Abstract
Genetic variation in fish stocks decreasing due to water pollution in the freshwater rivers, streams and canals. The objective of this study was to determine the genetic diversity and polymorphism in Oreochromis niloticus collected from the Wadi Hanefah Riyadh, Saudi Arabia by using RAPD-PCR. Total thirty fish specimens were harvested from each of four pre-determined locations of the reservoir which were designated as H1, H2, H3, and H4. Five random decamer primers were used to assess the diversity in the stock of O. niloticus. In this fish stock 48 bands were polymorphic and 12 were monomorphic. The maximum polymorphism (100%) was recorded in the fish samples procured from H4, followed by 88.75, 87.33 and 76.12% of the tilapia collected from H3, H2, and H4, respectively. Nei’s genetic distance value was ranged as 0.0005 to 0.1006. Maximum and minimum genetic distance was recorded as 0.1006 and 0.005 in tilapia harvested from H1 and H2 locations. Average heterozygosity was ranged from 0.3009 to 0.3744. This information about the genetic polymorphism of O. niloticus may be used by the concerned authorities to evolve strategies to conserve the diversity of tilapia in the country.
Keywords: Genetic diversity, Tilapia, Primers, RAPD
1. Introduction
The tilapia (Oreochromis niloticus) is widely cultured across the globe (Gottschalk et al., 2015). O. niloticus is categorized as a member of Cyprinidae (Nelson, 1994). The tilapia is cultured in every part of the world. Tilapia feeds on planktonic life to fulfill their metabolic requirements (Shair et al., 2011). The population of tilapia (O. niloticus) is gradually decreasing in water reservoirs due to anthropogenic activities, and water pollution and habitat degradation (Abder-Kader et al., 2013).
Genetic variation is necessary for natural stock of fish for the evolution purpose and to ensure their availability for the future generation (Xia et al., 2014, Xia et al., 2015). The decrease in genetic diversity in wild stock of freshwater fish species may influence on the adjustment of the fish into their changing environment (Arif and Khan, 2009, Mkare et al., 2017). The evolutionary variations are caused due to spontaneous mutation, migration, and genetic drift in the freshwater fish (Zhao et al., 2011). Genetic variation helps the s fish species to adjust in changing environment which is important for their survival. The information about the genetic structure is required to conserve the natural stocks (Carlson et al., 2015, Chauhan et al., 2007). The conservation of allelic variation is necessary for maintaining the genetic integrity and to conserve the natural stocks of freshwater fish (Perrier et al., 2011). The continuous monitoring of fish stocks in freshwater reservoirs is necessary to overcome their genetic decline (Alam et al., 2009, Islam et al., 2005). The information related about the diversity in natural stocks of fish is required to plan stocking program to observe the variation in genotype frequencies (Chupania et al., 2006, Frankham, 2010). Fisheries scientists are using different biotechnological techniques to explain the genetic diversity in fish culture (Okumus and Çiftci, 2003, Yousefian et al., 2011). DNA markers are the most commonly used tool for the conservation of fish stocks at a minimum level (Carlson et al., 2015).
The “randomly amplified polymorphic DNA (RAPD) technique is used for the investigation of genetic parameters, both in wild and inland fish stocks” (Alam and Islam, 2005). “Different biomarkers such as Microsatellites, RAPDs, AFLPs, RFLPs used to estimate the genetic variation to plan strategies for the conservation of fish population, sex determination, identification of disease carriers, and transgenesis (Figueras et al., 2016, Basavaraju et al., 2014)”. RAPDs is also in practice to determine the effect of contamination on genetic content (Liua and Cordes, 2004). The aim of this research work was to assess the genetic diversity and polymorphism in the tilapia stocks Wadi Haneffah, Riyadh, Saudi Arabia through RAPD analysis, and also to propose strategy for conservation of tilapia stocks in the freshwater reservoirs.
