Significance
Deciphering the mechanisms involved in the recognition of natural ligands by their G protein-coupled receptors (GPCRs) is a major issue in pharmacology, particularly for peptide-activated receptors, for which structural data are sparse. Here, we combined state-of-the-art biophysical methods to solve the structure of a peptide hormone, ghrelin, in its receptor-bound state. In addition of being a prototype for GPCR peptide ligands, ghrelin is a major player in physiology. The data emerging from this study clearly demonstrate that ligand conformational dynamics play a central role in receptor–peptide interactions. The results of this study will also aid in the future development of drugs targeting the ghrelinergic system.
Keywords: GPCR, ghrelin, acylation, NMR, coarse-grain modeling
Abstract
Ghrelin plays a central role in controlling major biological processes. As for other G protein-coupled receptor (GPCR) peptide agonists, the structure and dynamics of ghrelin bound to its receptor remain obscure. Using a combination of solution-state NMR and molecular modeling, we demonstrate that binding to the growth hormone secretagogue receptor is accompanied by a conformational change in ghrelin that structures its central region, involving the formation of a well-defined hydrophobic core. By comparing its acylated and nonacylated forms, we conclude that the ghrelin octanoyl chain is essential to form the hydrophobic core and promote access of ghrelin to the receptor ligand-binding pocket. The combination of coarse-grained molecular dynamics studies and NMR should prove useful in improving our mechanistic understanding of the complex conformational space explored by a natural peptide agonist when binding to its GPCR. Such information should also facilitate the design of new ghrelin receptor-selective drugs.
G protein-coupled receptors (GPCRs) compose the largest family of cell surface receptors and contribute to various central physiological processes (1). As such, they represent prominent therapeutic targets (2). Despite the wealth of structural information obtained over the last decade, few complexes between receptors and their natural agonists have been identified. This problem is particularly apparent for peptide ligands. Indeed, the structures of only a handful of all of the class A GPCRs activated by endogenous peptides or proteins have been elucidated (3, 4), often with constrained ligands and/or modified receptors (3). Although limited, the existing structural data nevertheless demonstrate that GPCR peptide ligands adopt very diverse and complex modes of binding (3).
Ghrelin is a 28-aa peptide hormone that exerts a wide range of biological effects, including controlling growth hormone secretion, food intake, glucose metabolism, and reward (5). These effects are all mediated by a class A GPCR, the growth hormone secretagogue receptor (GHSR) (6). A detailed description of the structure of ghrelin in its receptor-bound state is lacking and is urgently needed, given its primary physiological role and therapeutic potential (5). Intriguingly, ghrelin requires posttranslational octanoylation on the serine residue at position 3 for GHSR binding and activation, even though nonacylated ghrelin is the most abundant circulating form of the hormone (5). The role of the ghrelin octanoyl chain is currently unknown, although different roles have been proposed, such as partitioning into the membrane to increase the local concentration near the receptor or stabilizing the peptide conformation for optimal docking to GHSR. Here, by combining solution-state NMR with molecular modeling, we provide a detailed description of the structure and dynamics of ghrelin bound to its receptor and illuminate the role of the octanoyl chain in stabilizing the structure of the hormone and promoting its binding to GHSR.
Results
The Ghrelin-GHSR Complex.
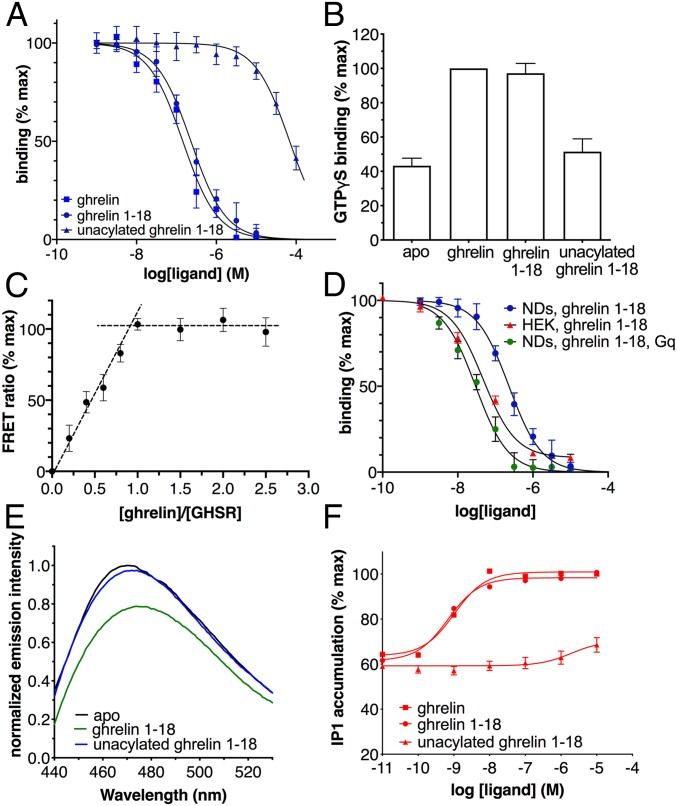
Wild-type human monomeric GHSR was expressed in Escherichia coli and assembled into POPC/POPG nanodiscs (7). Active GHSR was then purified on a ligand-immobilized affinity column (8) (SI Appendix). The receptor used in all NMR experiments was thus fully functional with regard to ghrelin binding and G protein activation (Fig. 1 A and B), with more than 90% of GHSR in our preparations competent to bind ligand, as assessed in a stoichiometric titration assay (Fig. 1C). Moreover, high-affinity ghrelin binding similar to that measured in HEK cells could be recovered by reconstituting the isolated receptor in nanodiscs with its cognate Gq protein (Fig. 1D). The effect of the G protein on agonist affinity provides a further measure of the functional properties of the purified receptor. It also indicates that the properties of ghrelin bound to isolated GHSR assessed in this work are signatures of the natural agonist bound to the low-affinity, G protein-uncoupled state of its GPCR.
Fig. 1.
