Significance
The nucleolus is an organelle in the nucleus that plays a key role in the transcription and processing of ribosomal RNA (rRNA). While many studies have highlighted the viscoelastic material state of the nucleolus, how the material properties of the nucleolus affect its function in rRNA biogenesis is not understood. Here we use an optogenetic protein system to tune the material state of the nucleolus. Our experiments show that above a certain concentration of light-activated protein, the nucleoli gel into a tightly linked meshwork and are no longer able to fuse. These material property changes lead to changes in rRNA processing, providing insight into how rRNA flux and processing may be natively tuned through the material state of nucleolus.
Keywords: nucleolus, material properties, transcription, liquid–liquid phase separation, condensate
Abstract
The nucleolus is a prominent nuclear condensate that plays a central role in ribosome biogenesis by facilitating the transcription and processing of nascent ribosomal RNA (rRNA). A number of studies have highlighted the active viscoelastic nature of the nucleolus, whose material properties and phase behavior are a consequence of underlying molecular interactions. However, the ways in which the material properties of the nucleolus impact its function in rRNA biogenesis are not understood. Here we utilize the Cry2olig optogenetic system to modulate the viscoelastic properties of the nucleolus. We show that above a threshold concentration of Cry2olig protein, the nucleolus can be gelled into a tightly linked, low mobility meshwork. Gelled nucleoli no longer coalesce and relax into spheres but nonetheless permit continued internal molecular mobility of small proteins. These changes in nucleolar material properties manifest in specific alterations in rRNA processing steps, including a buildup of larger rRNA precursors and a depletion of smaller rRNA precursors. We propose that the flux of processed rRNA may be actively tuned by the cell through modulating nucleolar material properties, which suggests the potential of materials-based approaches for therapeutic intervention in ribosomopathies.
Gene expression within the eukaryotic cell nucleus remains a fundamental and poorly understood problem. Membraneless subnuclear condensates are ubiquitous structures that are intimately associated with nuclear RNA, DNA, and proteins and play key roles in cellular processes. Biomolecular condensates such as the nucleolus, Cajal bodies, and speckles concentrate molecules in order to spatiotemporally facilitate the information flow from DNA to RNA (1). Condensates are biophysically interesting because while lacking an enclosing membrane their overall shape and size may be stable on the timescale of hours, even as their constituent biomolecules dynamically exchange over tens of seconds (2). Over the last decade, a host of studies have shown that these structures often exhibit behaviors consistent with condensed liquid phases and assemble above a concentration threshold, a hallmark of liquid–liquid phase separation (3–6).
The most prominent condensate in the nucleus is the nucleolus, a structure that despite being first described over 150 years ago (7) remains not fully understood (8). The nucleolus forms around regions of chromosomes containing stretches of tandem ribosomal DNA (rDNA) gene repeats, known as nucleolar organizer regions (NORs). The transcription of ribosomal RNA (rRNA) is comparable to the overall process of gene transcription elsewhere in the nucleus, with key differences arising from the demand for producing large quantities of the multisubunit ribosome complex. In most eukaryotes, a precursor 47S rRNA transcript is transcribed by RNA polymerase I (Pol I) and contains each of the cotranscribed 18S, 5.8S, and 28S rRNAs (9). By locally concentrating various processing factors, the nucleolus appears to facilitate the cleavage, chemical modifications, and key protein subunit assembly required to generate precursor ribosomal particles from the nascent rRNA transcript.
A host of recent studies have examined the nature and biomolecular origins of the rich phase behavior of the nucleolus (5, 10–14). Diploid human cells contain 10 NORs, but they are not all active at the same time. A human cell generally contains between 2 and 6 nucleoli and the number of nucleoli per cell tends to decrease over time due to fusion events, consistent with the liquid behavior of nucleoli (15, 16). As with other condensates, the phase behavior of nucleoli is largely driven by weak multivalent interactions, often involving segments of intrinsically disordered regions (IDRs) of proteins (regions that are able to sample a wide range of conformations) (4, 17). Two key nucleolar proteins that have received the most attention are nucleophosmin (B23/NPM1) and fibrillarin (FBL). NPM1 is a highly expressed nucleolar protein that is particularly enriched in the granular component (GC), the outer layer of the nucleolus that is associated with assembling ribosomal proteins together with processed rRNA into mature preribosomal particles. Purified NPM1 undergoes phase separation in vitro in an rRNA-dependent manner (18–20). FBL is a nucleolar protein that is enriched in the dense fibrillar component (DFC) of the nucleolus. The DFC is an internal core of proteins and RNAs that serves as an enzymatic crucible for rRNA modifications including methylation by FBL as part of the C/D box snoRNPs, and pseudouridylation by H/ACA box snoRNPs (21). FBL also undergoes phase separation in vitro, in a manner also promoted by the presence of RNA (10). NPM1 and FBL droplets have been shown to be relatively immiscible with one another, and their phase separation can recapitulate the core-shell architecture of the nucleolus in vitro (22).
The same weak molecular interactions that promote the condensation of nucleolar proteins also dictate nucleolar material properties. Coalescence and other liquid behavior of nucleoli, as well as microrheology and other direct measurements of nucleolar protein droplets formed in vitro, can be used to estimate values for its surface tension and viscosity (11, 22, 23). Interestingly, these material properties likely reflect nonequilibrium internal dynamics, as ATP depletion has been seen to increase the apparent viscosity, and purified nucleolar proteins exhibit time-dependent hardening. Indeed, the material state of the inner compartment of the nucleolus has been suggested to age in the intestinal nuclei of Caenorhabditis elegans worms (22). It is likely that the cell itself tunes molecular interaction parameters and thus nucleolar assembly and material properties through posttranslation modifications such as phosphorylation and methylation (24). Understanding the nucleolus as a material is key, since its material properties have been shown to play a central role in its internal organization (22). Furthermore, material properties are expected to impact internal diffusivity and thus rates of ribosome biogenesis and potential downstream biological function, particularly cell growth rates. However, direct tests of the links between the nucleolar material state and function have been lacking.
