Abstract
Hundred kernel weight is an important indicator for large-seeded genotype selection. A recombinant inbred line population was used to decipher the genetic architecture of seed size and three pod traits in cultivated groundnut based on the phenotypic data from six and three environments, respectively. The study revealed a consensus major QTL for HKW in B07 group that explained 10.5–23.9% phenotypic variation due to seed size. Further, two other minor QTLs were identified in B03 and B08 group for the seed size. Two minor QTLs for pod beak were positioned in B03 and A08. A minor QTL for pod reticulation was also mapped in the same map interval with the pod beak QTL in A08. Another minor QTL for pod constriction was co-mapped with the minor QTL for HKW in B08. The other minor QTL for pod constriction was placed in the neighboring map interval with the consensus QTL for seed size in B07 that suggests linkage of pod constriction with large seed trait. Analysis of the flanking markers profile in 71 cultivated groundnut genotypes revealed a strong association of pPGPseq_2E06 marker with large seed trait.
Electronic supplementary material
The online version of this article (10.1007/s13205-019-1881-7) contains supplementary material, which is available to authorized users.
Keywords: Arachis hypogaea L., Hundred kernel weight, Pod beak, Pod constriction, Pod reticulation, Quantitative trait loci
Introduction
Cultivated groundnut (Arachis hypogaea L.) is an important source of quality edible oil, protein and various bioactive compounds. Among the 100 groundnut-growing countries, India, China, USA and Nigeria hold more than 90% of the cultivated area under this crop. In India, it is grown on 5.30 million ha with a total production of 9.18 million tons (FAOSTAT 2017). It is a self-pollinated crop with allotetraploid (2n = 4x = 40, AABB) genome. It is mainly used for edible oil, various culinary purposes and preparation of various confectionary items. Cultivated groundnut originated in the Bolivian region (South Bolivia–Northwestern Argentina) (Krapovickas and Gregory 1994). It has two subspecies, hypogaea and fastigiata. The subspecies hypogaea is characterized by alternate branching and absence of flowers on the main stem. While fastigiata is recognized by sequential branching and presence of flowers on the main stem (Krapovickas 1969). In general, ssp. hypogaea includes genotypes with long duration, seed dormancy and large seed size. However, sustained breeding efforts through mutation and recombination breeding have generated large-seeded genotypes in subspecies fastigiata (Murty et al. 2004; Badigannavar and Mondal 2007).
Cultivated groundnut genotypes have sufficient diversity in morphological, physiological, and agronomic traits (Upadhyaya et al. 2005; Yol et al. 2018). Bhad et al. (2016) reported significant variation in seed size in cultivated groundnut genotypes that include varieties, mutants and newly evolved breeding lines. However, limited variation was found at the DNA level (Halward et al. 1991) owing to its evolution from a single natural polyploidization event between two diploid progenitor species, followed by chromosome duplication (Kochert et al. 1996; Bertioli et al. 2016). But the rapid development of whole genome sequencing, whole genome re-sequencing, availability of genome survey sequence and expressed sequence tag in Arachis hypogaea had prompted the development of a large number of simple sequence repeat (SSR) and single nucleotide polymorphism (SNP) markers that had been used widely for detecting sufficient genetic diversity in cultivated species (Ferguson et al. 2004; Mondal et al. 2012; Jiang et al. 2014; Pandey et al. 2014; Zhao et al. 2017). This genetic diversity information was later utilized to tag foliar disease-resistant traits (Mondal and Badigannavar 2018; Clevenger et al. 2018; Shirasawa et al. 2018), pod traits (Fonceka et al. 2012; Shirasawa et al. 2012), plant habits (Fonceka et al. 2012; Kayam et al. 2017) and various quantitative traits like pod yield (Faye et al. 2015), shelling percentage (Luo et al. 2018a), bruchid resistance (Mondal et al. 2014), and content of bioactive compounds (Mondal et al. 2015).
Seed size is an important parameter for enhancing yield of crop plants. In groundnut, seed size is usually being measured in terms of hundred kernel weight (HKW). Since yield is a complex trait which is governed by many genes, it is wise to improve its component characters towards the improvement of crop yield. Generally, seed-related traits like seed length, seed width, ratio of seed length to width and HKW significantly and directly impact the groundnut pod yield (Kale et al. 2000). For a practical breeding program, HKW is often used as a selection parameter for developing confectionary groundnut varieties (Venuprasad et al. 2011). Variable reports on genetics of seed size are available in literature. Balaiah et al. (1977) and Layrisse et al. (1980) reported that large seed is dominant over small. While others claimed small seed is dominant over large seed in groundnut (Cahaner 1978; Anderson et al. 1993). Pattanashetti et al. (2008) described trigenic inheritance of seed size. Most of the available literature on seed size claimed that it is controlled by polygenes with predominance of additive gene action (Garet 1976, Layrisse et al. 1980, Swe and Branch 1986, Anderson, et al. 1993). Hariprasanna et al. (2008) and Venuprasad et al. (2011) confirmed that seed size is mostly influenced by a combination of both maternal and nuclear genes. Genetic linkage mapping has detected numbers of quantitative trait loci (QTL) for seed length, seed width and HKW in various bi-parental mapping populations (Fonceka et al. 2012; Shirasawa et al. 2012; Huang et al. 2015; Chen et al. 2016; Hake et al. 2017). Pandey et al. (2014) and Zhao et al. (2017) have detected significant marker association with large phenotypic variance for seed length and HKW in multiple environments using association mapping approach. But often, this association mapping approach is cumbersome in polyploid crops due to lack of diverse germplasm, scoring of multiple bands during genotyping and complex analysis criteria used in data analysis. Thus, bi-parental immortal mapping population like recombinant inbred lines (RILs) is being used for detecting QTLs of various agronomically important quantitative traits in groundnut. Few reports are available on QTL information for pod beak, constriction and reticulation. Shirasawa et al. (2012) reported two QTLs for pod constriction in LG09 and single QTL for pod beak/shape of pod tip in LG03 of the reported genetic linkage map. By using an improved Axiom Arachis array, different SNPs were mapped and a single QTL was found for pod constriction in B07 chromosome (Patil et al. 2018). Here, we report detection of a consensus QTL region for HKW along with other QTLs for pod reticulation, beak and constriction in cultivated groundnut.
