Abstract
This article contains chemical characterization and biological activity data for a novel indirubin derivative, termed LDD-1819. The detailed synthesis procedure and associated NMR data are presented. The concentration-dependent inhibition data of two biological targets, glycogen synthase kinase-3 and aurora kinase A are described. The following biological data are also contained in this article: 1) the cellularization of skeletal muscle myotubes by LDD-1819 or two small molecule inhibitors of glycogen synthase kinase-3 and aurora kinase A (BIO and reversine) and gene expression data for the myoblast markers Pax-7 and Myf5, 2) Cell viability of hTERT human immortalized fibroblasts, colon cancer cells and breast cancer cells, and 3) Western blotting analysis of full length and cleaved caspse-7, and cleaved poly (ADP-ribose) polymerase (PARP) in hTERT fibroblasts treated with LDD-1819. A schematic diagram of the biological activities of LDD-1819 is also presented. Further interpretation and discussion of these data are provided in the associated research article ‘A novel indirubin derivative that increases somatic cell plasticity and inhibits tumorigenicity’ (Kim et al., 2019).
Keywords: Metastasis, Cell plasticity, Cellularization, Indirubin derivative, Glycogen synthase kinase-3 β, Aurora kinase A
Specifications Table
| Subject | Chemistry (General) |
| Specific subject area | Bioorganic and Medical Chemistry |
| Type of data | Images Graphs Figures |
| How data were acquired | NMR spectroscopy (JNM-ECX400P, JEOL), microscopy (Olympus's CLX41), western blot imaging system (LAS500, AB Biosciences), microplate reader (VersaMax, Molecular Devices). |
| Data format | Analyzed. |
| Parameters for data collection | Synthesis and cell plasticity/anti-cancer activity after treatment with a novel indirubin derivative, LDD-1819. |
| Description of data collection | The parameters were as follows: 1) 1H NMR spectra of the chemical structure of compound LDD-1819. 2) Cell viability after LDD-1819 treatment. 3) Microscopic observation of myotube cellularization. 4) Quantitative PCR analysis of gene expression related to myogenesis, apoptosis or cancer cell invasion. 5) Inhibition activity against glycogen synthase kinase-3 β and aurora kinase A. |
| Data source location | Gwangju Republic of Korea (South Korea). |
| Data accessibility | With the article |
| Related research article | Author's name: Kim WH, Jeong P, Kim SW, Cho H, Lee JM, Seo S, Shen H, Ahn Y, Jung DW, Kim YC, Williams DR. Title: A novel indirubin derivative that increases somatic cell plasticity and inhibits tumorigenicity Journal: Bioorganic & Medicinal Chemistry. https://doi.org/10.1016/j.bmc.2019.05.025 |
Value of the data
|
1. Data
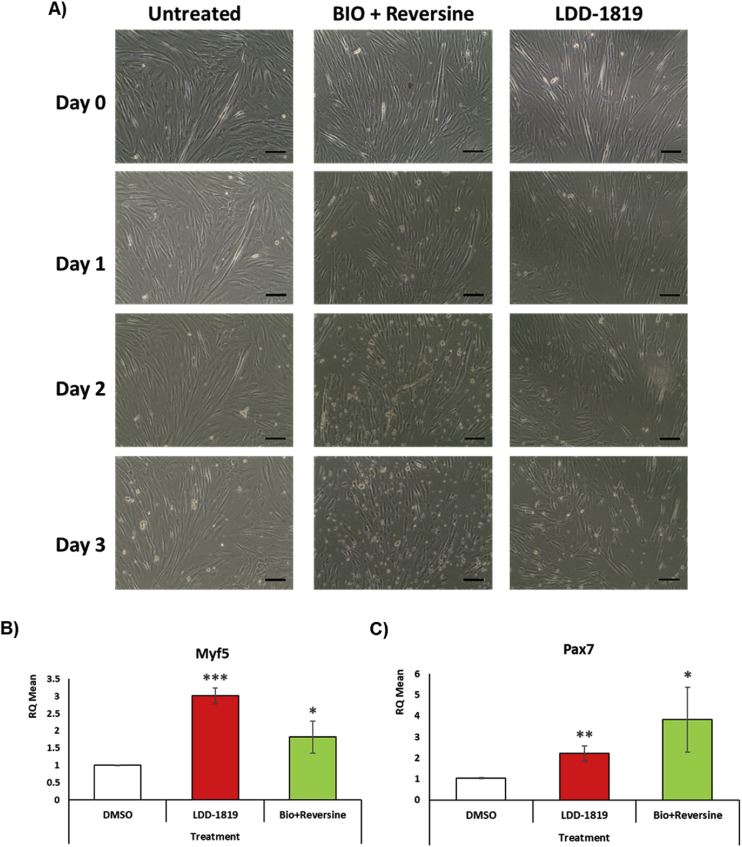
C2C12 myotube cultures were treated with LDD-1819, or the compounds BIO and reversine (a glycogen synthase kinase-3 (GSK-3) inhibitor and an aurora kinase A inhibitor) [2]. Using microscopic analysis, evidence of cellularization in the myotubes could be observed after treatment with LDD-1819 or BIO plus reversine (Fig. 1A). qPCR analysis was carried out for Pax-7 and Myf5 expression, which are genes expressed in undifferentiated myoblasts and downregulated during differentiation into multinucleated myotubes [[3], [4]]. Treatment with LDD-1819, or BIO plus reversine, increased Pax-7 and Myf5 expression, which supports the observation of cellularization and reversal of the multinucleated myotube differentiated state (Fig. 1B and C).
