioprocess
In the present study, agro residues were screened for the production of fibrinolytic enzyme from X. oryzae IND3. All selected substrates supported fibrinolytic enzyme production. However, the enzyme yield varied widely. The enzyme yield was 132 ± 12.1, 334 ± 8.4, 217 ± 6.9, 402 ± 11.3, and 419 ± 9.8 U/g for the substrates banana peel, green gram husk, rice bran, wheat bran and cow dung, respectively. Previously, various wastes were utilized for the production of proteolytic enzymes. The proteinaceous tannery wastes were utilized as the cheap substrate by Ravindran et al. (2011). Considering the availability of substrate, cow dung is available widely and is too cheap than other reported substrates elsewhere. The application of cow dung as the substrate for the production of enzyme in an industrial scale could reduce the environmental pollution. This is the first report on the production of fibrinolytic protease by Xanthomonas sp. using low cost substrate in SSF and optimization by statistical approach for industrial processes. Previously, cow dung was utilized for the production of fibrinolytic enzyme by Bacillus sp. IND7 and Paenibacillus sp. IND8 (Vijayaraghavan et al., 2016, Vijayaraghavan and Vincent, 2014b).
3.3. Optimization of fibrinolytic enzyme production by OVAT approach
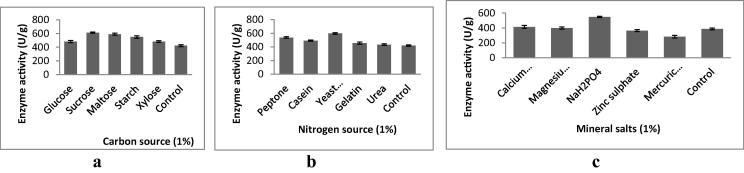
The screened X. oryzae IND3 utilized various carbon sources and these carbon sources induced enzyme production. Enzyme production was high when sucrose was used as the carbon source (481 ± 16.9 U/g) (Fig. 2a) and low in the substrate containing glucose. This low enzyme activity could be the result of catalytic repression. It was previously reported that the increased production of proteolytic enzymes by the supplementation of maltose (Tsuchiya et al., 1991), and sucrose (Phadatare et al., 1993) along with the major components. The complex nitrogen sources were previously reported to enhance the production of fibrinolytic enzymes. In our study, fibrinolytic protease yield was found to be high when yeast extract was supplemented with substrate (597 ± 12.6 U/g) (Fig. 2b). The present study revealed that yeast extract was found to be the appropriate nitrogen source for enzyme production. This result was in accordance with result on fibrinolytic protease production by Proteus penneri SP-20 (Jhample et al., 2015).
Fig. 2.
Effect of carbon source (a), nitrogen source (b) and mineral salts (c) on fibrinolytic protease production.
In the present study, supplementation of sodium dihydrogen phosphate (1%, w/w) with substrate enhanced the production of fibrinolytic proteases (548 ± 9.64 U/g) than control (Fig. 2c). Supplementation of sodium dihydrogen phosphate could responsible for buffering the culture medium. This could be the reason for the increased fibrinolytic activity in the culture medium in the presence of this salt. This result was in accordance the observations made earlier with Paenibacillus sp. IND8 (Vijayaraghavan and Vincent, 2014b). It was also described that the mineral salts, such as calcium, copper, cobalt, manganese, and magnesium, were very much required for the production of proteolytic enzymes. Moreover, the requirement of mineral salts for the production of proteolytic enzymes varies widely, for example, potassium phosphate enhanced fibrinolytic enzyme production in Bacillus licheniformis (Mao et al., 1992).