2. Materials and methods
2.1. Study area
“Wadi Hanefah is also known as Riyadh River/Riyadh Lake and has a length of 120 km (75 mi) from northwest to southeast, cutting through the city of Riyadh, Saudi Arabia. Riyadh City has a population of approximately 4 million. This water reservoir receives water treated by the Riyadh municipality’s sewerage system and untreated discharge from local industry and adjacent areas along the length of the river. The water of this reservoir is used for irrigation of various fruit farms and vegetables grown in adjacent areas” (Mahboob et al., 2014).
2.2. Sampling of fish
Oreochromis niloticus samples (120) was collected from four different sampling locations viz., “Wadi Labn, Wadi Ubayr, Wadi Liha, and Al-Hair”, which were designated as H1, H2, H3, and H4 in the Wadi Hanefah, Riyadh Saudi Arabia. Blood was collected from each fish and was stored in 95% ethyl alcohol at −20 °C. These samples were shifted to the Lab for extraction of DNA.
2.3. Isolation of DNA
Genomic DNA was isolated by the methodology of by Sambrook et al. (1989) with slight changes as explained by Yue and Orban (2005).
2.4. PCR amplification of RAPD loci
“Five random decamer primers (OPA-02, OPA-04, OPA-05, OPA-08, and OPA-09) purchased from Operon Technologies” were used in the estimation of polymorphisms. “Extracted genomic DNA was amplified by PCR” (Table 1). “The sequences of the primers were taken from the literature (Chandra et al., 2010) and oligonucleotides were custom synthesized by Eurofins genomics, Canada. Specific parental band profiles were generated by these five primers in at least three replicate PCRs. For non-denatured gel electrophoresis, 40% acrylamide gel was used”.
Table 1.
Random decamer primers with their Primer sequence, GC content, and annealing temperature.
| Sr. No | Locus | Primer sequence (5′-3′) | G + C (%) | Tm (C) |
|---|---|---|---|---|
| 1 | OPA-02 | TGCCGAGCTG | 60 | 32 °C |
| 2 | OPA-04 | AATCGGGCTG | 60 | 32 °C |
| 3 | OPA-05 | AGGGGTCTTG | 60 | 32 °C |
| 4 | OPA-08 | AGGGGTCTTG | 60 | 34 °C |
| 5 | OPA-09 | GTGACGTAGG | 60 | 34 °C |
2.5. Similarity analysis
“RAPD-ISSR data was used to generate a similarity matrix using the” Nei and Lei (1979) method.
2.6. Data analysis
“The genotypic data obtained from band counting were analyzed using the programs POPGENE and TFPGA. The genotype data for each locus were subjected to accurate analysis to estimate genetic diversity in the stock of O. niloticus. The banding patterns generated by RAPD markers were scored on the basis of presence or absence of visible, clear, and reproducible bands. The presence of the band was scored 1 and the absence of the band was scored 0. The RAPD loci were utilized for determination of genetic diversity, the number of polymorphic loci, and genetic distance. They were also used to construct an unweighted pair group method for the arithmetic mean for the UPGMA dendrogram for the population using Nei’s unbiased distance” (Ambak et al., 2006).
For every sample, “the proportion of polymorphic loci (P %), as well as the meaning of genetic diversity (H %) was calculated by POPGENE v.1.31. POPGENE software was utilized for analysis of polymorphic loci, genetic diversity within the population, genetic diversity between populations, and construction of a dendrogram based on Nei’s unbiased genetic distances. Genetic distances were estimated by utilizing TFPGA (Tool for population genetic analysis”).
3. Results
3.1. RAPD analysis
In this study 48 bands were polymorphic and 12 bands were monomorphic (Table 2) from four sampling sites. The highest bands were observed in the tilapia harvested from H4 location produced by primer OPA-04 and the minimum (10 bands) in O. niloticus procured from H1 by primer OPA-02.
Table 2.
