Abstract
Salmonella is an important pathogen for poultry production as well as for human due to zoonotic importance. It has more than 2600 identified serovars despite of this identification and classification of Salmonella isolates into different serovars is critical for study of incidence and surveillance. This study investigates the epidemiology and molecular characterization of Salmonella isolates in broiler chicks during 1st week of life. A total of (n = 1000) samples including liver, intestine, yolk sac, spleen and heart blood were collected from El-Gharbia, El-Behera, Kafr-Elshikh, Alexandria, Marsamatroh Provinces in Egypt and tested through bacteriological, biochemical, serological and molecular examinations. Incidence of Salmonella was demonstrated on 75 positive samples from 1000 samples and the predominance of Salmonella that isolated from internal organs of newly hatched chicks was highest from yolk sacs (10%), liver and intestines (9%) followed by the spleen (7.5%) then heart blood (2%). Serotyping of the isolated strains using slide agglutination test revealed that 24 isolates belonging to S. enteritidis (1,9,12 g.m 1,7), while, 14 isolates belonging to S. virchow (6,7 r 1,2), in addition to, 12 isolates belonging to S. typhimurium (1,4,5,12.i.1,2) and 8 isolates belonging to S. kentucky (6,8.I,z). Enterobacterial Repetitive Intergenic Consensus (ERIC) PCR revealed that two S. enteriditis isolates were identical and one isolate differ by 40%, while two S. typhimurium isolates were identical by 80% and one isolate was similar by 20% to the other two isolates, in addition, two S. virchow isolates were identical by 80% and the two S. kentucky isolates were different.
Keywords: Salmonella, Chicken, Bactreiolgical, ERIC-PCR, Serotypes
Introduction
Salmonella isolates are considered as the most circulating and frequent bacterial agents causing disease poultry and other avian species. It is associated with high economic losses because of high mortality, morbidity and impaired productions. It is considered as a major food-borne pathogen in most countries of the world especially in developing countries (Soultose et al. 2003; Carraminana et al. 2004). Salmonella contamination of poultry and poultry products are frequently occurred and can be transmitted to humans through transportation and consumption of undercooked poultry meat (Bailey and Cosby 2003; Kimura et al. 2004). Wide variations of Salmonella serovars commonly infect poultry and one serovar may be common in a country for number of years before it is substituted by another isolate. The serovars may vary geographically, but the most common serovars reported globally are S. typhimurium and S. enteritidis as reported by World Health Organization (2006). Salmonellosis has been associated with infection of broiler flocks that has ability of vertical transmission to progeny (Irshad et al. 2013). The predominant serotypes have been identified in Egyptian poultry farms are Salmonella enterica serovar Typhimurium and S. enterica serovar Enteritidis (Abd El-Ghany et al. 2012).
Serotyping is a basic biomarker to investigate the epidemiological situation of Salmonella infections and it is commonly used to trace back the contamination sources during outbreaks. White and Kauffmann developed the serotyping scheme on 1920 that was based on the flagella H, somatic O antigens and the observed phase-shift in flagella antigen (Molbak et al. 2006). This method is worldwide and it is considered as the standard method for Salmonella serotypes identification. The advantages of identifying Salmonella serotypes include providing information about the disease severity, contamination source and the resistance pattern (Molbak et al. 2006). Moreover, molecular techniques have been used to differentiate the strains of Salmonella isolates including pulsed field gel electrophoresis (PFGE), enterobacterial repetitive intergenic consensus (ERIC) PCR, Random Amplification of Polymorphic DNA (RAPD), Single Strand Conformation Polymorphism (SSCP), hybridization and ribotyping-PCR (Anjay et al. 2015). Due scarce knowledge available on conventional and molecular identification of Salmonella species, this investigation was designed to follow the epidemiology of Salmonella isolates through biochemical, serological and molecular methods.