Fig. 1.
(A) Microscopic indication of cellularization in myotubes treated with 1 μM LDD-1819, or 5 μM BIO and 100 nM reversine, for 72 h in differentiation media. Scale bar = 100 μm. (B) qPCR analysis of the expression of myoblast marker genes Myf5 and Pax7, which are down-regulated during differentiation, in myotubes treated with 5 μM BIO and 100 nM reversine, or 1 μM LDD-1819, for 72 h. *, **, or**** = p < 0.05, p < 0.01 or p < 0.001 compared to myotubes treated with 0.1% DMSO. RQ = mean expression level for the gene of interest.
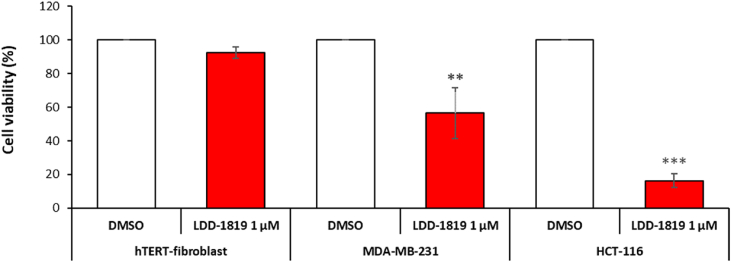
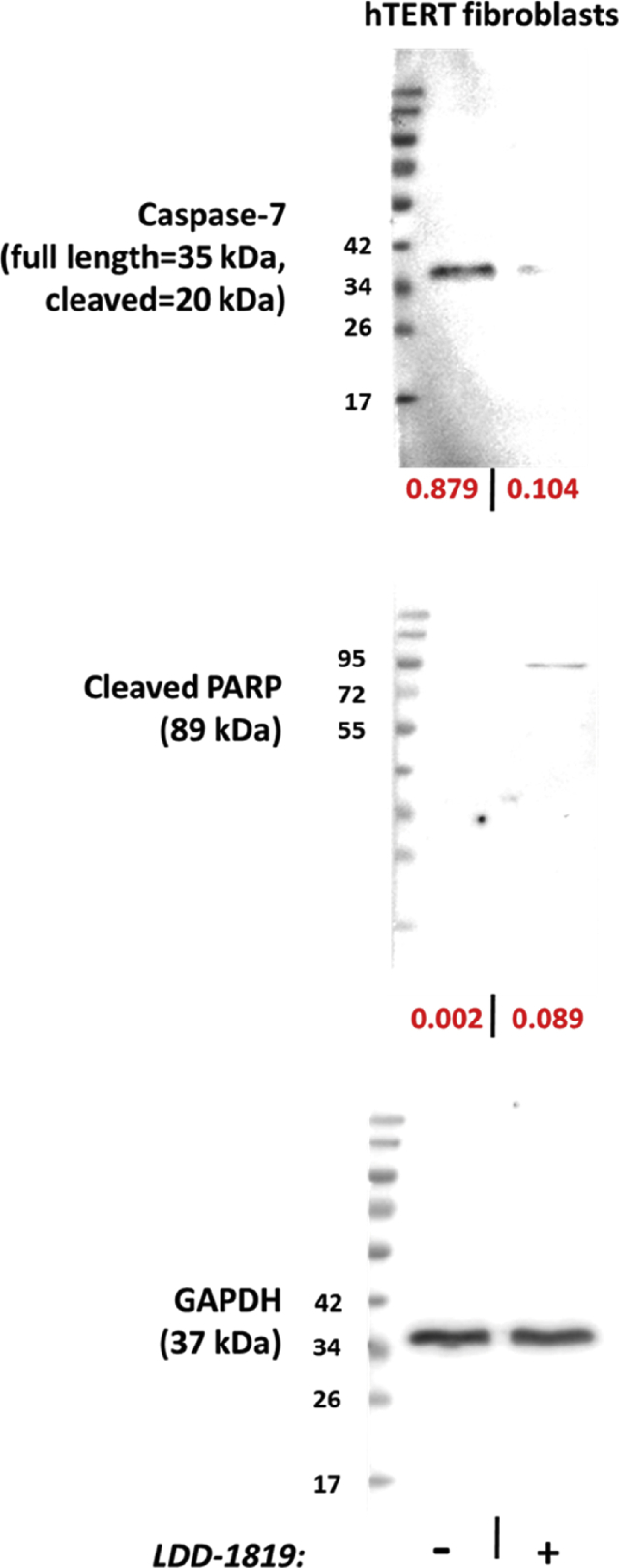
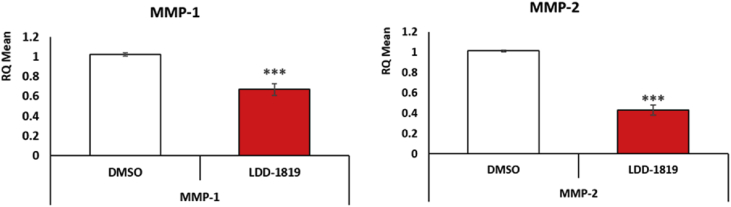
The anti-cancer activity was investigated in human, normal hTERT immortalized fibroblasts, and human breast and colon carcinoma cells (MDA-MB-231 and HCT116 cells). Cell viability data showed that LDD-1819 treatment was selectively cytotoxic for the cancer cells, with a greater toxic response in the colon carcinoma cells compared to the breast carcinoma cells (Fig. 2). The expression level of the pro-apoptosis proteins cleaved caspase-7 and cleaved Poly (ADP-ribose) polymerase (PARP) was measured in untreated and LDD-1819 treated hTERT fibroblasts. It was observed that LDD-1819 treatment did not produce detectable levels of cleaved caspase-7 and reduced the expression of full length cleaved caspase-7 (Fig. 3). Cleaved PARP was detected in hTERT fibroblasts after treatment with LDD-1819 (Fig. 3.). The expression level of two enzymes linked to cancer cell invasion (matrix metalloproteinase (MMP)-1 and MMP-2) was measured in LDD-1819 treated breast carcinoma cells using qPCR. LDD-1819 treatment reduced the expression of MMP-1 and MMP-2 in the cancer cells (Fig. 4).