3.4. Optimization of fibrinolytic protease production by two level full factorial designs
A two-level (25) full factorial design was used to screen all selected variables (moisture, pH, sucrose, yeast extract, and sodium dihydrogen phosphate) for fibrinolytic protease production. The response (Y) for two level full factorial design is shown in Table 1b. The two-level full factorial results revealed a wide variation on fibrinolytic enzyme yield. The designed 2 level full factorial statistical model was significant (p < 0.05). In this model, pH, sucrose, and yeast extract were most significant factors (p < 0.05). The first-order regression equation showed fibrinolytic protease activity, as a function of moisture, pH, sucrose, yeast extract, and sodium dihydrogen phosphate in terms of coded factors:
| Enzyme activity = +769.38 + 34.69A + 63.75B − 135.31C + 27.50D − 19.38E + 71.87AC + 67.81AD + 53.44AE + 54.06BC − 60.63BD + 90.00BE + 121.56CE − 60.00DE + 128.13ABC + 40.31ABD + 25.94ABE − 58.12ACD − 106.25ACE − 147.19ADE + 72.81BCD + 40.94BCE + 130.63BDE − 47.19CDE − 24.37ABCD + 112.50ABCE − 70.94ABDE + 98.75ACDE − 156.56BCDE + 108.75ABCDE |
where Y is the fibrinolytic protease activity (U/g), A, B, C, D, and E are moisture, pH, sucrose, yeast extract, and sodium di hydrogen phosphate, respectively.
The interaction between AC, AD, AE, BC, BE, CE, ABC, ABD, ABE, BCE, BDE, ABCE, ACDE, and ABCD had a positive interactive effect on enzyme production. The lack of fit of this model was non-significant, which confirmed that the designed model is good. According to the results, 80% moisture, pH 9.0, 0.1% sucrose, 0.1% yeast extract, and 0.1% NaH2PO4 significantly enhanced fibrinolytic protease production (2245 U/g) (Table 1c). Moisture content is one of the significant factors in SSF. However, in our study, the moisture content of the medium was not statistically significant (p > 0.05). Unlike other substrates, cow dung possesses high moisture holding capacity, and it supports the anchorage of bacterial cells in a wide range of moisture content. This could be the reason for non-significant moisture level in this experiment. This indicated that X. oryzae can produce enzyme at wide moisture content. The predicted R2 of this model was 0.8094, which was in reasonable agreement with the R2 of this model (0.993), and the adjusted R2 of this model was 0.988.
Table 1c.
Analysis of variance for 25 factorial experimental design.
| Source | Sum of squares | df | Mean square | F-value | p-value | |
|---|---|---|---|---|---|---|
| Model | 6.952 + 006 | 29 | 2.40E+05 | 92.54 | 0.0107 | Significant |
| A-Moisture | 3.85E+04 | 1 | 3.85E+04 | 14.86 | 0.0612 | |
| B-pH | 1.30E+05 | 1 | 1.30E+05 | 50.2 | 0.0193 | |
| C-Sucrose | 5.86E+05 | 1 | 5.86E+05 | 226.16 | 0.0044 | |
| Yeast extract | 2.42E+04 | 1 | 2.42E+04 | 9.34 | 0.0924 | |
| E- NaH2PO4 | 1.20E+04 | 1 | 1.20E+04 | 4.64 | 0.1644 | |
| AC | 1.65E+05 | 1 | 1.65E+05 | 63.81 | 0.0153 | |
| AD | 1.47E+05 | 1 | 1.47E+05 | 56.8 | 0.0172 | |
| AE | 1.29E+05 | 1 | 1.29E+05 | 49.71 | 0.0195 | |
| BC | 9.35E+04 | 1 | 9.35E+04 | 36.1 | 0.0266 | |
| BD | 1.18E+05 | 1 | 1.18E+05 | 45.4 | 0.0213 | |
| BE | 2.59E+05 | 1 | 2.59E+05 | 100.05 | 0.0098 | |
| CE | 4.73E+05 | 1 | 4.73E+05 | 182.53 | 0.0054 | |
| DE | 1.15E+05 | 1 | 1.15E+05 | 44.47 | 0.0218 | |
| ABC | 5.25E+05 | 1 | 5.25E+05 | 202.77 | 0.0049 | |
| ABD | 5.20E+04 | 1 | 5.20E+04 | 20.07 | 0.0464 | |
| ABE | 2.15E+04 | 1 | 2.15E+04 | 8.31 | 0.1022 | |
| ACD | 1.08E+05 | 1 | 1.08E+05 | 41.73 | 0.0231 | |