Polymorphic and Monomorphic bands.
| Locus | No. of polymorphic bands | No. of monomorphic bands |
|---|---|---|
| OPA-02 | 9 | 3 |
| OPA-04 | 15 | 4 |
| OPA-05 | 13 | 0 |
| OPA-08 | 13 | 0 |
| OPA-09 | 13 | 3 |
| Total | 50 | 10 |
3.2. Polymorphism among the primers
The polymorphism % in the O. niloticus was recorded as: OPA-05 (99.9%) > OPP-08 (88.75%) > OPA-02 (85.10%) > OPA-04 (83.52%) > OPA-09 (63.40%). Out of 5 primers the maximum and minimum polymorphism was observed at 99.9 and 63.40% through OPA-05 and OPA-09 primers (Table 3). The maximum (99.5%) polymorphism out of 5 primers was recorded in O. niloticus collected from H3 (Table 3).
Table 3.
Total number of amplified fragments, number of polymorphic bands, and percentage polymorphisms generated by PCR using five primers.
| Primers | Band pattern | Populations |
Total no. of bands | |||
|---|---|---|---|---|---|---|
| WH1 | WH2 | WH3 | WH4 | |||
| OPA-02 | P | 1 | 4 | 2 | 2 | 9 |
| M | 0 | 2 | 1 | 0 | 3 | |
| %P | 100.00 | 66.66 | 70.23 | 100.00 | 84.22% | |
| OPA-04 | P | 5 | 5 | 3 | 2 | 15 |
| M | 1 | 2 | 1 | 0 | 4 | |
| %P | 83.33 | 71.42 | 75.00 | 100.00 | 82.43% | |
| OPA-05 | P | 4 | 4 | 3 | 2 | 13 |
| M | 0 | 0 | 0 | 0 | 0 | |
| %P | 100.00 | 100.00* | 100.00 | 100.00 | 100.00% | |
| OPA-08 | P | 1 | 5 | 4 | 3 | 13 |
| M | 1 | 0 | 0 | 0 | 0 | |
| %P | 50.00 | 100.00 | 100.00 | 100.00 | 87.50% | |
| OPA-09 | P | 2 | 4 | 3 | 4 | 13 |
| M | 2 | 0 | 0 | 0 | 2 | |
| %P | 50.00 | 100.00 | 100.00 | 100* | 62.50% | |
| Average %P | 76.12% | 87.62% | 88.75% | 100% | ||
P - Polymorphic bands; M - Monomorphic bands.
Primer showing highest percent polymorphism**Primer showing lowest percent polymorphism.
3.3. Genetic diversity
The genetic variation in the natural stocks of O. niloticus harvested from the Wadi Haneffah showed a decline in the genetic variation due to water pollution and other human activities, genetic drift, and inbreeding.
3.4. Allelic frequency
“Allelic frequencies for each locus in the population of O. niloticus collected from four different locations is given” in Table 4. Null alleles were observed with a value of 1 in the fish harvested from H1, H2, H3 and H4.
Table 4.
Allele frequencies in four populations of Oreochromis niloticus at five loci.
| Locus Name | Populations |
|||||||
|---|---|---|---|---|---|---|---|---|
| WH1 |
WH2 |
WH3 |
WH4 |
|||||
| #Obs | Allele frequency | #Obs | Allele frequency | # Obs | allele frequency | # Obs | Allele frequency | |
| OPA-02 | 24 | 0.565 | 26 | 0.642 | 26 | 0.645 | 27 | 0.687 |
| 6 | 0.435 | 4 | 0.358 | 4 | 0.355 | 3 | 0.313 | |
| OPA-04 | 27 | 0.701 | 26 | 0.650 | 26 | 0.646 | 26 | 0.640 |
| 3 | 0.299 | 4 | 0.350 | 4 | 0.344 | 4 | 0.360 | |
| OPA-05 | 27 | 0.672 | 25 | 0.597 | 26 | 0.627 | 27 | 0.687 |
| 3 | 0.328 | 5 | 0.403 | 4 | 0.373 | 3 | 0.313 | |
| OPA-08 | 4 | 0.092 | 26 | 0.642 | 26 | 0.632 | 26 | 0.637 |
| 26 | 0.908 | 4 | 0.358 | 4 | 0.368 | 4 | 0.363 | |
| OPA-09 | 30 | 1.0000 | 30 | 1.0000 | 30 | 1.0000 | 30 | 1.0000 |
| 0 | 0.0000 | 0 | 0.0000 | 0 | 0.0000 | 0 | 0.0000 | |
| Null alleles | 1 | 1 | 1 | 1 | ||||
3.5. Heterozygosity
The heterozygosity was ranged from 0.380 to 0.475. The heterozygosity was 0.380, 0.468, 0.463, and 0.475 in O. niloticus collected from H1, H2, H3, and H4, respectively (Table 5).
