Abstract
We provide bacterial 16S rRNA community and hydrochemical data from water and sediments of Lake Neusiedl, Austria. The sediments were retrieved at 5 cm intervals from 30–40 cm push cores. The lake water community was recovered by filtration through a 3.0/0.2 µm filter sandwich. For 16S rRNA gene amplicon-based community profiling, DNA was extracted from the sediment and filters and the bacterial V3-V4 regions were amplified and sequenced using a MiSeq instrument (Illumina). The reads were quality-filtered and processed using open source bioinformatic tools, such as PEAR, cutadapt and VSEARCH. The taxonomy was assigned against the SILVA SSU NR 132 database. The bacterial community structure was visualised in relation to water and porewater chemistry data. The bacterial community in the water column is distinct from the sediment. The most abundant phyla in the sediment shift from Proteobacteria to Chloroflexota (formerly Chloroflexi). Ammonium and total alkalinity increase while sulphate concentrations in the porewater decrease. The provided data are of interest for studies targeting biogeochemical cycling in lake sediments.
Subject terms: Sequencing, Microbial ecology, Environmental microbiology
| Design Type(s) | source-based data analysis objective • biodiversity assessment objective |
| Measurement Type(s) | freshwater metagenome |
| Technology Type(s) | DNA sequencing |
| Factor Type(s) | Environment • depth |
| Sample Characteristic(s) | metagenome • Neusiedlersee • lake |
Machine-accessible metadata file describing the reported data (ISA-Tab format)
Background & Summary
Lake Neusiedl is the largest, seasonally evaporative lake in western Europe covering an area of approximately 315 km2 1. Its sediments show high contents of authigenic high magnesium calcite and poorly ordered dolomite, which have been the focus of multiple studies on sediment formation, geochemistry and water level1–4. There is a strong economic interest in the lake and the surrounding national parks due to their recreational value1,5. Thus, the lake’s water quality, including potential pathogenic microbes, is monitored on a regular basis6–9. Nevertheless, the bacterial community composition of water and sediment remains largely unexplored, particularly in relation to the lakes’ hydrochemistry.
Soft sediment push-cores were taken in the bay of Rust in August 2017 (Fig. 1a). Two 30–40 cm cores were used for bacterial community analysis and one for porewater extraction and analysis. The water (core supernatant) was filtered through a 3.0 and 0.2 µm filter sandwich. All samples for bacterial community analysis were stored in RNAprotect Bacteria Reagent (Qiagen, Hilden, Germany) for transport. The reagent was removed by centrifugation from the samples prior to storage at −80 °C. Metagenomic DNA was extracted from 0.25 g of sediment or one third of a filter. Subsequently, the V3-V4 region of bacterial 16S rRNA genes were amplified using primers described by Klindworth et al.10. After purification with magnetic beads, the amplicons were sequenced, yielding a total of 6,044,032 raw paired-end reads. Bioinformatic processing of the data included quality-filtering and base pair correction of overlapping regions (fastp), read-merging (PEAR), primer clipping (cutadapt), size-selection, dereplication, denoising and chimera removal (VSEARCH). After taxonomic assignment 2,263,812 high-quality 16S rRNA gene sequences remained in the dataset11. Amplicon sequence variants12 (ASVs) with 100% sequence identity were screened with BLASTn against the SILVA SSU 132 NR database for taxonomic assignment. The ASV abundance table13 was used for visualisation of the bacterial community. Total alkalinity (TA) was determined by titration. Major cations and anions were measured by ion-chromatography and ICP-MS was used to determine trace element content. Nutrient concentrations and total sulphide were assessed photometrically14. Porewater chemistry and bacterial community composition were analysed in intervals of 5 cm15 (Fig. 1c).
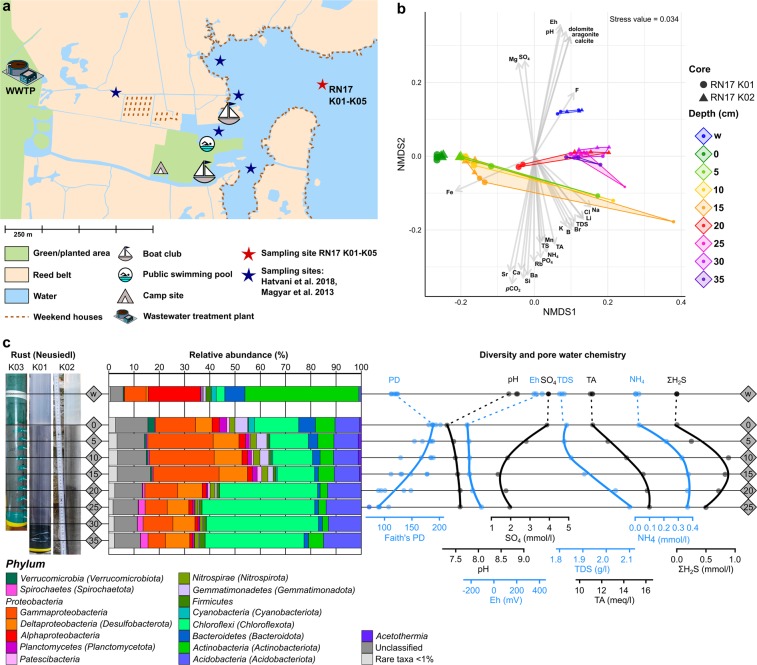
Fig. 1.