Materials and methods
Sample collection
A total of one thousand samples including liver, intestine, yolk sac, spleen and heart blood of newly hatched chicks during first week of life were collected aseptically from 25 poultry farms located in five different governorates in Egypt (El-Gharbia, El-Kafr-Elshikh, El-Behera, Alexandria and Matroh) with 10 chicks for each farm as shown in Table 1. The samples were collected in separate sterile plastic bags and immediately transported to the laboratory in ice box (4 °C).
Table 1.
History of examined farms
| Farm No. | Location | No. of chicks | Total farm No. | Age of chick (day) | Mortalities in week % | Antibiotic used at 1st 3 days of age |
|---|---|---|---|---|---|---|
| 1 | El-Gharbia | 40 | 5000 | 1 | 10 | Ciprofloxacin |
| 2 | El-Gharbia | 40 | 7000 | 5 | 15 | Colistine + tylosine |
| 3 | El-Gharbia | 40 | 5000 | 3 | 5 | Ciprofloxacin |
| 4 | El-Gharbia | 40 | 10,000 | 5 | 12 | Florfenicol |
| 5 | El-Gharbia | 40 | 12,000 | 1 | 18 | Ciprofloxacin |
| 6 | El-Behera | 40 | 15,000 | 3 | 15 | Enrofloxacin + colistine |
| 7 | El-Behera | 40 | 5000 | 1 | 2 | Enrofloxacin |
| 8 | El-Behera | 40 | 10,000 | 5 | 7 | Colistine + tylosine |
| 9 | El-Behera | 40 | 15,000 | 3 | 12 | Ciprofloxacin |
| 10 | El-Behera | 40 | 20,000 | 1 | 14 | Florfenicol |
| 11 | Kafr-Elshikh | 40 | 10,000 | 5 | 10 | Enrofloxacin |
| 12 | Kafr-Elshikh | 40 | 10,000 | 5 | 8 | Oxytetracyclin + tylosine |
| 13 | Kafr-Elshikh | 40 | 15,000 | 3 | 7 | Enrofloxacin + colistine |
| 14 | Kafr-Elshikh | 40 | 20,000 | 1 | 10 | Ciprofloxacin |
| 15 | Kafr-Elshikh | 40 | 10,000 | 3 | 5 | Florfenicol |
| 16 | Alexandria | 40 | 15,000 | 5 | 8 | Ciprofloxacin |
| 17 | Alexandria | 40 | 15,000 | 5 | 14 | Colistine + tylosine |
| 18 | Alexandria | 40 | 5000 | 1 | 5 | Ciprofloxacin |
| 19 | Alexandria | 40 | 5000 | 5 | 10 | Ciprofloxacin |
| 20 | Alexandria | 40 | 10,000 | 1 | 15 | Oxytetracyclin + tylosine |
| 21 | Marsamatroh | 40 | 5000 | 1 | 5 | Oxytetracyclin + tylosine |
| 22 | Marsamatroh | 40 | 5000 | 5 | 12 | Enrofloxacin |
| 23 | Marsamatroh | 40 | 10,000 | 1 | 20 | Ciprofloxacin |
| 24 | Marsamatroh | 40 | 5000 | 1 | 7 | Florfenicol |
| 25 | Marsamatroh | 40 | 5000 | 3 | 2 | Ciprofloxacin |
Bacterial isolation
The collected samples were cultured on 1% peptone broth then 1 ml selenite F. broth and incubated aerobically at 37 °C for 18 h then were subcultured to MacConkey, Salmonella shigella agar and/or XLD media. The cultured plates were incubated at 37 °C for 24 h. Suspected colonies were picked up, preserved into semi solid agar as stock medium and into slant agar for further biochemical and serological identification.
Biochemical identification
Dry heat fixed smears of suspected colonies were stained using Gram’s stain then were examined, revealing the presence of Gram negative bacilli. The suspected isolates were identified biochemically (Hossain et al. 2006) by applying catalase test, oxidase test and IMViC group of biochemical tests. The identified isolates as Salmonella species were cultivated on triple sugar iron agar (TSI).