Fig. 2.
MTT assay to measure cell viability in hTERT immortalized human fibroblasts (non-cancer cells), MDA-MB-231 human breast cancer cells and HCT-116 human colon cancer cells treated with LDD-1819 for 72 h. ** = p < 0.01 and *** = p < 0.001 compared to 0.1% DMSO treated cells.
Fig. 3.
Western blotting analysis of full length or cleaved caspase-7, and PARP expression, in hTERT fibroblasts treated with 500 nM LDD-1819 for 72 h. GAPDH was used as a housekeeping gene control. Fold-change in protein expression is shown as red numbers beneath each panel.
Fig. 4.
qPCR analysis of MMP-1 and MMP-2 expression in MDA-MB-231 breast cancer cells treated with 50 nM LDD-1819 for 24 h. *** = p < 0.001 compared to 0.1% DMSO treated cells.
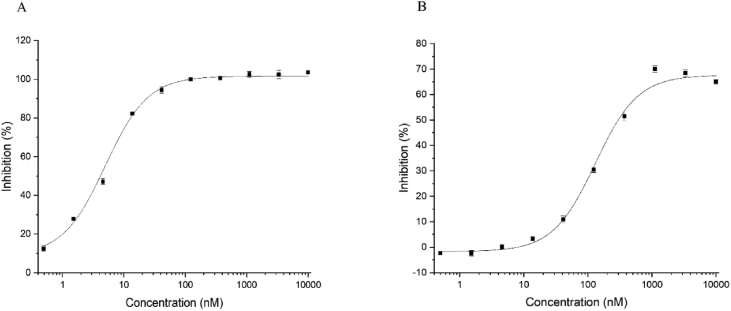
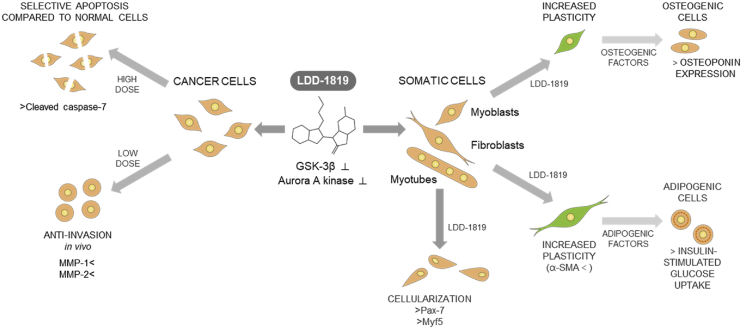
The inhibitory activity of LDD-1819 against two biological targets, GSK-3β and aurora kinase A, was measured in a concentration dependent experiment (Fig. 5). The IC50 was calculated as 5.46 nM for GSK-3β and 222 nM for aurora kinase A. A schematic diagram summarizing the biological effects of compound LDD-1819, based on the data herein and in the associated research article ‘A novel indirubin derivative that increases somatic cell plasticity and inhibits tumorigenicity’ [1] is shown in Fig. 6.