| ACE | 3.61E+05 | 1 | 3.61E+05 | 139.45 | 0.0071 | |
| ADE | 6.93E+05 | 1 | 6.93E+05 | 267.6 | 0.0037 | |
| BCD | 1.70E+05 | 1 | 1.70E+05 | 65.49 | 0.0149 | |
| BCE | 5.36E+04 | 1 | 5.36E+04 | 20.7 | 0.0451 | |
| BDE | 5.46E+05 | 1 | 5.46E+05 | 210.76 | 0.0047 | |
| CDE | 7.13E+04 | 1 | 7.13E+04 | 27.5 | 0.0345 | |
| ABCD | 1.90E+04 | 1 | 1.90E+04 | 7.34 | 0.1135 | |
| ABCE | 4.05E+05 | 1 | 4.05E+05 | 156.33 | 0.0063 | |
| ABDE | 1.61E+05 | 1 | 1.61E+05 | 62.16 | 0.0157 | |
| ACDE | 3.12E+05 | 1 | 3.12E+05 | 120.45 | 0.0082 | |
| BCDE | 7.84E+05 | 1 | 7.84E+05 | 302.78 | 0.0033 | |
| ABCDE | 3.79E+05 | 1 | 3.79E+05 | 146.08 | 0.0068 | |
| Residual | 5.18E+03 | 2 | 2.59E+03 | |||
| Cor Total | 6.96E+06 | 31 |
3.5. Central composite design and response surface methodology
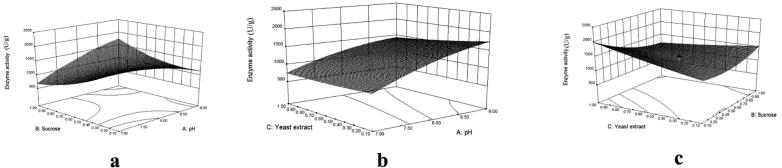
The concentrations of sucrose, yeast extract, and pH value were chosen for optimizing medium composition. Table 2b shows the experimental result of CCD. The designed model was well fit to the quadratic response surface model. The following regression equation of the second-order model provides the levels of fibrinolytic protease activity as a function of sucrose, yeast extract, and pH.
| Enzyme activity = +1282.03 + 100.01A − 134.94B + 92.92C + 428.38AB − 61.62AC − 268.12BC − 121.57A2 + 187.79B2 + 28.34C2 |
where Y is the fibrinolytic activity (U/g), and A, B, and C were sucrose, yeast extract, and pH, respectively.
The F-value of the CCD model was 97.55, and the corresponding p-value was <0.001 (Table 2c). In this model, p-value was < 0.05 for the variables pH, sucrose, and yeast extract. These suggested that the concentration of sucrose, yeast extract, and pH of the culture medium has significant impact on fibrinolytic enzyme production. The p-value of the lack of fit was 0.1081, which revealed that the lack of fit of this model was non-significant. Non-significant lack of fit is good. The coefficient of determination (R2) for this model was 0.9887. 3D response surface curves explained the interactive effects of variables on fibrinolytic enzyme production (Fig. 3a–c). The optimal levels of the important variables for the maximum fibrinolytic protease production for A. oryzae IND3 were as follows: pH 9.0, 0.96% sucrose, and 0.1% yeast extract. Fibrinolytic enzyme production was 2293 ± 12.8 U/g at these optimized concentration. The observed result (2293 ± 12.8 U/g) was close with the predicted response (2340 U/g). Results revealed that the developed model was very highly accurate and reliable for predicting the response.
Table 2c.
Analysis of variance for central composite design.
| Source | Sum of Squares | df | Mean Square | F-Value | p-Value | |
|---|---|---|---|---|---|---|
| Model | 3.38E+06 | 9 | 3.75E+05 | 97.55 | <0.0001 | Significant |
| A-pH | 1.37E+05 | 1 | 1.37E+05 | 35.54 | 0.0001 | |
| B-Sucrose | 2.49E+05 | 1 | 2.49E+05 | 64.62 | <0.0001 | |
| C-Yeast extract | 1.18E+05 | 1 | 1.18E+05 | 30.64 | 0.0002 | |
| AB | 1.47E+06 | 1 | 1.47E+06 | 381.46 | <0.0001 | |
| AC | 3.04E+04 | 1 | 3.04E+04 | 7.89 | 0.0185 | |
| BC | 5.75E+05 | 1 | 5.75E+05 | 149.44 | <0.0001 | |
| A2 | 2.13E+05 | 1 | 2.13E+05 | 55.34 | <0.0001 | |
| B2 | 5.08E+05 | 1 | 5.08E+05 | 132.06 | <0.0001 | |
| C2 | 1.16E+04 | 1 | 1.16E+04 | 3.01 | 0.1136 | |
| Residual | 3.85E+04 | 10 | 3848.45 | |||
| Lack of fit | 29530.99 | 5 | 5906.2 | 3.3 | 0.1081 | Not significant |
| Pure error | 8.95E+03 | 5 | 1790.7 | |||
| Cor total | 3.42E+06 | 19 |
Fig. 3.