Serological identification
Serogrouping of identified bacterial isolates was performed according to Kauffmann–White method (Aribam et al. 2015).
Molecular identification
Biochemically, identified Salmonella isolates were then serotyped and further characterization was done by using ERIC PCR for intra-serotyping of Salmonella isolates. DNA was extracted from studied isolates according to QIAamp DNA mini kit instructions and PCR Master Mix was prepared according to Emerald Amp GT PCR master mix (Tarkara) Code.No.RR310Akit using the following primer set ERIC-DG111-F with primers sequences ATG TAA GCT CCT GGG GAT TCA C and ERIC-DG112-R with primers sequences AAG TAA GTG ACT GGG GTG AGC G. Amplification of primers was done by using thermal cycling (Fendri et al. 2013). Briefly, denaturation at 94 °C for 2 min, annealing at 49 for and extention at 72 for 2 min followed by 35 cycles including 94 °C for 1 min, 56 °C for 1 min and 72 °C for 2 min and final extension at 72 °C for 5 min. After that the amplified product was loaded on 1.5% agrose gel using 100 bp gene ruler for 1 h at 5 V and the gel was visualized by chemical documentation (Bio Red).
Results
Morphological identification of the isolated organisms
Morphology revealed the 75 samples out of one thousands appeared on MacConkey agar, colorless and translucent, though they sometimes have dark centers. Gram’s stain smears from suspected colonies showed Gram-negative rod-shaped motile bacteria, or bacillus. On XLD, they were pink with or without black centers while, colonies on S.S agar media appeared as white colonies with black center.
Biochemical identification of the isolated organisms
The isolated micro-organisms were positive for methyl red, catalase, TSI, citrate utilization test, lysine iron agar, oxidase and christensen citrate while negative to indole, Phenol red, sucrose, and Voges-Proskauer. They ferment variety of sugar types but remain negative on KCN medium and ONPG reaction as illustrated in Table 2.
Table 2.
Biochemical identification of various organisms suspected to Salmonella isolates
| Biochemical tests | Salmonella isolates |
|---|---|
| Indole | −ve |
| Methyl red | +ve |
| Voges Proskauer | −ve |
| Citrate utilization test | +ve |
| TSI | K/A. + ve H2S |
| Lysine iron agar | +ve |
| Christensen citrate | +ve |
| Hydrolysis of urea | −ve |
| Gelatin liquefaction | −ve |
| Oxidase test | −ve |
| Ornithine decarboxylase | +ve |
| Mannitol | +ve |
| l-arabinose | +ve |
| Maltose | +ve |
| l-rhamnose | +ve |
| Glucose | +ve |
| KCN medium | −ve |
| ONPG-reaction | −ve |
| Catalase test | +ve |
Incidence of Salmonella in different organs
Revealing to traditional identification on media and biochemically identification the proportion of isolates result as Salmonella isolates from various organs of newly hatched chicks represented by 7.5% total distribution in various organ as shown in Table 3.
Table 3.
Incidence of Salmonella isolates in various organs of 1 week old chicks
| Organs | No. of examined organs | No of Salmonella +ve organs | Percentage (%) of isolation |
|---|---|---|---|
| Liver | 200 | 18 | 9 |
| Yolk sac | 200 | 20 | 10 |
| Intestine | 200 | 18 | 9 |
| Spleen | 200 | 15 | 7.5 |
| Heart blood | 200 | 4 | 2 |
| Total | 1000 | 75 | 7.5 |
Serological identification of the isolated organisms
The serotyping investigated the S. typhimurium, S. enteritidis, S. virchow and S. kentucky with O antigen are 4, 3, 2 and 2 while presence of H factor only in S. enteritidis. S. typhimurium and S. virchow as shown in Table 4.
Table 4.