Sampling site in the bay of Rust, NMDS and depth profiles of the bacterial community composition and porewater properties. (a) Sampling site of this study (red star) and previous studies (blue stars). Markers for anthropogenic influences, such as a wastewater treatment, holiday houses (brown dashed lines) and recreational sites (pool, boat club, camp site) are indicated by pictograms or dashed lines. (b) Non-metric multidimensional scaling (NMDS) of bacterial communities (n = 47) with the environmental fit (p < 0.01) of porewater properties (grey arrows) based on a weighted generalized UniFrac analysis using the vegan package incorporated into ampvis251,57. Depths are indicated in cm or w (water column) and triangles or circles indicate the sediment core. (c) Sampling depths of the sediment cores (Rust Neusiedl RN-K01 and RN-K02) for bacterial community analysis. Each bacterial phylum depicted here comprises more than 1% relative abundance of the bacterial community in at least one sample. All other amplicon sequence variants (ASVs) are summarized as rare taxa and those with a taxonomic match below 95% sequence identity were summarized as “Unclassified”. The phylum Proteobacteria is shown at class level (Alpha-, Gamma-, Deltaproteobacteria). Names in brackets indicate revised phylum classifications according to Parks et al.28. The phylogenetic diversity (Faith’s PD) was calculated based on the rarefied community (5,873 reads per sample) and a midpoint-rooted phylogenetic tree. Indicators for microbial activity in the porewater chemistry were selected and depicted as profiles of up to 25 cm depth.
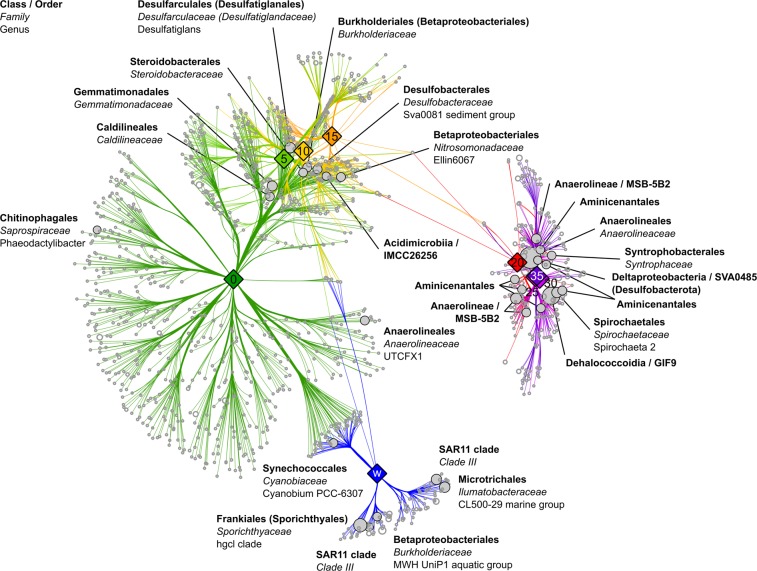
The bacterial community composition and diversity as well as the porewater chemistry of the sediment are distinct from the water column and change gradually with depth (Fig. 1b). The water column has a lower phylogenetic diversity than the top sediment layers (Fig. 1c) and is dominated by aquatic Actinobacteria (hgcl clade)16–20 and freshwater Alphaproteobacteria (SAR11 clade III)16,21–25 with relative abundances of more than 40% and up to 20% (Figs 1c and 2). The uppermost sediment layer is the most diverse and harbours the largest number of associated genera (Fig. 2). It shares community members of water and sediment, such as Synechococcus or the algae-associated Phaeodactylibacter26,27. The phylogenetic diversity (Fig. 1c) and associated genera (Fig. 2) in the sediment decrease gradually with depth until approximately 20 cm. Members of the Proteobacteria and Chloroflexota28 are dominant in the sediment community, which shifts from 15–35% Gammaproteobacteria in the top 15 cm to approximately 40% Chloroflexota below 15 cm. Notably, the upper sediment layers harbour sulphate-reducing bacteria, such as Desulfobacteraceae and Desulfarculaceae29–32 (Fig. 2). The decline in sulphate, increase in total sulphide (ΣH2S) and low redox potential also indicate sulphate reduction (Fig. 1c). Below 15 cm the bacterial community is associated with Anaerolineae, Aminicenantales and Dehalococcoidia (Fig. 2). Members of these taxa are known fermenters, organohalide respirators and hydrocarbon degraders33–35. Increasing degradative processes are indicated by the increase in ammonium and total alkalinity (Fig. 1c).