Response surface plot showing the effect of pH and sucrose concentration and their interactive effect on the production of fibrinolytic enzyme (a). Response surface plot showing the effect of pH and yeast extract concentration and their interactive effect on the production of fibrinolytic enzyme (b). Response surface plot showing the effect of sucrose and yeast extract concentration and their interactive effect on the production of fibrinolytic enzyme (c).
In industrial point of view, a statistical experimental design played a significant role in enzyme production. RSM is one of the valid such statistical tools for studying the interactive effect of variables and to determine the optimal concentration of selected factors for enzyme production. RSM allows rapid screening of variables with minimum effect. In RSM, 3D helps simple visualization on interactions among variables (Mullai et al., 2010). RSM has been used for the production of fibrinolytic enzyme from Bacillus cereus IND1 and Bacillus natto NLSSE using CCD and RSM (Vijayaraghavan and Vincent, 2014a, Liu et al., 2005). In this study, the designed CCD model was effective and found to be suitable to enhance the production of protease. After statistical approach, fibrinolytic protease yield was 4-fold higher than un-optimized medium.
3.6. Applications of fibrinolytic enzymes
In our study, fibrinolytic enzyme effectively hydrolyzed blood clot directly invitro. The blood clot digestion was 3.8 ± 2.1, 17.8 ± 3.2, 39 ± 3.2, 82.5 ± 3.8, 97 ± 2.9 and 100 ± 2.1% after 2, 4, 6, 8, 10 and 12 h, respectively. In our study, after 12-h incubation, the fibrinolytic enzyme digested blood clot completely and converted blood clot into soluble form. Najafi et al. (2005) previously used crude protease and studied blood clot lytic activity in vitro from Pseudomonas aeruginosa PD100. Because of its blood clot lytic activity of enzyme from X. oryzae IND3, this enzyme may have great applications to treat thrombosis. Europe and Asia are the two major consumers of meat by-products, including lamb and beef. These industries generate blood wastes and in recent years increased attentions have been made to utilize these wastes (Jayathilakan et al., 2012). Also, the application of protease on removal of blood clot was studied by Anwar and Saleemuddin, 1997). In our study, the proteolytic enzyme hydrolyzed waste milk and protein content of waste milk sample was initially 30.02 ± 0.54 mg/ml. About the application of enzyme, it hydrolyzed the available proteins. Results revealed that the milk proteins concentration decreased as 21.07 ± 0.31, 18.3 ± 0.94, 14.8 ± 0.26, 6.03 ± 0.09, 1.29 ± 0.41, and 0.003 ± 0.001 mg/ml, after 10, 20, 30, 40, 50, and 60 min, respectively. In the present study, crude enzyme was subjected to digest proteins from sewage water. The protein content of municipal wastewater was 64.8 ± 0.063 mg/ml. The protein content was decreased as 49 ± 0.38, 37.1 ± 0.29, 23.8 ± 0.27, 1.92 ± 0.16, and 0.027 ± 0.001 mg/ml after, 10, 20, 30, 40, and 50 min incubation with enzyme. After 60 min incubation, the fibrinolytic enzyme digested considerable amount of protein from sewage water. The SDS-PAGE analysis revealed decreased protein content from municipal waste at higher incubation time with enzyme (Fig. 4). The extracellular microbial enzymes could hydrolyze proteins into smaller units (Codoret et al., 2002).
Fig. 4.
