Abstract
Phosphorylation is an important event in cell signaling that is modulated by kinases and phosphatases. In Trypanosoma cruzi, the etiological agent of Chagas disease, approximately 2% of the protein-coding genes encode for protein kinases. This parasite has a heteroxenic life cycle with four different development stages. In the midgut of invertebrate vector, epimastigotes differentiate into metacyclic trypomastigotes in a process known as metacyclogenesis. This process can be reproduced in vitro by submitting parasites to nutritional stress (NS). Aiming to contribute to the elucidation of mechanisms that trigger metacyclogenesis, we applied super-SILAC (super-stable isotope labeling by amino acids in cell culture) and LC-MS/MS to analyze different points during NS. This analysis resulted in the identification of 4205 protein groups and 3643 phosphopeptides with the location of 4846 phosphorylation sites. Several phosphosites were considered modulated along NS and are present in proteins associated with various functions, such as fatty acid synthesis and the regulation of protein expression, reinforcing the importance of phosphorylation and signaling events to the parasite. These modulated sites may be triggers of metacyclogenesis.
Electronic supplementary material
The online version of this article (10.1007/s12192-019-01018-7) contains supplementary material, which is available to authorized users.
Keywords: Trypanosoma cruzi, Metacyclogenesis, Nutritional stress, Proteomics, Phosphoproteomics, Super-SILAC
Introduction
Reversible protein phosphorylation plays an important role in the regulation of cellular behaviors, such as proliferation, differentiation, migration, and apoptosis, in response to extracellular and intracellular stimuli (Tan 2011). This mechanism is regulated by the action of protein kinases and phosphatases. In Trypanosoma cruzi, the etiological agent of Chagas disease (Chagas 1909), the protein-coding genes encode for protein kinases (Parsons et al. 2005), and there are 86 protein phosphatases, among which the majority are serine/threonine phosphatases (Brenchley et al. 2007). In this parasite, the regulation of gene expression occurs mainly at the posttranscriptional level, involving translation control, trans-splicing, mRNA stability, and posttranslational modifications, such as phosphorylation (Martínez-Calvillo et al. 2010; De Gaudenzi et al. 2011; Smircich et al. 2015).
T. cruzi has a heteroxenic life cycle and presents multiple morphological forms, among them epimastigotes (Epi) and metacyclic trypomastigotes (Meta) in the triatomine vector and amastigotes and trypomastigotes in the human host (Tyler and Engman 2001). Therefore, during its life cycle, the parasite is submitted to different microenvironments and receives several external stimuli that can lead to parasite differentiation. The signaling by phosphorylation may modulate this process. In the midgut of invertebrate host, Epi differentiate into Meta in a process known as metacyclogenesis. The increase in the cell population and the low nutrient availability are associated with the differentiation of the parasite (Kollien and Schaub 2000; Figueiredo et al. 2000). This process can be reproduced in vitro by submitting Epi to NS through incubation in TAU (triatomine artificial urine) medium (Contreras et al. 1985), which mimics the urine of the invertebrate host, followed by incubation in TAU3AAG medium, which allows the adhesion of the parasites to the substrate and the subsequent release of Meta (Bonaldo et al. 1988).
NS has been proposed to trigger substrate adhesion and metacyclogenesis (Figueiredo et al. 2000). The glucose depletion in medium leads to the elongation of Epi flagella (Tyler and Engman 2000), which may be a prestage to adhesion or an adaptation that allows greater capture of nutrients from the medium by increasing the membrane area (Hernández et al. 2012). In addition, the reservosome, a structure where lipids and proteins are stored, decreases in size, indicating that this stored content is consumed during differentiation (Soares 1999). A more recent study has shown ultrastructural changes in parasites subjected to NS, with a slimmer appearance, alterations in the location and structure of mitochondrion-kinetoplast, and the accumulation of vesicles in the cytoplasm of the parasites (Pérez-Morales et al. 2017).
The lipid content of Epi subjected to NS is more similar to that of Meta when compared with Epi, with a degree of unsaturation, influencing the membrane permeability of the parasite (Esteves et al. 1989). Autophagy is another mechanism related to NS, as the degradation of proteins and organelles is important for remodeling during development and cell differentiation (Alvarez et al. 2008; Schoijet et al. 2017). The involvement of phosphorylation in T. cruzi differentiation has also been described: the increase in eukaryotic initiation factor 2α phosphorylation during NS culminates in the reduction of protein synthesis (Tonelli et al. 2011), and cAMP-PKA signaling stimulates differentiation when cAMP levels are elevated (Gonzales-Perdomo et al. 1988; Bao et al. 2008), in addition to other possible targets of the PKA, such as trans-sialidases and the proteins involved in the adhesion and invasion of host cells (Bao et al. 2010).
Many efforts have been made to understand the differentiation of the parasite as well as the role of phosphorylation in this process (Nakayasu et al. 2009; Marchini et al. 2011; Queiroz et al. 2014; Amorim et al. 2017), but none of these studies have conducted the analyses focusing on different points during the NS stage, an essential step in the process of parasite differentiation. The super-SILAC strategy (Geiger et al. 2010), employed in the present study, allows the quantification of the proteins and phosphosites by spiking each sample with a labeled reference, allowing the indirect comparison of intensities in the different experimental conditions.
Due to the importance of NS for differentiation and because it is a process of short duration, with the receipt of several external stimuli, the present study is a proteomics and phosphoproteomics analysis of the different experimental points along NS. By using mass spectrometry (MS)–based proteomics and the super-SILAC quantitative methodology, we aimed to identify proteins involved in metacyclogenesis and possible phosphosites that play roles as triggers of differentiation, contributing to the elucidation of the mechanisms involved in metacyclogenesis.
Methods
Sample preparation for proteome and phosphoproteome analyses
Each experiment was performed in two biological replicates. T. cruziDm28c (Contreras et al. 1988) epimastigotes were cultured in liver infusion tryptose medium (LITB) (Camargo and Silva 1964) supplemented with 10% fetal bovine serum without agitation at 28 °C. The parasites were collected after 5 days of culture in the early stationary phase (ST-0min). To obtain parasites in different times of stress, the parasites were harvested after 5 days of culture by centrifugation at 7000×g for 5 min at 20 °C, washed once with TAU medium (2 mM CaCl2, 17 mM KCl, 2 mM MgCl2, 190 mM NaCl, 8 mM phosphate buffer, pH 6.0), resuspended in TAU at a final concentration of 5 × 108 parasites/mL and incubated for 5 min (ST-5min), 15 min (ST-15min), 1 h (ST-1h), and 2 h (ST-2h).
For the super-SILAC standard, T. cruzi Dm28c epimastigotes were cultured in LM-14B medium (De Paula Lima et al. 2014), replacing lysine and arginine with lysine 13C615N2 (Lys-8) and arginine 13C615N4 (Arg-10) (Cambridge Isotope Laboratories), without agitation at 28 °C. Then, 3 × 109 cells were collected after 3 and 5 days of culture and at different times (5 min, 15 min, 1 h, and 2 h) of NS in vitro, which were mixed in the same proportion, generating the super-SILAC standard.