Functional properties of the isolated GHSR. (A) FRET-monitored competition assays of ghrelin, ghrelin 1–18, and unacylated ghrelin 1–18 for binding GHSR assembled into nanodiscs. (B) GTPγS binding to Gαqβ1γ2 catalyzed by GHSR-containing nanodiscs in the absence of ligand or in the presence of 10 µM ghrelin, 10 µM ghrelin 1–18, or 500 µM unacylated ghrelin 1–18. (C) FRET-monitored stoichiometric titration of a 20 µM fluorescent ghrelin solution with increasing concentrations of GHSR-containing nanodiscs. (D) FRET-monitored competition assays of ghrelin 1–18 for binding GHSR assembled into nanodiscs (NDs) in the absence or in the presence of Gαqβ1γ2 compared with the HTRF-monitored competition binding assay of ghrelin 1–18 on GHSR expressed in stable HEK293 cell lines. (E) Fluorescence emission spectra of bimane attached to C3047.34 of GHSR in nanodiscs in the absence of ligand or in the presence of 10 µM ghrelin or of 500 µM unacylated ghrelin. (F) Ghrelin, ghrelin 1–18, and unacylated ghrelin-induced IP1 production in GHSR-expressing HEK293 cells. Except for E, data are the mean ± SEM of 3 measurements.
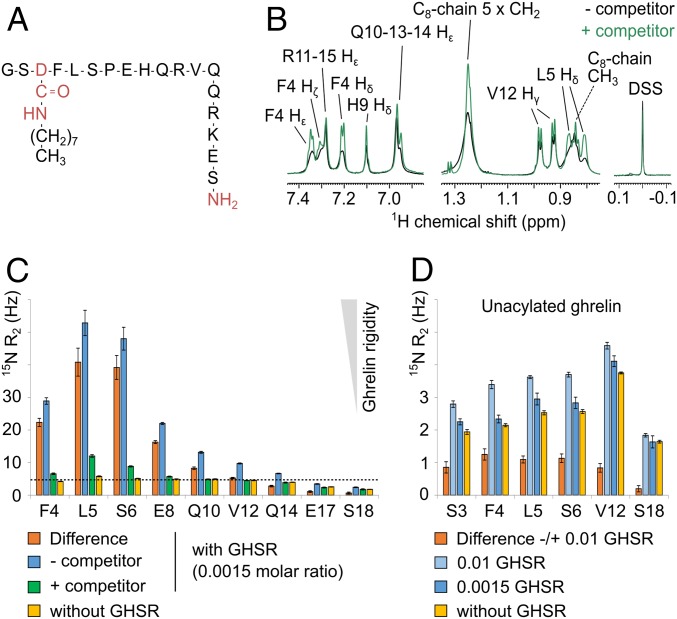
To facilitate NMR analyses, we used a ghrelin peptide containing residues 1 to 18 (ghrelin 1–18) instead of full-length ghrelin (Fig. 2A), because the N-terminal region of ghrelin has been shown to be sufficient for full GHSR binding and activation (9–12). Accordingly, this truncation did not have a major impact on either ghrelin affinity for GHSR or its ability to trigger Gq activation (Fig. 1 A and B) (9). In addition, the C8 fatty acid chain was attached to an Asp residue at position 3 instead of a Ser, to increase the stability of the octanoylated version of the peptide (Fig. 2A). This modification does not affect the pharmacologic profile of ghrelin (13), as the ester bond between the C8 chain and Ser3 is not indispensable for ghrelin activity (11). Ghrelin 1–18 was synthesized with a number of residues labeled with 15N on their amide bonds, spanning the entire peptide sequence (i.e., S2, F4, L5, S6, E8, Q10, V12, Q14, E17, and S18). Under our experimental conditions, ghrelin 1–18 was in fast exchange between its free and receptor-bound states on the chemical shift time scale (i.e., <10 ms). Indeed, we observed broadening of site-specific ghrelin signals in the presence of GHSR and linear chemical shift variations as a function of the bound fraction (Fig. 2B and SI Appendix, Fig. S1), as was previously observed for the dynorphin–κ-opioid receptor complex (14). We exploited these fast-exchange conditions to use a large excess of ghrelin compared with GHSR (0.0015 GHSR-to-ghrelin molar ratio). Under these fast-exchange conditions, 15N transverse relaxation rates and 1H cross-relaxation rates were weighted averages of their values in the free and bound states. Due to the GHSR nanodiscs’ slow tumbling motion, these values are 3 to 4 orders of magnitude higher in the bound state than in the free state. Thus, the former very likely dominated the measured averages, as confirmed by our results (Fig. 2C and SI Appendix, Fig. S2). To account for nonspecific binding, we systematically duplicated each NMR experiment in the presence of an orthosteric GHSR ligand, JMV5327. This nonpeptidic compound (15) dissociates slowly from the isolated receptor, which prevents ghrelin from binding to its orthosteric site (SI Appendix, Fig. S3).
Fig. 2.
Acylated and unacylated ghrelin structural dynamics in the bound state. (A) Primary structure of the ghrelin peptide studied in the present work. Differences from the human wild-type peptide (deletion of residues 19 to 28, amidification of the C terminus, and amidated aspartate at position 3) are shown in red. (B) 1D 1H spectrum of ghrelin with GHSR and with (green) or without (black) JMV5327. (C) 15N transverse relaxation rates (R2) of ghrelin backbone amides. Blue, apo GHSR (0.0015 GHSR:ghrelin molar ratio); green, GHSR loaded with JMV5327 (0.0015 GHSR:ghrelin molar ratio); orange, apo minus JMV5327-loaded difference; yellow, without GHSR. The dashed line corresponds to the 15N R2 of nonacylated ghrelin V12 with GHSR (0.01 molar ratio). The relaxation rate difference (in orange) is proportional to the bound fraction and to the order parameter S2, which describes the amplitudes of NH motions in the bound state. (D) 15N R2 of nonacylated ghrelin 1–18 backbone amides with apo GHSR (light blue, 0.01 GHSR:ghrelin molar ratio; blue, 0.0015 molar ratio), without GHSR (yellow), and the difference in 15N R2 (apo receptor at 0.01 GHSR:ghrelin molar ratio minus no receptor; orange).