Fig. 2.
Bacterial genera associated with the different depths of the sediment cores and water column. The association network was calculated with the indicspecies54 package in R and visualised in Cytoscape with an edge-weighted spring embedded layout. Branch lengths indicate the phi correlation coefficient. Each light grey circle indicates a bacterial genus associated (p < 0.001) with the depth it is connected to. The 30 most abundant genera are indicated by filled circles and named up to the point where the classification turns to uncultured. Revised names according to Parks et al.28 are indicated in brackets. Average relative abundance of each genus among all samples is indicated by the circle size. Each sampling depth is indicated by a filled diamond shape containing the depth in cm or w (water column).
The bacterial community of Lake Neusiedl has mainly been studied with regard to potential pathogens6,9. Here, Enterobacteriaceae, more specifically Escherichia/Shigella, but not Vibrionaceae were detected with a relative abundance of up to 10% at almost all depths in the sediment, but not in the water column. While they indicate an anthropogenic impact on the sediment, the bacteria detected are based on DNA amplification and may not be metabolically active. This data may contribute to studies identifying the sampling site as hotspot for faecal pollution6,7 (Fig. 1a). Further, this survey forms a basis for studies targeting biogeochemical cycling in alkaline lakes.
Methods
Sediment sampling at Lake Neusiedl, Austria
Three soft sediment push-cores (RN-K01/K02/K03) covering 30 to 40 cm depth were sampled in close lateral distance to each other at the bay of Rust (16°42′33.635″E, 47°48′12.929″N) at Lake Neusiedl, Austria in August of 2017. PVC coring tubes (Uwitec, Mondsee, Austria) of 60 cm length and 63 mm diameter (RN-K01/K03) or 100 cm and 50 mm diameter (RN-K02) were manually pushed into the sediment at the sampling site. A rubber plug was applied to the top of the coring tube to create a partial vacuum, which allowed retrieval of the sediment. After allowing the sediment to settle on cores RN-K01 and K02, 600 ml core supernatant (water column) was filtered through a 3.0 µm polycarbonate (Merck, Darmstadt, Germany) and 0.2 µm polyethersulfone (Sartorius, Göttingen, Germany) filter sandwich. Subsequently, filters were immediately stored in RNAprotect Bacteria Reagent (Qiagen, Hilden, Germany). Sampling of the sediment for community analysis occurred under exclusion of the outer 1 cm of sediment, which is in contact with the walls of the coring tubes. RN-K01/K02 were sampled in triplicate at every 5 cm of depth. RN-K02 was sampled at a higher resolution (every 2.5) as the sediment showed finer lamination. Every triplicate was immediately mixed with RNAprotect Bacteria Reagent (Qiagen, Hilden, Germany) and kept at ambient temperature in a cool box with freezer elements for transport. Before storage, samples were centrifuged at 3,220 × g for 15 min and the clear supernatant containing the RNAprotect Bacteria Reagent discarded. Samples were stored at −80 °C. Core RN-K03 and the core supernatant were stored in the cool and dark until analytical chemical analysis.
