Significance
Toxoplasma gondii infects a wide range of animals, including humans, where it can form chronic persistent infections for the life of the host. We demonstrate that the parasite T. gondii secretes an effector protein called TgIST that traffics to the host cell nucleus, binds to STAT1/STAT2 heterodimers, and blocks signaling through type I interferon (IFN). TgIST globally blocks induction of IFN-stimulated genes normally up-regulated by IFN-β, including those involved in innate defense pathways. Type I IFN is important for control of chronic infection by T. gondii in the central nervous system, and the ability of the parasite to modulate this pathway contributes to chronic persistence.
Keywords: interferon, central nervous system, inflammatory monocyte, NK cell, transcriptome
Abstract
In contrast to the importance of type II interferon-γ (IFN-γ) in control of toxoplasmosis, the role of type I IFN is less clear. We demonstrate here that TgIST, a secreted effector previously implicated in blocking type II IFN-γ signaling, also blocked IFN-β responses by inhibiting STAT1/STAT2-mediated transcription in infected cells. Consistent with a role for type I IFN in cell intrinsic control, ∆Tgist mutants were more susceptible to growth inhibition by murine and human macrophages activated with IFN-β. Additionally, type I IFN was important for production of IFN-γ by natural killer (NK) cells and recruitment of inflammatory monocytes at the site of infection. Mice lacking type I IFN receptors (Ifnar1−/−) showed increased mortality following infection with wild-type parasites and decreased virulence of ∆Tgist parasites was restored in Ifnar1−/− mice. The findings highlight the importance of type I IFN in control of toxoplasmosis and illuminate a parasite mechanism to counteract the effects of both type I and II IFN-mediated host defenses.
Signal Transducer and Activator of Transcription (STAT) factors control a number of host responses, including antimicrobial defense and inflammation (1, 2). Type I interferons (IFNs) (i.e., IFN-α, IFN-β, IFN-λ) induce JAK1 and TYK2 kinases to phosphorylate STAT1/STAT2 heterodimers that associate with IRF9 to activate genes that contain a canonical IFN-sensitive response element (ISRE) in their promoters (3). Type II IFN (IFN-γ) activates JAK1 and JAK2 to phosphorylate homodimers of STAT1 and induce expression of genes that contain a gamma-activated sequence (GAS) in their core promoters (4). There is a considerable overlap in the genes induced by type I and type II IFNs due to cross-talk between these signaling pathways (4). As well, both pathways activate transcription through STAT-independent means (3, 5). In addition to the strong activation that occurs through signaling via IFN receptors, some genes in this pathway continue to show low-level expression in the absence of added IFN, and these so-called tonic response genes also play key roles in host defense and inflammation (3, 4).
Toxoplasma gondii is an obligate intracellular parasite that infects a wide range of animals, including humans, where it is an opportunistic pathogen (6). Type II IFN-γ is crucial for control of infection in the mouse model, as shown by ablation of the Ifng gene (7) and specific antibody neutralization of IFN-γ (8). Studies using bone marrow chimeras have established that signaling is needed in both hematopoietic and nonhematopoietic cells, both of which express IFN-γ receptors (Ifngr1) (9). The requirement for IFN-γ signaling is thought to be due to up-regulation of IFN-stimulated genes (ISGs), including inducible nitric oxide synthase (iNOS, Nos2), NADPH oxidase (Nox1, Nox2), immunity-related guanosine triphosphatases, and guanylate binding proteins (GBPs), which collectively result in control of replication and elimination of the parasite (10). The importance of IFN-γ–mediated signaling is underscored by a number of virulence factors in the parasite that counteract these defenses (11). Although host effectors and parasite avoidance mechanisms differ in human cells, they also rely on IFN-γ to control intracellular replication of T. gondii (12).
In addition to blocking downstream effector mechanisms, preinfection with T. gondii blocks induction of IFN-γ–responsive genes in both mouse and human cells (13), thus preventing up-regulation of iNOS (14) and major histocompatibility complex (MHC) class II (15). In previously infected cells, activation with IFN-γ results in phosphorylation of STAT1 homodimers that are recruited to the nucleus and bind to the GAS-containing promoters, yet transcription is blocked (16, 17). Although STAT1 homodimers normally recycle through a process of dephosphorylation, in T. gondii-infected cells, activated phospho-Tyr701 STAT1 homodimers remain strongly associated with chromatin (16, 18, 19). The ability of T. gondii to block STAT1 signaling has been attributed to an effector called Inhibitor of STAT Transcription (TgIST), which blocks responses to IFN-γ (18, 19). TgIST is a dense granule protein that is released into the host cell, where it traffics to the host cell nucleus bound to phosphorylated dimers of STAT1, which, in turn, bind tightly to GASs in the promoters of ISGs. Paradoxically, even though activated STAT1 dimers are bound to the correct response regions of IFN-γ–activated genes, STAT1 transcription is blocked in T. gondii-infected cells. The block in transcription caused by TgIST may be related to recruitment of a nucleosome remodeling and repressive (NuRD) complex (18, 19), which is not normally a component of the STAT1 pathway but is known for its role in transcriptional repression during development (20, 21). Previous studies demonstrated that TgIST is both necessary and sufficient for this block, and that it imparts a protective role in cells that are infected before receiving the IFN-γ signal (19). The ability to block STAT1 signaling in naive macrophages is important during infection in vitro, as shown by the decreased expansion, dissemination, and lethality of ∆Tgist parasites (19).
In contrast to the dominant role of IFN-γ, type I IFNs appear to be less critical in control of T. gondii. Type I IFNs are primarily known for their role in combating viral infections, although they can also induce inflammation and create exacerbating pathology, especially in the context of coinfection with bacterial pathogens (22). In addition, type I IFNs can play protective roles during bacterial infection, although they can also counteract protective type II IFN pathways (22). Similarly, in parasitic infections, type I IFNs can play neutral or inhibitory roles (22) or be protective, as in the case of sporozoites of rodent malaria infecting the liver (23, 24). Although infection by most strains of T. gondii does not elicit strong type I IFN production (25), challenge with heat-killed lysates was able to potently induce such a response (26). Together, these findings suggest that the parasite possesses both the intrinsic capacity to induce the type I IFN pathway and also expresses mediators that block amplification of the response. Consistent with this finding, early studies demonstrated only modest control of T. gondii replication in human monocyte-derived macrophages treated with IFN-β (27, 28). As well, Ifnar1−/− knockout mice, which lack signaling through type I IFNs, show only a modest increase in susceptibility to infection (29). However, since infection with T. gondii blocks induction of genes regulated by type I IFN (16), and, overall, this pathway may be less important in controlling toxoplasmosis than induction of type II IFN, it is not surprising that loss of type I IFN signaling may not reveal a striking phenotype. Collectively these findings leave undefined whether type I IFN plays a meaningful role during infection with T. gondii.
Here, we reexamined the role of TgIST in modulating signaling by STAT1/STAT2 heterodimers in response to type I IFN. Treatment with IFN-β induced a set of type I IFN response genes that were strongly blocked by prior infection with T. gondii. This block was dependent on TgIST, which binds to the STAT1/STAT2 heterodimer and recruits the NuRD complex. Importantly, IFN-β treatment led to restricted growth of TgIST knockout parasites in both mouse and human macrophages. The ability to block type I IFN pathways was particularly important in the central nervous system (CNS), where loss of Ifnar1 receptors led to increased parasite load, encephalitis, and decreased survival. Our studies reveal roles for type I IFN in controlling chronic parasitic infection and for the evolution of a pleotropic effector capable of blocking both type I and type II IFNs.
Results
TgIST Suppresses Type I IFN Responses.
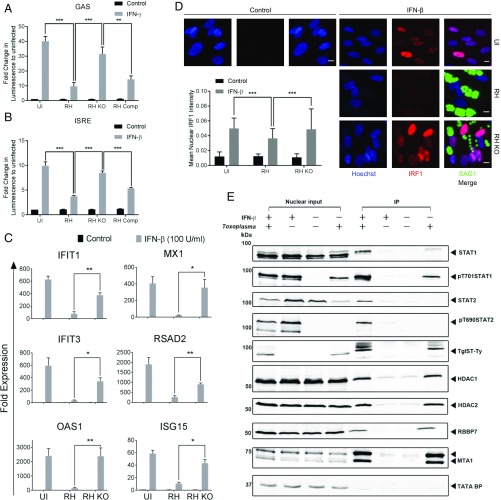
Although the secreted effector TgIST has previously been shown to block signaling from IFN-γ (18, 19), its role in modulating type I IFN signaling has not been reported. We tested the ability of TgIST to block both type I and type II IFN responses in HeLa cells stably expressing luciferase reporter downstream of GAS (γ-IFN–activated sequence) or ISRE sequences. The up-regulation of luciferase expression upon IFN-γ or IFN-β activation that was seen in uninfected cells was significantly abrogated in cells infected with either wild-type RH strain (i.e., RH) or TgIST-complemented parasites (i.e., RH Comp) (Fig. 1 A and B). In contrast, infection of cells with TgIST knockout (i.e., RH KO) parasites resulted in IFN-γ– or IFN-β–mediated luciferase expression similar to the levels in uninfected cells (Fig. 1 A and B). To examine the responsiveness of endogenous genes, we evaluated expression of genes normally up-regulated by IFN-β (e.g., IFIT1, MX1, IFIT3, RSAD2, OAS2, ISG15) in T. gondii-infected human foreskin fibroblast (HFF) cells. IFN-β strongly induced expression of these genes in uninfected cells, and this effect was significantly blocked in cells infected with wild-type RH parasites, but not when cells were infected with the TgIST knockout parasites (Fig. 1C). IRF1 is a transcription factor that is induced by IFN treatment, and following translocation to the nucleus, it further enhances expression of many of the IFN-activated genes in a feed-forward manner (4). Therefore, IRF1 expression and its translocation to the nucleus were also tested in HFFs upon IFN-β activation. Treatment with IFN-β induced translocation of IRF1 to the nucleus (Fig. 1D). IRF1 translocation to the nucleus was significantly higher in cells infected with TgIST knockout parasites compared with wild-type parasites upon IFN-β activation (Fig. 1D). In combination with previous studies, these results indicate that TgIST blocks both type I and type II IFN-induced responses in human cells.
Fig. 1.