Receptor-Bound Ghrelin Structure, Dynamics, and Interaction Mapping.
First, we measured amide nitrogen 15N transverse relaxation rates (15N R2), which are direct reporters of peptide internal mobility in its receptor-bound state (14). We carried out these measurements with a perdeuterated receptor, as perdeuteration is straightforward in the E. coli expression system that we used (16) and prevents spin diffusion in 15N relaxation and proton transferred nuclear Overhauser effect (trNOE) NMR experiments (17, 18). We observed significant differences in the 15N R2 of ghrelin between apo- and JMV5327-loaded GHSR (Fig. 2C). Specifically, we found considerably higher rates for residues F4 to E8 in the absence of the competing compound. This 15N transverse relaxation rate profile indicates that the N-terminal region encompassing the functionally important residues F4 (in part), L5, and S6 is rigidified in the GHSR-bound state while the peptide backbone becomes progressively more flexible (i.e., unfolded) toward the C terminus.
Complementary information on side chain dynamics was provided by 1D proton spectrum line shape analysis (Fig. 2B and SI Appendix, Fig. S1). We observed significantly broadened signals for ghrelin protons G1-Hα, S2-Hβ, F4 aromatic ring, L5 side chain methyl, S6-Hβ, and C8 chain in the presence of the receptor and in the absence of JMV5327 compared with signals from other residues, such as H9, V12, Q10, Q13, Q14, R11, and R15. Taken together, these data clearly demonstrate the specific formation of a hydrophobic core in the N-terminal part of ghrelin upon receptor binding, while the C-terminal region of the peptide encompassing residues from E8 to S18 remains flexible.
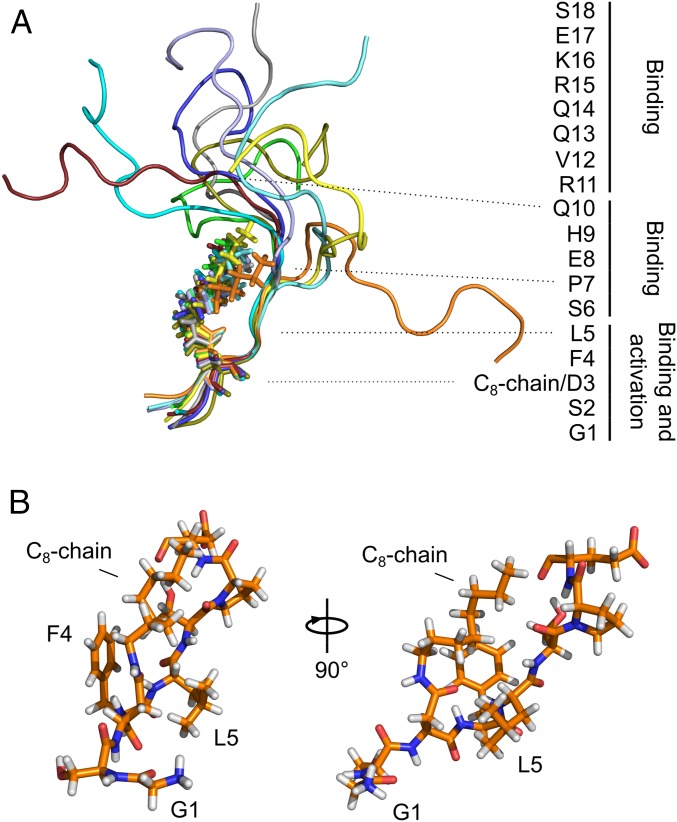
We then used trNOE experiments to determine the structure of ghrelin bound to GHSR (14, 17–20). We conducted nuclear Overhauser effect spectroscopy (NOESY) experiments at several mixing times with the ghrelin samples in the presence of perdeuterated GHSR and in the absence or presence of JMV5327. As was the case for 15N R2 relaxation rates, 1H-1H cross-relaxation rates leading to NOEs were dominated by the contributions of slow tumbling motion, reflecting the bound state (14). We fit the evolution of the NOESY cross-peak integrals with mixing time (build-up curves) using biexponential functions using the isolated spin pair approximation (21) (SI Appendix, Fig. S2). Interproton proximities in the bound state were distributed throughout the peptide sequence, with a higher density in the rigidified N-terminal part, where several medium-range NOE contacts were observed (SI Appendix, Fig. S2). Importantly, we observed several NOE contacts between the C8 chain terminal methyl and the peptide chain, namely at S6 and H9. We used distance restraints derived from the NOE build-up curves for structure calculation by simulated annealing with AMBER14 (22) (SI Appendix, Table S1). We calculated an ensemble of 10 receptor-bound ghrelin structures that were in best agreement with the experimental data (Fig. 3A; Protein Data Bank ID code 6H3E) (23). The C terminus peptide comprising residues S18 to R11 was disordered, while the peptide center from Q10 to S6 progressively adopted a more rigid structure. In the N-terminal region, critical residues L5, F4, and the D3-linked C8 chain formed a well-defined hydrophobic core (Fig. 3B). In this ensemble of structures, the C8 chain was aligned with the peptide backbone, pointing toward its C terminus, and was surrounded by the aromatic ring of F4 and the side chain of L5. Interestingly, this ensemble explains the 15N transverse relaxation rates; the profile of relative order parameters calculated from 15N R2 data is in good agreement with that calculated using the ensemble of NMR conformers (SI Appendix, Fig. S4). One exception is F4 NH, indicating some additional flexibility for this amide group. S2 and G1 remained more disordered than the peptide’s hydrophobic core in the ensemble of structures. Although we could not measure 15N relaxation rates for S2, as its NH was in fast exchange with water protons and thus not visible in the heteronuclear single quantum coherence (HSQC) spectra, evidence for the relative mobility of this residue came from the lack of intense NOEs and from the saturation transfer difference (STD) profile.
Fig. 3.