DNA extraction and amplification of bacterial 16S rRNA genes
DNA was extracted from 0.25 mg of sediment from each sample of RN-K01/K02 using the MoBio Power Soil Kit (MoBio, CA, USA) with minor modifications. For this purpose, sediments were thawed on ice and homogenized to disrupt any layering caused by the previous centrifugation step. Subsequently, 0.25 mg were transferred into bead-beating tubes supplied by the manufacturer. DNA from the water column (core supernatant) was extracted by cutting one third of the frozen filter sandwiches into small pieces in the bead-beating tubes. After the addition of SDS-containing Solution C1, cells were mechanically disrupted with a FastPrep (MP Biomedicals, Eschwege, Germany) at 6.5 m/s for 20 s. After disruption, the DNA was extracted according to manufacturer’s instructions. Subsequently, DNA was eluted twice in 50 µl of prewarmed DEPC-treated water36. Bacterial 16 S rRNA genes were amplified by PCR with forward and reverse primers published by Klindworth et al.10 and added adapters for MiSeq sequencing (underlined) (D-Bact-0341-b-S-17, TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG CCTACGGGNGGCWGCAG; S-D-Bact-0785-a-A-21, GTCTCGTGGGCTCGGAGATGTGTATAAGA GACAGGACTACHVGGGTATCTAATCC). PCR reactions were performed in a total volume of 50 µl containing 10 µl of five-fold GC Buffer (Thermo Scientific, Waltham, MA, USA), 5% DMSO, 0.2 mM of forward and reverse primer, 200 µM dNTPs, 0.2 mM MgCl2, 1 U Phusion High-Fidelity DNA polymerase (Thermo Scientific, Waltham, MA, USA) and 20–25 ng template DNA. The PCR mixture was denatured for 1 min at 98 °C and then subjected to 25 cycles at 98 °C for 45 s, 45 s at 60 °C, and 30 s at 72 °C, followed by a final extension at 72 °C for 5 min. Negative controls were prepared without template and positive controls with genomic E. coli DH5α DNA as template. PCR reactions for each sample were performed in triplicate. PCR triplicates were pooled in equal amounts in order to minimize amplification bias, concentrated and purified with MagSi-NGSPrep magnetic beads as recommended by the manufacturer (Steinbrenner, Wiesenbach, Germany). After the final washing step, the beads were air-dried and DNA eluted in 30 µl of elution buffer EB (Qiagen, Hilden, Germany). Purified PCR products were quantified and sequenced as described by Schneider et al.37 using a MiSeq instrument and v3 chemistry (Illumina, San Diego, CA, USA).
Bioinformatic processing of 16S rRNA gene amplicons
Paired-end sequencing data from the Illumina MiSeq were quality-filtered with fastp38 (version 0.19.4) using default settings with the addition of an increased per base phred score of 20, base pair corrections by overlap (-c), as well as 5′- and 3′-end read trimming with a sliding window of 4, a mean quality of 20 and minimum sequence size of 50 bp. After quality control, the paired-end reads were merged using PEAR39 (version 0.9.11) and primers clipped using cutadapt40 (version 1.18) with default settings. Sequences were then processed using VSEARCH41 (v2.9.1). This included sorting and size-filtering of the paired reads to ≥300 bp (--sortbylength --minseqlength 300), dereplication (--derep_fulllength). Dereplicated amplicon sequence variants (ASVs) were denoised with UNOISE3 using default settings (--cluster_unoise – minsize 8) and chimeras were removed (--uchime3_denovo). An additional reference-based chimera removal was performed (--uchime_ref) against the SILVA SSU NR database (version 132). Raw reads were mapped to ASVs (--usearch_global–id 0.97). The taxonomy was assigned using BLAST 2.7.1+42 against the SILVA SSU 132 NR database with an identity of at least 95% to the query sequence resulting in a total of 21,009 ASVs43.
Bacterial community analysis
For data evaluation all samples from the 5 cm intervals were analysed. Additional samples taken due to the finer lamination of one core were not considered in the presented analysis but are available in the dataset44. Sequences comprising extrinsic domains, eukaryotes and archaea were removed from the ASV table using grepl, a base R function (version 3.4.4). A phylogenetic tree was generated by aligning all sequences of the filtered dataset with MAFFT45 at a maximum of 100 iterations. The tree was calculated using FastTree 2.1.7 (OpenMP)46, saved in newick format and midpoint rooted using FigTree47 (version 1.4.4).
The dataset was analysed in R48 (version 3.4.4) and RStudio49 (version 1.1.456). Depth profiles in the form of bar and line charts were generated with ggplot250 (version 3.1.0) using standard R packages. Alphadiversity indices and species richness were calculated with the ampvis251 package (version 2.4.1) and Faith’s phylogenetic diversity with picante52 (version 1.7) and the midpoint-rooted tree15. For this purpose, 16 samples with a read count below 5,000 were excluded from the diversity analysis (RN17_K1_DNA_Bac_2a, 3a, 5a, 6a, RN17_K2_DNA_Bac_5a-c, 7a-c, 9a-c, 11a-c). All other samples were rarefied in ampvis2 to 5,873 reads. For the visualisation in bar charts, the mean of all replicates from both cores was used to account for the variance at the sampling sites. The non-metric multidimensional scaling (NMDS) matrix was calculated using the ASV table and phylogenetic tree in a weighted generalized UniFrac analysis using the ampvis2 package (version 2.4.1) including the package GUniFrac53 (version 1.1). Environmental fit of the metadata were also calculated and plotted onto the NMDS if p < 0.01. An association network of the bacterial community was calculated using the indicspecies54 package (version 1.7.6) with the multipatt function and the r.g species-site group association function for calculation of the association strength. The significance cut-off for the phi coefficient was set to p < 0.001. The network was visualised in Cytoscape (version 3.5.1) with an edge-weighted spring-embedded layout using weight as the force and average abundance as the circle size.