A primary factor hindering studies to date is a lack of tools for directly modulating material states within living cells. However, recent studies have begun to exploit optogenetics to tune molecular interactions and the resulting phase behavior in living cells (25–28). Cry2 is a photolyase homology region of Arabidopsis thaliana CRY2 protein, that upon blue light exposure exhibits enhanced interactions (29), and when combined with weak IDR-mediated interactions was shown to drive light-dependent phase transitions of “optoDroplets” (27). The material state of these optoDroplets appears to depend on the degree of supersaturation (i.e., the degree to which the intracellular concentration is driven to exceed the threshold for phase separation) (27). Moreover, a mutant Cry2olig has been shown to display disassociation times correlated with enhanced interaction strength (30), which can be utilized for tuning optoDroplet material properties. These optogenetic tools offer a set of approaches for dynamically tuning material properties of intracellular condensates and probing the link between material state and function.
Here we use an optogenetic gelation strategy to show that the material state of the nucleolus affects its ribosome biogenesis function. A Cry2olig optogenetic module is used to tune specific protein interactions in the nucleolus, thereby modulating its material properties. We show that by adding a light-tunable interaction domain to nucleolar molecules we can affect the material state of the nucleolus as well as its ability to process ribosomal RNA. The implications of these findings likely extend beyond the nucleolus to other condensates whose internal dynamics are tuned by cells to perform diverse biological functions.
Results
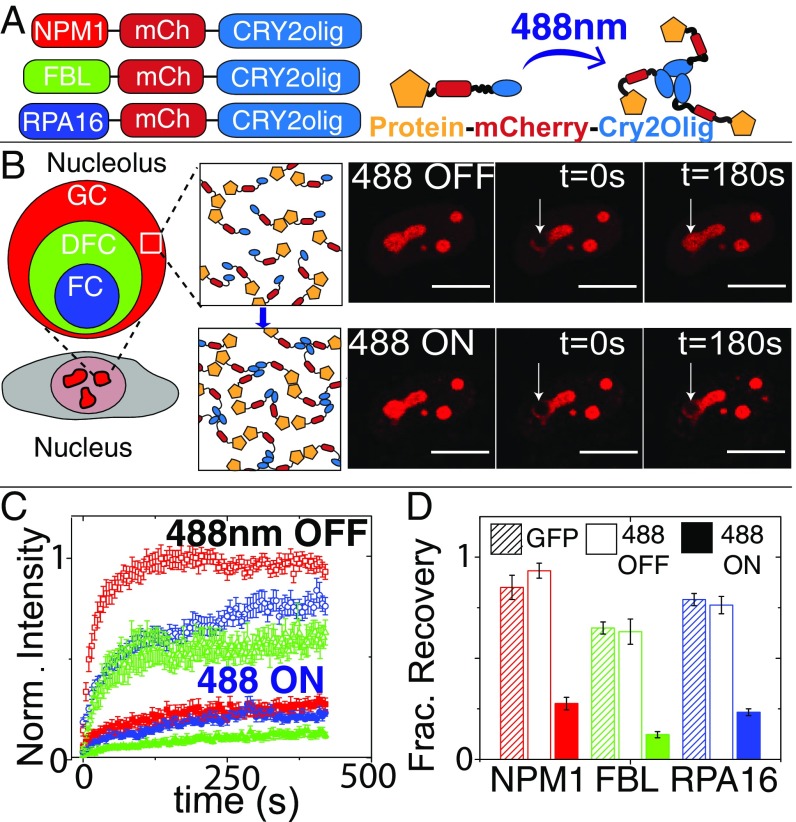
To modulate the interaction of proteins within the nucleolus, we fused proteins found in each nucleolar compartment (GC–NPM1, DFC–FIB1, and FC–RPA16) with Cry2olig; we refer to these variants as “opto-” (e.g. opto-NPM1) and expressed them in NIH3T3 cells at a range of concentrations using lentiviral transfection. The fused proteins localize to their respective nucleolar compartment (Fig. 1B and SI Appendix, Fig. S1), with colocalization and fluorescent recovery after photobleaching (FRAP) dynamics similar to green fluorescent protein (GFP)-tagged versions of the proteins in the absence of blue light (Fig. 1D). However, when these cells are exposed to blue light, FRAP results show that mobility of the Cry2olig-tagged protein is severely diminished, with a decrease in the mobile fraction and an increase in the recovery timescale (Fig. 1 B–D and SI Appendix, Fig. S1).
Fig. 1.
Fusing Cry2olig to nucleolar proteins: localization and FRAP dynamics. (A) Schematic of opto-nucleolar constructs. (B, Left) Schematic of three compartments of the nucleolus. (B, Right) FRAP images before, during, and after bleach for opto-NPM1 before and after blue light activation in the same cell. (Scale bars, 10 μm.) (C) Normalized FRAP curves for opto-nucleolar constructs before and after blue light activation. (D) Normalized fraction recovery at t = 300 s for nonopto GFP-tagged protein, unactivated opto-proteins (488 OFF), and blue light-activated (488 ON) opto-proteins. Error bars are SEM of 15 to 20 cells.