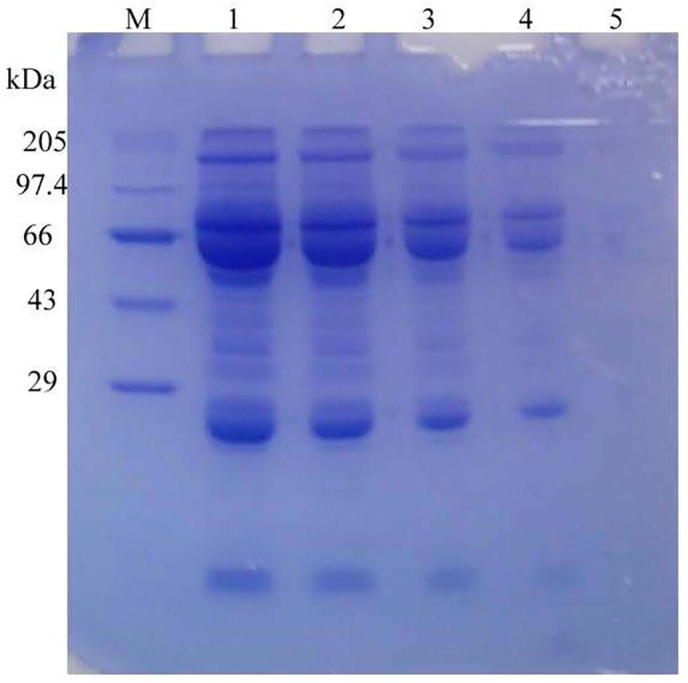
SDS–PAGE results of solubilization of proteins from the municipal wastewater (M: protein marker; Lane 1: protein profile of crude protein precipitated from slaughterhouse wastewater without incubation with enzyme (control); Lane 2: crude protein profile after 15 min digestion with enzyme; Lane 3: digestion after 30 min; Lane 4: digestion after 45 min; Lane 5: digestion after 60 min).
4. Conclusions
Fibrinolytic enzyme has potent application to treat cardiovascular diseases. This study reports the optimization of fibrinolytic protease by Xanthomonas oryzae IND3 using statistical approach in solid state fermentation. It completely digested Goat blood clot invitro after 6 h. Also, the crude extracellular enzyme effectively solubilizes proteins from waste milk from the processing plant and protein waste from municipal wastewater. Considering the efficiency of X. oryzae IND3 fibrinolytic enzyme, this enzyme could be useful in the treatment of cardiovascular diseases.
Conflict of Interest Statement
The authors declared no conflict of interest.
Acknowledgement
The authors would like to extend their sincere appreciation to the Deanship of Scientific Research at King Saud University for its funding of this research through the Research Group Project No. RG-1435-071.
Footnotes
Peer review under responsibility of King Saud University.
Contributor Information
Ponnuswamy Vijayaraghavan, Email: venzymes@gmail.com.
Mariadhas Valan Arasu, Email: mvalanarasu@ksu.edu.sa.
References
- Almalki M.A. Solid state fermentation of agro-residues for the production of amylase from Bacillus subtilis for industrial applications. Int. J. Curr. Microbiol. App. Sci. 2018;7(3):1341–1348. [Google Scholar]
- Amat D., Arora A., Nain L., Nain L. Biomass hydrolyzing enzymes from plant pathogen Xanthomonas axonopodis pv. punicae: optimizing production and characterization. Ann. Microbiol. 2014;64(1):267–274. [Google Scholar]
- Ansen M.L. The estimation of pepsin, trypsin, papain, and cathepsin with hemoglobin. J. Gen. Physiol. 1938;22:79–89. doi: 10.1085/jgp.22.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anwar A., Saleemuddin M. Alkaline-pH-acting digestive enzymes of the polyphagous insect pest Spilosoma obliqua: stability and potential as detergent additives. Biotechnol. Appl. Biochem. 1997;25(1):43–46. [Google Scholar]
- Astrup T., Mullertz S. The fibrin plate method for estimating fibrinolytic activity. Arch. Biochem. Biophys. 1952;40(2):346–351. doi: 10.1016/0003-9861(52)90121-5. [DOI] [PubMed] [Google Scholar]
- Bajaj B.K., Singh S., Khullar M., Singh K., Bhardwaj Optimization of fibrinolytic protease production from Bacillus subtilis I-2 using agro-residues. Braz. Arch. Biol. Technol. 2014;57(5):653–662. [Google Scholar]
- Cadoret A., Conrad A., Block J.C. Availability of low and high molecular weight substrates to extracellular enzymes in whole and dispersed activated sludges. Enzy. Microb. Technol. 2002;31:179–186. [Google Scholar]
- Collen D., Gold H.K. New developments in thrombolytic therapy. Adv. Exp. Med. Biol. 1990;281:333–354. [PubMed] [Google Scholar]
- Da Silva C.R., Silva M.L.C., Kamida H.M., Goes-Neto A., Koblitz M.G.B. Lytic enzyme production optimization using low-cost substrates and its application in the clarification of xanthan gum culture broth. Food Sci. Nut. 2014;2(4):299–307. doi: 10.1002/fsn3.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon G.S., Brar S.K., Kaur S., Valero J.R. Potential of apple pomace as a solid substrate for fungal cellulase and hemicellulase bioproduction through SSF. Ind. Crops Prod. 2012;38:6–13. [Google Scholar]