Results of serotyping of the isolated Salmonella strains
| Serial No. | Salmonella serotype | Group | Antigenic structure | ||
|---|---|---|---|---|---|
| O-antigen | H-factor | ||||
| Phase I | Phase II | ||||
| 1 | S. enteritidis | D | 1,9,12 | g,m | 1.7 |
| 2 | S. typhimurium | B | 1,4,5,12 | I | 1.2 |
| 3 | S. virchow | C1 | 6,7 | R | 1.2 |
| 4 | S. kentucky | C3 | 6,8 | L.z | – |
Strain wise distribution of Salmonella species
Serotyping revealed that the distribution of S. entritidis was comparatively higher than S. Virchow, S. typhimurium and S. Kentucky as 2.4, 1.4, 1.2 and 0.8% while 1.7% strains were untypable as illustrated in Table 5.
Table 5.
Strain wise distribution of isolated Salmonella species
| Salmonella serotype | No. of the isolated strains | % of the isolated strains |
|---|---|---|
| S. enteritidis | 24 | 2.4 |
| S. typhimurium | 12 | 1.2 |
| S. Virchow | 14 | 1.4 |
| S. Kentucky | 8 | 0.8 |
| Un typable | 17 | 1.7 |
| Total isolated strains | 75 | 7.5 |
ERIC-PCR revealed that two Salmonella enteritidis were found identical while one was different i-e; lane S. T1, S. T2 and S. T3 with 232, 235 and 235 bp respectively. Similarly, three S. typhimurium were identical i-e; lane S.E1, S.E2 and S.E3 166, 166 and 166 bp. Additionally, lane S.V1 and S.V2 i-e; 266 and 266 bp showed two S. virchow were identical to each other while lane S.K1 and S.K2 i-e; 149 and 151 bp revealed that two S. Kentucky differ from each other as shown in Table 6 and Fig. 1.
Table 6.
ERIC PCR for selected strain of investigated Salmonella
| PCR bands (bp) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| S.T1 | S.T2 | S.T3 | S.E1 | S.E2 | S.E3 | S.V1 | S.V2 | S.K1 | S.K2 |
| 1909 | 1968 | 1938 | 1200 | 1200 | 1200 | 1372 | 1357 | 958 | 948 |
| 1200 | 1214 | 1214 | 1070 | 1070 | 1070 | 1034 | 1034 | 576 | 581 |
| 700 | 700 | 700 | 708 | 700 | 700 | 760 | 760 | 378 | 378 |
| 391 | 395 | 395 | 450 | 445 | 445 | 445 | 445 | 277 | 280 |