Materials and methods
Plant materials and phenotyping
A recombinant inbred line population (consisting of 164 lines) derived from a cross between VG 9514 and TAG 24 was used for this study. The genotype VG 9514 was derived from a wide hybridization between Co 1 (A. hypogaea L.) X A. cardenasii (Varman 1999). It has foliar disease resistance, red testa and small seed size. TAG 24, a well-adapted cultivated groundnut variety developed at Bhabha Atomic Research Centre (BARC), Mumbai, India, has high yield potential, rose testa and moderate seed size (Patil et al. 1995). Another 71 active Trombay groundnut genotypes were used for validation of associated alleles of two flanking markers with seed size (Suppl. Table 1).
Phenotyping of RIL population
RILs along with parents were grown in randomly complete block design in two replicates for 3 years, 2008, 2009 and 2011. Phenotyping of these RILs for HKW was carried out at six environments based on IPGRI and ICRISAT (1992) descriptors. These six environments were summer season 2008 at Kehal (Keh-S08); summer 2008 at Gauribidanur (Gau-S08); rainy season 2008 at Gauribidanur (Gau-R08) and at Trombay (Tro-R08); rainy season 2009 at Trombay (Tro-R09) and rainy season 2011 at Gauribidanur (Gau-R11). For pod-related traits (beak, constriction and reticulation), the data were collected only from three environments (Tro-R08; Tro-R09 and Gau-R11) as per description in IPGRI and ICRISAT (1992). Descriptive statistics and normality test of all the phenotypic data were performed using PAST ver 3.11 (Hammer et al. 2001).
SSR marker development, DNA isolation and marker analysis
Based on our earlier effort on QTL mapping for seed size in our laboratory (unpublished data), a chromosomal portion (49.05 cM) between GM2073 and TE282 (Suppl. Figure 1) was retrieved from genomic sequence of A. ipaensis in PeanutBase (http://www.peanutbase.org). Sequence information from this retrieved B07 genome was used to identify di- (minimum six repeats), tri- (five repeats), tetra- (five repeats), penta- (four repeats), hexanucleotide (four repeats) and compound repeat motifs. The flanking sequences of the identified repeat loci were used for primer pair designing (Mondal and Badigannavar 2018). These chromosome-specific SSR markers were named as B07_1 to B07_110 (Suppl. Table 2).
Total genomic DNA was isolated from fresh young leaf tissues of described RILs by GenElute™ Plant Genomic DNA mini-prep kit (Sigma Aldrich, USA) following manufacturer’s recommendation. DNA samples were quantified using a NanoDrop™ spectrophotometer (Thermo Fisher Scientific, Waltham, USA) and adjusted to a final concentration of 10 ηg/µl. Each 10 µl PCR reaction volume contained 10 ηg genomic DNA, 1 × Go Taq buffer, 1.5 mM MgCl2, 0.2 µM of each primer, 0.20 mM of each dNTP and 0.5 U of Go Taq DNA polymerase (Promega, Madison, USA). SSR amplification was performed in a thermal cycler (Eppendorf, Hamburg, Germany). The reaction was carried out under the following temperature conditions: 94 °C for 5 min with 35 cycles of 94 °C for 30 s, 50–55 °C (depending upon annealing temperature of respective primer pair) for 30 s, 72 °C for 30 s and a final extension at 72 °C for 10 min. The amplified SSRs were size separated in high-resolution DNA Cartridge (Qiagen, Germany) fitted with an automated capillary gel electrophoresis system (Qiagen, Germany).
Linkage analysis
A χ2 test was performed to test the null hypothesis of 1:1 segregation of each new SSR marker in the RILs. The linkage analysis of polymorphic marker was performed using QTL IciMapping ver 4.1 (Wang et al. 2016). Mapping criteria and graphical representation of the linkage map were followed as mentioned in Mondal and Badigannavar (2018). The newly developed polymorphic SSR markers were also included in the existed genetic linkage map of Mondal et al. (2014) by following the above mapping protocol in QTL IciMapping ver 4.1.
QTL analysis for HKW and pod traits
Phenotypic data (HKW and pod traits) of the RILs in each season were entered along with the genotypic data of the RILs in inclusive composite interval mapping (ICIM) analysis (Li et al. 2007). To identify the main QTLs, analyses were performed with QTL IciMapping version 4.1 (Wang et al. 2016) using inclusive composite interval mapping of additive (ICIM-ADD). Additive QTLs were then detected using 1.0 cM speed in scanning by following a stepwise regression method. The probability used in stepwise regression for additive QTLs was 0.001. To claim a significant QTL, a LOD threshold of 2.5 was set for additive QTLs. Significant LOD thresholds were determined for each dataset by 1000 permutations with type I probability of 0.01 (Doerge 2002). Graphics for linkage map and different QTLs were generated through Mapchart version 2.1 (Voorrips 2002).