Receptor-bound ghrelin structure. (A) Overlay of the 10 best GHSR-bound ghrelin conformations. The C8 chains are shown in all-atom stick representation. The peptide sequence annotation summarizes structure-activity relationship data from the literature. (B) All-atom stick representation of the N-terminal region of the best ghrelin conformer. Only residues G1 to E8 are shown.
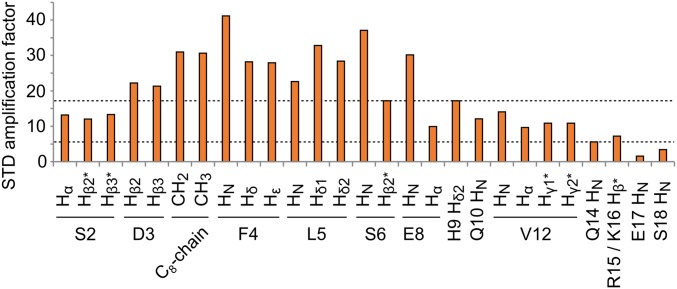
Finally, we obtained a complementary picture of the interaction of ghrelin with GHSR using STD epitope mapping experiments (24, 25), with a protonated receptor sample. STD amplification factors depend on both ligand-receptor contacts and ligand dynamics and thus provide an additional description of the ligand- binding pattern (25). We measured STD in the absence and presence of JMV5327 and analyzed the differences in STD amplification factors for signals spanning the whole peptide sequence (Fig. 4). Specifically, protons within the hydrophobic core—i.e., D3-Hβ, C8 chain methylenes and methyl, F4 aromatic ring, and L5 methyls—had the strongest STD signals, indicating spatial proximity with GHSR protons and confirming their involvement in a rigid domain. Consistent with the partial disorder of G1 and S2 in the ensemble of NMR structures, we observed medium-strength STD signals for G1-Hα, S2-Hα, and S2-Hβ protons. Overall, these results confirm and extend STD data obtained with GHSR in bicelles, where the residues S3 and F4 of ghrelin show strong STD signals (26), indicative of a tight interaction with the receptor.
Fig. 4.
Map of the area of interaction of ghrelin with its receptor. STD amplification factors of ghrelin bound to GHSR, with ASTD = (I0 − Isat)/I0 × [ghrelin]/[GHSR] (24). This number indicates the efficiency of magnetization transfer from the receptor to the peptide. It depends on the proximity of receptor and peptide protons, as well as on the internal dynamics of the complex. The difference in amplification factors in the absence and presence of JMV5327 is plotted here. Backbone amide protons and other protons signals were identified from 2D 15N-1H and 1D 1H STD experiments, respectively. An asterisk indicates signals for which stereospecific assignment was not performed. Dashed lines indicate the medium-intensity STD signal of S6 Hβ and the low-intensity STD signal of Q14 HN. The STD signal of G1-Hα is similar to the STD signals of S2-Hα and S2-Hβ; however, its amplification factor could not be measured precisely due to partial overlap with other signals in 1D proton spectra.
Ghrelin Binding Pathway.
To further delineate the possible ghrelin-binding pathway, we used a protocol previously validated with other peptide-binding GPCRs (27). This protocol, termed CG-REMD, combined a MARTINI coarse-grained (CG) force field with replica exchange molecular dynamics (REMD), starting from free ghrelin in the water phase and GHSR embedded in a lipid bilayer. Incorporating the full set of NMR restraints on the peptide at this initial stage prevented ghrelin from entering its binding pocket, possibly because it could not adopt the intermediate conformations required for navigating its binding pathway. This implies that the conformational dynamics of ghrelin are essential for GHSR binding. Thus, we considered only the restraints that maintained the octanoyl group along the peptide main chain. These restraints prevented insertion of the acyl chain into the membrane that might limit ghrelin–GHSR interaction events (SI Appendix, Fig. S5). At the end of the simulations, the ghrelin conformations in contact with the receptor were clustered according to their structural similarities (SI Appendix). Among the 10 most populated clusters, 8 were systematically found in each of the 3 independent CG-REMD simulations that we carried out, indicating a good convergence of sampling (SI Appendix, Fig. S6). Thus, we considered only these 8 clusters (SI Appendix, Table S2 and Fig. S7). Ghrelin was in close contact with the orthosteric pocket of the receptor in all but 1 of these 8 clusters (cluster 7), in which the hydrophobic core was embedded into the membrane. Among the remaining clusters, in only 1 (cluster 6) did ghrelin have its N terminus oriented toward the bottom of the receptor-binding pocket (SI Appendix, Fig. S7 and Movie S1), as expected based on previous pharmacologic studies (11), with a conformation closely resembling that of the NMR-derived ensemble of structures (Fig. 5A, purple curve). The conformation derived from this cluster was the only one consistent with most of the NOE restraints selected for CG modeling (SI Appendix, Figs. S8 and S9). The only exception to the NMR measurements involved F4, which was found to be shifted away from D3 and L5 in the modeling.
Fig. 5.