| 232 | 235 | 235 | 166 | 166 | 166 | 266 | 266 | 149 | 151 |
Fig. 1.
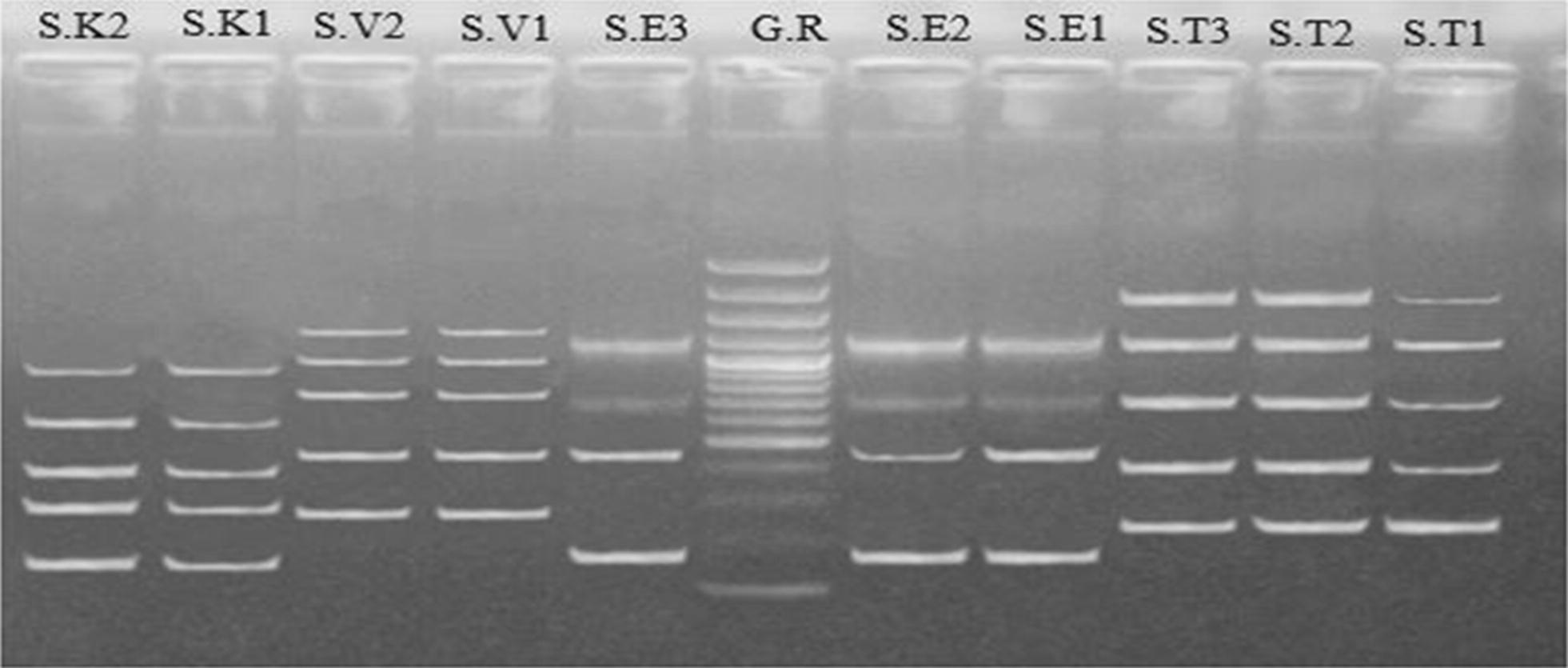
ERIC PCR of different isolates of Salmonella. lane S. T1, S. T2 and S. T3 are S. typhimurium, lane S.E1, S.E2 and S.E3 are S. enteritidis, lane S.V1 and S.V2 are S. virchow while lane S.K1 and S.K2 are S. Kentucky verified by using lane G.R 100 bp gene ruler
Discussion
Salmonella is considered as one of the major pathogenic agents which infect the variety of avian species specially poultry birds including layer as well as broiler reared in the modern intensive system with higher biosafety, biosecurity and standard management. Any contributions for elimination of Salmonella incidences and infection in birds could have a major influence in reducing the populations of the organism under natural conditions. One thousand samples were collected from different farms including liver, intestine, yolk sac, spleen and heart blood of newly hatched chicks at El-Gharbia, El-Behera, Kafr-Elshikh, Alexandria, Marsamatroh Provinces. The samples were examined bactriologically to isolate the Salmonella isolates.
In this study, 75 samples out of 1000 samples (7.5%) were found positive. The higher percentage of isolation from the internal organs from yolk sacs (10%) then from livers (9%) and from 20 intestines (9%) were the same, spleen (7.5%), and finally heart blood (2%) (Table 3). These results are in contrary to (Ahmed et al. 2008; Islam et al. 2016) who showed that the prevalence of avian Salmonellosis was highest in adult layer (53.25%) followed by brooding (14.55%) then growing (16.10%) and pullet (16.10%). The prevalence rate of Salmonella spp. in different poultry farm were different i-e; the 80 samples were tested from the clinically healthy birds showed 44 (55%) positive (Ahmed14). Moreover the samples from birds having diarrhea infection rate (66.67%) (Hossain et al. 2006).
