Significance
Ribosomally synthesized and posttranslationally modified peptides (RiPPs) are a large class of natural products with a range of bioactivities including antibiotic, anticancer, and antinociception. Several classes of RiPPs contain dehydro amino acids that are installed on peptide substrates by LanB enzymes through the glutamyl-tRNA–dependent dehydration of Ser and Thr residues. Despite extensive research efforts, little is known about how LanBs carry out these posttranslational modifications. Here, we have used a combination of synthetic chemistry, structural biology, and biochemistry to identify features of the LanB active sites. Our mechanistic insights will facilitate future synthetic biology efforts aimed at diversifying the products of LanB enzymes.
Keywords: dehydratase, X-ray crystallography, lantibiotic, nisin, mechanism
Abstract
The peptide natural product nisin has been used as a food preservative for 6 decades with minimal development of resistance. Nisin contains the unusual amino acids dehydroalanine and dehydrobutyrine, which are posttranslationally installed by class I lanthipeptide dehydratases (LanBs) on a linear peptide substrate through an unusual glutamyl-tRNA–dependent dehydration of Ser and Thr. To date, little is known about how LanBs catalyze the transfer of glutamate from charged tRNAGlu to the peptide substrate, or how they carry out the subsequent elimination of the peptide-glutamyl adducts to afford dehydro amino acids. Here, we describe the synthesis of inert analogs that mimic substrate glutamyl-tRNAGlu and the glutamylated peptide intermediate, and determine the crystal structures of 2 LanBs in complex with each of these compounds. Mutational studies were used to characterize the function of the glutamylation and glutamate elimination active-site residues identified through the structural analysis. These combined studies provide insights into the mechanisms of substrate recognition, glutamylation, and glutamate elimination by LanBs to effect a net dehydration reaction of Ser and Thr.
Ribosomally synthesized and posttranslationally modified peptides (RiPPs) are a major class of genetically encoded peptide natural products with a wide array of biological activities, including antimicrobial properties (1). RiPP natural products are biosynthesized from a genetically encoded precursor peptide that is composed of a C-terminal core peptide (CP), where the posttranslational modifications occur, and an N-terminal leader peptide (LP), which is often essential for recognition by the modification machinery and is ultimately excised in the mature final product (2). The class I lanthipeptide dehydratases (LanBs) are involved in the biosynthesis of several families of RiPPs, including lanthipeptides and thiopeptides (3).
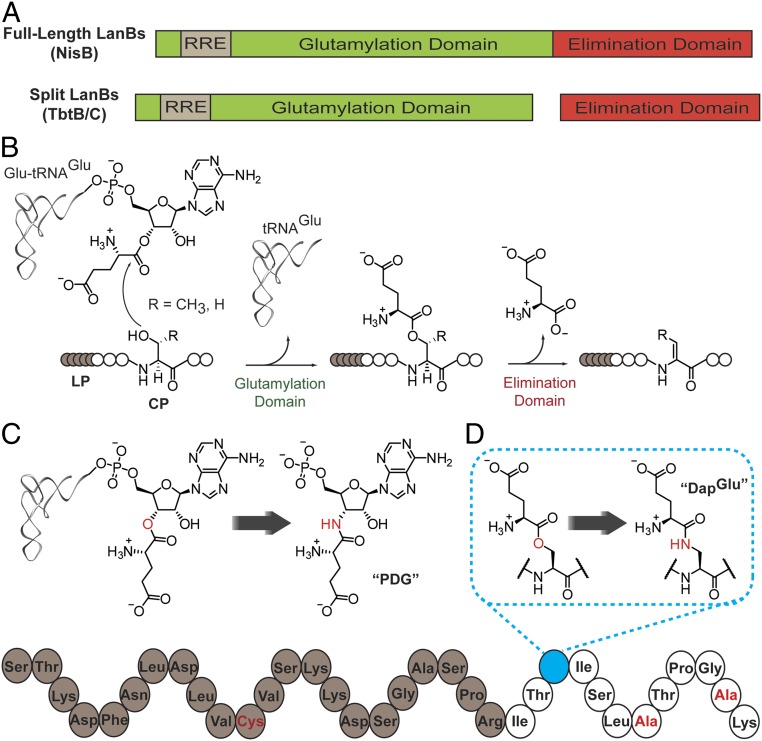
LanBs convert target serine and threonine residues in the core region of the cognate precursor peptide (LanA) to the dehydro amino acids dehydroalanine (Dha) and dehydrobutyrine (Dhb), respectively. These enzymes transfer glutamate from glutamyl-tRNAGlu to the side-chain hydroxyl groups of Ser/Thr residues in the LanA peptide substrate, and subsequently carry out glutamate elimination to yield Dha or Dhb (Fig. 1 A and B) (3, 4). LanBs have been categorized into subclasses that include full-length LanBs, split LanBs (in which glutamate transfer and elimination are catalyzed by 2 proteins), and small LanBs (which lack the elimination domain altogether) (3).
Fig. 1.