Then, 1.5 × 109 cells from the super-SILAC standard were combined with 1.5 × 109 cells from each experimental time point (ST-0min, ST-5min, ST-15min, ST-1h, ST-2h). The cells were washed twice with phosphate-buffered saline (PBS; 137 mM NaCl, 4.3 mM Na2HPO4, 2.7 mM KCl, 1.5 mM KH2HPO4, pH 7.2), centrifuged at 7000×g for 5 min at 20 °C, resuspended in 1.95 mL lysis buffer (4% SDS, 100 mM Tris-HCl pH 7.5, 100 mM DTT), vortexed, incubated at 95 °C for 5 min, and sonicated for approximately 1 h to remove viscosity. The cell lysates were then centrifuged at 20,000×g for 10 min at 20 °C, and the supernatants were transferred to new tubes and stored at − 80 °C until use. The cell lysates were digested using the filter-aided sample preparation (FASP) method (Wiśniewski et al. 2009) with some modifications, as previously described (Amorim et al. 2017).
After protein digestion, the peptides were fractionated by high pH reversed-phase chromatography using Sep Pack C18 130 mg columns (Waters) as previously described (Amorim et al. 2017). After each elution, the peptides were acidified with 0.5% trifluoroacetic acid (TFA). Approximately 50 μg of peptides was separated for the proteomics analysis, dried in a vacuum centrifuge, desalted, and stored in C18 Stage-Tips (Rappsilber et al. 2003) at 4 °C until analysis. The remaining volume was dried and processed for phosphopeptide enrichment. Fractions were resuspended in 1 mL of 80% MeCN, 2.5% TFA, and 70 mg/mL phthalic acid (Sigma) and incubated for 1 h by end-over-end rotation with 100 μL of microbead solution (80% MeCN, 2.5% TFA, 40 mg/mL TiO2 microbeads—GL Sciences), following a previously published protocol (Batista et al. 2017). After incubation, microbeads were spun down and washed twice with 500 μL of wash buffer 1 (80% MeCN, 2.5% TFA, and 70 mg/mL phthalic acid), twice with 500 μL of wash buffer 2 (80% MeCN, 0.1% TFA) and twice with 500 μL of wash buffer 3 (0.1% TFA). For peptide elution, the microbeads were incubated for 5 min with 50 μL of 0.3 M ammonium hydroxide followed by centrifugation at 1000×g for 1 min, and the supernatants were recovered into a fresh tube. This elution process was repeated three times, and the combined eluates were fully dried in a vacuum centrifuge, desalted, and stored in C18 Stage-Tips (Rappsilber et al. 2003).
Nano-LC-MS/MS analysis
After elution from C18 Stage-Tips, the peptide mixtures were separated by online C18 reversed-phase (RP) nanoscale capillary liquid chromatography (nano-UPLC) and analyzed by nanoelectrospray tandem mass spectrometry (nano-ESI MS/MS) in duplicate. The experiments were performed with an EASY-nLC 1000 chromatograph (Thermo Scientific) coupled to an LTQ Orbitrap XL ETD (Thermo Scientific) mass spectrometer (mass spectrometry facility RPT02H/Carlos Chagas Institute – Fiocruz Parana). Separation of the peptides was performed at a flow rate of 250 nL/min of the mobile phase (MeCN, 0.1% formic acid, 5% DMSO) with a linear gradient from 5 to 40% MeCN in 120 min for phosphoproteomics. For proteomics, the gradient ranged from 5 to 28% MeCN in 214 min followed by 28–40% MeCN in 26 min (total of 240 min), and two technical replicates were performed. The chromatography was performed on an analytical silica column of 30 cm, 75 μm ID and C18 particles with a diameter of 1.9 μM and heated to 60 °C. The peptides were ionized by nanoelectrospray (voltage 2.7 kV) and injected into the MS. The mode of acquisition was data-dependent analysis (DDA) conducted as follows: a preview full scan was acquired on the Orbitrap with a resolution of 15,000, followed by a selection of the 10 most intense ions, which were fragmented by CID and analyzed in the Ion trap. Parallel to the MS/MS, a full scan was acquired on the Orbitrap with a resolution of 60,000. When selecting ions for MS2, a dynamic exclusion list of 90 s was used. In phosphoproteomics runs, the isolation and fragmentation of the precursors containing phosphate groups were performed using the methods of neutral loss and multistage activation. The lock mass option was used at 401.922718 m/z to obtain better accuracy—error below 0.5 ppm—on a mass of tryptic peptides detected by MS. The mass spectrometry (phospho) proteomics data were deposited in the ProteomeXchange Consortium via the PRIDE (Vizcaíno et al. 2016) partner repository with the dataset identifier PDXD012113.
Data analysis
The comparison of data obtained in MS and MS/MS with database identification and quantification of peptides, proteins, and phosphorylation sites were carried out in MaxQuant [32] software version 1.5.2.8. For identification, we used the T. cruziDm28c database of the UniProt (www.UniProt.org), downloaded on April 3, 2016, containing 11,346 sequences. A decoy database containing the reverse protein sequence of each entry was also used to estimate the false discovery rate (FDR). The MaxQuant software was set with parameters: for MS spectra accuracy of 20 ppm (first search) and 4.5 ppm (second search); for MS/MS spectra accuracy of 0.5 Da; the options “match between runs” and “requantify” were enabled. In searches related to the phosphoproteomics analysis, the phosphorylation of serine, threonine, and tyrosine (STY) was included as a variable modification. For identification, peptides with at least 7 amino acids were required. An FDR of 1% for peptide and protein identification was independently applied. Similar protein sequences that did not permit distinction were clustered in groups of proteins. The proteins and phosphorylation sites were quantified by the difference in ratios between the pairs of “light” and “heavy” peptides. The tables generated by MaxQuant were analyzed by Perseus software version 1.5.3.2. The contaminants and reverse sequences were removed, and the normalized ratio of intensity heavy/light was converted to a −log2(x) scale. The Benjamini–Hochberg test (Benjamini and Hochberg 1995) and a q value ≤ 0.2 Student’s t test were applied as cutoffs to select the modulated proteins and phosphorylation sites. For phosphoproteomics data, we normalized the intensity heavy/light of phosphorylation sites containing 1, 2, or 3 phospho groups for each biological replicate by subtracting the heavy/light value of proteins (obtained in the proteomics analysis) from the heavy/light value of the corresponding phosphosites. The normalized phosphosite intensities resulted in modulation of the phosphorylation sites, regardless of the protein variation. The enriched GO terms were obtained from the TriTrypDB online tool at http://tritrypdb.org/tritrypdb/. The search for molecular function terms was performed in the T. cruzi background. Terms with p values < 0.02 were considered significant.