Result and discussion
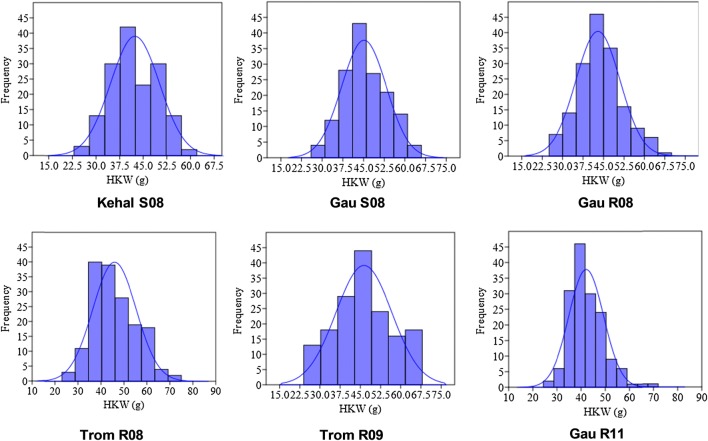
Field experiments in six different environments revealed that TAG 24 has medium-sized seeds with HKW of 52.0–56.5 g and VG 9514 has small seed type with HKW of 35.0–39.2 g. ANOVA analysis of HKW data of RILs over six environments detected significant difference among RILs and environments. There was also a significant genotype X environment interaction for the tested seed size trait in groundnut (Suppl. Table 3). The RIL population derived from these two parents showed wide distribution of seed size (in terms of HKW) ranging from 23.0 g to 75.0 g at Trombay rainy season 2008 (Table 1). Distribution of these seed phenotypes beyond the parental limit indicated the possible role of transgressive segregation and additive genes from both the parents (Anderson et al. 1993; Venuprasad et al. 2011). The frequency distribution in histogram for HKW in most of the tested environments followed normal distribution except in Gauribidanur rainy season 2011 (Fig. 1; Table 1). Skewness values showed that HKW in this RIL population was positively skewed. The low-to-medium positive skewed values indicated additive gene action for seed size in groundnut (Fisher et al. 1932). Four environments (Keh-S08; Gau-S08; Tro-R08; Tro-R09) showed negative kurtosis for HKW, but the distribution in Gauribidanur rainy season 2008 and 2011 had leptokurtic configurations. The negative kurtosis in most of the environments suggested that HKW trait is controlled by more number of genes without any gene interactions among them (Robson 1956). Frequency distribution of pod-related traits (beak, constriction and reticulation) revealed that these traits were not following normal distribution (Table 2; Suppl. Figure 2). All types of pod features were noticed in RILs. The parent VG 9514 has the highest score for beak and reticulation. TAG 24 has almost no beak and with moderate reticulation. Almost nil constriction was there in VG 9514 but moderate constriction was found in TAG 24 (Fig. 2). The score for pod beak and constriction followed negative skewness and positive kurtosis which means few numbers of genes interact among them and control these traits (Coffelt and Hammons 1974; Pattanashetti et al. 2008). The distribution of pod reticulation score was not skewed but had negative kurtosis. This suggests more number of genes is controlling pod reticulation without any possible gene interactions among them in groundnut.
Table 1.
Descriptive parameters of HKW of RILs at different six environments
| Environment | Number of RILs | Min HKW (g) | Max HKW (g) | Mean HKW (g) | Skewness | Kurtosis | Normality test | |
|---|---|---|---|---|---|---|---|---|
| Shapiro–Wilk statistic | Jarque–Bera statistic | |||||||
| Keh-S08 | 156 | 23 | 62 | 42.35 | 0.09 | − 0.55 |
0.99 (P = 0.31) |
2.35 (P = 0.31) |
| Gau-S08 | 153 | 26 | 66 | 45.16 | 0.15 | − 0.32 |
0.99 (P = 0.34) |
1.35 (P = 0.51) |
| Gau-R08 | 164 | 25 | 70 | 42.95 | 0.41 | 0.24 |
0.98 (P = 0.11) |
4.70 (P = 0.09) |
| Tro-R08 | 164 | 23 | 75 | 45.90 | 0.41 | − 0.12 |
0.98 (P = 0.05) |
4.62 (P = 0.09) |
| Tro-R09 | 162 | 24 | 68 | 46.30 | 0.06 | − 0.70 |
0.98 (P = 0.01) |
3.61 (P = 0.16) |
| Gau-R11 | 157 | 24.3 | 72 | 42.10 | 0.87 | 1.77 |
0.96 (NS) |
37.82 (NS) |
Keh-S08 Kehal, Maharashtra in summer (Jan to May) 2008, Gau-S08 Gauribidanur, Karnataka in summer 2008, Gau-R08 Gauribidanur in rainy season (June to September) 2008, Tro-R08 Trombay, Mumbai in rainy season 2008, Tro-R09 Trombay in rainy season 2009, Gau-R11 Gauribidanur in rainy season 2011
Fig. 1.
Frequency distribution of hundred kernel weight of RIL population at six different environments
Table 2.
Descriptive parameters of pod traits of RILs at different environments
| Environment | RILs | Min score | Max score | Mean score | Skewness | Kurtosis | Normality test | |
|---|---|---|---|---|---|---|---|---|
| Shapiro–Wilk statistic | Jarque–Bera statistic | |||||||
| Pod beak | ||||||||
| Tro-R08 | 164 | 0 | 9 | 5.76 | − 0.42 | 0.88 | 0.82 (NS) | 9.46 (NS) |
| Tro-R09 | 162 | 0 | 9 | 5.34 | − 0.51 | 0.36 | 0.88 (NS) | 7.79 (NS) |
| Gau-R11 | 164 | 0 | 9 | 5.76 | − 0.42 | 0.88 | 0.82 (NS) | 9.45 (NS) |
| Pod constriction | ||||||||
| Tro-R08 | 164 | 0 | 9 | 4.08 | − 0.11 | 0.62 | 0.84 (NS) |
2.51 (P = 0.28) |
| Tro-R09 | 162 | 0 | 9 | 4.44 | − 0.47 | 0.23 | 0.87 (NS) | 6.25 (NS) |
| Gau-R11 | 157 | 0 | 9 | 4.08 | − 0.10 | 0.62 | 0.84 (NS) |
2.50 (P 0.28) |
| Pod reticulation | ||||||||
| Tro-R08 | 164 | 0 | 9 | 5.77 | 0.05 | − 0.80 | 0.89 (NS) |
4.64 (P = 0.09) |
| Tro-R09 | 162 | 3 | 9 | 6.17 | 0.008 | − 0.84 | 0.88 (NS) |
4.95 (P = 0.08) |
| Gau-R11 | 157 | 0 | 9 | 5.77 | 0.05 | − 0.88 | 0.89 |
4.64 (P 0.09) |
Tro-R08 Trombay, Mumbai in rainy season (June to September) 2008, Tro-R09 Trombay in rainy season 2009, Gau-R11 Gauribidanur in rainy season 2011
Fig. 2.