Domain organization of and reactions catalyzed by lanthipeptide dehydratases. (A) General domain structure of full-length and split LanBs. (B) Glutamylation and glutamate elimination reactions that result in Ser and Thr dehydration. The leader peptide is designated LP; the core peptide, CP. (C) Glutamyl-tRNAGlu and an amide-linked 3′-glutamyl-AMP mimic, PDG. (D) glutamylated-NisA and an amide-linked stable mimic, NisA-Ser3DapGlu. Residues highlighted in red in the bottom structure represent additional amino acid substitutions that differ from the wild-type NisA peptide and that are described further in the text.
Insights into how dehydro amino acids are installed on a peptide substrate were initially derived from biochemical and crystallographic studies of NisB and MibB, which are full-length LanBs involved in the biosynthesis of the lanthipeptides nisin and microbisporicin (NAI-107), respectively (3, 5). Both proteins contain a ∼90-kDa N-terminal glutamylation domain that catalyzes the transesterification from glutamyl-tRNAGlu to Ser/Thr, and a smaller (∼30-kDa) C-terminal domain responsible for glutamate elimination from the glutamyl-LanA intermediates. The LanB N-terminal region contains an embedded subdomain (3), termed the RiPP recognition element (RRE) (6), which is often essential for binding to the LP region of the LanA prior to modification on the CP. The structure of a NisB-NisA complex was originally obtained with sufficient electron density to model a portion of the LP of NisA bound to the RRE (3), while the MibB structure was determined without bound substrate peptide (5).
To date no structures are available for any LanB enzyme with substrate or any ligand bound at either the glutamylation or elimination active site. Indeed, structures of RiPP biosynthetic enzymes with substrates or intermediates bound are rare, and this lack of structural information poses a critical hurdle for understanding the molecular details of these catalysts. As a result, current understanding of active-site substrate recognition and catalysis during lanthipeptide and thiopeptide biosynthesis is restricted to inference based on mutational analysis of conserved amino acids of the enzyme and nucleotides of the glutamyl-tRNAGlu cosubstrate (3–5).
Unlike the full-length LanBs that catalyze both glutamylation and glutamate elimination in lanthipeptide biosynthesis, the orthologous enzymes that install dehydro amino acids in thiopeptides are split into 2 polypeptides (Fig. 1A). The larger of these enzymes (termed B) also utilizes glutamyl-tRNAGlu to activate the Ser/Thr side chains of substrate peptides. For instance, TbtB from Thermobispora bispora involved in thiomuracin biosynthesis transfers the glutamate from glutamyl-tRNAGlu to Ser residues in the TbtA peptide that was previously acted upon by the TbtEFG proteins to convert six Cys to the corresponding thiazoles (7). The C enzymes (e.g., TbtC) carry out subsequent glutamate elimination from the modified Ser or Thr to form Dha or Dhb, respectively.
In order to better understand the mechanisms of both glutamylation and glutamate elimination reactions, we designed and carried out the chemical synthesis of nonreactive substrate mimics for each reaction. Substitution of amide linkages in place of esters in the intermediates of catalysis was expected to prevent transesterification by the glutamylation domain and β-elimination by the elimination domain (Fig. 1 C and D). We also report the structures of TbtB bound to a small-molecule aminoacyl-tRNA mimic and of NisB bound to a nonreactive glutamyl-NisA mimic in its elimination domain. Together with biochemical characterization of structure-derived active-site variants, these studies reveal key molecular features involved in substrate recognition and catalysis.
Results
Strategy for Crystallization of Substrate Analogs with LanB Enzymes.
Despite multiple crystallization attempts using several different orthologs of full-length LanBs, we were unable to visualize any ligand bound to the glutamylation active sites in any of the resultant structures. We reasoned that the presence of the elimination active site might hamper productive binding of substrates/analogs in the glutamylation active site. Therefore, we focused efforts on the split LanB systems found in thiopeptide biosynthetic clusters, in which the B enzyme carries out only the glutamylation reaction. Crystallization efforts with TbtB from the thiomuracin biosynthetic cluster were successful, and this enzyme bound nucleotide and substrate analogs with suitable affinity to allow further analysis. We did not encounter difficulties in capturing a glutamylated peptide mimic in the NisB elimination active site, using a peptide in which the leader sequence was covalently tethered to the RRE (8) as described below.
Overall Structure of TbtB.