Fig. 5.
Concentration dependent inhibition of GSK-3β (A) and aurora kinase A (B) by LDD-1819. IC50 for GSK-3β = 5.46 nM; IC50 for aurora kinase A inhibition = 222 nM.
Fig. 6.
Schematic diagram of the biological effects of compound LDD-1819 in cancer cells and somatic cells.
2. Experimental design, materials, and methods
2.1. Reagents and antibodies
BIO ((2′Z,3′E)-6-Bromoindirubin-3′-oxime), CelLytic™ M lysis buffer, (β-D-arabinofuranosyl) cytosine) (AraC), and TRI-Solution™ were purchased from Sigma-Aldrich. Reversine was purchased from Santa Cruz Biotechnology, Inc. An antibody for PARP (catalogue number #9541) was purchased from Cell Signaling Technology. An antibody recognizing both full length and cleaved caspase-7 (catalogue number #9492) was also purchased from Cell Signaling Technology. An antibody for GAPDH was purchased from Santa Cruz Biotechnology, Inc (catalogue number sc-365062).
2.2. Synthetic procedure of LDD-1819
To a solution of 5-hydroxyanthranilic acid (1) in 50 mL of methanol was added 0.5 mL of acetic acid, followed by ethyl glyoxalate and NaCNBH3. The reaction mixture was stirred for overnight at room temperature. Then the methanol was removed by evaporation. The residue was taken up in a solution of saturated NH4Cl in water and extracted with ethyl acetate (3 times). The combined extracts were dried over sodium sulfate, filtered and evaporated. The residue was purified by silica gel column chromatography (CHCl3:CH3OH = 30:1) to give the ester derivative (2) which was subsequently hydrolyzed in 1N NaOH(aq) and methanol. The reaction mixture was stirred for 1 hour at room temperature and the resulting solution was neutralized by 1N HCl to afford the solid product (3) which was filtered and washed with water. To the di-acid compound (3) was added acetic anhydride and Na2CO3. The reaction mixture was refluxed for 4 hours and the product was extracted with ethyl acetate (3 times) and washed with water. The combined extracts were dried over sodium sulfate, filtered and evaporated. The residue was purified by silica gel column chromatography (hexane: ethyl acetate = 2:1) to give diacetate compound (4). Indirubin oxime compound, 6 was synthesized from aldol condensation of the indoxyl-N,O-diacetate (4) with 5-nitroisatin under basic conditions to yield the compound (5), followed by the reaction with hydroxylamine in pyridine with reflux condition for 6 hour. The reaction mixture was acidified by 1N HCl, filtered and washed with water to obtain N-oxime compound (6) [5]. Next, a solution of 2-(Boc-amino)ethyl bromide, N-oxime analog (6) and potassium carbonate in DMF was stirred at ambient temperature for overnight. The resulting mixture was partitioned in saturated aq. NH4Cl solution and ethyl acetate (3 times). The organic phase was evaporated and the product (7) was purified by silica gel column chromatography. Finally, to a solution of compound 7 in THF, 4N HCl in 1,4-dioxane was added dropwise in ice-bath. After the completion of reaction, the precipitate was filtered and washed with DCM. The resulting product was dried in vacuum oven to afford the hydrochloride salt, LDD-1819.
2.3. Analytical data of the compound
Starting materials, reagents, and solvents were purchased from Aldrich Chemical Co. (Milwaukee, WI) and TCI (Tokyo) and used as supplied without further purification. Thin-layer chromatography (TLC) was performed on fluorescent silica gel plates (60 F254 from Merck) and visualized with either short-wave UV light. Chromatographic purifications were achieved using kieselgel 60 (Merck) 0.040–0.063 mm column chromatography. 1H NMR spectra were determined with a JEOL JNM-LA 300WB spectrometer at 300 MHz or JEOL JNM-ECX 400P spectrometer at 400 MHz, and spectra were taken in CDCl3 or DMSO-d6 or D2O. Unless otherwise noted, chemical shifts are expressed as ppm downfield from internal tetramethylsilane, or relative ppm from DMSO (2.5 ppm), H2O (4.79 ppm).
2.3.1. 2-((2-ethoxy-2-oxoethyl)amino)-5-hydroxybenzoic acid (2)
Yield: 89%, 1H NMR 1H NMR (400 MHz, DMSO-d6) δ ppm 8.72 (s, 1H), 7.22 (s, 1H), 6.86-6.83 (dd, J = 8.8, 2.8 Hz, 1H), 6.46-6.43 (d, J = 8.8 Hz, 1H), 4.14-4.09 (q, J = 7.2 Hz, 2H), 3.98 (s, 2H), 1.20-1.16 (t, J = 6.8 Hz, 3H).