Contrasting pod and seed features of TAG 24 and VG 9514
Previously, we have generated a genetic linkage map with 190 markers in cultivated groundnut (Mondal et al. 2014). Based on this genetic linkage information, a preliminary QTL analysis revealed that the consensus QTL peak for HKW lies in between GM2073 and TE282 marker in linkage group B07, but these two flanking markers were placed at 49.05 cM interval (Suppl. Figure 1). Searching of repeat motif in B07 (in between marker GM2073 and TE282) chromosomal sequence revealed 110 SSR repeat motifs with minimum repeat length of 14 bp. Of the 110 repeat motifs, 40 were with dinucleotide, 47 with trinucleotide, 3 with tetranucleotide, 2 with pentanucleotide, 2 with hexanucleotide and 16 with compound repeat motif (Suppl. Table 2). Of these 110 SSR primer pairs, six (5.45%) were found polymorphic between two parents. All these six new SSR markers were used for genotyping in 164 RILs for updating the existing genetic linkage map (mentioned in Mondal et al. 2014). Genotyping of 164 RILs revealed that four new markers (B07_53, B07_77, B07_80 and B07_109) have followed 1:1 ratio (P value = 0.87–1.00). Of the six B07 specific SSR markers, five were placed in B07 linkage group and the remaining one (B07_72) was unlinked. Placing of five new markers in the updated linkage map has increased map density from 22.3 cM/interval to 12.5 cM/interval in B07 group in the present study.
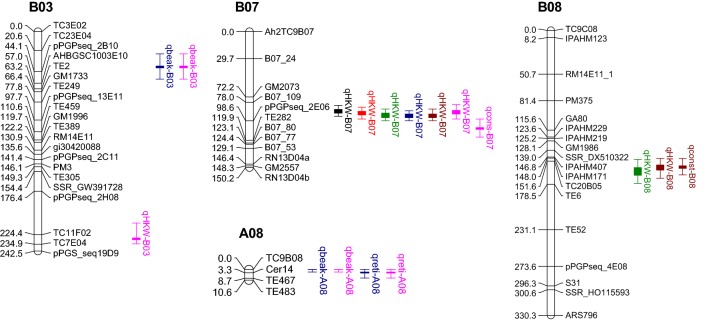
A total of three QTLs for seed size (in terms of HKW), two for pod beak, one for pod reticulation and two for pod constriction were identified in this study. QTL analysis for HKW revealed a major consensus QTL (qHKW-B07) in B07 linkage group in all the six analyzed environments. This major consensus QTL explained 10.50–23.88% phenotypic variance of HKW in RILs (Table 3). The significant QTL peak detected in between SSR markers B07_109 and pPGPseq_2E06 within a 20.6 cM map interval (Fig. 3). The additive effect of this major QTL was contributed by TAG 24 allele. Fonceka et al. (2012) identified two QTLs for HKW in chromosome A07 and B02. The closest marker pPGPseq_2E06 was placed in linkage group A07 in that report. In another report, Luo et al. (2018b) revealed three QTLs for hundred pod weight, pod length and pod width that were co-localized in a 5 cM interval (1.48 Mb in physical map) on chromosome A07. The present study detected two flanking markers pPGPseq_2E06 (genbank accession CC000271) and B07_109 (designed from chromosome 7 of A. ipaensis) for the consensus QTL for HKW in B07 linkage group. When we used CC000271 as query sequence in BLASTn in Peanutbase, we found that it has 99.86% sequence identity with 1,982,594–1,983,295 bp in Arahy.17 chromosome. Similarly the SSR loci portion of B07_109 has 100% identity with 4,645,102–4,645,410 bp in Arahy.17 chromosome. Two major QTLs for HKW were also reported in B02 and B03 chromosome by Huang et al. (2015) and the QTL in B03 chromosome was positioned distally at 124.2 cM. In the present study, another minor QTL was identified in B03 where it placed at the distal portion of the chromosome in between SSR marker TC11F02 and TC7E04 (Fig. 3). This QTL in B03 (qHKW-B03) appeared only in Gauribidanur rainy season 2011 and explained 8.88% of phenotypic variance (Table 3). The additive effect of this minor QTL in B03 was contributed by TAG 24 allele. Another minor QTL (qHKW-B08) was also identified in linkage group B08 (Table 3). The QTL in B08 was detected only in two environments (Gauribidanur rainy 2008 and Trombay rainy 2009) and it explained 6.71–7.97% phenotypic variance. This minor QTL in B08 group had additive effect that was contributed by the VG 9514 allele. Two flanking markers for the identified major QTL for HKW were later genotyped in 71 cultivated groundnut genotypes. Based on the single marker ANOVA, it was found that the SSR marker pPGPseq_2E06 was significantly associated with HKW and explained 17.94% phenotypic variation (R2 = 0.1794, P = 0.0005). But no association was detected for the other flanking marker B07_109.
Table 3.