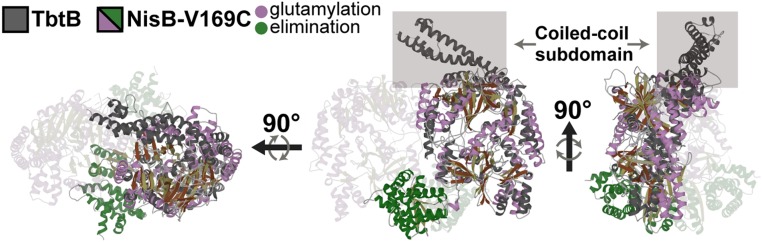
TbtB was crystallized in the absence of bound ligands, and its structure solved to 2.1-Å resolution using diffraction data from crystals soaked in thiomersal (PDB 6EC7). The overall structure of TbtB (Fig. 2) resembles the NisB and MibB glutamylation domains (NisB5–729, rmsd = 5.0 Å, over 2,889 atoms; MibB4–809, rmsd = 3.5 Å, over 2,586 atoms) with 1 notable exception. The N terminus of TbtB contains a distinguishing coiled-coil subdomain (residues 36–153; helices α2–α5), which is absent in the type I lanthipeptide dehydratases NisB and MibB (for a multiple sequence alignment, see SI Appendix, Fig. S1). This coiled-coil subdomain is integral to stand-alone LanB glutamylation enzymes and is involved in extensive interactions with helix α23 as well as surrounding portions of the core structure (total buried surface area ∼2,600 Å2; SI Appendix, Fig. S2). Genome neighborhood analysis using the Rapid Open Reading Frame Description & Evaluation Online (RODEO) bioinformatics portal (9) suggests this extended N-terminal subdomain is a feature common to split and small LanB-encoding biosynthetic clusters and is represented in equivalent enzymes from across diverse RiPP pathways (e.g., cyclothiazomycin, goadsporin, thiomuracin, pinensin, and 3-thiaglutamate; SI Appendix, Fig. S2). Although the overall structure of TbtB resembles the glutamylation domains of MibB and NisB, the electrostatic potential maps are notably different (SI Appendix, Fig. S3). The RRE domain in TbtB recapitulates the structural organization observed in NisB and MibB and encompasses strands β6–β7–β8 and helices α10–α12 (SI Appendix, Fig. S4). However, previous experiments demonstrated that TbtB (and TbtC) can carry out glutamylation (and glutamate elimination) independent of the leader peptide, suggesting that the RRE may be largely vestigial.
Fig. 2.
Apo TbtB superimposed on NisB-Val169Cys. β–strands are shown in brown for TbtB and in khaki for NisB-Val169Cys. One NisB-Val169Cys monomer is made 80% transparent for clarity. Helices of the glutamylation and elimination domains of NisB-Val169Cys are shown in purple and green, respectively. Two NisA-Ser3DapGlu molecules bound to the NisB-Val169Cys homodimer are not shown.
Cocrystal Structure of TbtB and 5′-Phosphoryl-Desmethylglutamycin.
TbtB cocrystallized with AMP showed clear electron density for the nucleotide in the putative glutamylation active site. Assuming that AMP occupies the position of the 3′-end of the tRNA, we prepared an aminoacyl-tRNA analog to better define key interactions and the mechanism of glutamate transfer. Inspired by prior work (10, 11) we designed 5ʹ-phosphoryl-desmethylglutamycin (PDG), a phosphorylated glutamate analog of puromycin (Fig. 1C), which was synthesized as shown in SI Appendix, Scheme S1.
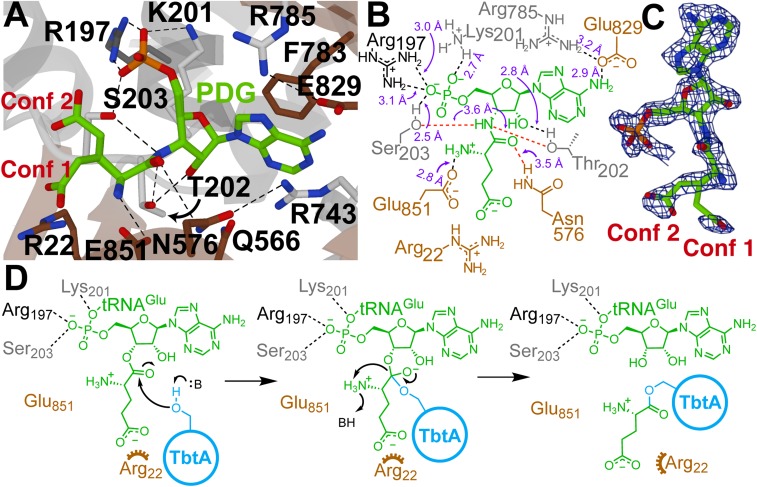
Diffraction analysis of ligand-free TbtB crystals soaked with PDG showed clear and near-continuous electron density for PDG in the glutamylation active site with the nucleotide oriented in a fashion similar to that of AMP. In the cocrystal structure, the adenine ring of PDG wedges between the phenyl side chain of Phe783 and the guanidinium of Arg743, forming a π-π-cation interaction (Fig. 3A). The exocyclic amine of the adenine ring hydrogen-bonds with the Glu829 carboxylate and backbone amide oxygen of Glu830. The 2′-hydroxyl of PDG and the Thr202 side chain engage in hydrogen bonding, and an electrostatic interaction is observed between the protonated α-amine of the glutamate moiety in PDG and the carboxylate of Glu851 in TbtB. Additional electrostatic interactions are present between the 5′-phosphate of the ligand and side chains of Arg197 and Lys201 (Fig. 3 A and B). The γ-carboxylate of the 3′-glutamate of PDG is disordered in the crystal structure, showing 2 orientations with 1 low-occupancy conformer having the carboxylate situated near Arg22 (Fig. 3C). Given the resolution of the current structure, it is not feasible to determine the occupancy of each conformer, but we conservatively assigned an occupancy of 10% to this conformer based on i) the electron density maps and ii) experimental data showing the importance of Arg22 (see below). Alternatively, it is plausible that Arg22 interacts with the γ-carboxylate of the glutamate following its transfer to the peptide substrate. Interestingly, Thr202 and Ser203 are close to the amide structure in PDG and suggest a possible ping-pong mechanism in which the glutamate is initially transferred from tRNA to the side chain of either of these residues, followed by tRNA dissociation and binding of the TbtA substrate that then accepts the glutamate from the acyl-enzyme intermediate (Fig. 3D). Ser203 is of particular interest given that mutation of the corresponding residue in NisB resulted in complete loss of activity (4).