2.3.2. 2-((carboxymethyl)amino)-5-hydroxybenzoic acid (3)
Yield: 90%, 1H NMR 1H NMR (400 MHz, DMSO-d6) δ ppm 8.73 (s, 1H), 7.22 (s, 1H), 6.86-6.83 (dd, J = 8.8, 2.8 Hz, 1H), 6.44-6.41 (d, J = 8.8 Hz, 1H), 3.89 (s, 2H).
2.3.3. 1-acetyl-1H-indole-3,5-diyl diacetate (4)
Yield: 32%, 1H NMR (300 MHz, CDCl3) δ ppm 8.36 (d, J = 9.3 Hz, 1H), 7.95(s, 1H), 7.31 (d, J = 2.4 Hz, 1H), 7.15-7.12(dd, J = 9.3, 2.4 Hz, 1H), 2.67(s, 3H), 2.36 (s, 3H), 2.31(s, 3H). ESI [M+H] = 214.16.
2.3.4. (Z)-5-hydroxy-5′-nitro-[2,3′-biindolinylidene]-2′,3-dione (5)
Yield: 32%, 1H NMR (300 MHz, DMSO-d6) δ ppm 11.78 (s, 1H), 11.51 (s, 1H), 9.59 (d, J = 2.4 Hz, 1H), 8.27-8.25 (d, J = 9.16 Hz, 1H), 8.14-8.11 (dd, J = 8.84, 2.44 Hz, 1H), 7.52-7.48 (m, 2H), 7.12-7.07 (m, 2H)
2.3.5. (2Z,3E)-5-hydroxy-3-(hydroxyimino)-5′-nitro-[2,3′-biindolinylidene]-2′-one (6)
Yield: 66%, 1H NMR (400 MHz, DMSO-d6) δ ppm 13.87 (s, 1H), 11.78 (s, 1H), 11.35 (s, 1H), 9.41 (d, J = 2.8 Hz, 1H), 9.32 (s, 1H), 8.05 (dd, J = 11.6, 2.8 Hz, 1H), 7.76 (d, J = 3.2 Hz, 1H), 7.29 (d, J = 11.6 Hz, 1H), 7.04 (d, J = 11.2 Hz, 1H) 6.86 (dd, J = 11.2, 3.2 Hz, 1H).
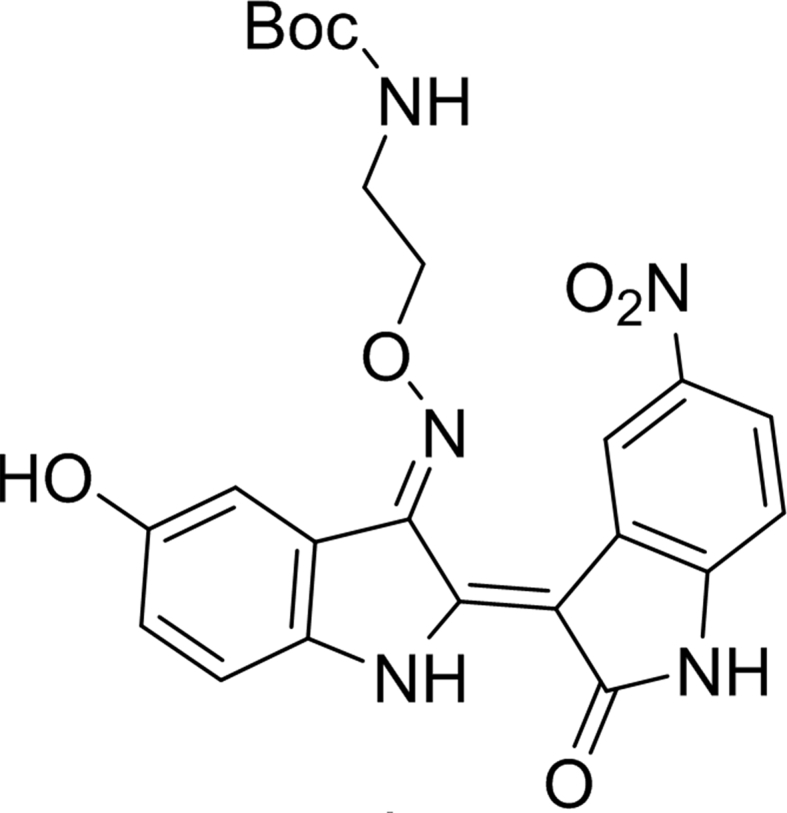
2.3.6. tert-butyl (2-((((2Z,3E)-5-hydroxy-5′-nitro-2′-oxo-[2,3′-biindolinylidene]-3-ylidene)amino)oxy)ethyl)carbamate (7)
Yield: 48%, 1H NMR(400 MHz, DMSO-d6) δ ppm 11.66 (s, 1H), 11.39 (s, 1H), 9.85 (s, 1H), 9.36 (s, 1H), 8.10-8.07 (dd, J = 8.8, 2.4 Hz, 1H), 7.65 (d, J = 2.4 Hz, 1H), 7.13–7.29 (d, J = 8.8 Hz, 1H), 7.07-7.03 (m, 2H), 6.92-6.89 (dd, J = 8.8, 2.4 Hz, 1H), 4.71-4.69 (t, J = 5.6 Hz, 2H), 3.55-3.54 (m, 2H), 1.31-1.24 (brs, 9H).