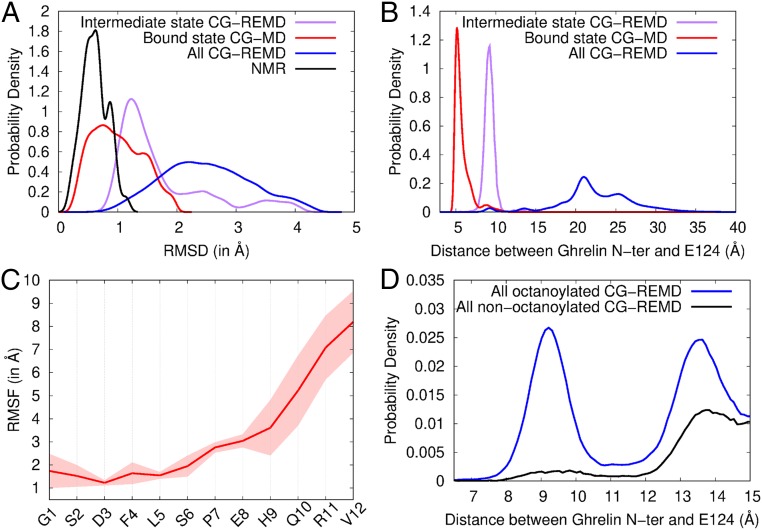
Conformation and flexibility of ghrelin bound to GHSR. (A) Root mean square deviation (RMSD) profiles of coarse-grained models from the ensemble of NMR structures (black), the bound state model (red), the intermediate state model (purple), and all models in contact with GHSR from CG-REMD simulations (blue). The RMSD is a measure of structural deviation from a reference structure, here the NMR structure ensemble. (B) Probability densities of the distance between the ghrelin N terminus and the side chain bead of E1243.33 in the different models. (C) Root mean square fluctuation (RMSF) profile of ghrelin backbone during restrained CG-MD simulations. The RMSF shows the variability in position of each residue along the trajectory (Movie S2). Conformations were aligned on the receptor pocket to account for the global flexibility of ghrelin inside its binding cavity. The SD inferred from the 2 CG-MD simulations is shown as a transparent area around the mean (red line). (D) Probability densities of the distance between octanoylated and nonoctanoylated ghrelin N termini and the side chain bead of E1243.33. Densities are shown from 6.5 Å to 15 Å for clarity. Complete density profiles are provided in SI Appendix, Fig. S11.
To further refine the ligand-receptor model, we performed an additional 30 µs of CG molecular dynamics (CG-MD) simulations starting from cluster 6 and incorporating the full set of NMR restraints. Over several nanoseconds, ghrelin explored a restricted conformational space encompassing the whole set of NMR conformers (Fig. 5A, red curve) and a deeper position in the receptor-binding pocket (Fig. 5B). This position could correspond to the final bound state of ghrelin (“bound state”), and thus cluster 6 could correspond to an intermediate site in the binding pathway (“intermediate state”).
Importantly, in the bound state, ghrelin maintained a certain degree of internal flexibility (Fig. 5C and Movie S2), in line with the dynamic features identified by NMR. To better describe this flexibility, we analyzed all possible interaction pairs between ghrelin and GHSR in this state (Fig. 6 and SI Appendix, Table S3). Despite its deep location in the receptor binding pocket, the G1 residue of ghrelin explored several interaction partners, in particular E1243.33 (with the superscript indicating Ballesteros–Weinstein numbering) (28) (Figs. 5B and 6), which has been identified as a major interaction spot for the terminal amine of ghrelin (29). Similarly, S2 showed different putative partners, in agreement with the relative degree of flexibility shown by our NMR data. The most stable peptide–receptor interactions involved the hydrophobic core, consistent with its rigidification in the GHSR-bound state. Finally, weaker interactions were found for S6 and P7, and no stable interactions were found for any of the other residues (E8, H9, Q10, R11, and V12), in agreement with our NMR ensemble of structures and the profile of order parameters.
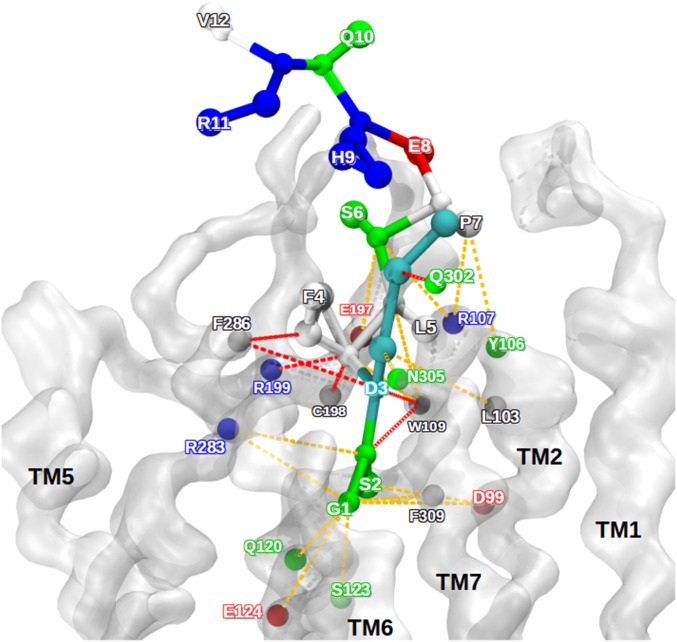
Fig. 6.
Interaction pattern between ghrelin and GHSR in the bound state model. Ghrelin is represented as balls and sticks. The receptor backbone is shown as a transparent white surface. The colors of the amino acids correspond to their physical and chemical properties: cyan, octanoylated aspartate; white, hydrophobic; green, polar; red, negatively charged; blue, positively charged. The dashed colored lines represent the contacts between the receptor and the ghrelin along the 2 trajectories. Red contacts represent strong persistent contacts, and orange contacts indicate transient and weaker contacts. This qualitative determination of contact used distances from the center of CG beads for all snapshots of the CG-MD trajectories.
Role of the Octanoyl Chain in Ghrelin Structure and Binding.
Removing the C8 chain induces a dramatic deterioration in ghrelin’s potency and affinity for GHSR (9, 11, 12). In agreement with this data, we found here that the nonacylated form of ghrelin 1–18 bound weakly to the purified receptor in nanodiscs (Fig. 1A). Absence of the octanoyl chain also impeded GHSR activation, based on the absence of fluorescence changes of bimane attached to the isolated receptor (Fig. 1E), the absence of any GHSR-catalyzed GTPγS binding to Gαq (Fig. 1B), and the very limited IP1 production in GHSR-transfected HEK cells (Fig. 1F). Overall, these data indicate that nonacylated ghrelin 1–18 is likely unable to stabilize an active GHSR conformation, although it can bind the receptor with a very low affinity (Ki = 34.4 µM, vs. 115.3 nM for the acylated version) (Fig. 1A).