Table 5.
Heterozygosity values in four populations of Oreochromis niloticus at five loci.
| Locus name | Populations |
|||||||
|---|---|---|---|---|---|---|---|---|
| WH1 |
WH2 |
WH3 |
WH4 |
|||||
| #hets | het freq | # hets | het freq | # hets | het freq | # hets | het freq | |
| OPA-02 | 15.31 | 0.495 | 13.96 | 0.465 | 12.95 | 0.436 | 13.94 | 0.467 |
| 15.31 | 0.495 | 13.96 | 0.465 | 12.95 | 0.436 | 13.94 | 0.467 | |
| OPA-04 | 13.33 | 0.431 | 13.97 | 0.467 | 13.93 | 0.466 | 13.93 | 0.469 |
| 13.33 | 0.431 | 13.97 | 0.467 | 13.93 | 0.466 | 13.93 | 0.469 | |
| OPA-05 | 13.30 | 0.430 | 13.98 | 0.469 | 12.99 | 0.437 | 14.54 | 0.487 |
| 13.30 | 0.430 | 13.98 | 0.469 | 12.99 | 0.437 | 14.54 | 0.487 | |
| OPA-08 | 4.83 | 0.158 | 13.97 | 0.468 | 13.94 | 0.469 | 13.95 | 0.470 |
| 4.83 | 0.158 | 13.97 | 0.468 | 13.94 | 0.469 | 13.95 | 0.470 | |
| OPA-09 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.0000 | |
| Average Heterozygosity | 0.378 | 0.467 | 0.452 | 0.473 | ||||
hets: heterozygosity; het freq: heterozygosis frequency.
3.6. UPGMA cluster analysis
The phylogenetic tree exhibited the presence of 3 clusters (Fig. 1). The first cluster, second and third cluster was formed between the H4 and H3, the H2 and H3 tilapia stocks, and the fish collected from H1 showed a separate cluster. Genetic distance was highest in the fish harvested from H1 and H3, which showed a heterozygous genotype. “The minimum heterozygosity recorded between the fish collected HM and HK, which showed a homogenate genotype. Node distance included population (1) 0.0005, 2, 3 (2) 0.0025 2, 3, 4, and (3) 0.0996, 1, 2, 3, 4.”
Fig. 1.
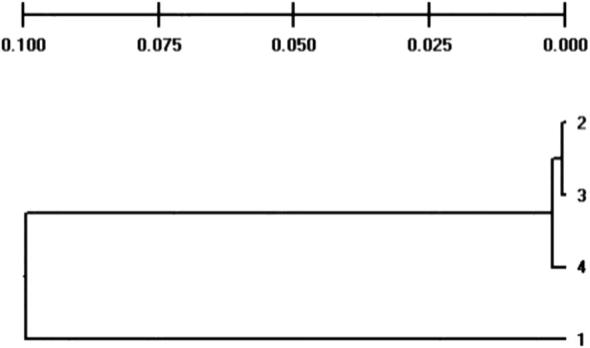
Dendrogram construction based on Nei’s genetic distance among Oreochromis niloticus populations.