Fig. 3.
Structural analysis of the active site of TbtB. (A) TbtB bound to PDG (2.15 Å; PDB 6EC8) with active-site residues targeted for mutagenesis represented as sticks (residues residing in an α-helix, β-strand, or random coil are shown in dark gray, brown, or light gray, respectively). (B) Two-dimensional representation of A with labeled interatomic distances (≤3.2 Å in black; ≥3.5 Å in red). The distances reflect crystallographic nonhydrogen interatomic distances, but the dashed lines are drawn to satisfy proposed hydrogen-bonding interactions. (C) Feature-enhanced map contoured at 2σ for the PDG ligand (30). PDG was modeled as 2 conformations [Conf 1 (10%) and Conf 2 (90%)] due to partial disorder of the Glu side-chain atoms. (D) Proposed mechanism of TbtB-catalyzed glutamylation of TbtA. An alternative ping-pong mechanism, described in the text, was ruled out based on site-directed mutagenesis studies.
Mutational Analysis of TbtB and Inhibition of LanB Activity by EF-Tu In Vitro.
To probe the relevance of these interactions to the enzyme-catalyzed glutamylation reaction, we created 14 structure-based alanine mutants of TbtB and assessed dehydration activity in vitro (SI Appendix, Figs. S4–S6). Reactions contained ATP and the T. bispora tRNAGlu/GluRS pair for in situ generation of glutamyl-tRNAGlu, as well as the elimination enzyme TbtC to allow for dehydration of a TbtA hexazole substrate peptide (7). End-point analysis by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) revealed a complete lack of dehydratase activity for Ala-variants of Arg22 and the 5′-phosphate–binding residues Arg197 and Lys201 (SI Appendix, Fig. S6), indicating that they are essential for TbtB catalysis. These findings are consistent with previous results for NisB, in which the corresponding mutations were also devoid of dehydratase activity (4) (SI Appendix, Tables S1 and S2). Mutation of Thr202, Ser203, Tyr564, and Glu851 to Ala resulted in partial dehydration of the substrate. The partial activities of the Thr202Ala and Ser203Ala mutants argue against a ping-pong mechanism. The partial activity of Glu851Ala is also of note given its strong conservation (SI Appendix, Table S1) and position directly next to the proposed site of transesterification (Fig. 3A). The high activities of the Gln566, Asn576, and Arg743 mutants are less surprising since these residues are not highly conserved between full-length and split/small LanBs (SI Appendix, Fig. S1). For completeness, we also generated 5 NisB glutamylation domain variants corresponding to mutated TbtB residues that were not previously examined for NisB (3, 4). They were tested for activity by in vivo coexpression with NisA in E. coli, providing results that were consistent with the outcomes of mutagenesis on TbtB, suggesting a similar glutamylation active site in NisB (SI Appendix, Fig. S7).
Concomitant with our structural and mutational studies of the glutamylation active site of LanBs, we also questioned how these enzymes acquire aminoacyl-tRNA from the intracellular pool. Given that aminoacylated tRNA is expected to be bound by elongation factor Tu (EF-Tu) within the cell (12–14), we examined the impact of EF-Tu on NisB activity in vitro. Addition of recombinant C-terminally His6-tagged EF-Tu from E. coli (15, 16) that had been preactivated with GTP resulted in a dose-dependent decrease in the efficiency of NisB-mediated dehydration of NisA (SI Appendix, Fig. S8). This observation suggests EF-Tu is capable, at least in vitro, of sequestering glutamyl-tRNAGlu from NisB, thereby limiting its activity. The implications of this effect for heterologous production of lanthipeptides and thiopeptides are under investigation.
Design and Synthesis of a Nonreactive Glutamyl-NisA Mimic.
We previously showed that full-length NisB could catalyze glutamate elimination on the exogenously provided glutamylated NisA peptide (3, 4). Thus, for structural analysis, we prepared a stable mimic of the glutamylated NisA peptide in which the glutamyl-ester was substituted with a less reactive amide functionality (Fig. 1D). The core peptide of the analog was truncated C-terminally to Lys12 to reduce conformational flexibility.