TgIST suppresses the type I IFN response. Luminescence is reported in HeLa cells expressing GAS (A) or ISRE (B) Gaussia Luciferase reporter constructs with and without (Control) incubation with IFN-γ (A) or IFN-β (B) at 100 units/mL for 6 h. The cells were left uninfected (UI) and infected with wild-type RH or RH KO (RH strain with knockout of TgIST) for 12 h. Luminescence is expressed as fold change ± SEM, compared with uninfected and control cells from 3 independent experiments done in triplicate. There were significant differences between the compared groups (**P = 0.0002 and ***P < 0.001 using an unpaired Student’s t test). (C) Real-time PCR showing fold induction of mRNA transcripts in HFFs infected with RH or RH KO for 12 h, followed by treatment with IFN-β (100 units/mL for 6 h). Comparative cycle threshold values were used to evaluate the fold change in transcripts using YWHAZ as an internal transcript control. Data are plotted as fold change ± SEM compared with UI and untreated (Control) cells from at least 3 independent experiments per gene. There were significant differences between the compared groups (*P < 0.05 and **P < 0.01 using a multiple Student’s t test with Holm–Sidak correction). (D) Representative images showing nuclear localization of IRF1 in HFFs infected with wild-type RH or RH KO for 6 h, followed by treatment with IFN-β (100 units/mL for 12 h). UI and untreated HFFs were used as controls. The cells were stained using a mAb against IRF1 (red) and Hoechst (100 ng/mL) to label the nuclei (blue), and parasites were detected using mAb DG52 (SAG1) (green), followed by secondary antibodies. (Scale bars, 10 μm.) The bar graph shows the mean of nuclear IRF1 intensity per image (arbitrary units) ± SD of at least 150 images per sample from a representative experiment. There was a significant difference between compared groups (***P < 0.0001 using 2-way ANOVA with Tukey’s multiple comparison test). (E) Western blot analysis of TgIST-Ty immunoprecipitated from nuclear lysates of HFF cells. Cells were either left uninfected or infected with Toxoplasma (RH or RH KO, each expressing Ty-tagged TgIST) for 16 h, followed by treatment with IFN-β (150 units/mL) for 1 h. Control cells were left untreated. Different proteins were probed for their relative enrichment across samples in the immunoprecipitated fraction. Equal amounts of nuclear lysates used for immunoprecipitation are loaded alongside as nuclear input controls. IP, immunoprecipitation.
TgIST Associates with STAT1/STAT2 Heterodimers and Recruits the NuRD Complex.
Previous studies have shown that TgIST binds to phosphorylated homodimers of STAT1 that form in response to IFN-γ, and that this complex remains tightly bound to chromatin (18, 19). Type I IFNs share the requirement of STAT1, which pairs with STAT2 to form an active heterodimer that, together with IRF9, drives expression of a distinct set of genes harboring ISREs in their promoters (30). Therefore, we examined whether TgIST binds to active, phosphorylated STAT1/STAT2 heterodimers in nuclear extracts of IFN-β–treated cells. HFF cells were infected with wild-type or TgIST knockout strains for 16 h, followed by IFN-β stimulation for 1 h. Expectedly, IFN-β stimulation led to phosphorylation and nuclear translocation of both STAT1 (Y701) and STAT2 (Y690) (Fig. 1E). TgIST was immunoprecipitated with both phosphorylated STAT1 and STAT2 from IFN-β–stimulated cells. However, unlike STAT1, STAT2 was not associated with TgIST in naive cells. In addition, components of the NuRD complex, namely, HDAC1, HDAC2, RBBP7, and MTA1, were also immunoprecipitated with TgIST in cells stimulated with IFN-β. Similar to HFFs, TgIST also immunoprecipitated the NuRD complex in IFN-β–stimulated U3A cells ectopically expressing STAT1 (SI Appendix, Fig. S1). These results strongly suggest that TgIST blocks STAT1/STAT2 heterodimer-mediated transcription by recruiting the Mi2/NuRD complex, similar to what has previously been reported for STAT1 homodimers in response to type II IFN (18, 19).
Global Transcriptomic Response to IFN-β in T. gondii Infection.
To further confirm TgIST-mediated inhibition of type I IFN responses, we performed next-generation messenger RNA (mRNA) sequencing of HFFs that were infected with wild-type or TgIST knockout RH parasites, followed by IFN-β stimulation. Single 50-base pair (bp) reads were sequenced from 3 biological replicates using a HiSeq 2500 Illumina platform and mapped to the human genome using CLC Genomics Workbench version 9.5.3 to analyze reads across all samples (Dataset S1). Hierarchical cluster analysis of genes expressed at significantly different levels (SI Appendix, Fig. S2 A and B and Dataset S2) showed that genes induced by IFN-β stimulation of uninfected cells in cluster 3 (comprising 437 differentially expressed genes) were inhibited in cells infected with wild-type RH, whereas cells infected with TgIST knockout parasites maintained their expression (SI Appendix, Fig. S2C). Ingenuity pathway analysis (IPA) showed that the majority of the genes in cluster 3 were involved with IFN signaling as the top canonical pathway (SI Appendix, Fig. S2D).
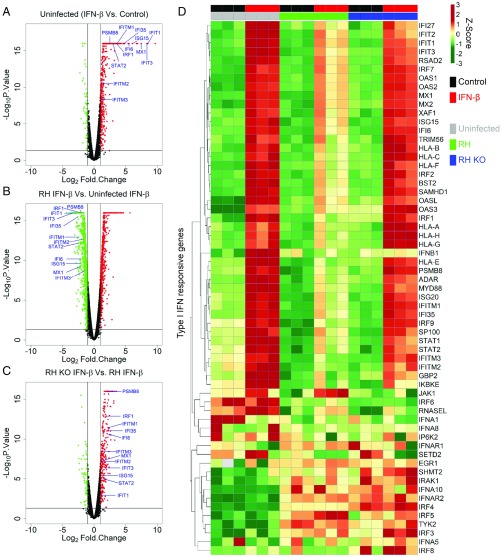
To further illustrate these differences, we compared control and IFN-β–treated samples for several pairs of samples. In uninfected cells, IFN-β treatment led to significant up-regulation of 514 genes, including many genes associated with type I IFN response (Fig. 2A). IPA of these differentially expressed genes also confirmed significant up-regulation of canonical pathways associated with IFN signaling (SI Appendix, Fig. S3A), as expected. However, infection with wild-type RH parasites led to suppression of most of the genes that were up-regulated by IFN-β in the uninfected control (Fig. 2B). In contrast, IFN-β activation of HFFs infected with TgIST knockout parasites compared with wild-type RH infection showed up-regulated expression of the ISGs seen in uninfected cells (Fig. 2C). IPA of the genes differentially regulated by knockout vs. wild-type parasites upon IFN-β activation also showed significant modulation of IFN signaling as the top canonical pathway, respectively (SI Appendix, Fig. S3 B and C). Further analysis of a subset of genes associated with type I IFN responses [Gene Ontology (GO) list: GO:0060337, GO:0071357, and GO:0004905 from QuickGO EMBL-EBI (31)] demonstrated that the majority of genes up-regulated by IFN-β were suppressed by infection with wild-type parasites and relieved in cells infected with the TgIST knockout parasites (Fig. 2D). The expression pattern of type I IFN-activated genes measured by qPCR (Fig. 1C) was also maintained in the RNA-sequencing dataset: These genes characteristically showed inhibition upon infection with wild-type parasites and rescue with TgIST knockout parasites (SI Appendix, Fig. S4A). Further upstream analysis of differentially expressed genes also showed that the vast majority (i.e., 63 of 77) of genes associated with activated STAT1 were up-regulated upon infection with TgIST knockout parasites compared with wild-type parasites in IFN-β–stimulated HFFs (SI Appendix, Table S1).
Fig. 2.
Global transcriptomic response to IFN-β in T. gondii infection. Volcano plots between fold change (Log2Fold.Change) of genes vs. significance of change (−Log10P.Value). (A) HFF cells treated with IFN-β (100 units/mL for 6 h) normalized to untreated (Control) cells. (B) HFF cells infected with wild-type RH and treated with IFN-β normalized to uninfected cells treated with IFN-β. (C) HFF cells infected with RH KO (TgIST knockout RH parasites) and treated with IFN-β normalized to cells infected with wild-type RH and treated with IFN-β. HFF cells in B and C were infected with RH or RH KO for 12 h, followed by IFN-β (100 units/mL) treatment for 6 h. Points in red and green represent significantly up-regulated (Log2Fold.Change > 1 and −Log10P.Value > 1.3) and down-regulated (Log2Fold.Change < −1 and −Log10P.Value > 1.3) genes, respectively. Blue text denotes genes responsive to type I IFNs. (D) Heatmap of gene expression across all samples with their 3 independent biological replicates was clustered using Euclidean distance and complete linkage on normalized Log2 (Total gene reads). Normalized Z-scores are color-scaled from green to red showing relative down-regulation to up-regulation. Uninfected, RH-infected, and RH KO-infected HFFs are represented in gray, green, and blue, respectively. Samples with control and IFN-β treatment are represented in black and red, respectively.
In order to further characterize genes that were modulated by TgIST, we also analyzed IFN-responsive genes that are known to have a role in host defense. TgIST also regulated expression of most of the genes that were up-regulated by IFN-β and associated with cellular defense against pathogens (32) (SI Appendix, Fig. S4B). Included in this set of genes are the GBPs that were induced by IFN-β stimulation of the HFFs, with GBP1 showing up-regulation of up to 15,000-fold compared with control (SI Appendix, Fig. S4B). In addition to GBPs, a number of the IFN regulatory factors (IRFs) and tripartite motif-containing (TRIM) proteins were also up-regulated by IFN-β stimulation.
Previous studies have emphasized the importance of tonic gene expression, which is detected in the type I IFN pathway even in the absence of added stimuli (3). We also examined the expression of genes in the type I pathway in the absence of IFN treatment to determine the effect of TgIST on tonic gene expression. IPA of all 315 differentially regulated transcripts also showed IFN signaling was the most enriched canonical pathway that was up-regulated in HFF cells infected with TgIST knockout parasites compared with wild-type parasites (SI Appendix, Fig. S3D). In the absence of added IFN-β, 17 of the 22 STAT1-associated transcripts were up-regulated upon infection with the TgIST knockout parasites compared with wild-type parasites (SI Appendix, Table S2). This finding suggests that infection with wild-type parasites suppresses these tonic genes, even in the absence of induced signaling. Taken together, these results demonstrate that TgIST suppresses tonic type I IFN-mediated transcription in addition to blocking the induction of gene expression following addition of IFN-β, thus affecting both basal pathways and those that are up-regulated for host defense. Although type I IFN-responsive genes comprise the majority of genes that were differentially regulated by TgIST, there were also genes associated with other pathways altered in the absence of this effector (SI Appendix, Fig. S3 C and D), suggesting it may modulate additional signaling pathways.