Results
Profile of the proteomics data
In this study, we applied LC-MS/MS associated with super-SILAC methodology to identify proteins and phosphosites regulated during NS that can play roles as triggers of differentiation. For this purpose, parasites were cultured in LIT-B complex medium and LM-14B defined medium containing lysine and arginine with heavy isotopes and then submitted to NS for different times. The super-SILAC standard showed enough labeled proteins (96.3%) to perform this experiment.
A total of 80 runs were performed and 2,873,119 MS/MS spectra were acquired. These mass spectra were compared with the database, and 35.5% were identified, corresponding to 34,341 nonredundant peptide sequences that were grouped into 4205 protein groups (Table A1), of which 98.7% could be quantified. Approximately 2465 were annotated as uncharacterized proteins (approximately 59% of total proteins identified). This number represents how several proteins have unknown functions in the T. cruzi genome, reinforcing the study and characterization of these proteins. The numbers of proteins and peptides identified and quantified in the two biological replicates were similar (Fig. A1A), and the samples showed a high correlation between biological replicates (Fig. A1B).
Amorim et al. (2017) also analyzed the experimental point ST-2h using another quantitative methodology, label-free quantification (LFQ). Considering only the data obtained at this experimental time point, 78% of the proteins were identified in both studies, with 392 identified only by Amorim et al. (2017) and 586 identified exclusively in the current study (Fig. A2). Regarding the quantification of these proteins, approximately 75% of the proteins were quantified in both studies, and the number of proteins quantified in the present study was higher than that of Amorim et al. (2017). Tebbe et al. (2015) demonstrates that LFQ allows greater coverage of the proteome; however, super-SILAC is more accurate for protein quantification.
Proteins differentially expressed during nutritional stress
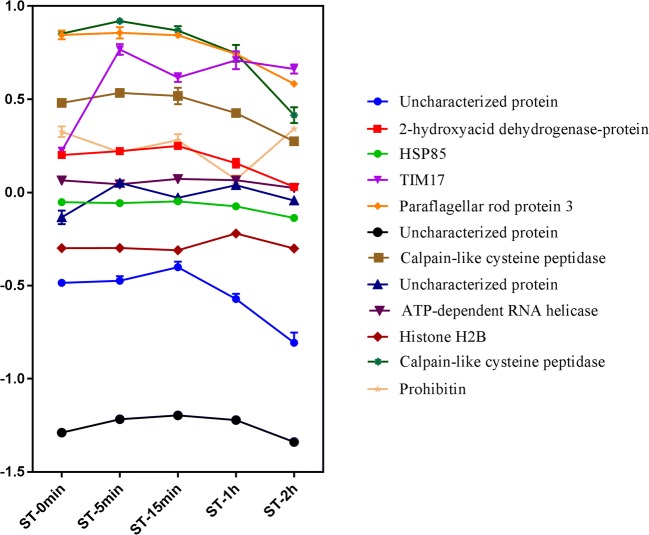
Using only proteins quantified at all experimental points (2751 proteins), with p value and q value < 0.2, few proteins were differentially expressed along NS, as listed in Table A2. Of these proteins, mitochondrial import inner membrane translocase subunit (TIM17) was upregulated in the first 5 min of NS, while five other proteins (two calpain-like cysteine peptidases, paraflagellar rod protein 3, D-isomer-specific 2-hydroxy acid dehydrogenase protein, and the uncharacterized protein V5B3E2) presented a downregulation pattern in the final phase of NS, but with a small variation in expression (Fig. 1). In the GO enrichment analysis (Table A3), the enrichment of terms associated with cysteine peptidases was observed.
Fig. 1.
Profile of the differentially expressed proteins (DEPs) during NS. Proteins were represented by ratio of intensities during different time points of NS. The proteins represented: Uncharacterized protein (TCDM_05191), 2-hydroxyacid dehydrogenase-protein (TCDM_03590), HSP85 (TCDM_06775), TIM17 (TCDM_14283), Paraflagellar rod protein 3 (TCDM_07792), Uncharacterized protein (TCDM_07431), Calpain-like cystein peptidase (TCDM_09423), Uncharacterized protein (TCDM_01051), ATP-dependent RNA helicase (TCDM_00483), Histone H2B (TCDM_14237), Calpain-like cysteine peptidase (TCDM_07700), Prohibitin (TCDM_03718)
Outline of phosphoproteomics data
After 40 runs, a total of 458,884 MS/MS spectra were obtained. Excluding contaminants and reverse sequences, 6436 peptides were identified, of which 3643 were phosphorylated. Most of these peptides (approximately 61%) were monophosphorylated (Fig. A3A), as expected based on the enrichment method used. A total of 4846 phosphorylation sites were located (Table A4), of which 3730 were located in serine, 1034 were located in threonine, and 42 were located in tyrosine (Fig. A3B). Among the phosphorylation sites located, 2685 showed more than 99% reliability in localization (Fig. A3C).
Phosphosites modulated during nutritional stress
To evaluate the phosphosites that are differentially phosphorylated along NS, we normalized the intensity of the phosphorylation site by subtracting the protein intensity obtained from the proteomics analysis, allowing the quantification of the variation of phosphosites, independent of protein oscillation. This approach has a limitation. In addition to the identification and quantification of the phosphorylation sites, it was also necessary for the identification and quantification of the protein in which this phosphosite is inserted in the proteomics analysis. In this way, it was possible to normalize 593 phosphosites (approximately 12% of the total sites identified). Among these phosphosites, using the p value and q value < 0.2 as cutoffs, 93 phosphosites were considered modulated. As normalization did not significantly alter most of the intensities in phosphosites, and because of the technical limitation of normalization, since not all phosphorylated peptides contain corresponding identification in the proteomics data, were obtained a low number of phosphosites to be analyzed. With this, we opted to analyze the non-normalized phosphorylation sites and created a final table containing all the phosphosites modulated during NS (Table A5). The GO enrichment analysis of proteins with modulated phosphosites is represented in Table A6, and terms associated with the activity of peptidases and phosphatases, among others, were enriched.
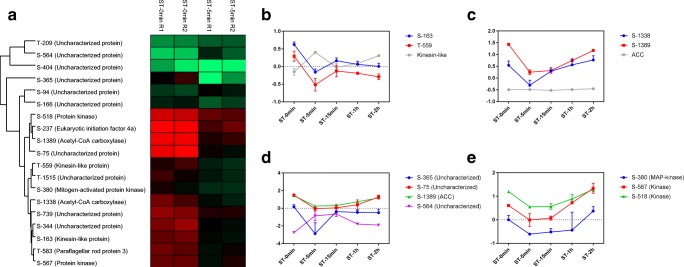
Figure 2a shows the phosphosites modulated more than 1.5-fold in the first 5 min of NS. The kinesin-like protein (Fig. 2b) and acetyl-CoA carboxylase (ACC) (Fig. 2c) had two phosphosites modulated in this first time point analyzed. Kinesin is a microtubule-dependent motor protein involved in microtubule dynamics. A decrease in phosphorylation was observed in both phosphosites represented in the first 5 min, while there was an increase in protein expression. ACC is associated with fatty acid synthesis. Both phosphosites showed a decrease in phosphorylation in the first 5 min, and phosphorylation increased again over time, whereas no significant change in the expression of the protein was observed. The phosphosites represented in Fig. 2d showed the greatest variation between ST-0min and ST-5min, with a difference of more than 2-fold. The phosphosite S-564 (uncharacterized protein) showed an increase in phosphorylation, while the phosphosites S-365 (uncharacterized protein), S-75 (uncharacterized protein), and S-1389 (acetyl-CoA carboxylase) showed a decrease in phosphorylation. In addition, three different kinases had modulated sites in the first 5 min of NS (Fig. 2e), indicating that phosphorylation has great relevance to this process.