- Dow J.M., Fan M.J., Newman M.A., Daniels M.J. Differential expression of conserved protease genes in crucifer-attacking pathovars of Xanthomonas campestris. Appl. Environ. Microbiol. 1993;59(12):3996–4003. doi: 10.1128/aem.59.12.3996-4003.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayathilakan K., Sultana K., Radhakrishna K., Bawa A.S. Utilization of byproducts and waste materials from meat, poultry and fish processing industries: A review. J. Food Sci. Technol. 2012;49:278–293. doi: 10.1007/s13197-011-0290-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhample S.B., Bhagwat P.K., Dandge P.B. Statistical media optimization for enhanced production of fibrinolytic enzyme from newly isolated Proteus penneri SP-20. Biocat. Agric. Biotechnol. 2015;4(3):370–379. [Google Scholar]
- Johnvesly B., Manjunath B.R., Naik G.R. Pigeon pea waste as a novel, inexpensive, substrate for production of a thermostable alkaline protease from thermoalkalophilic Bacillus sp. JB-99. Bioresour. Technol. 2002;82:61–64. doi: 10.1016/s0960-8524(01)00147-x. [DOI] [PubMed] [Google Scholar]
- Jung J., Xing X.H., Matsumoto K. Recoverability of protease released from disrupted excess sludge and its potential application to enhanced hydrolysis of proteins in wastewater. Biochem. Eng. J. 2002;10:67–72. [Google Scholar]
- Kalashnikova E.E., Chernyshova M.P., Ignatov V.V. The extracellular proteases of the phytopathogenic bacterium Xanthomonas campestris. Microbiology. 2003;72(4):443–447. [PubMed] [Google Scholar]
- Laemmli U.K. Cleavage of structural proteins during assembly of head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Liu J., Xing J., Chang T., Ma Z., Liu H. Optimization of nutritional conditions for nattokinase production by Bacillus natto NLSSE using statistical experimental methods. Process Biochem. 2005;40:2757–2762. [Google Scholar]
- Lowry O.H., Rosebrough N.J., Farr A.L., Randall R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Mao W., Pan R., Freedman D. High production of alkaline protease by Bacillus licheniformis in a fed-batch fermentation using a synthetic medium. J. Ind. Microbiol. 1992;11:1–6. [Google Scholar]
- Margesin R., Schinner F. Characterization of metalloprotease from psychrophilic Xanthomonas maltophilia. FEMS Microbiol. Lett. 1991;79(2–3):257–262. [Google Scholar]
- Mukherjee A.K., Rai S.K. A statistical approach for the enhanced production of alkaline protease showing fibrinolytic activity from a newly isolated Gram-negative Bacillus sp. strain AS-S20-1. New Biotechnol. 2011;28:182–189. doi: 10.1016/j.nbt.2010.11.003. [DOI] [PubMed] [Google Scholar]
- Mukherjee A.K., Adhikari H., Rai S.K. Production of alkaline protease by a thermophilic Bacillus subtilis under solidstate fermentation (SSF) condition using Imperata cylindrical grass and potato peel as low-cost medium: characterization and application of enzyme in detergent formulation. Biochem. Eng. J. 2008;39:353–361. [Google Scholar]
- Mullai P., Fathima N.S.A., Rene E.R. Statistical analysis of main and interaction effects to optimize xylanase production under submerged cultivation conditions. J. Agric. Sci. 2010;2:144–153. [Google Scholar]
- Najafi M.F., Deobagkar D., Deobagkar D. Potential application of protease isolated from Pseudomonas aeruginosa PD100. Electron. J. Biotechnol. 2005;8:197–203. [Google Scholar]