Type I IFN Suppresses Growth of TgIST-Deficient Parasites.
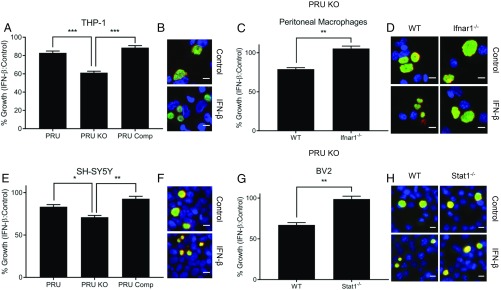
Having established that TgIST inhibits the type I IFN response, we then explored its importance for T. gondii infection. IFN-β has been shown to inhibit growth of T. gondii parasites in human retinal epithelial cells (33) and mouse macrophages (34), suggesting that the down-modulation of this pathway by TgIST may be important for parasite survival. To evaluate the potential for growth restriction, we used an assay that monitors the size of the parasitophorous vacuole (SAG1- and GRA7-positive vacuoles) using plate-based automated microscopy (SI Appendix, Fig. S5A). Vacuolar growth was measured as mean area of the parasitophorous vacuoles per image at 36 h postinfection with type I RH infection and 40 h postinfection with type II PRU infection (SI Appendix, Fig. S5 B–J). However, none of the wild-type, TgIST knockout, or TgIST-complemented strains on the RH background showed growth inhibition in human THP-1 macrophages activated with IFN-β (SI Appendix, Fig. S5B). Since type I strains are equipped with additional mechanisms to block downstream IFN-mediated effectors (11), we used a TgIST knockout mutant in the type II PRU strain, which was previously shown to have increased susceptibility to IFN-γ–mediated inhibition (19).
Human THP-1 macrophages showed significant growth inhibition of TgIST knockout parasites in this background compared with wild-type parasites and TgIST-complemented PRU parasites upon activation with human IFN-β (hIFN-β) plus human tumor necrosis factor-α (hTNF-α) (Fig. 3 A and B). Activation with mouse IFN-β (mIFN-β) plus mouse TNF-α (mTNF-α) also led to significant growth inhibition of TgIST knockout parasites in mouse peritoneal macrophages, and this effect was reversed in cells from mice lacking IFN-α/β receptors (Ifnar1−/−), confirming specificity of the type I IFN response in controlling parasite growth (Fig. 3 C and D). We also examined type I IFN control in human SH-SY5Y cells, which are often used to study human neuronal biology (35), and in mouse microglial BV2 cells, which are used for studying macrophage function in the CNS (36). Human SH-SY5Y neuroblastoma cells showed significant growth inhibition of TgIST knockout parasites compared with wild-type parasites and complemented parasites upon activation with hIFN-β plus hTNF-α (Fig. 3 E and F). Incubation with mIFN-β plus mTNF-α also led to significant growth inhibition of TgIST knockout parasites in BV2 cells, and this was abrogated in cells lacking Stat1 (Fig. 3 G and H). Importantly, phorbol 12-myristate 13-acetate (PMA)–differentiated THP-1 and SHSY-5Y cells treated with IFN-β plus TNF-α also showed growth inhibition of wild-type and complemented parasites, albeit to a lower level than that of TgIST knockout parasites (Fig. 3 A and C). In mouse cells, we did not observe a significant inhibition of replication of wild-type parasites treated with IFN-β; however, TgIST knockout parasites showed significant growth inhibition (SI Appendix, Fig. S5 G–J).
Fig. 3.
Type I IFN-mediated growth suppression of TgIST-deficient parasites. Comparison of vacuolar growth in control or IFN-β–treated cells of various types infected with type II wild-type parasites (PRU), ∆Tgist knockout (PRU KO), or complemented lines (PRU Comp). Differentiated THP-1 human macrophages (A and B) and SH-SY5Y human neuroblastoma cells (E and F) are shown. ∆Tgist knockout (PRU KO)-infected (C and D), wild-type (WT), Ifnar1−/− thioglycolate-elicited peritoneal macrophages (G and H), mouse microglial WT, and Stat1−/− BV2 cells are shown. Cells were infected for 2 h and then treated with TNF-α (10 ng/mL; Control) alone or in combination with IFN-β (100 units/mL) during 40 h of infection. Growth or mean vacuolar size per image of parasites upon IFN-β treatment relative to control is plotted as % Growth ± SEM of at least 3 independent replicates with at least 50 images per replicate and sample. There were significant differences between the compared samples (*P < 0.05, **P < 0.01, and ***P < 0.001 using 1-way ANOVA in A and E and an unpaired Student’s t test in C and G). Representative images of vacuolar growth of PRU KO parasites in THP-1, peritoneal macrophages, SH-SY5Y, and BV2 cells are shown in B, D, F, and H, respectively. Parasites were stained with mouse mAb anti-SAG1 (green), and the vacuolar membrane was stained with Pc rabbit anti-GRA7 (red) and detected with appropriately conjugated secondary antibodies. Nuclei were stained with Hoechst (100 ng/mL). (Scale bars, 10 μm.)
Taken together, our results further demonstrate that growth inhibition by IFN-β relies on Ifnar receptors and the STAT1 transcription factor, which are required for the type I IFN response. These findings are consistent with a role for type I IFN in restricting the growth of T. gondii in human and mouse cells and with TgIST abrogating this effect.
Type I IFN Controls In Vivo Expansion of Acute and Chronic Toxoplasma Infection.
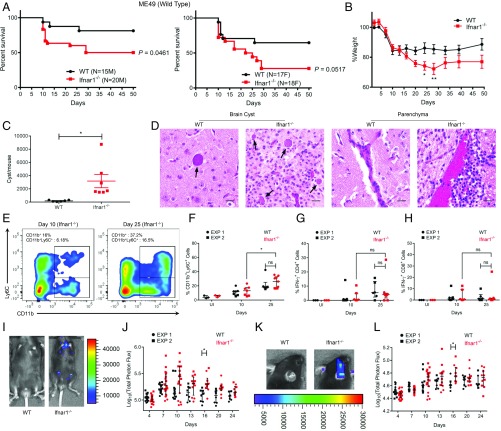
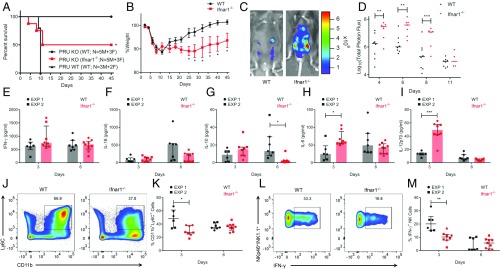
A previous study reported a modest role for type I IFN in controlling T. gondii infection in mice (29). However, the contribution of type I IFN is likely masked by the ability of TgIST to block the pathway, as indicated by the findings above. With this possibility in mind, we reexamined the role of type I IFN signaling in control of T. gondii infection using type II parasite strains of intermediate virulence, which allow comparison of both acute and chronic infection. We infected mice with ME49 tachyzoites by intraperitoneal (i.p.) inoculation and measured IFN-α and IFN-β mRNAs during the acute phase of infection. Peritoneal cells of infected mice showed significant up-regulation of IFN-β mRNA at 6 d postinfection, although only modest changes in IFN-α were observed (SI Appendix, Fig. S6A). Both male and female Ifnar1−/− mice on C57BL/6 background showed significantly higher susceptibility to oral challenge with tissue cysts of wild-type II ME49 strain parasites when compared with wild-type mice (Fig. 4A). During the acute phase of infection, Ifnar1−/− mice showed a significant increase in weight loss compared with wild-type mice, suggesting an overall lower ability to control T. gondii infection in the absence of type I IFN signaling (Fig. 4B). Mice that survived ME49 infection were killed 50 d postinfection, and their brains were harvested to measure cyst burden. Interestingly, survivors from the Ifnar1−/− group showed a significantly higher cyst burden than the wild-type group (Fig. 4C). Both IFN-α and IFN-β mRNAs were also up-regulated in brain tissue of wild-type and Ifnar1−/− mice infected with ME49 cysts compared with uninfected mice (SI Appendix, Fig. S6 B and C). Additionally, histopathological features in the surviving wild-type and Ifnar1−/− mice showed minimal to mild meningitis with perivascular accumulation of lymphocytes and macrophages. Both of the groups also showed perivascular cuffing by lymphocytes and macrophages in the brain consistent with chronic toxoplasmosis. However, the brain sections of Ifnar1−/− mice stained with hematoxylin and eosin showed a higher cyst burden and increased cellularity due to gliosis, with a higher presence of microglial nodules and focal necrosis. Ifnar1−/− mice also showed a higher density of inflammatory cell accumulation compared with wild-type mice in the brain parenchyma (Fig. 4D).
Fig. 4.
Type I IFN controls in vivo expansion of acute and chronic Toxoplasma infection. (A) Kaplan–Meier curve showing survival of wild-type (n = 15 male [M] and 17 female [F]) and Ifnar1−/− (n = 20 M and 18 F) mice infected orally with 5 cysts of the type II ME49 line expressing firefly luciferase (ME49/FLUC) strain. Cumulative data from 3 independent replicates are shown separately for M and F mice. Statistical difference between wild-type and Ifnar1−/− mice was calculated using a log-rank Mantel–Cox test. (B) Percent weight of mice infected compared with average weight of wild-type mice before infection (0 d) is plotted as mean ± SEM from 2 independent experiments (*P < 0.05, using a multiple Student’s t test with Holm–Sidak correction). (C) Cyst burden in surviving mice is plotted as mean ± SEM (*P = 0.0197, Student’s t test). (D) Histopathological fields showing cyst burden and brain parenchyma. Arrows denote tissue cysts. (Scale bars, 20 μm.) (E) Representative fluorescence-activated cell sorting plot showing CD11b+ and CD11b+Ly6C+ cells in Ifnar1−/− mice at 10 d and 25 d postinfection. (F) Percentages of CD11b+Ly6C+ cells after gating on CD45+CD19−CD3− (hematopoietic, non-B/T cells) brain mononuclear cells from 2 independent replicates. Each point represents 1 mouse. EXP, experiment; ns, not significant. Percentages of IFN-γ+ CD4+ T cells (G) and IFN-γ+ CD8+ T cells (H) are shown after gating on a parent population of live CD45+CD19−CD3+CD4/8+ brain mononuclear cells from uninfected or ME49 infected wild-type and Ifnar1−/− mice at 10 d and 25 d postinfection. Bioluminescent imaging of ME49/FLUC infection in the peritoneum (I) and cranium (K) of wild-type and Ifnar1−/− mice is shown. Representative images show radiance from respective sites at day 16. The scatter plot shows total peritoneal (J) and cranial (L) radiance in each mouse across different time points up to 24 d postinfection. Total photon flux is log-transformed and plotted as median ± interquartile range. There was a significant difference between compared groups (*P < 0.05 using a multiple Student’s t test with Holm–Sidak correction). The number of animals per group is indicated.