Water column and porewater analysis
For hydrochemical analysis, capped and tightly sealed sediment cores, including the supernatant water column above, were stored upright in the cool and dark until analytical investigation 5 days after sampling. Core supernatants were collected in 250 ml polyethylene (PE) bottles for anion, nutrient, and total alkalinity determination. For cation analysis, a 50 ml aliquot of the supernatants was filtered through 0.7 μm diameter membrane filters (Merck, Darmstadt, Germany) into a PE-bottle and acidified with 100 µl HNO3 (suprapure, Merck, Darmstadt, Germany). Physicochemical parameters of the core supernatants were measured using a WTW Multi 3430 device equipped with a WTW Tetracon 925 conductivity probe, a WTW FDO 925 probe for dissolved O2, a Schott Pt 61 redox electrode, and a WTW Sentix 940 electrode for temperature and pH, which was calibrated against standard pH-buffers 7.010 and 10.010 (HI6007 and HI6010, Hanna Instruments, Vöhringen, Germany). Total alkalinity (TA) was determined via titration using a hand-held titration device and 1.6 N H2SO4 cartridges (Hach, Loveland, CO, USA).
Redox potential (Eh) and pH gradients were measured through boreholes directly in the sediment core using a portable WTW 340i pH meter equipped with an Inlab Solids Pro pH-electrode (Mettler Toledo, Gießen, Germany) and a Pt 5900A redox electrode (SI Analytics, Mainz, Germany). Porewater was extracted from core RN-K03 using 5 cm CSS Rhizon samplers (Rhizosphere, Wageningen, Netherlands). Immediately after extraction, aliquots were fixed with Zn-acetate for determination of total sulphide or acidified with suprapure HNO3 for analysis of main cations and trace elements. Porewater alkalinity was immediately determined by titration with cartridges (Hach, Loveland, CO, USA) containing self-prepared 0.01 n HCl as titrant. An aliquot for determination of nutrients and anions was stored in the cool and dark until subsequent analysis. Total sulphide (ΣH2S) and nutrient concentrations (NH4, NO2, PO4, SiO2) were measured by photometric methods according to Grasshoff et al.14, using an SI Analytics Uviline 9400 spectrophotometer within a few days after extraction.
Major cation (Ca, Mg, Na, K and Li) and anion (Cl, F, Br and SO4) concentrations of all water samples (porewaters, water column) were analysed by ion chromatography with non-suppressed and suppressed conductivity detection, respectively (Metrohm 820 IC/Metrosep C3-250 analytical column, Metrohm 883 Basic IC/Metrohm ASupp5-250 analytical column). ICP-MS (ICAP-Q, Thermo Fisher, Waltham, MA, USA) was used to determine Sr, Ba, Fe, Mn, Rb and B, as well as control for the cation determination by ion chromatography. Total dissolved salts (TDS) were calculated as the sum of all measured cations and anions. The chemical analysis was completed within two weeks after extraction with the analytical accuracy of all methods exceeding 1.5%15.
All measured values were processed by the PHREEQC software package, version 355, using the phreeqc.dat and wateqf4.dat databases in order to calculate ion activities and pCO2 (partial pressure of CO2) of the water samples and mineral saturation states. The saturation indices of all mineral phases are given as log (IAP/KSO) where IAP denotes the ion activity product and KSO is the solubility product of the corresponding mineral (solid phase).
Data Records
The 16S rRNA gene paired-end raw reads were deposited to the National Center for Biotechnology Information Sequence Read Archive (SRA) and can be found under the accession number PRJNA507590 (Bio Project 507590/SRP171602). This BioProject contains 63 samples and 126 zipped FASTQ files, which were processed using the CASAVA software (Illumina, San Diego, CA, USA)44. The processing included demultiplexing and adapter removal from the sequences. The following files have been deposited at figshare: a FASTA file with the assigned ASV sequences after bioinformatic processing56; the ASV count table with taxonomic assignments13, the read statistics before, during and after bioinformatic processing11; the metadata, porewater chemical data and alphadiversity metrics of each sample15. The individual files may also be accessed through a figshare collection43.