The study revealed that pink colonies with or without black centers were typical for Salmonella on XLD. Many cultures of Salmonella spp. may produce large colonies with glossy black centers or may appear as almost completely black that is similar to (Ramya et al. 2012). Correspondingly, the characteristics of Salmonella spp. colonies are translucent, small round, smooth, black or colorless was observed on SSA, black colonies on TSI agar (Islam et al. 2016 and Sujatha et al. 2003). The isolated micro-organisms were Catalase-positive, oxidase, indole, Phenol red, sucrose, Voges-Proskauer and urease negative while methyl red, H2S production, citrate-positive and glucose positive. The current finding is similar Islam et al. (2016) who have found Salmonella isolates were MR test and citrate utilization test positive, ferment dextrose, maltose and mannitol but fail to ferment sucrose and lactose.
In the present study serological identification of the isolated bacteria revealed 24 isolates belonging to group D and identified as S. enteritidis (1,9,12. g,m 1,7) and 12 isolates belonging in the group B and identified as S. typhimurium (1,4,5,12.i.1,2) and 14 isolates belonging in the group C1 and identified as S. virchow (6,7.r,1,2) and 8 isolates belonging in the group C3 and identified as S. kentucky (6,8.I,z). Meanwhile, 17 isolates were untypable (Table 4). Moreover, 68 serotypes were identified among 75 Salmonella isolates, and 17 isolates were untypeable (Table 5). The most prevalent serovar detected in this study was S. enteritidis 2.4% followed by S. virchow 1.4%, S. typhimurium 1.2% and S. kentucky 0.8%. The most commonly isolated serotype from different organs was S. enteritidis the same results were recorded in Egypt by (Sujatha et al. 2003; Akeila et al.2013 and Rabie et al. 2012) who confirmed the prevalence of S. enteritidis and S. typhimurium by (58.33% and 41.66%), respectively from chickens. In addition, S. enteritidis and S. typhimurium were predominant in Saudi Arabia, by (55.56% and 22.22%, respectively) among the detected Salmonella serovars from chickens (Moussa et al. 2010). Im et al. (2015) reported that the most prevalent Salmonella serovars in the flocks were Salmonella bareilly 41.2%), Salmonella mbandaka (32.4%), and Salmonella rissen (17.6%).
Ten Salmonella isolates belonging to 4 serotypes produced ERIC PCR fingerprints that were distinct for each serotype (Table 6). ERIC PCR found that three S. enteriditis isolates (isolates 2 and 3 identical in 1200, 1070, 700, 445, 166 bands but isolate 1 different from it in 708,450 bands) so two S. enteriditis isolates were identical and one isolate was different from it by 40%. Three S. typhimurium isolates (isolate 1 belonging to 1909, 1200, 700, 391 and 232 bands it was different from isolates 2 and 3 while isolates 2 and 3 identical in 1214, 700, 395, 235 bands and the difference in 1968 and 1938 bands) so two S. typhimurium isolates were identical by 80% and one isolate was similar by 20% to the other two isolates two S. virchow isolates were identical in 1034, 760, 445, 266 bands and the difference in 1372 and 1357 bands) so two S. virchow isolates were identical by 80%. Two S. kentucky isolates (isolate 1 belonging to 958, 576, 378, 277, 149 bands and it was different from isolates 2 which belonging to 948,581,378,280,151 bands) so two S. kentucky isolates were not identical. ERIC-PCR is a useful and recent method for DNA typing for analysis and evaluation of fingerprinting. It is used in epidemiology of Salmonella enteritidis (Suh and Song 2006). Using specific ERIC primers, a total of 30 strains of Salmonella enteritidis of four main clusters had found 60% similarity.