Solid-phase peptide synthesis (SPPS) with an orthogonal deprotection strategy was used for the synthesis of the desired peptide, herein referred to as NisA-Ser3DapGlu (SI Appendix, Scheme S2). Incorporation of an amide-linked glutamate was accomplished by first substituting an orthogonally protected 2,3-diaminopropionic acid (Dap) for the Ser at position +3 of the CP; +/− refers to amino acid position in the precursor peptide relative to the natural proteolysis site between the LP and the CP. Selective removal of the protecting group followed by coupling of Boc-Glu(OtBu)-OH afforded the desired amide-linked adduct (SI Appendix, Scheme S2). The location of this Ser3Dap substitution was chosen based on the N to C directionality of NisB-catalyzed dehydration, wherein the enzyme exhibits a preference for serine and threonine residues toward the N-terminal side of the CP region (17, 18). A Ser(−12)Cys substitution was made in the LP region of the substrate mimic to form a disulfide bond with a reciprocal cysteine introduction (Val169Cys) in the leader-binding RRE of NisB (8). Lastly, Cys residues in the core peptide region were substituted with Ala to prevent intramolecular disulfide formation.
Biochemical Evaluation of Glutamyl-NisA Mimics In Vitro.
Preliminary biochemical analyses of the glutamyl-NisA mimic and its synthetic intermediate, NisA-Ser3DapAlloc (SI Appendix, Fig. S9), were carried out using wild-type (WT) NisB activity assays. The peptide containing the amide-linked glutamate adduct, NisA-Ser3DapGlu, was dehydrated by NisB at the remaining Ser/Thr residues of the CP, but the efficiency of dehydration was reduced in comparison to WT NisA (SI Appendix, Fig. S9). In contrast, NisA-Ser3DapAlloc was dehydrated more efficiently, suggesting that the structure of the glutamate adduct is an important factor in reducing the dehydration rate, presumably because the amide-linked glutamate adduct occupies the elimination active site, preventing efficient catalysis. The synthetic peptides competed with WT NisA for binding to NisB in fluorescence polarization measurements, with the competition using NisA-Ser3DapGlu yielding IC50 values that were four- to fivefold lower than the other peptides examined, indicating that the amide-linked glutamate adduct provides additional binding affinity and cooperates with the LP for NisB engagement (SI Appendix, Fig. S10). Mutation of the proposed catalytic base (see below) His961 to Ala revealed a similar binding effect. However, mutation of Arg786 to Ala abolished the observed additive-binding effect (SI Appendix, Fig. S10).
Structural Analysis of NisB Containing a Stable Glutamyl-NisA Mimic.
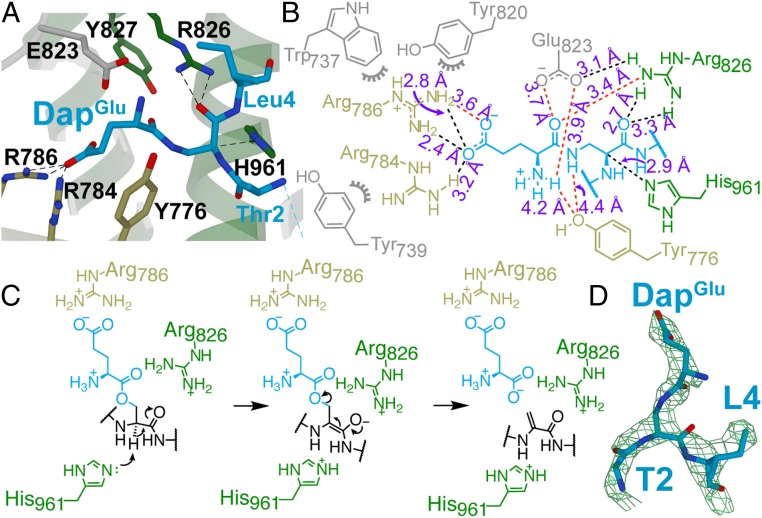
To better understand the mechanism of elimination, we next determined the structure of the covalent complex of NisB-Val169Cys and NisA-Ser3DapGlu-Ser(−12)Cys to 2.8 Å. The overall structure of NisB in this complex is highly similar to that of the previously determined structure of the noncovalent NisA-NisB complex (3) with an rmsd of 0.4 Å over 9,196 atoms. Continuous electron density is only observed for residues −21 to −9 of the leader peptide and residues 2–4 of the core peptide (Fig. 4 and SI Appendix, Fig. S11A). The LP interfaces as an antiparallel β-sheet with β7 of the NisB RRE, and clear electron density is observed for the engineered disulfide linkage between Cys169 of NisB and Cys(−12) of the peptide (SI Appendix, Fig. S11A). Residues 2–4 of the CP are visible in the elimination domain active site, with the side chain of the glutamate adduct inserted into a cavity containing residues previously shown to be important for catalysis (Fig. 4 A, B, and D) (4). The imidazole τ-nitrogen of His961 is situated 2.9 Å from the α-carbon of the glutamylated Dap residue and is poised for proton abstraction, suggesting that it plays the role of catalytic base in the glutamate elimination reaction (Fig. 4C). This α-proton is acidified by interaction of the carbonyl group of the Dap residue with the side chain of Arg826. In addition, the carboxylic acid of Glu823 is positioned to make a hydrogen bond to the carbonyl oxygen in the amide linkage between Dap and Glu. With the native substrate, this interaction may stabilize the negative charge that builds up on this oxygen during the elimination reaction. The Glu residue is further oriented by the phenol oxygen of Tyr776 that interacts with the amine group. Specificity for the Glu residue is provided by Arg786, which forms an electrostatic interaction with the carboxylate side chain of the glutamate attached to the Dap residue. The side chain of another Arg (Arg784) is positioned 3.2 Å from the side chain of the Glu attached to Dap.