Details of QTLs information of HKW in different environments
| QTL name | Environment | Chr. | Peak position (cM) | Left marker | Right marker | LOD value | PVE (%) | Additive value |
|---|---|---|---|---|---|---|---|---|
| qHKW-B07 | Keh-S08 | B07 | 88 | B07_109 | pPGPseq_2E06 | 8.14 | 20.55 | − 4.40 |
| qHKW-B07 | Gau-S08 | B07 | 90 | B07_109 | pPGPseq_2E06 | 6.47 | 17.80 | − 4.20 |
| qHKW-B07 | Gau-R08 | B07 | 94 | B07_109 | pPGPseq_2E06 | 4.35 | 10.50 | − 3.22 |
| qHKW-B08 | B08 | 163 | TC20B05 | TE6 | 2.89 | 7.97 | 2.82 | |
| qHKW-B07 | Tro-R08 | B07 | 93 | B07_109 | pPGPseq_2E06 | 9.73 | 23.88 | − 5.41 |
| qHKW-B07 | Tro-R09 | B07 | 93 | B07_109 | pPGPseq_2E06 | 4.71 | 11.87 | − 4.45 |
| qHKW-B08 | B08 | 159 | TC20B05 | TE6 | 2.58 | 6.71 | 3.35 | |
| qHKW-B07 | Gau-R11 | B07 | 89 | B07_109 | pPGPseq_2E06 | 6.08 | 18.13 | − 3.32 |
| qHKW-B03 | B03 | 229 | TC11F02 | TC7E04 | 3.64 | 8.88 | − 2.32 |
Keh-S08 Kehal, Maharashtra in summer (Jan to May) 2008, Gau-S08 Gauribidanur, Karnataka in summer 2008, Gau-R08 Garibidanur in rainy season (June to September) 2008, Tro-R08 Trombay, Mumbai in rainy season 2008, Tro-R09 Trombay in rainy season 2009, Gau-R11 Gauribidanur in rainy season 2011
Fig. 3.
QTLs for hundred kernel weight, pod beak, pod reticulation and pod constriction in the linkage map of cultivated groundnut. Color codes for QTLs are as follows: black = QTL region for Keh-S08, red = QTL region for Gau-S08, green = QTL region for Gau-R08, blue = QTL region for Tro-R08, maroon = QTL region for Tro-R09, and pink = QTL region for Gau-R11
Identification of QTLs for pod traits (beak, constriction, reticulation) is equally important, as these traits determine the consumer or market acceptability. Confectionary groundnut with desirable pod features normally fetches premium price (Kale et al. 2000). Thus, QTLs for both HKW and pod traits will be helpful in introgression of desirable alleles in breeding for large seed trait in groundnut. Analysis of QTL for pod-related traits revealed five minor QTLs (in total) for pod beak, constriction and reticulation (Table 4). Two minor QTLs for pod beak (qbeak-B03 and qbeak-A08) were detected in linkage group B03 and A08 that explained 8.73–8.89% and 6.31–6.78% of phenotypic variance, respectively. The QTL peak in B03 was present at 39 cM distance in between markers TC23E04 and pPGPseq_2B10. The other QTL peak in A08 was placed in between marker TC9B08 and Cer14 (Fig. 3). The same map interval in linkage group A08 had another minor QTL (qreti-A08) for pod reticulation. This minor QTL for pod reticulation was detected in two environments and it explained 7.30–7.80% phenotypic variance (Table 4). Further, a correlation analysis revealed that pod reticulation and pod beak are positively correlated (r = 0.30, P = 0.0007). An earlier report by Shirasawa et al. (2012) revealed a QTL for pod beak named qSTP03 in between marker AhTE0570 and AHGS1744 in A03. The same report also described two other QTLs for pod constriction in linkage group A09 and B09. Another two minor QTLs for pod constriction (qconst-B08 and qconst-B07) were detected in B08 (in Trombay rainy 2009) and B07 (in Gauribidanur rainy 2011) and explained 8.84% and 8.45% phenotypic variance, respectively (Table 4). Interestingly, both the qHKW-B08 and qconst-B08 were mapped in the same map interval (Fig. 3) and their additive effects were contributed by VG 9514 allele. Recently a single genomic region was found to be associated with pod constriction in B07 chromosome and the reported QTL explained 32% of the total phenotypic variation due to pod constriction in a mapping population (Patil et al. 2018). The identified minor QTL for pod constriction in this study is situated in between marker pPGPseq_2E06 and TE282 of B07 linkage group (Fig. 3). The same marker interval was closely spaced with the consensus QTL for HKW (qHKW-B07) in B07 linkage group in this study. The additive effect of both these QTLs (qHKW-B07 and qconst-B07) in B07 was contributed by TAG 24 alleles. Thus, to identify recombinant with large seed and less constriction, a large number of segregating plant materials has to be generated to identify desirable crossovers within these above QTLs. Future research may be directed to fine map this interesting loci that control both HKW and pod constriction in cultivated groundnut.
Table 4.
Details of QTLs information of pod traits in different environments
| QTL name | Environment | Chr. | Peak position (cM) | Left marker | Right marker | LOD value | PVE (%) | Additive value |
|---|---|---|---|---|---|---|---|---|
| qbeak-B03 | Tro-R08 | B03 | 39.0 | TC23E04 | pPGPseq_2B10 | 2.75 | 8.73 | − 0.43 |
| qbeak-A08 | A08 | 0.0 | TC9B08 | Cer14 | 2.63 | 6.31 | − 0.37 | |
| qbeak-B03 | Gau-R11 | B03 | 39.0 | TC23E04 | pPGPseq_2B10 | 2.81 | 8.89 | − 0.57 |
| qbeak-A08 | A08 | 0.0 | TC9B08 | Cer14 | 2.71 | 6.78 | − 0.41 | |
| qreti-A08 | Tro-R08 | A08 | 3.0 | TC9B08 | Cer14 | 2.58 | 7.30 | − 0.54 |
| qreti-A08 | Gau-R11 | A08 | 3.0 | TC9B08 | Cer14 | 2.71 | 7.80 | − 0.67 |
| qconst-B08 | Tro-R09 | B08 | 159 | TC20B05 | TE6 | 3.54 | 8.84 | 0.76 |
| qconst-B07 | Gau-R11 | B07 | 107.0 | pPGPseq_2E06 | TE282 | 3.22 | 8.45 | − 0.52 |
Tro-R08 Trombay, Mumbai in rainy season (June to September) 2008, Tro-R09 Trombay in rainy season 2009, Gau-R11 Gauribidanur in rainy season 2011
Electronic supplementary material
Below is the link to the electronic supplementary material.