We also examined the infiltration of leukocytes, and their production of cytokines, in the brain at 10 and 25 d postinfection. There was a significant increase in both the percentage and total number of CD45+ hematopoietic cells in the brain of infected mice at 25 d compared with 10 d postinfection (SI Appendix, Fig. S6 D–F). There was also a significant increase in the percentage and total number of CD11b+Ly6C+ inflammatory monocytes in brains of infected mice at 25 d compared with day 10 postinfection (Fig. 4E and SI Appendix, Fig. S6G). We also measured IFN-γ–secreting CD4+ and CD8+ T cells, which are important in controlling Toxoplasma infection during the chronic phase (37). Although higher cell numbers were consistently seen at later time points, there was no significant difference in the percentages (Fig. 4 G and H) or total numbers (SI Appendix, Fig. S6 H and I) of IFN-γ+ CD4+, or IFN-γ+ CD8+ T cells between wild-type and Ifnar1−/− mice at 10 d or 25 d postinfection. It is important to note that we did not observe any difference in the percentages or numbers of CD11b+Ly6C+ inflammatory monocytes, IFN-γ+ CD4+ cells, and IFN-γ+ CD8+ T cells between wild-type and Ifnar1−/−-infected mice at either time point. There was also no significant difference in serum levels of interleukin-10 (IL-10), IL-6, IL-18, and IL-12p70 apart from IFN-γ, which showed a decrease in Ifnar1−/− mice compared with wild-type mice at 10 d postinfection (SI Appendix, Fig. S6 J–N). In parallel studies, bioluminescence imaging of cyst infection resulted in significant increases in parasite burden at day 16 postinfection in Ifnar1−/− mice compared with wild-type mice (Fig. 4 I and J). Higher parasite numbers in Ifnar1−/− mice were also evident from images taken of the cranial region at day 16 postinfection (Fig. 4 K and L).
The ∆Tgist knockout in the type II background is attenuated in vivo, but at a higher inoculum, it can cause mortality in mice (18). To test the importance of TgIST in mediating the susceptibility to type I IFN in vivo, we reasoned that the loss of the type I pathway should partially offset the virulence defect of TgIST knockouts. To test this hypothesis, we used a high dose of ∆Tgist knockout PRU parasites to infect Ifnar1−/− mice by tachyzoite injection. Although avirulent in wild-type mice, infection with 105 ∆Tgist knockout PRU strain parasites led to a significant increase in the susceptibility of Ifnar1−/− mice (Fig. 5A). At the same dose, i.p. infection with PRU wild-type (WT) strain resulted in the death of all mice within 10 d (Fig. 5A). Ifnar1−/− mice challenged with ∆Tgist knockout parasites also showed increased weight loss compared with wild-type infection (Fig. 5B), suggesting a relatively higher parasite burden. Consistent with this prediction, bioluminescence imaging showed a significantly higher parasite (∆Tgist PRU KO parasites) burden during the initial phase of acute infection in Ifnar1−/− mice compared with wild-type mice (Fig. 5 C and D). Serum levels of different cytokines were measured to ascertain whether increased expansion of parasites in Ifnar1−/− mice correlates with changes in systemic immune responses. Somewhat surprisingly, there was no significant difference in serum levels of IFN-γ and IL-18 at day 3 or 6 postinfection with ∆Tgist knockout parasites (Fig. 5 E and F). However, there was a significant decrease in the serum levels of antiinflammatory IL-10 at day 6 postinfection in Ifnar1−/− mice compared with wild-type mice (Fig. 5G). Additionally, there was a significant increase in IL-6 and IL-12p70 serum levels at day 3 postinfection in Ifnar1−/− mice compared with wild-type mice (Fig. 5 H and I). These findings suggest that enhanced expansion of ∆Tgist knockout parasites in Ifnar1−/− mice is not due to a global impairment in immune responsiveness.
Fig. 5.
Susceptibility of Ifnar1−/− mice to TgIST knockout type II PRU T. gondii. Analysis of C57BL/6J wild-type (n = 8) and Ifnar1−/− (n = 8) mice infected i.p. with 105 PRU KO or PRU WT (n = 5) tachyzoites per mouse and monitored for 45 d. (A) Kaplan–Meier curve showing survival of both groups of mice. There was a significant difference at between PRU KO-infected wild-type and Ifnar1−/− mice (*P = 0.0256 using a log-rank Mantel–Cox test). (B) Percent weight of mice infected compared with average weight before infection (0 d). (C) Bioluminescence imaging of luciferase-expressing PRU KO tachyzoite infection in the peritoneum of wild-type (WT) and Ifnar1−/− mice. Representative images of the radiance from the peritoneum at day 6 are shown. (D) Scatter plot shows total peritoneal radiance in each mouse across different time points up to 11 d postinfection. Total photon flux is log-transformed, with the dash representing the median. (E–I) Serum levels of IFN-γ, IL-18, IL-10, IL-6, and IL-12p70 in wild-type and Ifnar1−/− mice at days 3 and 6 postinfection with PRU KO. EXP, experiment. (J) Representative plot of inflammatory monocytes (CD11b+Ly6C+) in wild-type and Ifnar1−/− mice at day 3 postinfection. (K) Percentages of CD11b+Ly6C+ cells after gating on CD45+CD19− (hematopoietic, non-B cells) peritoneal cells in wild-type and Ifnar1−/− mice at days 3 and 6 postinfection. (L) Representative plot of intracellular IFN-γ in NK cells (NK1.1+NKp46+) at 3 d postinfection. (M) Quantitative percentage of IFN-γ+ NK cells after gating on CD45+CD19−CD3− (hematopoietic, non-B/T cells) peritoneal cells in wild-type and Ifnar1−/− mice at days 3 and 6 postinfection. Cumulative data from 2 independent experiments are shown in E–I, K, and M. The data in E–I, K, and M are plotted as median ± interquartile range. (B–G) There were significant differences between compared groups (*P < 0.05, **P < 0.01, and ***P < 0.005) using a multiple Student’s t test with Holm–Sidak correction.
Previous studies have reported that type I IFN is required for optimal production of IFN-γ by natural killer (NK) cells in vitro (38), and this pathway is important for recruitment of protective inflammatory monocytes to the peritoneum (39). To determine whether the increase in parasitemia in the absence of type I IFN signaling might be due to a local defect in type II IFN production at the site of infection, we measured the recruitment of inflammatory monocytes to the site of infection. Ifnar1−/− mice showed significantly lower recruitment of CD11b+Ly6C+ inflammatory monocytes to the peritoneal cavity at 3 d postinfection with ∆Tgist PRU parasites compared with wild-type mice (Fig. 5 J and K and SI Appendix, Fig. S7A). Similarly, there was a significant decrease in recruitment of both IFN-γ–producing NK cells (Fig. 5 L and M and SI Appendix, Fig. S7B) and total NK cells (SI Appendix, Fig. S7C) at 3 d postinfection in Ifnar1−/− mice compared with wild-type mice. Concomitantly, there was an increase in neutrophils in Ifnar1−/− mice compared with wild-type mice at 3 and 6 d postinfection (SI Appendix, Fig. S7 D and E). There were no differences in recruitment of other IFN-γ–producing CD4+ and CD8+ T cells (SI Appendix, Fig. S7 F–I) or in CD11b+CD11c+MHCII+ dendritic cells and F4/80+Ly6C− resident macrophages (SI Appendix, Fig. S7 J and K). Collectively, these results suggest that impaired control of acute T. gondii infection in Ifnar1−/− mice is partially due to a local defect in induction of the normally protective type II IFN.
Discussion
Type I (α, β, and λ) (22) and type II (γ) (10) IFNs act through STAT factors (1, 2) to up-regulate expression of defense mechanisms that are important in the control of microbial pathogens. Previous studies have shown that T. gondii secretes an effector called TgIST that translocates to the host nucleus and blocks induction of IFN-γ–activated genes, thereby promoting parasite survival (18, 19). Because STAT1 is common to both type I and type II IFN signaling pathways, we sought evidence that TgIST might also control type I IFN responses, and we also investigated whether this pathway plays a role in controlling infection. Like IFN-γ–activated genes, TgIST was also responsible for globally suppressing genes normally up-regulated by type I IFN-β. TgIST was associated with both phosphorylated STAT1 and STAT2 in the nucleus and also recruited the NuRD complex in response to IFN-β stimulation, suggesting it acts by a common means to block both type I and type II IFNs. Addition of IFN-β stimulation led to growth suppression of TgIST knockout parasites in human and mouse macrophages. Ifnar1−/− mice that lack type I IFN responses were more susceptible to type II T. gondii infection during both acute and chronic phases, a result attributable to impaired type II IFN responses as well as loss of cell intrinsic control. Collectively, these studies reveal a common mechanism whereby T. gondii blocks STAT1-mediated type I and type II IFN signaling to enhance its survival, including in the CNS during chronic infection.