HRMS (FAB) m/z calculated for C23H23N5O7 [M+H]+ 482.16, found 482.1679.
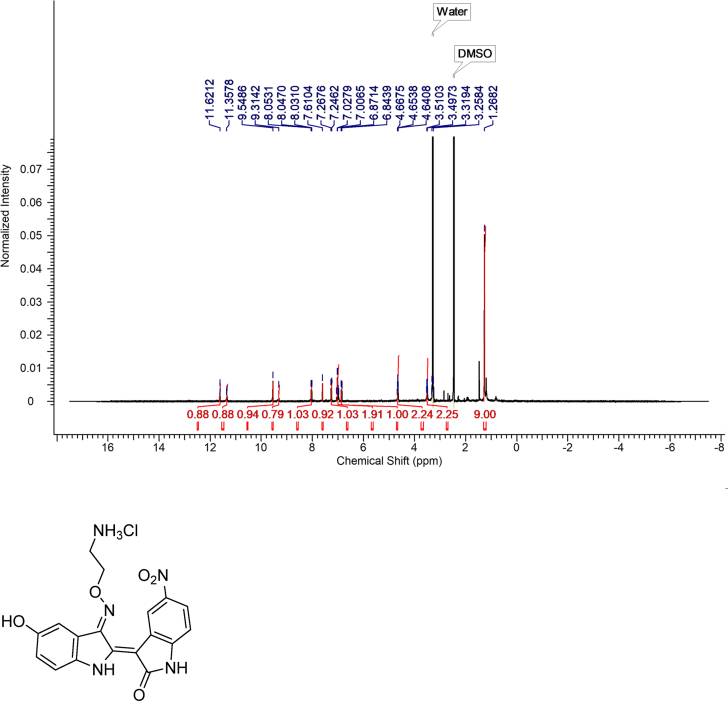
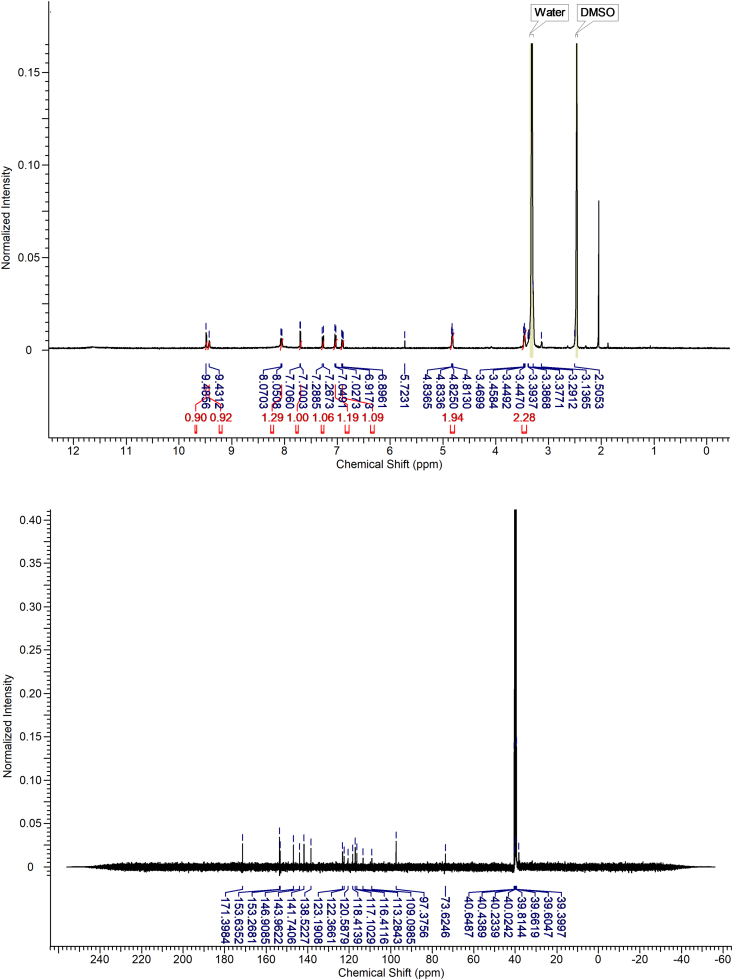
2.3.7. (2Z,3E)-3-((2-aminoethoxy)imino)-5-hydroxy-5′-nitro-[2,3′-biindolinylidene]-2′-one hydrochloride (LDD-1819)
Yield: 98%, 1H NMR(400 MHz, DMSO-d6) δ ppm 11.66(s, 1H), 11.47(s, 1H), 9.51(d, J = 2.4 Hz, 1H), 9.48(s, 1H), 8.24(brs, 3H), 8.09(dd, J = 8.4, 2.4 Hz, 1H), 7.78(d, J = 2.4 Hz, 1H), 7.30 (d, J = 8.4 Hz, 1H), 7.07 (d, J = 8.8 Hz, 1H), 6.95 (dd, J = 8.4, 2.4 Hz, 1H), 4.87 (t, J = 4.4 Hz, 2H), 3.49 (t, J = 4.4 Hz, 2H).