To understand how blue light exposure can alter molecular dynamics within the nucleolus, we examined the impact on the self-association strength of these proteins before and after blue light exposure. We used fluorescence correlation spectroscopy (FCS) to estimate the values of the apparent second virial coefficient B2 in the nucleoplasm of the mammalian cell. For concentrations that are below the condensed droplet concentrations, the diffusivity of a protein is influenced by interactions with other proteins (31, 32). A positive B2 is associated with repulsive protein interactions and can lead to larger effective protein diffusion coefficients. Conversely, a negative B2 is associated with attractive protein interactions and leads to smaller effective protein diffusion coefficients. Here, we estimated an effective second virial coefficient, which we refer to as β, by measuring protein diffusivity as a function of protein concentration and analyzing the data using the relation D ≈ D0(1 + βC), where D is protein diffusivity, D0 is the diffusivity at infinite dilution, and C is the protein concentration of the opto-construct (33).
In the absence of blue light, NPM1-mCherry-Cry2olig (opto-NPM1) diffusivity is weakly dependent on protein concentration, with a low, but nonzero, slope β (Fig. 2 A, black symbols). This slope is similar to that observed with a nonopto NPM1-mCherry, suggesting that in the dark state Cry2olig contributes little to the inherent NPM1-mediated interactions; consistent with this, dark state β values for mCh-Cry2olig are close to zero (Fig. 2 B, open red bar, 488 Off). However, in the presence of high-intensity blue light, the diffusivity of opto-NPM1 becomes more strongly dependent on protein concentration, yielding more strongly negative β values (Fig. 2A). These more negative β values are a direct consequence of light-dependent Cry2olig interactions, since β for NPM1-mCh exhibits no significant light dependence, while β for mCh-Cry2olig is significantly more negative upon blue light activation (Fig. 2 A and B). Taken together, these results show that we can successfully tune the molecular mobility of opto-NPM1 with the Cry2olig optogenetic system, by modulating light-dependent Cry2olig interactions.
Fig. 2.
Opto-NPM1’s average protein interactions are tunable by blue light. (A) Diffusion vs. concentration in the nucleoplasm measured by FCS for opto-NPM1 unactivated (black) and with increasing levels of [488] intensity (red is 70 nW and blue is 175 nW). Error bars are the SDs of 3 to 5 cells (binned by concentration). (B) β (effective second viral coefficient) measured for 488 Off (open bars) and 488 On (closed bars) opto-NPM1 (green), NPM1-mCh (blue), and mCh-Cry2olig (red). Error bars are error of the linear fit (i.e., dotted line in A). P values < 0.05 as indicated by asterisks.
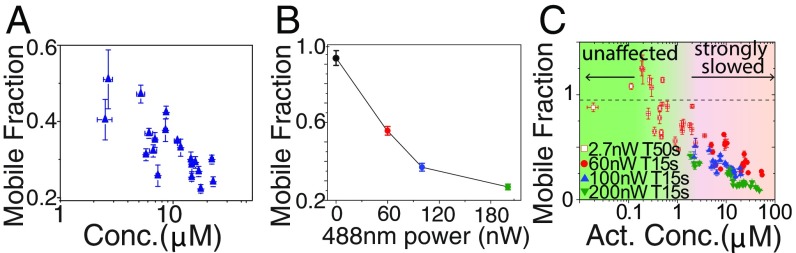
We next sought to determine how these light-dependent interactions give rise to collective changes in molecular dynamics within the nucleolus. As with other optogenetic proteins, Cry2olig switches between an active and inactive state. We reasoned that the observed molecular dynamics should strongly depend on the concentration of activated opto-proteins that results from this light-dependent conversion. Consistent with this expectation, for a given set of activation conditions we find that the normalized fraction recovery (mobile fraction) of photobleached opto-NPM1 exhibits a strong dependence on the total concentration of opto-NPM1 expression (Fig. 3A). Moreover, for cells with similar opto-NPM1 expression (∼5 to 10 μM), we find a strong dependence of the mobile fraction on blue light intensity (Fig. 3B). Indeed, cells activated with very low laser power, and at long intervals between activation, exhibit nearly complete recovery, suggesting only a small fraction of activated opto-NPM1 proteins contribute to decreased mobility (Fig. 3 C, open squares). To quantify these light-dependent dynamics, we use a simple kinetic model (27), with an overall activation rate proportional to light intensity, k1 = kact*[blue], and a light-independent deactivation rate, k2 (27). Cells were activated with different activation intervals (T) and blue light intensities (27) (SI Appendix, Fig. S2), yielding fitted values of kact = 1.1 ± 5.6 μW−1⋅s−1 and k2 = 0.002 ± 0.005 s−1, which are comparable to previous measurements (27). Consistent with model predictions, the fraction of inactivated molecules, Finact, increases with either longer time in between intervals (T) or weaker blue light intensity (SI Appendix, Fig. S3). Remarkably, FRAP recovery data for cells spanning 2 orders of magnitude in expression levels, and 4 different activation conditions (2.7, 60, 100, and 200 nW), can be collapsed by plotting the data against the calculated concentration of activated opto-NPM1 (Fig. 3C and SI Appendix, Fig. S3 for uncollapsed data).
Fig. 3.
Mobile fraction of Opto-NPM1 measured at 300 s after bleach as a function of (A) opto-NPM1 concentration and (B) 488-nm power. (C) Mobile fraction of Opto-NPM1 collapsed by plotting against activated Opto-NPM1 concentration. Error bars are SE of 5 to 10 cells.