STAT1 homodimers, which form in response to type II IFN-γ activation, and ISGF3 complexes, which form in response to type I IFN-α/β activation, translocate to the nucleus and bind to GASs containing the TTCN2–4GAA motif or ISREs containing the YAGTTTC(A/T)YTTTYCC motif, respectively (30). These 2 pathways induce the expression of a largely overlapping set of ISGs, and yet type I IFN is generally considered antiviral, while type II IFN acts primarily to restrict intracellular bacterial and protozoal pathogens (10, 22). Stimulation of T. gondii-infected cells with IFN-γ or IFN-β increases binding of phosphorylated STAT1 to chromatin, and yet the transcriptional activity from both pathways is blocked in infected cells (16). It has previously been shown that the parasite protein TgIST binds to activated STAT1 homodimers on the chromatin and blocks transcription induced by IFN-γ (18, 19). We have extended these findings by showing that TgIST also binds to STAT1/STAT2 heterodimers and blocks transcription induced by IFN-β. TgIST recruits a repressive complex called the NuRD complex, which is known for its role in chromatin modification and gene silencing (20, 21), to both transcription complexes, suggesting they share a common mechanism of inhibition. Although its precise role has not been delineated, the NuRD complex may act to repress transcription by modifying chromatin through its deacetylation and adenosinetriphosphatase activities.
TgIST inhibits STAT1-mediated transcription at a genome-wide level in HFF cells activated by IFN-γ (18). Similarly, our findings indicate that TgIST blocks STAT1/STAT2-mediated gene expression induced by IFN-β. This pattern of repression included canonical type I IFN genes like IFIT1, ISG-15, and MX1, as well as transcription enhancers like IRF1, IRF2, IRF7, and IRF9. Although, IRF1 induction is more commonly associated with IFN-γ stimulation of host cells, treatment with IFN-β has also been shown to induce its expression (40, 41). Our biochemical studies revealed that TgIST also binds to and enhances phospho-STAT1 and the NuRD complex components in naive cells without IFN-β stimulation. Consistent with this, TgIST was responsible for repression of STAT1-associated tonic transcription of type I responses in infected cells. Previous studies have emphasized the importance of tonic IFN expression for control of latent or chronic viral infection (3), which may be relevant to the observed increase in tissue cyst burden in Ifnar1−/− mice infected with T. gondii, as described below. Type III or λ-IFNs signal through the Ifnlr, which also activates STAT1/STAT2-dependent gene expression (42); hence, it will be interesting to determine whether TgIST also augments pathways controlled by these receptors.
IFN-β induces growth restriction of Toxoplasma in retinal epithelial cells (33), mouse peritoneal macrophages (34), and human monocyte-derived macrophages (27). In the majority of these studies, the degree of growth impairment achieved with treatment of IFN-β is much less substantial than that observed with IFN-γ, consistent with a major role for the latter pathway in control of infection in mice (7, 8). Our findings are consistent with this pattern and extend the repertoire of cell types to include mouse microglial cells that constitute tissue macrophages in the CNS, human THP-1 monocytes, and human neuroblastoma cells. Importantly, the growth-inhibitory effects of IFN-β were enhanced in ∆Tgist knockout parasites on the type II PRU background, consistent with the finding that TgIST suppresses the induction of genes normally activated by IFN-β. Although treatment with IFN-β was able to restrict growth in multiple cell types, it did not lead to vacuolar destruction or clearance of intracellular parasites, and thus differs from control mechanisms in mouse cells that are induced by IFN-γ. IFN-β induces many molecules known to be involved in controlling intracellular infection by viral pathogens (32, 43), although the roles of such effectors in control of intracellular growth of T. gondii have not been examined. Candidate ISGs that might explain the growth restriction in IFN-β–treated cells include effectors such as GBPs (44, 45), TRIM21 (46), and ISG-15 (47). Regardless of the exact mechanism, growth inhibition in neuronal cell types (i.e., BV2 cells, SH-SY5Y cells) suggests that IFN-β–mediated control of T. gondii infection may also be important in the CNS, which normally harbors the chronic cyst form of toxoplasmosis.
Initial studies with Toxoplasma infection in mice showed the protective effect of recombinant IFN-β treatment (28). Separately, another study showed that absence of type I IFN production by monocytes results in a modest increase in mortality of Ifnar1−/− male mice (29). In contrast, we observed significant increases in the mortality of both male and female Ifnar1−/− mice, indicating that type I IFN signaling is required for control of toxoplasmosis in both sexes. Another prior study (48) also failed to find a substantial role for type I IFN, but those investigators only focused on the acute phase of infection, where type II IFN predominates. Our finding that type I IFN is important in control of T. gondii may arise from the lower dose of ME49 cysts used in the present experiment, and hence lower mortality in wild-type mice during the acute phase of the infection, when compared with the prior study (29). The defect in Ifnar1−/− mice becomes apparent later in infection, consistent with its presentation in the CNS during chronic infection. The enhanced susceptibility of Ifnar1−/− mice may result from impaired growth restriction by infected cells, given that this phenotype is evident in multiple cell types, including macrophages from both the periphery and CNS.
Apart from direct growth-inhibitory effect on infected cells, type I IFN signaling in vivo has also been shown to affect recruitment of IFN-γ–secreting and/or immune cells that control infection. Previous in vitro studies have shown that NK cell production of IFN-γ is dependent on type I IFN signaling (38). The early activation of NK cells, as well as their production of IFN-γ, has also been linked to the recruitment and activation of inflammatory monocytes to the peritoneal cavity (39). Our studies extend these findings by demonstrating that in T. gondii-infected mice lacking type I IFN signaling, NK cell recruitment and induction of IFN-γ are impaired. Inflammatory monocytes, but not neutrophils, are critical to control acute infection with T. gondii (49, 50). Hence, the diminished production of IFN-γ by NK cells in Ifnar1−/− mice, and corresponding decrease in recruitment of inflammatory monocytes, likely contributes to decreased control at the local site of infection. The apparent decrease in Ly6C+CD11b+ monocytes in Ifnar1−/− mice might also reflect diminished expression of this receptor, which has been shown to be induced by type I IFN in T cells (51); however, the defect was only seen at day 3, and not beyond. Reports from other infection models also support the idea that recruitment of inflammatory monocytes is impaired in Ifnar1−/− mice (52, 53). Regardless, these differences in cellular recruitment did not result in a systemic decrease in IFN-γ levels, and proinflammatory cytokines IL-6 and IL-12 p70 actually increased in the serum of ∆Tgist-infected Ifnar1−/− mice compared with control mice. Taken together, these findings suggest that the decreased control of infection at later time points is not simply due to impaired immunity at early time points, but rather results from a defect within the CNS.
Following control of the acute phase, Toxoplasma differentiates from fast-replicating tachyzoites into slow-replicating bradyzoites, a process that occurs primarily in long-lived differentiated cells like those in the brain and skeletal muscles of the infected animal. Recently, neurons have been identified to be the primary target for Toxoplasma infection in the CNS (54). However, unlike macrophages and mouse embryonic fibroblasts, primary mouse neurons do not show growth inhibition or clearance of parasites upon IFN-γ stimulation in vitro (55). Mouse neurons show delayed or dampened responses to IFN-γ (56), which may be a protective response, given the propensity of type II IFN to inhibit cell growth, and even induce apoptosis in some cases (57). The ability of IFN-β to suppress growth in human SH-SY5Y neuroblastoma cells and mouse microglial BV2 cells in vitro suggests that local control within the CNS may rely instead on type I IFN. The higher CNS burden in Ifnar1−/− mice, despite normal systemic responses during acute infection, suggests that lack of type I IFN signaling in the CNS is responsible for increased susceptibility during the chronic stage of infection.
Our findings extend the role of type I IFNs in controlling growth of intracellular pathogens based on several lines of evidence. First, although type II IFN plays a major role in resistance to T. gondii, absence of type I IFN signaling leads to increased mortality and higher cyst burdens in the CNS. Second, direct treatment with IFN-β can induce restriction of T. gondii growth in multiple cell types from mice and humans. Whether this results from a unique type I ISG-mediated effect or common ISGs that are induced by both type I and type II IFN is uncertain. Regardless of the underlying mechanisms, our studies make clear that optimal control of infection requires expression of both type I and type II signaling pathways. Finally, the importance of the type I pathway is underscored by the fact that the parasite has evolved an efficient mechanism to block signaling mediated by IFN-β. The secretory protein TgIST binds to both STAT1 homodimers and STAT1/STAT2 heterodimers, recruits the NuRD complex, and suppresses gene expression normally induced by both pathways. Our studies elucidate the mechanism by which type I IFN responses contribute to antiparasitic activity against an intracellular pathogen and identify a strategy by which the parasite effectively evades this control mechanism.
Materials and Methods
Reagents and Antibodies.
d-Luciferin, PMA, and thioglycolate medium were obtained from Sigma-Aldrich. The hIFN-γ, hTNF-α, mTNF-α, hIFN-β, and mIFN-β were obtained from R&D Systems. T. gondii parasites were stained with mouse monoclonal antibody (mAb) DG52 against the surface antigen SAG1 (58). GRA7 was detected using a rabbit Pc serum described previously (59). Ty-tagged TgIST was immunoprecipitated with mouse mAb BB2 (60). Mouse monoclonal anti-HDAC1 (10E2, no. 5356) antibody; rabbit polyclonal antibodies against STAT1 (no. 9172), STAT2 (no. 4594), and RBB7 (no. 6882); and rabbit mAbs against MTA1 (D17G10; no. 5646), IRF1 (D5E4, no. 8478), phospho-STAT1(Tyr701) (58D6, no. 9167), and phospho-STAT2 (Tyr690) (D3P2P, no. 88410) were obtained from Cell Signaling Technologies. Rabbit polyclonal anti-HDAC2 (sc-7899) antibody was obtained from Santa Cruz Biotechnology. Goat polyclonal anti-TATA binding protein (TBP; ab134575) antibody was obtained from Abcam. Secondary anti-immunoglobulin G (IgG) conjugated to IRDye800 and IRDye700 were obtained from Li-Cor Biosciences. Hoechst, goat anti-mouse IgG, and goat anti-rabbit IgG secondary antibodies conjugated to Alexa 488 or Alexa 594 were obtained from Life Technologies Corporation. Fluorescein isothiocyanate (FITC)-conjugated Dolichos biflorus lectin (DBL) was obtained from Vector Laboratories, Inc. Flow cytometry antibodies were obtained from BioLegend, Inc. Anti–IFN-γ antibody and Brefeldin-A were obtained from Thermo Fisher Scientific.
Parasite Strains.