Suppl. Figure 1: Initial QTL mapping of seed size detected map interval in between GM2073 and TE282 (DOCX 27 kb)
Suppl. Figure 2: Frequency distribution of pod traits of RIL population at three environments. (DOCX 182 kb)
Suppl. Table 1: List of Trombay groundnut genotypes used for marker validation. (XLSX 15 kb)
Suppl. Table 2: Details of newly developed B07-specific SSR markers in the present study. (XLSX 17 kb)
Suppl. Table 3: Analysis of variance for seed size in RILs over six different environments (DOCX 11 kb)
Acknowledgements
Sincere thanks to Associate Director (A), Bioscience group and Head, NA&BTD, BARC for his encouragement and support. Authors duly acknowledged the efforts of T. Chalapathi and Sujit Tota during field experimentation.
Author contributions
SM and AMB developed the mapping population, evaluated it for seed size and pod traits in multi-environments. SM conducted laboratory experiments, analyzed data and wrote the first draft of the manuscript. Both the authors checked and edited the manuscript and prepared the final version.
Compliance with ethical standards
Conflict of interest
Authors declare that they have no conflict of interest.
References
- Anderson WF, Fitzner MS, Isleib TG, Wynne JC, Phillips TD. Combining ability for large pod and seed traits in peanut. Peanut Sci. 1993;20:49–52. doi: 10.3146/i0095-3679-20-1-13. [DOI] [Google Scholar]
- Badigannavar AM, Mondal S. Mutation experiments and recent accomplishments in Trombay groundnuts. IANCAS Bullet. 2007;6:308–318. [Google Scholar]
- Balaiah C, Reddy PS, Reddi MV. Genic analysis in groundnut. I. Inheritance studies on 18 morphological characters in crosses with Gujarat narrow leaf mutant. Proc Ind Acad Sci. 1977;85:340–350. [Google Scholar]
- Bertioli DJ, Cannon SB, Froenicke L, Huang G, Farmer AD, Cannon EK, et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat Genet. 2016;48:436–443. doi: 10.1038/ng.3517. [DOI] [PubMed] [Google Scholar]
- Bhad PG, Mondal S, Badigannavar AM. Genetic diversity in groundnut (Arachis hypogaea L.) genotypes and detection of marker trait associations for plant habit and seed size using genomic and genic SSRs. J Crop Sci Biotechnol. 2016;19(3):203–221. doi: 10.1007/s12892-016-0060-1. [DOI] [Google Scholar]
- Cahaner A (1978) The inheritance of yield components and plant conformation in peanut, Arachis hypogaea L. PhD thesis, The Hebrew University, Israel
- Chen W, Jiao Y, Cheng L, Huang L, Liao B, Tang M, Ren X, Zhou X, Chen Y, Jiang H. Quantitative trait locus analysis for pod-and kernel-related traits in the cultivated peanut (Arachis hypogaea L.) BMC Genet. 2016;17:25. doi: 10.1186/s12863-016-0337-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevenger J, Chu Y, Chavarro C, Botton S, Culbreath A, Isleib TG, Holbrook CC, Ozias-Atkin P. Mapping late leaf spot resistance in peanut (Arachis hypogaea) using QTL-seq reveals markers for marker-assisted selection. Front Plant Sci. 2018;9:83. doi: 10.3389/fpls.2018.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffelt TA, Hammons RO. Inheritance of an albino seedling character in Arachis hypogaea L. Crop Sci. 1974;11:753–755. doi: 10.2135/cropsci1971.0011183X001100050045x. [DOI] [Google Scholar]
- Doerge RW. Multifactorial genetics: mapping and analysis of quantitative trait loci in experimental populations. Nat Rev. 2002;3:43–52. doi: 10.1038/nrg703. [DOI] [PubMed] [Google Scholar]
- FAOSTAT (2017) Food and Agricultural Organization of the United Nations. Data available at http://www.fao.org/faostat/en/#data/QC. Accessed 7 Jan 2018
- Faye I, Pandey MK, Hamidou F, Rathore A, Ndoye O, Vadez V, Varshney RK. Identification of quantitative trait loci for yield and yield related traits in groundnut (Arachis hypogaea L.) under different water regimes in Niger and Senegal. Euphytica. 2015;206:631–647. doi: 10.1007/s10681-015-1472-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson M, Burow M, Schulze S, Bramel P, Paterson A, Kresovich S, Mitchell S. Microsatellite identification and characterization in peanut (Arachis hypogaea L.) Theor Appl Genet. 2004;108:1064–1070. doi: 10.1007/s00122-003-1535-2. [DOI] [PubMed] [Google Scholar]
- Fisher RA, Immer FR, Tedin O. The genetical interpretation of statistics of the third degree in the study of quantitative inheritance. Genetics. 1932;17:107–124. doi: 10.1093/genetics/17.2.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonceka D, Tossim HA, Rivallan R, Vignes H, Faye I, Ndoye O, Morezsohn MC, Bertioli DJ, Glaszmann JC, Courtois B, Rami JF. Fostered and left behind alleles in peanut: interspecific QTL mapping reveals footprints of domestication and useful natural variation for breeding. BMC Plant Biol. 2012;12:26. doi: 10.1186/1471-2229-12-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garet B. Heterosis and combining abilities in groundnut (Arachis hypogaea L.) Oleagineux. 1976;29:435–442. [Google Scholar]