Fig. 2.
Phosphosites modulated between ST-0min and ST-5min. a Heat map showing phosphosites with differences of more than 1.5-fold. Profile of phosphorylation in modulated phosphosites in b kinesin-like (TCDM_04106) and c acetyl-CoA carboxylase protein (TCDM_08578). d Phosphosites with greater alterations between the experimental points. The proteins represented: Uncharacterized protein (TCDM_07592), Uncharacterized protein (TCDM_02660), ACC (TCDM_08578), Uncharacterized protein (TCDM_10416). e Profile of phosphorylation in modulated phosphosites in different kinases. The proteins represented: MAP kinase (TCDM_07669), Kinase (TCDM_02063), Kinase (TCDM_08224)
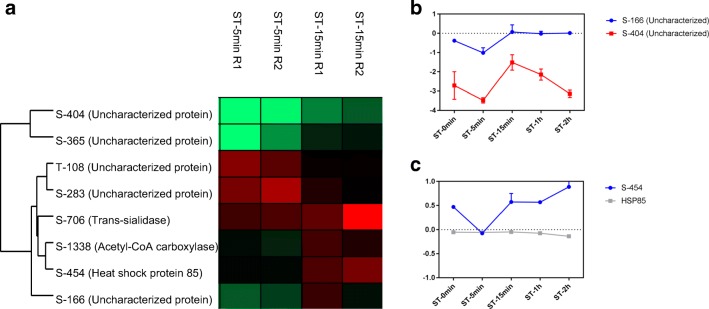
At the next point along NS, 15 min, other phosphosites presented more than 1.5-fold variations (Fig. 3a). The phosphosites S-166, S-404 (Fig. 3b), and S-365 (Fig. 2c), all in uncharacterized proteins, showed the highest variation between ST-5min and ST-15min, with a difference of more than 2-fold. The heat-shock protein (HSP) is associated with the stress response, and HSP-85 had a phosphosite S-454 modulated in the initial moments of NS (Fig. 2c). There was a decrease in phosphorylation at ST-5min, which was increased at ST-15min and ST-2h.
Fig. 3.
Phosphosites modulated between ST-5min and ST-15min. a Heat map showing phosphosites with differences of more than 1.5-fold. b Phosphosites with greater alterations between the experimental points. The proteins represented: Uncharacterized protein (TCDM_08527), Uncharacterized protein (TCDM_07220). c Profile of phosphorylation in modulated phosphosite in heat shock protein 85 (TCDM_06775)
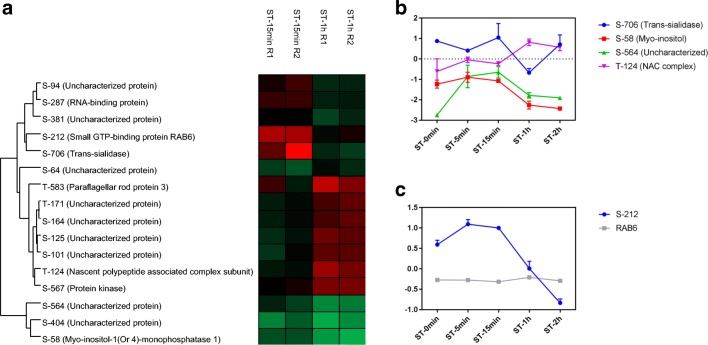
Comparing the points ST-15min and ST-1h, other phosphosites presented more than 1.5-fold variations (Fig. 4a), with an emphasis on the phosphosites S-706 (trans-sialidase), S-58 (myo-inositol-1(or 4)-monophosphatase-1), S-564 (uncharacterized protein), and T-124 (nascent polypeptide–associated complex (NAC)), with a difference of more than 2-fold (Fig. 4b). Trans-sialidase is an enzyme that transfers sialic acid from host glycoconjugates to the parasite; this protein showed a decrease in phosphorylation at ST-1h. Additionally, we observed a decrease in phosphorylation at phosphosites S-58 in myo-inositol-1(or 4)-monophosphatase-1, which is involved in inositol metabolism, and S-564, which is an uncharacterized protein. For the phosphosite T-124 of the NAC complex, an increase in phosphorylation was observed. A large decrease in the phosphorylation of S-212 in the RAB6 protein was observed starting at ST-15min and continuing until the end of NS (Fig. 3c). This protein is a GTPase involved in membrane traffic.
Fig. 4.
Phosphosites modulated between ST-15min and ST-1h. a Heat map showing phosphosites with differences of more than 1.5-fold. b Phosphosites with greater alteration between the experimental points. The proteins represented: Trans-sialidase (TCDM_12217), Myo-inositol (TCDM_03084), Uncharacterized protein (TCDM_10416), NAC complex (TCDM_05688). c Profile of phosphorylation in modulated phosphosite in RAB6 (TCDM_14273)
Finally, in the final moments of NS, some phosphosites showed variations of more than 1.5-fold when comparing ST-1h and ST-2h (Fig. 5a). A decrease in phosphorylation in the phosphosite S-582 in calmodulin was observed (Fig. 5b).
Fig. 5.
Phosphosites modulated between ST-1h and ST-2h. a Heat map showing phosphosites with differences of more than 1.5-fold. b Profile of phosphorylation in modulated phosphosite in calmodulin (TCDM_02086)
Discussion
In this study, we applied LC-MS/MS associated with a super-SILAC methodology to evaluate proteins and phosphorylation sites modulated during the nutritional stress of T. cruzi. NS is the initial stage of in vitro metacyclogenesis and has been proposed as the trigger that leads to the differentiation of the parasite (Figueiredo et al. 2000). The SILAC methodology was first employed in T. cruzi in 2016, using a semidefined medium for cultivation to identify the N-myristoylome of the parasite (Roberts and Fairlamb 2016). Under our conditions, continuous growth and sufficient metabolic protein labeling by SILAC of the parasite were only possible using the defined medium LM-14B (De Paula Lima et al. 2014).