4. Discussion
O. niloticus harvested from H1, H2, and H3 showed the maximum of genetic variation (99.99) caused by OPA-02, OPA-05, and OPA-08. The fish introgressive hybridization was due to discharge of untreated domestic and industrial waste. Isoenzyme analysis of O. niloticus specimens showed genetic diversity in the fish population from the same region and river (Kohlmann and Kersten, 1999). The maximum and minimum polymorphism was recorded as 99.99 and 63.40% by OPA-05 and OPA-09 (Table 2). The polymorphism was higher than the percentage obtained by Li and Chu-Wu (2006) by RAPD analysis. Out of 60 bands, 48 bands showed polymorphism. Basavaraju et al. (2007) reported 57.1% polymorphism in tilapia. Basavaraju et al. (2014) used 8 random primers to study genetic diversity in L. fimbriatus and observed polymorphic bands. We had recorded the highest genetic variation in O. niloticus collected from H4 location and lowest from H1, which indicates fish from H4 have more heterozygous genotypes. The similar findings were also reported by Chandra et al. (2010).
“Genetic distance ranged between 0.0005 and 0.0996. The highest and lowest genetic was recorded in the fish stocks obtained from H1 and H2, respectively”. Ji et al. (2014) studied the genetic distance in five populations of Megalobrama amblycephala and reported genetic variation. The highest heterozygosity in O. niloticus harvested from H1. A low level of genetic variation was observed in the fish stock collected from four different locations (Gopalakrishnan et al., 2009). These results were not consistent with findings of Kohlmann and Kersten (1999), they mentioned varying diversity in fish stocks. The cluster analysis exhibited the fish collected from H3 was resemble to H4, while the tilapia collected from H1 was genetically distant from the other stocks. Basavaraju et al. (2014) observed two cluster in a group of three different stock. Bartfai et al. (2003) reported no grouping in the stock of common carp. The loss in genetic variation in the fish stocks of O. niloticus in Wadi Haneffah, Riyadh, Saudi Arabia due to increased load of domestic and industrial waste. The increasing loss in the genetic variation in the tilapia may decrease its potential to overcome the habitat degradation due to anthropogenic activities (Milligan et al., 1994).
5. Conclusion
It has been concluded that RAPD technique is very useful to collect data about the genetic variation in the wild stock of fish population of the same geographic region. This information may be helpful to plan strategies for improvement in the breeding program by the fisheries biologist.
Acknowledgements
“The authors (SM, FAM and NAM) would like to express their sincere appreciation to the Deanship of Scientific Research at the King Saud University for its funding of this research through the Research Group Project No. RG-1435-012. The authors also thank the Deanship of Scientific Research and RSSU at King Saud University for their technical support”.
Footnotes
Peer review under responsibility of King Saud University.
References
- Abder-Kader A.M., Abdel-Hamid Z.G., Mahrous A.K.F. Genetic diversity among three species of tilapia in Egypt detected by Random Amplified Polymorphic DNA Marker. J. Appl. Biol. Sci. 2013;7(2):57–64. [Google Scholar]
- Alam M.S., Islam M.S. Population genetic structure of Catla catla (Hamilton) revealed by microsatellite DNA markers. Aquaculture. 2005;246:151–160. [Google Scholar]
- Alam Md.S., Maraya J., Hossain M.Md., Islam S. Population genetic structure of three major river populations of Rohu, Labeo rohita (Cyprinidae: Cypriniformes) using microsatellite DNA markers. Genes Genom. 2009;31:43–51. [Google Scholar]