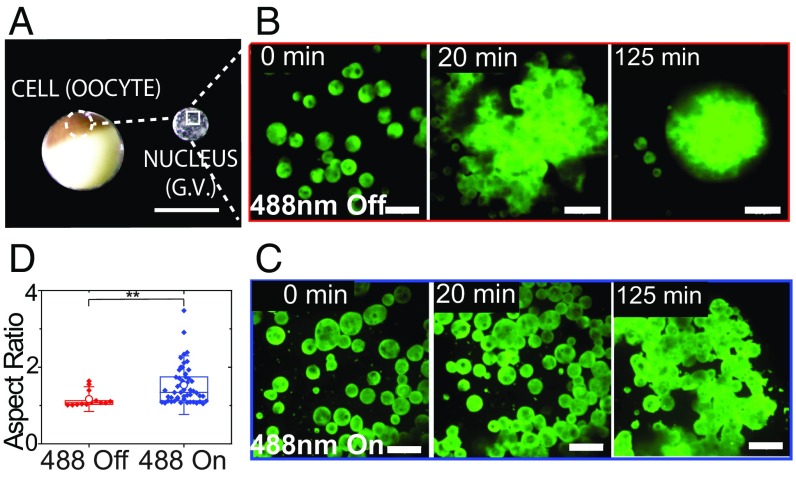
This ability to collapse the molecular mobility data (Fig. 3C) is consistent with a light-dependent oligomerization of Cry2olig inducing a transition in collective molecular dynamics within the nucleolus. To determine if these changes are associated with alterations in the mesoscale material properties of the nucleolus, we take advantage of mature Xenopus laevis oocytes, which contain a ∼500-µm-diameter nucleus (germinal vesicle) (Fig. 4A), whose large nuclear bodies have previously been exploited to examine nucleolar material properties (12, 15, 22, 34). As shown previously, we find that when actin is disrupted using latrunculin A (Lat-A) the nucleoli coalesce with one another and sediment due to gravity (15, 35) (Fig. 4 B and D, 488 Off). Without exposure to blue light, these nucleoli fuse and round up to a nearly perfect sphere, exhibiting an exponential relaxation to a single large droplet, with a ratio of droplet viscosity to surface tension, η/γ, of ∼30 s/μm, consistent with previous measurements (15, 22). Strikingly, in Lat-A–treated X. laevis nuclei expressing opto-NPM1, exposure to blue light results in nucleoli that no longer coalesce into spherical droplets (shown in Fig. 4C and SI Appendix, Fig. S4) but rather clump onto each other much like hard sticky beads (see aspect ratio, Fig. 4D); even after more than 2 h, these nucleoli are not able to relax and round up to a sphere (aspect ratio > 1). While an apparent η/γ cannot be reliably determined from these very slow dynamics, the coalescence timescale for opto-NPM1 nucleoli appears to be at least 10 times longer upon activation, implying that the value of η/γ is at least 10 times higher.
Fig. 4.
X. laevis nucleolar coalescence dynamics shows changes in bulk properties. (A) Images of the X. laevis oocyte and nucleus. (Scale bar, 1 mm.) (B) Nonactivated nucleolar fusion under actin disruption at 0, 20, and 125 min and (C) 488-nm activated nucleolar fusion under actin disruption at 0, 20, and 125 min. (Scale bars, 20 μm.) (D) Aspect ratios after 2 h of nucleoli in unactivated and activated nuclei expressing opto-NPM1. P value < 0.005.
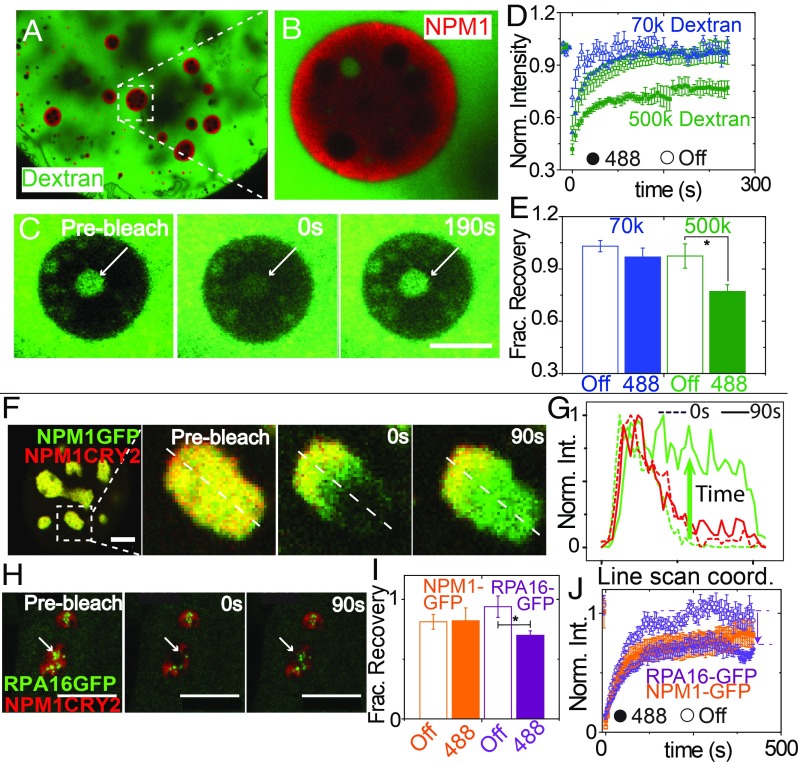
To examine how this light-dependent change in material state impacts mobility of other molecules within the nucleolus, we used dextran of varying molecular weight, injected into the nucleolus prior to blue light activation (Fig. 5 A–E). Dextrans ≥70 kDa are mostly excluded from nucleoli, while 10-kDa dextrans are able to partition into nucleoli, as described previously (33). The concentration of large dextrans is very low within the nucleolus, hindering reliable FRAP measurements on this population. However, we noticed that often there are nucleoplasmic pockets of dextrans that are trapped within the GC of nucleoli. By using FRAP on the population of dextran molecules within these nucleoplasmic pockets, we could examine the transport of dextran through the GC with both the opto-NPM1 construct and an NPM1-Cerulean control. For 70 kDa, we see the same relatively full recovery of dextran regardless of whether opto-NPM1 has been exposed to blue light or not (Fig. 5D), which suggests that small dextran can diffuse across the GC from the nucleoplasm and replenish the pool of molecules in the pocket. However, in the case of a very large dextran, 500 kDa, we find that for opto-NPM1–expressing nucleoli exposed to blue light (Fig. 5C) the final fraction of dextran is significantly lower (0.7 vs. ∼1, as shown in Fig. 5 D and E). Taken together, these data are consistent with an opto-NPM meshwork formed after exposure to blue light, with an effective mesh size that is sufficient to hinder the motion of other large biomolecules in the nucleolus.