Technical Validation
For microbial community analysis the layers (2.5–5 cm) of both soft sediment push-cores were sampled in three technical replicates to allow for the microbial heterogeneity at each depth. The PCR reactions were run in three technical replicates per sample and PCR products were pooled equimolar. Negative controls without DNA template and positive controls with genomic E. coli DH5α DNA as template were also performed. Correct amplicon size was determined on a 0.8% agarose gel. PCR triplicates per sample were pooled in equimolar amounts for amplicon sequencing to minimize possible PCR bias. Physiochemical data were measured with calibrated probes and ions and nutrients were measured against IC and nutrient standards from Merck (Darmstadt, Germany) and Honeywell Fluka (Charlotte, NC, USA). The analytical accuracy of all methods exceeded 1.5%.
ISA-Tab metadata file
Acknowledgements
This study was funded by the German research foundation (DFG) in the framework of the research unit “CHARON” (subproject TP7: DA 374/11-1, AR 335/8-1). We thank Prof. Dr. Patrick Meister, University of Vienna, for his support during the field work and his feedback. Anja Poehlein and Melanie Heinemann are hereby gratefully acknowledged for performing the amplicon sequencing and initial sequence processing after the MiSeq run. We acknowledge support by the Open Access Publication Funds of the University of Göttingen.
Author contributions
R.D., G.A., A.H. conceived the study. Experimental design and implementation of the bacterial community analysis was performed by A.H. supported by D.S. A.H., A.R., G.A., D.S. and D.F. participated in the sampling. Porewater chemistry was measured and analysed by D.F. and A.R., A.H. drafted the manuscript, which was revised by D.S. and R.D. All authors interpreted the results and contributed to the final version of the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
ISA-Tab metadata
is available for this paper at 10.1038/s41597-019-0172-9.
References
- 1.Soja G, Züger J, Knoflacher M, Kinner P, Soja A-M. Climate impacts on water balance of a shallow steppe lake in Eastern Austria (Lake Neusiedl) J. Hydrol. 2013;480:115–124. doi: 10.1016/j.jhydrol.2012.12.013. [DOI] [Google Scholar]
- 2.Kogelbauer I, Loiskandl W. Characterization of sediment layer composition in a shallow lake: from open water zones to reed belt areas. Hydrol. Earth Syst. Sci. 2015;19:1427–1438. doi: 10.5194/hess-19-1427-2015. [DOI] [Google Scholar]
- 3.Müller G, Irion G, Förstner U. Formation and diagenesis of inorganic Ca-Mg carbonates in the lacustrine environment. Naturwissenschaften. 1972;59:158–164. doi: 10.1007/BF00637354. [DOI] [Google Scholar]
- 4.Schroll E, Wieden P. Eine rezente Bildung von Dolomit im Schlamm des Neusiedler Sees. Tschermaks Mineral. und Petrogr. Mitteilungen. 1960;7:286–289. doi: 10.1007/BF01127917. [DOI] [Google Scholar]
- 5.Whitman RL, Nevers MB. Foreshore sand as a source of Escherichia coli in nearshore water of a Lake Michigan beach. Appl. Environ. Microbiol. 2003;69:5555–5562. doi: 10.1128/AEM.69.9.5555-5562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hatvani IG, Kirschner AKT, Farnleitner AH, Tanos P, Herzig A. Hotspots and main drivers of fecal pollution in Neusiedler See, a large shallow lake in Central Europe. Environ. Sci. Pollut. Res. 2018;25:28884–28898. doi: 10.1007/s11356-018-2783-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Magyar N, et al. Application of multivariate statistical methods in determining spatial changes in water quality in the Austrian part of Neusiedler See. Ecol. Eng. 2013;55:82–92. doi: 10.1016/j.ecoleng.2013.02.005. [DOI] [Google Scholar]
- 8.Jirsa F, Pirker D, Krachler R, Keppler BK. Total mercury in sediments, macrophytes, and fish from a shallow steppe lake in eastern Austria. Chem. Biodivers. 2014;11:1263–1275. doi: 10.1002/cbdv.201400172. [DOI] [PubMed] [Google Scholar]
- 9.Pretzer C, et al. High genetic diversity of Vibrio cholerae in the European lake Neusiedler See is associated with intensive recombination in the reed habitat and the long-distance transfer of strains. Environ. Microbiol. 2017;19:328–344. doi: 10.1111/1462-2920.13612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klindworth A, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:1–11. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.von Hoyningen-Huene A, 2019. 16S rRNA gene sequence processing statistics and biosample accession numbers. figshare. [DOI]
- 12.Callahan BJ, McMurdie PJ, Holmes SP. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017;11:2639–2643. doi: 10.1038/ismej.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.von Hoyningen-Huene A, 2019. ASV counts and taxonomic assignments PRJNA507590/SRP171602. figshare. [DOI]
- 14.Grasshoff, K., Kremling, K. & Ehrhardt, M. Methods of seawater analysis. (John Wiley & Sons, 2009).