Wild-type, TgIST knockout, and complemented lines on the type I RH and type II PRU backgrounds were generated previously (19). Parasites were grown as tachyzoites in HFFs (obtained from the laboratory of John Boothroyd, Stanford University, Stanford, CA), as described previously (61). Parasites were harvested shortly after natural egress and purified by passage through a 20-gauge needle and separated from host cell debris using 3.0-μm polycarbonate filters (Whatman). All strains and host cell lines were determined to be mycoplasma-negative using the e-Myco Plus Kit (Intron Biotechnology).
For generating tissue cysts for oral challenge, C57BL/6 or CBA/J mice were infected with ME49 (American Type Culture Collection [ATCC] 50611) or a ME49 line expressing firefly luciferase (ME49/FLUC) (62). The mice were then treated with 0.2 mg/L sulfadiazine in drinking water from day 6 to day 12 postinfection. The mice were then killed 4 to 6 wk postinfection, and cysts were isolated from infected brain as described previously (63).
Mammalian Culture.
HeLa cells stably expressing 5×-GAS-Gaussia Luciferase were generated and maintained as described previously (64). We generated an 11×-ISRE-Gaussia Luciferase reporter line by replacing the GAS tandem repeat element (TRE) of the pGLUC-5×-GAS-mCMV-neoR vector with an IFN-α/β–inducible ISRE TRE using the primers forward 5′-TAGTTTCACTTTCCCTAGTTTCACTTTCCCTAGTTTCACTTTCCCTAGTTTCACTTTCCCTAGTTTCACTTTCCCACTAGTTAGGCGTGTACGGT-3′ and reverse 5′-AGATCTCGATCCTCTACGCC-3′ in a PCR-based mutagenesis strategy (Q5 Site Directed Mutagenesis; New England Biolabs). After transfection into HeLa cells with Lipofectamine LTX (Thermo Fisher Scientific), stable colonies were selected under G418 (Sigma-Aldrich) and tested for activation of Gaussia Luciferase activity by type I IFN. Cells were maintained in Minimal Essential Medium (MEM) with 10% (vol/vol) fetal bovine serum (FBS) at 5% CO2 and 37 °C. U3A cells ectopically expressing STAT1 (U3A-STAT1) were generated and maintained as described previously (19). HFFs, THP-1 (ATCC TIB-202), and SH-SY5Y (ATCC CRL-2266) cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM), RPMI-1640, and a 1:1 mix of Eagle's MEM and F-12 medium, respectively, with 10% (vol/vol) FBS at 5% CO2 and 37 °C. THP-1 and SHSY-5Y cells were first differentiated with 50 nM PMA for 48 h. PMA was washed off, and cells were further incubated in their maintenance media for 24 h before infection with T. gondii tachyzoites. BV2 and Stat1−/− BV2 cells were maintained in DMEM with 10% (vol/vol) FBS at 5% CO2 and 37 °C, and were a kind gift from the laboratory of Herbert Virgin, Washington University School of Medicine in St. Louis. Thioglycolate-elicited peritoneal macrophages from wild-type and Ifnar1−/− mice were isolated and maintained as described previously (65).
IFN-Activated Luciferase Reporter Assay.
HeLa cells stably expressing 5×-GAS-Gaussia Luciferase or 11×-ISRE-Gaussia Luciferase reporters were infected with parasites for 12 h. The cells were then treated with 100 units/mL hIFN-γ or hIFN-β for another 6 h. The cells were then washed and assayed for expression of Gaussian Luciferase in cell lysates using a BioLux Gaussia Luciferase Assay Kit (New England BioLabs) as per the manufacturer’s instructions.
IRF1 Nuclear Translocation by Immunofluorescence.
HFFs were infected with parasites for 6 h. The cells were then treated with 100 units/mL hIFN-β for another 12 h. The cells were fixed and stained for parasites (anti-SAG1), host nuclei (Hoechst; 100 ng/mL), and IRF1 (anti-IRF1), followed by Alexa-conjugated secondary antibodies. Images were acquired at a magnification of 20× on a Cytation3 Multi-Mode plate-based imager (Biotek), and the IRF1-stained intensity per nuclei of all samples was determined using CellProfiler 2.1.1. Briefly, nuclei in each sample were identified as primary objects, and mean IRF1 intensity in those was measured as the nuclear IRF1 level. IRF1-stained images were first illumination-corrected before measurement of channel intensity in the nuclei.
Real-Time PCR.
Samples were lysed, and RNA was extracted using a QIAGEN RNeasy Mini Kit per the manufacturer’s instructions. For RNA isolation from brain tissues, uninfected controls and ME49-infected mice were first perfused with phosphate-buffered saline (PBS), followed by removal of half of the brain tissue. The tissue was then sectioned into slices <0.5 cm in width and kept in RNAlater reagents at −20 °C until processing using a QIAGEN RNeasy Midi Kit. Complementary DNA (cDNA) was prepared using a Bio-Rad iScript cDNA Synthesis Kit (Bio-Rad Laboratories, Inc.) as per the manufacturer’s instructions. Real-time PCR was performed using Clontech SYBR Advantage qPCR premix (Takara Bio USA, Inc.) as per the manufacturer’s instructions. Data acquisition was done in QuantStudio3 (Applied Biosystems) and analyzed using QuantStudio Design and Analysis Software (Applied Biosystems). Primers are listed in SI Appendix, Table S3. Comparative cycle threshold values were used to evaluate fold change in transcripts using the YWHAZ gene for human genes and β-Actin for mouse genes as an internal transcript control.
Immunoprecipitation and Western Blotting.
Immunoprecipitation experiments were performed as previously described (19). Briefly, HFF cells or U3A cells expressing STAT1 (U3A-STAT1) were left uninfected or infected with parasites for 16 h and treated with 150 units/mL IFN-β for 1 h. Control cells were left untreated. Nuclear extracts were prepared using a NE-PER Nuclear and Cytoplasmic Extraction Reagents Kit (Thermo Fisher Scientific). Anti-Ty antibody targeting to TgIST-Ty was added to nuclear extracts and incubated at 4 °C for 2 h. Prewashed Protein G Dynabeads (Thermo Fisher Scientific) were then added to the mix and incubated overnight at 4 °C. Samples were eluted after 3 washes with PBS, separated using 10 to 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis, and transferred onto a nitrocellulose membrane. The membrane was blocked using 5% nonfat dried milk diluted in Tris-buffered saline with 0.05% (vol/vol) Tween-20 (TBST) and probed with primary antibodies overnight at 4 °C, followed by 3 washes with TBST. The blot was then incubated with a 1:1,000 dilution of secondary antibodies conjugated to IRDye 700CW or IRDye 800CW for 1 h, followed by 3 washes with TBST. Blots were imaged using an ODYSSEY infrared imager.
Next-Generation mRNA Sequencing and Analysis.
HFFs were cultured without infection, infected with parasites for 12 h following treatment with hIFN-β (100 units/mL), or left untreated (control) for an additional 6 h. Samples were lysed, and RNA was extracted using a QIAGEN RNeasy Mini Kit (QIAGEN, Inc.) as per the manufacturer’s instructions. Total RNA was then submitted to the Genome Technology Access Center, Washington University School of Medicine in St. Louis, for next-generation mRNA sequencing. Briefly, mRNA was extracted from the total RNA using Oligo-dT beads binding poly-A-tail. The mRNA was then fragmented and reverse-transcribed to cDNA using random primers, followed by the addition of sample specific adaptors. A total of 544,234,059 read sequences were generated from 3 independent replicates sequenced across 2 lanes of a single flow cell on an Illumina HiSeq 2500 platform with 1 × 50-bp single-end reads. The base calling was performed with Illumina RTA version 1.18.64. Demultiplexing of the lane level fastq files into individual fastq files for each sample was performed with Illumina bcl2fastq2. The reads in fastq format were imported into CLC Genomics Workbench version 9.5.3 (QIAGEN Bioinformatics, Inc.) and were aligned to a Homo sapiens hg19 reference genome (downloaded from Ensembl via the CLC Genomics Workbench) that covered 57,773 genes and 173,446 transcripts in total. Gene expression and transcript expression tracks generated from read mapping were used to compare differential expression of all of the genes between different groups in the CLC Genomics Workbench. The expression data from the CLC Genomics Workbench were imported into R to generate volcano plots showing differentially expressed genes. The “DESeq2” R-package (66) was used to compare normalized total gene reads between all samples of certain genes known to be involved in host defense against Toxoplasma infection. The list of differentially expressed genes [|Fold Change| > 2 and −Log10 (P Value) > 1.3] was used for downstream IPA (QIAGEN Bioinformatics, Inc.). The Log2(Fold Change) of differentially expressed genes was used to identify direct and indirect relationships with the reference gene sets in the IPA knowledge base.
Vacuolar Size Assay.
Host cells were seeded in 96-well µCLEAR black plates (Greiner Bio-One International GmbH) at least 24 h prior to infection. Cells were infected at a multiplicity of infection of 0.5 for 30 min, followed by 3 PBS washes to remove extracellular parasites. At 2 h postinfection, cells were either treated with TNF-α (10 ng/mL, control) alone or in combination with IFN-β (100 units/mL) for the duration of the experiment. Cells were fixed at 36 h (RH infection) or 40 h (PRU infection) postinfection using 4% formaldehyde and stained to detect parasites (anti-SAG1) and the parasitophorous vacuole (anti-GRA7), followed by Alexa-conjugated secondary antibodies. Images were acquired at a magnification of 20× on a Cytation3 Cell Multi-Mode plate-based imager, and the size of parasitophorous vacuole-harboring parasites (SAG1-positive GRA7 vacuoles) was determined using CellProfiler 2.1.1 (SI Appendix, Fig. S5A). Data from at least 50 fields per sample and experiment were used to calculate the vacuolar size of parasites in different host cells.
Animals.
Mice were housed and bred locally at Washington University in Association for Assessment and Accreditation of Laboratory Animal Care-approved facilities. Animal studies were conducted according to the US Public Health Service policy on the humane care and use of laboratory animals as approved by the Institutional Animal Care and Use Committee at Washington University School of Medicine in St. Louis. All mice were on a C57BL/6 background. C57BL/6 wild-type mice were purchased from The Jackson Laboratory. Ifnar1−/− mice (67) were provided by M. S. Diamond, Washington University in St. Louis. For survival assays, 8- to 12-wk-old mice were challenged orally with 5 cysts of ME49 or ME49.Luciferase or i.p. with 105 PRU KO or PRU WT tachyzoites. The mice were monitored for weight loss and parasite burden during 45 to 50 d postinfection.