Few proteins were considered modulated along NS and showed small variations in their expression. Significant changes were observed in the expression profile of the proteins in the adhesion phase, the step subsequent to NS, during in vitro metacyclogenesis (Amorim et al. 2017), a more extensive process, which compared intermediate points of the adhesion process, in addition to two very different morphological forms of T. cruzi, Epi under NS and Meta. TIM17 is upregulated in the first 5 min of NS. This protein is part of a complex associated with the translocation of mitochondrial proteins synthesized in the cytosol. TbTIM17 is essential to procyclic forms of T. brucei, and its knockout reduces several nuclear-encoded mitochondrial proteins (Singha et al. 2008). In addition, the mitochondrial genome of T. cruzi has the potential to change gene expression in response to different stimuli, such as NS (Shaw et al. 2016). The upregulation of this protein may represent an initial mitochondrial response to NS stimulation.
Two calpain-like cysteine peptidases were downregulated in the final moments of NS. Proteinases play a key role in the differentiation of the parasite since proteases and proteinase blockers disrupt this process (Cardoso et al. 2008; Goldenberg and Avila 2011). The calpain-like cysteine-peptidase (TCDM_09423), named Tccalpx11, has already been characterized in T. cruzi (Giese et al. 2008). Tccalpx11 mRNA was 2.5 times more abundant in parasites under NS compared with Epi in culture (Giese et al. 2008), where it has been observed that this protein is downregulated in the final moments of NS, perhaps because culture does not encompass the Epi point. Additionally, Tccalpx11 was not expressed in Meta and amastigotes (Giese et al. 2008), indicating that this protein plays a fundamental role in differentiation.
Despite the small number of regulated proteins, several phosphosites were considered modulated along NS. ACC is associated with fatty acid synthesis, and the catalytic activity of this protein is regulated by phosphorylation. In this study, we identified a decrease in phosphorylation at sites S-1389 and S-1338 during the first 5 min of NS, and phosphorylation increased again at both sites during NS. In humans, phosphorylation at site S-80 of ACC alters its conformation and is not compatible with dimerization, which inactivates the catalytic domain of this protein (Cho et al. 2010). In Saccharomyces cerevisiae, substitution with alanine at another phosphorylation site in ACC, S-1157, resulted in increased catalytic activity (Wei et al. 2016). In T. brucei, ACC RNAi reduced fatty acid biosynthesis, resulting in growth inhibition (Vigueira and Paul 2011). Therefore, the decrease in phosphorylation identified in this study may result in altered protein activity and consequently stop cell growth. In addition, changes in fatty acid composition were observed in Epi and Meta, with Epi subjected to NS being more similar to Meta when compared with Epi, influencing the membrane permeability of the parasite (Esteves et al. 1989).
During differentiation, parasites undergo morphological changes. Paraflagellar rod protein 3 is located in the flagellum and is regulated during NS, showing a great increase in Meta (Amorim et al. 2017). These data suggest that flagellum structure modulation occurs throughout differentiation. In addition, two phosphosites were modulated in kinesin-like proteins. While there was a decrease in phosphorylation at both sites in ST-5min, an increase in protein expression was observed. The kinesin is a microtubule-dependent motor protein that participates in intracellular transport and the regulation of microtubule dynamics. TbKIN-D is required for maintaining the cell morphology of T. brucei (Hu et al. 2012). The RNAi of this protein disrupts the organization of the subpellicular microtubule and affects the cell morphology. Protein phosphorylation may represent a stimulus for a change in the organization of the microtubules, leading to the morphological changes observed in the differentiation. In fact, several kinesin-like proteins and other cytoskeletal proteins are regulated during metacyclogenesis (Amorim et al. 2017).
HSPs have several functions, including the stress response (Feder and Hofmann 1999; Ürményi et al. 2014). HSP85 is differentially expressed and has a phosphosite modulated during NS. In T. brucei, the HSP85 mRNA levels increase at 41.5 °C, but at 42.5 °C, there is a decrease in these transcripts, and it is suggested that there is a disruption of trans-splicing in these transcripts under severe heat shock conditions (Muhich et al. 1989). The modulation of HSP expression is the result of the activation of various intracellular signaling pathways (Feder and Hofmann 1999). We detected a decreasing peak in phosphorylation in phosphosite S-454 at ST-5min, which may be a factor that influences the expression of this protein. During metacyclogenesis, a decrease in the levels of this protein was observed at the end of adhesion and in Meta (Amorim et al. 2017). Another important heat shock protein for protozoa is HSP90, which has already been associated with the process of differentiation and invasion of host cells, and is modulated by posttranslational modifications (Roy et al. 2012; Schopf et al. 2017). In Toxoplasma gondii, HSP90 is associated with bradyzoite differentiation, host-cell invasion, and growth and virulence (Sun et al. 2017). The inactivation of HSP90 for drug geldanamycin or radicicol in Leishmania donovani leads to the differentiation of the promastigote form into the amastigote (Wiesgigl and Clos 2001). In T. cruzi, the use of geldanamycin does not lead to differentiation, but an increase in the level of other heat-shock proteins is observed (Graefe et al. 2002).
The phosphosite T-124 was modulated in the NAC. This complex interacts with ribosomes, promotes translation, and assists in the folding of nascent polypeptides (Kirstein-Miles et al. 2013). Under stress conditions, this protein forms aggregates, reducing the number of proteins available in the cytosol for translation (Kirstein-Miles et al. 2013). Also associated with protein synthesis, the phosphosite S-237 in protein eIF4A showed a decrease in phosphorylation. This protein is an RNA helicase and plays a role in the initiation of translation by unwinding secondary structure in the 5′-untranslated region of mRNA, which contributes to ribosomal subunit binding to mRNA and scanning of the initiation codon (Rogers et al. 2001).
A decrease in the S-706 phosphosite of a trans-sialidase was observed at ST-1h. Trans-sialidase family members are substrates of TcPKA and regulate intracellular trafficking, protein turnover, or enzymatic activities (Bao et al. 2010). In addition, trans-sialidase is a membrane-anchored enzyme that catalyzes the transfer of sialic acid from the host to a parasite, allowing the parasite to evade the host immune system (Miller III and Roitberg 2013). Additionally, a decrease in phosphosite S-58 of myo-inositol-1(or 4)-monophosphatase was observed in the final moments of NS. This enzyme participates in myo-inositol de novo synthesis, dephosphorylating the inositol 1-phosphatase produced by myo-inositol-1-phosphate (Martin and Smith 2005). Myo-inositol-1-phosphate is essential to the bloodstream form of T. brucei (Martin and Smith 2005). Myo-inositol can then be incorporated into phosphatidylinositol (PI), which is utilized in a variety of cellular functions, including the biosynthesis of GPI (glycosylphosphatidylinositol) anchors. In T. cruzi, d-myo-inositol-1,4,5-trisphosphate (IP3) is associated with Ca2+ signaling (Hashimoto et al. 2013). The phosphoinositide phospholipase C (PI-PLC) is involved in amastigogenesis, and the modulation mediated by IP3 contributes to differentiation (Okura et al. 2005). In addition, high levels of Ca2+ are required for efficient cell invasion (Hashimoto et al. 2013). Furthermore, the regulation of Ca2+ is involved in the metacyclogenesis and multiplication of Epi (Lammel et al. 1996). A decrease in the phosphorylation of phosphosite S-582 of calmodulin was also observed in the final stress stage. Calmodulin is an intracellular acceptor of Ca2+; therefore, it is also associated with Ca2+ signaling (Ogueta et al. 1996). The regulation of the proteins associated with Ca2+ signaling reinforces the importance of this process for the differentiation events in this parasite, as previously reported in both amastigogenesis (Okura et al. 2005) and metacyclogenesis (Lammel et al. 1996).