To examine the possible effects of the acyl chain on the structure of ghrelin that could be responsible for these pharmacologic features, we duplicated the 15N R2 measurements with ghrelin 1–18 devoid of the C8 chain. We found that 15N R2 values in the presence of GHSR (0.0015 GHSR:ghrelin molar ratio, i.e., the ratio used with acylated ghrelin) were almost identical to those of the peptide free in solution. Increasing the GHSR:ghrelin molar ratio to 0.01 yielded a small increase in R2 of ∼1 Hz, compared with the 40-Hz increase observed with acylated ghrelin at a 0.0015 molar ratio (Fig. 2D). The difference in 15N R2 values between samples in the absence and presence of GHSR exhibited a flat profile from S3 to V12. These data indicate that nonacylated ghrelin residues 3 to 12 remain largely disordered under our experimental conditions even though the peptide should be mostly bound to the receptor (>95%) at the concentration used (1.37 mM for the sample with a 0.0015 GHSR:ghrelin molar ratio, i.e., ∼40-fold greater than the measured Ki value). We also recorded 2D 1H-1H NOESY spectra of nonacylated ghrelin with mixing times of 100 and 300 ms in the presence of the receptor (0.0015 and 0.01 GHSR:ghrelin molar ratio). Consistent with the 15N R2 measurements, we did not observe any of the trNOEs important for the structure of acylated ghrelin, particularly the backbone “medium-range” (S3Hα-L5HN and S6Hα-E8HN) and hydrophobic core (S3Hα-F4Hδ, S3Hα-F4Hε, S3Hβ-F4Hδ, F4HN-L5Hδ1/2, F4Hα-L5Hδ1/2, F4Hδ-L5Hδ1/2, F4Hε-L5Hδ1/2, F4Hε-L5HN, and F4Hε-L5Hα) proton pairs (SI Appendix, Fig. S10). Overall, these results highlight the essential role of the C8 chain in the formation of the hydrophobic core of ghrelin and in the associated rigidity of the central region of the peptide after receptor binding.
To further assess the possible origin of these effects, we analyzed the binding of nonacylated ghrelin to GHSR using the CG modeling protocol described above. The conformations thus obtained were more uniformly distributed than those obtained for the acylated peptide (SI Appendix, Table S2). Five of the 8 clusters described for the octanoylated peptide were also observed with the nonacylated peptide, but the latter was close to the receptor-binding pocket in very few conformations (Fig. 5D and SI Appendix, Fig. S11). In particular, cluster 6 (“intermediate state”) was almost unpopulated (SI Appendix, Table S2). Overall, our simulations reveal that the nonacylated peptide can explore positions at the receptor surface common with acylated ghrelin, but the octanoyl chain strongly facilitates access to deeper binding pockets.
Discussion
By integrating solution-state NMR spectroscopy data with advanced molecular modeling, we provide here a detailed description of the conformational features of ghrelin bound to GHSR. The molecular picture obtained illustrates the complex conformational space that GPCR peptide ligands explore when navigating from their free state to their receptor-bound state.
Our data indicate a binding mode in which only the N-terminal region of ghrelin becomes rigid through a direct interaction with GHSR, with the formation of a well-defined hydrophobic core composed of the octanoyl moiety, F4 and L5, while the C-terminal portion of the peptide remains highly disordered. Interestingly, we observed the specific ordering of the N-terminal region in CG modeling, despite starting from a ghrelin peptide in solution with a minimal set of NMR-derived restraints. Associated with the absence of any well-defined structure in the absence of GHSR (30), this strongly indicates that rigidification of the N-terminal region of ghrelin and formation of the hydrophobic core are intimately associated with the binding process. This mode of interaction is fully consistent with previous structure-activity relationship studies. The specific rigidification of the N-terminal region of ghrelin on binding to GHSR, as well as the receptor-ligand contacts inferred from STD, correlate directly with the observation that the first 5 residues of ghrelin are sufficient to bind and activate GHSR (9–12). In the same way, the mobility in the C-terminal region of ghrelin is consistent with studies that found no change in binding affinity on deletion of residues 10 to 14 of ghrelin (9). Finally, the partial disorder at S2 and lack of persistent interactions between this residue and the receptor indicates that residues G1-S2 explore different positions relative to the hydrophobic core of ghrelin, which is consistent with the proposed role of this 2-residue segment as a spacer between the 2 important functional motifs of ghrelin, the N-terminal amine and the hydrophobic core (9, 11). Ghrelin rigidity after binding to its receptor was also proposed in the ghrelin-GHSR model inferred from Rosetta modeling based on solid-state NMR chemical shifts (26). However, this model proposed a helical conformation for residues 9 to 18 of ghrelin, while our results indicate that this region is disordered. Helical propensity in this region was also proposed in a model of membrane-associated ghrelin obtained through a similar approach (30). Whether this difference arises from the distinct lipid models used (bicelles vs. nanodiscs) or from the experimental conditions (e.g., −30 °C for the solid-state NMR experiments vs. buffered solution at 7 °C) is an open question.
Our combined NMR and CG modeling data with nonacylated ghrelin highlight the paramount role of the octanoyl chain in ghrelin structure and binding to GHSR. Specifically, the C8 chain appears to be necessary for folding of the ghrelin hydrophobic motif. Moreover, its presence is necessary for ghrelin to access its final orthosteric binding site in GHSR, as well as for negotiating certain intermediate positions within the transmembrane bundle. This strongly suggests that the C8 chain acts as a dynamic, apolar structural hub that stabilizes the peptide hormone in a specific receptor-bound conformation. Based on our data, we propose a multistep model for ghrelin binding: initial interactions at the receptor surface occurs with and without the octanoyl chain, while the latter is required for engaging deeper binding sites. The role of the acyl chain in structuring a hydrophobic core rather than in mediating a highly specific receptor-ligand interaction agrees with the observation that unspecific C8 chain bulky hydrophobic substitutions preserve ghrelin binding and activity (9, 11). The preference for an 8-carbon fatty acid most likely arises from the acyl chain selectivity of the modifying enzyme (31). In contrast, polar or charged modifications within the C8 chain are strongly detrimental to both affinity and potency (9), likely because they disturb the ghrelin hydrophobic core. C8 chain attachment to S6 also significantly affects the affinity and potency of the ghrelin peptide (9). Our structure, in which the C8 chain interacts with the side chains of F4 and L5 and points toward the peptide C terminus, indicates that displacing the C8 chain toward the C-terminal end should prevent its interaction with these side chains, which in turn should inhibit formation of the hydrophobic core. Overall, this represents an original mechanism to explain the crucial role of this unique modification of the ghrelin peptide on the binding process and energetics.