Fig. 5.
Light-activated nucleolar mesh size effect in X. laevis oocytes and in tissue culture cells. (A) Nucleus of X. laevis oocyte expressing opto-NPM1 and injected with FITC-500-kDA dextran. (B) Zoom-in of nucleolus, showing dextran-rich nucleoplasmic compartment. (C) Prebleach, bleach, and recovery of 500-kDA dextran in nucleoplasmic buffer pocket of a representative nucleolus. (Scale bar, 10 μm.) Normalized FRAP curves (D) and normalized fraction recovery at 180 s (E) for 70-kDA (blue) and 500-kDA (green) dextrans in 5 to 10 nonopto (open) and activated opto-NPM1 nucleoli (solid). (F) Mammalian nuclei expressing NPM1-GFP and opto-NPM1 before bleach, during bleach, and after bleach. (G) Line scan through nucleolus from F during bleach (dotted lines) and 90 s after bleach (solid lines). (H) Mammalian nuclei expressing RPA16-GFP and opto-NPM1 before bleach, during bleach, and after bleach. (I) Normalized fraction recovery at 300 s after bleach and normalized FRAP curves (J) for NPM1-GFP (orange) and RPA16-GFP (purple) with (solid) and without (open) opto-NPM1 of 8 to 10 nucleoli. (Scale bars, 10 μm.) Asterisk denotes P value < 0.05.
To interrogate this mesh size effect in tissue culture cells, we activate opto-NPM1 and examine whether non–Cry2olig-tagged proteins are able to diffuse freely. Interestingly, we find that the mobility of NPM1-GFP within opto-NPM1 nucleoli is essentially unchanged before and after blue light activation (Fig. 5F). Moreover, other proteins of comparable size exhibit a similar activation-independent mobility, even in the presence of a variety of different opto-constructs (SI Appendix, Fig. S5). However, we find that the diffusivity of RPA16, a subunit of the large [∼550 kDa (36)] protein complex RNA Pol I, is significantly affected by activation of a range of different opto-constructs (Fig. 5 F–J and SI Appendix, Fig. S5). These data suggest that the presence of a blue light-dependent meshwork gives rise to an effective gelation of the nucleolus, with molecular-size–dependent changes in internal dynamics.
We next asked whether the size-dependent mesh of the optogenetically gelled nucleolus impacts processing of precursor ribosomal RNA subunits. Pre-rRNAs undergo processing in a directional manner, from the innermost DFC region to the outermost GC region (Fig. 6A). In budding yeast, early precursor ribosomes are ∼30 nm (37) but are expected to be even larger in human cells due to the large number of processing factors. These particles are thus similar in size or even larger than RNA Pol I, and thus may be larger than the apparent mesh size of opto-activated nucleoli. We thus reasoned that nucleolar gelation could lead to changes in their mobility and processing.
Fig. 6.
Opto-nucleolus gelling leads to specific inhibitions of early pre-rRNA processing. (A) Mouse pre-rRNA processing pathway. The probes used are shown. (B) Quantification of 6 early pre-RNA species as a function of time exposed to blue light (0, 24, 48, and 72 h). Raw band intensities of NPM1-CRY2 cells were first normalized to the initial time (t = 0) for each transcript and then to control 3T3 cells (Materials and Methods). Schematic of native, nonactivated (C) and blue light-activated (D) opto-NPM1 effect on rRNA processing.
To test the impact on rRNA processing, NIH 3T3 cells with mCh-Cry2olig tagged at the endogenous C-terminal locus of NPM1 were generated using CRISPR-Cas9 and exposed to blue light for 0, 24, 48, and 72 h. Total RNA was extracted, separated on denaturing gels, and analyzed by Northern blotting using probes that detect major pre-rRNA intermediates (Fig. 6A). We first examined NIH 3T3 control cells exposed to blue light, in which we found a general decrease in transcript level, likely due to mild toxicity associated with prolonged blue light exposure (SI Appendix, Fig. S6). By contrast, CRISPR NPM1-Cry2olig display specific processing inhibitions upon blue light exposure. Notably, they strongly accumulate the 46S and 45S pre-rRNAs, which is clearly seen in quantification normalized to controls (Fig. 6B and SI Appendix, Fig. S8). The 46S and 45S accumulation indicates inhibitions at cleavage sites 6 (in 3′-ETS), A0 (in 5′-ETS) and 2c (in ITS1). Consequently, several downstream species appear to be also impacted including 43S, 41S, 36S, 34S, and 29S (Fig. 6B and SI Appendix, Fig. S7). These data indicate that the flux of rRNA through the nucleolus can be controlled by altering its material properties, with functional consequences on pre-rRNA processing and ribosome biogenesis.