Conclusion
In the current study, we evaluated (phospho) proteomic regulation during NS using LC-MS/MS and super-SILAC approaches. This is a first approach that aims at a survey of possible targets significantly expressed and modulated by phosphorylation during NS, and other experiments are necessary to reinforce the results presented here, since the differences observed among the experimental points do not have high levels of statistical significance. Despite the large number of proteins identified during NS, few were considered regulated among the five experimental time points analyzed in the present study. In contrast, among the identified phosphorylation sites, several were considered to be modulated along NS. Regulated phosphosites are present in proteins associated with various functions, such as fatty acid synthesis and the regulation of protein expression, structural components, stress response, and intracellular signaling, indicating that several proteins are phosphorylated or dephosphorylated along NS, which is the beginning of the stimulus that leads to differentiation. Many of these sites are present in proteins with no unknown function, reinforcing the importance of the study and characterization of hypothetical T. cruzi proteins. Lastly, possible phosphosites that trigger metacyclogenesis were identified, and it is necessary to characterize these sites and these proteins to determine their functions.
Electronic supplementary material
Numbers obtained in proteomic analysis. (A) The number of proteins and peptides identified in total and at each experimental point (ST-0min, ST-5min, ST-15min, ST-1h, ST-2h). R1 and R2 represent different biological replicates. (B) Correlation of protein intensity between independent biological replicates. Pearson correlation value is presented in blue. (PNG 839 kb)
(TIF 58 kb)
(TIF 40 kb)
Numbers obtained in phosphoproteomic data. (A) Number of phosphosites per peptide. (B) Phosphosite distribution on serine, threonine, and tyrosine residues of phosphopeptides. (C) Localization probability of phosphogroups within a peptide based on the total probability score. (PNG 266 kb)
(TIF 43 kb)
- Protein Groups identified during NS (XLSX 9697 kb)
- Proteins modulated during NS (XLSX 12 kb)
- GO enrichment of proteins modulated during NS (XLSX 11 kb)
- Phosphosites identified during NS (XLSX 7130 kb)
- Phosphosites modulated during NS (XLSX 38 kb)
- GO of phosphosites modulated during NS (XLSX 10 kb)
Compliance with ethical standards
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Alvarez VE, Kosec G, Sant’Anna C, et al. Autophagy is involved in nutritional stress response and differentiation in Trypanosoma cruzi. J Biol Chem. 2008;283:3454–3464. doi: 10.1074/jbc.M708474200. [DOI] [PubMed] [Google Scholar]
- Amorim JC, Batista M, da Cunha ES, Lucena ACR, Lima CVP, Sousa K, Krieger MA, Marchini FK. Quantitative proteome and phosphoproteome analyses highlight the adherent population during Trypanosoma cruzi metacyclogenesis. Sci Rep. 2017;7:9899. doi: 10.1038/s41598-017-10292-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y, Weiss LM, Braunstein VL, Huang H. Role of protein kinase A in Trypanosoma cruzi. Infect Immun. 2008;76:4757–4763. doi: 10.1128/IAI.00527-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y, Weiss LM, Ma YF, et al. Protein kinase A catalytic subunit interacts and phosphorylates members of trans-sialidase super-family in Trypanosoma cruzi. Microbes Infect. 2010;12:716–726. doi: 10.1016/j.micinf.2010.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista M, Kugeratski FG, de Paula Lima CV, Probst CM, Kessler RL, de Godoy LM, Krieger MA, Marchini FK. The MAP kinase MAPKLK1 is essential to Trypanosoma brucei proliferation and regulates proteins involved in mRNA metabolism. J Proteome. 2017;154:118–127. doi: 10.1016/j.jprot.2016.12.011. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc. 1995;57:289–300. [Google Scholar]
- Bonaldo MC, Souto-Padron T, de Souza W, Goldenberg S. Cell-substrate adhesion during Trypanosoma cruzi differentiation. J Cell Biol. 1988;106:1349–1358. doi: 10.1083/jcb.106.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley R, Tariq H, McElhinney H, Szoor B, Huxley-Jones J, Stevens R, Matthews K, Tabernero L. The TriTryp phosphatome: analysis of the protein phosphatase catalytic domains. BMC Genomics. 2007;8:434. doi: 10.1186/1471-2164-8-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo EP, Silva LHP. Growth and differentiation in Trypanosoma cruzi. I. Origin of metacyclic trypanosomes in liquid media. Rev Inst Med Trop Sao Paulo. 1964;12:93–100. [PubMed] [Google Scholar]
- Cardoso J, Soares MJ, Menna-Barreto RFS, Bloas RL, Sotomaior V, Goldenberg S, Krieger MA. Inhibition of proteasome activity blocks Trypanosoma cruzi growth and metacyclogenesis. Parasitol Res. 2008;103:941–951. doi: 10.1007/s00436-008-1081-6. [DOI] [PubMed] [Google Scholar]
- Chagas C. Nova tripanosomiaze humana: estudos sobre a morfolojia e o ciclo evolutivo do Schizotrypanum cruzi n. gen., n. sp., ajente etiolojico de nova entidade morbida do homem. Mem Inst Oswaldo Cruz. 1909;1:159–218. doi: 10.1590/S0074-02761909000200008. [DOI] [Google Scholar]
- Cho YS, Il LJ, Shin D, et al. Molecular mechanism for the regulation of human ACC2 through phosphorylation by AMPK. Biochem Biophys Res Commun. 2010;391:187–192. doi: 10.1016/j.bbrc.2009.11.029. [DOI] [PubMed] [Google Scholar]
- Contreras VT, Salles JM, Thomas N, Morel CM, Goldenberg S. In vitro differentiation of Trypanosoma cruzi under chemically defined conditions. Mol Biochem Parasitol. 1985;16:315–327. doi: 10.1016/0166-6851(85)90073-8. [DOI] [PubMed] [Google Scholar]
- Contreras VT, Araujo-Jorge TC, Bonaldo MC, et al. Biological aspects of the Dm 28c clone of Trypanosoma cruzi after metacyclogenesis in chemically defined media. Mem Inst Oswaldo Cruz. 1988;83:123–133. doi: 10.1590/S0074-02761988000100016. [DOI] [PubMed] [Google Scholar]
- De Gaudenzi JG, Noé G, Campo VA, Frasch AC. Gene expression regulation in trypanosomatids. Essays Biochem. 2011;51:31–46. doi: 10.1042/bse0510031. [DOI] [PubMed] [Google Scholar]