- Hake AA, Shirasawa K, Yadawad A, Sukruth M, Patil M, Nayak SN, Lingaraju S, Patil PV, Nadaf HL, Gowda MVC, Bhat RS. Mapping of important taxonomic and productivity traits using genic and non-genic transposable element markers in peanut (Arachis hypogaea L.) PLoS One. 2017;12(10):e0186113. doi: 10.1371/journal.pone.0186113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halward TM, Stalker HT, Kochert G. Use of single primer DNA amplification in genetic studies of peanut (Arachis hypogaea L.) Plant Mol Biol. 1991;18:315–325. doi: 10.1007/BF00034958. [DOI] [PubMed] [Google Scholar]
- Hammer Ø, Harper DAT, Ryan PD (2001) PAST: Paleontological statistics software package for education and data analysis. Palaeontologia Electronica, 4(1). http://folk.uio.no/ohammer/past. Accessed 10 Dec 2018
- Hariprasanna K, Lal C, Radhakrishnan T, Gor HK, Chikani BM. Analysis of diallel cross for some physical-quality traits in peanut (Arachis hypogaea L.) Euphytica. 2008;160:49–57. doi: 10.1007/s10681-007-9553-9. [DOI] [Google Scholar]
- Huang L, He H, Chen W, Ren X, Chen Y, Zhou X, Xia Y, Wang X, Jiang X, Liao B, Jiang H. Quantitative trait locus analysis of agronomic and quality-related traits in cultivated peanut (Arachis hypogaea L.) Theor Appl Genet. 2015;128:1103–1115. doi: 10.1007/s00122-015-2493-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IPGRI, ICRISAT . Descriptors for peanut. Rome: International Crop Research Institute for the Semi-Arid Tropics, Patancheru, India; International Board for Plant Genetic Resources; 1992. p. 125. [Google Scholar]
- Jiang H, Huang L, Ren X, Chen Y, Zhou X, Xia Y, Huang J, Lei Y, Yan L, Wan L, Liao B. Diversity characterization and association analysis of agronomic traits in a Chinese peanut (Arachis hypogaea L.) mini-core collection. J Integr Plant Biol. 2014;56:159–169. doi: 10.1111/jipb.12132. [DOI] [PubMed] [Google Scholar]
- Kale DM, Badigannavar AM, Murty GSS. Development of new large pod Trombay groundnut (Arachis hypogaea) selections. Indian J Agric Sci. 2000;70(6):365–369. [Google Scholar]
- Kayam G, Brand Y, Faigenboim-Doron A, Patil A, Hedvat I, Hovav R. Fine-mapping the branching habit trait in cultivated peanut by combining bulked segregant analysis and high throughput sequencing. Front Plant Sci. 2017;8:467. doi: 10.3389/fpls2017.00467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochert G, Stalker HT, Gimenes M, Galgaro L, Lopes CR, Moore K. RFLP and cytogenetic evidence on the origin and evolution of allotetraploid domesticated peanut (Arachis hypogaea (Leguminosae) Am J Bot. 1996;83:1282–1291. doi: 10.2307/2446112. [DOI] [Google Scholar]
- Krapovickas A. The origin, variability and spread of the peanut (Arachis hypogaea) In: Ucko PJ, Dimbledy GW, editors. The domestication and exploitation of plant and animals. London: Gerald Duckworth Co Ltd; 1969. pp. 427–441. [Google Scholar]
- Krapovickas A, Gregory WC. Taxonomy of genus Arachis (Leguminosae) Bonplandia. 1994;8:1–186. [Google Scholar]
- Layrisse A, Wynne JC, Isleib TG. Combining ability for yield, protein and oil of peanut lines from South American centres of diversity. Euphytica. 1980;29:561–570. doi: 10.1007/BF00023203. [DOI] [Google Scholar]
- Li H, Ye G, Wang J. A modified algorithm for the improvement of composite interval mapping. Genetics. 2007;175:361–374. doi: 10.1534/genetics.106.066811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Pandey MK, Khan AW, Guo J, Wu B, Cai Y, Huang L, Zhou X, Chen Y, Chen W, Liu N, Lei Y, Liao B, Varshney RK, Jiang H. Discovery of genomic regions and candidate genes controlling shelling percentage using QTL-seq approach in cultivated peanut (Arachis hypogaea L.) Plant Biotechnol J. 2018 doi: 10.1111/pbi.13050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Guo J, Ren X, Chen W, Hunag L, Zhou X, Chen Y, Liu N, Xiong F, Lei Y, Liao B, Jiang H. Chromosomes A07 and A05 associated with stable and major QTLs for pod weight and size in cultivated peanut (Arachis hypogaea L.) Theor Appl Genet. 2018;131:267–282. doi: 10.1007/s00122-017-3000-7. [DOI] [PubMed] [Google Scholar]
- Mondal S, Badigannavar AM. Mapping of a dominant rust resistance gene revealed two R genes around the major Rust_QTL in cultivated peanut (Arachis hypogaea L.) Theor Appl Genet. 2018;131:1671–1681. doi: 10.1007/s00122-018-3106-6. [DOI] [PubMed] [Google Scholar]
- Mondal S, Badigannavar AM, D’Souza SF. Developement of genic molecular markers linked to a rust resistance gene in cultivated groundnut (Arachis hypogaea L.) Euphytica. 2012;188:163–173. doi: 10.1007/s10681-011-0619-3. [DOI] [Google Scholar]
- Mondal S, Hadapad AB, Hande P, Badigannavar AM. Identification of quantitative trait loci for bruchid (Caryedon serratus Olivier) resistance components in cultivated groundnut (Arachis hypogaea L.) Mol Breed. 2014;33:961–973. doi: 10.1007/s11032-013-0011-1. [DOI] [Google Scholar]
- Mondal S, Phadke RR, Badigannavar AM. Genetic variability of total phenolics, flavonoids and antioxidant activity of testaless seeds of a peanut recombinant inbred line population and identification of their controlling QTLs. Euphytica. 2015;204:311–321. doi: 10.1007/s10681-014-1324-9. [DOI] [Google Scholar]