HRMS (FAB) m/z calculated for C18H15N5O57 [M-Cl]+ 381.11, found 382.1152.
2.4. Cell culture
C2C12 murine skeletal myoblasts, HCT116 human colon cancer cells and MDA-MB-231 human breast cancer cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS), 100 units/mL penicillin and 100 μg/mL streptomycin (PenStrep). Confluent cultures of C2C12 myoblasts were induced to undergo differentiation into myotubes by incubation with differentiation media (DMEM containing 2% horse serum and PenStrep) for 5 d. The differentiation media was replenished every 24 h. To remove undifferentiated myoblasts, the cultures were treated with 50 μM AraC for 3 d (2d after the onset of differentiation), using the previously published protocol [6].
hTERT expressing immortalized human fibroblasts (hTERT fibroblasts) were a generous gift from Professor Jin Kim, Department of Oral Pathology, Yonsei University, Republic of Korea. hTERT fibroblasts were cultured with F-media (DMEM + F-12 media at a 3:1 ratio) supplemented with 10% FBS and 1% PenStrep.
2.5. Cell toxicity assay
Cytotoxicity was measured using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, a tetrazole (MTT) assay, as previously described [7] using a VersaMax plate reader (Molecular Devices). Cell viability was assessed after 72 h compound treatment.
2.6. Western blotting analysis
Proteins were separated using 10% SDS-PAGE and transferred onto 0.2 μm nitrocellulose (Bio-Rad). Densitometry analysis of band intensity was carried out with Image J 1.54s software (National Institutes of Health, USA).
2.7. Quantitative PCR (qPCR) analysis of Pax7, Myf5, MMP-1 and MMP-2 expression
RNA was harvested using the TRIzol reagent (Thermo Fisher Scientific, MA, USA) following the manufacturer's instructions. For qPCR, the transcript level of the gene of interest was measured using the StepOnePlus Real Time PCR System (Applied Biosystems, UK). Total RNA was reverse transcribed into cDNA using the AccuPower® RT PreMix (Bioneer Corporation) and used for real-time PCR in accordance with the manufacturer's guidelines, and PCR was performed in triplicate in a total volume of 20 μL TOPrealTM qPCR 2X PreMIX (SYBR Green with high ROX) (Enzynomics co. Ltd., South Korea) containing 250 nM final concentration of the specific primer and 1 μL cDNA. The mixture was incubated for 10 min at 95 °C prior to PCR amplification, which consisted of 40 cycles of denaturation, annealing and extension. Denaturation was carried out at 15 s at 95 °C, annealing was carried out at 1 min at 60 °C and extension was carried out at 72 °C for 20 s with fluorescence detection at 72 °C after each cycle. When the final cycle was completed, melting-point analysis was performed within the range of 60–95 °C and continuous fluorescence detection. A specific cDNA sample was included in each run and used as a reference for comparison between runs. The final mRNA expression in each experiment was calculated as the average value of three independent sets of experiments. The primer sequence for murine Myf5 was AGCTGGGCAGAATACGTGCTT (forward) and AGAACAGGCAGAGGAGAATCCA (reverse; product size 112 b.p.) and the primer sequence for murine Pax7 was CACAGAGGCAGAGCTGATTGC (forward) and CCAATTGAGGAGAGTGACAGGTT (reverse; product size 157 b.p.). The primer sequence for mouse GAPDH was CTCCACTCACGGCAAATTCA (forward) and GCCTCACCCCATTTGATGTT (reverse; product size 120 b.p.) The primer sequence for human MMP-1 was GGACCTGGAGGAAATCTTGCT (forward) and TCCAAGAGAATGGCCGAGTT (reverse; product size 150 b.p.) and the primer sequence for human MMP-2 was CGTCTGTCCCAGGATGACATC (forward) and CTGTTTGCAGATCTCAGGAGTGA (reverse; product size 120 b.p.). The primer sequence for human GAPDH was AAGGTCGGAGTCAACGGATTT (forward) and TTGACGGTGCCATGGAATTT (reverse; product size 170 b.p.).
2.8. Western blotting analysis
Proteins were separated by 10% SDS-PAGE and transferred onto 0.2 μm nitrocellulose (Bio-Rad). Densitometry analysis of band intensity was carried out using the Image J 1.54s software (National Institutes of Health, USA).