Fig. 4.
Structural analysis of the elimination active site of NisB. (A) NisB-Val169Cys bound to NisA-Ser3DapGlu (2.79 Å; PDB 6M7Y) showing elimination domain active-site residues as sticks (residues residing in an α-helix, β-strand, or random coil are shown in green, khaki, or light gray, respectively). (B) Two-dimensional representation of A with labeled interatomic distances (≤3.3 Å in black; ≥3.4 Å in red). The distances reflect crystallographic nonhydrogen interatomic distances, but the dashed lines are drawn to satisfy proposed hydrogen-bonding interactions where appropriate. (C) Proposed mechanism of NisB-catalyzed elimination of glutamylated NisA. (D) Feature-enhanced map contoured at 1.8σ for residues 2–4 of the CP (blue sticks) containing the DapGlu adduct.
Mutational Analysis of the NisB Elimination Domain.
Previous mutations of highly conserved residues showed that the NisB mutants Arg786Ala, Arg826Ala, and His961Ala resulted in buildup of glutamylated NisA (4), suggesting that these residues are important for glutamate elimination. Mutation of Glu823 to Ala led to less efficient dehydration but not abolishment of activity (4). Guided by the crystallographic evidence presented here, additional mutations were generated in the elimination domain of NisB. These mutations were primarily directed toward the conserved aromatic residues surrounding the glutamate adduct on NisA. Mutant enzymes were coexpressed with NisA in E. coli as described previously (4), and activity was ascertained by MALDI-TOF MS analysis of the purified NisA–derived products. Alanine substitutions at Glu344, Asp973, Phe772, Tyr776, and Phe840 of NisB did not result in notable differences in activity compared to WT (SI Appendix, Fig. S7). Alanine substitution at Trp737, Tyr739, and Tyr820, however, led to diminished dehydration activity and buildup of glutamylated NisA. Given that Trp737, Tyr739, and Tyr820 are directly adjacent to Arg786 (SI Appendix, Fig. S11B), it seems likely that they contribute to glutamate-adduct recognition by orienting the side chain of Arg786 and/or Arg784 toward the glutamylated intermediate. Lastly, mutation of Tyr827 resulted in a minor decrease in dehydration activity. Interestingly, however, this decrease in activity was not accompanied by a buildup of glutamylated peptide.
Discussion
Combining current structural data with sequence homology and previous findings, a hypothesis can be formulated as to how the glutamylation domain of LanB enzymes engages its glutamyl-tRNAGlu cosubstrate. Residues Arg83 and Arg87 were previously shown to be critical for NisB activity; however, the reasons were unclear (4). Structure-guided mutagenesis of the corresponding Arg197 and Lys201 residues in TbtB reveals that these conserved basic residues are required for engaging the phosphate preceding the terminal 3′-adenosine of tRNA (i.e., the phosphodiester group linking cytidine and adenosine in the typical CCA sequence at the 3′-end of tRNAs). In addition, Arg22 (Arg14 in NisB) is essential for catalysis, presumably because of the interaction with the γ-carboxylate of glutamyl-tRNAGlu based on its proximity in the structure, although our data show only low occupancy for the PDG γ-carboxylate in this orientation. This observation may be a result of the incorporation of an amide bond in the PDG mimic, which could distort the normal orientation of the aminoacyl bond. Alternatively, it is possible that a well-defined orientation of Glu is only attained upon binding of the peptide cosubstrate.
The possibility of a transesterification step producing an acyl enzyme intermediate with glutamate covalently bound to Thr202 or Ser203 is unlikely since mutations at these positions did not inactivate the enzyme, as would be expected for such a mechanism. For similar reasons, Tyr80 in NisB, which was reported to be indispensable for catalysis (19), does not appear to be a possible site for covalent attachment of the glutamate group given that it is distant (∼15 Å) from the aminoacyl bond in the TbtB structure with PDG. The strong decrease in dehydration activity observed with the Tyr80Ala mutant relative to WT does suggest, however, that Tyr80 may be involved in either engagement of additional features of the tRNA or structural maintenance of the active site. Interestingly, mutation of Glu851 in TbtB (Asp700 in NisB) to Ala did not dramatically impair glutamylation activity despite universal conservation of an acidic residue at this position and its location adjacent to the aminoacyl moiety of PDG. Similar effects were observed when Phe783 (Tyr643 in NisB) was mutated (SI Appendix, Tables S1 and S2).