- Ambak M.A., Bolong A.A., Ismail P., Tam B.M. Genetic variation of Snakehead fish (Channa striata) populations using random amplified polymorphic DNA. Biotechnology. 2006;5:104–110. [Google Scholar]
- Arif I.A., Khan H.A. Molecular markers for biodiversity analysis of wildlife animals: a brief review. Anim. Biodiv. Conserv. 2009;32:9–17. [Google Scholar]
- Bartfai R., Sandor E., Yue G., Kovacs B., Urbanyi G., Tamas L.H., Orban L. Genetic analysis of two common carp brood stocks by RAPD and microsatellite markers. Aquaculture. 2003;219:157–167. [Google Scholar]
- Basavaraju Y., Narasimha A., Reddy K., Rajanna B., Chethan N. Random amplified polymorphic DNA (RAPD) analysis of three stocks of Labeo fimbriatus from Indian peninsula. Global J. Biosci. Biotechnol. 2014;3:278–283. [Google Scholar]
- Basavaraju Y., Prasad D.T., Rani K., Kumar S.P., Naika U.D., Jahageerdar S., Srivastava P.P., Penman D.J., Mair G.C. Genetic diversity in common carp stocks assayed by random amplified polymorphic DNA markers. Aquacul. Res. 2007;38:147–155. [Google Scholar]
- Carlson S.M., Cunningham C.J., Westley P.A.H. Evolutionary rescue in a changing world. Trends Ecol. Evol. 2015;29:521–530. doi: 10.1016/j.tree.2014.06.005. [DOI] [PubMed] [Google Scholar]
- Chandra G., Saxena A., Barat Genetic diversity of two riverine populations of Eutropiichthys vacha (Hamilton, 1822) using RAPD markers and implications for its conservation. J. Cell and Mol. Biol. 2010;8(2):77–85. [Google Scholar]
- Chauhan T., Lal K.K., Mohindra V., Singh R.K., Punia P., Gopalakrishnan A., Sharma P.C., Lakra W.S. Evaluating genetic differentiation in wild populations of the Indian major carp, Cirrhinus mrigala (Hamilton-Buchanan, 1882): evidence from allozyme and microsatellite markers. Aquaculture. 2007;269:135–149. [Google Scholar]
- Chupania L., Niksirata H., Velíšeka J., Staráa A., Hradilováb S., Kolaříkb J., Zuskováa, El-Zaeem S.Y., Ahmed M.M.M. Genetic differentiation between sex reversal and normal of full-sib Nile Tilapia (Oreochromis niloticus) based on DNA finger printing. Res. J. Fish. Hydrobiol. 2006;1:1–5. [Google Scholar]
- Figueras A., Robledo D., Corvelo A., Hermida M., Pereiro P., Rubiolo J.A., Gómez-Garrido J., Carreté L., Bello X., Gut M., Gut I.G., Marcet-Houben M., Forn-Cuní G., Galán B., García J.L., Abal-Fabeiro J.L., Pardo B.G., Taboada X., Fernández C., Vlasova A., Hermoso-Pulido A., Guigó R., Álvarez-Dios J.A., Gómez-Tato A., Viñas A., Maside X., Gabaldón T., Novoa B., Bouza C., Alioto T., Martínez P. Whole genome sequencing of turbot (Scophthalmus maximus; Pleuronectiformes): a fish adapted to demersal life. DNA Res. 2016 doi: 10.1093/dnares/dsw007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankham R. Challenges and opportunities of genetic approaches to biological conservation. Biol. Conserv. 2010;143:1919–1927. [Google Scholar]
- Gopalakrishnan A., Musammilu K.K., Basheer V.S., John L., Padmakumar K.G., Lal K.K., Mohindra V., Punia P., Dinesh K., Manjebrayakath H., Ponniah K.K., Lakra W.S. Low genetic differentiation in the populations of the Malabar carp, Labeo dussumier as revealed by Allozymes, Microsatellites and RAPD. J. Fish. Hydrobiol. 2009;22:359–391. [Google Scholar]
- Gottschalk F., Lassen C., Kjoelholt J., Christensen F., Nowack B. Modeling flows and concentrations of nine engineered nanomaterials in the Danish environment. Int. J. Environ. Res. Public Health. 2015;12:5581–5602. doi: 10.3390/ijerph120505581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M.S., Ahmed A.S.I., Azam M.S., Alam M.S. Genetic analysis of three river populations of Catla (Hamilton) using randomly amplified polymorphic DNA markers. Asian-Austr. J. Anim. Sci. 2005;18:453–457. [Google Scholar]