Discussion
The molecular interactions which dictate the material properties of biocondensates and the way those material properties and associated molecular dynamics in turn determine biological function have received much recent attention (22, 38–44). Previous studies have illustrated how the cell may modulate protein interaction strength or interaction valency through posttranslational modifications (e.g., phosphorylation) or through oligomerization domains (e.g., NPM1 pentamerization), respectively (13, 19, 20, 45). However, these studies have relied on correlative changes between material properties and biological function, and our understanding of the causal relationships remains poor. In this paper, we used an optogenetics approach to tune the strength of protein–protein interactions and thus the material state of the nucleolus with light. FCS and kinetic modeling allowed us to examine the way in which strong light-meditated interactions control the fraction of activated molecules, which allow tuning of the bulk material properties of the nucleolus. This gives rise to strong functional perturbations, in particular hindering the flux of newly transcribed and partially processed rRNA molecules.
These changes to the material properties and molecular mobility depend strongly on the concentration of light-oligomerized molecules. Our FRAP data show that only when the nucleolar concentration of activated opto-NPM1 molecules becomes greater than ∼1 μM is there a significant decrease in molecular mobility (Fig. 3). This threshold concentration is significantly smaller than reported cellular concentration of endogenous (untagged) NPM1 (46), suggesting that the transition does not simply reflect an excess of activated opto-NPM1 compared with endogenous NPM1. If activation caused isolated opto-NPM1 clusters to form, they could potentially decrease the collective mobility within the nucleolus. However, in soft matter, true gelation is known to occur only beyond a “percolation” threshold, which describes the critical level of molecular connectivity required for solid-like material response (47); a percolated network scaffolded through opto-NPM1 activation likely underlies our findings of a strong concentration dependence of internal mobilities, and associated impact on bulk material properties. While here we have focused on controlling interaction strength, the geometry of interactions is likely also to be used by the cell to control condensate material state. For instance, NPM1 pentamerization leads to a radial set of interactions, while oligomerization on nucleic acid scaffolds could yield more extended interaction networks. Future work is needed to explore the interplay between interaction strength, geometry, and the connectivity transitions underlying gelation.
These changes in the mobility of opto-nucleolar components manifest as changes in the material state of the nucleolus. Fusion experiments showed slowed fusion tendency, which is expected as the effective viscoelasticity of droplets is increased (17, 22). However, if these changes can be understood as a light-induced nucleolar gelation, the gel must exhibit a large effective mesh size. Indeed, while our opto-tagged nucleolar proteins are strongly impacted, such that their apparent mobile fraction is decreased to as low as 20%, many nontagged nucleolar proteins are relatively unaffected (Fig. 5). The high mobility of GFP-tagged NPM1 in activated opto-NPM1–expressing cells is surprising and suggests that mixed NPM1 hetero-pentamerization may not be occurring with these tagged constructs, possibly due to interference from the tag. In any case, while opto-components appear to be forming a partially connected network, the network is capable of allowing relatively unhindered mobility of smaller nontagged proteins. We interpret this as proteins smaller than the pore size of the mesh diffusing through unhindered, while proteins/complexes larger than the pore size of the mesh are hindered. This appears to explain our findings that RPA16, a protein component of the large (∼10 nm) multisubunit complex of RNA pol I, is significantly hindered in gelled nucleoli. While this is interpreted as a size effect, we cannot rule out possible interactions of RNA pol I with the opto-NPM1 mesh, which could potentially also contribute to hindered mobility. However, the size dependence of nucleolar proteins is consistent with the hindered diffusion we measure for 500-kDa dextran, whose radius of gyration is expected to be similarly large, ∼15 nm (48). These findings are reminiscent of the apparent mesh size determined for other phase-separated condensates (33, 34).
The primary function of the nucleolus is in facilitating ribosome biogenesis, through the processing and assembly of rRNA into preribosomal particles. This is a multistep process, which occurs through a steady-state flux of newly transcribed 47S rRNA, through steps which involve a series of cleavage and chemical modifications that result in production of mature segments of 18S, 5.8S, and 28S rRNA (49). We found that by using light to gel the nucleolus the flux of rRNA was significantly altered. Notably, we found a buildup of 46S and 45S rRNA (Fig. 6) and a depletion of downstream processing intermediates (e.g., 43S and 36S). The 46S and 45S rRNAs are two of the very first precursors produced from the 47S transcript and thus appear to be sufficiently large that their mobility is hindered by nucleolar gelation, leading to their buildup and associated decrease in downstream products. Future work may utilize approaches such as those commonly employed to understand metabolic flux, to more precisely link these spatiotemporal changes in pre-RNA flux with altered nucleolar material properties and internal mobility.