2.9. Measurement of aurora kinase A inhibition
Inhibitory activity against Aurora kinase A was measured using Z′-LYTE® Biochemical assay based on the differential sensitivity of phosphorylated and non-phosphorylated peptides to proteolytic cleavage. The inhibitory activity of compounds was evaluated on the ATP (Km value), Aurora kinase A (0.91 - 10 ng) and Synthetic FRET peptide (2 μM) concentration. The kinase reaction buffer consists of 50 mM HEPES pH 7.5, 0.01% BRIJ-35, 10 mM MgCl2, 1 mM EGTA. The total reaction volume was 10 μL and serial dilutions of compounds, FRET peptide and ATP were incubated. Kinase reactions were performed for 30 min at room temperature in standard 384-well plates, and then 5 μL of the development reagent solution was treated to the reaction plates 1 h before reading the plates. After reagents were added for detection, the multi-label reader was set at 445 nm for excitation, and 520 nm for emission. The FRET emission ratio was represented as ratio F445/F520 nm. The IC50 values were calculated with nonlinear regression analysis using OriginPro 9.1 software (OriginLab, Northampton, MA).
2.10. Measurement of GSK-3β kinase inhibition
Inhibitory activity against GSK-3β was measured using Z′-LYTE® Biochemical assay based on the differential sensitivity of phosphorylated and non-phosphorylated peptides to proteolytic cleavage. The inhibitory activity of compounds was evaluated on the ATP (Km value), GSK-3β (0.22 - 0.92 ng) and Synthetic FRET peptide (2 μM) concentration. The kinase reaction buffer consists of 50 mM HEPES pH 7.5, 0.01% BRIJ-35, 10 mM MgCl2, 1 mM EGTA. The total reaction volume was 10 μL and serial dilutions of compounds, FRET peptide and ATP were incubated. Kinase reactions were performed for 30 min at room temperature in standard 384-well plates, and then 5 μL of the Development Reagent solution was treated to the reaction plates 1 h before reading the plates. After reagents were added for detection, the multi-label reader was set at 445 nm for excitation, and 520 nm for emission. The FRET emission ratio was represented as ratio F445/F520 nm. The IC50 values were calculated with nonlinear regression analysis using OriginPro 9.1 software (OriginLab, Northampton, MA).
2.11. Statistical analysis
Statistical significance was determined using the Student's t-test (Microsoft Excel, 2016). A p value < 0.05 was deemed to be significant. Unless otherwise stated, all data shown are representative of three experimental repeats and the error bars for the graphs are standard deviation.
Acknowledgments
This work was partly supported by Institute for Information and communications Technology Promotion(IITP) grant funded by the Korea government(MSIP) (No. 2019-0-00567, Development of Intelligent SW systems for uncovering genetic variation and developing personalized medicine for cancer patients with unknown molecular genetic mechanisms). This work was also supported by (1) Basic Science Research Program through the NRF funded by the Korean government, MSIP (2019R1A2C1006331), (2) Bio & Medical Technology Development Program of the NRF funded by the Korean government, MSIP (NRF-2015M3A9C6030838). This work was also supported by the GIST Research Institute (GRI) in 2019. We also acknowledge Eun-Ryeong Lee for designing the schematic diagram in Fig. 6.
Contributor Information
Da-Woon Jung, Email: jung@gist.ac.kr.
Yong-Chul Kim, Email: yongchul@gist.ac.kr.
Darren R. Williams, Email: darren@gist.ac.kr.
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Kim W.H. A novel indirubin derivative that increases somatic cell plasticity and inhibits tumorigenicity. Bioorg. Med. Chem. 2019;27(13):2923–2934. doi: 10.1016/j.bmc.2019.05.025. [DOI] [PubMed] [Google Scholar]
- 2.Meijer L. GSK-3-selective inhibitors derived from Tyrian purple indirubins. Chem. Biol. 2003;10(12):1255–1266. doi: 10.1016/j.chembiol.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 3.Ustanina S. The myogenic factor Myf5 supports efficient skeletal muscle regeneration by enabling transient myoblast amplification. Stem Cells. 2007;25(8):2006–2016. doi: 10.1634/stemcells.2006-0736. [DOI] [PubMed] [Google Scholar]
- 4.Olguin H.C., Pisconti A. Marking the tempo for myogenesis: Pax7 and the regulation of muscle stem cell fate decisions. J. Cell Mol. Med. 2012;16(5):1013–1025. doi: 10.1111/j.1582-4934.2011.01348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi S.J. 5,5'-substituted indirubin-3'-oxime derivatives as potent cyclin-dependent kinase inhibitors with anticancer activity. J. Med. Chem. 2010;53(9):3696–3706. doi: 10.1021/jm100080z. [DOI] [PubMed] [Google Scholar]
- 6.Kim W.H. Small molecules that recapitulate the early steps of urodele amphibian limb regeneration and confer multipotency. ACS Chem. Biol. 2012;7(4):732–743. doi: 10.1021/cb200532v. [DOI] [PubMed] [Google Scholar]
- 7.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods. 1983;65(1-2):55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]