- Peng Y., Huang Q., Zhang R.H., Zhang Y.Z. Purification and characterization of a fibrinolytic enzyme produced by Bacillus amyloliquefaciens DC-4 screened from douche, a traditional Chinese soybean food. Comp. Biochem. Physiol. Part B. 2003;134:45–52. doi: 10.1016/s1096-4959(02)00183-5. [DOI] [PubMed] [Google Scholar]
- Phadatare S.V., Srinivasan M.C., Deshpande V.V. High activity alkaline protease from Conidiobolus coronatus (NCL 86. 8. 20); enzyme production and compatibility with commercial detergents. Enzy. Microb. Technol. 1993;15:72–76. [Google Scholar]
- Ramkumar A., Sivakumar N., Victor R. Fish waste-potential low cost substrate for bacterial protease production: a brief review. Open Biotechnol. J. 2016;10:335–341. [Google Scholar]
- Ravindran B., Ganesh Kumar A., Aruna Bhavani P.S., Sekaran G. Solid-state fermentation for the production of alkaline protease by Bacillus cereus 1173900 using proteinaceous tannery solid waste. Curr. Sci. 2011;100(5):726–730. [Google Scholar]
- Sadh P.K., Duhan S., Duhan J.S. Agro-industrial wastes and their utilization using solid state fermentation: a review. Bioresour. Bioprocess. 2018;2018(5):1. [Google Scholar]
- Silva C.R., Silva M.L.C., Kamida H.M., Goes-Neto A., Koblitz M.G.B. Lytic enzyme production optimization using low-cost substrates and its application in the clarification of xanthan gum culture broth. Food Sci. Nut. 2014;2(4):299–307. doi: 10.1002/fsn3.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S., Bajaj B.K. Bioprocess optimization for production of thermoalkali-stable protease from Bacillus subtilis K-1 under solid-state fermentation. Prep. Biochem. Biotech. 2016;46(7:717–724. doi: 10.1080/10826068.2015.1135455. [DOI] [PubMed] [Google Scholar]
- Sole M., Scheibner F., Hoffmeister A.K., Hartmann H., Hause G., Rother A., Jordan M., Lautier M., Arlat M., Buttner D. Xanthomonas campestris pv. vesicatoria Secretes Proteases and Xylanases via the Xps Type II Secretion System and Outer Membrane Vesicles. J. Bacteriol. 2015;197(17):2879–2893. doi: 10.1128/JB.00322-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X.P. North Carolina State University; Raleigh: 1991. Isolation and characterization of an extracellular protease produced by Xanthomonas campestris pv. zinniae and its role in pathogenesis of bacterial spot of zinnia (Zinnia elegans Jacq.). Dissertation (Ph.D.) p. 78. [Google Scholar]
- Tao S., Peng L., Beihui L., Deming L., Zuohu L. Solid state fermentation of rice chaff for fibrinolytic enzyme production by Fusarium oxysporum. Biotechnol. Lett. 1997;19:465–467. [Google Scholar]
- Tsuchiya K., Sakashita H., Nakamura Y., Kimura T. Production of thermostable alkaline protease by alkalophilic Thermoactinomyces sp. HS682. Agric. Biol. Chem. 1991;55:3125–3127. [Google Scholar]
- Vijayaraghavan P., Vincent S.G.P. Statistical optimization of fibrinolytic enzyme production using agroresidues by Bacillus cereus IND1 and Its thrombolytic activity in vitro. BioMed Res. Int. 2014:11. doi: 10.1155/2014/725064. Article ID 725064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayaraghavan P., Vincent S.G.P. Medium optimization for the production of fibrinolytic enzyme by Paenibacillus sp. IND8 using response surface methodology. Sci. World J. 2014:9. doi: 10.1155/2014/276942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayaraghavan P., Arun A., Vincent S.G.P., Arasu M.V., Al-Dhabi N.A. Cow dung is a novel feedstock for fibrinolytic enzyme production from newly isolated Bacillus sp. IND7 and its application in in vitro clot lysis. Front. Microbiol. 2016;7:361. doi: 10.3389/fmicb.2016.00361. [DOI] [PMC free article] [PubMed] [Google Scholar]