Various diseases of ribosome biogenesis, known as ribosomopathies, are associated with a number of different genetic mutations (50). In many cases, these defects could reflect an unbalanced flux of rRNA out of the nucleolus that arises from changes in the viscoelasticity of the nucleolus. Indeed, similar perturbations in rRNA flux could also be at play in repeat expansion diseases such as amyotrophic lateral sclerosis caused by C9orf72 trinucleotide expansion, which results in dipeptide repeats that bind to NPM1, localize to the nucleolus, and lead to slowed mobility (51). Another example where rRNA flux may be at play is in premature aging disorders such as Hutchinson–Gilford progeria syndrome where ribosome biogenesis is elevated and nucleoli are enlarged (52). Indeed, it has been suggested that smaller nucleoli can be a hallmark of longevity in flies, mice, and humans (53). The development of optogenetic approaches to engineer the assembly of de novo condensates (25–28), and now to modulate the properties of endogenous ones, promises to have an enormous impact on our understanding of these emerging links between the material properties and (dys)function of the nucleolus. These insights will suggest new therapeutic approaches for tackling ribosome disorders by modulating nucleolar assembly and material properties, adding novel options to the developing arsenal targeting the nucleolus for anticancer intervention (54).
Our finding that altering the material properties of the nucleolus impacts the flux of nascent rRNA processing is likely relevant beyond this specific aspect of transcription. Indeed, we propose a similar causal link is at play throughout the genome, where the material properties of nanoscale transcriptional condensates likely dictate subsequent downstream RNA modifications. Moreover, condensates comparable in size to the nucleolus, such as nuclear speckles and Cajal bodies, which play key roles in splicing and other RNA processing steps, have functions likely to be tightly linked to their material properties. Future studies will build on the present study as well as other recently developed optogenetic approaches (25–28) to quantitatively map intracellular phase behavior and its link to underlying molecular function and associated phenotype.
Materials and Methods
Preparation of Mammalian and X. laevis Nucleoli.
Mammalian NIH 3T3 cell lines were maintained and imaged at 37 °C using standard conditions. Frog experiments were approved by the Princeton Institutional Review Board and nuclei were injected with plasmid constructs, manually dissected in mineral oil, and subsequently imaged. For actin disruption and coarsening in X. laevis, to disrupt actin Lat-A (Sigma) treatment was performed for 1 to 2 h. Nuclei were then dissected in mineral oil and imaged. For RNA analysis, cells were exposed to blue light (Teleopto LED array system, LED array model LEDA-X with driver LAD-1, setting 7.05) for 1, 2, or 3 d. Total RNA was extracted and Northern blot was performed as in ref. 55. Experiment was done in a 37 °C incubator. See SI Appendix, Supplementary Information Text for FRAP, actin disruption, FCS, dextran experiments, and RNA methods in detail.
Supplementary Material
Acknowledgments
We thank David Sanders, Amy Strom, Josh Riback, and other members of the C.P.B. laboratory for help with experiments and comments on the manuscript. This work was supported by the Howard Hughes Medical Institute and grants from the NIH 4D Nucleome Program (U01 DA040601), the Princeton Center for Complex Materials, the NSF-supported Materials Research Science and Engineering Center (DMR 1420541), an NSF CAREER award (1253035), NSF Grant PHY-1607612, and the NSF Graduate Research Fellowship Program (DCE-1656466). D.L.J.L. and L.W. were supported by Fonds de la Recherche Scientifique, Fonds Jean Brachet, Région Wallonne (DGO6), and the Université Libre de Bruxelles.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1903870116/-/DCSupplemental.
References
- 1.Zhu L., Brangwynne C. P., Nuclear bodies: The emerging biophysics of nucleoplasmic phases. Curr. Opin. Cell Biol. 34, 23–30 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Phair R. D., Misteli T., High mobility of proteins in the mammalian cell nucleus. Nature 404, 604–609 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Molliex A., et al. , Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nott T. J., et al. , Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell 57, 936–947 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weber S. C., Brangwynne C. P., Inverse size scaling of the nucleolus by a concentration-dependent phase transition. Curr. Biol. 25, 641–646 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wippich F., et al. , Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell 152, 791–805 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Wagner R., Einige bemerkungen und fragen über das keimbläschen (vesicular germinativa). Müller’s Archiv. Anat. Physiol. Wissenschaft. Med., 373–377 (1835). [Google Scholar]
- 8.Pederson T., The nucleolus. Cold Spring Harb. Perspect. Biol. 3, a000638 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boisvert F. M., van Koningsbruggen S., Navascués J., Lamond A. I., The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 8, 574–585 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Berry J., Weber S. C., Vaidya N., Haataja M., Brangwynne C. P., RNA transcription modulates phase transition-driven nuclear body assembly. Proc. Natl. Acad. Sci. U.S.A. 112, E5237–E5245 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brangwynne C. P., Soft active aggregates: Mechanics, dynamics and self-assembly of liquid-like intracellular protein bodies. Soft Matter 7, 3052–3059 (2011). [Google Scholar]
- 12.Feric M., Brangwynne C. P., A nuclear F-actin scaffold stabilizes RNP droplets against gravity in large cells. Nat. Cell Biol. 15, 1253–1259 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitrea D. M., et al. , Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. eLife 5, e13571 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falahati H., Wieschaus E., Independent active and thermodynamic processes govern the nucleolus assembly in vivo. Proc. Natl. Acad. Sci. U.S.A. 114, 1335–1340 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brangwynne C. P., Mitchison T. J., Hyman A. A., Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl. Acad. Sci. U.S.A. 108, 4334–4339 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amenta P. S., Fusion of nucleoli in cells cultured from the heart of Triturus viridescens. Anat. Rec. 139, 155–165 (1961). [DOI] [PubMed] [Google Scholar]