- 15.von Hoyningen-Huene A, 2019. Lake Neusiedl sample metadata and porewater properties. figshare. [DOI]
- 16.Aguilar P, Dorador C, Vila I, Sommaruga R. Bacterioplankton composition in tropical high-elevation lakes of the Andean plateau. FEMS Microbiol. Ecol. 2018;94:1–9. doi: 10.1093/femsec/fiy004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warnecke F, Amann R, Pernthaler J. Actinobacterial 16S rRNA genes from freshwater habitats cluster in four distinct lineages. Environ. Microbiol. 2004;6:242–253. doi: 10.1111/j.1462-2920.2004.00561.x. [DOI] [PubMed] [Google Scholar]
- 18.Tandon K, et al. Bacterial community in water and air of two sub-alpine lakes in Taiwan. Microbes Environ. 2018;33:120–126. doi: 10.1264/jsme2.ME17148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newton RJ, Jones SE, Eiler A, McMahon KD, Bertilsson S. A guide to the natural history of freshwater lake bacteria. Microbiol. Mol. Biol. Rev. 2011;75:14–49. doi: 10.1128/MMBR.00028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neuenschwander SM, Ghai R, Pernthaler J, Salcher MM. Microdiversification in genome-streamlined ubiquitous freshwater Actinobacteria. ISME J. 2018;12:185–198. doi: 10.1038/ismej.2017.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herlemann DPR, Woelk J, Labrenz M, Jürgens K. Diversity and abundance of “Pelagibacterales” (SAR11) in the Baltic Sea salinity gradient. Syst. Appl. Microbiol. 2014;37:601–604. doi: 10.1016/j.syapm.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 22.Brown MV, et al. Global biogeography of SAR11 marine bacteria. Mol. Syst. Biol. 2012;8:1–13. doi: 10.1038/msb.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Zhao Z, Dai M, Jiao N, Herndl GJ. Drivers shaping the diversity and biogeography of total and active bacterial communities in the South China Sea. Mol. Ecol. 2014;23:2260–2274. doi: 10.1111/mec.12739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Llirós M, et al. Bacterial community composition in three freshwater reservoirs of different alkalinity and trophic status. PLoS One. 2014;9:e116145. doi: 10.1371/journal.pone.0116145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eiler A, et al. Tuning fresh: radiation through rewiring of central metabolism in streamlined bacteria. ISME J. 2016;10:1902–1914. doi: 10.1038/ismej.2015.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flombaum P, et al. Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. PNAS. 2013;110:9824–9829. doi: 10.1073/pnas.1307701110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lei X, et al. Phaeodactylibacter luteus sp. nov., isolated from the oleaginous microalga Picochlorum sp. Int. J. Syst. Evol. Microbiol. 2015;65:2666–2670. doi: 10.1099/ijs.0.000321. [DOI] [PubMed] [Google Scholar]
- 28.Parks DH, et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 2018;36:996–1004. doi: 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 29.Schneider D, Arp G, Reimer A, Reitner J, Daniel R. Phylogenetic Analysis of a Microbialite-Forming Microbial Mat from a Hypersaline Lake of the Kiritimati Atoll, Central Pacific. PLoS One. 2013;8:e66662. doi: 10.1371/journal.pone.0066662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diaz MR, et al. Functional gene diversity of oolitic sands from Great Bahama Bank. Geobiology. 2014;12:231–249. doi: 10.1111/gbi.12079. [DOI] [PubMed] [Google Scholar]
- 31.Broman E, Sjöstedt J, Pinhassi J, Dopson M. Shifts in coastal sediment oxygenation cause pronounced changes in microbial community composition and associated metabolism. Microbiome. 2017;5:1–18. doi: 10.1186/s40168-017-0311-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reyes Carolina, Dellwig Olaf, Dähnke Kirstin, Gehre Matthias, Noriega-Ortega Beatriz E., Böttcher Michael E., Meister Patrick, Friedrich Michael W. Bacterial communities potentially involved in iron-cycling in Baltic Sea and North Sea sediments revealed by pyrosequencing. FEMS Microbiology Ecology. 2016;92(4):fiw054. doi: 10.1093/femsec/fiw054. [DOI] [PubMed] [Google Scholar]