- De Paula Lima CV, Batista M, Kugeratski FG, et al. LM14 defined medium enables continuous growth of Trypanosoma cruzi. BMC Microbiol. 2014;14:238. doi: 10.1186/s12866-014-0238-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteves MG, Gonzales-Perdomo M, Alviano CS, Angluster J, Goldenberg S. Changes in fatty acid composition associated with differentiation of Trypanosoma cruzi. FEMS Microbiol Lett. 1989;59:31–34. doi: 10.1111/j.1574-6968.1989.tb03077.x. [DOI] [PubMed] [Google Scholar]
- Feder ME, Hofmann GE. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol. 1999;61:243–282. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- Figueiredo RC, Rosa DS, Soares MJ. Differentiation of Trypanosoma cruzi epimastigotes: metacyclogenesis and adhesion to substrate are triggered by nutritional stress. J Parasitol. 2000;86:1213–1218. doi: 10.1645/0022-3395(2000)086[1213:DOTCEM]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Geiger T, Cox J, Ostasiewicz P, Wisniewski JR, Mann M. Super-SILAC mix for quantitative proteomics of human tumor tissue. Nat Methods. 2010;7:383–385. doi: 10.1038/nmeth.1446. [DOI] [PubMed] [Google Scholar]
- Giese V, Dallagiovanna B, Marchini FK, Pavoni DP, Krieger MA, Goldenberg S. Trypanosoma cruzi: a stage-specific calpain-like protein is induced after various kinds of stress. Mem Inst Oswaldo Cruz. 2008;103:598–601. doi: 10.1590/S0074-02762008000600015. [DOI] [PubMed] [Google Scholar]
- Goldenberg S, Avila AR. Aspects of Trypanosoma cruzi stage differentiation. In: Weiss LM, Tanowitz HB, Kirchhoff LV, editors. Advances in parasitology. Burlington: Academic Press; 2011. pp. 285–305. [DOI] [PubMed] [Google Scholar]
- Gonzales-Perdomo M, Romero P, Goldenberg S. Cyclic AMP and adenylate cyclase activators stimulate Trypanosoma cruzi differentiation. Exp Parasitol. 1988;66:205–212. doi: 10.1016/0014-4894(88)90092-6. [DOI] [PubMed] [Google Scholar]
- Graefe SEB, Wiesgigl M, Gaworski I, Macdonald A, Clos J. Inhibition of HSP90 in Trypanosoma cruzi induces a stress response but no stage differentiation. Eukaryot Cell. 2002;1:936–943. doi: 10.1128/EC.1.6.936-943.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Enomoto M, Morales J, Kurebayashi N, Sakurai T, Hashimoto T, Nara T, Mikoshiba K. Inositol 1,4,5-trisphosphate receptor regulates replication, differentiation, infectivity and virulence of the parasitic protist Trypanosoma cruzi. Mol Microbiol. 2013;87:1133–1150. doi: 10.1111/mmi.12155. [DOI] [PubMed] [Google Scholar]
- Hernández R, Cevallos AM, Nepomuceno-Mejía T, López-Villaseñor I. Stationary phase in Trypanosoma cruzi epimastigotes as a preadaptive stage for metacyclogenesis. Parasitol Res. 2012;111:509–514. doi: 10.1007/s00436-012-2974-y. [DOI] [PubMed] [Google Scholar]
- Hu H, Hu L, Yu Z, Chasse AE, Chu F, Li Z. An orphan kinesin in trypanosomes cooperates with a kinetoplastid-specific kinesin to maintain cell morphology by regulating subpellicular microtubules. J Cell Sci. 2012;125:4126–4136. doi: 10.1242/jcs.106534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirstein-Miles J, Scior A, Deuerling E, Morimoto RI. The nascent polypeptide-associated complex is a key regulator of proteostasis. EMBO J. 2013;32:1451–1468. doi: 10.1038/emboj.2013.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollien A, Schaub G. The development of Trypanosoma cruzi in Triatominae. Parasitol Today. 2000;16:381–387. doi: 10.1016/S0169-4758(00)01724-5. [DOI] [PubMed] [Google Scholar]
- Lammel E, Barbieri M, Wilkowsky S, Berrtini F, Isola E. Trypanosoma cruzi: involvement of intracellular calcium in multiplication. Exp Parasitol. 1996;83:240–249. doi: 10.1006/expr.1996.0070. [DOI] [PubMed] [Google Scholar]
- Marchini FK, de Godoy LMF, Rampazzo RCP, Pavoni DP, Probst CM, Gnad F, Mann M, Krieger MA. Profiling the Trypanosoma cruzi phosphoproteome. PLoS One. 2011;6:e25381. doi: 10.1371/journal.pone.0025381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KL, Smith TK. The myo-inositol-1-phosphate synthase gene is essential in Trypanosoma brucei. Biochem Soc Trans. 2005;33:983–985. doi: 10.1042/BST0330983. [DOI] [PubMed] [Google Scholar]
- Martínez-Calvillo S, Vizuet-de-Rueda JC, Florencio-Martínez LE, Manning-Cela RG, Figueroa-Angulo EE. Gene expression in trypanosomatid parasites. J Biomed Biotechnol. 2010;2010:525241. doi: 10.1155/2010/525241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BR, III, Roitberg AE. Trypanosoma cruzi trans-sialidase as a drug target against Chagas disease (American trypanosomiasis) Future Med Chem. 2013;5:1889–1900. doi: 10.4155/fmc.13.129. [DOI] [PubMed] [Google Scholar]
- Muhich ML, Hsu MP, Boothroyd JC. Heat-shock disruption of trans-splicing in trypanosomes: effect on Hsp70, Hsp85 and tubulin mRNA synthesis. Gene. 1989;82:169–175. doi: 10.1016/0378-1119(89)90042-5. [DOI] [PubMed] [Google Scholar]
- Nakayasu ES, Gaynor MR, Sobreira TJP, Ross JA, Almeida IC. Phosphoproteomic analysis of the human pathogen. Proteomics. 2009;9:3489–3506. doi: 10.1002/pmic.200800874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogueta S, Intosh GM, Téllez-Iñon MT. Regulation of Ca2+/calmodulin-dependent protein kinase from Trypanosoma cruzi. Mol Biochem Parasitol. 1996;78:171–183. doi: 10.1016/S0166-6851(96)02622-9. [DOI] [PubMed] [Google Scholar]
- Okura M, Fang J, Salto ML, Singer RS, Docampo R, Moreno SNJ. A lipid-modified phosphoinositide-specific phospholipase C (TcPI-PLC) is involved in differentiation of trypomastigotes to amastigotes of Trypanosoma cruzi. J Biol Chem. 2005;280:16235–16243. doi: 10.1074/jbc.M414535200. [DOI] [PubMed] [Google Scholar]