Our data not only provide a detailed description of the structural features of ghrelin bound to GHSR, but also illustrate an interesting mechanism in which ligand conformational dynamics could represent a central component in the binding of endogenous peptide ligands to their GPCR. Our data also directly demonstrate some degree of conformational and positional local dynamics of the peptide even after it has reached its ligand-binding pocket. It is then tempting to speculate that the degree of ligand dynamics in the bound state is intricately linked to the conformational flexibility of the receptors themselves, which explore a complex conformational landscape with multiple states (32–35). This would extend the concept of conformational multiplicity of GPCRs to ligand-receptor concerted dynamics, and even to a global picture of concerted dynamics within the full signaling complex (36), if one considers the proposed conformational dynamics of G proteins associated with their receptor (37–39). This dynamic picture of the interaction between GPCRs and conformationally labile ligands and its relationship to signaling output requires relevant experimental strategies, such as that used here, that can uncover these dynamics.
Materials and Methods
More details of the experimental methods are provided in SI Appendix.
Receptor Production, Ligand Synthesis, and Labeling.
Monomeric GHSR was produced as described previously (7). All peptides were produced by solid-phase peptide synthesis.
NMR Experiments.
NMR data were acquired with the peptides and GHSR in buffer containing 20 mM perdeuterated MES pH 6.5, 100 mM KCl, 100 µM DSS, and 10% vol/vol 2H2O at 280 K on a Bruker Avance III HD 700 MHz spectrometer (1H Larmor frequency). Experiments on GHSR-bound octanoylated ghrelin were carried out at a 0.0015 GHSR:ghrelin molar ratio. Each experiment was performed with 2 independent receptor samples, apo- and JMV5327-loaded GHSR, to remove nonspecific binding contributions.
NMR Data Analysis.
15N transverse relaxation rates (R2) were extracted by monoexponential decay fitting from R2 CPMG experiments. For each labeled ghrelin amide group, the difference in R2 (apo minus JMV5327-loaded GHSR) is directly proportional to the receptor-bound order parameter S2 (14). Integrals of 2D 1H-15N STD-HSQC signals and isolated 1D 1H STD signals were extracted, and amplification factors, ASTD = (I0 – Isat)/I0 × [ghrelin]/[GHSR] (24), were calculated. Signals from 2D 1H-1H NOESY and 3D 1H-1H-15N NOESY-HMQC spectra were assigned and integrated for each mixing time. The build-up curves were obtained by biexponential fitting (SI Appendix, Fig. S2) (14, 21). Structure calculations were performed by simulated annealing under distance restraints derived from build-up curves.
Coarse-Grained Modeling of the Ghrelin-GHSR Complex.
Coarse-grained modeling of the complex was carried out as described previously starting from a GHSR homology model based on the crystal structure of NTSR1 in complex with neurotensin (Protein Data Bank ID code 4GRV) (40) and a MARTINI representation of the ghrelin peptide obtained by converting the all-atom model, including the octanoyl chain. The parameters of this chain were assigned by analogy with a C8 lipid chain from the MARTINI force field, with 2 elastic bonds added to keep the octanoyl group in close contact with the peptide main chain (SI Appendix, Fig. S12).
Supplementary Material
Acknowledgments
We thank Centre Informatique National de l’Enseignement Supérieur (Project A0020707530) and Partnership for Advanced Computing in Europe (Project 2017174234) for calculation resources and A. Atkinson (Kings College London) for a critical reading of the manuscript. This work was realized with the support of the High Performance Computing Platform MESO@LR, financed by the Occitanie/Pyrénées-Méditerranée Region, Montpellier Mediterranean Metropole and the University of Montpellier. This work was supported by CNRS, Université Montpellier, Agence Nationale de la Recherche (ANR-17-CE11-0011). The Integrated Screening Platform of Toulouse was funded by CNRS, Université Paul Sabatier, Infrastructures en Biologie Santé et Agronomie European Structural Funds, and the Midi-Pyrénées Region. Financial support for P.R. was provided by Chemistry of Molecular and Interfacial Systems (CheMISyst) Labex, and support for B.D. was provided by Université de Montpellier.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 6H3E).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1905105116/-/DCSupplemental.