This study found that the Serotyping of the isolated strains revealed that 24 isolates belonging to S. enteritidis (1,9,12 g.m 1,7), while, 14 isolates belonging to S. virchow (6,7 r 1,2), in addition to, 12 isolates belonging to S. typhimurium (1,4,5,12.i.1,2) and 8 isolates belonging to S. kentucky (6,8.I,z). ERIC-PCR revealed that two S. enteriditis isolates were identical and one isolate was different from it by 40%, while two S. typhimurium isolates were identical by 80%and one isolate was similar by 20% to the other two isolates, in addition to, two S. virchow isolates were identical by 80% and the two S. kentucky isolates were not identical. This study will help future researchers to uncover new and critical methods that should be used to improve diagnosis and control measures for prevention zoonotic infections of Salmonella species.
Acknowledgements
The authors extended their appreciation to the Deanship of Scientific Research at King Saud University for funding this work through the College of Food and Agriculture Sciences Research Center.
Authors’ contributions
All authors participated in making the design, performing the experiment, analyses of the data, and writing the paper.
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
This trial was performed strictly according to the recommendations and guidelines of the committee on the ethics of animal experiments of Alexandria University, Egypt. All efforts were made to minimize suffering.
Consent for publication
All authors gave their informed consent prior to their inclusion in the study.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mahmoud E. Sedeik, Email: seddeeklab@yahoo.com
Nahed A. El-shall, Email: nahed.elshall@gmail.com, Email: nahed.abdelgawad@alexu.edu.eg
Ashraf M. Awad, Email: dr.ashrafawad@gmail.com
Sally M. Elfeky, Email: s.elfeky@gmail.com
Mohamed E. Abd El-Hack, Email: m.ezzat@zu.edu.eg, Email: dr.mohamed.e.abdalhaq@gmail.com
Elsayed O. S. Hussein, Email: shessin@ksu.edu.sa
Ayman A. Swelum, Email: aswelum@ksu.edu.sa
References
- Abd El-Ghany W, El-Shafii S, Hatem M. A survey on Salmonella species isolated from chicken flocks in Egypt. Asian J Anim Vet Adv. 2012;7:489–501. doi: 10.3923/ajava.2012.489.501. [DOI] [Google Scholar]
- Ahmed AKM, Islam MT, Haider MG, Hossain MM. Sero prevalence and pathology of naturally infected Salmonellosis in poultry with isolation and identification of causal agents. J Bangladesh Agric Univ. 2008;6:327–334. doi: 10.3329/jbau.v6i2.4830. [DOI] [Google Scholar]
- Akeila MA, Ellakany HF, Sedeik ME, Behar HM. Characterization and plasmid profiling of Salmonella enteritidis isolated from broiler chickens. Alex J Vet Sci. 2013;39:105–111. [Google Scholar]
- Anjay AK, Agarwal RK, Ramees TP, Dubal ZB, Kaushik P, Kumar MS, Dudhe NC, Milton AAP, Abhishek BK, Shagufta B. Molecular typing of Salmonella typhimurium and S. enteritidis serovars from diverse origin by ERIC-PCR. J Pure Appl Microbiol. 2015;9:2627–2634. [Google Scholar]
- Aribam SD, Elsheimer-Matulova M, Matsui H, Hirota J, Shiraiwa K, Ogawa Y, Hikono H, Shimoji Y, Eguchi M. Variation in antigen-antibody affinity among serotypes of Salmonella O4 serogroup, determined using specific antisera. FEMS Microbiol Let. 2015;362:362–368. doi: 10.1093/femsle/fnv168. [DOI] [PubMed] [Google Scholar]
- Bailey JS, Cosby DE. Detection of Salmonellae from chicken rinses and chicken hot dogs with automated Bax PCR system. J Food Protect. 2003;66:2138–2140. doi: 10.4315/0362-028X-66.11.2138. [DOI] [PubMed] [Google Scholar]