We had hoped that covalently linking the leader peptide to NisB and providing a nonreactive mimic of the glutamylated NisA substrate would result in electron density for the entire peptide such that we could trace its conformation from the leader peptide–binding site to the elimination active site, but this was not the observed outcome. As noted previously (20, 21), it appears that the substrate peptide sequence between the leader peptide binding site and the residues in the CP that are modified does not make specific interactions with the enzyme and merely functions as a spacer to allow the CP to reach the active sites of the enzyme. Similarly to the previous noncovalent cocrystal structure of dehydrated NisA and NisB (3), and the previous evaluation of a covalent link between the LP of NisA and NisB (8), the stoichiometry observed in the cocrystal structure in this study is 2 glutamylated NisA mimics per NisB dimer. This contrasts with small-angle X-ray scattering measurements suggesting that, in the presence of the cyclase NisC, a NisB2NisCNisA complex is formed with only 1 substrate per NisB dimer (22). It is possible that the presence of NisC leads to half-site activity as is observed in other enzymatic systems (23, 24).
Our structural data strongly suggest that the conserved His961 serves as the catalytic base needed for deprotonation at the α-position of the glutamylated residue to initiate Glu elimination. Arg826 most likely aids in this process through enolate stabilization (Fig. 4C). In addition, Arg784, Arg786, and Glu823 all appear to be involved in recognition or orientation of the glutamate adduct in the active site based on current structural data and previous mutagenesis studies. Further mutational analysis conducted in this work reveals that some aromatic residues (Trp737, Tyr739, and Tyr820) are also important for elimination activity, presumably by providing a scaffold for the glutamyl recognition pocket and/or orienting Arg784 and Arg786 toward the γ-carboxylate of the adduct.
Our finding that EF-Tu negatively impacts NisB activity in vitro raises interesting questions regarding how LanBs are able to access glutamyl-tRNAGlu. While it has been reported that EF-Tu concentrations diminish within the cell as growth rate slows (12), which is when most RiPPs are produced, this decrease is generally accompanied by a decrease in tRNA concentration (25) resulting in a net ratio of EF-Tu to total tRNA that is fairly consistent. Furthermore, despite being one of the most abundant tRNAs in Escherichia coli, tRNAGlu levels have been shown to decrease in lockstep with other tRNAs as growth rate slows (26). A better understanding of the interplay between the use of glutamyl-tRNA by the translational machinery and the dehydratases of class I lanthipeptides/thiopeptides may help improve heterologous expression systems and thereby genome-mining efforts.
In summary, we report the application of nonreactive substrate mimics to probe the active-site structure and mechanism of class I lanthipeptide and thiopeptide dehydratases. Based on structural data, we speculate that these domains may be amenable to bioengineering approaches, such as “bump-and-hole” or orthogonal expression strategies in heterologous hosts that would not interfere with existing pathways that rely upon aminoacyl-tRNA (27, 28). If possible, such systems could facilitate the production of RiPPs with academic or pharmaceutical value.
Materials and Methods
Plasmids containing WT tbtB or site-directed mutants were constructed using the pET hexahis (His6) small ubiquitin-like modifier (SUMO) tobacco etch virus (TEV) protease ligation-independent cloning (LIC) vector (2S-T), a gift from Scott Gradia (Addgene plasmid #29711). Alanine mutants in the glutamylation and elimination domains of NisB were generated using site-directed, ligation-independent mutagenesis (SLIM) on a pRSF-Duet1 template containing His6-tagged NisA in multiple cloning site (MCS) I and NisB in MCS II (29). Detailed procedures for all mutagenesis studies are provided in the SI Appendix. Escherichia coli JM109/pKECA-Tu (15, 16) producing C-terminally His6-tagged EF-Tu was a generous gift from the laboratory of Dr. Michael Ibba (Ohio State University, Columbus, OH). All protein purification methods and synthetic details are provided in the SI Appendix.
Supplementary Material
Acknowledgments
We thank I. Rahman and S. Biswas for constructive discussions and assistance with instrumentation. We thank K. Brister and colleagues at Life Sciences Collaborative Access Team (ID-21; Argonne National Laboratory) for facilitating synchrotron data collection. This work was supported by the National Institutes of Health (GM058822 to W.A.v.d.D, GM079038 to S.K.N., and F32 GM117765 to I.R.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors reported in this paper have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 6EC8, 6M7Y, and 6EC7).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1905240116/-/DCSupplemental.