- Parsons M, Worthey EA, Ward PN, Mottram JC. Comparative analysis of the kinomes of three pathogenic trypanosomatids: Leishmania major, Trypanosoma brucei and Trypanosoma cruzi. BMC Genomics. 2005;6:127. doi: 10.1186/1471-2164-6-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Morales D, Hernández KDR, Martínez I, Agredano-Moreno LT, Jiménez-García LF, Espinoza B. Ultrastructural and physiological changes induced by different stress conditions on the human parasite Trypanosoma cruzi. Cell Stress Chaperones. 2017;22:15–27. doi: 10.1007/s12192-016-0736-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queiroz RML, Charneau S, Mandacaru SC, Schwämmle V, Lima BD, Roepstorff P, Ricart CAO. Quantitative proteomic and phosphoproteomic analysis of Trypanosoma cruzi amastigogenesis. Mol Cell Proteomics. 2014;13:3457–3472. doi: 10.1074/mcp.M114.040329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 2003;75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- Roberts AJ, Fairlamb AH. The N-myristoylome of Trypanosoma cruzi. Sci Rep. 2016;6:1–11. doi: 10.1038/s41598-016-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GW, Richter NJ, Lima WF, Merrick WC. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J Biol Chem. 2001;276:30914–30922. doi: 10.1074/jbc.M100157200. [DOI] [PubMed] [Google Scholar]
- Roy N, Nageshan RK, Ranade S, Tatu U. Heat shock protein 90 from neglected protozoan parasites. Biochim Biophys Acta. 2012;1823:707–711. doi: 10.1016/j.bbamcr.2011.12.003. [DOI] [PubMed] [Google Scholar]
- Schoijet AC, Sternlieb T, Alonso GD. The phosphatidylinositol 3-kinase class III complex containing TcVps15 and TcVps34 participates in autophagy in Trypanosoma cruzi. J Eukaryot Microbiol. 2017;64:308–321. doi: 10.1111/jeu.12367. [DOI] [PubMed] [Google Scholar]
- Schopf FH, Biebl MM, Buchner J. The HSP90 chaperone machinery. Parasite. 2017;18:345–360. doi: 10.1038/nrm.2017.20. [DOI] [PubMed] [Google Scholar]
- Shaw AK, Kalem MC, Zimmer SL. Mitochondrial gene expression is responsive to starvation stress and developmental transition in Trypanosoma cruzi. Mol Biol Phisiol. 2016;1:e00051–e00016. doi: 10.1128/mSphere.00051-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singha UK, Peprah E, Williams S, Walker R, Saha L, Chaudhuri M. Characterization of the mitochondrial inner membrane protein translocator Tim17 from Trypanosoma brucei. Mol Biochem Parasitol. 2008;159:30–43. doi: 10.1016/j.molbiopara.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smircich P, Eastman G, Bispo S, Duhagon MA, Guerra-Slompo EP, Garat B, Goldenberg S, Munroe DJ, Dallagiovanna B, Holetz F, Sotelo-Silveira JR. Ribosome profiling reveals translation control as a key mechanism generating differential gene expression in Trypanosoma cruzi. BMC Genomics. 2015;16:443. doi: 10.1186/s12864-015-1563-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares MJ. The reservosome of Trypanosoma cruzi epimastigotes: an organelle of the endocytic pathway with a role on metacyclogenesis. Mem Inst Oswaldo Cruz. 1999;94(Suppl 1):139–141. doi: 10.1590/S0074-02761999000700015. [DOI] [PubMed] [Google Scholar]
- Sun H, Zhuo X, Zhao X, Yang Y, Chen X, Yao C, du A. The heat shock protein 90 of Toxoplasma gondii is essential for invasion of host cells and tachyzoite growth. Parasite. 2017;24:1–11. doi: 10.1051/parasite/2017001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan CSH. Sequence, structure, and network evolution of protein phosphorylation. Sci Signal. 2011;4:mr6–mr6. doi: 10.1126/scisignal.2002093. [DOI] [PubMed] [Google Scholar]
- Tebbe A, Klammer M, Sighart S, Schaab C, Daub H. Systematic evaluation of label-free and super-SILAC quantification for proteome expression analysis. Rapid Commun Mass Spectrom. 2015;29:795–801. doi: 10.1002/rcm.7160. [DOI] [PubMed] [Google Scholar]
- Tonelli RR, Augusto LS, Castilho BA, Schenkman S. Protein synthesis attenuation by phosphorylation of eIF2α IS required for the differentiation of Trypanosoma cruzi into infective forms. PLoS One. 2011;6:1–10. doi: 10.1371/journal.pone.0027904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler KM, Engman DM. Flagellar elongation induced by glucose limitation is preadaptive for Trypanosoma cruzi differentiation. Cell Motil Cytoskeleton. 2000;278:269–278. doi: 10.1002/1097-0169(200008)46:4<269::AID-CM4>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Tyler KM, Engman DM. The life cycle of Trypanosoma cruzi revisited. Int J Parasitol. 2001;31:472–481. doi: 10.1016/S0020-7519(01)00153-9. [DOI] [PubMed] [Google Scholar]
- Ürményi TP, Silva R, Rondinelli E, et al. The heat shock protein of Trypanosoma cruzi. In: Santos ALS, et al., editors. Proteins and proteomics of Leishmania and Trypanosoma. 2014. pp. 119–135. [DOI] [PubMed] [Google Scholar]
- Vigueira PA, Paul KS. Requirement for acetyl-CoA carboxylases in Trypanosoma brucei is dependent upon the growth environment. Mol Microbiol. 2011;80:117–132. doi: 10.1111/j.1365-2958.2011.07563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno JA, Csordas A, Del-Toro N, et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Zhang Y, Yu T, et al. A unified molecular mechanism for the regulation of acetyl-CoA carboxylase by phosphorylation. Nat Publ Gr. 2016;80:1–12. doi: 10.1038/celldisc.2016.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesgigl M, Clos J. Heat shock protein 90 homeostasis controls stage differentiation in Leishmania donovani. Mol Biol Cell. 2001;12:3307–3316. doi: 10.1091/mbc.12.11.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Numbers obtained in proteomic analysis. (A) The number of proteins and peptides identified in total and at each experimental point (ST-0min, ST-5min, ST-15min, ST-1h, ST-2h). R1 and R2 represent different biological replicates. (B) Correlation of protein intensity between independent biological replicates. Pearson correlation value is presented in blue. (PNG 839 kb)
(TIF 58 kb)
(TIF 40 kb)
Numbers obtained in phosphoproteomic data. (A) Number of phosphosites per peptide. (B) Phosphosite distribution on serine, threonine, and tyrosine residues of phosphopeptides. (C) Localization probability of phosphogroups within a peptide based on the total probability score. (PNG 266 kb)
(TIF 43 kb)
- Protein Groups identified during NS (XLSX 9697 kb)
- Proteins modulated during NS (XLSX 12 kb)
- GO enrichment of proteins modulated during NS (XLSX 11 kb)
- Phosphosites identified during NS (XLSX 7130 kb)
- Phosphosites modulated during NS (XLSX 38 kb)
- GO of phosphosites modulated during NS (XLSX 10 kb)