- Ji W., Zhang G.R., Ran W., Gardner J.P.A., Wei K.J., Wang W.M., Zou W. Genetic diversity of and differentiation among five populations of blunt snout bream (Megalobrama amblycephala) revealed by SRAP markers: implications for conservation and management. PLoS ONE. 2014;9:e108967. doi: 10.1371/journal.pone.0108967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlmann K., Kersten P. Genetic variability of German and foreign common carp (Cyprinus carpio L.) populations. Aquaculture. 1999;173:435–445. [Google Scholar]
- Li L., Chu-Wu L. Genetic diversity and molecular markers of five snapper species. J. Agricul. Biotechnol. 2006;14:349–355. [Google Scholar]
- Liua Z.J., Cordes J.F. DNA marker technologies and their applications in aquaculture genetics. Aquaculture. 2004;238:1–37. [Google Scholar]
- Mahboob S., Al-Balwai H.F.A., Al-Misned F., Al-Ghanim K.A., Ahmad Z. A study on the accumulation of nine heavy metals in some important fish species from a natural reservoir in Riyadh, Saudi Arabia. Toxicol. Environ. Chem. 2014;96:783–798. [Google Scholar]
- Milligan B.G., Leebens-Mack J., Strand A.E. Conservation genetics: beyond the maintenance of marker diversity. Mol. Ecol. 1994;3:423–435. [Google Scholar]
- Mkare T.K., Jansen van Vuuren J., Teske P.R. Conservation implications of significant population differentiation in an endangered estuarine seahorse. Biodiv. Conserv. 2017;26:1275–1293. [Google Scholar]
- Nei M., Lei W.H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proceed Nat. Acad. Sci. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson J. third ed. Wiley; New York: 1994. Fishes of the World. [Google Scholar]
- Okumus I., Çiftci Y. Fish population genetics and molecular markers: II-molecular markers and their applications in fisheries and aquaculture. Turk. J. Fish. Aqua. Sci. 2003;3:51–79. [Google Scholar]
- Perrier C., Rene G., Bagliere J., Evanno G. Determinants of hierarchical genetic structure in Atlantic salmon populations: environmental factors vs. anthropogenic influences. Mol. Ecol. 2011;20:4231–4245. doi: 10.1111/j.1365-294X.2011.05266.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. second ed. Cold Spring Harbor Laboratory Press; New York: 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- Shair M.O.A., Al-Ssum R.M., Bahkhali A.H. Genetic variation investigation of tilapia growth under Saudi Arabian controlled environment. Amm. J. Biochem. Mol. Biol. 2011;1:89–94. [Google Scholar]
- Xia J.H., Wan Z.Y., Ng Z.L., Wang L., Fu G.H., Lin G., Feng L., Gen H.Y. Genome-wide discovery and in silico mapping of gene-associated SNPs in Nile tilapia. Aquaculture. 2014;432:67–73. [Google Scholar]
- Xia J.H., Bai Z., Meng Z., Zhang Y., Wang L., Liu F., Jing W., Wan Z.Y., Li J., Lin H., Yue G.H. Signatures of selection in tilapia revealed by whole genome resequencing. Sci. Rep. 2015;5:14168. doi: 10.1038/srep14168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefian M., Sharifrohani M., Sahafi H.H., Laloei F., Makhdoomi C. Heritability estimation for growth-related traits in juvenile wild common carp (Cyprinus carpio L.) in the south of Caspian Sea. Iranian J. Fish. Sci. 2011;10:740–748. [Google Scholar]
- Yue G.H., Orban L. A simple and affordable method for high-throughput DNA extraction from animal tissues for polymerase chain reaction. Electrophoresis. 2005;26(16):3081–3083. doi: 10.1002/elps.200410411. [DOI] [PubMed] [Google Scholar]
- Zhao K., Duan Z., Peng Z., Gan X., Zhang R., He S., Zhao X. Phylogeography of the endemic Gymnocypris chilianensis (Cyprinidae): sequential westward colonization followed by allopatric evolution in response to cyclical Pleistocene glaciations on the Tibetan Plateau. Mol. Phylogen. Evol. 2011;59:303–310. doi: 10.1016/j.ympev.2011.02.001. [DOI] [PubMed] [Google Scholar]