References
- 1.Arnison P. G., et al. , Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30, 108–160 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oman T. J., van der Donk W. A., Follow the leader: The use of leader peptides to guide natural product biosynthesis. Nat. Chem. Biol. 6, 9–18 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ortega M. A., et al. , Structure and mechanism of the tRNA-dependent lantibiotic dehydratase NisB. Nature 517, 509–512 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garg N., Salazar-Ocampo L. M., van der Donk W. A., In vitro activity of the nisin dehydratase NisB. Proc. Natl. Acad. Sci. U.S.A. 110, 7258–7263 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ortega M. A., et al. , Structure and tRNA specificity of MibB, a lantibiotic dehydratase from Actinobacteria involved in NAI-107 biosynthesis. Cell Chem. Biol. 23, 370–380 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burkhart B. J., Hudson G. A., Dunbar K. L., Mitchell D. A., A prevalent peptide-binding domain guides ribosomal natural product biosynthesis. Nat. Chem. Biol. 11, 564–570 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hudson G. A., Zhang Z., Tietz J. I., Mitchell D. A., van der Donk W. A., In vitro biosynthesis of the core scaffold of the thiopeptide thiomuracin. J. Am. Chem. Soc. 137, 16012–16015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Repka L. M., Hetrick K. J., Chee S. H., van der Donk W. A., Characterization of leader peptide binding during catalysis by the nisin dehydratase NisB. J. Am. Chem. Soc. 140, 4200–4203 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tietz J. I., et al. , A new genome-mining tool redefines the lasso peptide biosynthetic landscape. Nat. Chem. Biol. 13, 470–478 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang B., Zhang L., Sun L., Cui Z., Synthesis of pCpCpA-3′-NH-phenylalanine as a ribosomal substrate. Org. Lett. 4, 3615–3618 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Balg C., Huot J. L., Lapointe J., Chênevert R., Inhibition of Helicobacter pylori aminoacyl-tRNA amidotransferase by puromycin analogues. J. Am. Chem. Soc. 130, 3264–3265 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Furano A. V., Content of elongation factor Tu in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 72, 4780–4784 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Louie A., Ribeiro N. S., Reid B. R., Jurnak F., Relative affinities of all Escherichia coli aminoacyl-tRNAs for elongation factor Tu-GTP. J. Biol. Chem. 259, 5010–5016 (1984). [PubMed] [Google Scholar]
- 14.Schrader J. M., Uhlenbeck O. C., Is the sequence-specific binding of aminoacyl-tRNAs by EF-Tu universal among bacteria? Nucleic Acids Res. 39, 9746–9758 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vorstenbosch E. L., Potapov A. P., de Graaf J. M., Kraal B., The effect of mutations in EF-Tu on its affinity for tRNA as measured by two novel and independent methods of general applicability. J. Biochem. Biophys. Methods 42, 1–14 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Ling J., Yadavalli S. S., Ibba M., Phenylalanyl-tRNA synthetase editing defects result in efficient mistranslation of phenylalanine codons as tyrosine. RNA 13, 1881–1886 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lubelski J., Khusainov R., Kuipers O. P., Directionality and coordination of dehydration and ring formation during biosynthesis of the lantibiotic nisin. J. Biol. Chem. 284, 25962–25972 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Q., et al. , Structural investigation of ribosomally synthesized natural products by hypothetical structure enumeration and evaluation using tandem MS. Proc. Natl. Acad. Sci. U.S.A. 111, 12031–12036 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khusainov R., van Heel A. J., Lubelski J., Moll G. N., Kuipers O. P., Identification of essential amino acid residues in the nisin dehydratase NisB. Front. Microbiol. 6, 102 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Repka L. M., Chekan J. R., Nair S. K., van der Donk W. A., Mechanistic understanding of lanthipeptide biosynthetic enzymes. Chem. Rev. 117, 5457–5520 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plat A., Kluskens L. D., Kuipers A., Rink R., Moll G. N., Requirements of the engineered leader peptide of nisin for inducing modification, export, and cleavage. Appl. Environ. Microbiol. 77, 604–611 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reiners J., Abts A., Clemens R., Smits S. H., Schmitt L., Stoichiometry and structure of a lantibiotic maturation complex. Sci. Rep. 7, 42163 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seydoux F., Malhotra O. P., Bernhard S. A., Half-site reactivity. CRC Crit. Rev. Biochem. 2, 227–257 (1974). [DOI] [PubMed] [Google Scholar]
- 24.Wielgus-Kutrowska B., Grycuk T., Bzowska A., Part-of-the-sites binding and reactivity in the homooligomeric enzymes–facts and artifacts. Arch. Biochem. Biophys. 642, 31–45 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Rosset R., Julien J., Monier R., Ribonucleic acid composition of bacteria as a function of growth rate. J. Mol. Biol. 18, 308–320 (1966). [DOI] [PubMed] [Google Scholar]
- 26.Dong H., Nilsson L., Kurland C. G., Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol. 260, 649–663 (1996). [DOI] [PubMed] [Google Scholar]
- 27.Bishop A., et al. , Unnatural ligands for engineered proteins: New tools for chemical genetics. Annu. Rev. Biophys. Biomol. Struct. 29, 577–606 (2000). [DOI] [PubMed] [Google Scholar]
- 28.Liu C. C., Jewett M. C., Chin J. W., Voigt C. A., Toward an orthogonal central dogma. Nat. Chem. Biol. 14, 103–106 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chiu J., March P. E., Lee R., Tillett D., Site-directed, ligase-independent mutagenesis (SLIM): A single-tube methodology approaching 100% efficiency in 4 h. Nucleic Acids Res. 32, e174 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Afonine P. V., et al. , FEM: Feature-enhanced map. Acta Crystallogr. D Biol. Crystallogr. 71, 646–666 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.