Bioluminescence Imaging.
Infected wild-type and Ifnar1−/− mice were imaged at different days postinfection for estimation of parasite burden. Mice were injected i.p. with 150 μg of d-luciferin per gram of body weight and imaged using a Xenogen IVIS 200 In Vivo imaging system with continuous administration of 2.5% isoflurane via nose cone. Images were acquired and analyzed using Xenogen Living Image software (Caliper Life Sciences).
Cyst Burden Estimation.
Mice surviving to chronic infection were killed, and half of their brains was harvested in 1 mL of 6% (vol/vol) formaldehyde and 0.25% (vol/vol) Triton X-100 in cold PBS. Cyst burden in each sample was estimated as described previously (68). Cysts were stained with 20 μg/mL FITC-conjugated DBL in 10% goat serum. Each sample was counted 3 times in aliquots of 12.5 μL on a fluorescence microscope in the FITC emission channel. The other half of their brains was fixed in 10% buffered neutral formalin, processed, embedded in paraffin wax, and cut into 4-μm sagittal sections, followed by staining with hematoxylin and eosin. Anonymously labeled samples were submitted for histopathology analysis to the Veterinary Pathology Service, Division of Comparative Animal Medicine, Washington University in St. Louis.
Cytokine Measurements.
Serum samples from wild-type and Ifnar1−/− mice at 3 and 6 d postinfection with PRU KO (∆Tgist) and at 10 and 25 d postinfection with ME49 cysts in 2 independent experiments were collected and stored at −80 °C until used. A ProcartaPlex immunoassay kit (Thermo Fisher Scientific) was used to measure serum levels of IFN-γ, IL-18, IL-10, IL-6, and IL-12p70 in the samples on the Luminex FLEXMAP 3-dimensional platform at The Bursky Centre for Human Immunology and Immunotherapy Programs, Washington University School of Medicine in St. Louis.
Flow Cytometry.
Eight- to 12-wk-old wild-type and Ifnar1−/− mice were infected with 105 PRU KO parasites i.p. or 5 ME49 cysts orally. Mice were killed 3 and 6 d postinfection with PRU KO, followed by isolation of the peritoneal cells as described previously (68). In ME49 cyst infection, the mice were first perfused with PBS and half of the brain was removed and kept in RPMI. Brain mononuclear cells were then isolated as described previously (69). For intracellular IFN-γ staining, cells were incubated with 5 μg/mL Brefeldin-A for 4 h at 37 °C. Cells were then stained with Fixable Vitality Dye eFluor 450 (Thermo Fisher Scientific) at a 1:500 dilution for 15 min at 4 °C before cell surface staining. For staining of peritoneal monocytes, neutrophils, macrophages, and dendritic cells, cells were treated with 2.4G2 hybridoma supernatant (ATCC), stained with BV650-Anti-CD45.2 (1:200), Alexa700-Anti-MHCII (1:200), Peridinin–chlorophyll–protein complex (PerCP)-conjugated Anti-CD11b (1:200), Allophycocyanin: Cy-7 tandem (APCCy7)-conjugated Anti-CD11c (1:100), APC-Anti-F4/80 (1:100), Phycoerythrin (PE)-conjugated Anti-Ly6G (1:250), PacBlue-Anti-Ly6C (1:250), and PECy7-Anti-CD19 (1:200) in staining buffer (2% [vol/vol] FBS, 1 mM ethylenediaminetetraacetic acid, and 0.002% [wt/vol] sodium azide in PBS) for 30 min at 4 °C. For detection of intracellular IFN-γ, Brefeldin A-treated cells were treated with 2.4G2 hybridoma supernatant and stained with APCCy7-Anti-CD45.2 (1:200), PE-Anti-CD3 (1:200), PECy7-Anti-CD19 (1:200), Alexa700-Anti-CD4 (1:250), APC-Anti-CD8 (1:250), BV650-Anti-NK1.1 (1:200), and PerCPCy710-Anti-NKp46 (1:50) in staining buffer for 30 min at 4 °C. For staining of brain monocytes and T cells, Brefeldin-A–treated brain mononuclear cells were treated with 2.4G2 hybridoma supernatant and stained with BV650-Anti-CD45.2 (1:200), PerCP-Anti-CD11b (1:200), PE-Anti-CD3 (1:200), PECy7-Anti-CD19 (1:200), Alexa700-Anti-CD4 (1:250), APC-Anti-CD8 (1:250), and FITC-Anti-Ly6C (1:200). Cells were then fixed with BD CytoFix (BD Biosciences) for 15 min, and stained with PacBlue-Anti-IFN-γ (1:100) in BD CytoPerm buffer (BD Biosciences) for 30 min at 4 °C. The data were acquired using FACSCanto (BD Biosciences) and analyzed using FlowJo (FlowJo LLC). Live cells gated on a CD45+CD19− (hematopoietic non-B cells) parent population were used to identify inflammatory monocytes (CD11b+Ly6C+), neutrophils (CD11b+Ly6G+), dendritic cells (CD11b+CD11c+MHCII+), and resident macrophages (F4/80+Ly6C−). Live cells gated on a CD45+CD19−CD3− (hematopoietic non-B/T cells) parent population were used to identify NK cells (NK1.1+NKp46+). Live cells gated on a CD45+CD19−CD3+ parent population were used identify CD4+ and CD8+ T cells. The total numbers of each specific cell type were calculated by normalizing their respective levels in each parent gate to the total yield of peritoneal and brain mononuclear cells per mouse.
Statistical Analysis.
An unpaired 2-tailed Student’s t test or multiple t test with Holm–Sidak correction was used for comparison between experiments with normally distributed data using Prism (GraphPad). For bioluminescent imaging experiments, total photon flux was log-transformed before the test. Ordinary 1-way ANOVA was used to test difference in relative growth of parasites in IFN-β–treated THP-1 and SH-SY5Y cells compared with untreated controls. Ordinary 1-way ANOVA was also used to test the difference in flow cytometric staining of brain mononuclear cell populations across different samples and serum cytokine levels from uninfected and ME49 cyst-infected wild-type and Ifnar1−/− mice. A nonparametric Kruskal–Wallis test was used to compare the fold differences in gene expression in brain tissues of mice. Two-way ANOVA with Turkey’s multiple comparison test was used to compare relative intranuclear IRF1 immunofluorescence. Survival statistics were compared using the log-rank Mantel–Cox test in Prism (GraphPad).
Supplementary Material
Acknowledgments
We thank Jennifer Barks for assistance with cell culture, Dr. Suellen Greco for histopathology analyses, and Dr. Diane Bender (The Bursky Center for Human Immunology, Alvin J. Siteman Cancer Center at Washington University School of Medicine, and Barnes-Jewish Hospital in St. Louis, MO) for Luminex cytokine analysis. The Siteman Cancer Center is supported in part by an NCI Cancer Center Support Grant P30 CA091842. This study was supported by a grant from the NIH (National Institute of Allergy and Infectious Diseases Grant AI118426).
Footnotes
The authors declare no conflict of interest.
Data deposition: The RNA sequencing data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE125066). Higher quality figures available on figshare (DOI: 10.6084/m9.figshare.9401234.v1).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1904637116/-/DCSupplemental.
References
- 1.Stark G. R., Darnell J. E. Jr, The JAK-STAT pathway at twenty. Immunity 36, 503–514 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reich N. C., Liu L., Tracking STAT nuclear traffic. Nat. Rev. Immunol. 6, 602–612 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Mostafavi S., et al. ; Immunological Genome Project Consortium , Parsing the interferon transcriptional network and its disease associations. Cell 164, 564–578 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schroder K., Hertzog P. J., Ravasi T., Hume D. A., Interferon-gamma: An overview of signals, mechanisms and functions. J. Leukoc. Biol. 75, 163–189 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Ramana C. V., Chatterjee-Kishore M., Nguyen H., Stark G. R., Complex roles of Stat1 in regulating gene expression. Oncogene 19, 2619–2627 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Dubey J. P., Toxoplasmosis of Animals and Humans (CRC Press, Boca Raton, 2010). [Google Scholar]
- 7.Scharton-Kersten T. M., et al. , In the absence of endogenous IFN-gamma, mice develop unimpaired IL-12 responses to Toxoplasma gondii while failing to control acute infection. J. Immunol. 157, 4045–4054 (1996). [PubMed] [Google Scholar]
- 8.Suzuki Y., Orellana M. A., Schreiber R. D., Remington J. S., Interferon-gamma: The major mediator of resistance against Toxoplasma gondii. Science 240, 516–518 (1988). [DOI] [PubMed] [Google Scholar]
- 9.Yap G. S., Sher A., Effector cells of both nonhemopoietic and hemopoietic origin are required for interferon (IFN)-gamma- and tumor necrosis factor (TNF)-alpha-dependent host resistance to the intracellular pathogen, Toxoplasma gondii. J. Exp. Med. 189, 1083–1092 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacMicking J. D., Interferon-inducible effector mechanisms in cell-autonomous immunity. Nat. Rev. Immunol. 12, 367–382 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hunter C. A., Sibley L. D., Modulation of innate immunity by Toxoplasma gondii virulence effectors. Nat. Rev. Microbiol. 10, 766–778 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saeij J. P., Frickel E. M., Exposing Toxoplasma gondii hiding inside the vacuole: A role for GBPs, autophagy and host cell death. Curr. Opin. Microbiol. 40, 72–80 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim S. K., Fouts A. E., Boothroyd J. C., Toxoplasma gondii dysregulates IFN-gamma-inducible gene expression in human fibroblasts: Insights from a genome-wide transcriptional profiling. J. Immunol. 178, 5154–5165 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Lüder C. G., Algner M., Lang C., Bleicher N., Gross U., Reduced expression of the inducible nitric oxide synthase after infection with Toxoplasma gondii facilitates parasite replication in activated murine macrophages. Int. J. Parasitol. 33, 833–844 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Lüder C. G., Walter W., Beuerle B., Maeurer M. J., Gross U., Toxoplasma gondii down-regulates MHC class II gene expression and antigen presentation by murine macrophages via interference with nuclear translocation of STAT1alpha. Eur. J. Immunol. 31, 1475–1484 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Rosowski E. E., Nguyen Q. P., Camejo A., Spooner E., Saeij J. P., Toxoplasma gondii inhibits gamma interferon (IFN-γ)- and IFN-β-induced host cell STAT1 transcriptional activity by increasing the association of STAT1 with DNA. Infect. Immun. 82, 706–719 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneider A. G., Abi Abdallah D. S., Butcher B. A., Denkers E. Y., Toxoplasma gondii triggers phosphorylation and nuclear translocation of dendritic cell STAT1 while simultaneously blocking IFNγ-induced STAT1 transcriptional activity. PLoS One 8, e60215 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gay G., et al. , Toxoplasma gondii TgIST co-opts host chromatin repressors dampening STAT1-dependent gene regulation and IFN-γ-mediated host defenses. J. Exp. Med. 213, 1779–1798 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olias P., Etheridge R. D., Zhang Y., Holtzman M. J., Sibley L. D., Toxoplasma effector recruits the Mi-2/NuRD complex to repress STAT1 transcription and block IFN-γ-dependent gene expression. Cell Host Microbe 20, 72–82 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bowen N. J., Fujita N., Kajita M., Wade P. A., Mi-2/NuRD: Multiple complexes for many purposes. Biochim. Biophys. Acta 1677, 52–57 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Denslow S. A., Wade P. A., The human Mi-2/NuRD complex and gene regulation. Oncogene 26, 5433–5438 (2007). [DOI] [PubMed] [Google Scholar]