References
- 1.Lagerström M. C., Schiöth H. B., Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat. Rev. Drug Discov. 7, 339–357 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Hauser A. S., Attwood M. M., Rask-Andersen M., Schiöth H. B., Gloriam D. E., Trends in GPCR drug discovery: New agents, targets and indications. Nat. Rev. Drug Discov. 16, 829–842 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu F., Song G., de Graaf C., Stevens R. C., Structure and function of peptide-binding G protein-coupled receptors. J. Mol. Biol. 429, 2726–2745 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Koehl A., et al. , Structure of the µ-opioid receptor-Gi protein complex. Nature 558, 547–552 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Müller T. D., et al. , Ghrelin. Mol. Metab. 4, 437–460 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Howard A. D., et al. , A receptor in pituitary and hypothalamus that functions in growth hormone release. Science 273, 974–977 (1996). [DOI] [PubMed] [Google Scholar]
- 7.Damian M., et al. , High constitutive activity is an intrinsic feature of ghrelin receptor protein: A study with a functional monomeric GHS-R1a receptor reconstituted in lipid discs. J. Biol. Chem. 287, 3630–3641 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Damian M., et al. , Ghrelin receptor conformational dynamics regulate the transition from a preassembled to an active receptor:Gq complex. Proc. Natl. Acad. Sci. U.S.A. 112, 1601–1606 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bednarek M. A., et al. , Structure-function studies on the new growth hormone-releasing peptide, ghrelin: Minimal sequence of ghrelin necessary for activation of growth hormone secretagogue receptor 1a. J. Med. Chem. 43, 4370–4376 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Matsumoto M., et al. , Structural similarity of ghrelin derivatives to peptidyl growth hormone secretagogues. Biochem. Biophys. Res. Commun. 284, 655–659 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Matsumoto M., et al. , Structure-activity relationship of ghrelin: Pharmacological study of ghrelin peptides. Biochem. Biophys. Res. Commun. 287, 142–146 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Van Craenenbroeck M., Gregoire F., De Neef P., Robberecht P., Perret J., Ala-scan of ghrelin (1-14): Interaction with the recombinant human ghrelin receptor. Peptides 25, 959–965 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Leyris J. P., et al. , Homogeneous time-resolved fluorescence-based assay to screen for ligands targeting the growth hormone secretagogue receptor type 1a. Anal. Biochem. 408, 253–262 (2011). [DOI] [PubMed] [Google Scholar]
- 14.O’Connor C., et al. , NMR structure and dynamics of the agonist dynorphin peptide bound to the human kappa opioid receptor. Proc. Natl. Acad. Sci. U.S.A. 112, 11852–11857 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maingot M., et al. , New ligands of the ghrelin receptor based on the 1,2,4-triazole scaffold by introduction of a second chiral center. Bioorg. Med. Chem. Lett. 26, 2408–2412 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Casiraghi M., Damian M., Lescop E., Banères J. L., Catoire L. J., Illuminating the energy landscape of GPCRs: The key contribution of solution-state NMR associated with Escherichia coli as an expression host. Biochemistry 57, 2297–2307 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Milon A., Miyazawa T., Higashijima T., Transferred nuclear overhauser effect analyses of membrane-bound enkephalin analogues by 1H nuclear magnetic resonance: Correlation between activities and membrane-bound conformations. Biochemistry 29, 65–75 (1990). [DOI] [PubMed] [Google Scholar]
- 18.Catoire L. J., et al. , Structure of a GPCR ligand in its receptor-bound state: Leukotriene B4 adopts a highly constrained conformation when associated to human BLT2. J. Am. Chem. Soc. 132, 9049–9057 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Bersch B., Koehl P., Nakatani Y., Ourisson G., Milon A., 1H nuclear magnetic resonance determination of the membrane-bound conformation of senktide, a highly selective neurokinin B agonist. J. Biomol. NMR 3, 443–461 (1993). [DOI] [PubMed] [Google Scholar]
- 20.Clore G. M., Gronenborn A. M., Theory of the time-dependent transferred nuclear overhauser effect—Applications to structural analysis of ligand-protein complexes in solution. J. Magn. Reson. 53, 423–442 (1983). [Google Scholar]
- 21.Vögeli B., The nuclear Overhauser effect from a quantitative perspective. Prog. Nucl. Magn. Reson. Spectrosc. 78, 1–46 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Case D. A., et al. , AMBER 14 (University of California, San Francisco, 2014). [Google Scholar]
- 23.Ferré G., et al. , Receptor-bound ghrelin conformation. Protein Data Bank. http://www.rcsb.org/structure/6H3E. Deposited 18 July 2018.
- 24.Mayer M., Meyer B., Group epitope mapping by saturation transfer difference NMR to identify segments of a ligand in direct contact with a protein receptor. J. Am. Chem. Soc. 123, 6108–6117 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Yong K. J., et al. , Determinants of ligand subtype-selectivity at α1A-adrenoceptor revealed using saturation transfer difference (STD) NMR. ACS Chem. Biol. 13, 1090–1102 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Bender B. J., et al. , Structural model of ghrelin bound to its G protein-coupled receptor. Structure 27, 537–544.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delort B., et al. , Coarse-grained prediction of peptide binding to G-protein coupled receptors. J. Chem. Inf. Model. 57, 562–571 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Ballesteros J. A., Weinstein H., “Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors” in Methods in Neurosciences, Sealfon S. C., ed. (Academic Press, 1995), pp. 366–428. [Google Scholar]
- 29.Holst B., et al. , Overlapping binding site for the endogenous agonist, small-molecule agonists, and ago-allosteric modulators on the ghrelin receptor. Mol. Pharmacol. 75, 44–59 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Vortmeier G., et al. , Integrating solid-state NMR and computational modeling to investigate the structure and dynamics of membrane-associated ghrelin. PLoS One 10, e0122444 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Darling J. E., et al. , Structure-activity analysis of human ghrelin O-acyltransferase reveals chemical determinants of ghrelin selectivity and acyl group recognition. Biochemistry 54, 1100–1110 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Casiraghi M., et al. , Functional modulation of a G protein-coupled receptor conformational landscape in a lipid bilayer. J. Am. Chem. Soc. 138, 11170–11175 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Nygaard R., et al. , The dynamic process of β(2)-adrenergic receptor activation. Cell 152, 532–542 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manglik A., et al. , Structural insights into the dynamic process of β2-adrenergic receptor signaling. Cell 161, 1101–1111 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okude J., et al. , Identification of a conformational equilibrium that determines the efficacy and functional selectivity of the μ-opioid receptor. Angew. Chem. Int. Ed. Engl. 54, 15771–15776 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hilger D., Masureel M., Kobilka B. K., Structure and dynamics of GPCR signaling complexes. Nat. Struct. Mol. Biol. 25, 4–12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Furness S. G. B., et al. , Ligand-dependent modulation of G protein conformation alters drug efficacy. Cell 167, 739–749.e11 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Gregorio G. G., et al. , Single-molecule analysis of ligand efficacy in β2AR-G-protein activation. Nature 547, 68–73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Damian M., et al. , GHSR-D2R heteromerization modulates dopamine signaling through an effect on G protein conformation. Proc. Natl. Acad. Sci. U.S.A. 115, 4501–4506 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.White J. F., et al. , Structure of the agonist-bound neurotensin receptor. Nature 490, 508–513 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.