- Murty GSS, Badigannavar AM, Mondal S, Kale DM. Research and impact of groundnut mutation breeding in India. In: Basu MS, Singh NB, editors. Groundnut Research in India. Junagadh: NRCG (now, Directorate of Groundnut Research); 2004. pp. 57–69. [Google Scholar]
- Pandey MK, Upadhyaya HD, Rathore A, Vadez V, Sheshshayee M, Sriswathi M, Govil M, Kumar A, Gowda MVC, Sharma S, Hamidou F, Kumar VA, Khera P, Bhat RS, Khan AW, Singh S, Li H, Monyo E, Nadaf HL, Mukri G, Jackson SA, Guo B, Liang X, Varshney RK. Genome wide association studies for 50 agronomic traits in peanut using ‘reference set’ comprising 300 genotypes from 48 countries of the semi-arid topics of the world. PLoS One. 2014;9:e105228. doi: 10.1371/journal.pone.0105228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil SH, Kale DM, Deshmukh SN, Fulzele GR, Weginwar BG. Semi-dwarf, early maturing and high yielding new groundnut variety, TAG 24. J Oilseed Res. 1995;12:254–257. [Google Scholar]
- Patil AS, Popvsky S, Levy Y, Chu Y, Clevenger J, Ozias-Atkins P, Hovav R. Genetic insight and mapping of the pod constriction trait in Virginia-type peanut. BMC Genet. 2018;19:93. doi: 10.1186/s12863-018-0674-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanashetti SK, Gowda MVC, Girija Inheritance of morphological traits and pod features in groundnut (Arachis hypogaea L.) Indian J Genet Plant Breed. 2008;68:157–162. [Google Scholar]
- Robson DS. Application of K4 statistics to genetic variance component analysis. Biometrics. 1956;12:433–444. doi: 10.2307/3001682. [DOI] [Google Scholar]
- Shirasawa K, Koilkonda P, Aoki K, Hirakawa H, Tabata S, Watanabe M, Hasegawa M, Kiyoshima H, Suzuki S, Kuwata C, Naito Y, Kuboyama T, Nakaya A, Sasamoto S, Watanabe A, Kato M, Kawashima K, Kishida Y, Kohara M, Kurabayashi A, Takahashi C, Tsuruoka H, Wada T, Isobe S. In silico polymorphism analysis for the development of simple sequence repeat and transposon markers and construction of linkage map in cultivated peanut. BMC Plant Biol. 2012;12:80. doi: 10.1186/1471-2229-12-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasawa K, Bhat RS, Khedikar Y, Sujay V, Kolekar RM, Yeri SB, Sukruth M, Cholin S, Asha B, Pandey MK, Varshney RK, Gowda MVC. Sequencing analysis of genetic loci for resistance for late leaf spot and rust in peanut (Arachis hypogaea L.) Front Plant Sci. 2018;9:1727. doi: 10.3389/fpls.2018.01727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swe SY, Branch WD. Estimates of combining ability and heterosis among peanut cultivars. Peanut Sci. 1986;13:70–74. doi: 10.3146/i0095-3679-13-2-7. [DOI] [Google Scholar]
- Upadhyaya HD, Swamy BPM, Goudar PVK, Kullaiswamy BY, Singh S. Identification of diverse groundnut germplasm through multi-environment evaluation of a core collection for Asia. Field Crops Res. 2005;93:293–299. doi: 10.1016/j.fcr.2004.10.007. [DOI] [Google Scholar]
- Varman PV. A foliar disease resistant line developed through interspecific hybridization in groundnut (Arachis hypogaea L.) Indian J Agric Sci. 1999;69:67–68. [Google Scholar]
- Venuprasad R, Aruna R, Nigam SN. Inheritance of traits associated with seed size in groundnut (Arachis hypogaea L.) Euphytica. 2011;181:169–177. doi: 10.1007/s10681-011-0390-5. [DOI] [Google Scholar]
- Voorrips RE. Mapchart: software for the graphical presentation of linkage map and QTL. J Heredity. 2002;93:77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- Wang J, Li H, Zhang L, Meng L (2016) Users’ Manual of QTL IciMapping. The Quantitative Genetics Group, Institute of Crop Science, Chinese Academy of Agricultural Sciences (CAAS), Beijing 100081, China, and Genetic Resources Program, International Maize and Wheat Improvement Centre (CIMMYT), Apdo. Postal 6-641, 06600 Mexico, D.F., Mexico
- Yol E, Furat S, Upadhyaya H, Uzun B. Characterization of groundnut (Arachis hypogaea L.) collection using quantitative and qualitative traits in the Mediterranean basin. J Integr Agric. 2018;17:63–75. doi: 10.1016/S2095-3119(17)61675-7. [DOI] [Google Scholar]
- Zhao J, Huang L, Ren X, Pandey MK, Wu B, Chen Y, Zhou X, Chen W, Xia Y, Li Z, Luo H, Lei Y, Varshney RK, Liao B, Jiang H. Genetic variation and association mapping of seed-related traits in cultivated peanut (Arachis hypogaea L.) using single-locus simple sequence repeat markers. Front Plant Sci. 2017;8:2105. doi: 10.3389/fpls.2017.02105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suppl. Figure 1: Initial QTL mapping of seed size detected map interval in between GM2073 and TE282 (DOCX 27 kb)
Suppl. Figure 2: Frequency distribution of pod traits of RIL population at three environments. (DOCX 182 kb)
Suppl. Table 1: List of Trombay groundnut genotypes used for marker validation. (XLSX 15 kb)
Suppl. Table 2: Details of newly developed B07-specific SSR markers in the present study. (XLSX 17 kb)
Suppl. Table 3: Analysis of variance for seed size in RILs over six different environments (DOCX 11 kb)