- 33.Hug LA, et al. Community genomic analyses constrain the distribution of metabolic traits across the Chloroflexi phylum and indicate roles in sediment carbon cycling. Microbiome. 2013;1:1–17. doi: 10.1186/2049-2618-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dombrowski N, Seitz KW, Teske AP, Baker BJ. Genomic insights into potential interdependencies in microbial hydrocarbon and nutrient cycling in hydrothermal sediments. Microbiome. 2017;5:1–13. doi: 10.1186/s40168-017-0322-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kadnikov VV, Mardanov AV, Beletsky AV, Karnachuk OV, Ravin NV. Genome of the candidate phylum Aminicenantes bacterium from a deep subsurface thermal aquifer revealed its fermentative saccharolytic lifestyle. Extremophiles. 2019;23:189–200. doi: 10.1007/s00792-018-01073-5. [DOI] [PubMed] [Google Scholar]
- 36.Schneider, D., Wemheuer, F., Pfeiffer, B. & Wemheuer, B. Extraction of total DNA and RNA from marine filter samples and generation of a cDNA as universal template for marker gene studies. In Metagenomics: Methods and Protocols (eds Streit, W. R. & Daniel, R.) Ch.2 (Humana Press, 2017). [DOI] [PubMed]
- 37.Schneider D, et al. Gut bacterial communities of diarrheic patients with indications of Clostridioides difficile infection. Sci. Data. 2017;4:170152. doi: 10.1038/sdata.2017.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: a fast and accurate illumina paired-end read merger. Bioinformatics. 2014;30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. journal. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 41.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altschul SF, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 43.von Hoyningen-Huene A, 2019. Bacterial succession along a sediment porewater gradient at Lake Neusiedl in Austria. figshare. [DOI] [PMC free article] [PubMed]
- 44.2019. NCBI Sequence Read Archive. SRP171602
- 45.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Price MN, Dehal PS, Arkin AP. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rambaut, A. FigTree - tree figure drawing tool. Institute of Evolutionary Biology, University of Edinburgh, http://tree.bio.ed.ac.uk/software/figtree/ (2018).
- 48.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, https://www.R-project.org/ (2019).
- 49.RStudio Team. RStudio: integrated development for R. RStudio Inc., http://www.rstudio.com/ (2016).
- 50.Wickham, H. ggplot2: elegant graphics for data analysis. (Springer-Verlag, 2016).
- 51.Andersen, S. K., Kirkegaard, R. H., Karst, S. M. & Albertsen, M. ampvis2: an R package to analyse and visualise 16S rRNA amplicon data. bioRxiv 1–2, 10.1101/299537 (2018).
- 52.Kembel SW, et al. Picante: R tools for integrating phylogenies and ecology. Bioinformatics. 2010;26:1463–1464. doi: 10.1093/bioinformatics/btq166. [DOI] [PubMed] [Google Scholar]
- 53.Chen J, et al. Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics. 2012;28:2106–2113. doi: 10.1093/bioinformatics/bts342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De Cáceres, M. & Legendre, P. Associations between species and groups of sites: indices and statistical inference. Ecology 90 3566–3572 (2009). [DOI] [PubMed]
- 55.Parkhurst, D. L. & Appelo, C. A. J. Description of input and examples for PHREEQC version 3—a computer program for speciation, batch-reaction, one-dimensional transport, and inverse geochemical calculations. In U.S. Geological Survey Techniques and Methods Book 6 Ch. A43, https://pubs.usgs.gov/tm/06/a43/ (2013).
- 56.von Hoyningen-Huene A, 2019. ASV sequences PRJNA507590/SRP171602. figshare. [DOI]
- 57.Oksanen, J. et al. vegan: community ecology package, https://cran.r-project.org/package=vegan (2018).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- von Hoyningen-Huene A, 2019. 16S rRNA gene sequence processing statistics and biosample accession numbers. figshare. [DOI]
- von Hoyningen-Huene A, 2019. ASV counts and taxonomic assignments PRJNA507590/SRP171602. figshare. [DOI]
- von Hoyningen-Huene A, 2019. Lake Neusiedl sample metadata and porewater properties. figshare. [DOI]
- von Hoyningen-Huene A, 2019. Bacterial succession along a sediment porewater gradient at Lake Neusiedl in Austria. figshare. [DOI] [PMC free article] [PubMed]
- 2019. NCBI Sequence Read Archive. SRP171602
- von Hoyningen-Huene A, 2019. ASV sequences PRJNA507590/SRP171602. figshare. [DOI]