- Carraminana JJ, Agustin I, Herrera A. High prevalence of multiple resistance to antibiotics in Salmonellae serovars isolated from a poultry slaughterhouse in Spain. Vet Microbiol. 2004;104:133–139. doi: 10.1016/j.vetmic.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Fendri I, Hassena AB, Grosset N, Barkallah M, Khannous L, Chuat V, Gautier M, Gdoura R. Genetic diversity of food-isolated Salmonella strains through Pulsed Field Gel Electrophoresis (PFGE) and Enterobacterial Repetitive Intergenic Consensus (ERIC-PCR) PLoS ONE. 2013;8:e81315. doi: 10.1371/journal.pone.0081315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MA, Aalbaek B, Christensen JP, Elisabeth H, Islam MA, Pankaj K. Observations on experimental infection of Salmonella gallinarum in Fayoumi and Hyline layer chickens. Bangladesh J Prog Agric. 2006;14:85–89. [Google Scholar]
- Im MC, Jeong JS, Kwon Y, Jeong O, Lee YL. Prevalence and characteristics of Salmonella spp. isolated from commercial layer farms in Korea. Animal and Plant Quarantine Agency: Republic of Korea; 2015. [DOI] [PubMed] [Google Scholar]
- Irshad B, Asad AA, Iftikhar M. The prevalence of Salmonellosis in poultry farms in and around district Kasur, Pakistan. Sci Int. 2013;25(3):603–604. [Google Scholar]
- Islam JAT, Mahbub-E-Elahi AT, Kamrul H. Isolation and identification of Salmonella spp. from broiler and their antibiogram study in Sylhet, Bangladesh. J Appl Biol Biotechnol. 2016;4:046–051. [Google Scholar]
- Kimura AC, Reddy V, Marcus R. Chicken consumption is a newly identified risk factor for sporadic Salmonellae enteric serotype enteritidis infections in the United States. Clin Infect Dis. 2004;38:244–252. doi: 10.1086/381576. [DOI] [PubMed] [Google Scholar]
- Molbak K, Olsen J, Wegener H (2006) Salmonella Infections. In Reimann H, Cliver D (eds) Food borne infections and intoxications. Academic press, pp 55–115.
- Moussa IM, Gassem MA, AlDoss AA, Mahmoud WA, Abdel Mawgood AL. Using molecular techniques for rapid detection of Salmonella serovars in frozen chicken and chicken products collected from Riyadh, Saudi Arabia. Afr J Biotechnol. 2010;9:612–619. doi: 10.5897/AJB09.1761. [DOI] [Google Scholar]
- Rabie N, Nashwa K, Mervat ER, Jehan F. Epidemiological and molecular studies of Salmonella isolates from chicken, chicken meat and human in Toukh. Egypt. Glob Vet. 2012;8:128–132. [Google Scholar]
- Ramya P, Madhavarao T, Venkateswara Rao L. Study on the incidence of Salmonella enteritidis in poultry and meat samples by cultural and PCR methods. Vet World. 2012;5:541–545. doi: 10.5455/vetworld.2012.541-545. [DOI] [Google Scholar]
- Soultose N, Koidis P, Madden RH. Prevalence of Listeria and Salmonellae in retail chicken in Northern Ireland. Appl Microbiol. 2003;37:421–423. doi: 10.1046/j.1472-765X.2003.01423.x. [DOI] [PubMed] [Google Scholar]
- Suh DK, Song JC. Analysis of Salmonella enterica serotype Enteritidis isolated from human and chickens by repetitive sequence-PCR fingerprinting, antibiotic resistance and profiles. J Vet Sci. 2006;7:37–41. doi: 10.4142/jvs.2006.7.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sujatha K, Dhanalakshmi K, Rao AS. Isolation and characterization of Salmonella gallinarum from chicken. Indian Vet J. 2003;80:473–474. [Google Scholar]
- World Health Organization . Global Salmonella-Survey Progress Report (2000–2005): building capacity for laboratory-based foodborne disease surveillance and outbreak detection and response. Geneva: World Health Organization; 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