- 17.Elbaum-Garfinkle S., et al. , The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. U.S.A. 112, 7189–7194 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrolino M. C., Mitrea D. M., Michael J. R., Kriwacki R. W., Compositional adaptability in NPM1-SURF6 scaffolding networks enabled by dynamic switching of phase separation mechanisms. Nat. Commun. 9, 5064 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitrea D. M., et al. , Self-interaction of NPM1 modulates multiple mechanisms of liquid-liquid phase separation. Nat. Commun. 9, 842 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitrea D. M., et al. , Structural polymorphism in the N-terminal oligomerization domain of NPM1. Proc. Natl. Acad. Sci. U.S.A. 111, 4466–4471 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodriguez-Corona U., Sobol M., Rodriguez-Zapata L. C., Hozak P., Castano E., Fibrillarin from archaea to human. Biol. Cell 107, 159–174 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Feric M., et al. , Coexisting liquid phases underlie nucleolar subcompartments. Cell 165, 1686–1697 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caragine C. M., Haley S. C., Zidovska A., Surface fluctuations and coalescence of nucleolar droplets in the human cell nucleus. Phys. Rev. Lett. 121, 148101 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leary D. J., Huang S., Regulation of ribosome biogenesis within the nucleolus. FEBS Lett. 509, 145–150 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Bracha D., et al. , Mapping local and global liquid phase behavior in living cells using photo-oligomerizable seeds. Cell 175, 1467–1480.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dine E., Gil A. A., Uribe G., Brangwynne C. P., Toettcher J. E., Protein phase separation provides long-term memory of transient spatial stimuli. Cell Syst. 6, 655–663.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shin Y., et al. , Spatiotemporal control of intracellular phase transitions using light-activated optoDroplets. Cell 168, 159–171.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shin Y., et al. , Liquid nuclear condensates mechanically sense and restructure the genome. Cell 175, 1481–1491.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bugaj L. J., Choksi A. T., Mesuda C. K., Kane R. S., Schaffer D. V., Optogenetic protein clustering and signaling activation in mammalian cells. Nat. Methods 10, 249–252 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Taslimi A., et al. , An optimized optogenetic clustering tool for probing protein interaction and function. Nat. Commun. 5, 4925 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harding S. E., Johnson P., The concentration-dependence of macromolecular parameters. Biochem. J. 231, 543–547 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tirado-Miranda M., Haro-Pérez C., Quesada-Pérez M., Callejas-Fernández J., Hidalgo-Alvarez R., Effective charges of colloidal particles obtained from collective diffusion experiments. J. Colloid Interface Sci. 263, 74–79 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Wei M. T., et al. , Phase behaviour of disordered proteins underlying low density and high permeability of liquid organelles. Nat. Chem. 9, 1118–1125 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Handwerger K. E., Cordero J. A., Gall J. G., Cajal bodies, nucleoli, and speckles in the Xenopus oocyte nucleus have a low-density, sponge-like structure. Mol. Biol. Cell 16, 202–211 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feric M., Brangwynne C. P., A nuclear F-actin scaffold stabilizes ribonucleoprotein droplets against gravity in large cells. Nat. Cell Biol. 15, 1253–1259 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hannan R. D., et al. , Affinity purification of mammalian RNA polymerase I. Identification of an associated kinase. J. Biol. Chem. 273, 1257–1267 (1998). [DOI] [PubMed] [Google Scholar]
- 37.Chaker-Margot M., Barandun J., Hunziker M., Klinge S., Architecture of the yeast small subunit processome. Science 355, eaal1880 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Patel A., et al. , A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Shin Y., Brangwynne C. P., Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Kato M., et al. , Cell-free formation of RNA granules: Low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weber S. C., Brangwynne C. P., Getting RNA and protein in phase. Cell 149, 1188–1191 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Zhang H., et al. , RNA controls PolyQ protein phase transitions. Mol. Cell 60, 220–230 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin Y., Protter D. S., Rosen M. K., Parker R., Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell 60, 208–219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elbaum-Garfinkle S., Brangwynne C. P., Liquids, fibers, and gels: The many phases of neurodegeneration. Dev. Cell 35, 531–532 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Wang J. T., et al. , Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically disordered proteins in C. elegans. eLife 3, e04591 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hein M. Y., et al. , A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell 163, 712–723 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Stauffer D., Aharony A., Introduction to Percolation Theory (Taylor & Francis, London, ed. 2, 1994). [Google Scholar]
- 48.Armstrong J. K., Wenby R. B., Meiselman H. J., Fisher T. C., The hydrodynamic radii of macromolecules and their effect on red blood cell aggregation. Biophys. J. 87, 4259–4270 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lafontaine D. L., Noncoding RNAs in eukaryotic ribosome biogenesis and function. Nat. Struct. Mol. Biol. 22, 11–19 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Narla A., Ebert B. L., Ribosomopathies: Human disorders of ribosome dysfunction. Blood 115, 3196–3205 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kwon I., et al. , Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science 345, 1139–1145 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Buchwalter A., Hetzer M. W., Nucleolar expansion and elevated protein translation in premature aging. Nat. Commun. 8, 328 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tiku V., et al. , Small nucleoli are a cellular hallmark of longevity. Nat. Commun. 8, 16083 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Catez F., et al. , Ribosome biogenesis: An emerging druggable pathway for cancer therapeutics. Biochem. Pharmacol. 159, 74–81 (2019). [DOI] [PubMed] [Google Scholar]
- 55.Tafforeau L., et al. , The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of Pre-rRNA processing factors. Mol. Cell 51, 539–551 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.