- 22.McNab F., Mayer-Barber K., Sher A., Wack A., O’Garra A., Type I interferons in infectious disease. Nat. Rev. Immunol. 15, 87–103 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liehl P., et al. , Host-cell sensors for Plasmodium activate innate immunity against liver-stage infection. Nat. Med. 20, 47–53 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller J. L., Sack B. K., Baldwin M., Vaughan A. M., Kappe S. H. I., Interferon-mediated innate immune responses against malaria parasite liver stages. Cell Rep. 7, 436–447 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Melo M. B., et al. , Transcriptional analysis of murine macrophages infected with different Toxoplasma strains identifies novel regulation of host signaling pathways. PLoS Pathog. 9, e1003779 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beiting D. P., et al. , Differential induction of TLR3-dependent innate immune signaling by closely related parasite species. PLoS One 9, e88398 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmitz J. L., Carlin J. M., Borden E. C., Byrne G. I., Beta interferon inhibits Toxoplasma gondii growth in human monocyte-derived macrophages. Infect. Immun. 57, 3254–3256 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orellana M. A., Suzuki Y., Araujo F., Remington J. S., Role of beta interferon in resistance to Toxoplasma gondii infection. Infect. Immun. 59, 3287–3290 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han S. J., et al. , Internalization and TLR-dependent type I interferon production by monocytes in response to Toxoplasma gondii. Immunol. Cell Biol. 92, 872–881 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Platanias L. C., Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 5, 375–386 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Huntley R. P., et al. , The GOA database: Gene Ontology annotation updates for 2015. Nucleic Acids Res. 43, D1057–D1063 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Randow F., MacMicking J. D., James L. C., Cellular self-defense: How cell-autonomous immunity protects against pathogens. Science 340, 701–706 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagineni C. N., Pardhasaradhi K., Martins M. C., Detrick B., Hooks J. J., Mechanisms of interferon-induced inhibition of Toxoplasma gondii replication in human retinal pigment epithelial cells. Infect. Immun. 64, 4188–4196 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mahmoud M. E., Ui F., Salman D., Nishimura M., Nishikawa Y., Mechanisms of interferon-beta-induced inhibition of Toxoplasma gondii growth in murine macrophages and embryonic fibroblasts: Role of immunity-related GTPase M1. Cell. Microbiol. 17, 1069–1083 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Kovalevich J., Langford D., Considerations for the use of SH-SY5Y neuroblastoma cells in neurobiology. Methods Mol. Biol. 1078, 9–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stansley B., Post J., Hensley K., A comparative review of cell culture systems for the study of microglial biology in Alzheimer’s disease. J. Neuroinflammation 9, 115 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gazzinelli R., Xu Y., Hieny S., Cheever A., Sher A., Simultaneous depletion of CD4+ and CD8+ T lymphocytes is required to reactivate chronic infection with Toxoplasma gondii. J. Immunol. 149, 175–180 (1992). [PubMed] [Google Scholar]
- 38.Hunter C. A., Gabriel K. E., Radzanowski T., Neyer L. E., Remington J. S., Type I interferons enhance production of IFN-gamma by NK cells. Immunol. Lett. 59, 1–5 (1997). [DOI] [PubMed] [Google Scholar]
- 39.Goldszmid R. S., et al. , NK cell-derived interferon-γ orchestrates cellular dynamics and the differentiation of monocytes into dendritic cells at the site of infection. Immunity 36, 1047–1059 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fujita T., et al. , Induction of the transcription factor IRF-1 and interferon-beta mRNAs by cytokines and activators of second-messenger pathways. Proc. Natl. Acad. Sci. U.S.A. 86, 9936–9940 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Su Z. Z., Sarkar D., Emdad L., Barral P. M., Fisher P. B., Central role of interferon regulatory factor-1 (IRF-1) in controlling retinoic acid inducible gene-I (RIG-I) expression. J. Cell. Physiol. 213, 502–510 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Zhou Z., et al. , Type III interferon (IFN) induces a type I IFN-like response in a restricted subset of cells through signaling pathways involving both the Jak-STAT pathway and the mitogen-activated protein kinases. J. Virol. 81, 7749–7758 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mitchell G., Isberg R. R., Innate immunity to intracellular pathogens: Balancing microbial elimination and inflammation. Cell Host Microbe 22, 166–175 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Selleck E. M., et al. , Guanylate-binding protein 1 (Gbp1) contributes to cell-autonomous immunity against Toxoplasma gondii. PLoS Pathog. 9, e1003320 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnston A. C., et al. , Human GBP1 does not localize to pathogen vacuoles but restricts Toxoplasma gondii. Cell. Microbiol. 18, 1056–1064 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Foltz C., et al. , TRIM21 is critical for survival of Toxoplasma gondii infection and localises to GBP-positive parasite vacuoles. Sci. Rep. 7, 5209 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Napolitano A., et al. , Cysteine-reactive free ISG15 generates IL-1β-producing CD8α+ dendritic cells at the site of infection. J. Immunol. 201, 604–614 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lieberman L. A., Banica M., Reiner S. L., Hunter C. A., STAT1 plays a critical role in the regulation of antimicrobial effector mechanisms, but not in the development of Th1-type responses during toxoplasmosis. J. Immunol. 172, 457–463 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Dunay I. R., et al. , Gr1(+) inflammatory monocytes are required for mucosal resistance to the pathogen Toxoplasma gondii. Immunity 29, 306–317 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dunay I. R., Fuchs A., Sibley L. D., Inflammatory monocytes but not neutrophils are necessary to control infection with Toxoplasma gondii in mice. Infect. Immun. 78, 1564–1570 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DeLong J. H., et al. , Cytokine- and TCR-mediated regulation of T cell expression of Ly6C and Sca-1. J. Immunol. 200, 1761–1770 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee P. Y., et al. , Type I interferon modulates monocyte recruitment and maturation in chronic inflammation. Am. J. Pathol. 175, 2023–2033 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seo S. U., et al. , Type I interferon signaling regulates Ly6C(hi) monocytes and neutrophils during acute viral pneumonia in mice. PLoS Pathog. 7, e1001304 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cabral C. M., et al. , Neurons are the primary target cell for the brain-tropic intracellular parasite Toxoplasma gondii. PLoS Pathog. 12, e1005447 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schlüter D., Deckert M., Hof H., Frei K., Toxoplasma gondii infection of neurons induces neuronal cytokine and chemokine production, but gamma interferon- and tumor necrosis factor-stimulated neurons fail to inhibit the invasion and growth of T. gondii. Infect. Immun. 69, 7889–7893 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rose R. W., Vorobyeva A. G., Skipworth J. D., Nicolas E., Rall G. F., Altered levels of STAT1 and STAT3 influence the neuronal response to interferon gamma. J. Neuroimmunol. 192, 145–156 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maher S. G., Romero-Weaver A. L., Scarzello A. J., Gamero A. M., Interferon: Cellular executioner or white knight? Curr. Med. Chem. 14, 1279–1289 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Burg J. L., Perelman D., Kasper L. H., Ware P. L., Boothroyd J. C., Molecular analysis of the gene encoding the major surface antigen of Toxoplasma gondii. J. Immunol. 141, 3584–3591 (1988). [PubMed] [Google Scholar]
- 59.Alaganan A., Fentress S. J., Tang K., Wang Q., Sibley L. D., Toxoplasma GRA7 effector increases turnover of immunity-related GTPases and contributes to acute virulence in the mouse. Proc. Natl. Acad. Sci. U.S.A. 111, 1126–1131 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bastin P., Bagherzadeh Z., Matthews K. R., Gull K., A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei. Mol. Biochem. Parasitol. 77, 235–239 (1996). [DOI] [PubMed] [Google Scholar]
- 61.Su C., Howe D. K., Dubey J. P., Ajioka J. W., Sibley L. D., Identification of quantitative trait loci controlling acute virulence in Toxoplasma gondii. Proc. Natl. Acad. Sci. U.S.A. 99, 10753–10758 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tobin C. M., Knoll L. J., A patatin-like protein protects Toxoplasma gondii from degradation in a nitric oxide-dependent manner. Infect. Immun. 80, 55–61 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fux B., et al. , Toxoplasma gondii strains defective in oral transmission are also defective in developmental stage differentiation. Infect. Immun. 75, 2580–2590 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Olias P., Sibley L. D., Functional analysis of the role of Toxoplasma gondii nucleoside triphosphate hydrolases I and II in acute mouse virulence and immune suppression. Infect. Immun. 84, 1994–2001 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang X., Goncalves R., Mosser D. M., The isolation and characterization of murine macrophages. Curr. Protoc. Immunol. Chapter 14, Unit 14.1 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Daffis S., et al. , The naturally attenuated Kunjin strain of West Nile virus shows enhanced sensitivity to the host type I interferon response. J. Virol. 85, 5664–5668 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Matta S. K., et al. , NADPH oxidase and guanylate binding protein 5 restrict survival of avirulent type III strains of Toxoplasma gondii in naive macrophages. MBio 9, e01393-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pino P. A., Cardona A. E., Isolation of brain and spinal cord mononuclear cells using percoll gradients. J. Vis. Exp., 10.3791/2348 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.